Abstract
Seed oil melting point is an adaptive, quantitative trait determined by the relative proportions of the fatty acids that compose the oil. Micro- and macro-evolutionary evidence suggests selection has changed the melting point of seed oils to covary with germination temperatures because of a trade-off between total energy stores and the rate of energy acquisition during germination under competition. The seed oil compositions of 391 natural accessions of Arabidopsis thaliana, grown under common-garden conditions, were used to assess whether seed oil melting point within a species varied with germination temperature. In support of the adaptive explanation, long-term monthly spring and fall field temperatures of the accession collection sites significantly predicted their seed oil melting points. In addition, a genome-wide association study (GWAS) was performed to determine which genes were most likely responsible for the natural variation in seed oil melting point. The GWAS found a single highly significant association within the coding region of FAD2, which encodes a fatty acid desaturase central to the oil biosynthesis pathway. In a separate analysis of 15 a priori oil synthesis candidate genes, 2 (FAD2 and FATB) were located near significant SNPs associated with seed oil melting point. These results comport with others’ molecular work showing that lines with alterations in these genes affect seed oil melting point as expected. Our results suggest natural selection has acted on a small number of loci to alter a quantitative trait in response to local environmental conditions.
Key Words: adaptation, evolution, fatty acid composition, GWAS.
Evolution requires the presence of heritable genetic variation in traits, and most traits are quantitative. Therefore, to understand adaptive evolution, we must understand the genetic architecture of complex trait variation. Understanding the genetic basis of adaptive evolution of quantitative traits, one of the major areas of evolutionary research, has become attainable due to recent genotyping advances and methods for analyzing large genetic data sets (Brachi et al. 2010; Childs et al. 2010; Chan et al. 2011; Hancock et al. 2011). This field also has practical applications, such as identifying genes underlying human disease (Stein and Elston 2009) and those important for crop improvement (Rafalski 2010).
Evidence supports adaptive evolution of seed oil composition, the types and relative proportions of the fatty acids in seed oil, in angiosperms that produce oil seeds (Linder 2000). In these species, seed oil supplies the energy for germination and establishment prior to photosynthesis (Harwood 1980). Seed oil composition determines the total amount of energy stored in the seed (when oil content is held constant) and also determines the melting point of the oil. Seed oil melting point tends to decrease with increasing fatty acid desaturation and with decreasing fatty acid chain length (Eckey 1954; Hilditch and Williams 1964). Per unit of change, desaturation has a stronger effect on melting point than decreasing chain length. On the other hand, saturated fatty acids cost less to synthesize and provide more energy per carbon atom when metabolized than unsaturated fatty acids (Lehninger 1993). Therefore, increased relative amounts of unsaturated fatty acids decrease seed oil melting point at the expense of total energy stores. In locations with cooler germination temperatures, seeds with higher proportions of unsaturated fatty acids are expected to be able to germinate earlier and grow more rapidly prior to photosynthesis, which would be advantageous under competition. Under warmer germination temperatures, these advantages decrease, and seeds with a higher amount of total energy stores, i.e., proportionately more saturated fatty acids, may be favored. Therefore, the adaptive evolution of seed oil composition is thought to be a trade-off between total energy stores and the rate those stores can be used, with germination temperature as the selective agent (Linder 2000).
Based on this adaptive explanation and seed oil biosynthesis, genes encoding enzymes that desaturate fatty acids would be expected to play an important role in modulating seed oil melting point. Most of the enzymes involved in seed oil biosynthesis are well understood and their functions validated in mutant and molecular studies (reviewed in Li-Beisson et al. 2013). Still, relatively little is known about how lipid synthesis is regulated to produce variation in fatty acid composition in nature (Li-Beisson et al. 2013). In addition to the genes in the oil synthesis pathway, genes encoding trans-acting regulatory factors such as kinases and transcription factors could act to control proportions of fatty acids produced and, therefore, be important in modifying the seed oil melting point (Ruuska et al. 2002).
The adaptive evolution of seed oil composition, seen at the family and genus levels (Linder 2000), would also be expected within a species with a broad geographic distribution. Previous work has shown extensive variation in seed oil composition in natural accessions of A. thaliana (Millar and Kunst 1999; O’Neill et al. 2003), and this variation follows the same pattern as seen among species (Sanyal and Linder 2011).
To date, the study of the quantitative genetics of seed oil composition has been confined to quantitative trait loci (QTL) studies (Hobbs et al. 2004; O’Neill et al. 2011), and only one of these has examined the quantitative genetics of the melting point of seed oil (Sanyal and Linder 2011). In the Sanyal and Linder study, the QTL peaks covered large genomic regions spanning dozens to hundreds of genes.
With a world-wide distribution and the availability of abundant genetic resources—particularly dense SNP data sets—A. thaliana is an excellent system to determine the fine-scale genetic basis of adaptive evolution. Hundreds of thousands of markers are available, allowing associations to be resolved to the genic level in most instances (Bergelson and Roux 2010; Weigel 2012). Further, genome-wide association studies (GWAS) in species with a high degree of population structure, like A. thaliana, are now possible because of new statistical tests that control for relatedness (Yu et al. 2006; Kang et al. 2008; Kang et al. 2010; Zhang et al. 2010; Zhou and Stephens 2012). These tests reduce the false positive rate caused by markers confounded with the population structure but also can lead to an increase in false negatives, especially for traits that have been selected along the same geographic pattern as the population structure (Brachi et al. 2010).
The goals of this study were 3-fold: 1) to determine whether natural variation in the seed oil melting point in A. thaliana varied with temperature; 2) to identify the genetic basis of observed natural variation in the seed oil melting point of A. thaliana; and 3) to assess whether the genes associated with melting point shed any light on the adaptive evolution of seed oil melting point. We were particularly interested in assessing whether any genes from a list of 15 a priori candidate genes for melting point (Sanyal and Linder 2011) would be implicated, and, if not, what new candidate genes would be identified. Using the fatty acid compositions and estimated seed oil melting points from 391 natural accessions of A. thaliana, a study was performed on the a priori candidate genes and a GWAS was performed to identify a posteriori candidate genes.
Materials and Methods
Accessions and Seed Production
Three hundred ninety-one accessions (Figure 1) from across the geographic range of A. thaliana were used to encompass the extent of seed oil melting point variation. Three hundred fifty-six were within the native range of A. thaliana (348 European, 6 Asian, 1 from the border between Europe and Asia, and 1 African), and 35 were recent introductions outside of its native range (United States 28, Japan 3, Cape Verde Islands 1, Canada 2, and New Zealand 1). Seeds for each accession were acquired from either the Arabidopsis Biological Resource Center (ABRC) or the Juenger lab at the University of Texas at Austin.
Figure 1.
Geographic distribution of the 391 A. thaliana accessions used in this study. Gray circles represent collection locations of: (A) native accessions (N = 356) across Europe, mainland Asia, and northern Africa, (B) non-native accessions from the United States of America (N = 31), and (C) the remaining non-native accessions (N = 4).
Growth conditions and seed production for the accessions were as in Branham et al. (2015). Briefly, seeds for 3 replicates of each accession were cold-stratified in water at 4 °C for 6 days and subsequently grown in a completely randomized design in a glasshouse under common garden conditions. Seeds from each plant were harvested at maturity and stored at room temperature in coin envelopes before seed oil compositions were determined.
Melting Point Analysis
Fatty acid compositions of the accessions were determined using gas chromatography of fatty acid methyl esters (FAMEs) as described in Branham et al. (2015). Two extractions per plant were performed and their average was used for all analyses. Using the relative proportions of the 9 principle fatty acids in A. thaliana seed oil (16:0, 18:0, 18:1, 18:2, 18:3, 20:0, 20:1, 20:2, and 22:1) (Branham et al. 2015), seed oil melting point (MP) was estimated as the average of the melting points of the individual fatty acids weighted by their relative proportions (Malkin 1954).
Proc Mixed in SASTM software (SAS Institute, Inc., Cary, NC) was used to calculate the broad-sense heritability (H2) of seed oil melting point (untransformed) with accession as the class variable (coded as random). Because the raw data were not normally distributed, the analysis was also performed on the box-cox transformed data and with tray as a random variable in the model.
Relative Effect of Fatty Acids on Seed Oil Melting Point
The effect of each fatty acid on the seed oil melting point is dependent not only on the relative proportion of that fatty acid but also the degree to which its individual melting point differs from the overall seed oil melting point. To assess which fatty acids had the greatest potential to alter the seed oil melting point, the effect of each fatty acid on melting point (MP) was calculated as follows: effect = [(fatty acid MP − seed MP)/seed MP] × fatty acid proportion. We refer to this value as the melting point effect metric.
Climate and Geographic Regressions
The geographic origins of 63 of the accessions used in this study may have been misidentified (Anastasio et al. 2011). Therefore, to assess the relationship between environmental temperatures and seed oil melting point, as would be expected if the melting point is adaptive, we used only the 328 accessions whose geographic origins are not in question. Linear regressions of the boxcox-transformed melting points were performed in R (R core team 2014) for each of the following monthly average temperature values as independent variables: minimum, mean, and maximum (Hijmans et al. 2005). The monthly temperature data from worldclim.org specific to the accessions was compiled and formatted to a user-friendly version in a previous study (Lasky et al. 2012). To determine whether MP varied along the same geographic lines as population structure, linear regressions were also performed (R core team 2014) using latitude, longitude, and altitude as independent variables.
Because A. thaliana was introduced to non-European and non-Asian regions fairly recently (Sharbel et al. 2000; Beck et al. 2008), the non-native accessions in this study may not be locally adapted. We, therefore, also performed the climate and geographic regressions on just the native (N = 299) accessions whose locations are not in question.
Association Mapping
Of 214 051 SNPs available for A. thaliana (http://walnut.usc.edu/2010/SNPs, v3.06), 13 748 with allele frequencies of 5% or less were removed from the dataset to control for the effect of minor alleles on the analysis (Kang et al. 2008; Atwell et al. 2010).
GWAS was performed for seed oil melting point as in Branham et al. (2015) using all 3 replicates of the 391 accessions to increase the power to detect associations (Kang et al. 2008). Seed oil melting point was Box-Cox transformed using the car package in R (Box and Cox 1964; Fox and Weisberg 2011; R core team 2014). SNP associations were identified using the R implementation EMMA v. 1.1.2 for Linux (Kang et al. 2008). FDR corrected P values were generated using the p.adjust package in R (Benjamini and Hochberg 1995; R core team 2014). SNPs were considered significant at FDR < 0.10. Gene functions and GO terms of a posteriori candidate genes were obtained from TAIR (www.arabidopsis.org). GWAS was redone using highly significant SNPs (FDR < 0.05) as covariates in the mixed model to determine whether the remaining significant SNPs were independently associated with MP. Linkage disequilibrium (LD) between highly significant SNPs and all surrounding SNPs up to ±1Mb was explored to search for a posteriori candidate genes. LD was calculated using TASSEL v5.2.16 (Bradbury et al. 2007).
A priori Candidate Genes
Previously, a set of 15 a priori candidate genes known to encode fatty acid synthesis enzymes was chosen as likely to affect seed oil melting point (Sanyal and Linder 2011). A separate FDR correction was applied to the P values of SNPs that fell within the 200kb window of the coding regions of these genes to provide more power to determine whether they were associated with MP.
Data Archiving
To fulfill data archiving requirements (Baker 2013), accession phenotypes, P values of significant SNPs from all models, results of the LD analyses, and genes in significant regions with their annotations have been deposited in Dryad.org.
Results
Seed Oil Melting Point and Heritability
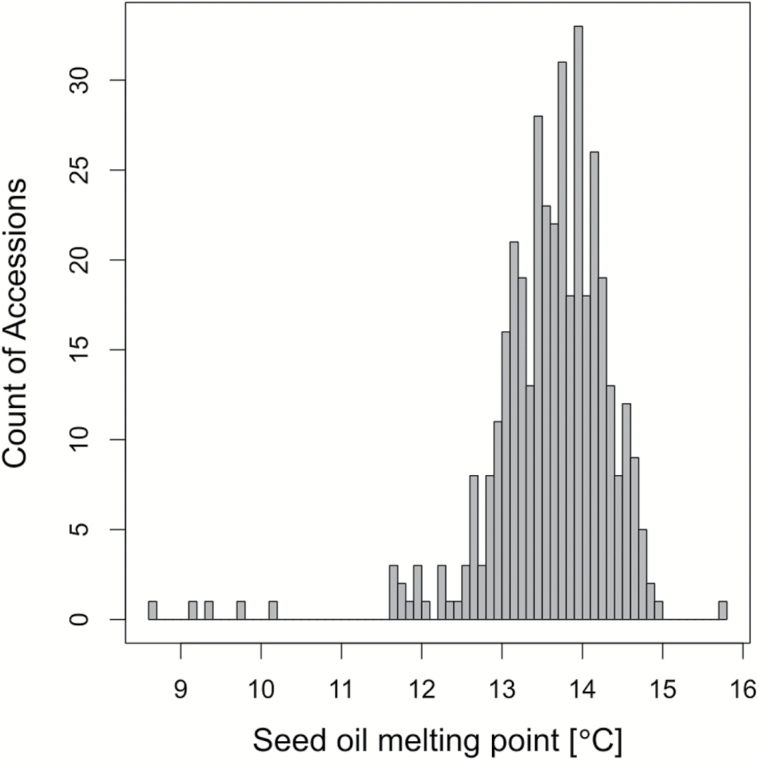
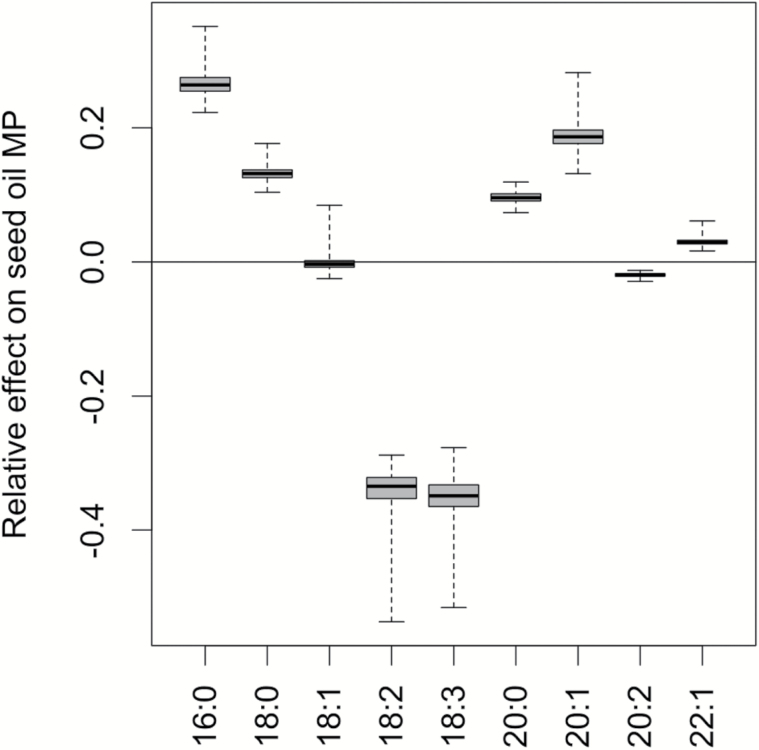
Seed oil melting point (MP) varied across the 391 accessions by 7.1 °C, from 8.7 °C to 15.8 °C with a mean of 13.6 °C (Figure 2). The distribution of melting points was mostly normal with a small number of low melting point outliers. The MP mean and range did not vary substantially between the All (N = 391) and the Native (N = 356) sets. Of the 4 fatty acids with the highest values for the seed oil MP effect metric (Figure 3), 3 (18:2, 18:3, and 20:1) constituted the highest average proportions in the seed oil (Branham et al. 2015), while the proportionately low 16:0 had a large effect because of its comparatively high melting point (62.9 °C) relative to that of the seed oil (mean = 13.6 °C). 18:2 and 18:3 most strongly decreased the melting point, while 16:0 had the strongest increase on MP followed by 20:1 (Figure 3).
Figure 2.
Distribution of seed oil melting points. Histogram of the average melting points for the accessions in the All dataset (N = 391).
Figure 3.
Relative effect of each fatty acid on seed oil melting point. Boxplots (minimum, lower hinge, median, upper hinge, maximum) of the effect each fatty acid had on average seed oil melting point among the 391 accessions. A zero value means the fatty acid has no effect on seed oil melting point because its melting point is equivalent to the seed oil melting point.
The broad-sense heritability of seed oil melting point was high whether estimated on untransformed (H 2 = 0.94) or boxcox-transformed (H 2 = 0.90) data. Although this value is likely to be inflated relative to what would be seen in nature, due to our common-garden growth conditions for generating the seeds, its high values indicate it is likely also a high heritability in nature. Inclusion of tray did not improve the log likelihood of the model and was therefore excluded.
Climate and Geographic Regressions
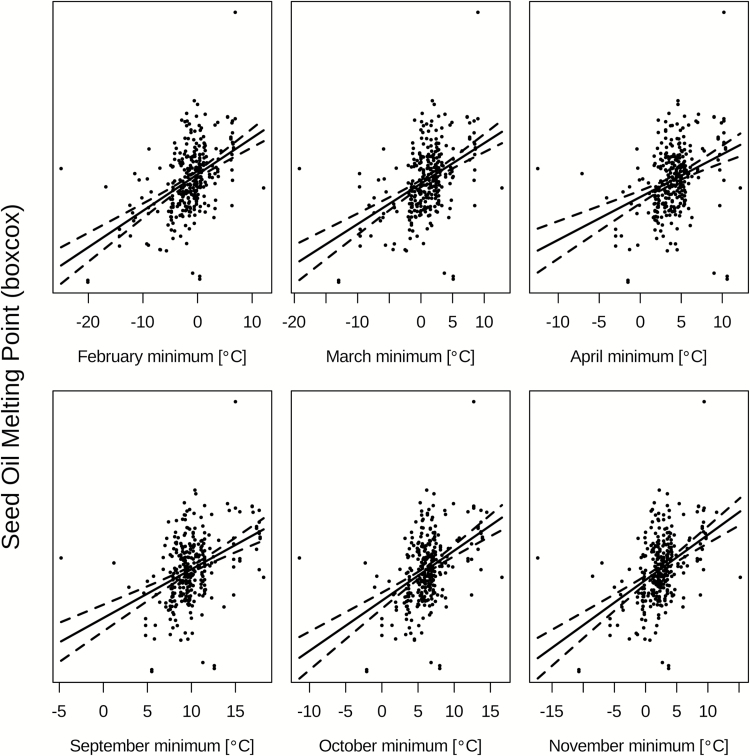
For the geographically validated Native accessions, monthly average minimum, maximum and mean temperatures for each of the months of September through April significantly predicted seed oil MP (Table 1), with average minimum explaining the most variation for all months. Further, average minimum temperature is significant all months of the year, and explains the largest amount of MP variation February through April and September through November (r 2 = 0.22–0.23; Figure 4).
Table 1.
Climate and geographic regressions with boxcox transformed MP as the dependent variable
| X | P valueb | r 2 |
|---|---|---|
| Latitude | 2.3e−6*** | 0.069 |
| Longitude | <2e−16*** | 0.22 |
| Altitude | 0.20 | 2.3e−3 |
| January min temperature | <2e−16*** | 0.22 |
| January mean temperature | <2e−16*** | 0.22 |
| January max temperature | <2e−16*** | 0.22 |
| February min temperaturea | <2e−16*** | 0.23 |
| February mean temperature | <2e−16*** | 0.22 |
| February max temperature | <2e−16*** | 0.21 |
| March min temperature | 2.6e−16*** | 0.20 |
| March mean temperature | 2.4e−15*** | 0.19 |
| March max temperature | 1.0e−13*** | 0.17 |
| April min temperature | 5.5e−10*** | 0.12 |
| April mean temperature | 1.8e−7*** | 0.084 |
| April max temperature | 4.2e−5*** | 0.052 |
| May min temperature | 2.0e−5*** | 0.056 |
| May mean temperature | 7.3e−3** | 0.021 |
| May max temperature | 0.26 | 1.0e−3 |
| June min temperature | 1.0e−3** | 0.032 |
| June mean temperature | 0.085 | 6.6e−3 |
| June max temperature | 0.67 | −2.7e−3 |
| July min temperature | 3.7e−4*** | 0.039 |
| July mean temperature | 0.021* | 0.015 |
| July max temperature | 0.25 | 1.1e−3 |
| August min temperature | 1.2e−6*** | 0.073 |
| August mean temperature | 5.9e−4*** | 0.036 |
| August max temperature | 0.039* | 0.011 |
| September min temperature | 9.7e−13*** | 0.15 |
| September mean temperature | 6.9e−9*** | 0.10 |
| September max temperature | 2.9e−5*** | 0.054 |
| October min temperature | 1.0e−15*** | 0.19 |
| October mean temperature | 2.3e−13*** | 0.16 |
| October max temperature | 3.1e−10*** | 0.12 |
| November min temperature | <2.2e−16*** | 0.20 |
| November mean temperature | 7.1e−16*** | 0.19 |
| November max temperature | 1.1e−14*** | 0.18 |
| December min temperature | <2.2e−16*** | 0.21 |
| December mean temperature | <2.2e−16*** | 0.21 |
| December max temperature | 3.2e−16*** | 0.20 |
aThe most significant predictor of seed oil melting point.
b*P < 0.05, **P < 0.01, ***P < 0.001.
Figure 4.
Boxcox transformed melting point regressed on the average minimum temperatures for the months of February through April and September through November. Best fit lines and 95% confidence intervals are shown.
February average minimum temperature remained the strongest predictor of MP when using all 328 geographically validated accessions (Native and Non-native) in the regressions. While there were no major deviations in rank order of the significant variables when regressions were performed with all accessions, the r 2 values were all weaker, as would be expected if non-native accessions were less well adapted due to their recent introduction. Of the geographic variables, longitude and latitude were significant predictors of MP (Table 1), with longitude explaining the most variation (r 2 = 0.22) by a large margin.
Association Analysis of A Priori Candidate Genes
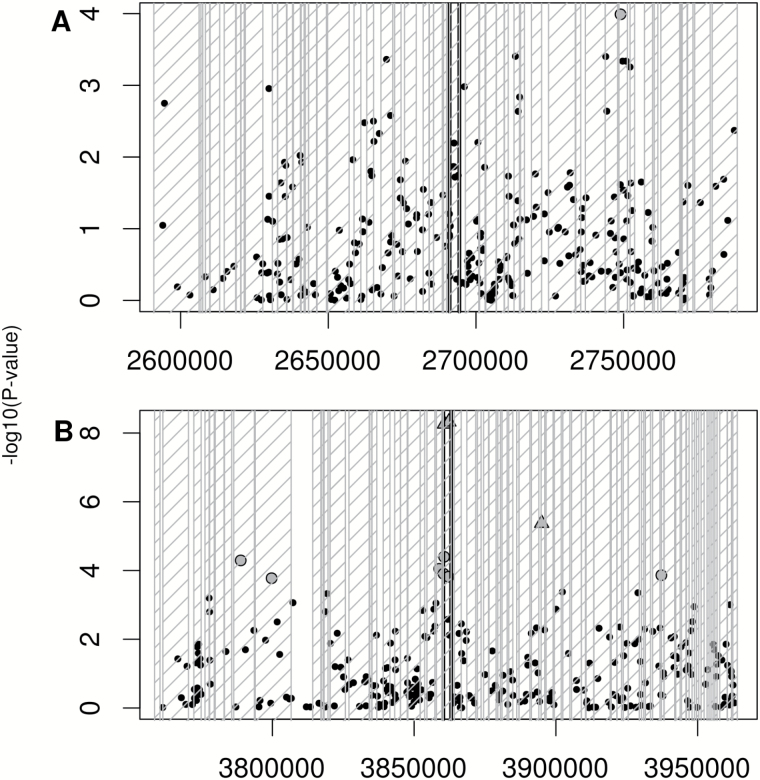
Of the 5318 SNPs found within ±100kb of the 15 a priori candidate genes, 11 were significantly associated with MP variation (uncorrected P = 1.7×10−4 to 4.7×10−9) and were located near 2 genes (FAD2 and FATB) (Figure 5). Ten SNPs were within 74kb of FAD2 (AT3G12120; a fatty acid desaturase) and one was less than 55kb from the thioesterase FATB (AT1G08510). The strongest associations were with SNPs linked to FAD2.
Figure 5.
Scatterplots of the significant associations in the a priori analysis. Negative log P values of all SNPs found within 100kb upstream of the start codon and 100kb downstream of the stop codon of the significant a priori candidate genes (bounded by vertical black lines). Gene locations in each region are represented by striped gray boxes. Significant SNPs are shape coded. Gray triangle = FDR < 0.05, gray circle = FDR < 0.1. (A) Region on chromosome 1 surrounding FATB (AT1G08510), (B) Region on chromosome 3 surrounding FAD2 (AT3G12120).
Genome-Wide Association Study
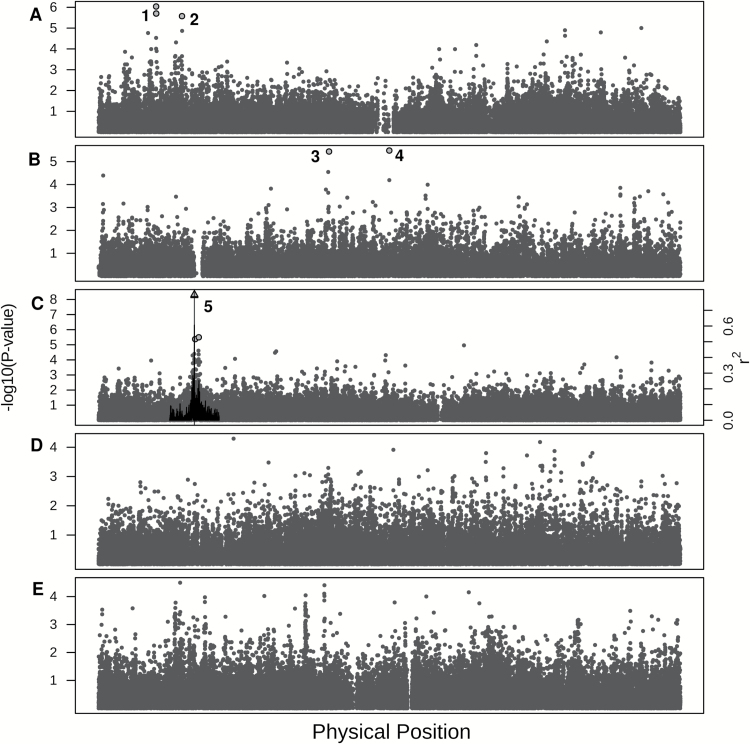
The GWAS revealed a single area of highly significant association (FDR < 0.05) with MP and 4 less significant areas (0.05 < FDR < 0.1), with 10 total significant SNPs. (Figure 6). The area of strongest association consisted of 3 significant SNPs within a 34-kb region on chromosome 3 and a fourth significant SNP 144kb downstream (Region 5 in Figure 6). The 4 less-significant regions contained 7 SNPs spread across chromosomes 1 and 2 (Figure 6). Region 1 contained 3 significant SNPs within 6kb on chromosome 1 (FDR = 0.06–0.08; P = 9×10−7 to 2×10−6). Regions 2 through 4 contained only a single significant SNP each (P = 2.7×10−6 to 3.6×10−6).
Figure 6.
GWAS results for boxcox-transformed seed oil melting point in A. thaliana using the EMMA package in R. Scatterplot of the negative log P values for each chromosome. Significant SNPs are shape coded. Gray triangle = FDR < 0.05, gray circle = FDR < 0.1. The vertical black line indicates the position of the a priori candidate gene FAD2 (AT3G12120). Regions of significant association are labeled 1–5. Linkage disequilibrium (r 2) in a 200kb window around the most significant SNP in the genome is shown with a black line graph. (A) Chromosome 1, (B) Chromosome 2, (C) Chromosome 3, (D) Chromosome 4, and (E) Chromosome 5.
Two SNPs had FDR values 2 orders of magnitude smaller than any other SNPs in the genome (P = 4.8×10−9 and 5.5×10−9). The 2 SNPs are in LD with one another (r 2 = 0.61) but not with any of the other significant SNPs (r 2 = 7.8×10−5 to 0.026). When either or both of these SNPs are used as a covariate in the GWAS model, all other associations become insignificant. These 2 highly significant SNPs are located in an exon and intron of the fatty acid desaturase gene, FAD2. LD remained above background levels (r 2 > 0.1) for approximately 150kb upstream and 250kb downstream of the significant SNPs. There were 131 genes within the 400kb window around these 2 SNPs, but the area of strongest association centered on FAD2 with a gradual decrease in strength with distance from this gene (Figure 5). While FAD2 is the most likely candidate gene, a fatty acid reductase (FAR2), 7 transcription factors, and 4 kinases were also found in this region and represent potential a posteriori candidate genes.
Discussion
We found support for the hypothesis that seed oil melting point has been under selection to change with germination temperature as previously hypothesized (Linder 2000). Specifically, fall and spring average minimum temperatures are most predictive of seed oil melting point in the native accessions. The low r 2 values (r 2 = 0.22–0.23) of these associations indicate that factors other than temperature are also involved in determining seed oil melting point. This pattern makes sense since the minimum temperatures experienced by newly germinated seeds are likely to be the best indication of whether the seedling will be in an environment that will produce freezing damage. Temporal variation in germination timing can be attributed to both seasonal life-history variation in the timing of germination (leading to summer, fall and winter annuals) and to climatic variation across the range of A. thaliana (Mitchell-Olds and Schmitt 2006). While few field studies of A. thaliana germination have been reported, natural populations in Europe have been observed germinating in September through November with a spring flush in March and a summer cohort in Northern Europe in June and July (Fournier-Level et al. 2013). In field trials of seeds sewn in November in Rhode Island, germination was observed throughout the winter and spring (Donohue et al. 2005). Climatic variation across Eurasia follows both longitudinal and latitudinal clines and both of these variables were predictive of seed oil melting point variation in the native accessions.
A Priori Candidate Genes
With respect to the genetic basis of the naturally occurring variation in seed oil MP, the a priori-specific analyses linked 2 genes (FAD2 and FATB) to significant SNPs. Both genes either do or could change the ratios of the fatty acids with the greatest potential to alter melting point (16:0 and 20:1 to increase melting point, and 18:2 and 18:3 to decrease melting point). This suggests natural selection has acted most strongly on the genes that can most readily affect MP, making it relatively easy for populations to optimize their MP as they migrate to environments with novel germination temperature regimens. Due to the most significant SNPs in the genome being located within the coding region of FAD2 and also being identified in the GWAS analysis, it is likely this is the key gene for modifying MP with FATB playing a more minor role.
Molecular studies of FAD2 and FATB variants confirm that they can influence the quantities of the fatty acids with the greatest potential effects on MP. FAD2 catalyzes the desaturation of 18:1 to 18:2 (Okuley et al. 1994). The rate of activity of FAD2 could influence seed oil melting point directly by changing the proportion of 18:2, but also indirectly because 18:2 is the substrate for 18:3. 18:2 and 18:3 have the greatest effects on lowering seed oil melting point. The importance of FAD2 for seed oil melting point is also supported by characterization of the seed oil composition of the fad2-1 (CS8041) ethyl methane sulfonate mutant (Lemieux et al. 1990), which showed a 9.6 ºC increase its seed oil melting point (MP = 22.5 ºC) relative to the wild-type (MP = 12.9 ºC) due to a 38% increase in 18:1, a 29% decrease in 18:2, a 15% decrease in 18:3, and a 10% decrease in 20:1.
FATB, an acyl-acyl carrier protein thioesterase which hydrolyzes acyl-ACP to produce free fatty acids has a substrate preference for 16:0-ACP (Salas and Ohlrogge 2002) but will also hydrolyze 18:1-ACP (24% less activity) and 18:0-ACP (64% less activity). Because of this substrate preference for 16:0 and because 16:0 has the strongest influence on increasing seed oil melting point, down regulation of FATB or protein variants of FATB could play a significant role in lowering A. thaliana’s melting point by decreasing the amount of 16:0 exported to the ER to be incorporated into seed oils. As expected, a T-DNA insertion mutation in FATB (CS6525) (Sussman et al. 2000) reduced seed oil melting point by 3 ºC as compared to wild-type. The MP reduction was primarily produced by a 56% reduction of 16:0 in seeds and a 30% reduction of 18:0 (Bonaventure et al. 2003).
In the future, it would be valuable to characterize the naturally occurring variation in FAD2 and FATB to assess whether protein sequence variation or expression variation is most commonly used to affect fatty acid production and incorporation into TAGs.
Although only 2 of the 15 a priori candidate genes were located near significant SNPs, many of the a priori candidate genes are part of gene families that might have redundant functions, making their individual influences difficult to identify through association mapping. There are 5 stearoyl-ACP desaturases (ST1-5), and they have high sequence similarity to FAB2 (AT2G43710) and DES6 (AT1G43800). All 7 of these genes likely contribute to production of 18:1 (Kachroo et al. 2007). Similarly, there are four 3-ketoacyl-CoA synthases (FAE1.1–1.4), which perform elongation of the 18-carbon FAs to 20 and 22 carbons, and they are also functionally related to 20 additional elongases with high sequence similarity (Blacklock and Jaworski 2006). Finally, FATA (AT3G25110) has 2 copies (Kachroo et al. 2007).
A Posteriori Candidate Genes
GWAS confirmed the single major QTL centered on the coding region of FAD2. While the initial GWAS identified 10 significant SNPs, these associations disappeared when the FAD2 SNPs were included as covariates in the model and are therefore likely spurious. The only previous mapping study to investigate seed oil melting point identified 9 QTLs using 4 RIL populations (Sanyal and Linder 2011 One of these QTL collocates with FAD2. GWAS can fail to detect true associations due to complex genetic interactions such as epistasis (Rowe et al. 2008; Chan et al. 2010) and genetic heterogeneity (Bergelson and Roux 2010).
Conclusion
We showed seed oil melting point in wild accessions of A. thaliana varies with local minimum monthly temperatures. In addition, we identified 2 candidate genes (FAD2 and FATB) that may have influenced the adaptive evolution of seed oil composition to optimize seed oil melting point relative to germination temperatures in nature. Variants of these genes were significantly associated with MP and have been shown to alter seed oil melting point in other studies. Characterization of the seed oil composition of up- and down-regulated transgenics and knockouts of both candidate genes should be done to show whether they affect seed oil melting point in the predicted manner. To test whether these genes are adaptive, fitness assays should be conducted comparing the relative fitness of the allelic variants in the same genetic background (either using NILs or transgenics) under cold and warm germination temperatures (Mackay 2001; Wright and Gaut 2005; Mitchell-Olds and Schmitt 2006; Shindo et al. 2007).
Funding
This work was supported by a National Science Foundation doctoral dissertation improvement grant (DEB-1011609) to (S.E.P., C.R.L.).
Data Availability
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.sh2mb
Acknowledgments
Thank you to T. Juenger for providing accession seed. We are grateful for analysis advice from J. Lasky and T. Nakov. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing high performance computing resources that have contributed to the research results reported within this paper. http://www.tacc.utexas.edu.
References
- Anastasio AE, Platt A, Horton M, Grotewold E, Scholl R, Borevitz JO, Nordborg M, Bergelson J. 2011. Source verification of mis-identified Arabidopsis thaliana accessions. Plant J. 67:554–566. [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 465:627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Beck JB, Schmuths H, Schaal BA. 2008. Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Mol Ecol. 17:902–915. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 57:289–300. [Google Scholar]
- Bergelson J, Roux F. 2010. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana . Nat Rev Genet. 11:867–879. [DOI] [PubMed] [Google Scholar]
- Blacklock BJ, Jaworski JG. 2006. Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem Biophys Res Commun. 346:583–590. [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Salas JJ, Pollard MR, Ohlrogge JB. 2003. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell. 15:1020–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box G, Cox D. 1964. An analysis of transformations. J R Stat Soc Ser B. 26:211–252. [Google Scholar]
- Brachi B, Faure N, Horton M, Flahauw E, Vazquez A, Nordborg M, Bergelson J, Cuguen J, Roux F. 2010. Linkage and association mapping of Arabidopsis thaliana flowering time in nature. PLoS Genet. 6:e1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 23:2633–2635. [DOI] [PubMed] [Google Scholar]
- Branham SE, Wright SJ, Reba A, Linder CR. 2015. Genome-wide association study of Arabidopsis thaliana Identifies determinants of natural variation in seed oil composition. J Hered. 106, doi:10.1093/jhered/esv100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ. 2011. Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana . PLoS Biol. 9:e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Kliebenstein DJ. 2010. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics. 185:991–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs LH, Witucka-Wall H, Günther T, Sulpice R, Korff MV, Stitt M, Walther D, Schmid KJ, Altmann T. 2010. Single feature polymorphism (SFP)-based selective sweep identification and association mapping of growth-related metabolic traits in Arabidopsis thaliana . BMC Genomics. 11:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K, Dorn L, Griffith C, Kim E, Aguilera A, Polisetty CR, Schmitt J. 2005. Environmental and genetic influences on the germination of Arabidopsis thaliana in the field. Evolution. 59:740–757. [PubMed] [Google Scholar]
- Eckey EW. 1954. Vegetable fats and oils. New York: Reinhold. [Google Scholar]
- Fournier-Level A, Wilczek AM, Cooper MD, Roe JL, Anderson J, Eaton D, Moyers BT, Petipas RH, Schaeffer RN, Pieper B, et al. 2013. Paths to selection on life history loci in different natural environments across the native range of Arabidopsis thaliana . Mol Ecol. 22:3552–3566. [DOI] [PubMed] [Google Scholar]
- Fox J, Weisberg S. 2011. An {R} Companion to Applied Regression, 2nd Edition Thousand Oaks: (CA: ): Sage; http://socserv.socsci.mcmaster.ca/jfox/Books/Companion. [Google Scholar]
- Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, Roux F, Bergelson J. 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science. 334:83–86. [DOI] [PubMed] [Google Scholar]
- Harwood JL. 1980. Plant acyl lipids: Structure, distribution, and analysis. In: Stumpf PK, editor. The biochemistry of plants: a comprehensive treatise, vol. 4. New York: Academic Press. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 25:1965–1978. [Google Scholar]
- Hilditch TP, Williams PN. 1964. The chemical constitution of natural fats. New York: Wiley. [Google Scholar]
- Hobbs D., Flintham J., Hills M. 2004. Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiol. 136: 3341–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Shanklin J, Whittle E, Lapchyk L, Hildebrand D, Kachroo P. 2007. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol Biol. 63:257–271. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. 2008. Efficient control of population structure in model organism association mapping. Genetics. 178:1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, et al. 2010. Variance component model to accound for sample structure in genome-wide association studies. Nat Genet. 42:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky JR, Des Marais DL, McKay JK, Richards JH, Juenger TE, Keitt TH. 2012. Characterizing genomic variation of Arabidopsis thaliana: the roles of geography and climate. Mol Ecol. 21:5512–5529. [DOI] [PubMed] [Google Scholar]
- Lehninger AI. 1993. Biochemistry. New York: Worth. [Google Scholar]
- Lemieux B, Miquel M, Somerville C, Browse J. 1990. Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor Appl Genet. 80:234–240. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. 2013. Acyl-lipid metabolism. Arabidopsis Book. 11:e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder C. 2000. Adaptive evolution of seed oils in plants: accounting for the biogeographic distribution of saturated and unsaturated fatty acids in seed oils. Am Nat. 156:442–458. [DOI] [PubMed] [Google Scholar]
- Mackay TF. 2001. Quantitative trait loci in Drosophila. Nat Rev Genet. 2:11–20. [DOI] [PubMed] [Google Scholar]
- Malkin T. 1954. The polymorphism of glycerides. In Holman RT, Lundberg WO, editors. Progress in the chemistry of fats and other lipids. Pergamon, NY. [Google Scholar]
- Millar AA, Kunst L. 1999. The natural genetic variation of the fatty-acyl composition of seed oils in different ecotypes of Arabidopsis thaliana . Phytochemistry. 52:1029–1033. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. 2006. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 441:947–952. [DOI] [PubMed] [Google Scholar]
- O’Neill CM, Gill S, Hobbs D, Morgan C, Bancroft I. 2003. Natural variation for seed oil composition in Arabidopsis thaliana . Phytochemistry. 64:1077–1090. [DOI] [PubMed] [Google Scholar]
- O’Neill CM, Morgan C, Hattori C, Brennan M, Rosas U, Tschoep H, Deng PX, Baker D, Wells R, Bancroft I. 2012. Towards the genetic architecture of seed lipid biosynthesis and accumulation in Arabidopsis thaliana . Heredity (Edinb). 108:115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. 1994. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell. 6:147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski JA. 2010. Association genetics in crop improvement. Curr Opin Plant Biol. 13:174–180. [DOI] [PubMed] [Google Scholar]
- R Core Team 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. 2008. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell. 20:1199–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska S, Girke T, Benning C, Ohlrogge JB. 2002. Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell. 14:1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas JJ, Ohlrogge JB. 2002. Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch Biochem Biophys. 403:25–34. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Randal Linder C. 2012. Quantitative trait loci involved in regulating seed oil composition in Arabidopsis thaliana and their evolutionary implications. Theor Appl Genet. 124:723–738. [DOI] [PubMed] [Google Scholar]
- Sharbel TF, Haubold B, Mitchell-Olds T. 2000. Genetic isolation by distance in Arabidopsis thaliana: biogeography and postglacial colonization of Europe. Mol Ecol. 9:2109–2118. [DOI] [PubMed] [Google Scholar]
- Shindo C, Bernasconi G, Hardtke CS. 2007. Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology. Ann Bot. 99:1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CM, Elston RC. 2009. Finding genes underlying human disease. Clin Genet. 75:101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S. 2000. The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124:1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. 2012. Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol. 158:2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Gaut BS. 2005. Molecular population genetics and the search for adaptive evolution in plants. Mol Biol Evol. 22:506–519. [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, et al. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 38:203–208. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, et al. 2010. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 42:355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. 2012. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 44:821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.sh2mb