Abstract
Activation of G protein-coupled receptors results in multiple waves of signaling that are mediated by heterotrimeric G proteins and the scaffolding proteins β-arrestin 1/2. Ligands can elicit full or subsets of cellular responses, a concept defined as ligand bias or functional selectivity. However, our current understanding of β-arrestin-mediated signaling is still very limited. Here we provide a comprehensive view of β-arrestin-mediated signaling from the cannabinoid 1 receptor (CB1R). By using a signaling biased receptor, we define the cascades, specific receptor kinases, and molecular mechanism underlying β-arrestin-mediated signaling: We identify the interaction kinetics of CB1R and β-arrestin 1 during their endocytic trafficking as directly proportional to its efficacy. Finally, we demonstrate that signaling results in the control of genes clustered around prosurvival and proapoptotic functions among others. Together, these studies constitute a comprehensive description of β-arrestin-mediated signaling from CB1Rs and suggest modulation of receptor endocytic trafficking as a therapeutic approach to control β-arrestin-mediated signaling.
Introduction
Ligand-induced signaling from G protein-coupled receptors (GPCRs) was initially conceptualized as a linear series of sequential steps leading to specific biologic outcomes. Research over the last 20 years has shown that many ligands can differentially stabilize receptors into multiple signaling conformations resulting in pluridimensional efficacies, a concept defined as functional selectivity or biased signaling (Urban et al., 2007; Kenakin, 2011; Shenoy and Lefkowitz, 2011). This complexity at the signaling level has significantly changed our understanding of GPCRs function and provides new challenges and opportunities for drug discovery (Kenakin, 2007; Atwood et al., 2012; Chang and Bruchas, 2014). Upon ligand activation, GPCRs undergo conformational changes leading to activation of heterotrimeric G proteins and their effectors such as adenylyl cyclase among others. These conformations are detected by G protein-coupled receptor kinases (GRKs) and they differentially phosphorylate GPCRs, generating specific patterns or barcodes depending on the ligand (Liggett, 2011; Nobles et al., 2011). These barcodes are subsequently recognized by β-arrestins, which are recruited to the plasma membrane sterically hindering G protein-receptor interactions and terminating the first wave, while initiating the second wave, of receptor signaling (Pierce et al., 2002; Shenoy and Lefkowitz, 2011). More recently a third wave has been described, in which some GPCRs can re-engage in G protein signaling after internalization in specific intracellular compartments (Irannejad and von Zastrow, 2014).
β-Arrestins have two major roles—as negative regulators of heterotrimeric G protein signaling during receptor desensitization and internalization and as signaling scaffolds (Gainetdinov et al., 2004; Schmid and Bohn, 2009; Tzingounis et al., 2010). As negative regulators of receptor activity, β-arrestins block G protein signaling and recruit components of the endocytic machinery to initiate receptor endocytosis (Goodman et al., 1996; Claing et al., 2002). As a signaling scaffold molecule, the focus has been placed on their role during the activation of selected downstream effectors such as mitogen-activated protein kinase (DeFea, 2011). At the mechanistic level, the kinetics of interaction between receptors and β-arrestins during endocytosis have been suggested as a mechanism to control β-arrestin-mediated signaling efficacy (Shenoy et al., 2009; Flores-Otero et al., 2014). In this scenario, ligand activation results in specific phosphorylation barcodes, that control the recruitment and kinetics of receptor-arrestin interactions and β-arrestin-mediated signaling (Shenoy and Lefkowitz, 2011; Reiter et al., 2012). However, our understanding of β-arrestin-mediated signaling is still rudimentary and limited to selected well-studied signaling pathways without much information on their roles or the mechanisms controlling them; yet β-arrestin-mediated pathways have been proposed as therapeutic targets in several disorders (Allen et al., 2011; Gurevich, 2014; Urs et al., 2015).
To delineate the complex mechanisms, physiologic roles, and therapeutic potential of β-arrestin-mediated signaling, a more comprehensive approach including cell networks analysis and transcriptomics is needed (Maudsley et al., 2013). To achieve this goal, we sought to investigate the dynamic mechanisms of β-arrestin-mediated signaling downstream from the cannabinoid 1 receptor (CB1R), one of the most abundant GPCRs in the central nervous system and the target of ∆9-tetrahydrocannabinol, the main psychoactive ingredient in marijuana. Our results distinguish β-arrestin-mediated signaling from G protein signaling, illustrate the pharmacological complexity and mechanisms contributing to this pathway, and propose modulation of the interaction between receptor and β-arrestin as a mechanism to control signaling output. Furthermore, these results suggest that therapeutic drugs developed to control β-arrestin-mediated signaling for the CB1R could present a novel approach to target the long-term effects of CB1R activation. Finally, our work also provides a framework to investigate β-arrestin signaling from other GPCRs and suggests endocytic dwell times as new biomarkers for identifying β-arrestin biased compounds.
Materials and Methods
Cell Culture and Transfections.
Human embryonic kidney (HEK) 293 cells (ATCC, Manassas, VA) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 3.5 mg/ml glucose at 37°C in 5% CO2. SEP-CB1R cDNA in the pcDNA3.1 vector was a generous gift from Andrew Irving (University of Dundee, Scotland, UK) and the SEP-S426A/S430A construct was generated by site-directed mutagenesis (QuickChange, Agilent Technologies, Santa Clara, CA) using the SEP-CB1R construct as a template. The plasmids pcDNA3.1-3xHA-CB1Rs encoding the N-terminal 3xHA epitope-tagged CB1Rs were generated by PCR using the HA-CB1 plasmid (Daigle et al., 2008). For transfection, HEK 293 cells were seeded in a 6-well plate (300,000 cells per well) and transfected with a total of 2 µg of plasmids using lipofectamine (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. Twenty-four hours posttransfection, cells were incubated for an additional 16 hours in serum-free growth media before agonist treatment. siRNA (Qiagen, Valencia, CA) transfection was carried out as previously described (Roman-Vendrell et al., 2012; Flores-Otero et al., 2014). GRK expression was targeted using the following siRNA sequences: GRK2, 5′-CCGGGAGATCTTCGACTCATA-3′; GRK3, 5′-AAGATGTTCAGTGTTGGGTAA-3′; GRK4, 5′-CCGGGTGTTTCAAAGACATCA-3′; GRK5, 5′-AGCGTCATAACTAGAACTGAA-3′, GRK6, 5′-AAGGATGTTCTGGACATTGAA-3′. Silencing of GRK expression was assessed by immunoblotting using anti-GRK2 antibody (1: 2000; Cell Signaling, Cell Signaling Technology, Danvers, MA), anti-GRK3, anti-GRK5, and anti-GRK6 antibodies (1:3000, 1: 6000, and 1:3000, respectively; Abcam, Cambridge, MA) and anti-GRK4 antibody (1: 3000; Sigma-Aldrich, St. Louis, MO).
Immunoblotting Studies.
After serum starvation for 16 hours, cells expressing SEP-CB1R were washed twice and exposed to either 1 μM WIN or 10 μM 2-AG diluted in Dubecco’s modified Eagle’s medium for 5, 10, 15, and 30 minutes at 37°C. To observe the effect of pertussis toxin (PTX) on ERK1/2 phosphorylation, cells were pretreated with 10 ng/ml for 16 hours at 37°C. For gene silencing experiments, siRNA transfection for β-arrestin 1 and β-arrestin 2 was carried out as previously described(Flores-Otero et al., 2014). Cells were then washed with ice-cold phosphate-buffered saline and lysed in ice-cold lysis buffer consisting of 150 mM NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 7.5, and a protease inhibitor cocktail 4-(2-aminoethyl)benzenesulfonyl fluoride, pepstatin A, E-64, bestatin, leupeptin, and aprotinin; Sigma-Aldrich). Cell lysates were cleared by centrifugation at 18,500 g for 15 minutes at 4°C. 13 μg of total proteins were resolved by SDS-PAGE gel electrophoresis in 10% gels and transferred onto polyvinylidene fluoride membrane. After incubating with blocking reagent (Fisher Scientific, Pittsburgh, PA), the membrane was incubated for 1 hour at room temperature with the primary antibody (1:4000 phospho-p44/42 and p44/42 antibodies; Cell Signaling Technology, Danvers, MA). After incubation with anti-rabbit peroxidase-conjugated secondary antibody (1:6000; Cell Signaling Technology) for 50 minutes at room temperature, the specific immunoreactivity was visualized using the SuperSignal West Femto Chemiluminescent Substrate System (Thermo Fisher Scientific, Rockford, IL). Immunoreactive bands of phospho-ERK1/2 were quantified by densitometric analysis using the ImageJ program (http://rsb.info.nih.gov/ij/) and normalized to the intensity of total-ERK1/2. Data are expressed as a fold increase above the basal level of phosphorylation.
Coimmunoprecipitation.
HEK293 cells were cotransfected with HA-CB1R and either β-arrestin 1-RFP or β-arrestin 2-RFP and treated with WIN and 2-AG as described above. Cells were lysed in a buffer containing 1% n-dodecyl-β-d-maltoside, 10% glycerol, 250 mM NaCl, 50 mM Tris (pH 8), 0.5 mM EDTA, and protease inhibitor cocktail (Sigma-Aldrich). The precleared supernatant fraction (approximately 400 μg) was incubated overnight at 4°C with 2 μg of anti-HA antibody (Roche, Indianapolis, IN) and Protein A/G Plus-Agarose (Santa Cruz Biotechnology, Dallas, TX). The beads were washed four times in lysis buffer and elution was performed in 40 μl of reducing 1× Laemmli buffer at 37°C for 30 minutes. Samples were separated by SDS-PAGE gel electrophoresis in 10% gels and subjected to Western blotting using anti-RFP and anti-HA antibodies as described above.
Total Surface Fluorescence.
Receptor expression levels were compared between wild-type and mutant receptors by total surface fluorescence. HEK293 cells transiently expressing SEP-CB1R or SEP-CB1R S426A/S430A 72 hours after transfection were analyzed with a Cellometer Vision from Nexcelom Bioscience (Lawrence, MA) following manufacturer protocol (Chan et al., 2011). Briefly, cells were harvested and centrifuged for 5 minutes at 1500 rpm. Cells were resuspended in phosphate-buffered saline and transferred to imaging chambers. Captured brightfield and fluorescence images using the GFP filter set (optic module 1: VB535-402) were saved and analyzed using the equipment software. Acquisition exposure was set to 7.5 seconds.
TIRF and Confocal Microscopy.
TIRF microscopy was performed as previously described(Yudowski and von Zastrow, 2011; Roman-Vendrell and Yudowski, 2015). Briefly, HEK293 cells transiently expressing SEP-CB1R or SEP-CB1R S426A/S430A were imaged using a Motorized Nikon Ti-E inverted microscope with a CFI-Apo ×100 1.49 oil TIRF objective lens and a motorized stage with perfect focus (Melville, NY). Light sources were 488- and 561-nm Coherent sapphire lasers (Coherent Inc., Santa Clara, CA) with 50 and 100 mW, respectively. The microscope was coupled to an iXonEM + DU897 back illuminated EMCCD camera (Andor, Belfast, UK). Imaging settings were kept constant throughout our imaging: readout speed: 10 Hz, exposure time: 100 ms every 3 seconds, EM gain 300, binning: 1 × 1, image: 512 × 512, BitDepth = 14 bits, temperature: −75, and laser power: 10%. Cells were kept at 37°C with a Stable Z stage and objective warmer (Bioptechs, Butler, PA). Cells were gently rinsed three times with OptiMem supplemented with 20 mM HEPES (Life Technologies) and kept in the incubator for 10–30 minutes to acclimate before imaging. TIRF microscopy recording was conducted in the same imaging media for 1–3 minutes under basal condition (without any treatment) and was followed by bath application of selected ligand using a custom-built perfusion chamber as previously described (Flores-Otero et al., 2014; Roman-Vendrell and Yudowski, 2015). Total time of live-imaging visualization and recording was less than 30 minutes. Confocal microscopy was performed using a Zeiss LSM 5 Pascal laser-scanning confocal microscope equipped with Ar 488 nm and HeNe 543 nm laser-lines using a 63x Plan-APOCHROMAT oil-immersion objective (NA = 1.4) with the following emission filters: BP 505-530 and BP 560-615, respectively. Images were collected at 1024 × 1024 pixel resolution using sequential scanning mode.
Image Processing and Analysis.
Analysis was performed using the public domain National Institutes of Health Image program ImageJ/FIJI software, which is freely available at http://fiji.sc/Fiji as described before(Yudowski and von Zastrow, 2011; Flores-Otero et al., 2014; Roman-Vendrell et al., 2014). Briefly, raw images were first background-subtracted and flat field-corrected. Individual endocytic events were quantified by an observer blinded to experimental details multiple times manually and using the particle tracking algorithm two-dimensional spot tracker. Event location, time, and fluorescence profile were logged and recorded. Individual endocytic events were identified and scored according to the following criteria: 1) individual events appeared and disappeared within the time series; 2) endocytic events displayed limited movement in the x and y axes as described for clathrin endocytic pits during their maturation; and 3) the events did not collide or merge with other structures. Dwell times were calculated as the time between the first frame where spot tracker detected an event above background fluorescence levels and the last. Because the fluorescence from individual events can fluctuate and the algorithm from the tracking software can misinterpret endocytic events, we manually verify all individual events after automated analysis. To analyze statistical significance between groups, we counted the number of events in each independent experiment (i.e., each separate imaging session and different dish of cultured cells were treated as independent experiments), analyzed their normality by D’Agostino and Pearson test, and used unpaired two-tailed Student’s t tests to test for statistical significance. All data are expressed as means ± S.E.M. unless stated. Statistical analyses between dwell times were calculated using the GraphPad Prism Software (La Jolla, CA). Box and whiskers plot represent minimum and maximum values, the box extends from 25 to 75% with the mean value.
Phospho-Kinase Array Analysis.
The phosphor-kinase array was performed using the Proteome Profiler Human Phospho-Kinase Array Kit (R&D Systems, Minneapolis, MN). Briefly, HEK293 cells expressing the CB1R wild-type or S426/430A mutant receptor at similar expression levels (Supplemental Fig. 1) were treated with to either 1 μM WIN or 10 μM 2-AG for 5 and 15 minutes. PTX treatment and siRNA transfection for β-arrestins were carried out as described above. Cells were lysed with lysis buffer (R&D Systems, Minneapolis, MN) and agitated for 30 minutes at 4°C. Cell lysates were subjected to protein assay. Preblocked nitrocellulose membranes spotted with antibodies for 43 kinases were incubated with 400 μg of the lysates overnight at 4°C on a rocking platform. The membranes were incubated with a biotinylated detection antibody cocktail and then streptavidin-horseradish peroxidase. Chemiluminescent detection reagents were applied to detect spot densities. Array images were analyzed quantified by densitometric analysis using the ImageJ program (http://rsb.info.nih.gov/ij/). Every spot was subtracted by the averaged background level from negative control spots. The phospho-antibody array experiment was repeated three times. Data of duplicated spots from three array results were expressed with the relative fold change over the basal level (in the absence of agonist). Statistical analysis was performed using one-way analysis of variance followed by Bonferroni’s post hoc test.
RNA-seq Experiments and Transcriptome Analysis.
HEK293 cells expressing the HA-tagged rat wild-type or S426/430A mutant receptors were treated with 1 μM WIN for 2 hours. After extensive washing, total RNA was isolated using TRIzol reagent (Life Technologies). The RNA samples were further cleaned up using RNeasy Mini Kit (Qiagen), with purity and quantity assessed spectrophotometrically. RNA was polyA enriched and libraries constructed. Sequencing (50 nt, single strand) was performed using the Illumina HiSeq2000 sequencing system (CGB, Indiana University). Each sample gave ∼10,000,000 reads/sample with >90% of reads uniquely mapped using Tophat2 ver 2.0.10 and using GRCh38 as the reference genome. Differential expression was examined using DESeq2 (Bioconductor) at a 5% false discovery rate.
Results
β-Arrestin-Mediated Signaling Is Enhanced in the CB1R S426A/S430A Receptor.
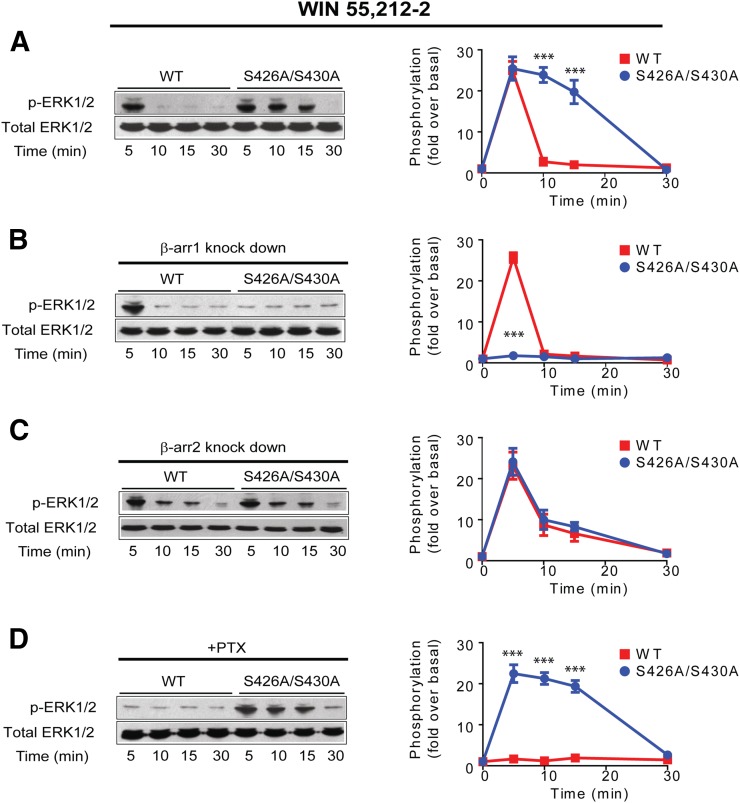
To define the signaling cascades mediated by β-arrestins downstream from the CB1R, we took advantage of the wild-type and S426A/S430A receptors. These serines have been shown to be phosphorylated in previous mass spectrographic analysis, and our previous work revealed that mutation of the putative GRK phosphorylation sites S426A/S430A to alanines resulted in lower levels of receptor internalization, reduced desensitization, and persistent activation of ERK1/2 independently from Gi/Go, suggestive of a β-arrestin-mediated pathway (Daigle et al., 2008; Huttlin et al., 2010; Wiśniewski et al., 2010; Trinidad et al., 2012; Morgan et al., 2014). This persistent ERK1/2 activation was identical to the response we observed when prolonging receptor-arrestin interactions at the endocytic pit, a mechanism controlling β-arrestin signaling (Flores-Otero et al., 2014). Sustained ERK1/2 activation by the S426A/S430A receptor led us to the hypothesis that signaling mediated by β-arrestins could be enhanced in this receptor and used as a tool to investigate this type of signaling. To test this hypothesis, first we analyzed ERK1/2 phosphorylation in HEK293 cells expressing the wild-type CB1R or S426A/S430A receptor exposed to maximal concentrations of the synthetic CB1R agonist WIN 55,212-2 (WIN) (Fig. 1A). Second, we assessed the role of β-arrestins during the sustained phosphorylation of ERK1/2 by using small interfering RNA (siRNA) against β-arrestin 1 and 2. Silencing β-arrestin 1 resulted in a complete reduction of ERK1/2 phosphorylation for the S426A/S430A receptor, whereas no significant effect was observed for the wild-type receptor. The effectiveness of siRNAs for β-arrestins 1 and 2 was confirmed by Western blots (Supplemental Fig. 2). In contrast, reduction of β-arrestin 2 expression did not show a substantial effect at 5 minutes (Fig. 1, B and C). Interestingly, however, both wild-type and S426A/S430A receptors show a smaller but sustained level of ERK1/2 phosphorylation at the later time points (10 and 15 minutes). Collectively, our data suggest that β-arrestin 2 may not have a primary role in ERK1/2 signaling. Rather, as our previous studies indicate, β-arrestin 2 is critical for receptor internalization, and its removal may impair receptor internalization (Ahn et al., 2013; Flores-Otero et al., 2014). The effectiveness of siRNAs for β-arrestins 1 and 2 was confirmed by Western blots (Supplemental Fig. 2). Finally, consistent with previous findings, PTX completely blocked ERK1/2 phosphorylation for the wild-type receptor (Daigle et al., 2008; Flores-Otero et al., 2014), although it had no effect on the mutant receptor (Fig. 1D). Together these results suggest that WIN induced prolonged activation of ERK1/2 in the mutant receptor that is solely mediated by β-arrestin 1.
Fig. 1.
WIN induced signaling from CB1R S426A/S430A is biased to β-arrestins. (A) HEK293 cells expressing SEP-CB1Rs or S426A/S430A were exposed to 1 μM WIN 55212-2 for 5, 10, 15, and 30 minutes. Cell lysates were analyzed using Western blots with phospho-ERK1/2 (p-ERK1/2, top panel) or total ERK1/2 (bottom panel). Representative Western blot images and analysis of ERK1/2 phosphorylation are shown. Time course showing ERK1/2 phosphorylation levels in the wild-type (red) and S426A/S430A (blue) receptors. (B) HEK293 cells expressing SEP-CB1Rs or the S426A/S430A receptor were cotransfected with β-arrestin 1 siRNA and exposed to WIN. The quantified time course shows complete abrogation of signal in the mutant receptor. (C) HEK293 cells expressing SEP-CB1Rs or S426A/S430A were cotransfected with β-arrestin 2 siRNA and exposed to WIN. Graph provide quantified time course for the wild-type and mutant receptor. (D) HEK293 cells expressing SEP-CB1Rs or S426A/S430A were pretreated with PTX (16 hours). Quantified time course indicates complete inhibition of phosphorylation in the wild-type receptor. Data represent the mean ± S.E.M. of at least three independent experiments. Statistically significant differences between the wild-type and mutant receptor were assessed using one-way analysis of variance and Bonferroni’s post hoc test. ***P < 0.005.
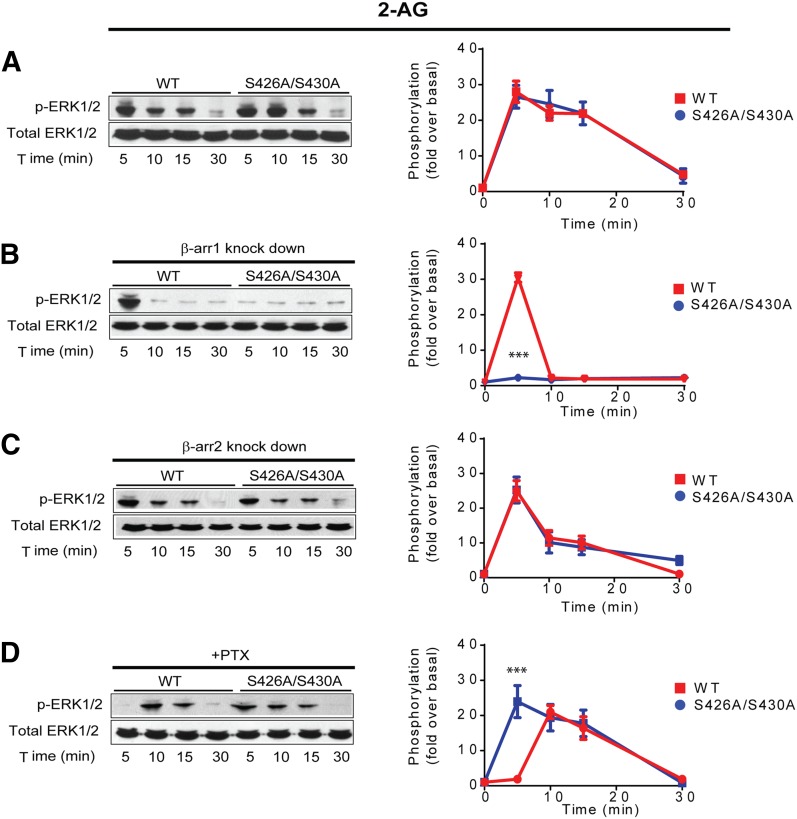
To further characterize signaling from the S426A/S430A receptor, we evaluated the phosphorylation of ERK1/2 elicited by the endogenous CB1R agonist 2-AG. Because we previously reported for cells expressing the wild-type receptor (Flores-Otero et al., 2014), 10 μM 2-AG induced a peak at 5 minutes followed by a slow decay in ERK1/2 phosphorylation. A similar response was observed with the S426A/S430A receptor (Fig. 2A). However, only in the latter case, silencing of β-arrestin isoform 1 and not 2 resulted in a complete reduction of ERK1/2 phosphorylation at 5 minutes (Fig. 2, B and C). On the other hand, preincubation with PTX fully abrogated the peak at 5 minutes in the wild-type receptor, but had no effect on the S426A/S430A receptor (Fig. 2D). Interestingly, removal of β-arrestin 2 also increased β-arrestin 1 signaling as in Fig. 1. These results together with data from WIN (Fig. 1) support the idea that activation of the S426A/S430A receptor leads to increased β-arrestin 1-mediated signaling, probably by reducing β-arrestin 2-induced internalization.
Fig. 2.
2-AG induced signaling from CB1R S426/430 is biased to β-arrestins. (A) HEK293 cells expressing SEP-CB1Rs or the S426A/S430A receptor were exposed to 10 μM 2-AG for 5, 10, 15, and 30 minutes. Cell lysates were analyzed using Western blots with phospho-ERK1/2 (p-ERK1/2, top panel) or total ERK1/2 (bottom panel). Representative Western blot images and analysis to quantify ERK1/2 phosphorylation are shown. Time course showing ERK1/2 phosphorylation levels in the wild-type (red) and S426A/S430A (blue) receptor. (B) HEK293 cells expressing SEP-CB1Rs and S426A/S430A were cotransfected with β-arrestin 1 siRNA and exposed to 2-AG as indicated. Quantified time course shows complete abrogation of signal in the mutant receptor. (C) HEK293 cells expressing SEP-CB1Rs or the S426A/S430A receptor were cotransfected with β-arrestin 2 siRNA and exposed to WIN. Graphs provide quantified time courses for the wild-type and mutant receptor. (D) HEK293 cells expressing SEP-CB1Rs or the S426A/S430A receptor were pretreated with PTX (16 hours). Quantified time courses indicate inhibition of phosphorylation only at 5 minutes in the wild-type receptor. Data represent the mean ± S.E.M. of at least three independent experiments. Statistically significant differences between the wild-type and mutant at each time point were assessed using one-way analysis of variance and Bonferroni’s post hoc test. ***P < 0.005.
The Kinome Downstream from CB1R/β-Arrestins.
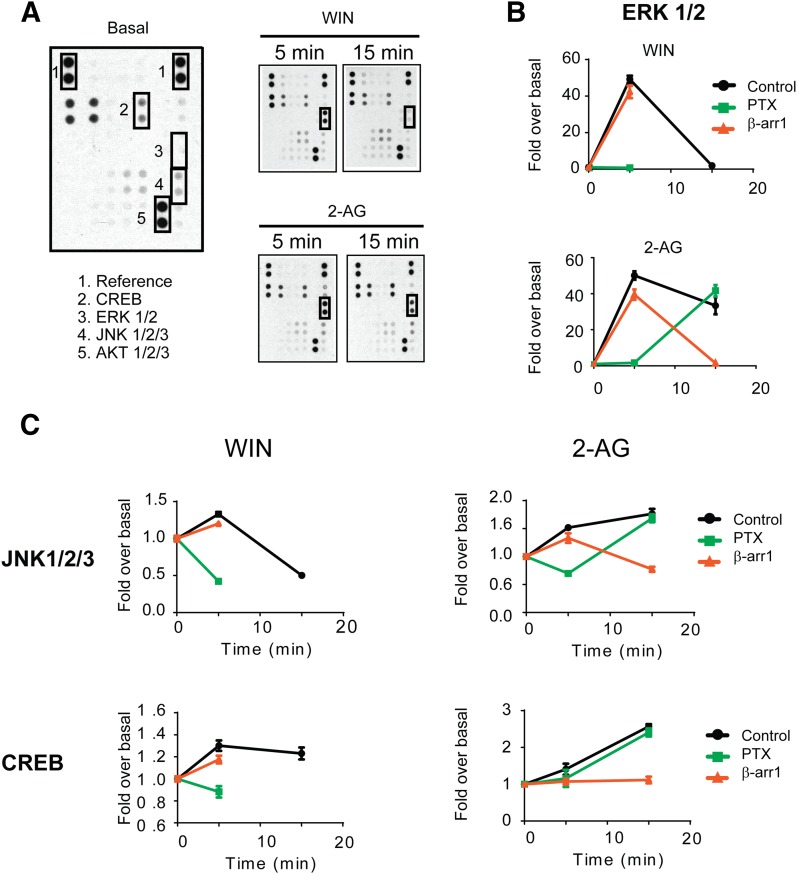
Beta-arrestins have been shown to coordinate multiple signaling networks downstream from many GPCRs (Maudsley et al., 2013). To identify additional pathways regulated by β-arrestins downstream from CB1Rs we applied an unbiased screen to detect the phosphorylation of kinases upon CB1R activation. We used a human phospho-kinase antibody array to simultaneously identify the phosphorylation of 43 different kinases and two related proteins, including ERK and CREB, whose activation was previously shown to be mediated by CB1Rs (Flores-Otero et al., 2014; Laprairie et al., 2014). Cells expressing the wild-type receptor were incubated with 10 μM 2-AG for 5 and 15 minutes (Fig. 3A). The effects of siRNA against β-arrestin 1 and preincubation with PTX were also evaluated to resolve pathways from β-arrestin 1 and Gi/Go proteins, respectively (Supplemental Fig. 3). Consistent with Fig. 2 and our prior findings (Flores-Otero et al., 2014), ERK1/2 phosphorylation elicited by 2-AG was mediated by PTX-sensitive G proteins at 5 minutes and exclusively by β-arrestin 1 at 15 minutes (Fig. 3, A and B). Significant increases in phosphorylation levels upon 2-AG treatment were also observed in JNK1/2/3 and CREB, whereas there is a decrease in the phosphorylation of HSP60. The levels of CREB and HSP60 phosphorylation were specifically affected by β-arrestin 1 siRNA and PTX treatment, respectively (Fig. 3C and Supplemental Fig. 3). In contrast, JNK1/2/3 phosphorylation was abolished by PTX treatment and β-arrestin 1 siRNA at 5 and 15 minutes, respectively. Interestingly, these pathways were previously described as regulated by CB1Rs, further validating our approach, but the involvement of β-arrestins was previously unknown (Rueda et al., 2000; Derkinderen et al., 2001; Hart et al., 2004). These results suggest that prolonged stimulation of CB1Rs with the endocannabinoid 2-AG leads to a β-arrestin 1-dependent activation of multiple signaling pathways. We did not observe changes in p-AKT 1/2/3 as previously reported (Laprairie et al., 2014), although phosphorylation levels were significantly elevated under basal conditions, possibly masking any changes (Figs. 3A and 4A). These differences could also be explained by quantitative relationship between receptors and signaling proteins or sensitivity issues in our detection methods. For the wild-type receptor upon WIN treatment, results were comparable to those of 2-AG treatment of 5 minutes (Fig. 3, B and C). In contrast, no further phosphorylation of these kinases was observed at 15 minutes.
Fig. 3.
Signaling networks elicited from CB1R activation. (A) Representative dot blots evaluated by profiling phosphorylation of 43 human kinases. HEK293 cells expressing SEP-CB1R were incubated with 1 μM WIN and 10 μM 2-AG for 5 and 15 minutes. Cell lysates from untreated and treated cells were applied to a nitrocellulose membrane spotted with the antibodies for 43 kinases. Three kinases (CREB, ERK1/2, and JNK1/2/3) with significant changes in phosphorylation level after agonist treatment were highlighted with boxes. AKT1/2/3 and reference spots (loading control) are shown for comparison. (B) Graphs provide the quantified time course showing ERK1/2 phosphorylation induced by 1 μM WIN and 10 μM 2-AG. Data are expressed as the fold change over the basal (no compound) level for each compound. The effects of β-arrestin 1 knockdown (red) and PTX treatment (green) are shown. (C) Graphs provide the quantified time course showing JNK1/2/3 and CREB phosphorylation induced by 1 μM WIN and 10 μM 2-AG. Data are expressed as the fold change over the basal level for each compound. Statistical significance of agonist-induced phosphorylation compared with basal (0 minute) was assessed using one-way analysis of variance and Bonferroni’s post hoc test.
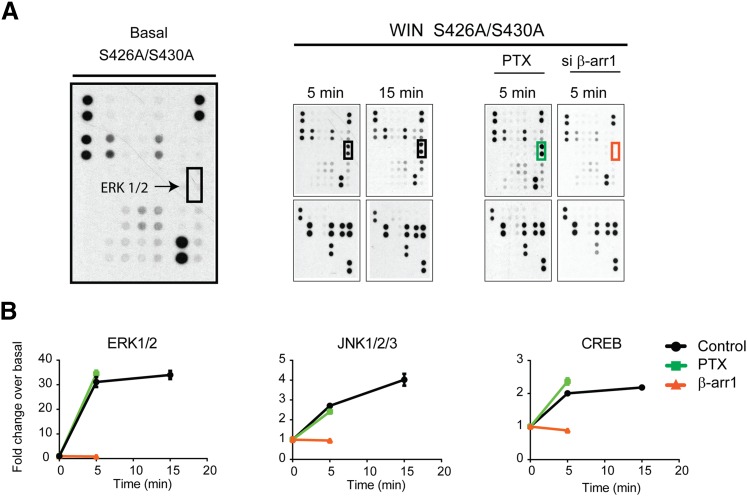
Fig. 4.
CB1R S426A/S430A is a β-arrestin 1 biased receptor. (A) Representative dot blots evaluated by profiling phosphorylation of 43 human kinases. Lysates from untreated and agonist-treated (1 μM WIN for 5 and 15 minutes) cells expressing the S426A/S430A receptor were applied to a nitrocellulose membrane spotted with antibodies for 43 kinases along with control antibodies. ERK1/2 phosphorylation levels were highlighted with boxes. The effects of β-arrestin 1 knockdown (red) and PTX treatment (green) are shown. (B) Graphs provide the quantified time course showing ERK1/2, JNK1/2/3, and CREB phosphorylation induced by 1 μM WIN. Data are expressed as the fold change over basal level for each compound. The effect of β-arrestin 1 knockdown (red) and PTX treatments are included. Statistical significance of the differences compared with basal (0 minute) was assessed using one-way analysis of variance and Bonferroni’s post hoc test.
Next, because β-arrestin-mediated signaling is enhanced in the S426A/S430A receptor, we used the phospho-kinase antibody array to investigate the pathways downstream from CB1R/β-arrestin 1 (Fig. 4A). Although phosphorylation levels for kinases were similar to those described for the wild-type receptor exposed to WIN at 5 minutes (Fig. 3B), key differences were identified. For example, unlike the wild-type receptor, ERK1/2 phosphorylation for the mutant receptor at 5 minutes is β-arrestin 1 mediated, and the phosphorylation level at 15 minutes remained increased and all were dependent on β-arrestin 1 not Gi/Go proteins (Fig. 4, A and B). Significant increases in phosphorylation upon WIN treatment were also observed for JNK1/2/3, CREB, and EGFR (Fig. 4B and Supplemental Fig. 3). These were specifically abolished by β-arrestin 1 siRNA, further suggesting that the mutation S426A/S430A resulted in enhanced β-arrestin-mediated signaling. Taken together these results indicate that this mutant receptor is an ideal tool to investigate β-arrestin-mediated signaling.
The Differential Interaction between Receptor and β-Arrestin Isoforms.
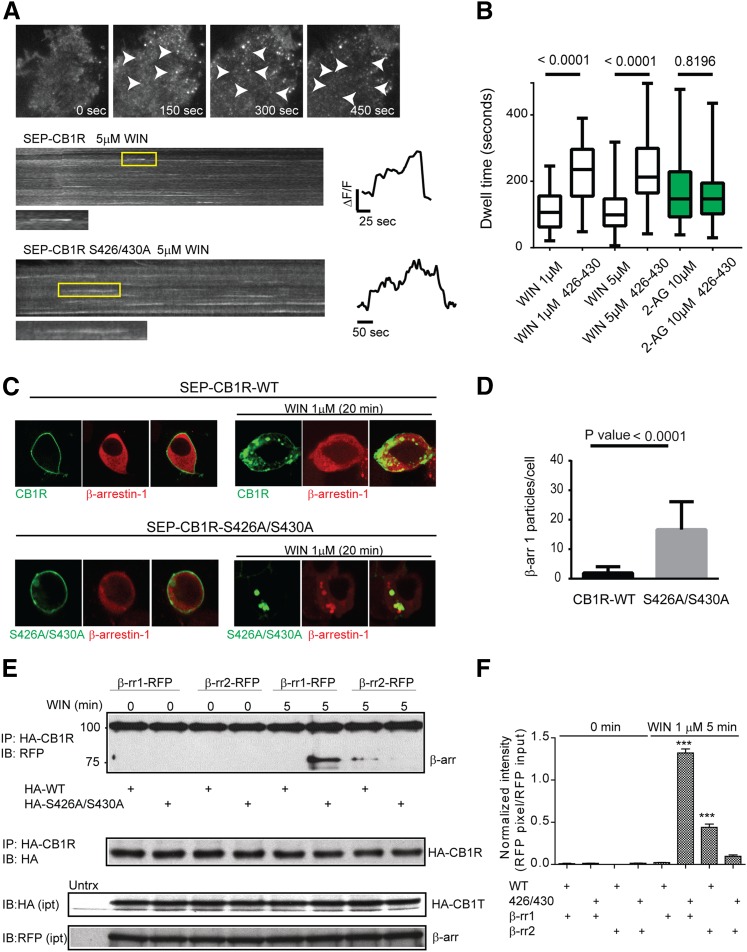
Because activation of the S426A/S430A receptor resulted in enhanced β-arrestin, we used this receptor to investigate the molecular mechanisms by which receptor activation translates into β-arrestin-mediated signaling. Previously, we proposed ligand-specific endocytic dwell times, the time during which receptors and β-arrestins are clustered at the cell surface during the endocytic process, as a possible mechanism modulating β-arrestin-mediated signaling (Flores-Otero et al., 2014). This work suggested that ligands inducing prolonged dwell times (>140 seconds) were more efficacious at β-arrestin-mediated signaling than ligands inducing short dwell times (<120 seconds). Here we sought to compare the endocytic dwell times of the wild-type and the mutant S426A/S430A receptor in the presence of WIN and 2-AG using total internal reflection microscopy (TIRFM) (Fig. 5A). Upon treatment, receptors clustered into individual endocytic events as previously described (Fig. 5A, top). By analyzing individual endocytic events, we found that dwell times of the S426A/S430A mutant receptor were prolonged compared with wild-type receptors in the presence of 5 μM WIN (Fig. 5A, kymographs and individual traces). Analysis of multiple endocytic dwell times indicated that mutation of S426A/S430A significantly prolonged dwell times elicited by 1 and 5 μM WIN (Fig. 5B), supporting the enhanced β-arrestin mediated signaling observed with the mutant receptor. However, no changes were observed in the wild-type versus the mutant in the presence of 10 μM 2-AG (Fig. 5B). These prolonged dwell times correlate and support their role as a predictor of β-arrestin-mediated signaling (Flores-Otero et al., 2014). Next, we investigated if the prolonged interaction between S426A/S430A receptors and β-arrestins during the endocytic process at the cell surface extends to intracellular compartments. HEK293 cells expressing the wild-type or mutant receptors were transfected with β-arrestin 1-RFP, incubated with 1 μM WIN for 20 minutes and imaged by live-cell confocal microscopy. After 20 minutes, wild-type receptors were internalized and localized into intracellular vesicles, whereas β-arrestin 1 was homogenously distributed in the cytoplasm (Fig. 5C, middle). Interestingly, S426A/S430A receptors and β-arrestin 1 were highly localized in intracellular clusters (Fig. 5 C, bottom). These clusters were frequently observed with the S426A/S430A receptor after WIN treatment but not with the wild-type receptor (Fig. 5D). To test the idea that the interaction between β-arrestin 1 and the mutant receptor is enhanced, we performed coimmunoprecipitations. β-Arrestins are notoriously difficult to immunoprecipitate; however, we observed a major band corresponding to β-arrestin 1-RFP when the S426A/S430A receptor was immunoprecipitated after incubation with 1 μM WIN (Fig. 5E). In marked contrast, no β-arrestin 1 band was observed upon WIN treatment of the wild-type receptor. We observed a weaker band for RFP-β-arrestin 2 after 1 μM WIN with immunoprecipitation of either the wild-type or the mutant receptors though the level of the latter was substantially reduced (Fig. 5F). Finally, we investigated the recruitment kinetics of β-arrestins to the cell surface in cells expressing either CB1R or S426A/S430A by TIRFM (Supplemental Fig. 4). Interestingly, β-arrestin 2 recruitment to the plasma membrane was severely impaired in the mutant receptor, strongly supporting the roles of S426/430 on β-arrestin 2 recruitment (Supplemental Fig. 4C). However, no significant difference was observed on the recruitment kinetics of β-arrestin 1 (Supplemental Fig. 4B)
Fig. 5.
CB1R S426A/S430A interacts with β-arrestin 1 more strongly than CB1R. (A) Total internal reflection images showing HEK293 cell expressing SEP-CB1Rs before and after 1 μM WIN (time indicated in seconds) showing individual endocytic events (arrowheads). Kymographs from HEK293 cell expressing either SEP-CB1Rs (top panel) or S426A/S430A (bottom panel) in the presence of 1 μM WIN. Individual endocytic events are indicated by the yellow rectangle. Intensity measurements from indicated events are represented to the right. (B) Box and whiskers plot (mean values with min/max range) from endocytic dwell times were analyzed for the indicated concentrations and receptors (n = 10-19 cells). (C) Live HEK293 cells expressing either SEP-CB1Rs (top panel) or SEP-CB1R S426A/S430A (bottom panel) were imaged by confocal before and after 20 minutes incubation with 1 μM WIN. (D) Intracellular β-arrestin 1 particles were quantified after treatments (n = 7–10 cells). (E) HEK293 cells were cotransfected with HA-CB1R and either β-arrestin 1-RFP or β-arrestin 2-RFP and treated with 1 μM WIN for 5 minutes. Lysates were immunoprecipitated with anti-HA antibody and subjected to immunoblot analysis using anti-RFP and anti-HA antibodies. Representative blot images show substantial β-arrestin 1 pull-down was observed with the HA-S426A/S430A mutant receptor, but not the HA-wild-type receptor after 5-minute treatment. The lower panels show the input levels of HA-CB1Rs and β-arrestins-RFP, respectively. (F) The bar graph provides quantification of β-arrestin-RFP shown in (E). Data are the mean ± S.E.M. from three independent experiments. The statistical significance of the differences compared with control (0 minute) was assessed using one-way analysis of variance and Bonferroni’s post hoc test. ***p < 0.001.
Taken together these results indicate that strong physical interactions between the receptor and β-arrestin 1 correlate with β-arrestin-mediated signaling. This interaction is initiated at the cell surface (i.e., prolonged dwell times) and continues after internalization in intracellular compartments, suggesting prolonged interaction as a mechanism to engage sustained β-arrestin-mediated signaling.
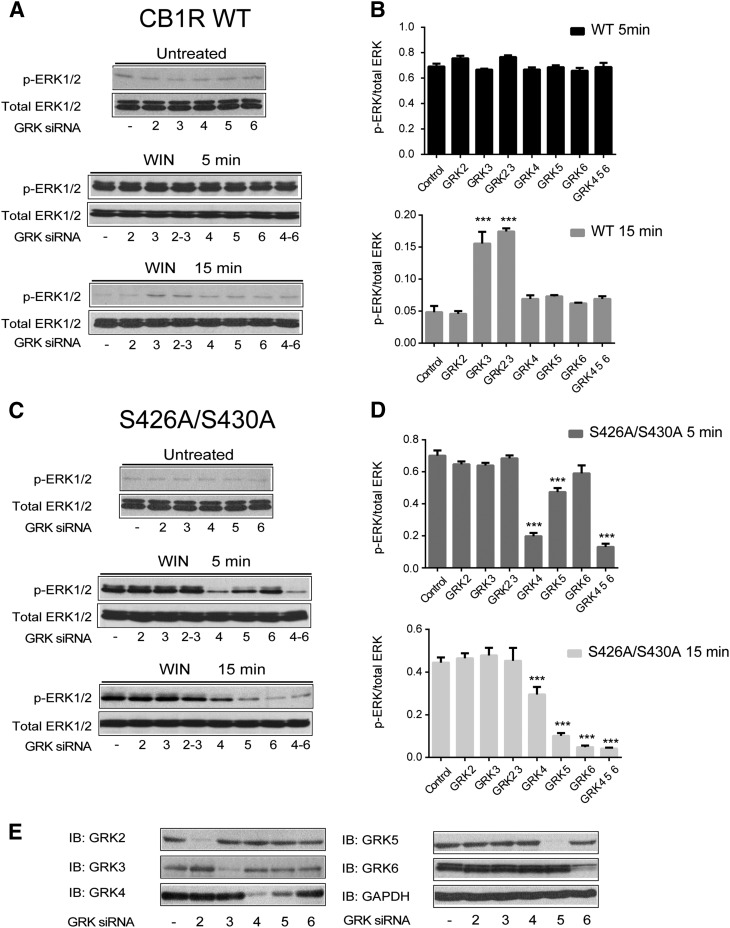
Specific GRK Isoforms Are Responsible for β-Arrestin-Mediated Signaling from the CB1R.
GRKs have been proposed to recognize different ligand-induced receptor conformations and generate phosphorylation barcodes that lead to G protein- or β-arrestin-mediated signaling (Zidar et al., 2009; Liggett, 2011; Nobles et al., 2011, 2012). To examine the mechanisms controlling β-arrestin-mediated signaling, we sought to investigate the roles of specific GRKs in the signaling of the wild-type and mutant receptor. We used siRNA technology to specifically remove GRK isoforms 2–6 and investigated ligand induced ERK1/2 phosphorylation at 5 and 15 minutes by the two receptors (Fig. 6). Consistent with Figs. 1 and 3, incubation with WIN induced robust ERK1/2 phosphorylation at 5 minutes in the wild-type receptor. This phosphorylation was independent of GRKs because siRNAs had no effect, consistent with a G protein-mediated mechanism (Bouaboula et al., 1995; Howlett, 2005) (Fig. 6, A and B). However, when GRK3 was knocked down, a small but significant increase in ERK1/2 phosphorylation was observed for the wild-type receptor upon WIN treatment of 15 minutes (Fig. 6, A and B, bottom). These data are consistent with results suggesting that GRK3 may play a key role in CB1R internalization, and thus in its absence, the receptor remains at the cell surface signaling slightly longer via G protein (Jin et al., 1999). Interestingly, in cells expressing the S426A/S430A receptor, removal of GRK 4 or 5 significantly reduced ERK activation at 5 minutes and the knockdown of GRKs 5 and 6 reduced ERK1/2 phosphorylation further at 15 minutes (Fig. 6C). These results indicate that GRKs are not involved in the phosphorylation of ERK1/2 at 5 minutes in the wild-type receptor but control phosphorylation at 5 and 15 minutes in the S426A/S430A receptor, strongly suggesting a β-arrestin-dependent mechanism at these time points. To control for the effectiveness of each siRNA, we analyzed the expression levels of endogenously expressed GRKs in HEK293 cells (Fig. 6E). Taken together the reductions in phosphorylation in the mutant receptor strengthen the barcode hypothesis where specific GRKs mediate β-arrestin-mediated signaling (GRKs 4, 5, 6, but not 3) versus G protein activation, even when other GRKs are present in HEK293 cells (Atwood et al., 2011) and indicate that β-arrestin-mediated signaling is controlled by a specific subsets of GRKs.
Fig. 6.
GRK subtype specific regulation of β-arrestin 1 signaling. (A–D). Cells expressing either the wild-type (A) or S426A/S430A (C) receptors were cotransfected with control, GRK2, GRK3, GRK4, GRK5, or GRK6 siRNAs as described in Materials and Methods. After exposure to 1 μM WIN for 5 and 15 minutes, cells were lysed and the lysates were separated by SDS-PAGE and blotted with anti-phospho-ERK1/2 and anti-total ERK1/2 antibodies. Representative Western blot images are shown. Graphs provide the quantified levels of ERK1/2 phosphorylation for the wild-type (B) and S416/430A (D) receptors. The statistical significance of the differences compared with control was assessed using one-way analysis of variance and Bonferroni’s post hoc test. ***P < 0.001. (E) Representative Western blot bands demonstrates specific knockdown of GRKs endogenously expressed in HEK293 cells.
Genes Specifically Controlled by β-Arrestin-Mediated Signaling.
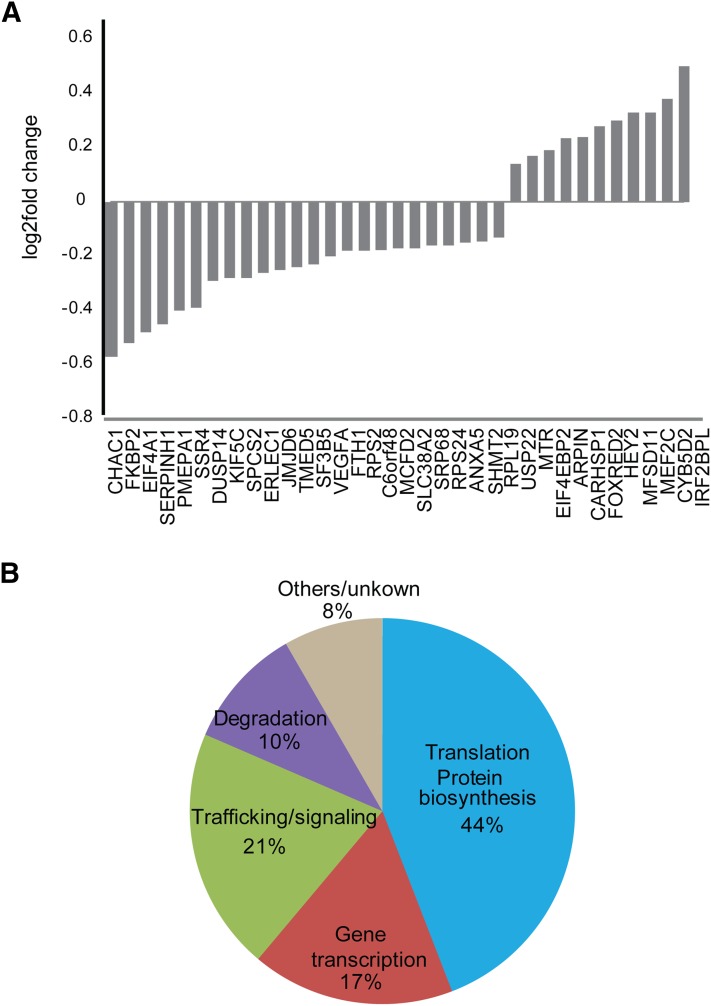
β-Arrestin mediated signaling has been shown to regulate protein synthesis and gene transcription (DeWire et al., 2008; Maudsley et al., 2015). Interestingly, activation of the δ opioid receptor has been shown to translocate β-arrestin 1 to the nucleus and affect histone modification and gene transcription (Kang et al., 2005). We took advantage of the enhanced β-arrestin signaling of the S426A/S430A receptor to explore the transcriptional profile of β-arrestin-mediated signaling from the CB1R. We extracted mRNA and performed RNA sequencing from HEK293 cells expressing these receptors with and without 1 μM WIN treatment of 2 hours. We examined differentially expressed genes using a false discovery rate of 5% to compare the activation by WIN of the mutant S426A/S430A receptor versus activation of the wild-type receptor (Fig. 7A and Supplemental Figs. 6 and 7). Several differentially regulated genes reflected activation of CB1R and induction of kinase stimulation as described in Supplemental Figs. 6 and 7, further validating our results. The phosphatases DUSP1, 5, and 16 are all significantly upregulated in S426A/S430A cells after prolonged treatment with WIN (Supplemental Fig. 6). These phosphatases play roles as negative regulation of ERK, JNK, and P38 and suggest crosstalk downstream from β-arrestin-mediated signaling and the dampening MAP kinase- and cAMP-dependent pathways.
Fig. 7.
Transcripts selectively regulated by β-arrestin 1 signaling. HEK293 cells expressing either the wild-type or S426A/S430A receptors were stimulated with 1 μM WIN for 2 hours. RNA was isolated and sequenced and results analyzed as described in Materials and Methods. (A) Transcripts that were differentially expressed in S426A/S430A expressing cells treated with WIN. Shown as log2 fold change. (B) Varied function of the differentially regulated genes. The majority of differentially regulated genes are involved in transcription and protein synthesis or degradation.
Interestingly, the E3 ubiquitin ligase MDM2, which regulates ubiquitination of β-arrestin 2 and signaling from the β2 adrenergic receptor (Shenoy et al., 2009) was also upregulated selectively in S426A/S430A expressing cells (Supplemental Fig. 6). Finally, we investigated the transcripts specifically regulated by WIN in the mutant receptor versus the wild-type receptor (Fig. 7A and Supplemental Fig. 7). From this list, ∼70% of the genes are involved in gene regulation, mRNA processing, and protein translation and degradation (Fig. 7 B), underlining the significant role of β-arrestin-mediated signaling in the long-term effects of CB1R activation. Remarkably, some of the genes downstream of β-arrestin include VEGFA, GH1, and ADAMTS1, which have been involved in cancer growth and neurodegeneration.
Discussion
Our understanding of GPCR signaling has dramatically changed over the last 15 years. Initially described as on and off switches, GPCRs are now likened to microprocessors where their activation can lead to multiple active states and cellular responses (Kenakin, 2006, 2011). This current understanding of GPCR pharmacology is integrated into the concepts of functional selectivity and biased signaling (Urban et al., 2007; Kenakin, 2011). In this paradigm, ligands can be biased to activate selected signaling pathways from the full signaling repertoire available to individual GPCRs. G protein-dependent pathways have been extensively described from the functional to the structural level (Pierce et al., 2002; Rosenbaum et al., 2009; Venkatakrishnan et al., 2013). However, despite its immense therapeutic potential, our general understanding of β-arrestin-mediated signaling is currently very limited (Shenoy and Lefkowitz, 2011; Maudsley et al., 2013; Venkatakrishnan et al., 2013; Srivastava et al., 2015).
Here we sought to apply a comprehensive approach to investigate the β-arrestin “signalosome,” the repertoire of cascades elicited downstream from the CB1R/ β-arrestin. To achieve this goal, first we identified a CB1R with enhanced β-arrestin signaling. Mutation of the putative GRK phosphorylation sites S426/S430 to alanines resulted in lower levels of receptor internalization (Supplemental Fig. 1), reduced desensitization, and persistent activation of ERK1/2 independently from Gi/Go and reduced β-arrestin 2 recruitment to receptors (Supplemental Fig. 4). Reduced internalization rates could give the receptors more opportunity to interact with β-arrestin 1 and increase their signaling (Supplemental Fig. 1) (Daigle et al., 2008; Morgan et al., 2014). Interestingly, mice with these mutations have an increased response to agonist, supporting the idea of impaired desensitization. Next, we compared the kinases activated by G proteins and β-arrestin using a human phospho-antibody array. This approach in combination with siRNA technology and toxin treatments allowed us to simultaneously screen for signaling pathways specifically mediated by β-arrestins. Consistent with our previous work and the work of others (Flores-Otero et al., 2014; Laprairie et al., 2014), signaling from the wild-type receptor elicited by the endogenous 2-AG was mediated by G proteins at 5 minutes and it was completely replaced with β-arrestin-mediated signaling at the later time point. In addition, the β-arrestin-mediated signaling from the S426A/S430A receptor showed significant switch from G protein to β-arrestin pathways. Results from this receptor strengthens the idea that this pathway is involved in the regulation of long-term cellular events such as protein translation, gene transcription, and epigenetic regulation (Ma and Pei, 2007; Maudsley et al., 2013). Activation of ERK 1/2, JNK1/2/3, CREB, and EGFR RTK was dependent of β-arrestin-1, strengthening the specific role of this molecule during CB1R signaling (Supplemental Figs. 1, 3, and 4)(Ahn et al., 2013; Srivastava et al., 2015). These cascades have been previously associated to CB1R signaling in different cellular backgrounds, but not all of the cascades were shown to be mediated by β-arrestins (Derkinderen et al., 2001, 2003; Dalton and Howlett, 2012; Hoffman and Lupica, 2013). Interestingly, not all previously described cascades mediated by β-arrestins such as AKT were observed in our model (Gómez del Pulgar et al., 2000; Ozaita et al., 2007; Trazzi et al., 2010). This lack in response could be explained by differences in the quantitative relationship between receptor and signaling proteins, detection issues, differential basal kinase activity levels in these cells, or by the possibility that β-arrestin-mediated signaling activates only a subset of the complete signalosome available to CB1Rs in these particular cells. However, receptor expression levels and their cellular localization were not significantly difference between wild-type and mutants (Supplemental Fig. 1). Nevertheless, is interesting to note that downstream cascades are in general very conserved among different cellular environments as previously observed for the PTH receptor (Maudsley et al., 2015).
How is GPCR activation translated into β-arrestin-mediated signaling? Our previous work (Flores-Otero et al., 2014) with TIRFM suggested that a prolonged interaction between receptors and β-arrestins at endocytic pits could play a role in β-arrestin-mediated signaling. Supporting this, chemical or genetic prolongation of the dwell times of receptors/β-arrestins at endocytic pits dramatically increased β-arrestin-mediated signaling (Flores-Otero et al., 2014). Ubiquitination has been also proposed as a molecular mechanisms to control this interaction and signaling (Shenoy et al., 2009). We explored the interaction of CB1Rs with β-arrestins at the cell surface and in intracellular compartments by microscopy and immunoprecipitations (Fig. 5). WIN, which induced little or no β-arrestin-mediated signaling, elicited short endocytic dwell times in cells expressing the wild-type receptor (Flores-Otero et al., 2014). Remarkably, WIN elicited prolonged dwell times in the S426A/S430A receptor, similar to the dwell times obtained with 2-AG, which show a strong correlation with β-arrestin signaling. Next, we explored the intracellular localization of CB1R and β-arrestin 1. Surprisingly, we observed receptor-β-arrestin 1 clusters in intracellular compartments after incubation with WIN only with the mutant receptor, suggesting a “stronger” interaction that is maintained after internalization. We tested this interaction biochemically by immunoprecipitation. Supporting the idea of a stronger (perhaps also a prolonged) interaction as a mechanism to control β-arrestin-mediated signaling, the S426A/S430A receptor successfully precipitated β-arrestin 1 after WIN treatment. Little or no β-arrestins were coprecipitated with the wild-type receptor. Finally, we looked into the hypothesis that specific GRKs are required for β-arrestin-mediated signaling. By using siRNA technology, we observed that GRKs were not necessary for the early activation of ERK (G protein dependent). However, GRKs 4–6 were necessary for ERK1/2 phosphorylation by the S426A/S430A receptor. Taken together this suggests that ligands eliciting β-arrestin-mediated signaling induce conformations that are recognized by specific GRKs, leading to a stronger interaction between receptors and β-arrestins during the endocytic trafficking. This prolonged interaction results in enhanced β-arrestin-mediated signaling. Interestingly, these results indicate that a high throughput approach to analyze this interaction could be used to screen for β-arrestin biased compounds, providing new tools for drug discovery.
Because β-arrestin-mediated signaling activates kinases that control gene expression, we used transcriptomics to identify genes targeted by the CB1R/β-arrestin pathways. Interestingly, ∼70% of the genes specifically regulated by β-arrestin-mediated signaling control gene transcription and protein synthesis, suggesting a significant role of β-arrestin-mediated signaling on the long-term effects of CB1R activation.
MDM2, an E3 ubiquitin ligase previously implicated in β-arrestin-mediated signaling from the β2 adrenergic receptor, was upregulated selectively in S426A/S430A expressing cells, suggesting a role for MDM2 in the regulation of CB1R signaling (Shenoy et al., 2009). We additionally noted upregulation of several genes of the unfolded protein response pathway (ATF4, ASNS, MTHFD2, HERPUD1, ATF3, TRIB3, EIF2AK3, GADD45B, HSPA5, XBP1, GADD45A, and PPP1R15A). Comparing activation of the ER-stress pathway genes in cells expressing the S426A/S430A receptor we noted important differences. Although the components of ER-stress pathway were also activated in these cells, we noted the ATF4 arm of the ER-stress pathway was induced to a lower extent. Two robust transcriptional targets of ATF4, CHAC1 and TRIB3, were among the highest differentially expressed genes noted comparing agonist stimulated wild-type versus β-arrestin selective receptor expressing cells (Fig. 7B and Supplemental data). Previous work has shown that the proapoptotic and antitumor activity of cannabinoids is partially dependent on TRIB3 and the Akt/mTORC1 pathway (Salazar et al., 2009). Taken together these data indicate that β-arrestin could mediate the and antitumor action of CB1Rs (Velasco et al., 2012). Remarkably, the vascular endothelial growth factor A (VEGFA) gene and the growth hormone GH1 were downregulated by β-arrestin-mediated signaling, hinting at a molecular link between cannabinoids and cancer (Blázquez et al., 2004). The transcription factor IRF2BPL, also known as EAP1, was among the highest genes upregulated specifically by β-arrestin-mediated signaling. Although little information is available on this gene and its function, it has been proposed to modulate proenkephalin expression, suggesting a new link between the cannabinoid system and pain (Heger et al., 2007).
The complexity and specificity of GPCR signaling networks controlled by β-arrestins provides the opportunity to develop new therapeutic compounds with desired bias and reduced side effects (Violin et al., 2010; Kenakin and Christopoulos, 2013; Luttrell, 2014). Our work together with others suggest identification and screening of β-arrestin signatures as a rational approach to the development of biased drugs and propose that some of the positive effects associated with cannabis use may be dependent on β-arrestin-mediated signaling.
Supplementary Material
Acknowledgments
The authors thank John Allen for suggesting the use of kinome arrays and Julieta Gleiser and the members of the G.A.Y. laboratory for valuable discussions and Andrew Irving (Dundee, UK) for critical reagents.
Abbreviations
- CB1R
cannabinoid 1 receptor
- CREB
cAMP response element-binding protein
- ERK1/2
extracellular signal-regulated protein kinases 1 and 2
- GPCRs
G protein-coupled receptors
- GRK
G protein-coupled receptor kinases
- HEK
human embryonic kidney
- PTX
pertussis toxin
- RFP
monomeric red fluorescent protein
- SEP
super-ecliptic phluorin
- siRNA
small interfering RNA
- TIRFM
total internal reflection microscopy
- 2-AG
2-arachidonoylglycerol
- WIN
WIN55,212-2
Authorship contributions
Participated in research design: Delgado-Peraza, Ahn, Nogueras-Ortiz, Mackie, Kendall, and Yudowski.
Conducted experiments: Delgado-Peraza, Ahn, Nogueras-Ortiz, Mackie, and Yudowski.
Performed data analysis: Delgado-Peraza, Nogueras-Ortiz, Mungrue, Mackie, Kendall, Ahn, and Yudowski.
Wrote or contributed to the writing of the manuscript: Mungrue, Mackie, Kendall, and Yudowski.
Footnotes
This work was supported by research grants from the National Institutes of Health National Institute of Drug Abuse to G.A.Y. and C.N.O. [Grants DA023444 and R01DA037924]; National Institute of Minority Health and Health Disparities to G.A.Y and F.D.P. [Grants NIMHD 8G12-MD007600], and from the National Science Foundation [DBI 0115825]; D.A.K. was supported by the National Institute of Drug Abuse [Grant DA020763]; I.N.M was supported by the National Heart, Lung, and Blood Institute [Grants HL094709 and P30GM106392] and the National Science Foundation [Grant 1359140]; K.M. was supported by the National Institute of Drug Abuse [Grants DA011322 and DA021696]. G.A.Y. received further support from the Puerto Rico Science Trust.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Ahn KH, Mahmoud MM, Shim J-Y, Kendall DA. (2013) Distinct roles of β-arrestin 1 and β-arrestin 2 in ORG27569-induced biased signaling and internalization of the cannabinoid receptor 1 (CB1). J Biol Chem 288:9790–9800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JA, Yost JM, Setola V, Chen X, Sassano MF, Chen M, Peterson S, Yadav PN, Huang XP, Feng B, et al. (2011) Discovery of β-arrestin-biased dopamine D2 ligands for probing signal transduction pathways essential for antipsychotic efficacy. Proc Natl Acad Sci USA 108:18488–18493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood BK, Lopez J, Wager-Miller J, Mackie K, Straiker A. (2011) Expression of G protein-coupled receptors and related proteins in HEK293, AtT20, BV2, and N18 cell lines as revealed by microarray analysis. BMC Genomics 12:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood BK, Wager-Miller J, Haskins C, Straiker A, Mackie K. (2012) Functional selectivity in CB(2) cannabinoid receptor signaling and regulation: implications for the therapeutic potential of CB(2) ligands. Mol Pharmacol 81:250–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez C, González-Feria L, Alvarez L, Haro A, Casanova ML, Guzmán M. (2004) Cannabinoids inhibit the vascular endothelial growth factor pathway in gliomas. Cancer Res 64:5617–5623. [DOI] [PubMed] [Google Scholar]
- Bouaboula M, Poinot-Chazel C, Bourrié B, Canat X, Calandra B, Rinaldi-Carmona M, Le Fur G, Casellas P. (1995) Activation of mitogen-activated protein kinases by stimulation of the central cannabinoid receptor CB1. Biochem J 312:637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan LL, Lyettefi EJ, Pirani A, Smith T, Qiu J, Lin B. (2011) Direct concentration and viability measurement of yeast in corn mash using a novel imaging cytometry method. J Ind Microbiol Biotechnol 38:1109–1115. [DOI] [PubMed] [Google Scholar]
- Chang SD, Bruchas MR. (2014) Functional selectivity at GPCRs: new opportunities in psychiatric drug discovery. Neuropsychopharmacology 39:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claing A, Laporte SA, Caron MG, Lefkowitz RJ. (2002) Endocytosis of G protein-coupled receptors: roles of G protein-coupled receptor kinases and beta-arrestin proteins. Prog Neurobiol 66:61–79. [DOI] [PubMed] [Google Scholar]
- Daigle TL, Kearn CS, Mackie K. (2008) Rapid CB1 cannabinoid receptor desensitization defines the time course of ERK1/2 MAP kinase signaling. Neuropharmacology 54:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton GD, Howlett AC. (2012) Cannabinoid CB1 receptors transactivate multiple receptor tyrosine kinases and regulate serine/threonine kinases to activate ERK in neuronal cells. Br J Pharmacol 165:2497–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFea KA. (2011) Beta-arrestins as regulators of signal termination and transduction: how do they determine what to scaffold? Cell Signal 23:621–629. [DOI] [PubMed] [Google Scholar]
- Derkinderen P, Ledent C, Parmentier M, Girault JA. (2001) Cannabinoids activate p38 mitogen-activated protein kinases through CB1 receptors in hippocampus. J Neurochem 77:957–960. [DOI] [PubMed] [Google Scholar]
- Derkinderen P, Valjent E, Toutant M, Corvol J-C, Enslen H, Ledent C, Trzaskos J, Caboche J, Girault J-A. (2003) Regulation of extracellular signal-regulated kinase by cannabinoids in hippocampus. J Neurosci 23:2371–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, Kim J, Whalen EJ, Ahn S, Chen M, Lefkowitz RJ. (2008) Beta-arrestin-mediated signaling regulates protein synthesis. J Biol Chem 283:10611–10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Otero J, Ahn KH, Delgado-Peraza F, Mackie K, Kendall DA, Yudowski GA. (2014) Ligand-specific endocytic dwell times control functional selectivity of the cannabinoid receptor 1. Nat Commun 5:4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. (2004) Desensitization of G protein-coupled receptors and neuronal functions. Annu Rev Neurosci 27:107–144. [DOI] [PubMed] [Google Scholar]
- Gómez del Pulgar T, Velasco G, Guzmán M. (2000) The CB1 cannabinoid receptor is coupled to the activation of protein kinase B/Akt. Biochem J 347:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. (1996) Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature 383:447–450. [DOI] [PubMed] [Google Scholar]
- Gurevich VV. (2014) Arrestins - Pharmacology and Therapeutic Potential, Springer Berlin Heidelberg, Berlin, Heidelberg. [Google Scholar]
- Hart S, Fischer OM, Ullrich A. (2004) Cannabinoids induce cancer cell proliferation via tumor necrosis factor alpha-converting enzyme (TACE/ADAM17)-mediated transactivation of the epidermal growth factor receptor. Cancer Res 64:1943–1950. [DOI] [PubMed] [Google Scholar]
- Heger S, Mastronardi C, Dissen GA, Lomniczi A, Cabrera R, Roth CL, Jung H, Galimi F, Sippell W, Ojeda SR. (2007) Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J Clin Invest 117:2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman AF, Lupica CR. (2013) Synaptic targets of Δ9-tetrahydrocannabinol in the central nervous system. Cold Spring Harb Perspect Med 3:a012237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett AC. (2005) Cannabinoid receptor signaling. Handb Exp Pharmacol 168:53–79. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143:1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irannejad R, von Zastrow M. (2014) GPCR signaling along the endocytic pathway. Curr Opin Cell Biol 27:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Brown S, Roche JP, Hsieh C, Celver JP, Kovoor A, Chavkin C, Mackie K. (1999) Distinct domains of the CB1 cannabinoid receptor mediate desensitization and internalization. J Neurosci 19:3773–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Shi Y, Xiang B, Qu B, Su W, Zhu M, Zhang M, Bao G, Wang F, Zhang X, et al. (2005) A nuclear function of β-arrestin1 in GPCR signaling: regulation of histone acetylation and gene transcription. Cell 123:833–847. [DOI] [PubMed] [Google Scholar]
- Kenakin T. (2006) Receptors as microprocessors: pharmacological nuance on metabotropic glutamate receptors 1alpha. Sci STKE 2006:pe29. [DOI] [PubMed] [Google Scholar]
- Kenakin T. (2007) Collateral efficacy in drug discovery: taking advantage of the good (allosteric) nature of 7TM receptors. Trends Pharmacol Sci 28:407–415. [DOI] [PubMed] [Google Scholar]
- Kenakin T. (2011) Functional selectivity and biased receptor signaling. J Pharmacol Exp Ther 336:296–302. [DOI] [PubMed] [Google Scholar]
- Kenakin T, Christopoulos A. (2013) Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov 12:205–216. [DOI] [PubMed] [Google Scholar]
- Laprairie RB, Bagher AM, Kelly MEM, Dupré DJ, Denovan-Wright EM. (2014) Type 1 cannabinoid receptor ligands display functional selectivity in a cell culture model of striatal medium spiny projection neurons. J Biol Chem 289:24845–24862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggett SB. (2011) Phosphorylation barcoding as a mechanism of directing GPCR signaling. Sci Signal 4:pe36. [DOI] [PubMed] [Google Scholar]
- Luttrell LM. (2014) Minireview: More than just a hammer: ligand “bias” and pharmaceutical discovery. Mol Endocrinol 28:281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Pei G. (2007) Beta-arrestin signaling and regulation of transcription. J Cell Sci 120:213–218. [DOI] [PubMed] [Google Scholar]
- Maudsley S, Martin B, Gesty-Palmer D, Cheung H, Johnson C, Patel S, Becker KG, Wood WH, 3rd, Zhang Y, Lehrmann E, et al. (2015) Delineation of a conserved arrestin-biased signaling repertoire in vivo. Mol Pharmacol 87:706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudsley S, Siddiqui S, Martin B. (2013) Systems analysis of arrestin pathway functions. Prog Mol Biol Transl Sci 118:431–467. [DOI] [PubMed] [Google Scholar]
- Morgan DJ, Davis BJ, Kearn CS, Marcus D, Cook AJ, Wager-Miller J, Straiker A, Myoga MH, Karduck J, Leishman E, et al. (2014) Mutation of putative GRK phosphorylation sites in the cannabinoid receptor 1 (CB1R) confers resistance to cannabinoid tolerance and hypersensitivity to cannabinoids in mice. J Neurosci 34:5152–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles KN, Xiao K, Ahn S, Shukla AK, Christopher M, Rajagopal S, Strachan RT, Huang T, Bressler EA, Hara MR, et al. (2012). Distinct phosphorylation sites on the β2 -adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci Signal 4(185):ra51. [DOI] [PMC free article] [PubMed]
- Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, Rajagopal S, Strachan RT, Huang T-Y, Bressler EA, Hara MR, et al. (2011) Distinct phosphorylation sites on the β(2)-adrenergic receptor establish a barcode that encodes differential functions of β-arrestin. Sci Signal 4:ra51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaita A, Puighermanal E, Maldonado R. (2007) Regulation of PI3K/Akt/GSK-3 pathway by cannabinoids in the brain. J Neurochem 102:1105–1114. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. (2002) Seven-transmembrane receptors. Nat Rev Mol Cell Biol 3:639–650. [DOI] [PubMed] [Google Scholar]
- Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. (2012) Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol 52:179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Vendrell C, Chevalier M, Acevedo-Canabal AM, Delgado-Peraza F, Flores-Otero J, Yudowski GA. (2014) Imaging of kiss-and-run exocytosis of surface receptors in neuronal cultures. Front Cell Neurosci 8:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Vendrell C, Yu YJ, Yudowski GA. (2012) Fast modulation of μ-opioid receptor (MOR) recycling is mediated by receptor agonists. J Biol Chem 287:14782–14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Vendrell C, Yudowski GA. (2015) Real-time imaging of mu opioid receptors by total internal reflection fluorescence microscopy. Methods Mol Biol 1230:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SGF, Kobilka BK. (2009) The structure and function of G-protein-coupled receptors. Nature 459:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda D, Galve-Roperh I, Haro A, Guzmán M. (2000) The CB(1) cannabinoid receptor is coupled to the activation of c-Jun N-terminal kinase. Mol Pharmacol 58:814–820. [DOI] [PubMed] [Google Scholar]
- Salazar M, Carracedo A, Salanueva ÍJ, Hernández-Tiedra S, Lorente M, Egia A, Vázquez P, Blázquez C, Torres S, García S, et al. (2009) Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest 119:1359–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid CL, Bohn LM. (2009) Physiological and pharmacological implications of beta-arrestin regulation. Pharmacol Ther 121:285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK, Lefkowitz RJ. (2011) β-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol Sci 32:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK, Modi AS, Shukla AK, Xiao K, Berthouze M, Ahn S, Wilkinson KD, Miller WE, Lefkowitz RJ. (2009) Beta-arrestin-dependent signaling and trafficking of 7-transmembrane receptors is reciprocally regulated by the deubiquitinase USP33 and the E3 ligase Mdm2. Proc Natl Acad Sci USA 106:6650–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Gupta B, Gupta C, Shukla AK. (2015) Emerging functional divergence of β-arrestin isoforms in GPCR function. Trends Endocrinol Metab 26:628–642. [DOI] [PubMed] [Google Scholar]
- Trazzi S, Steger M, Mitrugno VM, Bartesaghi R, Ciani E. (2010) CB1 cannabinoid receptors increase neuronal precursor proliferation through AKT/glycogen synthase kinase-3beta/beta-catenin signaling. J Biol Chem 285:10098–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, Burlingame AL. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics 11:215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzingounis AV, von Zastrow M, Yudowski GA. (2010) Beta-blocker drugs mediate calcium signaling in native central nervous system neurons by beta-arrestin-biased agonism. Proc Natl Acad Sci USA 107:21028–21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JD, Clarke WP, von Zastrow M, Nichols DE, Kobilka B, Weinstein H, Javitch JA, Roth BL, Christopoulos A, Sexton PM, et al. (2007) Functional selectivity and classical concepts of quantitative pharmacology. J Pharmacol Exp Ther 320:1–13. [DOI] [PubMed] [Google Scholar]
- Urs NM, Bido S, Peterson SM, Daigle TL, Bass CE, Gainetdinov RR, Bezard E, Caron MG. (2015) Targeting β-arrestin2 in the treatment of L-DOPA-induced dyskinesia in Parkinson’s disease. Proc Natl Acad Sci USA 112:E2517–E2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco G, Sánchez C, Guzmán M. (2012) Towards the use of cannabinoids as antitumour agents. Nat Rev Cancer 12:436–444. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. (2013) Molecular signatures of G-protein-coupled receptors. Nature 494:185–194. [DOI] [PubMed] [Google Scholar]
- Violin JD, DeWire SM, Yamashita D, Rominger DH, Nguyen L, Schiller K, Whalen EJ, Gowen M, Lark MW. (2010) Selectively engaging β-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J Pharmacol Exp Ther 335:572–579. [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Nagaraj N, Zougman A, Gnad F, Mann M. (2010) Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J Proteome Res 9:3280–3289. [DOI] [PubMed] [Google Scholar]
- Yudowski GA, von Zastrow M. (2011) Investigating G protein-coupled receptor endocytosis and trafficking by TIR-FM. Methods Mol Biol 756:325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidar DA, Violin JD, Whalen EJ, Lefkowitz RJ. (2009) Selective engagement of G protein coupled receptor kinases (GRKs) encodes distinct functions of biased ligands. Proc Natl Acad Sci USA 106:9649–9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.