Abstract
Lynch syndrome (LS) is an autosomal-dominant inherited disorder mainly caused by a germline mutation in the DNA mismatch repair (MMR) genes (MLH1, MSH2, MSH6, and PMS2) and is associated with increased risk for various cancers, particularly colorectal cancer and endometrial cancer (EC). Women with LS account for 2% to 6% of EC patients; it is clinically important to identify LS in such individuals for predicting and/or preventing additional LS-associated cancers. PMS2 germline mutation (PMS2-LS) is the rarest contribution to LS etiology among the 4 LS-associated MMR germline mutations, and its detection is complicated. Therefore, prudent screening for PMS2-LS is important as it leads to an efficient LS identification strategy. Immunohistochemistry is recommended as a screening method for LS in EC. Isolated loss of PMS2 (IL-PMS2) expression is caused not only by PMS2-LS but also by MLH1 germline mutation or MLH1 promoter hypermethylation (MLH-PHM). This study aimed to determine the association between MLH1-PHM and IL-PMS2 to avoid inappropriate genetic analysis. We performed MLH1 methylation analysis and MLH1/PMS2 germline mutation testing on the IL-PMS2 cases. By performing MMR-immunohistochemistry on 360 unselected ECs, we could select 8 (2.2%) cases as IL-PMS2. Heterogenous MLH1 staining and MLH1-PHM were detected in 4 of 8 (50%) IL-PMS2 tumors. Of the 5 IL-PMS2 patients who underwent genetic analysis, 1 had PMS2 germline mutation with normal MLH1 expression (without MLH1-PHM), and no MLH1 germline mutation was detected. We suggest that MLH1 promoter methylation analysis for IL-PMS2 EC should be performed to exclude sporadic cases before further PMS2 genetic testing.
Key Words: Lynch syndrome, endometrial cancer, PMS2, MLH1 promoter hypermethylation, heterogenous
Among endometrial cancer (EC) patients, Lynch syndrome (LS) accounts for approximately 2% to 6% of cases.1–5 LS is an autosomal-dominant inherited syndrome mainly caused by germline mutations in the DNA mismatch repair (MMR) genes MLH1, MSH2, MSH6, and PMS2.6 Mutation carriers have an increased lifetime risk of developing colorectal cancer (CRC, 40% to 80%), EC (33% to 61%), ovarian cancer (9% to 12%), and other LS-associated cancers.7 Thus, it is clinically relevant to identify LS women among EC patients to predict and prevent the development of other LS-associated cancers. It would also provide blood relatives an opportunity for genetic analysis and surveillance for LS-associated cancers. Each of the 4 MMR germline mutations lead to distinct molecular pathologies,8 and thus individuals carrying different mutations should not be regarded as suffering from the same disease. PMS2 germline mutation is associated with later onset, weaker family history, and a lower risk for cancer compared with other MMR germline mutations.9,10 Indeed, PMS2 germline mutation is the rarest genetic alteration among the 4 LS-associated MMR germline mutations, and its detection is more complicated than that of other MMR germline mutations because of the presence of a large family of highly homologous PMS2 pseudogenes.11
Immunohistochemistry (IHC) is recommended as a primary screen for LS in patients with newly diagnosed EC,12,13 as it can rapidly detect loss of MMR protein expression. In predicting MMR germline mutation, the sensitivity of IHC using a panel of 4 MMR antibodies (against MLH1, MSH2, MSH6, and PMS2) is as high as that of microsatellite instability (MSI) testing,13,14 which has been also used as a screening tool for LS. IHC is simple and fast, cost-effective, and practical in many institutions. It can also be used to predict corresponding germline mutations and is more suited for detection of MSH6 germline mutation compared with MSI testing.12,14 In general, the presence of nuclear staining in tumor cells is good evidence of retained MMR protein, even if it is focal and weak staining.14 This has led to neglect of staining pattern interpretation, with the exception of cases that show complete absence of nuclear staining. However, variable staining patterns are very confusing to interpret, as they present as heterogenous staining, weak staining, or cytoplasmic staining (CS).14–16 These variabilities are commonly seen in MLH1, and some studies have reported that MLH1 germline mutation may underlie weak MLH1 staining.17
The major reason for loss of MLH1 expression in sporadic cancers is MLH1 promoter hypermethylation (MLH1-PHM).2 This phenomenon is seen in 15% to 20% of CRCs and 20% to 30% of ECs.18 Performing MLH1 promoter methylation analysis to determine the cause of MLH1 loss would avoid unnecessary MLH1 germline mutation testing. MLH1-PHM is unevenly distributed in tumors, and there are some reports that this correlates with heterogenous MLH1/PMS2 staining.15,19 Therefore, MLH1-PHM can occasionally lead to unclear staining in IHC.16
MLH1 and PMS2 proteins form functional heterodimer complexes.20 MLH1 is obligatory for PMS2 protein stability, and its dysfunction leads to degradation and/or loss of PMS2.20 The converse is not true, because MLH1 can also bind to other MMR proteins.20 In contrast, some MLH1 germline mutations induce only loss of PMS2 protein and yet MLH1 antigenicity is retained.16,21,22 Thus, in cases of isolated loss of PMS2 (IL-PMS2) expression, MLH1 disorders cannot be excluded.21,22 Guidelines from The National Society of Genetic Counselors and the Collaborative Group of the Americas on Inherited Colorectal Cancer recommend MLH1 germline mutation testing in IL-PMS2 cases in which PMS2 germline mutations are absent.13 The National Comprehensive Cancer Network guidelines list PMS2 and MLH1 germline mutations as plausible etiologies in IL-PMS2.23 These guidelines (and some additional studies21,22) mention MLH1 germline mutation in IL-PMS2; yet, few studies have investigated MLH1 promoter methylation in IL-PMS2. Moreover, all of the previous studies focused on CRC, and there is no adequate consensus on the genetic alterations that predispose individuals to EC.
In an LS identification strategy that adopted universal MMR-IHC screening, “Lynch like (LL)” patients who had MMR-IHC deficiency without germline mutations formed a distinct subgroup.24–26 In families of LL CRC patients, the incidence of CRC was lower than that in families of confirmed LS cases and higher than that in families of sporadic cases.27 From this trend, both unknown hereditary cancers and sporadic cancers are likely to be intermixed in the LL CRC group.24–26 It has been suggested that LL CRC patients and their relatives should undergo the same management as LS patients.28 However, little is known regarding the clinical features of LL patients in EC.
In a previous study, we proposed a screening strategy for LS in 360 newly diagnosed EC patients with lenient triage (original criteria) using selective IHC and optional MLH1 promoter methylation analysis.29 We performed IHC on samples from all 360 of these participants and detected 10 cases (2.8%) of IL-PMS2. Most of them were accompanied by MLH1 IHC abnormalities (such as heterogenous or weak staining). On the basis of these results and existing knowledge, we hypothesized that MLH1-PHM might exist in some IL-PMS2 cases. Clarifying the MLH1-PHM status in IL-PMS2 cases would avoid unnecessary genetic analysis; moreover, it would spare individuals and relatives from uncomfortable clinical diagnostic interventions. With this in mind, we designed the current study to determine the association between MLH1-PHM and IL-PMS2.
MATERIALS AND METHODS
Study Population and Procedures
A total of 360 EC patients who were diagnosed at Akita University Hospital between January 2003 and December 2013 were identified retrospectively (Fig. 1). All of the patients were Asians living in Japan. The patients’ clinical data, such as age, personal medical history, and family history, were collected from medical records. We designed criteria, named “APF criteria” (our original criteria for selection according to Age of onset below 50 years and/or Personal/Family history of Lynch-associated cancer), and applied it to unselected EC patients. The cases satisfying one or more of the 3 criteria were considered eligible. We performed MMR-IHC on the tumor of patients who met our criteria in our previous study.29 Additional IHC was performed on the tumor of patients who did not meet our criteria in this study. Performing the MLH1 methylation assay and MMR germline mutation testing on cases with IL-PMS2, we investigated the association between MLH1-PHM and IL-PMS2. All study participants provided written informed consent in the prescribed document. The Institutional Review Board of Akita University approved our study design.
FIGURE 1.
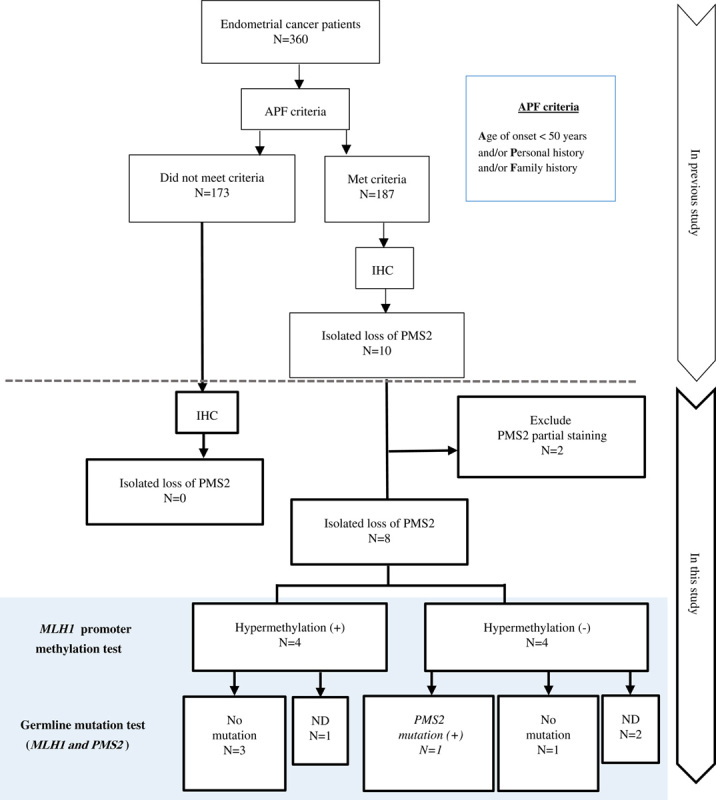
Summary of this study. The MLH1 promoter methylation test and germline mutation test for MLH1 and PMS2 were performed for IL-PMS2 cases. APF criteria, our original criteria for selection according to Age of onset below 50 years and/or Personal/Family history of Lynch-associated cancer. IHC analysis for MLH1, MSH2, MSH6, and PMS2. ND indicates not done germline mutation test.
IHC Staining for DNA MMR Proteins (MMR-IHC)
MMR-IHC was performed on tumors of all 360 EC patients to assess MMR protein (MLH1, MSH2, MSH6, and PMS2) expression, according to standard procedure. An appropriate paraffin-embedded tissue was cut at 4 μm thickness. The tissue sections were deparaffinized in xylene and rehydrated in graded alcohol. Subsequently, antigen retrieval was performed in 10 mmol/L Tris-EDTA buffer (pH 9.0) in a microwave oven for 20 minutes. These sections were allowed to cool at room temperature. Thereafter, the primary antibodies were applied overnight at 4°C. The primary antibodies were MLH1 (clone ES05, dilution 1:50; Dako), MSH2 (clone FE11, dilution 1:50; Dako), MSH6 (clone EP49, dilution 1:50; Dako), and PMS2 (clone EP51, dilution 1:40; Dako). The antigen-antibody reaction was visualized with the Envision kit (Dako). The slides were counterstained with hematoxylin. Adjacent normal endometrium and lymphocytes in the slides were used as an internal positive control. We judged the complete absence of nuclear staining in the tumor cells as loss of MMR protein expression.
MLH1 Promoter Methylation Analysis
In all 8 IL-PMS2 cases, we performed MLH1 promoter methylation analysis. The tumor DNA was extracted from mapped formalin-fixed, paraffin-embedded tissue sections to provide tumor samples for the assay. The SALSA MS-MLPA kit ME011 MMR genes (MRC-Holland, Amsterdam, The Netherlands) were used to detect aberrant CpG island methylation in the promoter of MMR genes, including 5 probes for MLH1. The MS-MLPA assay was performed as described by the manufacturer. We focused on the promoter C region (probe 3), which provides the best correlation with MLH1 expression.30 On the basis of a previous study associated with gene silencing,31 the dichotomization threshold to distinguish hypermethylated versus nonmethylated samples was set at 15%.
Germline Genetic Testing
Five of 8 IL-PMS2 cases underwent the genetic analysis for this study. Germline mutation testing of MLH1 and PMS2 was performed on genomic DNA isolated from peripheral blood leukocytes. Detection of point mutations was conducted using exon-by-exon polymerase chain reaction and direct sequencing of the whole coding sequence in and intron-exon boundaries for each gene. Large rearrangements (deletions and/or insertions) in the MMR gene were screened by MLPA according to the manufacturer’s protocols (SALSA MLPA kits P003, P008).
RESULTS
By performing MMR-IHC, we finally identified 8 (2.2%) cases as IL-PMS2 out of unselected 360 EC patients. We had originally recognized 10 cases as IL-PMS2 in the previous report, but we excluded 2 cases with weak PMS2 expression from the IL-PMS2 in this inspection. All 8 IL-PMS2 cases met the original APF criteria (Fig. 1). The clinical and pathologic characteristics of the IL-PMS2 cases are shown in Table 1. No cases of IL-PMS2 met the Amsterdam Criteria II, and 3 (37.5%) met the SGO 5-10% criteria. MLH1-PHM was detected in 4 (50%) of 8 IL-PMS2 cases (Table 1). In 4 cases with MLH1-PHM, between 25% and 65% of the MLH1 promoter C region was hypermethylated. All cases with MLH1-PHM were accompanied with MLH1 heterogenous staining and cytoplasmic immunoreactivity.
TABLE 1.
Clinical and Molecular Feature of Cases With IL-PMS2 Expression
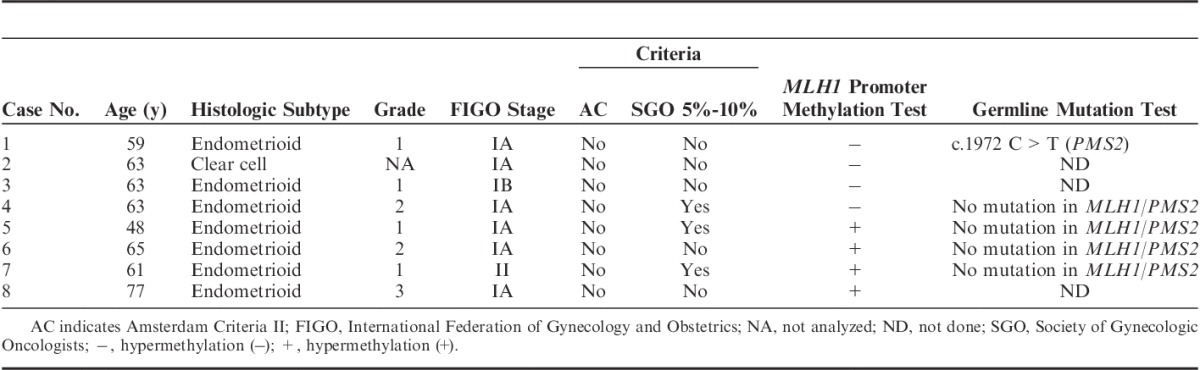
Of the 8 IL-PMS2 cases, PMS2 and MLH1 germline mutation tests were performed in 5 (63%) who donated blood samples. Of the 5 IL-PMS2 patients who underwent genetic testing, 1 had PMS2 germline mutation with normal MLH1 expression (without MLH1-PHM), and no MLH1 germline mutation was detected (Table 1).
DISCUSSION
PMS2 germline mutation is a rare cause of LS in EC, and the risk for LS-associated cancers is considerably lower with this genetic lesion than with those in the other 3 MMR genes.9,10 Thus, the benefits of screening for PMS2 germline mutation carriers (PMS2-LS) are less clear than those obtained by screening for other MMR germline mutation carriers. For developing the best screening strategy for LS in EC, exclusion of non-PMS2-LS is critical to avoid unnecessary PMS2 germline mutation testing.
In this study, we detected MLH1-PHM in half of the cases with IL-PMS2. In the MLH1-PHM cases, no PMS2 or MLH1 germline mutations were found, and we thus considered these as instances of sporadic EC. In cases without MLH1-PHM, 1 PMS2 germline mutation was detected, but no MLH1 germline mutations were found. In all MLH1-PHM cases, MLH1 expression was heterogenous. In contrast, we did not observe heterogenous MLH1 staining in non-MLH1-PHM cases (Tables 1, 2).
TABLE 2.
Staining Patterns for MMR Proteins With IL-PMS2 Cases
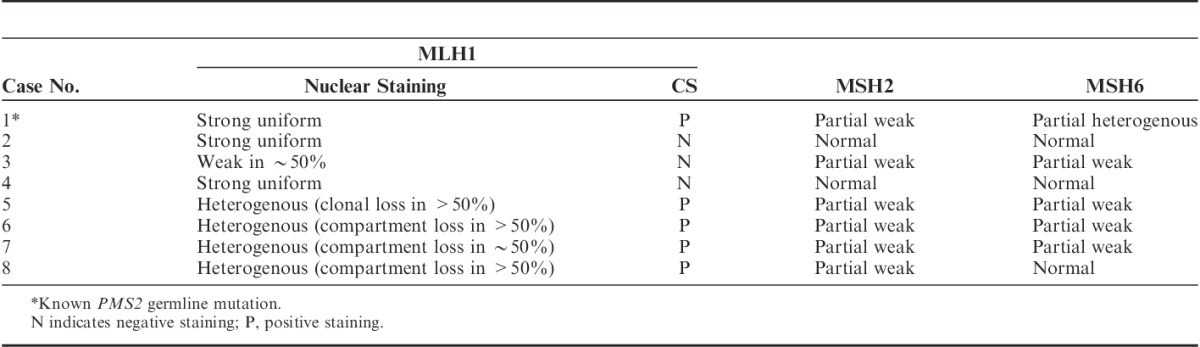
Previous studies have focused on areas with heterogenous MLH1 and PMS2 expression; areas with loss or retention of MLH1/PMS2 expression were assessed for MLH1 promoter methylation separately.15,19 Pai et al15 described 6 cases of heterogenous MLH1/PMS2 staining in EC. MLH1-PHM was detected in all of these cases, and focal MLH1-PHM (limited to the areas with MLH1/PMS2 loss) was reported in 2 cases.15 Joost et al19 reported 3 cases of heterogenous MLH1/PMS2 expression in CRC and performed methylation analysis in 2 of these cases. Both cases showed MLH1-PHM in only the area with loss of MLH1/PMS2 expression.19 These reports indicate that the heterogenous MLH1/PMS2 expression was most likely attributable to MLH1-PHM. In our study, heterogenous expression was detected only in MLH1 IHC; this expression pattern suggests that nonuniform hypermethylation was present. There were 2 patterns in MLH1 heterogenous staining: “compartmental,” which was defined as retained/lost staining in large areas of the tumor, and “clonal,” which was defined as retained/lost staining in whole glands or groups of glands (Table 2 and Fig. 2). Joost and colleagues also identified these patterns and suggested that they may be attributed to multiple causes, including variable epitope expression, second hit mutation or methylation in select tumors, or the influence of conditions in the tumor microenvironment, such as hypoxia and oxidative stress.19 Additional studies are required to fully determine the meaning of the heterogenous staining pattern.
FIGURE 2.
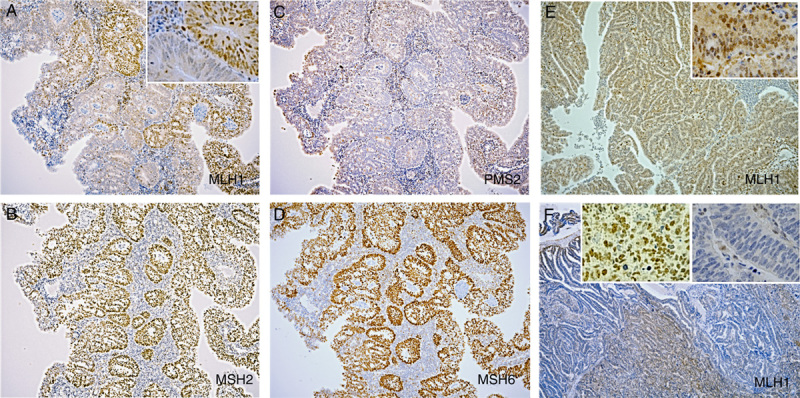
Examples of IHC staining for MMR protein. A, MLH1 heterogenous staining (clonal loss) in case 5. B, Normal MSH2 staining in case 5. C, Complete loss of PMS2 staining in case 5. D, Normal MSH6 staining in case 5. E, MLH1 staining with CS in case 6. F, MLH1 heterogenous staining (compartment loss) in case 8. Inset: 2A, E, F raised the magnification, ×10 to ×40. 2A, Heterogeneous staining part; 2E, CS; 2F, right side: staining area; left side: staining loss area.
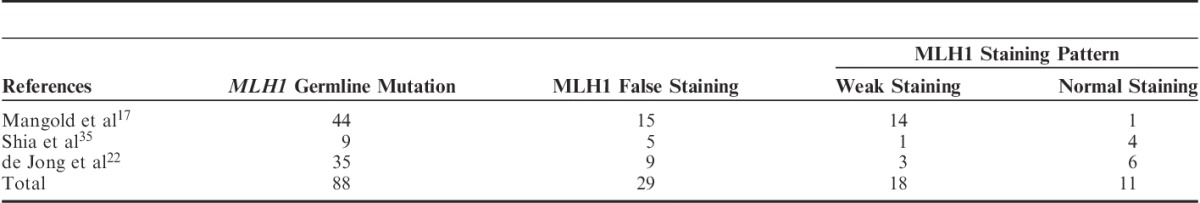
In the case with PMS2 germline mutation in our study, MLH1 expression in the tumor area was normal, whereas in the MLH1-PHM cases MLH1 expression in the tumor area was heterogenous. Further PMS2 genetic testing could be avoided in IL-PMS2 cases with abnormal MLH1 expression patterns (such as heterogenous and weak staining). Dudley et al21 reported 4 MLH1 germline mutations in 31 cases of IL-PMS2, and weak MLH1 staining was observed in 2 of those cases. As per the recent reports summarized in Table 3, weak MLH1 staining, as revealed by IHC, has been observed in 20% (18/88) of cases where a MLH1 germline mutation was present. Watson et al32 reported MLH1 germline mutation in cases with MLH1 heterogenous staining. Moreover, normal MLH1 staining was retained in 13% (11/88) of the MLH1 germline mutation cases (Table 3). Therefore, MLH1 weak staining, heterogenous staining, or even normal staining might be a result of false nuclear staining. In such cases, the possibility of an MLH1 germline mutation cannot be completely excluded. Shia et al16 reported that weak MLH1 staining in IL-PMS2 cases may suggest MLH1 genetic abnormalities. This is because some pathogenic MLH1 missense mutations functionally inactivate MLH1 protein and yet preserve its antigenicity.16,33,34
TABLE 3.
False Staining for MLH1 According to MLH1 Germline Mutation
CS is one of the most confusing patterns associated with aberrant MLH1 expression in IHC. MLH1 CS was observed in all MLH1-PHM cases and was sometimes seen locally in non-MLH1-PHM cases. In cases with MLH1 CS, it is challenging to determine whether the MLH1 protein is completely absent. Shia et al35 evaluated CS in CRC patients and found that CS extended to >30% of the tumor sample in 11% (12/105) of MLH1 IHC tests. However, the presence of CS was not correlated with MSI-H or germline mutation.35 There are many difficulties associated with the interpretation of MLH1-IHC; these include confounding variables such as MLH1 germline mutation, MLH1-PHM, CS, and other nonspecific reactions. We suggest that IL-PMS2 cases should include not only PMS2-LS but also MLH1-LS and MLH1-PHM subtypes.
On performing MLH1 promoter methylation analysis to exclude sporadic cases, the following types of LS might go undetected: those in which MLH1 germline mutation coexists with MLH1-PHM,34 those with coexisting PMS2 germline mutation and MLH1-PHM,10 and those with autosomal-dominant inherited MLH1-PHM (also known as constitutional MLH1 epimutation).36 These cases are rare, but their identification is clinically significant, particularly if individuals have a strong family history and/or present with young onset of LS-associated cancers. Methylation analysis cannot completely confirm that tumors are sporadic. Thus, the first 2 types listed above can be excluded with a MMR germline mutation test, whereas autosomal-dominant inherited MLH1-PHM cannot.
MLH1 can interact with MLH3 or PMS1 instead of PMS2 to form a heterodimer that functionally compensates for the absence of MutLα (MLH1+PMS2), thereby delaying disease onset.37 MLH1 germline mutation tends to result in the typical form of LS, whereas PMS2 germline mutation leads to an attenuated form of the disease.37 The MLH1 germline mutation–associated risk for CRC up to 70 years of age is considerably higher than the PMS2 germline mutation–associated risk (40% to 80% and 15% to 20%, respectively).10,38,39 Similarly, EC risk up to 70 years of age in individuals with MLH1 germline mutation is higher than that of PMS2 germline mutations (25% to 60% and 15%, respectively).10,38,39 The National Comprehensive Cancer Network guidelines recommend separate surveillance for MLH1 and PMS2 germline mutation carriers.23 Thus, verification of MLH1 and PMS2 germline mutation is important in the surveillance for individuals and their relatives. In the 5 IL-PMS2 cases in the current study, we did not find MLH1 mutation carriers. However, according to previous reports, MLH1 germline mutation was identified in 23% to 25% of IL-PMS2 cases.21,22 When PMS2 expression is absent, the possibility of a MLH1 germline mutation should not be excluded without additional information. This is true independent of the MLH1 expression status.
The spread of universal MMR-IHC screening for LS in EC would identify more LL (as well as LS) patients than classical selective screening.26 Buchanan et al26 reviewed LL cases and reported that 52% (52/101) of MMR-deficient EC cases were classified as LL. MMR-IHC deficiency in LL tumors is due to unidentified germline MMR gene mutations, biallelic somatic gene inactivation, and other rare causes.26 Haraldsdottir et al40 reported that almost 70% of LL tumors had somatic mutations in the MMR gene, and the majority of LL cases were considered nonhereditary. Distinction between LL tumor and sporadic EC may have considerable influence on the management of LL patients and their relatives.
In the current study we showed that 57% (4/7) of IL-PMS2 cases were misclassified as LL, and this error could be corrected by incorporating the MLH1 promoter methylation test.
In conclusion, we found that 50% of IL-PMS2 EC patients had MLH1-PHM. These MLH1-PHM cases did not have MMR germline mutation and were thus determined to be sporadic EC. MLH1 promoter methylation analysis for IL-PMS2 EC should be performed to exclude sporadic cases before further PMS2 genetic testing.
Footnotes
Conflicts of Interest and Source of Funding: The authors have disclosed that they have no significant relationships with, or financial interest in, any commercial companies pertaining to this article.
REFERENCES
- 1.Ferguson SE, Aronson M, Pollett A, et al. Performance characteristics of screening strategies for Lynch syndrome in unselected women with newly diagnosed endometrial cancer who have undergone universal germline mutation testing. Cancer. 2014;120:3932–3939. [DOI] [PubMed] [Google Scholar]
- 2.Buchanan DD, Tan YY, Walsh MD, et al. Tumor mismatch repair immunohistochemistry and DNA MLH1 methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level germline mismatch repair gene mutation testing. J Clin Oncol. 2014;32:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egoavil C, Alenda C, Castillejo A, et al. Prevalence of Lynch syndrome among patients with newly diagnosed endometrial cancers. PLoS One. 2013;8:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leenen CHM, Van Lier MGF, Van Doorn HC, et al. Prospective evaluation of molecular screening for Lynch syndrome in patients with endometrial cancer≤70 years. Gynecol Oncol. 2012;125:414–420. [DOI] [PubMed] [Google Scholar]
- 5.Hampel H, Frankel W, Panescu J, et al. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006;66:7810–7817. [DOI] [PubMed] [Google Scholar]
- 6.Lynch HT, Snyder CL, Shaw TG, et al. Milestones of Lynch syndrome: 1895-2015. Nat Rev Cancer. 2015;15:181–194. [DOI] [PubMed] [Google Scholar]
- 7.Barrow E, Hill J. Gareth ED. Cancer risk in Lynch syndrome. Fam Cancer. 2013;12:229–240. [DOI] [PubMed] [Google Scholar]
- 8.Cohen SA, Leininger A. The genetic basis of Lynch syndrome and its implications for clinical practice and risk management. Appl Clin Genet. 2014;7:147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanne W, Brohet RM, Tops CM, et al. Lynch syndrome caused by germline PMS2 mutations: delineating the cancer risk. J Clin Oncol. 2014;33:319–325. [DOI] [PubMed] [Google Scholar]
- 10.Senter L, Clendenning M, Sotamaa K, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clendenning M, Hampel H, LaJeunesse J, et al. Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat. 2006;27:490–495. [DOI] [PubMed] [Google Scholar]
- 12.Resnick KE, Hampel H, Fishel R, et al. Current and emerging trends in Lynch syndrome identification in women with endometrial cancer. Gynecol Oncol. 2009;114:128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weissman SM, Burt R, Church J, et al. Identification of individuals at risk for Lynch syndrome using targeted evaluations and genetic testing: National Society of Genetic Counselors and the Collaborative Group of the Americas on Inherited Colorectal Cancer joint practice guideline. J Genet Couns. 2012;21:484–493. [DOI] [PubMed] [Google Scholar]
- 14.Shia J. Immunohistochemistry versus microsatellite instability testing for screening colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome. Part I. The utility of immunohistochemistry. J Mol Diagn. 2008;10:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pai RK, Plesec TP, Abdul-Karim FW, et al. Abrupt loss of MLH1 and PMS2 expression in endometrial carcinoma: molecular and morphologic analysis of 6 cases. Am J Surg Pathol. 2015;39:993–999. [DOI] [PubMed] [Google Scholar]
- 16.Shia J, Holck S, Depetris G, et al. Lynch syndrome-associated neoplasms: a discussion on histopathology and immunohistochemistry. Fam Cancer. 2013;12:241–260. [DOI] [PubMed] [Google Scholar]
- 17.Mangold E, Pagenstecher C, Friedl W, et al. Tumours from MSH2 mutation carriers show loss of MSH2 expression but many tumours from MLH1 mutation carriers exhibit weak positive MLH1 staining. J Pathol. 2005;207:385–395. [DOI] [PubMed] [Google Scholar]
- 18.Society of Gynecologic Oncology/The American College of Obstetricians and Gynecologists. Practice Bulletin: Lynch syndrome No. 147, November 2014. Available at: http://www.sgo.or. Accessed September 17, 2015.
- 19.Joost P, Veurink N, Holck S, et al. Heterogenous mismatch-repair status in colorectal cancer. Diagn Pathol. 2014;9:126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shia J, Tang LH, Vakiani E, et al. Immunohistochemistry as first-line screening for detecting colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome: a 2-antibody panel may be as predictive as a 4-antibody panel. Am J Surg Pathol. 2009;33:1639–1645. [DOI] [PubMed] [Google Scholar]
- 21.Dudley B, Brand RE, Thull D, et al. Germline MLH1 mutations are frequently identified in Lynch syndrome patients with colorectal and endometrial carcinoma demonstrating isolated loss of PMS2 immunohistochemical expression. Am J Surg Pathol. 2015;39:1114–1120. [DOI] [PubMed] [Google Scholar]
- 22.de Jong AE, van Puijenbroek M, Hendriks Y, et al. Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:972–980. [DOI] [PubMed] [Google Scholar]
- 23.The NCCN Clinical Practice Guidelines in Oncology Genetic/Familial High-Risk Assessment. Colorectal Version 1.2015 National Comprehensive Cancer Network. Available at: http://www.nccn.org. Accessed September 17, 2015.
- 24.Mas-Moya J, Dudley B, Brand RE, et al. Clinicopathological comparison of colorectal and endometrial carcinomas in patients with Lynch-like syndrome versus patients with Lynch syndrome. Hum Pathol. 2015;46:1616–1625. [DOI] [PubMed] [Google Scholar]
- 25.Mills AM, Sloan EA, Thomas M, et al. Clinicopathologic comparison of Lynch syndrome-associated and” Lynch-like” endometrial carcinomas identified on universal screening using mismatch repair protein immunohistochemistry. Am J Surg Pathol. 2015. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 26.Buchanan DD, Rosty C, Clendenning M, et al. Clinical problems of colorectal cancer and endometrial cancer cases with unknown cause of tumor mismatch repair deficiency (suspected Lynch syndrome). Appl Clin Genet. 2014;7:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez–Soler M, Pérez–Carbonell L, Guarinos C, et al. Risk of cancer in cases of suspected lynch syndrome without germline mutation. Gastroenterology. 2013;144:926–932. [DOI] [PubMed] [Google Scholar]
- 28.Pérez-Carbonell L, Ruiz-Ponte C, Guarinos C, et al. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut. 2012;61:865–872. [DOI] [PubMed] [Google Scholar]
- 29.Sugawara T, Sato N, Shimizu D, et al. Efficient screening strategy for Lynch syndrome in Japanese endometrial cancer. Tohoku J Exp Med. 2015;235:117–125. [DOI] [PubMed] [Google Scholar]
- 30.Deng G, Chen A, Hong J, et al. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029–2033. [PubMed] [Google Scholar]
- 31.Joensuu EI, Abdel-Rahman WM, Ollikainen M, et al. Epigenetic signatures of familial cancer are characteristic of tumor type and family category. Cancer Res. 2008;68:4597–4605. [DOI] [PubMed] [Google Scholar]
- 32.Watson N, Grieu F, Morris M, et al. Heterogeneous staining for mismatch repair proteins during population-based prescreening for hereditary nonpolyposis colorectal cancer. J Mol Diagn. 2007;9:472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition—update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salahshor S, Koelble K, Rubio C, et al. Microsatellite instability and hMLH1 and hMSH2 colorectal cancer. Lab Invest. 2001;81:535–541. [DOI] [PubMed] [Google Scholar]
- 35.Shia J, Klimstra DS, Nafa K, et al. Value of immunohistochemical detection of DNA mismatch repair proteins in predicting germline mutation in hereditary colorectal neoplasms. Am J Surg Pathol. 2005;29:96–104. [DOI] [PubMed] [Google Scholar]
- 36.Hitchins MP, Lynch HT. Dawning of the epigenetic era in hereditary cancer. Clin Genet. 2014;85:413–416. [DOI] [PubMed] [Google Scholar]
- 37.Boland CR, Koi M, Chang DK, et al. The biochemical basis of microsatellite instability and abnormal immunohistochemistry and clinical behavior in Lynch syndrome: from bench to bedside. Fam Cancer. 2008;7:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohlmann W, Gruber SB. Lynch syndrome. In: GeneReviews at Gene Tests: Medical Genetics Information Resource (database online). Copyright, University of Washington, Seattle. 1993-2014. Available at: http://www.genetsets.org. Accessed September 17, 2015.
- 39.Bonadona V, Bonaïti B, Olschwang S, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 2011;305:2304–2310. [DOI] [PubMed] [Google Scholar]
- 40.Haraldsdottir S, Hampel H, Tomsic J, et al. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology. 2014;147:1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]


