Abstract
Oncogenic mutations in isocitrate dehydrogenase 1 and 2 (IDH1/2) occur in several types of cancer, but the metabolic consequences of these genetic changes are not fully understood. In this study, we performed 13C metabolic flux analysis on a panel of isogenic cell lines containing heterozygous IDH1/2 mutations. We observed that under hypoxic conditions, IDH1-mutant cells exhibited increased oxidative tricarboxylic acid metabolism along with decreased reductive glutamine metabolism, but not IDH2-mutant cells. However, selective inhibition of mutant IDH1 enzyme function could not reverse the defect in reductive carboxylation activity. Furthermore, this metabolic reprogramming increased the sensitivity of IDH1-mutant cells to hypoxia or electron transport chain inhibition in vitro. Lastly, IDH1-mutant cells also grew poorly as subcutaneous xenografts within a hypoxic in vivo microenvironment. Together, our results suggest therapeutic opportunities to exploit the metabolic vulnerabilities specific to IDH1 mutation.
Introduction
Mutations in the metabolic enzymes isocitrate dehydrogenase 1 and 2 (IDH1/2) have been identified in a variety of tumor types, including acute myelogenous leukemia (AML), gliomas, cholangiocarcinomas, and chondrosarcomas (1–9). These mutations are almost exclusively heterozygous point mutations that occur in specific residues within the catalytic pocket, and are suggestive of activating, oncogenic mutations. Although IDH mutants are no longer capable of efficiently carrying out the normal oxidative reaction [converting isocitrate and NADP+ to α-ketoglutarate (αKG), CO2, and NADPH), IDH mutations result in a novel gain-of-function involving the reductive, NADPH-dependent conversion of αKG to (D)2-hydroxyglutarate (2-HG; refs. 10, 11). 2-HG is not typically present at high levels in normal cells but accumulates considerably in cells with IDH1/2 mutations as well as in the tumors of patients with IDH1/2 mutations, and has thus been termed an “oncometabolite” (10–12).
Research into the oncogenic function of mutant IDH1/2 has focused in large part on the effects of 2-HG. Numerous reports have linked 2-HG accumulation to epigenetic changes, which are thought to contribute to alterations in cellular differentiation status (13–22). Additional mutant IDH phenotypes have also been reported, including changes in collagen maturation and hypoxia inducible factor-1α (HIF1α) stabilization (21, 23). These changes likely occur through inhibition of αKG-dependent dioxygenase activity by high levels of 2-HG. However, the diverse roles that αKG-dependent dioxygenases play in the cell and the numerous phenotypes associated with mutant IDH and 2-HG suggest that the phenotypes downstream of 2-HG induction could be cell type- or context-specific. We hypothesize that metabolic alterations induced by IDH mutations may also be present and might be a general phenotype that offers additional approaches to target these tumors. Previous work suggests that overexpression of mutant IDH alters the levels of several metabolites (24) and leads to increased sensitivity to glutaminase inhibitors (25). Studies by Leonardi and colleagues have indicated that the IDH1-mutant enzymes compromise the ability of this enzyme to catalyze the reductive carboxylation reaction (26). However, it is unclear how IDH mutations affect central carbon metabolism in the heterozygous cellular setting, and further exploration into how these metabolic differences could be therapeutically exploited is warranted. An important distinction between IDH1 and IDH2 is their localization in the cytosol/peroxisome and mitochondria, respectively, which may influence the ultimate metabolic phenotype of tumor cells with mutations in either enzyme.
Systems-based approaches that use stable isotope tracers (e.g., [13C]glutamine), mass spectrometry, and network modeling to estimate metabolic fluxes offer a unique means of characterizing intracellular metabolism (27). To understand the metabolic impact of heterozygous IDH mutation in vitro, we have applied 13C metabolic flux analysis (MFA) to a panel of cell lines that differ only with respect to their IDH1- and IDH2-mutant status. Using this approach, we have characterized how cells with wild-type (WT) and mutant IDH1/2 respond to hypoxia and pharmacologic induction of mitochondrial dysfunction.
Materials and Methods
Cell culture
HCT116 and MCF-10A isogenic clones were obtained from Horizon Discovery Ltd and IDH1/2 mutational status was verified by sequencing (28). HCT116 cells were cultured in McCoy’s 5A Modified Medium with 10% FBS. D-2-HG treatments were done at 10 mmol/L and replenished every 48 hours. MCF-10A cells were cultured as described previously (28). HT-1080, SW1353, A549, and 143B cells were cultured in DMEM supplemented with 10% FBS. HT-1080 and SW1353 cells were obtained from the ATCC, and cells were tested and authenticated by single-nucleotide polymorphism fingerprinting. A549 cells were obtained from ATCC and were not further tested or authenticated. 143B cells were kindly provided by Dr. Leonard Guarente (Massachusetts Institute of Technology, Cambridge, MA) and were not further tested or authenticated. Cells were routinely cultured in normoxia (21% O2) and then moved to hypoxia (1%–3% O2, as indicated in the figure legends) for 48 to 72 hours where indicated. Generation of the ρ0 cells is described in the Supplementary Methods. Xenograft assays are described in Supplementary Methods.
Steady-state labeling of organic, amino, and fatty acids was accomplished by culturing subconfluent cells in triplicate in tracer medium for 72 hours in a 6-well plate. Labeling studies of HCT116, SW1353, HT1080, A549, and 143B cells were performed in glucose or glutamine-free DMEM containing 10% FBS and 17.5 mmol/L [1,2-13C2]glucose, 4 mmol/L [U-13C5]glutamine, or 4 mmol/L [5-13C]glutamine. For the HCT116 isogenic cells, the initial seeding density was 150,000 cells per well except for the IDH1 R132H/+ 2H1, IDH1 R132C/+ 2A9, and IDH1 R32C/+ 3A4, which were 250,000 cells per well. Labeling studies of the MCF-10A cells were done in glutamine-free DMEM containing 4 mmol.mg/L [U-13C5]glutamine, 5% horse serum, 20 ng/mL EGF, 10 μg/mL insulin, 0.5 μg/mL hydrocortisone, and 100 ng/mL cholera toxin. Gas chromatography mass spectrometry (GC-MS) analysis is described in Supplementary Methods.
MFA
13C MFA was conducted using INCA, a software package based on the EMU framework (http://mfa.vueinnovations.com; ref. 29). Intracellular concentrations of free metabolites and intra- and extracellular fluxes were assumed to be constant over the course of the tracing experiment. Fluxes through a metabolic network comprising of glycolysis, the pentose phosphate pathway, the TCA cycle, biomass synthesis, and fatty acid synthesis were estimated by minimizing the sum of squared residuals between experimental and simulated MIDs and extracellular fluxes using nonlinear least squares regression (30). The best global fit was found after estimating 100 times using random initial guesses for all fiuxes in the network. A χ2 statistical test was applied to assess the goodness-of-fit using α of 0.01. The 95% confidence intervals for all fluxes in the network were estimated by evaluating the sensitivity of the sum of squared residuals to flux variations (30). Isotopomer Spectral Analysis was performed as previously described (31). See Supplementary Methods for further details of MFA.
Reagents
The following reagents were used at the doses indicated and as described in the text/figure legends: [1,2-13C2]glucose, [3-13C]glucose, [U-13C5]glutamine, [
3C]glutamine, and [5-13C]glutamine (all from Cambridge Isotope Laboratories); IDH-C277 (Xcessbio); HIF1α antibody (610958, BD Bioscinces). Synthesis of IDH1i A is described in Supplementary Methods.
Determination of oxygen consumption
HCT 116 cells were grown at either normoxia or hypoxia (3% O2), and respiration was measured using an XFe96 analyzer (Seahorse Bioscience). Cell growth and assays at 3% O2 were conducted using the Coy Dual Hypoxic Chambers for Seahorse XFe Analyzer (Coy Laboratory Products, Inc.) as described in Supplementary Methods.
Proliferation assays
To calculate doubling time, cells were trypsinized and viable cells were quantified on a ViCell (Beckman-Coulter). Doubling times are presented as the average of three or more independent experiments.
To generate longer-term growth curves, cells were plated at 3,000 cells per well in a 96-well plate in triplicate. Twenty-four hours later, the indicated treatment was started and confluency measurements were taken every 12 hours for 108 to 216 hours using an Incucyte Kinetic Imaging System (Essen BioScience). Confluency data were modeled using a generalized logistic growth equation (equation 1), and the maximum growth rate was estimated using nonlinear regression:
| (1) |
where U and L represent upper and lower asymptotes, t0 represents the time at which cell confluency reaches 50%, and μmax represents the maximum growth rate per hour.
Pharmacologic profiling of the CCLE was performed as previously described (32). The growth inhibition assays are described in Supplementary Methods.
Statistical analysis
All results shown as averages of multiple independent experiments are presented as mean ± SE; results shown as averages of technical replicates are presented as mean ± SD. P values were calculated using a Student two-tailed t test; *, P value between 0.005 and 0.05; **, P value between 0.001 and 0.005; ***, P value <0.001. All errors associated with MFA and ISA of lipogenesis are 95% confidence intervals determined via sensitivity analysis.
Results
Mutant IDH1 compromises metabolic reprogramming under hypoxia
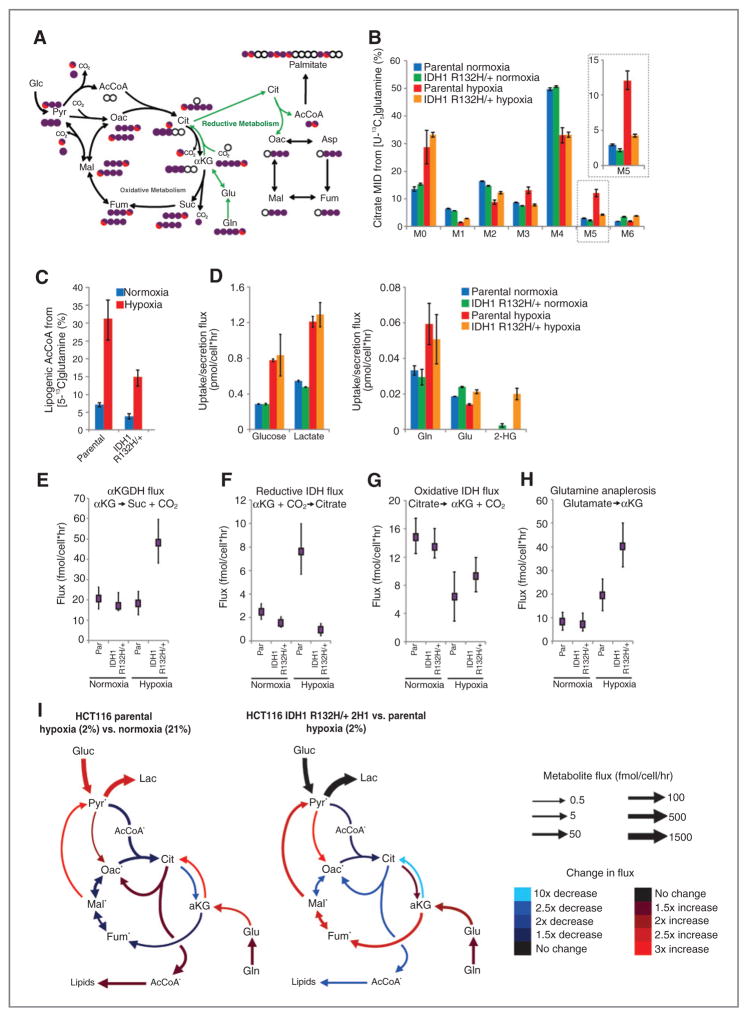
We and others have previously demonstrated that tricarboxylic acid (TCA) metabolism is reprogrammed under hypoxia, and flux through WT IDH1 and/or IDH2 become critical in these contexts (31, 33–37). Oncogenic mutations in IDH1 and IDH2 mitigate these enzymes’ WT function and, in particular, the ability to catalyze the reductive carboxylation reaction while inducing a neomorphic activity that results in the accumulation of D-2-HG (11, 26). Therefore, we hypothesized that cancer cells harboring mutations in either IDH1 or IDH2 may be compromised in their ability to modulate metabolism under low oxygen tensions. To identify metabolic liabilities induced by IDH1 mutations, we applied 13C MFA to isogenic HCT116 colon cancer cells with WT IDH1/2 (parental) or a heterozygous IDH1 mutation, IDH1 R132H/+ (clone 2H1). The IDH1-mutant, but not WT, cells have previously been shown to produce high levels of 2-HG (28). To gauge relative flux through the TCA cycle, each cell line was cultured in the presence of [U-13C5]glutamine (uniformly 13C-labeled glutamine) under normoxic or hypoxic (2% oxygen) conditions for 72 hours, and isotope enrichment in various metabolites was determined via mass spectrometry (Fig. 1A). Both cell lines displayed decreased oxidative TCA flux (as evidenced by decreased M3 αKG) in hypoxia (Supplementary Fig. S1A). Although minimal changes in labeling were detected when comparing the mass isotopomer distribution (MID) of citrate in each cell type grown in normoxia (designated as M0, M1, M2, etc. mass isotopomers, corresponding to ion fragments containing zero, one, or two 13C, respectively), more significant deviations occurred in cells proliferating under hypoxia (Fig. 1B). Parental cells under hypoxia exhibited increased M5 labeling indicative of reductive carboxylation (Fig. 1B), as has been seen previously for many WT IDH1/2 cell lines (31, 33, 35). In contrast, the IDH1 R132H/+ cells showed only a slight increase in the abundance of this mass isotopomer under hypoxia (Fig. 1B). M5 citrate can also be produced via M6 citrate and glutaminolysis (38); however, no increase in the low basal levels of M6 citrate was observed in hypoxia (Fig. 1B). We observed similar changes in the labeling of other TCA metabolites, including M3 fumarate, malate, and aspartate (derived from oxaloacetate), further supporting our finding that IDH1-mutant cells display compromised reductive glutamine metabolism in hypoxia (Fig. 1A and Supplementary Fig. S1B–S1D). To further quantify the metabolism of glutamine through the reductive carboxylation pathway in these cells, we determined the contribution of [5-13C]glutamine to palmitate synthesis using isotopomer spectral analysis (ISA), as this tracer specifically labels acetyl coenzyme A (AcCoA) through the reductive carboxylation pathway (Fig. 1A; ref. 39). Although parental cells increased the contribution of glutamine to lipogenic AcCoA almost 5-fold under hypoxia, cells with a mutant IDH1 allele were compromised in their ability to increase this reductive carboxylation flux (Fig. 1C).
Figure 1.
Isogenic IDH1 mutation compromises metabolic reprogramming under hypoxia. A, carbon atom (represented by circles) transitions and tracers used to detect changes in flux: [U-13C5]glutamine (purple circles) or [5-13C]glutamine (circle with red). The fifth carbon is lost during oxidative TCA metabolism but is retained on citrate, AcCoA, and palmitate in the reductive pathway (green arrows). B, citrate MID labeling from [U-13C5]glutamine from HCT116 parental and HCT116 IDH1 R132H/+ clone 2H1 cells cultured in normoxia or hypoxia (2% oxygen) for 72 hours. Data, representative of more than three independent experiments. Inset highlights changes in % M5 citrate. C, contribution of [5-13C]glutamine to lipogenic AcCoA from cells cultured as in Fig. 1B. D, uptake and secretion fluxes for cells cultured as in Fig. 1B. E–H, α-Ketoglutarate dehydrogenase (E), reductive IDH (F), oxidative IDH (G), and glutamine anaplerosis (H) flux estimates and 95% confidence intervals by the 13C MFA model. I, graphical representation of fluxes determined via MFA. Arrow thickness, level of flux in HCT116 cells cultured in hypoxia. Color, fold difference between hypoxic and normoxic parental HCT116 cells (left) or between hypoxic HCT116 IDH1 R132H/+ 2H1 cells and hypoxic HCT116 parental cells (right).*, metabolites that were modeled as existing in separate mitochondrial and cytosolic pools. aKG, α-Ketoglutarate; cit, citrate; fum, fumarate; gluc, glucose; glu, glutamate; gln, glutamine; lac, lactate; mal, malate; oac, oxaloacetate; pyr, pyruvate. See also Supplementary Methods, Supplementary Fig. S2, and Supplementary Tables S1–S4 for details of MFA model, results, and data.
To characterize the metabolic phenotype of HCT116 cells with WT or IDH1 R132H/+ in a more comprehensive and unbiased manner, we incorporated uptake/secretion fluxes and mass isotopomer data into a network of central carbon metabolism. This model included glycolysis, the pentose phosphate pathway (PPP), TCA metabolism, and various biosynthetic fluxes using [U-13C5]glutamine and [1,2-13C2]glucose (for the oxidative PPP bifurcation), as these tracers provide optimal flux resolution throughout central carbon metabolism (40). An elementary metabolite unit (EMU)–based algorithm was used to estimate fluxes and associated confidence intervals in the network (41, 30), and a detailed description of the model, assumptions, and the complete data set are included as Supplementary Material. As expected, glucose and lactate fluxes were significantly increased by hypoxia in both cell lines, and significant 2-HG secretion occurred only in IDH1 R132H/+ cells (Fig. 1D). Notably, 2-HG secretion was elevated under hypoxia, consistent with previous observations of 2-HG accumulation at low oxygen tension (33).
The modeling data comparing parental HCT116 cells grown in normoxia and hypoxia highlight some of the important metabolic changes that occur at low oxygen tensions (Fig. 1E–I, Supplementary Fig. S2, and Supplementary Tables S1–S4). In the HCT116 parental cells, pyruvate dehydrogenase and oxidative TCA metabolism are decreased under hypoxia, whereas pyruvate cycling through malic enzyme (ME) and pyruvate carboxylase (PC) are elevated under these conditions. Parental cells increased reductive IDH flux several fold, such that net IDH flux slightly favored the direction of reductive carboxylation (Fig. 1F, G, and I). As with the MID changes (Fig. 1B), only modest changes in metabolism were detected when comparing parental HCT116 cells with IDH1 R132H/+ 2H1 grown under normoxia (Fig. 1E–H and Supplementary Tables S1–S4). However, mutant cells maintained high oxidative IDH and αKG-dehydrogenase (αKGDH) fluxes and were unable to induce reductive carboxylation under hypoxia relative to the parental cells (Fig. 1E–G, I, and Supplementary Tables S1–S4). This oxidative TCA flux was maintained by increased glutamine anaplerosis and flux through ME and PC (Fig. 1H and I and Supplementary Tables S1–S4). Overall, these results demonstrate that significant reprogramming of TCA metabolism occurs in cells at 2% oxygen, and expression of IDH1 R132H/+ abrogates the cells’ ability to respond appropriately to hypoxic stress.
Compromised reductive TCA metabolism is specific to cells with mutant IDH1
The MFA results above suggest that heterozygous IDH1 mutations disrupt the metabolic response of cells to hypoxic stress. To determine whether this metabolic deficiency is common to cells with either IDH1 or IDH2 mutations, we interrogated a panel of previously reported IDH1- and IDH2-mutant isogenic cells (28) and measured the ability of each to initiate reductive carboxylation under 2% oxygen. With the exception of IDH2 R140Q/+cells, these mutant cell lines exhibit a ≥25-fold increase in 2-HG compared with parental HCT116 cells (28). We cultured each clone with [U-13C5]glutamine under normoxia and hypoxia, quantifying M5 citrate abundance (see Fig. 1A) to determine the relative extent of reductive carboxylation induction. All but one of the IDH1-mutant clones were consistently compromised in their ability to increase reductive carboxylation activity under hypoxia (Fig. 2A). The one exception being the IDH1 R132H/+ 2C11 clone, which showed a weaker phenotype relative to the other IDH1-mutant clones (Fig. 2A). This is likely explained by a lower level of IDH1 R132H protein (Fig. 2B) and lower level of 2-HG than the IDH1 R132H/+ 2H1 clone (28).
Figure 2.
Compromised reductive TCA metabolism is specific to cells with mutant IDH1. A, relative level of reductive glutamine metabolism, determined by M5 labeling of citrate from [U-13C5] glutamine in HCT 116 parental cells with or without 10 mmol/LD-2-HG, or HCT116 IDH1/2-mutant isogenic cells cultured in normoxia orhypoxia (2% oxygen). Percentage of M5 citrate levels are normalized to HCT116 parental cells in normoxia for each experiment. B, Western blot of HCT116 parental, HCT116 IDH1 R132H/+ clone 2C11, and HCT116 IDH1 R132H/+ clone 2H1 showing levels of IDH1 R132H and total levels of IDH1. C, ratio of α-ketoglutarate to citrate from cells cultured as in Fig. 2A. D, contribution of [U-13C5]glutamine to lipogenic AcCoA from cells cultured as in Fig. 2A. Four independent experiments are shown. E, correlation of reductive glutamine metabolism in 2% oxygen (as measured by % M5 citrate from [U-13C5]glutamine) to 2-HG ng/1 million cells. Note that HCT116 IDH2 R172K/+ 45G9 and 59F10 are not included in this figure. F, relative level of reductive glutamine metabolism, determined by M5 labeling of citrate from [U-13C5]glutamine in HCT116 parental cells, which were stably infected with doxycycline-inducible shRNA’s targeting IDH1 or IDH2, or nontargeting control (NTC). Cells were cultured for 48 hours in normoxia or hypoxia (1% oxygen) in the presence of 100 ng/mL doxycycline. Citrate labeling was determined via liquid chromatography/mass spectrometry. G, relative level of reductive glutamine metabolism, determined by M5 labeling of citrate from [U-13C5]glutamine in MCF-10A cells cultured in normoxia or hypoxia (2% oxygen) for 72 hours. H, contribution of [U-13C5]glutamine to lipogenic AcCoA from cells cultured as in Fig. 2G.
Unlike mutant IDH1 cells, HCT116 cells with IDH2 mutations exhibited levels of M5 citrate under hypoxia that were comparable with the parental cells (Fig. 2A). Addition of 10 mmol/L D-2-HG to the culture media also had a minimal effect on TCA metabolism (Fig. 2A). These data suggest that high 2-HG levels alone are unable to inhibit reductive carboxylation activity, even though this dose of exogenous D-2-HG is sufficient to induce the 2-HG–dependent epithelial–mesenchymal transition (EMT) phenotype previously described in these cell lines (28). Consistent changes were observed when normalizing M5 citrate to M5 glutamate (Supplementary Fig. S3A), and levels of M6 citrate were not high in any of the clones (Supplementary Fig. S3B), providing evidence that such changes are specific to the IDH/aconitase node of metabolism. Similar trends were also observed when measuring the ratio of αKG/citrate under normoxia and hypoxia (Fig. 2C), another metric that describes the extent of reductive versus oxidative IDH flux (36, 42). Finally, the contribution of glutamine to lipogenic AcCoA under hypoxia was significantly lower in cells with IDH1 mutations but not those with IDH2 mutations or exogenous 2-HG (Fig. 2D). Overall, the extent that each IDH1-mutant cell line produced 2-HG correlated with their ability to activate reductive carboxylation flux under hypoxia (Fig. 2E), whereas IDH2-mutant cells did not adhere to this trend.
We next conducted shRNA-mediated knockdown of IDH1 and IDH2 in parental HCT116 cells to examine the roles of WT IDH1 and IDH2 in mediating reductive glutamine metabolism (Supplementary Fig. S3C and S3D). Knockdown of IDH1 decreased levels of M5 citrate in cell populations cultured with [U-13C5]glutamine, whereas cells expressing shRNAs targeting IDH2 exhibited the same or higher M5 citrate (Fig. 2F). These results are consistent with previous studies in other cell lines (31), highlighting the importance of WT IDH1 expression in reprogramming TCA metabolism under hypoxia, and further suggest that IDH1 mutation selectively impedes WT IDH1 function in these cells.
To determine whether this mutant IDH1-induced metabolic deficiency is specific to the HCT116 genetic background or more broadly applicable to cells of different tissue origin, we performed similar analyses using MCF-10A immortalized mammary epithelial cells with heterozygous IDH1 mutations (28). When cultured for 3 days under normoxia or hypoxia, two distinct IDH1 R132H/+ clones were compromised in their ability to generate M5 citrate or lipogenic AcCoA from [U-13C5]glutamine (Fig. 2G and H and Supplementary Fig. S3E). Thus, cells with heterozygous IDH1 mutations, but not IDH2 mutations or exogenous 2-HG, are compromised for glutamine reductive carboxylation under hypoxia.
Cells with endogenous IDH1 and IDH2 mutations respond differently to mitochondrial stress
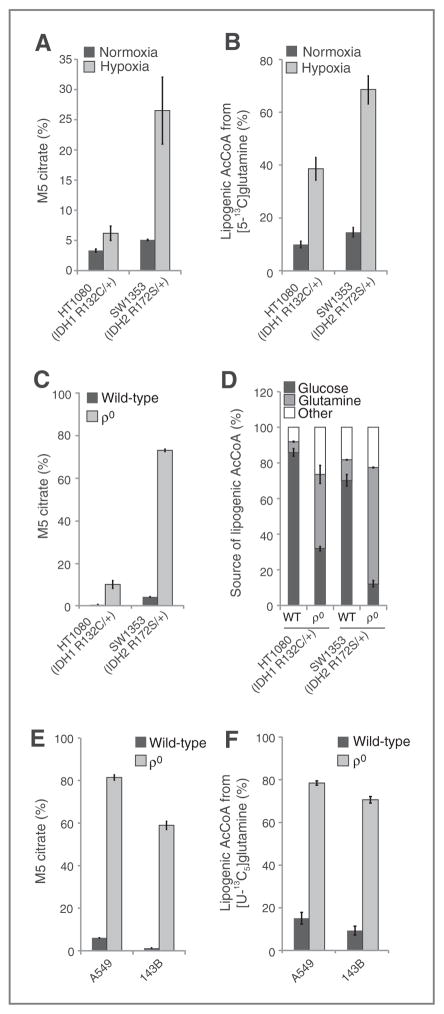
To examine whether these trends are observed in cell lines from cancers with endogenous IDH mutations, we evaluated the ability of two cell lines harboring IDH1 or IDH2 mutations to activate reductive carboxylation under conditions of mitochondrial stress. When switched to hyp-oxic growth, HT1080 IDH1 R132C/+ fibrosarcoma cells exhibited a significantly decreased ability to induce reductive glutamine metabolism in comparison with SW1353 IDH2 R172S/+ chondrosarcoma cells (Fig. 3A and Supplementary Fig. S4A). HT-1080 cells also used less glutamine for de novo lipogenesis than the SW1353 cells in 1% oxygen tension (Fig. 3B). Thus, a cell line with an endogenous mutation in IDH1, but not IDH2, displays compromised reductive glutamine metabolism in hypoxia.
Figure 3.
Cells with endogenous IDH1 and IDH2 mutations respond differently to mitochondrial stress. A, relative level of reductive glutamine metabolism, determined by M5 labeling of citrate from [U-13C5]glutamine in HT-1080 and SW1353 cells cultured in normoxia or hypoxia (2% oxygen) for 72 hours. B, contribution of [5-13C]glutamine to lipogenic AcCoA from HT-1080 and SW1353 cells cultured in normoxia or hypoxia (1% oxygen) for 72 hours. C, relative level of reductive glutamine metabolism, determined by M5 labeling of citrate from [U-13C5]glutamine in HT-1080 and SW1353 WT or ρ0 cells. D, contribution of [U-13C5]glutamine and [U-13C6]glucose to lipogenic AcCoA from cells cultured as in Fig. 3C. E, relative level of reductive glutamine metabolism, determined by M5 labeling of citrate from [U-13C5]glutamine in A549 and 143B WT or ρ0 cells. F, contribution of [U-13C5]glutamine to lipogenic AcCoA from cells cultured as in Fig. 3E.
In addition to low oxygen tensions, an alternative means of inducing reductive TCA metabolism is through the inhibition of oxidative phosphorylation (OXPHOS; refs. 34, 42). To compromise OXPHOS, we generated ρ0 cells that lack a functional electron transport chain (ETC) from various cell lines using established methods (43). As expected, oxidative mitochondrial metabolism was virtually extinguished, as evidenced by M3 and M3/M5 labeling of αKG in ρ0 cells generated from IDH-mutant cells (HT1080 and SW1353) or other cancer cell lines with WT IDH1/2 (143B osteosarcoma, A549 non–small cell lung cancer; Supplementary Fig. S4B–S4E). However, HT1080 IDH1 R132C/+ ρ0 cells were compromised in their ability to generate citrate and palmitate through reductive glutamine metabolism, whereas SW1353 and both of the IDH1/2 WT ρ0 cell lines were able to efficiently induce reductive carboxylation and use glutamine for lipid synthesis (Fig. 3C–F). Both the HT1080 and SW1353 ρ0 cells continued to use glucose for lipid synthesis, and this contribution was higher in the HT1080 ρ0 cells (Fig. 3D). The increased glucose utilization in HT1080 ρ0 cells compared with SW1353 ρ0 cells was facilitated by anaplerosis through pyruvate carboxylase, as demonstrated by increased labeling in TCA intermediates from [3-13C]glucose (Supplementary Fig. S4F and S4G). These results provide evidence that hypoxia and mitochondrial dysfunction drive reprogramming of the TCA cycle, and cells with spontaneously acquired IDH1 mutations are unable to efficiently reprogram metabolism to induce reductive glutamine carboxylation.
Mutant IDH1 affects TCA metabolism in vivo
The metabolic deficiencies of IDH1-mutant cells occur at oxygen tensions that are likely to occur in solid tumors and some normal tissues (44). To determine whether these metabolic phenotypes arise in vivo, we generated subcutaneous xenografts using parental, IDH1 R132H/+ 2H1, and IDH1 R132C/+ 2A9 HCT116 cells. After tumors achieved a minimum diameter of 0.8 cm, mice were infused with [1-13C]glutamine for 6 hours to achieve steady-state isotope enrichment in plasma and tumor (Fig. 4A and B; ref. 45). A targeted metabolomic analysis was performed on plasma and tumor extracts to quantify isotope enrichment and metabolite abundances. Significant 2-HG was detected only in the IDH1-mutant tumors (Fig. 4C). Insufficient isotope enrichment was achieved in plasma and intratumoral glutamine/αKG to detect label on citrate via reductive carboxylation (Fig. 4A, B, and D). However, in agreement with the results obtained from in vitro studies, the αKG/citrate ratio was significantly lower in IDH1-mutant tumors compared with those generated using parental HCT116 cells (Fig. 4E), indicating that TCA metabolism may also be perturbed in tumors comprised of IDH1-mutant cells. In addition, the contribution of glutamine anaplerosis to the αKG pool was significantly elevated in IDH1 R132H/+ and IDH1 R132C/+ tumors (Fig. 4F), which was also observed in our MFA results (Fig. 1H and I). Thus, the available data are consistent with our in vitro MFA results and provide evidence that TCA metabolism is similarly compromised by IDH1 mutations in vivo.
Figure 4.
Mutant IDH1 affects TCA metabolism in vivo. A and B, box–whisker plots showing [1-13C]glutamine enrichment in plasma (A) and tumor (B) from mice with xenografts derived from the indicated cell lines. Black dot, mean; center line, median; box, interquartile range; whiskers, maximum and minimum values. C, relative 2-HG levels from xenografts, normalized to HCT116 parental xenografts. D, percentage of M1 αKG derived from [1-13C]glutamine in tumor xenografts. E, nMols αKG/nMols citrate ratio from xenografts grown from the indicated HCT116 cells. F, glutamine anaplerosis, as determined by M1 labeling of αKG relative to M1 labeling of glutamine from xenografts grown from the indicated HCT116 cells.
Inhibition of mutant IDH1 does not rescue reprogramming of TCA metabolism
One possible explanation for the decrease in reductive carboxylation flux in IDH1-mutant cells is that localized substrate (αKG and NADPH) consumption by the mutant enzyme for production of 2-HG compromises this activity. Therefore, we examined whether pharmacologic inhibition of mutant IDH1 could increase reductive carboxylation activity and rescue the ability of cells to use this pathway for growth under hypoxia. To test this hypothesis, we treated IDH1 R132H/+ 2H1 or IDH1 R132C/+ 2A9 cells with a mutant IDH1 inhibitor (IDH1i A) similar to a previously described structural class (Fig. 5A; refs. 22, 46). Doses of 10 μmol/L were able to decrease 2-HG levels more than 25-fold in both clones (Fig. 5B). As would be expected from an engineered cell line that does not exhibit growth dependence on mutant IDH1 or 2-HG, 10 μmol/L of IDH1i A had no appreciable effect on the growth rate of either cell line (Fig. 5C). Both short-term (3 day) and long-term (31 day) treatment with 10 μmol/L IDH1i A induced minimal changes in metabolite abundances beyond 2-HG (Fig. 5D) and effectively reversed the mutant IDH-dependent EMT phenotype exhibited by these cells (Supplementary Fig. S5A).
Figure 5.
Inhibition of mutant IDH1 does not rescue reprogramming of TCA metabolism. A, structure and biochemical data for mutant IDH1 inhibitor, IDH1i A. Note that this is the (S) enantiomer. B, 2-HG levels in HCT116 IDH1 R132H/+ 2H1 and HCT116 IDH1 R132C/+ 2A9 cell lines treated with the indicated concentrations of IDH1i A for 3 days. C, doubling time of cells cultured with or without 10 μmol/L of IDH1i A. D, change in total metabolite levels of HCT116 IDH1 R132H/+ 2H1 and HCT116 IDH1 R132C/+ 2A9 cells cultured in the presence or absence of 10 μmol/L of IDH1i A for 3 or 31 days, the final 72 hours of which the cells are grown in hypoxia (2% oxygen). E, citrate MID labeling from [U-13C5]glutamine from cells cultured as in Fig. 5D. Dotted line over M5 citrate represents average % M5 citrate observed in HCT116 parental cells cultured in hypoxia. F, αKG/citrate ratio from cells cultured as in Fig. 5D. G, contribution of [U-13C5]glutamine to lipogenic AcCoA from cells cultured as in Fig. 5D. H, 2-HG levels in HCT116 IDH1 R132H/+ 2H1 and HCT116 IDH1 R132C/+ 2A9 cell lines treated with the indicated concentration of IDH1-C227 for 3 days. I, relative level of reductive glutamine metabolism, determined by M5 labeling of citrate from [U-13C5]glutamine in HCT116 IDH1 R132H/+ 2H1 and HCT116 IDH1 R132C/+ 2A9 cell lines treated with 1 μmol/L of IDH1-C227 for 3 or 12 days, the final 72 hours of which the cells are grown in hypoxia (2% oxygen). J, αKG/citrate ratio from cells cultured as in Fig. 5I. K, contribution of [U-13C5]glutamine to lipogenic AcCoA from cells cultured as in Fig. 5I.
IDH1i A did not rescue the ability of cells to initiate reductive TCA metabolism under hypoxia, as labeling of citrate (Fig. 5E) and other metabolites (Supplementary Fig. S5B–S5D) from [U-13C5]glutamine was not increased compared with vehicle treatment. Other indices of reductive TCA metabolism, including the ratio of αKG/citrate and contribution of glutamine to lipid biosynthesis, also indicated that IDH1i A failed to rescue reductive carboxylation flux in these cells (Fig. 5F and G). At 10 μmol/L, the dose that showed maximal 2-HG inhibition, IDH1i A mildly inhibited reductive carboxylation in the WT parental cells (Supplementary Fig. S5E and S5F), potentially due to off-target effects on WT IDH1 at high concentrations. To further address this issue, we also treated the IDH1-mutant cells with an additional inhibitor of mutant IDH1 at more moderate concentrations (Fig. 5H). We again observed no rescue in reductive glutamine metabolism (Fig. 5I–K), providing evidence that inhibition of IDH1-mutant activity may be insufficient to remove the block in metabolic reprogramming in response to hypoxic stress.
Cells expressing mutant IDH1 are sensitive to pharmacologic inhibition of mitochondrial oxidative metabolism
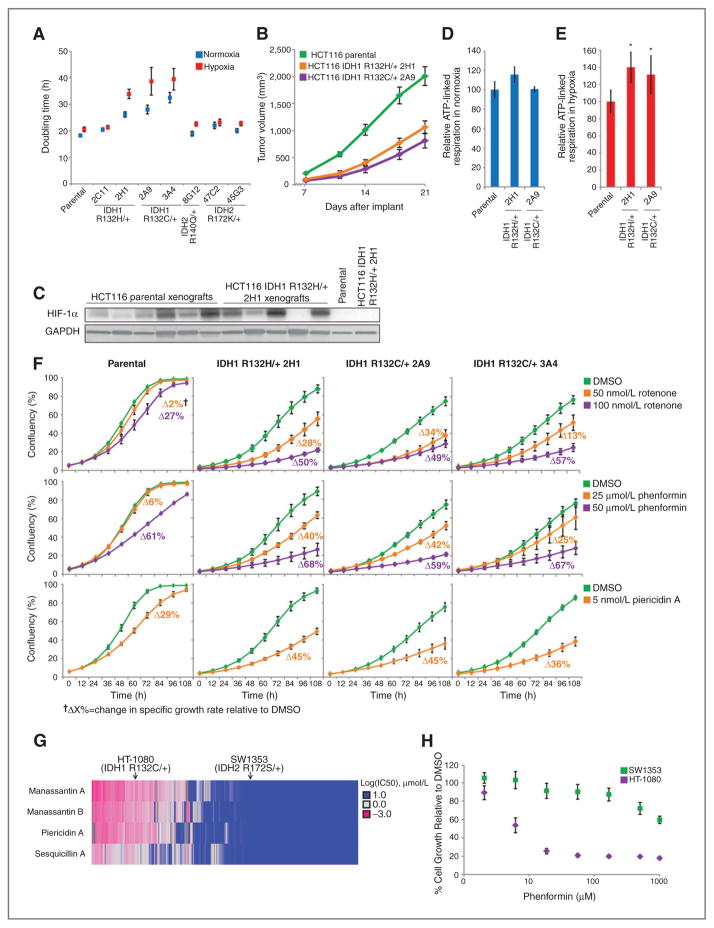
In comparing the growth rates of the HCT116 panel under normoxia and hypoxia, we observed that mutant IDH1 cells grew more poorly under conditions of low oxygen tension than parental cells or those expressing mutant IDH2 (Fig. 6A). HCT116 IDH1-mutant xenografts also grew at a significantly slower rate than the HCT116 parental cells (Fig. 6B), conditions that exhibited significant stabilization of HIF1α in both parental and IDH1-mutant tumors (Fig. 6C). The growth rate of IDH2-mutant cells as xenografts was also significantly decreased relative to the parental cells (Supplementary Fig. S6A), though our available data do not yet support a hypothesis for how IDH2 mutations affect in vivo growth.
Figure 6.
Cells expressing mutant IDH1 are sensitive to pharmacologic inhibition of mitochondrial oxidative metabolism. A, doubling times of HCT116 cells cultured in normoxia or hypoxia (2% oxygen) for 72 hours. B, growth curves for HCT116 parental and IDH1-mutant xenografts. C, Western blot showing HIF1α expression in HCT116 parental and HCT116 IDH1 R132H/+ 2H1 cells grown in normoxia in cell culture (last two lanes) or as xenografts. D, ATP-linked oxygen consumption for the indicated cell lines grown in normoxia. E, ATP-linked oxygen consumption for the indicated cell lines grown in hypoxia (3% O2). F, growth charts from cells cultured as indicated. Images were acquired every 12 hours to measure confluency. Change in growth relative to DMSO treatment (ΔX%) was calculated using a generalized logistics growth model for batch culture and represents the change in the specific growth rate relative to the DMSO treatment for the indicated cell line. G, heatmap displaying IC50 values to four inhibitors of mitochondrial metabolism for more than 500 cancer cell lines; HT-1080 and SW1353 cells are indicated. H, growth of HT-1080 and SW1353 cells under the indicated concentration of phenformin for 48 hours.
The slow growth of IDH1-mutant cells in the xenograft model suggests that altered TCA metabolism may contribute to the slower growth of these cells under the decreased oxygen levels in vivo. IDH1-mutant cells also exhibited increased oxidative TCA metabolism under hypoxia compared with parental cells (Fig. 1E and I), providing evidence that they are more dependent on OXPHOS. To further confirm this phenotype, we measured oxygen consumption in parental and IDH1-mutant cells under both normoxia and hypoxia. Consistent with our MFA results, basal respiration was not significantly altered in parental and IDH1-mutant cells under normoxic conditions, though uncoupled respiration was decreased in the IDH1-mutant cells (Supplementary Fig. S6B–S6E). Notably, mutant IDH1 cells exhibited significantly higher oligomycin-sensitive oxygen consumption under hypoxia compared with parental HCT116 cells, an effect not reproduced under normoxia (Fig. 6D and E and Supplementary Fig. S6F and S6G). Therefore, we hypothesized that, as with growth in hypoxia, cells harboring IDH1 mutations may be more susceptible to inhibition of oxidative mitochondrial metabolism than cells with WT IDH1/2 or mutant IDH2.
To address this question, we cultured parental HCT116 cells and three IDH1-mutant clones in the presence of several compounds that inhibit Complex I of the ETC and OXPHOS. Confluency measurements were taken every 12 hours, and the maximum specific growth rate of each cell was determined using a generalized logistic growth model and compared with vehicle treatment for each cell line (Fig. 6F). The proliferation rate of cells with mutant IDH1 was significantly more affected than that of parental HCT116 cells in response to discreet dose ranges of Complex I inhibitors. On the other hand, the IDH2 R172K/+ cells displayed no such increased sensitivity with the same treatments (Supplementary Fig. S6H). This altered sensitivity is not due to differences in target modulation, as 100 nmol/L rotenone effectively shut down oxidative TCA cycle flux in all cells tested (Supplementary Fig. S6I). Treatment of parental HCT116 cells with 100 nmol/L rotenone also induced reductive carboxylation, whereas R132H/+ 2H1 and R132C/+ 2A9 HCT116 cells were less able to increase flux through this pathway (Supplementary Fig. S6J and S6K). Treatment with Antimycin A, an inhibitor of Complex III of the ETC, also inhibited oxidative TCA metabolism (Supplementary Fig. S6L). Induction of reductive carboxylation by Antimycin A was observed in the parental, but not IDH1-mutant, cells (Supplementary Fig. S6M and S6N). However, this compound had differential effects on succinate labeling compared with rote-none (Supplementary Fig. S6O). Notably, IDH1-mutant cells did not exhibit increased sensitivity to Antimycin A (Supplementary Fig. S6P), suggesting that Complex III inhibition suppresses growth through distinct mechanisms compared with Complex I inhibitors (e.g., reactive oxygen species generation, and pyrimidine synthesis; ref. 43). Thus, these data indicate that IDH1-, but not IDH2-, mutant cells are selectively sensitive to Complex I inhibitors.
To determine whether IDH1 mutants are generally more sensitive to other treatments, we examined the effect of the cell-cycle inhibitor flavopyridol in the HCT116 panel of cells. IDH1-mutant cells did not display increased sensitivity in comparison with parental or IDH2-mutant cells (Supplementary Fig. S6Q), further suggesting that the differential sensitivity we observe is specific to inhibitors of mitochondrial metabolism. These results indicate that oncogenic IDH1 mutations induce cells to rely more heavily on Complex I of the ETC, rendering these cancer cells more susceptible to inhibition of this pathway compared with cells with WT IDH1/2 or mutant IDH2 alleles.
Finally, to evaluate whether these results are relevant to other cells, we interrogated the Cancer Cell Line Encyclopedia (CCLE; ref. 32), which contains compound sensitivity data across more than 500 cell lines for four ETC inhibitors (Fig. 6G; ref. 47). Cell lines clustered well into sensitive and insensitive groups, suggesting these compounds show consistent behavior across a wide panel of cell lines. Notably, IDH1-mutant HT1080 cells fell into the sensitive group, whereas IDH2-mutant SW1353 cells fell into the insensitive group (Fig. 6G). The differential effects of mitochondrial metabolism inhibitors were not likely due to HT1080 being generally more sensitive to compound treatments, as the sensitivities of HT1080 and SW1353 to a broad array of more than 1,300 compounds were within one SD of each other (Supplementary Fig. S6R). Furthermore, HT1080 cells were significantly more sensitive than SW1353 cells to phen-formin treatment, a compound that was not included in the CCLE screening set (Fig. 6H). Together, these data indicate that IDH1 mutation may substantially sensitize cells to inducers of mitochondrial stress.
Discussion
Since the discovery of oncogenic mutations in IDH1 and IDH2, significant efforts have been made to elucidate the mechanisms driving tumorigenesis in these cancers. Owing to the accumulation of D-2-HG in these tumors, researchers have focused on the role of this oncometabolite in regulating the phenotype of IDH1/2-mutant cancer cells. For example, high D-2-HG levels and other metabolites regulate the activity of αKG-dependent dioxygenases that control many distinct cellular processes (12). However, the diverse roles of these enzymes in mediating activities ranging from collagen hydroxylation and HIF stabilization to epigenetics regulation complicate identification of the specific process(es) driving tumor-igenesis in each tumor type.
Despite the central role of these enzymes in cellular metabolism, surprisingly few investigations have addressed the metabolic changes that occur as a result of these genetic modifications. Here, we find that IDH1 mutations cause cells to increase flux through the oxidative TCA cycle, increase respiration, and compromise the conversion of glutamine to citrate, AcCoA, and fatty acids under hypoxia compared with those with WT IDH1 (Fig. 7). Others have previously shown that IDH1-mutant proteins are biochemically compromised with respect to this latter functionality, suggesting that cells harboring such mutations may be similarly defective under certain conditions (26). However, the cellular consequences of this effect have not been well characterized within intact, heterozygous, IDH-mutant cells. Proliferating cells must double their membrane lipids to successfully complete cell division, and evidence suggests that tumors may rely more on de novo lipogenesis than do nonneoplastic tissues, and inhibition of lipid synthesis decreases tumor growth in vivo (48, 49). In addition, AcCoA is an important precursor for a number of other molecules, including cholesterol, phospholipids, amino acid modifications, and histone acetylation (50). Interestingly, previous studies have also found that overexpression of mutant IDH leads to a decrease in N-acetyl amino acids, and these changes were also observed when comparing WT human glioma tissue with that of tumors with mutant IDH1 (24). This suggests that other AcCoA-dependent molecules may be similarly perturbed in the IDH1-mutant setting. Our application of MFA to IDH1-mutant cells builds upon these results by addressing the functional consequences of heterozygous IDH1 mutations and in particular the metabolic limitations that arise in tumors cells with these genetic modifications. Given the importance of each of these AcCoA-dependent processes for cellular homeostasis and proliferation, we speculate that the reduced metabolic flexibility of these cells contributes to the decrease in growth rate that we observe in the IDH1-mutant cells under conditions of decreased oxidative mitochondrial metabolism.
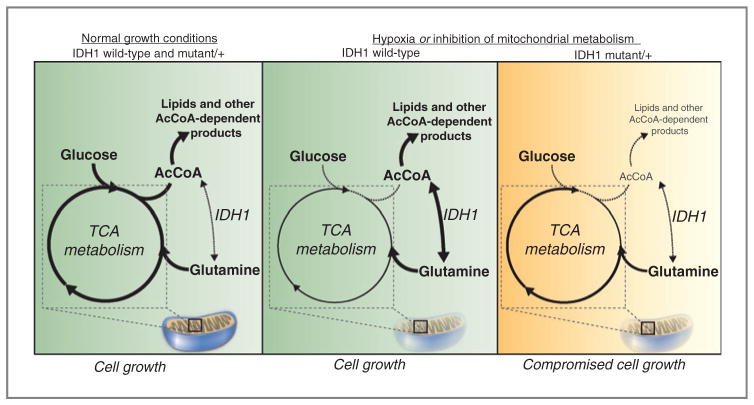
Figure 7.
Mutant IDH1 sensitizes cells to inhibition of oxidative mitochondrial metabolism. Left, under normal growth conditions, glucose is metabolized oxidatively in the mitochondria, and AcCoA and lipids are derived mainly from glucose carbons. Middle, in IDH1 WT cells, inhibition of oxidative mitochondrial metabolism (induced by growth in hypoxia or pharmacologic inhibitors of the ETC) limits glucose flux to the mitochondria, and cells instead rely on reductive glutamine metabolism via IDH1 to provide carbons for AcCoA generation and lipid synthesis. Right, when oxidative mitochondrial metabolism is inhibited, cells with a mutant IDH1 allele are unable to fully induce reductive glutamine metabolism and are thus compromised for AcCoA and lipid production, leading to decreased cell growth.
Although 2-HG–mediated control of αKG-dependent dioxygenase activity clearly plays a role in tumorigenesis driven by IDH mutations (13–18, 20–23), our results provide insights into therapeutic strategies that exploit the metabolic vulnerabilities caused by partial loss of WT IDH1 function. Interestingly, we observe that IDH1-mutant cells do not exhibit pronounced metabolic differences in normoxia; however, growth in low oxygen tensions or with pharmacologic inhibitors of mitochondrial metabolism results in the emergence of dramatic metabolic changes. Our MFA results identify several enzymes and pathways that are altered under hypoxia and in particular in the context of IDH1 mutations. Although compartment-specific IDH fluxes cannot be resolved with these data, these findings further highlight the importance of WT IDH1 activity in mediating reductive glutamine metabolism. Importantly, our results provide evidence that IDH1 mutations functionally compromise cellular metabolism under conditions of low oxygen levels, with the most pronounced effects being increased dependence on oxidative mitochondrial metabolism and an inability to induce reductive glutamine metabolism. We artificially induced such stresses using pharmacologic inhibitors of Complex I or manipulation of the oxygen tensions, and observed selective growth rate reductions in several IDH1-mutant cells, but not in parental or IDH2-mutant cancer cells. Other recent studies have also highlighted the importance of oxidative mitochondrial metabolism for tumor cell growth and survival both in vitro and in vivo (45, 51).
These results suggest that compromised IDH1 function may affect the proliferative capacity of tumor cells and furthermore that IDH1-mutant tumors may be sensitive to inhibitors that perturb mitochondrial metabolism. When comparing the metabolic phenotype of tumor xenografts derived from parental or IDH1-mutant cells with our in vitro results, similar changes were detected, including increased glutamine anaplerosis and a decreased αKG to citrate ratio. The increase in glutamine anaplerosis we observe in the IDH1-mutant cells is in agreement with previous findings, which suggest that IDH1-mutant cells display an increased sensitivity to glutaminase inhibitors (25). The similar metabolic changes that could be reliably measured in vivo suggest that the altered sensitivity we observe to inhibitors of mitochondrial metabolism in vitro may also be true in vivo. Additional studies are required to determine if cellular proliferation in the tumor microenvironment alone can drive hypoxia and induce reductive glutamine metabolism. Regardless, tumors would still be expected to increase their reliance on WT IDH1 (or cytosolic TCA) activity when treated with phenformin or other inhibitors of mitochondrial metabolism, suggesting that these strategies could be efficacious in IDH1-mutant cancers. As such, this increased susceptibility of cultured IDH1-mutant cells relative to parental cells or IDH2-mutant cells provides intriguing evidence of a potential therapeutic strategy associated with IDH1 mutational status and warrants further investigation in preclinical models.
We find that inhibition of mutant IDH1 is unable to reverse the observed metabolic phenotype. The DNA hypermethylator phenotype, which is highly associated with IDH mutation, is also not entirely reverted by a mutant IDH1 inhibitor (22), providing further evidence that some, but not all, mutant IDH-dependent phenotypes may be reversed by inhibitors targeting 2-HG production. Mechanistically, this result also suggests that the metabolic defect we observe may be independent of 2-HG production. A previous study used biochemical assays to quantify the effects of IDH1 mutations on reductive carboxylation activity, and, in agreement with our findings here, demonstrated that the mutant enzymes are unable to catalyze the conversion of αKG and CO2 to isocitrate (26). This study concluded that the subunits in a WT/mutant heterodimer function independently; however, our modeling data indicate that heterozygous IDH1 mutations lead to a much greater than 50% inhibition of reductive glutamine metabolism (Fig. 1F), suggesting a possible dominant effect of the mutant protein in cells or alternatively global metabolic reprogramming in response to the compromised cytosolic IDH1 activity caused by these mutations.
Importantly, as small molecules capable of inhibiting mutant IDH1 enzymatic activity and preventing D-2-HG accumulation fail to rescue mutant cell metabolism under hypoxia, this suggests that combinatorial therapeutic strategies that block oncogenic D-2-HG production (e.g., via a mutant-selective inhibitor of enzyme function), while simultaneously targeting mutant IDH1-induced metabolic liabilities, may be a viable option for therapy. Such an approach could involve IDH1-mutant inhibitor treatment to attenuate any prosurvival or dedifferentiation effects of D-2-HG, while increasing the tumor’s reliance on WT IDH1 activity through an inhibitor of oxidative mitochondrial metabolism. As drugs that could target the mutant IDH1 metabolic phenotype are already in the clinic (metformin, phenformin) and inhibitors of mutant IDH1 are currently being developed (22, 46), it is hoped that this hypothesis will be tested in the clinic in the near future.
Acknowledgments
The authors thank Nicholas Keen, William Sellers, Jonathan Coloff, Julian Levell, Charles Stiles, Juliet Williams, Brant Firestone, Wenlin Shao, Jonathan Solomon, Frank Stegmeier, and the Novartis Postdoctoral Fellows for helpful discussions of this work.
Grant Support
This work was supported by Novartis Institutes for BioMedical Research, American Cancer Society Institutional Research Grant 70-002 (C.M. Metallo), the DOD Lung Cancer Research Program Grant W81XWH-13-1-0105 (C.M. Metallo), and a UC Cancer Research Coordinating Committee grant (C.M. Metallo). S.J. Parker is supported by the NIH/National Institute of Biomedical Imaging and Bioengineering Interfaces Training Grant. A.R. Grassian is the recipient of a presidential postdoctoral fellowship from Novartis Institutes for Biomedical Research. M.G. Vander Heiden and S.M. Davidson acknowledge support from NIH grants R01CA168653 and 5-P30-CA14051-39, and support from the Koch Institute/DFHCC Bridge Project, the Bur-rough’s Wellcome Fund, the Smith Family, the Ludwig Foundation, and the Damon Runyon Cancer Research Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest
A.S. Divakaruni is a consultant/advisory board member of Seahorse Bioscience. M.G. Vander Heiden has ownership interest and is a consultant/advisory board member of Agios Pharmaceuticals. A.N. Murphy received a commercial research grant and is a consultant/advisory board member of Seahorse Bioscience. C. Straub, J.D. Growney, and R. Pagliarini have ownership interest in Novartis. C.M. Metallo has honoraria from the speakers’ bureau of Agios Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors’ Contributions
Conception and design: A.R. Grassian, S.J. Parker, X. Zhang, F. Lin, M.G. Vander Heiden, A.N. Murphy, R. Pagliarini, C.M. Metallo
Development of methodology: A.R. Grassian, S.J. Parker, S.M. Davidson, A.S. Divakaruni, X. Zhang, C. Vickers, C. Joud-Caldwell, F. Chung, H. Yin
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A.R. Grassian, S.J. Parker, S.M. Davidson, A.S. Divakaruni, C.R. Green, X. Zhang, K.L. Slocum, M. Pu, F. Lin, F. Chung, H. Yin, E.D. Handly, C. Straub, J.D. Growney, A.N. Murphy, C.M. Metallo
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A.R. Grassian, S.J. Parker, S.M. Davidson, A.S. Divakaruni, C.R. Green, X. Zhang, M. Pu, F. Lin, C. Joud-Caldwell, H. Yin, J.D. Growney, A.N. Murphy, R. Pagliarini, C.M. Metallo
Writing, review, and/or revision of the manuscript: A.R. Grassian, S.J. Parker, S.M. Davidson, C.R. Green, X. Zhang, F. Lin, F. Chung, C. Straub, M.G. Vander Heiden, R. Pagliarini, C.M. Metallo
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): A.R. Grassian, S.J. Parker, M. Pu, R. Pagliarini
Study supervision: A.R. Grassian, S.J. Parker, M.G. Vander Heiden, A.N. Murphy, R. Pagliarini, C.M. Metallo
References
- 1.Arai M, Nobusawa S, Ikota H, Takemura S, Nakazato Y. Frequent IDH1/2 mutations in intracranial chondrosarcoma: a possible diagnostic clue for its differentiation from chordoma. Brain Tumor Pathol. 2012;29:201–6. doi: 10.1007/s10014-012-0085-1. [DOI] [PubMed] [Google Scholar]
- 2.Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119:1901–3. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayden JT, Fruhwald MC, Hasselblatt M, Ellison DW, Bailey S, Clifford SC. Frequent IDH1 mutations in supratentorial primitive neuroectodermal tumors (sPNET) of adults but not children. Cell Cycle. 2009;8:1806–7. doi: 10.4161/cc.8.11.8594. [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–43. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 8.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–9. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, Jeck WR, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2012:1–10. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yen KE, Schenkein DP. Cancer-associated isocitrate dehydrogenase mutations. Oncologist. 2012;17:5–8. doi: 10.1634/theoncologist.2011-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–9. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–9. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–6. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 20.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–8. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio II, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26:2038–49. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci U S A. 2011;108:3270–5. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–7. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonardi R, Subramanian C, Jackowski S, Rock CO. Cancer-associated isocitrate dehydrogenase mutations inactivate NADPH-dependent reductive carboxylation. J Biol Chem. 2012;287:14615–20. doi: 10.1074/jbc.C112.353946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamboni N. 13C metabolic flux analysis in complex systems. Curr Opin Biotechnol. 2011;22:103–8. doi: 10.1016/j.copbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Grassian AR, Lin F, Barrett R, Liu Y, Jiang W, Korpal M, et al. Isocitrate dehydrogenase (IDH) mutations promote a reversible ZEB1/microRNA (miR)-200-dependent epithelial-mesenchymal transition (EMT) J Biol Chem. 2012;287:42180–94. doi: 10.1074/jbc.M112.417832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Elementary metabolite units (EMU): a novel framework for modeling isotopic distributions. Metab Eng. 2007;9:68–86. doi: 10.1016/j.ymben.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–37. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–6. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–8. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, et al. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J Biol Chem. 2011;286:42626–34. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, et al. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gameiro PA, Yang J, Metelo AM, Perez-Carro R, Baker R, Wang Z, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17:372–85. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–21. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Mol Cell. 2013;49:388–98. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metallo CM, Walther JL, Stephanopoulos G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J Biotechnol. 2009;144:167–74. doi: 10.1016/j.jbiotec.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young JD, Walther JL, Antoniewicz MR, Yoo H, Stephanopoulos G. An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol Bioeng. 2008;99:686–99. doi: 10.1002/bit.21632. [DOI] [PubMed] [Google Scholar]
- 42.Fendt SM, Bell EL, Keibler MA, Davidson SM, Wirth GJ, Fiske B, et al. Metformin decreases glucose oxidation and increases the dependency of prostate cancer cells on reductive glutamine metabolism. Cancer Res. 2013;73:4429–38. doi: 10.1158/0008-5472.CAN-13-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McClintock DS, Santore MT, Lee VY, Brunelle J, Budinger GR, Zong WX, et al. Bcl-2 family members and functional electron transport chain regulate oxygen deprivation-induced cell death. Mol Cell Biol. 2002;22:94–104. doi: 10.1128/MCB.22.1.94-104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hockel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–76. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 45.Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang XL, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012;15:827–37. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popovici-Muller J, Saunders JO, Salituro FG, Travins JM, Yan SQ, Zhao F, et al. Discovery of the first potent inhibitors of mutant IDH1 that lower tumor 2-HG in vivo. ACS Med Chem Lett. 2012;3:850–5. doi: 10.1021/ml300225h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai K, Selinger DW, Solomon JM, Wu H, Schmitt E, Serluca FC, et al. Integrated compound profiling screens identify the mitochondrial electron transport chain as the molecular target of the natural products manassantin, sesquicillin, and arctigenin. ACS Chem Biol. 2013;8:257–67. doi: 10.1021/cb300495e. [DOI] [PubMed] [Google Scholar]
- 48.Medes G, Thomas A, Weinhouse S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 1953;13:27–9. [PubMed] [Google Scholar]
- 49.Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–21. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 50.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grassian AR, Metallo CM, Coloff JL, Stephanopoulos G, Brugge JS. Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. Genes Dev. 2011;25:1716–33. doi: 10.1101/gad.16771811. [DOI] [PMC free article] [PubMed] [Google Scholar]