Abstract
Macroautophagy (hereafter autophagy) is an evolutionarily-ancient mechanism by which cellular material is delivered to lysosomes for degradation. Autophagy and cell death are intimately linked. For example, both processes often use the same molecular machinery and recent work suggests that autophagy has great influence over a cell’s decision to live or die. However, this decision making is complicated by the fact that autophagy’s role in determining whether a cell should live or die goes both ways—autophagy inhibition can result in more or less cell death depending on the death stimulus, cell type, or context. Autophagy may also differentially affect different types of cell death. Here we discuss recent literature that helps make sense of this seemingly inconsistent role of autophagy in influencing a cell to live or die.
Keywords: Macroautophagy, autophagy, apoptosis, necroptosis, necrosis, autosis, MOMP, caspases, BH3 only proteins, PUMA, FAP-1
Introduction
As we consider how autophagy affects cell death, it is important to note that autophagy can determine whether or not cells die without directly affecting the cell death machinery. For example, it is well established that autophagy is critical for preventing cell death due to amino acid starvation. But this doesn’t necessarily mean that the apoptosis and necrosis mechanisms that cause amino acid starved cells to die are controlled by autophagy. Instead, autophagy can be protective here simply because autophagy degrades proteins, thereby providing the cell with the amino acids it needs to avoid activating those cell death mechanisms in the first place. Similarly, in mice genetic inactivation of autophagy in neurons leads to accumulation of aggregated proteins and eventual neuronal cell loss [1, 2]; however, this doesn’t mean that autophagy was directly controlling the activity of the cell death machinery in those neurons. Instead, in this case, it is thought that autophagy is removing the toxic protein aggregates so that a death signal was never activated. In this review, we ignore this type of indirect mechanism to focus on recent findings that relate to more direct autophagy control of distinct programmed cell death pathways.
Another important point should be considered too. In the literature, one often reads statements to the effect of “although autophagy can sometimes promote cell death, the primary function of autophagy is to protect cells”. This has led to the view that autophagy promotion of cell death is controversial or only occurs under special circumstances. In fact, we arguably have better evidence for autophagy promoting cell death under physiological conditions than we do for a general protective effect. There are numerous examples in development and under normal physiological conditions where autophagy is required for cell death [3]. And, in such contexts, there are identified mechanisms that show direct regulation of the cell death apparatus by autophagy to promote death– e.g. during Drosophila oogenesis, nurse cells die because autophagic degradation of dBruce, an anti-apoptotic protein, makes them more likely to activate caspases [4]. This physiological cell death depends on autophagy because deletion of core components of the autophagy machinery causes the nurse cells to persist. Examples like this indicate that autophagy directly modulates the cell death machinery in a positive way. Surprisingly, given the widespread view that autophagy generally protects cells from dying, mechanistic evidence supporting a generally protective role is less clear (although, as we will discuss, this is improving). Indeed, unlike regulators of the core apoptosis machinery, such as anti-apoptotic BCL- family members where genetic inactivation leads to widespread spontaneous cell death during development [5], animals with genetic inactivation of core autophagy regulators show little evidence of widespread cell loss during development [6]. One might expect this result if autophagy functions to generally tamp down the cell death machinery. Instead, as noted above, what we tend to see is evidence that autophagy protection against cell death is indirect and due to removal of a toxic signal rather than through direct modulation of the cell death apparatus. Building on recent findings, we think it is now becoming clear that autophagy does in fact directly regulate the cell death machinery both positively and negatively depending on the death stimulus, cell type, and context. Autophagy can also influence different types of cell death [7].We aim to emphasize three important points: (1) Programmed cell death mechanisms and autophagy are indeed intimately linked. (2) The basis for autophagy influencing cell-fate decisions—whether to live or die—often depends on which proteins are recruited to autophagosomes for lysosomal degradation. (3) Where and when the autophagy pathway—early versus late stages, before or after a death stimulus—is manipulated by genetic changes or pharmacological agents should be considered when trying to influence survival or cell death. Understanding the cellular mechanisms by which autophagy regulates cell fate is critical for knowing how autophagy alteration might be beneficial in diseases where too much or too little cell death takes place e.g. cancer and neurodegenerative diseases.
Autophagy regulation
Autophagy degrades cytoplasmic substrates to obtain energy and metabolic building blocks, especially when the cell is subjected to nutrient deprivation [8]. Autophagy is also necessary for the homeostasis of organelle integrity, clearance of aggregated proteins, tissue remodeling during development, and infection resistance. The signaling associated with autophagy is best understood in the context of coping with nutritional stress and maintaining cellular fitness. Many studies have concentrated on amino acid and insulin-dependent signaling involving mTOR signaling. Upstream of mTOR, the 5’ AMP-activated protein kinase (AMPK) phosphorylates TSC2 in response to a low ATP/AMP ratio, leading to mTOR inactivation and autophagy induction. Other important complexes include the ULK1 initiation complex and the Beclin1-VPS34 PI3 Kinase complex, which are important for nucleation of the membrane that will eventually form the autophagosome [8]. In response to such signaling, coordinated actions of more than 30 ATG (autophagy-related genes) proteins promote the nucleation and elongation of an isolation membrane for engulfing cytosolic substrates into an autophagosome. Two ubiquitin-like conjugation systems cause elongation of the membrane and formation of the autophagosome. One of these systems is the covalent conjugation of ATG12 to ATG5, which interacts with ATG16L to form the ATG5-ATG12-ATG16L complex. The second system conjugates a lipid, phosphatidylethanolamine, to LC3 (and various family members) resulting in the autophagosome-associated LC3-II protein, which is commonly monitored as a measure of autophagic activity in cells. At this stage, cellular material including proteins and organelles are either specifically recruited to the autophagosomal structures or, in the case of bulk non-specific autophagy, randomly captured. After fusion of the membranes to make an intact vesicle containing the cargos that will eventually be degraded, the resulting double-membrane vesicle fuses with a lysosome to form an autolysosome. Then, lysosomal enzymes hydrolyze the proteins, lipids and nucleic acids contained in the autophagosomes. The completion of this process is termed autophagic flux, and the magnitude of this flux can be measured on western blots by the amount of lipidated autophagosome-associated protein LC3-II that accumulates with or without blockade of the lysosomal fusion step. Alternatively, fluorescent labeling of LC3 can allow for tracking the redistribution of LC3 to autophagosomes. By using fluorescent tags with different pH sensitivities, it is possible to assess both the formation of autophagosomes and their fusion with lysosomes. Autophagy substrates are often recruited to autophagosomes by ubiquitylation, and recognized by adaptor proteins such as SQSTM1 (p62), which contain an LC3-interacting region (LIR). These adaptor proteins are degraded along with the cargo, and their turnover can therefore also be used to measure autophagic flux. In this review, when we discuss “early” steps in the autophagy process, we are referring to the activities of ATG proteins and signaling complexes necessary for autophagosomal nucleation, elongation, and maturation but not fusion with the lysosome and degradation of the cargo. For example, inhibition of early autophagy could be achieved by pharmacologically inhibiting the phosphatidylinositol 3-kinase (VPS34) with a kinase inhibitor, or the genetic knock down of ATG7 thus preventing the protein conjugations necessary for autophagosomal membrane elongation. Conversely, when we refer to “late” steps in autophagy, we mean the degradation of the autophagosome and its cargo by the lysosome. Late autophagy can be inhibited with agents that prevent the acidification of the lysosome, like chloroquine or bafilomycin A1. As will be discussed below, interference with autophagy at these different steps may sometimes have quite different effects on the cell fate decision. Since we are already using drugs like chloroquine and hydroxychloroquine to try to inhibit autophagy in people during cancer therapy and drugs that block early steps in the process are also being developed to treat cancer, such differences could be very important.
Autophagy regulation of canonical apoptosis
Caspase-dependent apoptosis is by far the best understood mode of programmed cell death and recent studies have identified different ways that autophagy can influence apoptosis (Fig. 1). We usually think of apoptosis as being driven by either intrinsic or extrinsic pathways. Extrinsic apoptosis occurs through ligand binding to cell surface receptors such as CD95, also known as Fas, and tumor necrosis factor-related apoptosis inducing ligand (TRAIL). Activation of these death receptors causes formation of various protein complexes that ultimately serve to aggregate an initiator caspase (caspase-8). In some cells, this occurs efficiently enough to activate sufficient effector caspase (e.g. caspase-3) activity to kill the cell. However, in most cells, the active caspase-8 causes cleavage of the BH3-only protein BID to activate the so-called “intrinsic” apoptosis pathway and this is needed to induce enough caspase activity to cause apoptosis. The intrinsic pathway is also activated by apoptosis stimuli that do not work through death receptors.
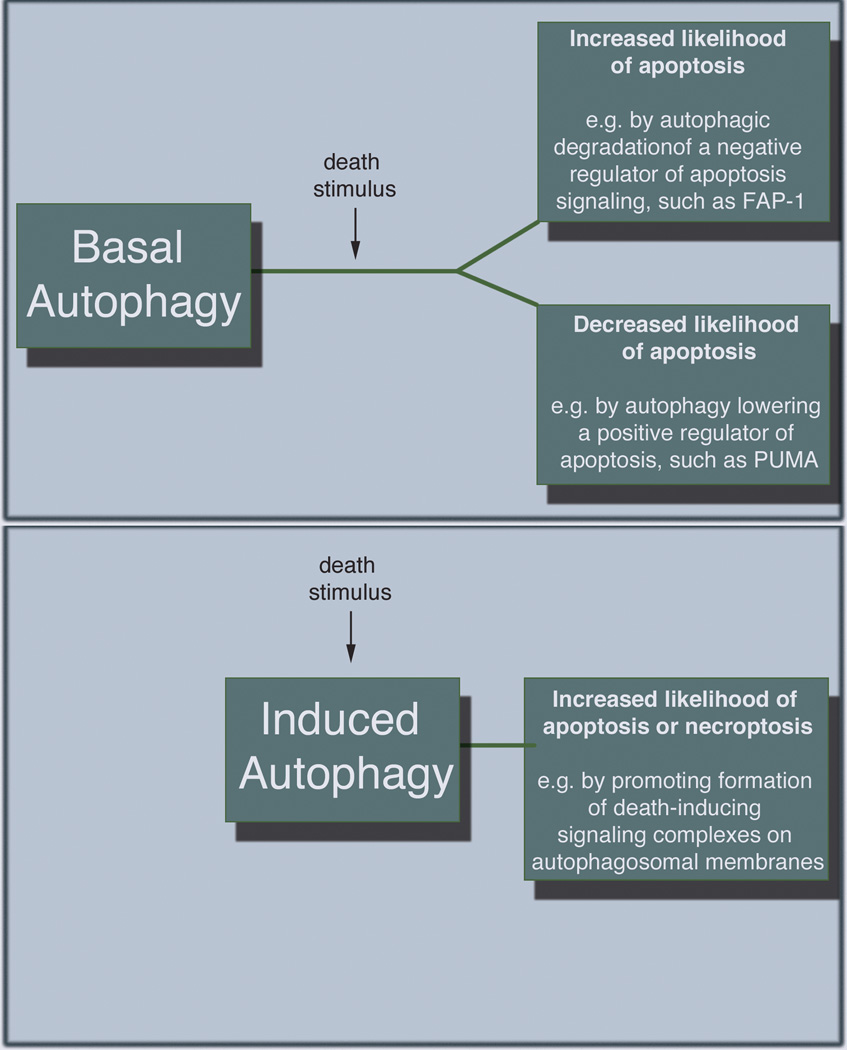
Figure 1.
Autophagy regulation of canonical apoptosis. Autophagy may both promote and inhibit apoptosis by degrading by altering the levels of key apoptosis regulators. This activity (panel A) can be governed by basal autophagy that takes place prior to any apoptotic signal being activated. Alternatively (panel B), autophagic structures can serve as scaffolds for efficient formation of death inducing complexes; this occurs after induction of the death signal.
Intrinsic apoptosis is characterized by the summation of pro- and anti-death signals converging at mitochondrial membranes that become permeabilized (MOMP—mitochondrial outer membrane permeabilization), leading to the release of mitochondrial intermembrane proteins. The key proteins include cytochrome c, which interacts with Apaf1 to form a scaffold for the activation of caspase-9 called the apoptosome, and Smac/Diablo, an inhibitor of caspase inhibitors that normally keep caspases in check. MOMP is followed by rapid cell death because the combination of caspase activation on the apopotosome along with simultaneous block of caspase inhibitors starts a cascade of active caspases that cleave hundreds of cellular substrates to cause cellular demise. Bcl-2 family proteins control MOMP. Under basal conditions, the anti-apoptotic BCL-2 proteins Bcl-2, Bcl-xL, and Mcl-1 prevent activation of pro-apoptotic family members Bax and Bak, which form the pores in the mitochondrial membrane that defines MOMP. In most cases, accumulation of cellular stress generates a synchronized effort by BH3-only members of the family that inhibit the Bcl-2 proteins, or directly activate Bax and Bak to promote MOMP.
MOMP is commonly thought of as a ‘point of no return’ that defines the cell’s commitment to apoptosis. However, new evidence questions this dogma by demonstrating that a small population of mitochondria in the cell may actually undergo MOMP, yet not result in the death of the cell. This has been termed “Incomplete” [9] or “Minority” MOMP [10]. Autophagy affects this process, since cells with higher autophagy are more likely to display delayed and incomplete MOMP than cells where autophagy is inhibited by knockdown of essential ATGs [11]. These effects whereby a cell activates incomplete MOMP and then recovers can have profound biological effects. Indeed, it was recently demonstrated that minority MOMP and caspase activation is sufficient to promote pro-tumorigenic effects in cells [10, 12]. Moreover, bursts of caspase activity that are not sufficient to cause the cell to die occur during development [13], suggesting that these effects may be important in normal biological processes. Thus complete MOMP causes enough caspase activation to cleave sufficient caspase substrates that the cell is destined to die with the morphology associated with canonical apoptosis, like plasma membrane blebbing and nuclear condensation etc. However, incomplete MOMP allows cells to recover after activating an insufficient level of caspases to cause death, and appears to be more likely when autophagy is high [11].
BCL family proteins regulate autophagy initiation, as well as control the balance of pro- and anti-apoptotic signals. One of the best examples of this regulation is the ability of BH3 (BCL-2 homology 3)-only proteins to neutralize BCL-2 anti-apoptotic proteins, stimulate pro-apoptotic proteins, and displace protein interactions that put the brake on autophagy induction. The subcellular localization and phosphorylation status of these BH3-only proteins dictate the direction toward survival or undergoing apoptosis, and the tendency to promote autophagy. BH3-only proteins, like PUMA, NOXA, NIX, BID, and BNIP3, disrupt inhibitory interactions between the BCL-2 proteins (Bcl-2, Bcl-XL, Mcl-1) and the autophagy regulator Beclin 1 (BECN1) [14]. Relieving the inhibition of BECN1 by Bcl-2 or Bcl-XL displacement allows for the activation of a PI3 Kinase called vacuolar protein sorting 34 (VPS34), resulting in the nucleation of an isolation membrane. This regulation of Bcl-2 protein and subsequent displacement from BECN1 is also controlled by phosphorylation, e.g. by the stress-activated c-Jun N-terminal protein kinase 1 (JNK1) [15].
The previous example shows how BCL family proteins (i.e. proteins that are best known as apoptosis regulators) can regulate autophagy. We recently described a mechanism for the converse– i.e. autophagy control of apoptosis– whereby autophagy regulates a specific BH3 protein to regulate the timing and extent of MOMP, subsequently regulating the timing and efficiency of apoptosis [11]. We found that as compared to autophagy competent cells, autophagy deficient cells are highly sensitized to Tumor Necrosis Factor-Related Apoptosis Inducing Ligand (TRAIL)-induced apoptosis. This indicates that autophagy is providing protection from TRAIL through the ability of autophagy to regulate constitutive levels of PUMA, a BH3-only pro-apoptotic protein. PUMA makes MOMP occur more efficiently, reducing the likelihood of incomplete MOMP and subsequent cellular recovery. Importantly, the majority of autophagy’s ability to inhibit apoptosis in this context was due to this mechanism because PUMA knockdown was sufficient to avoid sensitization to apoptosis when autophagy is inhibited. This example defines a mechanism by which autophagy can protect cells from a canonical apoptotic stimulus by controlling the level of a key regulator that makes the rate limiting step in apoptosis– i.e. MOMP– work more efficiently. As noted above, efficient versus inefficient/incomplete MOMP leading to some caspase activity, but not enough to kill the cell, could have important biological consequences; it is not known if this explains some of autophagy’s biological effects.
We recently reported [16] that in a population of cells, there are transient cell-to-cell variations in basal autophagy flux that dictates cell fate in both a cell-type-specific and stimulus-specific fashion. For example, distinct populations of cells with high or low autophagy flux respond very differently to death receptor activation. Fas ligand and TRAIL act in similar and well-understood ways, utilizing many of the same pathway components to elicit apoptosis. So, it was surprising when cell sorting to isolate cells that were undergoing high or low levels of autophagy, and treating these with Fas or TRAIL produced completely opposite results. High autophagy cells were sensitized to Fas ligand-induced apoptosis, but resistant to TRAIL treatment. As explained above, the protective effect of autophagy on TRAIL-induced apoptosis can be largely explained by PUMA being lower when autophagy is high, thus making MOMP more efficient. What explains the opposite effect when the apoptotic stimulus is the very similar death agonist Fas Ligand?
The answer was that the differential responses to two canonical death ligands is a result of what the high autophagy cells are or are not degrading. The key was the selective degradation of a phosphatase, FAP-1, which is a negative regulator of FAS signaling, but does not affect TRAIL signaling. This degradation of a negative regulator of FAS signaling was sufficient and necessary to explain how high autophagy can promote Fas-induced apoptosis. And, since FAP-1 has no effect on TRAIL receptors, this also explains why only autophagy’s protective effects were seen with TRAIL treated cells in this case. This example demonstrates how autophagy can both positively and negatively influence cell fate. The degradation of a negative apoptosis regulator results in more death in high autophagy cells, while autophagy regulation of a positive regulator of apoptosis leads to less death in high autophagy cells. This mechanism of apoptosis promotion by autophagy is conceptually similar to that seen in the Drosophila nurse cells described above–apoptosis is promoted by autophagic degradation of a negative regulator that controls the apoptosis machinery. The difference here is that the FAP-1 mechanism affects only apoptosis induced by Fas Ligand, whereas the dBruce degradation in the nurse cells removes an inhibitor of a caspase inhibitor, sensitizing them to any stimulus that activates caspases.
Above, we discussed how autophagy influences cell death by selective degradation of pro- or anti-apoptotic proteins. An intriguing story where autophagosomal membranes serve as platforms for death signaling complexes offers another mechanistically distinct example of how the autophagy machinery can influence cell fate. Experiments with the sphingosine kinase inhibitor (SKI-I), which promotes cell death through the suppression of sphingosine 1-phosphate, showed caspase-dependent cell death occurred only in the presence of functional autophagy [17]. Further, SKI-I was demonstrated to promote the translocation of caspase-8 homocomplex and Fas-associated protein with death domain (FADD) to ATG5-positive autophagosomal membranes, forming a scaffold for the efficient formation of an intracellular death-inducing signaling complex (iDISC). This death was abrogated by ATG5 depletion, which resulted in lower activation of mitochondrial amplification loop that is initiated by caspase-8. Overall, these studies show a mechanism of apoptosis that is dependent on autophagosomal structures but not necessarily the whole process of autophagy whereby the material sequestered in the autophagosome is degraded. It should also be noted that there are other examples where autophagosomal structures are proposed to serve as signaling scaffolds, [18] so there may be many instances where interference with the autophagy machinery could affect signaling events through such mechanisms.
This mechanism of alteration by autophagy is very different from the mechanisms described above where autophagy regulation of apoptosis is achieved by degrading specific proteins. Two important implications of this mechanism whereby the autophagy machinery alters signaling by serving as a scaffold should be considered. First, there may be different effects on the outcome of the cell depending on whether basal autophagy or stimulus-induced autophagy is manipulated. For the anti- and pro-apoptotic effects that are ultimately due to altering levels of apoptosis regulators such as PUMA, dBruce or FAP-1, autophagy’s effects must be mediated prior to the initiation of the death signal if they are to alter the likelihood that the apoptosis stimulus will be sufficient to cause the cells to die. Indeed, when we separated high and low autophagy cells from a population we found that this predicted their likelihood to undergo apoptosis that wasn’t even initiated until several hours after the separation of the cells into the two groups [16]. This is a case whereby the effects of basal autophagy that occur before the apoptosis signal are critical for deciding whether or not cells will die at a later time. Not surprisingly then, manipulations that affect such basal autophagy can influence these mechanisms [19]. When autophagic structures are working by forming a scaffold for efficient formation of death inducing protein complexes as in the case with the intracellular DISC formed after SK1 inhibitor treatment, autophagy is having its effects after initiation of the death stimulus. These kinds of effects are, therefore, more likely to be influenced not by the basal autophagy that occurred before the death stimulus, but instead by the induced autophagy that occurs in response to the stimulus. Since many apoptotic stimuli (e.g. drugs that kill cells as well as pure apoptosis inducers like the death receptor agonists) also induce autophagy, it has been assumed that this induced autophagy is doing the same things to the cell death machinery as the ongoing basal autophagy does. That’s not necessarily true. To date, it has been impossible to test such ideas because we usually design experiments to determine how autophagy affects cell death by inhibiting autophagy using genetic approaches (knockout or knockdown of essential ATGs, expression of dominant negatives etc.) that take several days to become effective [20]. Technical advances will be needed to address these questions because one needs a way to very rapidly, selectively and reversibly target stimulus-induced autophagy without affecting basal autophagy in order to test if both types of autophagy have the same effects, or not.
Second, interference with autophagy at different steps in the process may sometimes have the same effects or different effects on a cell’s likelihood of dying. If the important mechanism is regulation of proteins like FAP-1 or PUMA, it most likely doesn’t matter how autophagy is blocked—if you block formation upstream of autophagosomal structures or degradation of cargo, similar effects are expected. Indeed, that’s what we saw with FAP-1– genetic inhibition of autophagosome formation gave similar effects to blocking lysosomal function. However, if autophagy’s role in regulating apoptosis is by providing scaffolding structures, one might expect that inhibiting formation of autophagosomal structures would block cell death, while inhibiting their degradation (e.g. by blocking the lysosome) might promote these effects by causing more scaffolding. There are hints in the literature [21] where apoptosis inducing anti-cancer drugs are affected oppositely depending on whether autophagosome formation versus degradation are targeted. Do these kinds of mechanistic differences explain such effects? The answer to this question is important because although we are already treating cancer patients with lysosomal inhibitors, other autophagy inhibitors that affect early steps in the process are being developed [22–25]. For example, the VPS34 kinase inhibitor, SAR405, is a potent (KD=1.5 nM) and selective compound targeting the ATP binding cleft. SAR405 was shown to inhibit starvation-induced autophagy by limiting the formation of lipidated LC3, reducing GFP-LC3 puncta, and causing the accumulation of a receptor protein p62 [25]. Compounds like SAR405 that target the initiation of autophagy may prove to be efficacious in clinical trials as independent therapies, or useful alongside late-stage inhibitors like chloroquine. One could also imagine a situation in which these drugs might have the opposite effect that is hoped for (i.e. inhibit rather than promote tumor cell death) e.g. by interfering with scaffolding activities and this might be highly context dependent.
How Apoptosis can influence autophagy
Apoptosis also directly influences autophagy. For example, Beclin 1 is a substrate for cleavage by caspase-3. Two caspase cleavage sites in Beclin 1 were discovered [26], resulting in reduced affinity for Bcl-2, a reduction in autophagy, and an enhancement of apoptosis in HeLa cells. The cleavage of Beclin 1 may be a way to help the cell fully commit to apoptosis by putting the brake on autophagy induction, reducing the likelihood of cellular recovery. Other essential autophagy proteins are also targeted by caspases. The autophagosomal biogenesis regulatory protein ATG3 is a substrate for caspase-8 cleavage following the activation of extrinsic apoptosis [27]. This cleavage site in ATG3 is conserved in humans, mice, rats, zebra fish, frogs, worms, and baker’s yeast suggesting a conserved regulatory role. Cleavage of ATG3 following a death receptor signal inactivated autophagy, but the expression of a non-cleavable ATG3 could reestablish autophagic activity. Further, the autophagy protein AMBRA1 (activating molecule in BECN1-regulated autophagy) is degraded by calpains and caspases, and expressing a cleavage-resistant mutant leads to apoptosis avoidance by prolonging autophagy induction [28].
Autophagic cell death—autophagy as a direct cause of cell death
The mechanisms described above involve autophagy interplay with apoptosis; however, autophagy may also regulate non-apoptotic cell death mechanisms where there is no need for caspase activation in order for the cell to die. For example, it was shown that acute expression of oncogenic H-RAS can cause caspase-independent death with characteristics of autophagy [29]. This autophagic cell death was mediated by and dependent on Ras-induced expression of the pro-apoptotic protein, Noxa, and Beclin-1. These findings provided the first unequivocal genetic evidence for autophagy-dependent death in mammalian cells in response to a specific signaling event and although seen in an artificially controlled system, suggest that autophagic cell death may, under some circumstances, provide a mechanism to limit the oncogenic potential of dysregulated RAS signaling. However, in other contexts the opposite effect holds sway and tumor cells with RAS mutations are highly dependent upon autophagy in order to survive and grow [30–33]. Added complexity comes from the fact that autophagy inhibition can have opposing consequences in RAS-mutant tumors. These effects may be based on factors such as p53 status [34] or the cell line being studied [35], an could be unrelated to autophagy regulation of death pathways. Instead, as in the case of autophagy-addicted RAS-transformed cells, these effects may be connected to the regulation of metabolism [30–33].
A requirement for functional autophagy for programmed cell death has been most clearly shown in vivo in developmental systems that are genetically tractable, such as Drosophila [3]. For example, the first clear genetic evidence that autophagy is required for “physiological” cell death in vivo came from the demonstration that autophagy is induced just before salivary gland cell death, and salivary glands are not properly degraded in ATG mutants [36]. During salivary gland degradation, autophagic cell death is thought to take place alongside caspase-dependent apoptosis. Conversely, as mentioned above, during Drosophila oogenesis, autophagy controls cell death by promoting caspase activation and subsequent apoptosis [4]. In other contexts, developmental cell death in Drosophila can be shown to be autophagy-dependent but independent of caspases [37]. These examples show that developmentally programmed cell deaths can involve autophagy working alongside apoptosis, autophagy controlling apoptosis and autophagy working on its own with no involvement of the apoptosis machinery.
Another example of a morphologically distinct form of autophagy-dependent cell death was recently identified called “autosis” [38], which was activated by an autophagy-inducing peptide and is modulated by widely used cardiac glycosides that target the Na+/K+-ATPase. This type of death may also occur in response to physiological signals because autosis was demonstrated to occur in a small population of cells (~1%) during nutrient starvation conditions and in vivo during hypoxic-ischemic injury in neonatal rats. The morphological features of autosis include nuclear convolution, increased autophagosomes, nuclear shrinkage, and focal perinuclear swelling. Key experiments demonstrated that the inhibition of early autophagy abrogated Tat-Beclin 1-mediated cell death, and that this death does not require the canonical apoptosis or necroptotsis machinery. Interestingly, blocking autophagosome/lysosomal fusion with a late-stage autophagy inhibitor, bafilomycin A1, did not reduce the Tat-Beclin 1 death, suggesting that only the early steps of autophagy are required for this cell death mode. These sorts of distinctions—death modalities requiring early or late stage autophagy machinery—require consideration when developing autophagy inhibitors for patient use.
Necrosis and autophagy
For many years, necrosis was regarded as an “accidental” process, but it is now understood that necrosis is often highly regulated and an intentional programmed mechanism. The most extensively studied form of programmed necrosis is called necroptosis, which is a form of programmed cell death dependent on receptor-interacting Ser/Thr protein kinase 1 (RIPK1), RIPK3 and the pseudokinase MLKL. Necroptosis is best characterized by the stimulation of the TNFR1 (Tumor Necrosis Factor Receptor 1) by TNF resulting in the formation of different signaling complexes that create a “switch” leading to cell survival, apoptosis or necroptosis. The ubiquitin-editing system and initiator caspases dictate the response to TNF ligand binding to TNFR. Complex I formation upon TNF binding consists of TNFR-associated death domain (TRADD), RIPK1, cellular inhibitor of apoptosis 1 (cIAP1), cIAP2, TNFR-associated factor 2 (TRAF2) and TRAF5, leading to cell survival. Conversely, the internalization of the TNFR1 and deubiquitylation of RIPK1 by the deubiquitylating enzyme cylindromatosis results in the formation of complex II, promoting either apoptosis or necroptosis. This complex consists of RIPK1, RIPK3, TRADD, caspase-8, and FAS-associated protein with a death domain (FADD). One result of complex II formation is the proteolytic cleavage of RIPK1 and RIPK3 by caspase-8, generating a pro-apoptotic caspase activation cascade. However, when caspase-8 is deleted or inhibited, complex II cannot produce apoptotic signals. Instead, RIPK1 and RIPK3 phosphorylate each other, and aggregate in a complexe called the necrosome. These autophosphorylation and transphosphorylation events recruit the mixed-lineage kinase domain-like (MLKL), which is itself phosphorylated by RIPK3. MLKL is then responsible for the permeabilization of the plasma membrane and death of the cell [39, 40].
Little is known about how autophagy is intertwined with necroptosis. However, some of the first evidence to show that autophagy could promote cell death came from a system that has gone on to become the best understood necroptosis pathway [41] and in this case, autophagy was shown to modulate these effects by selectively degrading the reactive oxygen species (ROS) scavenger enzyme, catalase [42]. In another example, as with formation of an apoptosis-inducing scaffold described above, one piece of evidence points to autophagosomal membranes acting as platforms for necrosome assembly, and serving as key sites to mediate necroptosis. Obatoclax, or GX15-070) is an indole bipyrrole compound that antagonizes Bcl-2, Bcl-XL, Bcl-w, and Mcl-1, and has been shown to activate autophagy and elicit non-apoptotic cell death in rhabdomyosarcoma cells. GX15-070-stimulated autophagy was linked to the assembly of the necrosome—i.e. a complex involving FADD, RIPK1, and RIPK3—on autophagosomal membranes. ATG5 or ATG7 silencing mitigated this cell death, and co-immunoprecipitation studies suggest that GX15-070 stimulates an interaction between ATG5 and necrosome components. Further, RIPK1 knockdown or pharmacological inhibition of RIPK1 with necrostatin-1 blocked death by GX15-070 [43]. Together, these data point to the formation of autophagosomes as key mediators for achieving efficient necrosome formation, resulting in necroptotic cell death by GX15-070.
Bray et al provided another example of the coordination between necroptosis and autophagy [44]. They found that concurrent mTOR inhibition by CCI-779 and inhibiting autophagosome maturation with chloroquine lead to the accumulation of autophagosomes that induced RIPK3-dependent and ROS-dependent necroptosis in renal cell carcinoma lines. Some evidence also implies that RIPK1 might be degraded by autophagy. Overall, the above examples support the notion that autophagy can influence the fate of cells treated with compounds that induce necroptosis. However, while these examples are suggestive of important interactions between autophagy and the necroptosis machinery, more work is needed to uncover the mechanistic ties and to work out how these processes are controlled.
Conclusions
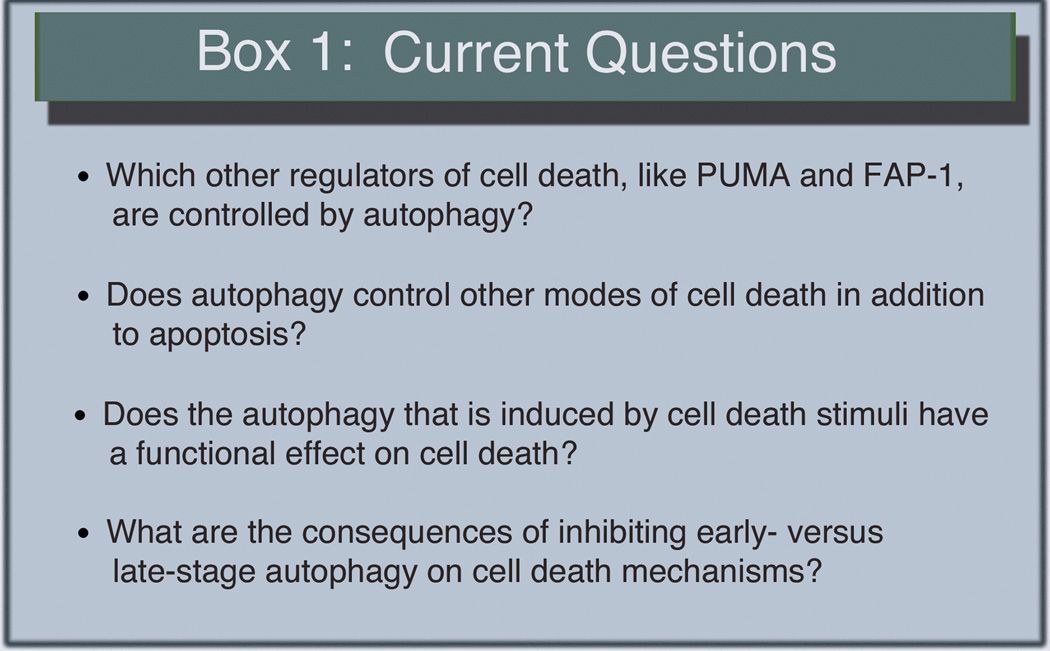
Although autophagy has been widely thought to be intimately involved in cell death regulation for many years, it is only recently that we have started to understand the underlying mechanisms and there is clearly much more that needs to be done and many remaining questions to be answered (Fig. 2). We now have examples such as the FAP-1 story described above that start to explain how autophagy can promote apoptosis but only for some stimuli. And, we are starting to see clear evidence that non-apoptotic forms of cell death are also affected by autophagy. However, even with our current limited understanding of these processes, it is already clear that these mechanisms are complicated and highly intertwined. Many of the past mechanistic studies involving autophagy and cell death have relied on in vitro validation in transformed cell lines that have evolved under conditions where cell death regulation is awry. Thus, while it is possible that some of the effects seen are specific to cancer cells and are of little relevance when it comes to understanding the interplay of autophagy and cell death in normal cell physiology, such differences may also provide an avenue for manipulation of processes that are cancer cell specific as a way to improve cancer therapy. Future studies aiming to uncover mechanistic ties between cell death and autophagy should be performed in a diverse selection of both transformed and primary cells, as well as in vivo. Efforts to do so will make for more robust conclusions about the implications of cell death and autophagy relationships in whole organisms. As we improve our understanding of the basic mechanisms, it will hopefully become clearer how (or if) we should try to manipulate these processes to improve treatment of the numerous diseases where excessive or too little cell death is the ultimate cause.
Figure 2.
Some unanswered questions.
Acknowledgments
Work in our lab is supported by NIH grants 2RO1CA150925 and 1RO1CA190170.
Abbreviations
- ATG
autophagy-related gene
- TRAIL
tumor necrosis factor-related apoptosis inducing ligand
- FADD
Fas-associated protein with death domain
- MOMP
mitochondrial outer membrane permeabilization
- PUMA
p53 upregulated modulator of apoptosis
- iDISC
intracellular death-inducing signaling complex
REFERENCES
- 1.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 2.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 3.Nelson C, Baehrecke EH. Eaten to death. The FEBS journal. 2014;281:5411–5417. doi: 10.1111/febs.13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjørkøy G, Johansen T, Rusten TE, Brech A, Baehrecke EH, Stenmark H. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010;190:523–531. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motoyama N, Wang F, Roth KA, Sawa H, Nakayama K, Nakayama K, Negishi I, Senju S, Zhang Q, Fujii S. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science. 1995;267:1506–1510. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- 6.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 7.Tait SW, Ichim G, Green DR. Die another way--non-apoptotic mechanisms of cell death. J Cell Sci. 2014;127:2135–2144. doi: 10.1242/jcs.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 9.Tait SWG, Parsons MJ, Llambi F, Bouchier-Hayes L, Connell S, Muñoz-Pinedo C, Green DR. Resistance to caspase-independent cell death requires persistence of intact mitochondria. Dev Cell. 2010;18:802–813. doi: 10.1016/j.devcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichim G, Lopez J, Ahmed SU, Muthalagu N, Giampazolias E, Delgado ME, Haller M, Riley JS, Mason SM, Athineos D, Parsons MJ, van de Kooij B, Bouchier-Hayes L, Chalmers AJ, Rooswinkel RW, Oberst A, Blyth K, Rehm M, Murphy DJ, Tait SW. Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol Cell. 2015;57:860–872. doi: 10.1016/j.molcel.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorburn J, Andrysik Z, Staskiewicz L, Gump J, Maycotte P, Oberst A, Green DR, Espinosa JM, Thorburn A. Autophagy controls the kinetics and extent of mitochondrial apoptosis by regulating PUMA levels. Cell Rep. 2014;7:45–52. doi: 10.1016/j.celrep.2014.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, He Y, Li F, Huang Q, Kato TA, Hall RP, Li C-Y. Caspase-3 promotes genetic instability and carcinogenesis. Molecular Cell. 2015;58:284–296. doi: 10.1016/j.molcel.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang HL, Tang HM, Fung MC, Hardwick JM. In vivo CaspaseTracker biosensor system for detecting anastasis and non-apoptotic caspase activity. Scientific reports. 2015;5:9015. doi: 10.1038/srep09015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gump JM, Staskiewicz L, Morgan MJ, Bamberg A, Riches DWH, Thorburn A. Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1. Nat Cell Biol. 2014;16:47–54. doi: 10.1038/ncb2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J, Sharma AK, Amin S, Hu C-D, Zhang J, Kester M, Wang H-G. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287:12455–12468. doi: 10.1074/jbc.M111.309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Lopez N, Athonvarangkul D, Mishall P, Sahu S, Singh R. Autophagy proteins regulate ERK phosphorylation. Nat Commun. 2013;4:2799. doi: 10.1038/ncomms3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yonekawa T, Gamez G, Kim J, Tan AC, Thorburn J, Gump J, Thorburn A, Morgan MJ. RIP1 negatively regulates basal autophagic flux through TFEB to control sensitivity to apoptosis. EMBO reports. 2015 doi: 10.15252/embr.201439496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staskiewicz L, Thorburn J, Morgan MJ, Thorburn A. Inhibiting autophagy by shRNA knockdown: Cautions and recommendations. Autophagy. 2013;9 doi: 10.4161/auto.24895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Y, Hamed HA, Cruickshanks N, Fisher PB, Grant S, Dent P. Obatoclax and lapatinib interact to induce toxic autophagy through NOXA. Mol Pharmacol. 2012;81:527–540. doi: 10.1124/mol.111.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egan DF, Chun MGH, Vamos M, Zou H, Rong J, Miller CJ, Lou HJ, Raveendra-Panickar D, Yang C-C, Sheffler DJ, Teriete P, Asara JM, Turk BE, Cosford NDP, Shaw RJ. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Molecular Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shpiro N, Ward R, Cross D, Ganley IG, Alessi DR. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. The Biochemical journal. 2014;463:413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, Cantwell J, Luu C, Cornella-Taracido I, Harrington E, Fekkes P, Lei H, Fang Q, Digan ME, Burdick D, Powers AF, Helliwell SB, D'Aquin S, Bastien J, Wang H, Wiederschain D, Kuerth J, Bergman P, Schwalb D, Thomas J, Ugwonali S, Harbinski F, Tallarico J, Wilson CJ, Myer VE, Porter JA, Bussiere DE, Finan PM, Labow MA, Mao X, Hamann LG, Manning BD, Valdez RA, Nicholson T, Schirle M, Knapp MS, Keaney EP, Murphy LO. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nature cell biology. 2014 doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 25.Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot M-F, Lamberton A, Mathieu M, Bertrand T, Marquette J-P, El-Ahmad Y, Filoche-Romme B, Schio L, Garcia-Echeverria C, Goulaouic H, Pasquier B. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, Zhao L, Liu L, Gao P, Tian W, Wang X, Jin H, Xu H, Chen Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell. 2010;1:468–477. doi: 10.1007/s13238-010-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, Gozuacik D. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis. 2012;17:810–820. doi: 10.1007/s10495-012-0735-0. [DOI] [PubMed] [Google Scholar]
- 28.Gu W, Wan D, Qian Q, Yi B, He Z, Gu Y, Wang L, He S. Ambra1 is an essential regulator of autophagy and apoptosis in SW620 cells: pro-survival role of Ambra1. PLoS One. 2014;9:e90151. doi: 10.1371/journal.pone.0090151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic ras-induced expression of noxa and beclin-1 promotes autophagic cell death and limits clonogenic survival. Molecular Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Guo JY, Chen H-Y, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JMS, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, Snyder E, Santanam U, Dipaola RS, Jacks T, Rabinowitz JD, White E. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, White E. Autophagy is Required for Glucose Homeostasis and Lung Tumor Maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell'antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, Kimmelman AC. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, Mackay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, Adams PD, Anderson KI, Gottlieb E, Sansom OJ, Ryan KM. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 35.Morgan MJ, Gamez G, Menke C, Hernandez A, Thorburn J, Gidan F, Staskiewicz L, Morgan S, Cummings C, Maycotte P, Thorburn A. Regulation of autophagy and chloroquine sensitivity by oncogenic RAS in vitro is context-dependent. Autophagy. 2014;10:1814–1826. doi: 10.4161/auto.32135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Shoji-Kawata S, Sumpter RM, Jr, Wei Y, Ginet V, Zhang L, Posner B, Tran KA, Green DR, Xavier RJ, Shaw SY, Clarke PG, Puyal J, Levine B. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuchs Y, Steller H. Live to die another way: modes of programmed cell death and the signals emanating from dying cells. Nat Rev Mol Cell Biol. 2015;16:329–344. doi: 10.1038/nrm3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol. 2010;11:700–714. doi: 10.1038/nrm2970. [DOI] [PubMed] [Google Scholar]
- 41.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 Program of Autophagic Cell Death by Caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 42.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basit F, Cristofanon S, Fulda S. Obatoclax (GX15-070) triggers necroptosis by promoting the assembly of the necrosome on autophagosomal membranes. Cell Death Differ. 2013;20:1161–1173. doi: 10.1038/cdd.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bray K, Mathew R, Lau A, Kamphorst JJ, Fan J, Chen J, Chen H-Y, Ghavami A, Stein M, Dipaola RS, Zhang D, Rabinowitz JD, White E. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PloS one. 2012;7:e41831. doi: 10.1371/journal.pone.0041831. [DOI] [PMC free article] [PubMed] [Google Scholar]