Abstract
Treatments that successfully modulate anti-cancer immunity have significantly improved outcomes for advanced stage malignancies and sparked intense study of the cellular mechanisms governing therapy response and resistance. These responses are governed by an evolving milieu of cancer and immune cell subpopulations that can be a rich source of biomarkers and biological insight, but it is only recently that research tools have developed to comprehensively characterize this level of cellular complexity. Mass cytometry is particularly well suited to tracking cells in complex tissues because 35+ measurements can be made on each of hundreds of thousands of cells per sample, allowing all cells detected in a sample to be characterized for cell type, signaling activity, and functional outcome. This review focuses on mass cytometry as an example of systems level characterization of cancer and immune cells in human tissues, including blood, bone marrow, lymph nodes, and primary tumors. This review also discusses the state of the art in single cell tumor immunology, including tissue collection, technical and biological quality controls, computational analysis, and integration of different experimental and clinical data types. Ex vivo analysis of human tumor cells complements both in vivo monitoring, which generally measures far fewer features or lacks single cell resolution, and laboratory models, which incur cell type losses, signaling alterations, and genomic changes during establishment. Mass cytometry is on the leading edge of a new generation of cytomic tools that work with small tissue samples, such as a fine needle aspirates or blood draws, to monitor changes in rare or unexpected cell subsets during cancer therapy. This approach holds great promise for dissecting cellular microenvironments, monitoring how treatments affect tissues, revealing cellular biomarkers and effector mechanisms, and creating new treatments that productively engage the immune system to fight cancer and other diseases.
Keywords: Systems immunology, tumor immunology, immunotherapy, mass cytometry, human immune monitoring
Introduction
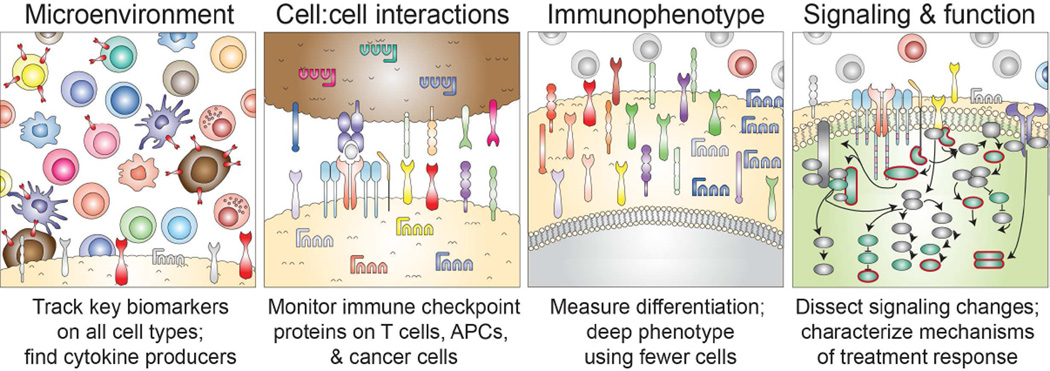
The immune system is a complex network comprised of localized and specialized tissue sites connected by circulating immune cells. Traditional immunological techniques and approaches have provided a depth of knowledge within each compartment, but struggle to comprehensively dissect the network and its interactions as a whole. In addition to system-wide complexity, each cell subset is itself a “system within a system”, possessing its own hierarchies and heterogeneity. As cancer and immune system cells compete in a complex and continuously evolving cycle (1), understanding the complex rules governing anti-cancer immune responses poses a challenge. Multiple subsets of immune cells are implicated as promoters or inhibitors of the anti-tumor immune responses (2, 3). To dissect and predict anti-cancer immune responses, it is crucial to not only monitor the cellular milieu of peripheral blood, tumor sites, and draining lymph nodes, but also to monitor the cell surface molecules responsible for cell:cell interactions, the deep immunophenotype of cell subsets of special interest, and intracellular signaling events including post translational protein modifications, proliferation, cytokine production, and other functional capabilities (Figure 1).
Figure 1. Focal single cell areas in systems cancer immunology.
Mass cytometry and other multidimensional single cell tools can be focused to resolve key biomarkers and mechanisms at different layers of cellular interaction. Most commonly, mass cytometry is used to provide cytomic resolution, meaning that all the different cell types present in a tissue are quantified and phenotyped. As this can generally be achieved with 10 markers on a typical mass cytometry panel, this leaves at least 25 mass channels available for detection of cell interaction markers, immunophenotype, and intracellular signaling (4). As nearly any cellular property can now be quantified at the single cell level (5), multidimensional cytometry enables biomarkers with complex expression patterns that can vary with cell type and activation state – such as PD-L1 (6) – to be broadly monitored. Another advantage of cytomic approaches is that cells with unusual and unexpected phenotypes present in a patient’s tissue sample do not escape detection due to expert bias or overly focused analysis strategies. These advantages of mass cytometry address ongoing needs in cancer and immune biomarker development (7).
Keeping Track of Complex Immune Networks
Milieu
Each step of the cancer-immunity cycle includes the potential for competition between effector and regulatory cells, and nearly every immune cell subset has been implicated in the anti-cancer immune response (8). Dendritic cells presenting tumor antigen are required to activate a specific anti-cancer adaptive immune response (1, 9). Effectors like CD8+ and CD4+ T cells, NK cells, and tumor specific antibodies participate in direct killing of tumor cells (10–13). Controlling these effectors are cells and signaling mechanisms that can check or attenuate immune responses, including regulatory and suppressive cells arising from the T cell (14), myeloid (15), and B cell lineages (16). These effector and regulatory cells are diverse in phenotype and variable in abundance (17). While some cell subsets comprise a substantial proportion of the total leukocyte pool (e.g. ~2–11% cytotoxic T cells), others, such as Tregs or memory B cells, can contain critical information while comprising <5% of total leukocytes (18–21). Although small in number, regulatory cell subsets can drastically impact the anti-tumor immune response. Thus, the ability of mass cytometry to characterize rare cells comprising as few as 1 in 10,000 cells is a key advantage for evaluating the state of a patient’s immune system (22). This capacity is highlighted in detection of minimal residual disease (MRD) in leukemia, a crucial metric for monitoring progress of patients on therapy (23). Recently mass cytometry has demonstrated proof of concept efficacy in working with small samples (22) and detecting MRD (24). While detecting MRD, mass cytometry also allows for the detection of multiple other cell subsets.
Deep phenotype
In addition to detecting rare cellular subsets, high dimensional single cell technologies are also capable of revealing cells with unusual or unexpected phenotypes. With single cell analysis, it is possible to not only resolve rare cell subsets or subtle changes in phenotype, but also to distinguish cancer cells from healthy non-hematopoietic cells, and immune cells (5, 25). Small phenotypic shifts, such as the slight downregulation of antigen receptors by activated T cells (26), may provide important information about the state of a patient’s immune system. At present, the effort is to track the status of cells and identify markers and mechanisms that indicate status can cell type, including “poised to attack cancer cells”, “in need of priming”, “held in check by regulation”, or “lacking key effector subsets”. Given sufficient examples, it may be possible to discern the signaling rules that govern cell identity and to use this information to precisely modulate the in vivo activity of target cell subsets.
Cell: Cell interactions
The behavior of effector immune cells is directly affected by the engagement of cell surface receptors. While some ligands are soluble, like many cytokines and chemokines, many are bound to the surface of antigen presenting cells and even cancer cells themselves. A long list cancer cell bound ligands are known to modulate and suppress the behavior of cytotoxic T cells within the tumor microenvironment (27). Although it is well known that cancer cells express molecules like PD-L1 that modulate the immune response, the surface phenotype of cancer cells is less well characterized than immune cells. For instance, when expressed on melanoma cells, HLA-DR, an MHC class II molecule generally restricted to professional antigen presenting cells, was shown to have predictive value in determining which patients will respond to PD-1 blockade (28). Including key markers of cancer cell type in immune classification panels makes it possible to localize biomarker and cytokine expression to cancer and immune cells, which is especially important given how cancer cells aberrantly express molecules from outside their lineage of origin.
Function and Signaling
Distinguishing “cancer” versus “healthy” cells and then attributing genotype and phenotype characteristics is especially critical when developing novel therapeutics expected to have selective activity on cancer cells. Analysis of non-cancer cells can provide information on off-target effects of therapeutics. For example, small molecule inhibitors have varied functional impacts across immune cell populations (29, 30). Preserving viability and effector function of healthy immune cells during cancer therapy is crucial for maintaining an effective anti-tumor response (31).
The Need for Longitudinal Cytomic Monitoring in Clinical Trails
The immune system is constantly in flux with cells undergoing stimulation, suppression, expansion and death resulting in phenotypic changes (32). Static snapshots of tissue and tumor resident or peripheral immune cells provide a wealth of information about overall immune function and tumor immune response, although lack key information about dynamic changes. Serial acquisition of healthy donor peripheral blood revealed low intra-donor variability but high inter-donor variability (33). The analysis of blood from melanoma patients, treated with anti-CTLA-4 or anti-PD-1, revealed small, but critical changes in certain cell subsets, such as the upregulation of activation marker HLA-DR on CD4 T cells (34). With the combination of high, single-cell resolution and primary patient samples over the course of therapy, it is possible to create an in-depth picture of the immune network as therapy progresses. By connecting this detailed picture with the clinical outcome of the patients, it may be possible to 1) identify a marker that predicts whether patients will respond to therapy and 2) identify a specific cell type or group of cells that are responsible for successful therapy.
Mass Cytometry: Optimized for Human Immune Monitoring
Multidimensional flow cytometry has been a gold standard of single cell biology, both within basic research labs and the clinic. Flow cytometry is routinely used in the clinic for analysis and diagnosis of leukemia and lymphoma, identification of lymphocyte subsets in HIV infection, monitoring solid organ transplantation matches, and detection of immunodeficiency. In traditional flow cytometry, single cell suspensions are stained with a cocktail of antibodies tagged with different fluorophores (35). Stained cells are then run through a flow cytometer in a single cell stream, passing by lasers that excite the fluorophores conjugated to the antibodies bound to the cell surface. The emissions of the excited fluorophores are then recorded by the machine’s detector (36).
Like fluorescence flow cytometry, mass cytometry utilizes single cell suspensions stained with antibody cocktails. However, instead of fluorophores, antibodies used in mass cytometry are coupled to pure metal isotopes rarely found in nature. Stained single cells are atomized and ionized by argon plasma. The resulting ion clouds are then resolved and quantified by time-of-flight mass spectrometry. Mass cytometry largely eliminates spectral overlap issues that can confound quantitative fluorescence cytometry and has been used to measure over 40 parameters simultaneously at the single cell level (37). The use of metal isotopes and inductively-coupled mass spectrometer (ICP-MS) allows for precise quantitation (38). The capacity to measure so many features per cell allows combined detection of surface proteins, intracellular phospho-proteins, transcription factors, and functional markers, such as cleaved caspases, within a single panel. While monitoring of these features in live cells is not possible, by fixing cells at multiple times following stimulation, a kinetic analysis of specific populations within a heterogeneous sample is obtained (25, 29, 30).
While mass cytometry is not the only quantitative, high-dimensional technique, it is especially effective for monitoring immune responses in patients undergoing immunotherapy. In particular, mass cytometry allows for high-dimensional, single cell analysis at a relatively high throughput of around 500 cells per second. Other multidimensional techniques may play a role in immune monitoring as well. Polychromatic fluorescence flow cytometry has contributed immensely to the field of tumor immunology and, unlike CyTOF, can be utilized for fluorescence associated cell sorting (FACS). Multidimensional fluorescence and mass based imaging techniques can provide information about cellular positioning and cell-cell contacts (30, 39, 40), but typically measure several orders of magnitude fewer cells per sample than either fluorescence or mass cytometry. These techniques make excellent use of widely-available formalin-fixed paraffin-embedded (FFPE) blocks of tumor tissue. Recent work indicates that fixed cells can be released for analysis by mass cytometry (41).
In addition to protein, it is possible to measure RNA transcripts at the single cell level. Single cell RNA sequencing (scRNAseq) measures the transcriptome quantitatively at the single cell level (42). Despite the high dimensional, single-cell capabilities, scRNAseq typically measures only tens to hundreds of cells per sample and detection of transcripts can be confounded by issues including cell size, cell cycle changes, cell death, and allele dropout (43). Additionally, scRNAseq is restricted to measuring RNA transcripts and does not provide information about which genetic material is ultimately translated into protein or how these proteins are post-translationally modified. Quantitative PCR can also measure DNA, but is generally focused on hundreds of targets. Polychromatic fluorescence cytometry, imaging cytometry, scRNAseq, qPCR, and many other high dimensional technologies have produced great advances in the field of tumor immunology and will continue to play key roles in discoveries aimed to improve patient care. However, mass cytometry’s ability to measure surface protein expression, post-translational modifications, transcription factors, and functional outcomes (e.g. cytokine production, apoptosis, cell cycle) for millions of cells at the single cell level (5) makes it uniquely adapted for monitoring immune responses in cancer patients undergoing immunotherapy.
Dissecting Tissue Microenvironments in the Cloud
The increasing dimensionality of mass and fluorescence cytometry has dramatically increased the robustness and complexity of cytometry data (44). Analysis of data from a single mass cytometry panel containing 30 antibodies would require hundreds of traditional, biaxial plots (4). The use of hundreds of biaxial plots to analyze high dimensional, single cell data is not only impractical, but also insufficient in its ability to characterize complex cellular phenotypes and overly reliant on prior knowledge of the cell of interest. Thus, the growing use of high-dimensional cytometry has necessitated the development of novel data analysis tools. These advances have ushered in an ‘information age’ of single cell biology (45) by providing researchers with access to machine learning tools for dimensionality reduction, clustering, and model building. Machine learning algorithms can be designed to learn and improve performance based on previous experience (46). Recently, tools from machine learning have been adapted to visualize and cluster cells. One group of tools uses dimensionality reduction to reduce data from the original dimensions (about 40 for mass cytometry) to a few dimensions (47) that might be interpreted by a human expert or further analyzed by clustering algorithms that group phenotypically similar cells (44). These algorithms thus facilitate interpretation of highly complex, multidimensional data.
The current need for analysis by human experts in clinical diagnostic and experimental cytometry introduces problems of bias and limitations on the speed, quantitative accuracy, and reproducibility that might be overcome by computational analysis. However, it is only recently that the “big data” being generated is comprehensive enough for the discovery of predictive correlations that might match or exceed the skill of experts. What is needed now are use cases that show the systems biology approach not only captures standard clinical readouts quickly and accurately, but that it goes beyond what is known to also reveals clinically useful information from unexpected cellular sources (45). For example, we recently used mass cytometry immune monitoring to reveal and characterize unexpected myelodysplastic syndrome (MDS) blasts in the blood of a melanoma patient treated with α-PD1 (Greenplate et al., in press). This case showed not only the potential for in depth characterization of changes within major immune cell populations during immune therapy, but also the ability to aid in diagnosis of potential pathological changes within human tissue. Ultimately, we expect the practical challenges of mass cytometry will be addressed and lead to increasing costs of not implementing high dimensional single cell tools and automated analysis. Critically, these approaches have been developed in a way to minimally impact current standards of care, such that the significant increase in actionable information should not be learned at a high cost.
Machine learning and computational analysis make it possible to analyze large data sets in a reasonable amount of time. The technological advances in flow cytometry have enabled the field to measure over 30 parameters simultaneously, either through mass cytometry or polychromatic fluorescence cytometry. Analysis via traditional biaxial gates would result in hundreds of biaxial gates, and these gates would still fail to show high-dimensional co-expression of measured proteins (4). Computational analysis tools like viSNE and SPADE reduce multi-dimensional data into two dimensions while retaining high dimensional phenotype (48, 49). This allows the user to comprehensively analyze co-expression of all measured proteins. In addition to comprehensively analyzing and displaying data, computational methods are able to reduce bias. Analyzing all the data allows for the identification of cells with unusual or novel phenotypes that might have been overlooked otherwise (45). Using mass cytometry and computational analysis tools, Becher et al identified a previously unknown tissue-resident CD11blo NK cells (50). Work flows have been developed to streamline cellular discovery, identifying both broad populations and cell subsets and characterizing individual cell phenotype within those subsets (44, 51). Computational tools, while critical for analysis of big data, still face several challenges. Currently, an “expert” is needed for unstructured problem solving. In other words, computational tools fail in solving problems or analyzing data in systems where the rules do not currently exist.
Clinical Impact of Systems Immunology
Several recent studies have demonstrated that overcoming the practical challenges of mass cytometry data acquisition and analysis in longitudinal immune monitoring can unlock biologic and clinical insights in tumor immunology (34, 52, 53). A comprehensive and dynamic picture of anti-tumor immunity can be achieved by using cutting edge, single cell tools on samples collected longitudinally, over the course of therapy, or after specific perturbation. This approach has had success in the study of CD8+ T cell types (17), vaccine responses (54), surgery outcomes (53), and NK cell diversity (55). Serial sample collection provides personalized response profiles that could be used to guide treatment in real time. Specifically, high resolution analysis immunophenotyping and single cell signaling profiles have the potential to uncover prognostic markers, biomarkers, and possible mechanisms of response. For example, external stimulation through the B cell receptor of follicular lymphoma revealed a population insensitive to BCR stimulation and was prognostic of a poor outcome (4) and potentiated cytokine signaling has characterizes clinically relevant leukemia cells (56, 57). Establishing a response to stimuli, whether through phenotypic changes, post-translational modifications, or genetic changes, can elucidate the behavior and function of each cell beyond what is possible with a static view (5).
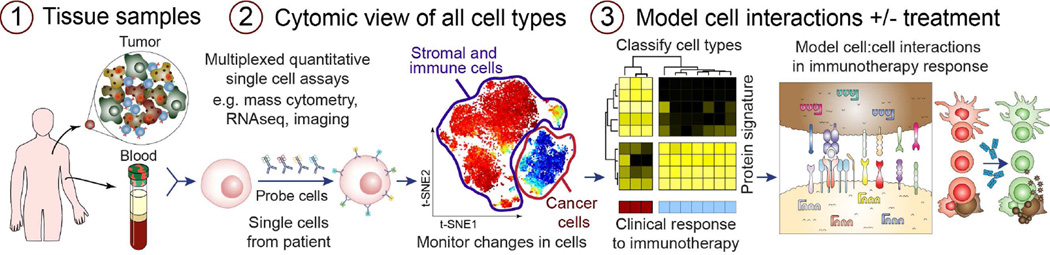
Cytomic response profiles are accelerating the transition of immune “monitoring” into an in vivo tool that identifies biomarkers of response and mechanisms of action for cancer patients undergoing immunotherapy (Figure 2). An early proof of concept of immune monitoring with mass cytometry was recently published: using cytomic response profiles of patients undergoing hip replacements with the goal of identifying immune signatures of recovery from surgery. Peripheral blood from patients was analyzed before and at several time points after surgery. Strong correlates were found between clinical parameters of surgical recovery and signaling responses in a subset of monocytes (53). Likewise, cytomic response profiling of melanoma patients undergoing immune checkpoint blockade therapy revealed that anti-CTLA-4, but not anti-PD-1, induced activation in a subset of CD4+ transitional memory T cells (7). More recently immune cells in breast cancer and acute myeloid leukemia were profiled by mass cytometry (57, 60, 61). Although these examples demonstrate the potential of HD immune monitoring in clinical practice, applying these principals to anti-tumor immunity remains relatively new and much work remains to identify standard procedures for experiments and data analysis.
Figure 2. Systems immune monitoring in cancer therapy.
1) Acquisition of high quality tissue samples pre- and post-treatment is a critical element of human immune monitoring. Following processing into single cell suspension, smaller samples of 100,000 to 2 million cells are generally best analyzed immediately, whereas larger samples are typically cryopreserved as aliquots (58). 2) Next, single cells must be detected using a quantitative technique. This review focuses on mass cytometry, but many other flow, imaging, and sequencing based approaches now yield quantitative single cell information with sufficient cellular throughput. Critical to the analysis are software tools that cope with high dimensional data and provide human-readable single cell views (44, 59). 3) Finally, statistical models are derived that correlate cell subsets and biomarkers with clinical outcomes. This information can be used to develop new mechanistic models of cell to cell interactions and the impact of treatment on signaling networks within and between cells.
Conclusion: Toward Clinical Cytomic Monitoring
Ultimately, it may be possible to bring cytomic profiling into the clinical setting such that a rapid assessment of cellular biomarkers could guide treatment. However, the current data analysis workflow remains driven by human experts and accounts for nearly half of the time spent going from sample to useful data. Expert driven manual analysis, the current gold standard, of high dimensional data is much too time consuming for practical clinical use. Computational analysis tools are not only able to dramatically speed up analysis, but also reduce bias and facilitate the discovery of unexpected cell types or phenotypes. By using an unsupervised computational workflow (44), it is now possible to automatically analyze cellular populations, subsets, and single-cell phenotype for millions of cells within minutes of data collection. In time, computational tools are expected to play an increasing role in guiding diagnosis and treatment selection.
Mass cytometry could play several potential roles in cancer therapy. First, identifying markers that predict response to immune checkpoint inhibitors is a major unmet need. Pre-treatment or early-on-treatment peripheral blood or tumor biopsies could be profiled to identify immune cell subsets that correlate with response to treatment or even severe toxicities. Second, with the advent of immune therapy combinations, dissecting the individual and collective effects of each agent will be critical. For example, a combination partner with immune therapy that dampens cytotoxic T cell proliferation or signaling may compromise rather than enhance the anti-tumor immune response. Mass cytometry could be used, therefore, in clinical trials or even in a high-throughput fashion to screen novel combinations. Finally, immune monitoring for patients who have responded to treatment may be useful. Particular cellular populations may herald durable responses, or conversely, impending relapses. Studies by our group and others are addressing these clinical challenges.
A critical element is the acquisition of high quality samples of human tissue. As a field, there is a need for standardized procedures and consistent support for sample acquisition as part of clinical trials, especially early phase trials. We recently published detailed step by step mass cytometry focused protocols for human tissue acquisition (58) and unsupervised data analysis (44). Still missing, however, are studies testing the impact of sample acquisition and preparation conditions on a wide number of endpoint assays and standard protocols for data quality assessment that include protocols for data analysis. This problem is especially significant in the realm of multi-center trials, where differences in capacity and experience among the centers leads to inconsistent practices that jeopardize data integrity. However, publication and agreement on standards of practice can ameliorate these inconsistencies. The field is actively meeting these needs as improvement in mass cytometry data normalization (62) consistent online sharing of annotated data can improve data quality and accuracy(59, 63). As mass cytometry and other single cell immune monitoring techniques become more widely adopted, it will be valuable for the field to further develop and implement standardized procedures for immune monitoring, from sample collection to cytometric protocols to data analysis.
bullet-point highlights.
A network of dynamic cells drives anti-cancer immune responses
Mass cytometry enables cancer immune monitoring in human blood and tissue biopsies
Cytomic profiling reveals known and novel cell types in complex tissues
Systems cancer immunology may soon help guide clinical decision making
Acknowledgments
Funding: NIH/NCI R00 CA143231 (J.M.I.), F31 CA199993 (A.R.G.), and K12 CA090625 (D.B.J. and P.B.F.) - provided salary or fellowship support for authors of this review
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict-of-interest disclosure: Co-founder and board member at Cytobank Inc. (JMI); research support from Incyte Corporation (JMI); advisory board for Bristol Myers Squibb, Genoptix (DBJ).
Bibliography
- 1.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12(4):298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 3.Giraldo NA, Becht E, Remark R, Damotte D, Sautes-Fridman C, Fridman WH. The immune contexture of primary and metastatic human tumours. Curr Opin Immunol. 2014;27:8–15. doi: 10.1016/j.coi.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler's guide to cytometry. Trends Immunol. 2012;33(7):323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irish JM, Doxie DB. High-dimensional single-cell cancer biology. Curr Top Microbiol Immunol. 2014;377:1–21. doi: 10.1007/82_2014_367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14(4):847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 7.Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 8.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scarlett UK, Rutkowski MR, Rauwerdink AM, Fields J, Escovar-Fadul X, Baird J, et al. Ovarian cancer progression is controlled by phenotypic changes in dendritic cells. J Exp Med. 2012;209(3):495–506. doi: 10.1084/jem.20111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smyth MJ, Crowe NY, Godfrey DI. NK cells and NKT cells collaborate in host protection from methylcholanthrene-induced fibrosarcoma. Int Immunol. 2001;13(4):459–463. doi: 10.1093/intimm/13.4.459. [DOI] [PubMed] [Google Scholar]
- 11.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410(6832):1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 12.Qin Z, Blankenstein T. CD4+ T cell--mediated tumor rejection involves inhibition of angiogenesis that is dependent on IFN gamma receptor expression by nonhematopoietic cells. Immunity. 2000;12(6):677–686. doi: 10.1016/s1074-7613(00)80218-6. [DOI] [PubMed] [Google Scholar]
- 13.Nimmerjahn F, Ravetch JV. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 2005;310(5753):1510–1512. doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- 14.Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer C, Sevko A, Ramacher M, Bazhin AV, Falk CS, Osen W, et al. Chronic inflammation promotes myeloid-derived suppressor cell activation blocking antitumor immunity in transgenic mouse melanoma model. Proc Natl Acad Sci U S A. 2011;108(41):17111–17116. doi: 10.1073/pnas.1108121108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balkwill F, Montfort A, Capasso M. B regulatory cells in cancer. Trends Immunol. 2013;34(4):169–173. doi: 10.1016/j.it.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36(1):142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strauss L, Bergmann C, Gooding W, Johnson JT, Whiteside TL. The frequency and suppressor function of CD4+CD25highFoxp3+ T cells in the circulation of patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 2007;13(21):6301–6311. doi: 10.1158/1078-0432.CCR-07-1403. [DOI] [PubMed] [Google Scholar]
- 19.Wolf AM, Wolf D, Steurer M, Gastl G, Gunsilius E, Grubeck-Loebenstein B. Increase of regulatory T cells in the peripheral blood of cancer patients. Clin Cancer Res. 2003;9(2):606–612. [PubMed] [Google Scholar]
- 20.Ling KL, Pratap SE, Bates GJ, Singh B, Mortensen NJ, George BD, et al. Increased frequency of regulatory T cells in peripheral blood and tumour infiltrating lymphocytes in colorectal cancer patients. Cancer Immun. 2007;7:7. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Min Z, Zhang D, Wang W, Marincola F, Wang X. Enhanced frequency and potential mechanism of B regulatory cells in patients with lung cancer. J Transl Med. 2014;12:304. doi: 10.1186/s12967-014-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao Y, Liu R, Shin MS, Trentalange M, Allore H, Nassar A, et al. CyTOF supports efficient detection of immune cell subsets from small samples. J Immunol Methods. 2014;415:1–5. doi: 10.1016/j.jim.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campana D, Coustan-Smith E. Minimal residual disease studies by flow cytometry in acute leukemia. Acta Haematol. 2004;112(1–2):8–15. doi: 10.1159/000077554. [DOI] [PubMed] [Google Scholar]
- 24.Amir E-aD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irish JM, Czerwinski DK, Nolan GP, Levy R. Altered B-cell receptor signaling kinetics distinguish human follicular lymphoma B cells from tumor-infiltrating nonmalignant B cells. Blood. 2006;108(9):3135–3142. doi: 10.1182/blood-2006-02-003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valitutti S, Muller S, Salio M, Lanzavecchia A. Degradation of T cell receptor (TCR)-CD3-zeta complexes after antigenic stimulation. J Exp Med. 1997;185(10):1859–1864. doi: 10.1084/jem.185.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson DB, Estrada MV, Salgado R, Sanchez V, Doxie DB, Opalenik SR, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun. 2016;7:10582. doi: 10.1038/ncomms10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bodenmiller B, Zunder ER, Finck R, Chen TJ, Savig ES, Bruggner RV, et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat Biotechnol. 2012;30(9):858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12(4):269–281. doi: 10.1038/nri3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholas KJ, Greenplate AR, Flaherty DK, Matlock BK, Juan JS, Smith RM, et al. Multiparameter analysis of stimulated human peripheral blood mononuclear cells: A comparison of mass and fluorescence cytometry. Cytometry A. 2015 doi: 10.1002/cyto.a.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carr EJ, Dooley J, Garcia-Perez JE, Lagou V, Lee JC, Wouters C, et al. The cellular composition of the human immune system is shaped by age and cohabitation. Nat Immunol. 2016 doi: 10.1038/ni.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das R, Verma R, Sznol M, Boddupalli CS, Gettinger SN, Kluger H, et al. Combination therapy with anti-CTLA-4 and anti-PD-1 leads to distinct immunologic changes in vivo. J Immunol. 2015;194(3):950–959. doi: 10.4049/jimmunol.1401686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4(8):648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 36.Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the Human Immunology Project. Nat Rev Immunol. 2012;12(3):191–200. doi: 10.1038/nri3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bendall SC, Davis KL, Amir el AD, Tadmor MD, Simonds EF, Chen TJ, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157(3):714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361(1–2):1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 39.Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med. 2014;20(4):436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A. 2013;110(29):11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmons AJ, Banerjee A, McKinley ET, Scurrah CR, Herring CA, Gewin LS, et al. Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-α-induced apoptosis in vivo. Mol Syst Biol. 2015;11(10):835. doi: 10.15252/msb.20156282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11(1):41–46. doi: 10.1038/nmeth.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16(3):133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 44.Diggins KE, Ferrell PB, Jr, Irish JM. Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods. 2015;82:55–63. doi: 10.1016/j.ymeth.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Irish JM. Beyond the age of cellular discovery. Nat Immunol. 2014;15(12):1095–1097. doi: 10.1038/ni.3034. [DOI] [PubMed] [Google Scholar]
- 46.Mjolsness E, DeCoste D. Machine learning for science: state of the art and future prospects. Science. 2001;293(5537):2051–2055. doi: 10.1126/science.293.5537.2051. [DOI] [PubMed] [Google Scholar]
- 47.Cheng Y, Wong MT, van der Maaten L, Newell EW. Categorical Analysis of Human T Cell Heterogeneity with One-Dimensional Soli-Expression by Nonlinear Stochastic Embedding. J Immunol. 2016;196(2):924–932. doi: 10.4049/jimmunol.1501928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amir el AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31(6):545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29(10):886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW, et al. High-dimensional analysis of the murine myeloid cell system. 2014;15(12):1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 51.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci U S A. 2014;111(26):E2770–E2777. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansmann L, Blum L, Ju CH, Liedtke M, Robinson WH, Davis MM. Mass cytometry analysis shows that a novel memory phenotype B cell is expanded in multiple myeloma. Cancer Immunol Res. 2015;3(6):650–660. doi: 10.1158/2326-6066.CIR-14-0236-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaudilliere B, Fragiadakis GK, Bruggner RV, Nicolau M, Finck R, Tingle M, et al. Clinical recovery from surgery correlates with single-cell immune signatures. Sci Transl Med. 2014;6(255):255ra131. doi: 10.1126/scitranslmed.3009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sen N, Mukherjee G, Sen A, Bendall SC, Sung P, Nolan GP, et al. Single-cell mass cytometry analysis of human tonsil T cell remodeling by varicella zoster virus. Cell Rep. 2014;8(2):633–645. doi: 10.1016/j.celrep.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, et al. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med. 2015;7(297):297ra115. doi: 10.1126/scitranslmed.aac5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118(2):217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 57.Levine JH, Simonds EF, Bendall SC, Davis KL, Amir el AD, Tadmor MD, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162(1):184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leelatian N, Diggins KE, Irish JM. Characterizing Phenotypes and Signaling Networks of Single Human Cells by Mass Cytometry. Methods Mol Biol. 2015;1346:99–113. doi: 10.1007/978-1-4939-2987-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010 doi: 10.1002/0471142956.cy1017s53. Chapter 10:Unit10 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 61.Gibbs KD, Jr, Jager A, Crespo O, Goltsev Y, Trejo A, Richard CE, et al. Decoupling of tumor-initiating activity from stable immunophenotype in HoxA9-Meis1-driven AML. Cell Stem Cell. 2012;10(2):210–217. doi: 10.1016/j.stem.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83(5):483–494. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spidlen J, Breuer K, Rosenberg C, Kotecha N, Brinkman RR. FlowRepository: a resource of annotated flow cytometry datasets associated with peer-reviewed publications. Cytometry A. 2012;81(9):727–731. doi: 10.1002/cyto.a.22106. [DOI] [PubMed] [Google Scholar]