Abstract
The gut microbiota is a key player in many physiological and pathological processes occurring in humans. Recent investigations suggest that the efficacy of some clinical approaches depends on the action of commensal bacteria. Antibiotics are invaluable weapons to fight infectious diseases. However, by altering the composition and functions of the microbiota, they can also produce long-lasting deleterious effects for the host. The emergence of multidrug-resistant pathogens raises concerns about the common, and at times inappropriate, use of antimicrobial agents. Here we review the most recently discovered connections between host pathophysiology, microbiota, and antibiotics highlighting technological platforms, mechanistic insights, and clinical strategies to enhance resistance to diseases by preserving the beneficial functions of the microbiota.
Keywords: Antibiotics, Gut Microbiota, Immunity, Disease, Antibiotic Resistance
In the past two decades, the gut microbiota has been recognized as a fundamental player orchestrating host physiology and pathology (Box 1). Trillions of bacteria inhabit the gastrointestinal (GI) tract of complex metazoans including humans, greatly expanding the host genetic repertoire [1]. This translates into the possibility for the host to perform functions that are not encoded by its own genome: commensals protect from pathogen invasion, extract additional energy from food, synthesize key molecules for tissue development, in a way that is highly specialized with respect to their location along the GI tract [2-4].
Box 1. The Gut Microbiota: a Structural Overview.
The human gut microbiota consists of an estimated 100 trillion bacteria, belonging to several hundreds of different species [129]. These fall into 4 major phyla covering more than 90% of the total bacterial population, namely Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, and many additional minor phyla, including Verrucomicrobia and Fusobacteria. The representation of these groups changes along the gastrointestinal tract, influenced by distinct microenvironments and nutrient availability [2]. The Firmicutes phylum is composed mainly by Gram+, aerobic and anaerobic bacteria. Prominent members are Clostridia strains, whose activities range from beneficial and protective (e.g. C. scindens, cluster IV-XIVa) to pathogenic (e.g. C. difficile, C. perfrigens). Potentially pathogenic streptococci, enterococci and staphylococci are also Firmicutes. Bacteroidetes are Gram− bacteria that are extremely well adapted to the intestinal environment. Here, they ferment otherwise indigestible carbohydrates producing SCFAs, molecules that have been implicated in a plethora of important processes. Actinobacteria are Gram+ bacteria generally considered beneficial, such as the Bifidobacterium genus, which are included in many probiotic preparations. The Proteobacteria phylum contains Gram-bacteria, most notably the family Enterobacteriaceae, including E. coli and K. pneumoniae. These are not very abundant under normal conditions, but tend to expand upon dysbiosis.
Notably, the majority of the studies of the microbiota have been performed in mice, even though human and mouse microbiota differ in genus representation [130]. Some genuses such as Prevotella, Faecalibacterium and Ruminococcus are abundant in humans, while others, namely Lactobacillus, Alistipes and Turicibacter are highly prevalent in mice [131]. However, a core of common taxa can be identified, and mouse and human intestinal metagenomes appear to be remarkably similar if analyzed from a functional perspective (i.e. representation of KEGG pathways, which depict the overall metabolic potential of a community) [132]. Most importantly, GF animals can be efficiently reconstituted with microbial communities isolated from other species, including humans, reproducing the effects that were observed in a donor, on a recipient host [133]. Reconstitution of GF mice with stool samples from obese or malnourished subjects is sufficient to phenocopy patient defects in energy harvest or growth [55, 65, 134, 135], demonstrating that despite inter-species divergences, the mouse model is a valuable tool to study the human microbiota.
Although the physiology of virtually all organs is influenced by the microbiota [5, 6], the intestinal mucosa and its immune components, are most affected by this symbiosis [7]. Here we first review recent findings elucidating the impact of the microbiota on the immune system. Second we discuss the involvement of gut commensals in the pathogenesis of disease. Third, we examine the role of antibiotics in perturbing or driving these processes. And finally, we discuss the mechanisms of antibiotic resistance development and spread, as well as the proposed approaches to overcome the drawbacks of antibiotic therapy.
Beneficial Roles of the Microbiota
The gut microbiota exerts many beneficial functions for the host, to a level that it can be considered an additional organ [8]. For example, commensal bacteria convert primary bile acids into secondary bile acids, thus allowing lipid adsorption. They also produce vitamins of the B and K groups and ferment otherwise indigestible plant-derived fibers producing short chain fatty acids (SCFAs, see Glossary) that feed enterocytes and modulate immune functions [2, 3]. Furthermore, the microbiota drives intestinal development by promoting vascularization, villus thickening, mucosal surface widening, mucus production, cellular proliferation and maintenance of epithelial junctions [9-11]. Notably, the influence of the microbiota is not limited to the intestine, and affects the physiology of most host organs, even the brain [9, 12-15].
One of the most prominent roles of the gut microbiota is to promote the development and education of the immune system, both locally and systemically, as described below.
Education of the Immune System
The close proximity of dense microbial populations to host tissues poses risks of invasion and the immune system must thoroughly monitor bacteria present in the gut lumen (Box 2). Nonetheless, the microbiota is allowed to prosper on the surface of the intestinal mucosa, orchestrating the overall physiology of the tissue lying underneath. This concept was established with the observation that antibiotic-treatment worsens the severity of DSS-induced colitis in mice, by depleting microbial ligands that normally signal through Toll-like receptors (TLRs) and function to ensure expression of tissue homeostasis and repair mediators [16] (Figure 1, Figure 2).
Box 2. Microbiota and the Immune System: Strategies to Ensure a Tricky Coexistence.
As a first challenge, the host must maintain in the gut lumen beneficial strains without allowing overproliferation of non-beneficial rapid growers. The epithelium has been proposed to act as a selectivity amplifier, causing major rearrangements in the composition of the microbial community through secretion of nutrients and reduced quantities of AMPs, and acting mainly on microbes that are closer to the mucosa [136]. Provision of nutrients is maintained at all costs. For instance, upon starvation-- a natural reaction of animals following infection --, a pathway signaling through DCs, ILCs and IECs induces massive fucosylation of the intestinal epithelium in mice, thus providing nutrients to commensal bacteria and maintaining colonization resistance to C. rodentium infection [137].
As a second challenge, immune recognition of commensal antigens and cues must be blunted. The luminal bacterial load, a few micrometers in distance, is as high as 1012 CFU/ml, and the host immune system is well-suited to react. Indeed, breakdown of the intestinal barrier upon infection or DSS treatment of mice has been shown to result in a complete effector and memory immune response against commensal flagellin [138 [80].
Under normal physiological conditions, mucus creates a physical barrier that keeps most bacteria a safe distance from the epithelium, mainly through diffusion of AMPs and IgA in the small intestine and formation of a tight, impenetrable structure in the large intestine [139]. At steady state, commensal microbes provide signals that dampen the activity of CX3CR1+CD11c+ mononuclear phagocytes in the intestine, thus reducing transport of bacterial antigens to the MLNs and preventing triggering of adaptive immune responses against luminal antigens, a tolerogenic mechanism that can be broken by antibiotic treatment [140]. Moreover, type 3 ILCs in the intestine blunt the immune response against the microbiota by inducing apoptosis in commensal-specific T cells, through an MHCII-dependent antigen presentation process that resembles thymic negative selection [141, 142]. Deletion of the MHCII gene on ILCs has been shown to result in a T cell-dependent inflammatory disease modeling IBD colitis, which could be ameliorated by antibiotic treatment [141, 142]. Of note, human ILC3s have also been found to express MHCII. ILC3s from pediatric patients with Crohn’s disease (IBD) were found to present reduced HLA-DR expression, and HLA-DR levels negatively correlated with the number of Th17 cells in colon biopsies from these patients [142].
Finally, commensals can also adopt strategies to “hide”. Some abundant strains of Bifidobacteria alternatively produce two variants of cell surface-associated exopolysaccharide, decreasing their chances of being recognized by the immune system [143]. B. thetaiotaomicron reduces the negative charge of LPS through phosphatase activity, thus acquiring resistance to positively-charged AMPs [99].
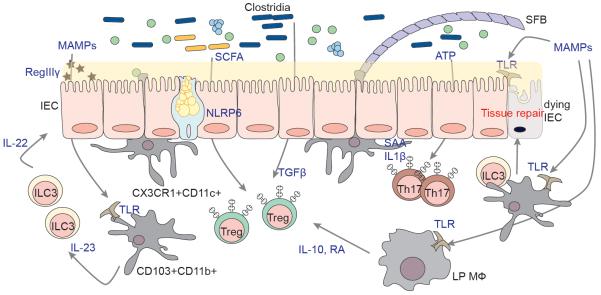
Figure 1. Roles of the Microbiota in the Development and Maintenance of the Intestinal Immune System.
The gut microbiota is separated from the intestinal epithelium by a thin layer of mucus, secreted by Goblet cells in a microbiota- and NLRP6-dependent manner. The mucus layer has a different structure in small and large intestine (not depicted in the figure). Microbial-associated molecular patterns (MAMPS) can be sensed by IECs as well as by myeloid cells in the lamina propria and induce a variety of effects, including tissue repair, and production of antimicrobial peptides such as RegIIIγ in intestinal epithelial and Paneth cells through a DC-ILC axis. Luminal ATP and SAA/IL1β produced by IECs and DCs in response to adhesion of segmented filamentous bacteria (SFB) promote Th17 development. Antigens presented during this process are largely derived from SFB. Treg induction is also regulated by bacterial cues. Clostridia of the IV and XIVa groups induce Tregs in a TGFβ-dependent manner. Short chain fatty acids (SCFA) promote Treg differentiation by acting directly on T cells and indirectly on DCs. Macrophages in the lamina propria are involved in a pathway that includes also ILCs (not shown) resulting in production of IL-10 and Retinoic Acid (RA), and also sustaining expansion of Tregs. All pathways illustrated in the figure were shown to be affected by the use of antibiotics, leading to a lack of homeostasis, an increased sensitivity to infection and an increase in the severity of various conditions, as in the case of allergy.
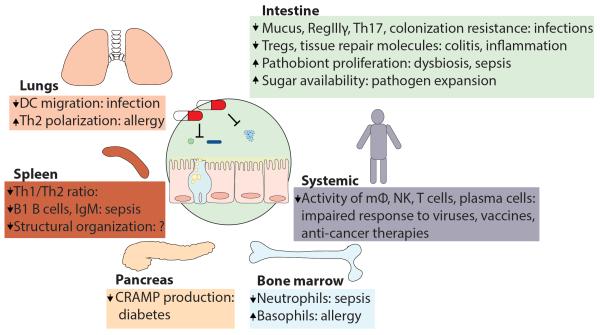
Figure 2. Antibiotic-Mediated Microbiota Depletion Causes Disease in Multiple Organs.
Antibiotics act on the gut microbiota by decreasing its density and modifying its composition in a long-lasting fashion. This causes reduced signaling to the intestinal mucosa and peripheral organs, which results in impaired functioning of the immune system. Depicted are examples of diseases that were shown to arise or be worsened as a consequence of antibiotic treatment in mouse models (see main text).
All branches of the immune system rely on this tonic signaling to properly function (Figure 1). Microbiota-derived LPS maintains basal level expression of RegIII-γ (a bactericidal C-type lectin) in intestinal epithelial cells (IECs) and Paneth cells. RegIII-γ is not detected in germ-free (GF) mice [17], and even short-term antibiotic treatment impairs its expression, rendering mice susceptible to vancomycin-resistant enterococcus (VRE) infection, a defect that can be reverted by oral administration of LPS [18]. Similarly, commensal flagellin sensing by TLR5 on CD103+CD11b+ lamina propria (LP) dendritic cells (DCs) contributes to maintenance of RegIII-γ expression. Upon TLR5 signaling, DCs produce IL-23, promoting IL-22 release by innate lymphoid cells (ILCs) and therefore, RegIII-γ expression in intestinal epithelial cells (IECs) [19, 20].
Granulocytes also receive commensal cues, which they sense while residing in the bone marrow (BM). NOD1-mediated sensing of meso-diaminopimelic acid (DAP) promotes neutrophil-mediated killing of pathogens such as Staphylococcus aureus and Staphylococcus pneumoniae [21, 22]. In GF mice reconstituted with Escherichia coli, DAP was detected in the blood and BM over the course of three days, showing that bacterial ligands from the intestinal lumen can have systemic distribution and therefore, a systemic effect [22]. Moreover, perinatal antibiotic exposure alters neutrophil number and functions by impairing G-CSF and IL-17 production, predisposing neonates to increased risk of E. coli or Klebsiella pneumonia-induced sepsis [23]. Similarly, antibiotic-treatment perturbs the basophil compartment in the blood and BM of mice by acting through a Th2-IL-4-IgE pathway, which leads to exacerbated allergic syndromes [24].
Professional antigen-presenting cells (APCs) also rely on the microbiota to orchestrate the immune response. For instance, DC migration and IL-1β/IL-18 production have been shown to be impaired in antibiotic-treated mice infected with influenza virus in an inflammasome-dependent manner [25]. Immunoglobulin levels, T cell numbers and IFN-γ production were consequently affected, resulting in increased viral titers. However, rectal stimulation with TLR agonists restored lung immunity in this study, indicating that microbial signals from the intestine can modulate and re-establish systemic immunity.
Antibiotics have been shown to impair immunity against lymphocytic choriomeningitis virus (LCMV) in mice, by lowering the expression of antiviral genes such as Ifnb and Irf7 in lung macrophages, resulting in defects in CD8+ T cell expansion, as well as in IFN-γ/TNF-α and IgG production [26]. Another study reported that splenic macrophages from GF and antibiotic-treated mice also failed to prime NK cells because of reduced chromatin accessibility in the promoter region of genes encoding for cytokines such as type I IFNs (α, β), IL6 and TNF-α [27]. As a result, antibiotic-treated mice failed to control infection with mouse cytomegalovirus (MCMV) [27].
With regard to adaptive immune cells, the activity of T lymphocytes is severely impaired by disruption of the dialogue between the immune system and the microbiota, as in the case of antibiotic exposure.
GF mice exhibit aberrant spleen architecture, decreased numbers of CD4+ T lymphocytes in lymphoid organs, and an increased Th2/Th1 ratio [28]. Reconstitution of mice with Bacteroides fragilis, an abundant commensal, reverts these defects via immunogenic presentation of the zwitterionic polysaccharide PSA and priming of splenic Th1 cells [28]. The biology of Th17 cells is particularly interconnected with the activity of luminal bacteria. Indeed, Th17 differentiation in the small intestine (SI) LP depends on the microbiota; GF mice have a reduced Th17 compartment that is restored by introduction of fecal material from conventional mice [29]. Of note, ATP, likely of commensal origin, contributes to the conversion of naïve T cells into Th17 in the intestinal LP [30]. Prominent inducers of Th17 differentiation are segmented filamentous bacteria (SFB), spore-forming anaerobic Gram+ bacilli residing in the terminal ileum of some mouse strains [31, 32]. Accordingly, treatment with Gram+-targeting antibiotics impairs Th17 development in mice [29]. Mice reconstituted with SFB-lacking microbiota develop a reduced intestinal Th17 compartment and are therefore more susceptible to Clostridium rodentium challenge, a model for human enterohemorrhagic infections [29].
Commensal bacteria are detected by T cells, and most mouse LP Th17 T cells recognize SFB-derived antigens [21]. Physical interactions between SFB and enterocytes, to which the bacteria are tightly anchored, are required for the induction of Th17-polarizing molecules (SAA, IL-1β) [33, 34].
Regulatory T cells (Tregs) have a fundamental protective role against autoimmune and chronic inflammatory diseases, such as IBD. Commensal sensing by LP-resident phagocytes promotes release of retinoic acid (RA) and IL-10 in mice, leading to the generation and expansion of Tregs, a mechanism that is important in establishing tolerance to food antigens [35].
Commensal Clostridia belonging to clusters IV and XIVa have been shown to promote accumulation of Tregs in the colon by increasing TGF-β production in IECs, thus protecting mice from DSS-induced colitis and strengthening oral tolerance [36]. A follow up study described a protective consortium of 17 clostridia strains isolated from a human fecal sample, a community with therapeutic potential for allergic or inflammatory diseases such as colitis [37]. Recently, three different laboratories have identified microbiota-derived SCFAs (particularly propionate and butyrate), to be responsible for Treg differentiation/accumulation [38-40]. Indeed, SCFAs were present at reduced concentrations in the fecal pellets of GF or antibiotic-treated mice, and oral administration of SCFAs protected mice from T cell-induced colitis by inducing immunosuppressive Tregs [38-40]. SCFAs were found to act directly on T cells via the receptor GPCR43, thus enhancing FoxP3 expression through DNA acetylation, and on DCs, conferring a higher capacity to drive naïve T cell differentiation into Tregs [38-40]. Notably, microbiota-produced SCFAs can also promote tolerance in non-intestinal tissue. SCFAs were reported to drive cathelicidin-related antimicrobial peptide (CRAMP) production in pancreatic β cells in mice, promoting a regulatory fate in macrophages, which enhanced the priming of Tregs [41]. In this study, CRAMP treatment of NOD mice ameliorated diabetes, while antibiotic treatment favored priming of diabetogenic T cells, thus promoting disease progression. Female NOD mice displayed lower SCFA production than in male NOD mice, and their CRAMP levels could be restored by conventionalization with feces from males, resulting in protection from diabetes. This falls in line with findings from a previous report, showing that microbiota from males could partially protect NOD females from diabetes by inducing high levels of circulating testosterone, glycerophospholipids and sphingolipids [42]. Androgens are in fact known to limit diabetes [43], possibly by reducing T cell proliferation and polarization towards a Th1 cell fate [44, 45].
Finally, SCFAs can also promote T cell tolerance in allergic diseases models. Mice fed a high fiber diet have shown increased levels of circulating SCFAs in a model of intranasal challenge with house dust mite (HDM) extract; the SCFAsprevented lung allergic inflammation by acting on the propionate receptor GPR41, and promoting the development of phagocytes with reduced Th2-polarizing capacity [46]. Collectively, these murine studies show that commensal bacteria contribute to the maintenance of immune tolerance in the host, principally through induction of regulatory T cells.
B cell-mediated immunity is also affected by perturbation of the microbiota. In fact, immunoglobulin titers are decreased in mice treated with antibiotics prior to viral infection [25, 26]. Furthermore, sensing of commensal-derived LPS has been shown to be necessary for the development of B1 cells in mouse spleen and for the maintenance of basal levels of circulating IgM, which exert a protective role in the cecal ligation and puncture (CLP)-induced sepsis model [47].
Surprisingly, gut microbes are capable of modulating vaccination outcomes. In a recent study, TIV (influenza trivalent inactivated) and other unadjuvanted vaccines were reported to induce IgG and IgM production in a TLR5-dependent manner through the microbiota in mice and, possibly, humans, as suggested by the correlation between leukocyte TLR5 expression levels and anti-influenza antibody titers in cohorts of vaccinees [48]. TIV-challenged antibiotic-treated (and TLR5-deficient, TLR5−/−) mice exhibited defective antibody production, but administration of the TLR5-agonist flagellin, restored the humoral response. TLR5 signaling had an effect on short-lived plasma cells, enhancing survival/differentiation and antibody production, as well as on lymph node (LN) medullary cord macrophages, leading to the production of plasma cell-sustaining factors such as TNF-α, IL-6, and APRIL.
The described studies provide substantial demonstration of a strong influence of the gut microbiota on the immune response. This connection is currently being investigated in the context of different pathological conditions and therapeutic approaches. In this regard, it is of particular interest that the gut microbiota has been recently proposed to modulate the efficacy of anti-cancer therapies (Box 3), rapidly becoming an area of active research.
Box 3. The Gut Microbiota can Modulate the Efficacy of Anticancer Therapeutics.
Recent publications have suggested an unexpected role for commensal bacteria in modulating the effectiveness of anticancer drug-based and antibody-based therapies. For instance, antibiotic treatment has been shown to impair responses to intratumoral CpG-oligodeoxynucleotides (ODN) injection or Oxaliplatin administration in mice, by reducing induction of necrosis (via TNF-α), the magnitude of CD8+ T cell responses, as well as the production of ROS by myeloid cells [144]. Commensal taxa have been positively (e.g. Alistipes and Ruminocuccus) or negatively (e.g. Lactobacillus) associated with anticancer responses in this model.
Along the same lines, the antitumor alkylating agent Cyclophosphamide (CTX) has been shown to cause intestinal damage and augmented epithelial permeability in mice, thus promoting translocation of commensals to MLNs and spleen (particularly L. johnsonii and E. hirae) [145]. In this study, IFN-γ producing Th17 cells (pTh17) responsive to these bacteria were generated in CTX-treated (but not control) mice. Transfer of these cells rescued immune responses in vancomycin-treated CTX-treated mice that were otherwise unable to arrest tumor growth, suggesting that commensal-specific pTh17 generated as a consequence of CTX treatment were largely responsible for the anti-cancer effect of this drug.
In another study, checkpoint blockade anti-PDL1 therapeutic efficacy was improved by the presence or transfer of Bifidobacterium (but not Lactobacillus) in a mouse model of melanoma [146]. In this model, bacterial translocation was not observed, but the commensal bacteria improved the functionality of DCs, upregulating the expression of cytokines as well as T cell activation-related genes (e.g. antigen presentation, cross-presentation and co-stimulatory molecules), thus inducing a more potent anti-tumor CD8+ T cell response.
Similar to PDL1, the efficacy of CTLA4 blockade (ipilimumab), was strongly impaired in GF or antibiotic-treated mice, with reduced T cell proliferation and IFN-γ/TNF-α production in several cancer models (melanoma, sarcoma, colon carcinoma) [147]. Anti-CTLA4 treatment induced T cell-mediated intestinal damage and altered the microbiota by increasing B. thataiotaomicron, B. uniformis and Burkholderiaceae representation [147]. GF mice reconstituted with these but not other species (E. coli, L. plantarum, B. diastonis., E. hirae) recovered full responses to anti-CTLA4 administration, owing to the generation of bacterium-specific IFN-γ-producing T cells. Notably, T cells with the same bacterial specificity could be recovered from patients, and melanoma tumor-ridden mice reconstituted with human stool enriched in protective species exhibited a high incidence of tumor regression.
Of note, development of colitis upon ipilimumab administration, a common adverse effect observed in cancer patients undergoing treatment, was reduced in tumor-bearing mice reconstituted with B. fragilis and B. crepacia [147]. Accordingly, a recent analysis of a cohort of patients undergoing ipilimumab treatment identified members of the Bacteroidetes phylum as being significantly increased in subjects who did not develop colitis [148]. Polyamine transport and B vitamin biosynthesis modules were also enriched in these subjects, and were successfully used as high-sensitivity and -specificity molecular markers to predict a patient’s risk for developing colitis. This analysis thus provided a potentially useful microbiota-based diagnostic tool [148].
Overall, these works demonstrate that the microbiota contributes to the efficacy of anticancer treatment and the development of adverse reactions. However, whether antibiotic use in the context of cancer treatment affects outcome, remains as of yet, an important albeit unanswered, question.
Colonization Resistance
The microbiota can confer protection against pathogens, a phenomenon referred to as colonization resistance, which can be severely impaired by antibiotic treatment [4]. Colonization resistance takes place through direct (not requiring host involvement) or indirect mechanisms (ultimately mediated by the host response, as mentioned above for the modulation of AMPs) [18, 19]. Mucus production is another mediator of indirect colonization resistance. Treatment with metronidazole, which selectively depletes anaerobes, but not streptomycin, reduced the thickness of the inner mucus layer in the large intestine (LI) – a process that depends on NLRP6 – and increased susceptibility to oral C. rodentium infection in mice [49, 50].
Another example of colonization resistance is that mediated by commensal anaerobes such as Blautia producta and B. thetaiothaomicron, that confer transkingdom resistance to C. albicans in a mouse model of oral infection by inducing expression of the hypoxic factor HIF1-α and production of the antimicrobial peptide CRAMP (LL37) [51].
Bifidobacterium longum has been shown to protect mice from enterohemorrhagic E. coli O157 infection through the production of SCFAs, that prevent IEC apoptosis and inflammation, thus preserving gut epithelium integrity, and reducing the spread of Stx2 toxin into the bloodstream, ultimately improving host survival [52].
In humans, recurrent infection with C. difficile occurs in hospitalized patients, in particular those undergoing antibiotic treatment, causing diseases ranging from mild diarrhea to deadly toxic megacolon. Colonization resistance against C. difficile presents itself in healthy individuals, and transfer of healthy microbiota is currently being performed to treat patients with recurrent infections [53]. Of note, the commensal bacterium C. scindens can inhibit growth of C. difficile through the generation of secondary bile acids deoxycholic acid (DCA) and lithocholic acid (LCA) [54]. Reconstitution with C. scindens alone or within a bacterial consortium, protected antibiotic-treated mice from C. difficile intestinal colonization. in this study, C. scindens was also associated to protection from C. difficile colonization in patients receiving allogeneic haematopoietic stem-cell transplantation [54].
By combining the study of microbiota recovery in human samples from infected patients, and GF mouse reconstitution, Ruminococcus obeum was found to provide direct colonization resistance against Vibrium cholerae (responsible for cholera disease). Through the LuxS-based AI-2 quorum sensing signaling system, whose expression is enhanced in the presence of V. cholerae, R. obeum induces down-regulation of virulence and colonization factors in the pathogen, promoting its clearance [55]. Moreover, enterococci carrying a plasmid encoding for bacteriocin bac-21 were shown to outcompete established VRE colonization upon continuous administration of the pathogen in drinking water to mice harboring an intact microbiota [56].
Thus, taken from these examples, it is clear that commensals can directly or indirectly protect from pathogens, and antibiotic use severely impairs this function, which in turn increases the risk of host infection.
Gut Microbiota and Pathogenesis
The microbiota can contribute to a variety of diseases through different mechanisms, including production of noxious catabolites, a capacity to overgrow, to sustain inflammation, or to provide support for pathogens. Many factors, such as diet, underlying inflammation and dysbiosis, can modulate such pathogenic potential. In the next section, we discuss recent literature on the role of gut commensal bacteria in disease pathogenesis .
The Microbiota in Diet-Promoted Disease
Diet profoundly influences gut microbiota composition and functions [57], an effect that can promote disease. Consumption of artificial sweeteners in mice and humans has been shown to lead to deregulation of the intestinal microbiota, with altered glycan degradation capacity promoting decreased glucose tolerance, a pre-diabetic condition [58]. An important role for the microbiome in modulating glycemic responses has been recently demonstrated, with the development of an effective machine-learning algorithm to predicti postprandial glucose levels in humans based on several personal as well biochemical parameters, including the composition of the microbiota in healthy individuals[59]. In this study, personalized dietary interventions adopted on the basis of algorithm predictions, significantly altered microbiota composition while improving glycemic control, suggesting that modulation of microbiome configuration through diet could be used to prevent diabetes-predisposing conditions.
A correlation between certain bacterial taxa and type 2 diabetes has been proposed, based on analyses of human gut metagenomes [60, 61]. However, a recent study suggested that patient treatment with the drug metformin, a variable that subjects had not been stratified for previously, constituted the main determinant of microbial signatures in diabetic cohorts [62]. This finding demonstrates that multiple variables can influence gut microbiota composition in disease [63].
Gut microbiota plays a key role in the pathogenesis of obesity. Indeed, experiments have shown that the condition can be transferred from obese mice or humans to healthy mice via conventionalization (i.e. microbiota transfer by feeding fecal material) [64, 65]. These studies strongly suggest that diet is responsible for the selection of commensal strains with enhanced energy-harvesting capacity.
Obese mice harbor an altered microbiota enriched in Clostridia that produces high quantities of DCA, contributing to liver inflammation, and predisposing the host to an increased risk of developing hepatic cancer [66]. Low fat diet, antibiotic treatment or pharmacological inhibition of microbial conversion of primary to secondary bile acids has had a protective effect in this model. Along similar lines, in a mouse model of colorectal cancer, microbiota-derived butyrate enhanced intestinal epithelial cell proliferation, as well as the occurrence of intestinal polyps and tumor development [67]. In this report, low carbohydrate diet or treatment with selected antibiotics could prevent polyp formation.
Furthermore, a series of recent studies [68-70] revealed a link between microbiota and atherogenesis. Dietary phosphatydilcholine (PC), a lipid broadly present in foods, and L-carnitine (LC), a molecule abundant in red meat, are transformed by the microbiota into pro-atherogenic trimethylamine (TMA). LC- or PC- supplemented diets were found to induce higher atheroma formation and wider aortic lesions in ApoE- mice, and disease was ameliorated by antibiotic treatment [68, 69]. Also in humans, PC or LC administration augmented TMA plasma levels, and increased the risk for cardiovascular disease; notably, antibiotic treatment dramatically reduced TMA levels in these subjects [68, 70]. Vegan or vegetarian subjects exhibited a reduced or non-existent capacity to produce TMA upon LC supplementation, and it was proposed that bacteria, presumably selected through the host’s diet, were incapable of performing this biochemical conversion [68]. Overall these studies convincingly demonstrate a prominent role for the gut microbiota in diet-driven pathogenesis of atherosclerosis. Interestingly, a follow-up study showed that feeding mice an analogue of choline, which inhibits TMA production by blocking microbial TMA lyases, could prevent atherosclerosis in mice [71]. Thus, the microbiota represent a promising target for pharmacological therapeutic intervention in atherosclerosis.
Commensal bacteria can also impair drug activity. Eggerthella lenta, a human commensal, reduces digoxin, a drug used to prevent heart failure and arrhythmia, thus destroying its biological activity. This activity is enhanced in the presence of other commensals, but is repressed by arginine, which can be supplemented through a high-protein diet [72].
The Microbiota in Infection, Inflammation and Aberrant Immunity
A strong link between the microbiota and intestinal inflammation has been established. For instance, it has been shown that NLRP6-deficient mice harbor a microbiota that is more prone to induce colitis upon DSS treatment, a phenotype that could be transferred to healthy WT mice upon conventionalization [73].
Upon dysbiosis or inflammation, pathobionts lodged among healthy commensal species can promote disease. For instance, like other enterobacteriaceae, E. coli can utilize nitrate as an electron acceptor for respiration, which confers a selective growth advantage following iNOS gene activation upon DSS-induced inflammation in mice [74]. This has been proposed to be a leading cause of inflammation-related dysbiosis [74]. As another example, DSS-induced intestinal damage has been demonstrated to allow the human and mouse commensal Proteus mirabilis to engage the NLRP3 inflammasome on mouse CCR2+Ly6Chi monocytes via release of the Hemolysin A toxin [75]. Consequently, NLRP3 activation leads to production of IL1β, fostering inflammation and inducing colitis which can be ameliorated by antibiotic treatment. By contrast, antibiotic treatment has itself been reported to promote sepsis in mice in at least one model of DSS-induced colitis, by inducing overgrowth of the antibiotic resistant commensal strain of E. coli O21:H+, which carries a virulence gene cluster [76]. Upon expansion and DSS-induced epithelial damage, this pathobiont could enter the circulation and induce IL1-β production by macrophages, thus promoting sepsis [76].
The microbiota can favor or amplify the activity of pathogens, and antibiotics play an important role in this process. Enterohemorrhagic E. coli carries a fucose-sensing system (FusKR) that regulates virulence gene expression and therefore competition with commensals [77]. In vitro, B. thetaiotaomicron can cleave off fucose from mucin and enhance virulence of E. coli. In vivo, E. coli lacking FusKR is outcompeted by a wild type strain [77]. These data suggest that in vivo, B. thetaiotaomicron liberates fucose, thus fueling pathogenic E. coli growth. In addition, B. thetaiotaomicron also carries sialidases that can cleave sialic acid from mucus, although the bacterium lacks the enzymatic machinery necessary to catabolize such sugar [78]. In mono-associated gnotobiotic mice, B. thetaiothaomicron-liberated sialic acid can support the expansion of pathogens such as C. difficile or S. enterica upon oral infection. Accordingly, pathogen burden is reduced upon infection of mice monocolonized with a sialidase-deficient B. thetaiothaomicron strain [78]. In conventional mice, antibiotics reduce the amount of commensals competing for sugars liberated prior to treatment, increasing nutrient availability and favoring pathogen expansion througha limited time-window (peaking at day 1 and ending at day 3 post-antibiotic treatment for this study) [78].
Commensal bacteria have also been implicated in long-term sequelae from resolved Yersinia pseudotuberculosis infections in conventional mice, such as those resulting in leakage of gut-draining lymphatic vessels and deviated trafficking of CD11b+CD103+ DCs to adipose tissues rather than to mesenteric LNs (MLNs) [79].. This “immunological scar” has been found to impair mucosal immune responses as well as oral tolerance to food antigens. DC migration was not impaired in GF mice, and was restored in conventional mice upon antibiotic treatment, implicating a role for the microbiota in promoting scar formation [79].
Finally, the gut microbiota has also been associated with the induction of autoimmunity. For instance, R161H mice bear transgenic T cells that recognize the retinal photoreceptor RBP3 and spontaneously develop autoimmune uveitis over time. However, as RBP3 is confined to the eye, which is an immune-privileged site, it has been unclear how auto-reactive T cells could encounter the cognate antigen and be activated prior to migrating to the eye and promoting inflammation. Recent data have emerged to suggest that cross-reactivity to unidentified commensal antigens triggers an autoimmune response in this model [80]. Indeed, effector auto-reactive T cells could be detected in R161H mice lacking RBP3 as well, especially in the intestinal LP, suggesting a nonendogenous source for cognate peptide. Autoreactive T cells responded to stimulation with cecal content. Importantly, the onset of uveitis in R161H mice could be delayed upon antibiotic treatment [80].
Collectively, the discussed data demonstrate that the gut microbiota, especially with dysbiosis, can promote a variety of pathological conditions. In some of these cases, targeted treatments to selectively deplete the involved pathogenic species may be considered as a promising therapeutic approach.
Antibiotic Treatment Induces Long-Lasting Changes in the Microbiota Correlating with Disease
Numerous studies have confirmed that antibiotics have a tremendous impact on the composition and functionality of the human microbiota.
One study documented that healthy volunteers treated for 1 week or less with antibiotics, reported effects on their bacterial flora that persisted six months to two years after treatment, including a dramatic loss in diversity as well as in representation of specific taxa, insurgence of antibiotic resistant strains, and up-regulation of antibiotic resistance genes (ARGs) [81]. Antibiotic treatment in mice recapitulates the impact and long-term shifts in human gut communities. For example, a single dose of Clindamycin has been shown to induce profound changes in mouse microbiota composition and, consequently, to confer long-lasting susceptibility to C. difficile infection [82].
One potential explanation for the magnitude and duration of antibiotic effects in vivo, is the remarkable interdependence of different bacterial taxa. For instance, Gram− commensals can be depleted by vancomycin, which is a Gram+-targeting drug [83]. As a consequence of its profound effect on gut autochthonous (native) communities, vancomycin has been shown to cause long-lasting susceptibility to a variety of secondary infections in both humans and mice [84].
Microbiota development during early life stages of humans and mice is critical, and its perturbation predisposes to disease in later infancy or adulthood, particularly in the case of allergic and metabolic syndromes.
For instance, while vaginally delivered neonates acquire bacteria through the birth canal, the microbiota of C-section delivered infants resembles that populating the skin of adults, with Streptococci being a dominant species [85]. It has been posited that this could represent a factor predisposing individuals to subsequent infections [85]. Notably, swabbing C-section-delivered newborns with gauze incubated in the vagina has been shown to partially restore a normal microbiota, albeit with effects that remain to be ascertained [86].
Surveys on thousands of children have highlighted a link between the use of antibiotics during the first year of life and development of asthma by the sixth-seventh year [87, 88]. Early use of macrolides in Finnish children was found to generate a distinct microbial profile characterized by a loss of Actinobacteriaceae and an increase in Bacteroidetes and Proteobacteria, an induction of ARGs, and a decrease in bile salt hydrolases. Such profile positively correlated with either a later development of asthma, or an increase in body mass index [89]. Concordant with these findings, studies in mice have shown that neonatal (from pre-birth) but not adult exposure to antibiotics, resulted in exacerbated asthma following intranasal challenge with ovalbumin [90]. Along similar lines, low dose penicillin (LDP) administration was shown to induce stronger physiological alterations if administered from the beginning of gestation, rather than at weaning, confirming a critical role for vertical acquisition of microbiota [91]. In particular, following early LDP administration, body mass and fat content were increased in adulthood, while the expression of intestinal immune genes coding for proteins such asRegIIIγ, β-defensins, and IL-17 was decreased.
Therefore, antibiotic exposure, even for short periods of time and especially during infancy, has long-lasting effects on the microbiota, which can predispose the host to a variety of diseases, some of which remain to be potentially identified. This evidently represents a matter of critical importance for public health.
Generation of Antibiotic Resistance: the Driving Forces
Antibiotic-resistant pathogens are a major public health burden. However, ARGs are highly represented not only in such pathogens, but also among human commensal bacteria. An early survey suggested that a sizable fraction of the anaerobe compartment within the microbiota of healthy subjects is resistant to one or multiple antibiotics, with the proportion of such bacteria increasing following antibiotic treatment [92]. A more recent metagenomic analysis of the gut microbiota obtained from two healthy subjects estimated that multidrug-resistant species accounted for 20% of total bacteria [93]. As discussed below, exogenous antibiotics can contribute considerably to such accumulation of ARGs. However, antimicrobial molecules and resistance mechanisms are abundant in any bacterial community, and play an important evolutionary and regulatory role. Accordingly, ARGs were identified in microbiota from an 11th century AD mummy [94], in 30000 year-old permafrost sediment [95], as well as in a cave in New Mexico that had been isolated for millions of years [96]; none of these environments could have been influenced by the presence of modern day drugs. Surprisingly, both the DNA sequences and the structural organization within AR operons identified in these ancient bacteria were found to exhibit high similarity to those carried by currently circulating microbes [95]. Furthermore, antibiotic resistance to multiple classes (up to 14) of antibiotics, even to semi-synthetic molecules, was also identified in these prehistoric samples [96].
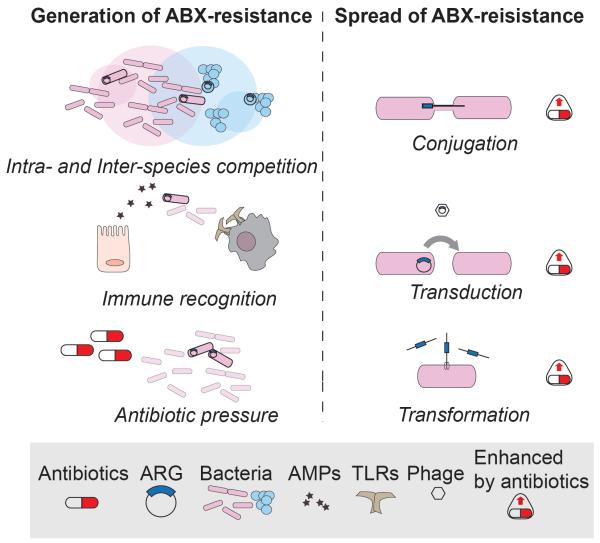
It is recognized that at least 3 major forces drive the development and spread of antibiotic resistance in the host or in the environment: i) immune recognition and response; ii) bacterial competition within communities; iii) exogenous antibiotic pressure [97] (Figure 3).
Figure 3. Generation and Spread of Antibiotic Resistance.
Left panel: antibiotic resistance is generated by mutations that can be induced by several driving forces. From the top: competition of bacteria (inter- or intra- species, here depicted as intersections between circles representing population niches) mainly mediated by bacteriocins, induces a selective pressure that favors development of resistance. Some TLRs or host-derived antimicrobial peptides (AMPs) target bacterial molecules that can undergo mutations and provide resistance to clinically-relevant antibiotics. Exogenous antibiotic pressure through medical or industrial practices (e.g.. antibiotic use in livestock) promotes the generation and selection of resistant strains that can rapidly diffuse. Right panel: antibiotic resistance genes can be exchanged among bacteria also of different species (not shown) through horizontal gene transfer, i.e. conjugation, transduction or transformation. Notably, these three mechanisms are all enhanced upon antibiotic exposure, resulting in a faster and more efficient spread of ARGs in the gut as well as in the environment.
Antibiotic resistance can arise in bacteria that inhabit animal body sites as a means to escape host defense strategies. For instance, erythromycin resistance gained by mutation of the 23s rRNA sequence was found to concurrently confer resistance to recognition by TLR13, which also happens to bind the same molecular target, and is likely to exert selective pressure in vivo [98]. The commensal B. thethaiotaomicron is highly resistant to the cationic peptide polymyxin B, which is considered a model for mammalian AMPs. Transposon mutation libraries in B. thethaiotaomicron have led to the identification of the bacterial gene lpxF, which encodes a phosphatase acting on LPS to diminish its negative charge and consequently, impair polymyxin B ligation to the bacterium [99]. Notably, upon DSS- or C. rodentium-induced inflammation, lpxF-deficient bacteria were outcompeted by WT strains in mouse intestines, suggesting that bacterial resistance can be both induced and maintained by the immune response [99].
The development of antibiotic resistance in natural environments, due to competition among different bacterial taxa, is a well-documented event [95, 96]. Intra-species competition, however, is sufficient to drive acquisition of resistance [100]. Growth of methycillin-resistant S. aureus (MRSA) in a high Mg2+ concentration medium which favors biofilm formation, has been shown to induce rapid generation of a mutated strain (W) that attempts to outcompete the WT strain (O) through secretion of surfactant molecules and the bacteriocin Bsa [100]. Increased Bsa concentrations drive in turn, the selection and expansion of a resistant S. aureus strain (Y), and as Bsa and vancomycin bind to a common target within the cell wall, the Y strain acquires resistance to the antibiotic, resulting in greater virulence in infected mice [100].
Finally, and most importantly, exogenous administration of antibiotics to a host can induce rapid expansion of ARGs (Box 4). In one study, pigs were fed a diet supplemented with a growth-enhancing antibiotic cocktail for two weeks. Subsequently, their microbiota showed increased expression of ARGs, conferring tolerance even to drugs that were not administered during the study [101]. That type of occurrence is of particular concern since drug-resistance genes can be horizontally transferred from soil bacteria, to pathogens [102]. Shotgun sequence analysis through an ARG-dedicated platform (PARFuMS) has in fact led to the identification of DNA cassettes conferring resistance to 7 different classes of antibiotics in soil bacteria, exhibiting perfect identity in coding and non-coding regions to genes encoded by common human pathogens, such as Pseudomonas aeruginosa and Salmonella enterica. Some cassettes were flanked by transposable elements, strongly suggesting horizontal gene transfer between soil-resident and pathogenic bacteria [102].
Box 4. Broad Use and Misuse of Antibiotics.
Since the beginning of their commercial distribution in the 40’s, antibiotics have proven an invaluable weapon against infectious agents, saving millions of lives. However, their use has broadened to a level that raises many concerns. In fact, it is estimated that a sizable proportion of the human population (1-3%) makes use of antibiotics everyday [110]. The overall administration of medical antibiotics has increased by more than 30% in the decade 2001-2011, and last-resort antibiotic use is alarmingly frequent. This has ignited a vicious circle in which the augmented use of antibiotics and development of antibiotic resistance fuel one another, leading to the return of previously well-controlled threats such as gonorrhea and Enterobacteriaceae infection [116].
Antibiotics are broadly employed in farming practices at low doses with the main aim of enhancing animal growth [149-151]. Livestock account for the vast majority of antibiotic production and use in the US (80% in 2013), resulting in the selection of resistant bacteria that can either infect humans or horizontally transfer resistance to pathogens. The use of antibiotics for livestock has been widely criticized and the FDA has responded by introducing Guidance#213 in 2013, which recommends avoidance of unnecessary antibiotics. This remains, however, a voluntary policy. Of note, it has been calculated that the ban of this procedure would not significantly increase costs for producers or consumers [150].
Interestingly, a correlation was suggested between the increased use of tetracycline in Danish livestock and the prevalence of tetracycline-resistance genes in Danish human subject microbiomes, despite β-lactams being the most widely prescribed antibiotic to patients in the same period and region [103].
A detailed analysis of dozens of metagenomic datasets obtained from hosts (humans, animals) or environments (water, soil, etc) has identified housekeeping alleles of genes such as rpsL, rpoB, gyrA conferring resistance to three classes of broad spectrum antibiotics, segregating at high frequency [104]. Of note, nearly 40% of host-associated bacteria carried quinolone-resistance genes, even in subjects that had never been exposed clinically. The “resistome” is defined as the pool of all ARGs present in a given microbiome, and appears early in life, detectable in infants as early as two months after birth [105]. Interestingly, twins display remarkably greater resistome similarity to each other than to unrelated infants or their mothers, suggesting that vertical transmission does not play a crucial role in its establishment. Moreover, the resistome expands over time, as the number of ARGs within the human microbiome positively correlate with age [106].
Altogether, these data provide evidence that antibiotic-resistance is naturally present in bacterial communities, including the gut microbiota, where it can be induced by several driving forces. However, exposure to exogenous antibiotics via medical or industrial practices has a marked capacity to enhance the insurgence and spread of ARGs.
Antibiotic Resistance Generation: De Novo Mutations
As previously discussed, antibiotics exert a selective pressure that drives rapid development of resistant strains. This process generally requires multiple DNA mutations. To understand how such mutations are acquired, in one study, E. coli cultures were challenged with increasing doses of 3 different antibiotics for 20 days in vitro[107]. Antibiotic resistance was found to arise following similar or identical mutational patterns in replicate experiments;. mutations affecting the same, or functionally-analogous genes, consistently emerged in bacterial cultures treated with a given drug [107]. Mutations also occurred in multidrug resistance genes. Furthermore antibiotics with a common molecular target also induced resistance to one another. Thus, it appears that there is an evolutionary trajectory in the acquisition of mutations conferring antibiotic resistance [107].
Importantly, within a clonal bacterial population under antibiotic pressure, not all cells acquire resistance. “Bacterial charity work” has been described as a mechanism to enhance resistance of the whole community with respect to a single resistant clone in an initially homogeneous population [108]. This study documented that within an E. coli culture exposed to norfloxacin or gentamicin, only a few cells rapidly acquired resistance-conferring mutations. Surprisingly, the antibiotic-resistant cells did not outcompete the antibiotic-sensitive members of the community, but rather, kept them alive. In fact, cells acquiring resistance to the drug also gained the ability (normally inhibited by antibiotics) to secrete indole, which acted on neighboring cells by activating defense mechanisms such as drug efflux pumps and oxidative-stress responses, ultimately allowing the survival of drug-sensitive bacteria. This mechanism resulted in higher growth rates of the entire population when compared to those of isolated resistant clones, which displayed lower proliferative capacity due to the metabolic cost of acquired mutations [108]. This study highlighted how acquisition of antibiotic resistance modifies ecological interactions within a homogeneous bacterial community, contributing to its own well-being. In well-mixed environments were different microbial taxa coexist in close proximity, distinct strains generally have the capacity to produce and to inactivate specific sets of anti-bacterial molecules. In this scenario, it has been proposed that leaky protection conferred by an antibiotic-degrading species to other surrounding bacteria is of public benefit [109]. In fact, ecological modeling has shown that in a bacterial consortium where every bacterial strain protects only itself from the action of a given antibiotic produced by a competing species (“rock, paper, scissors” model), fluctuations in the composition of the consortium increase, giving rise to a very unstable community that is ultimately dominated by one strain. On the contrary, if protection against a bactericidal molecule is provided in a leaky manner to surrounding species, -- by secretion of antibiotic-degrading enzymes in outer spaces, for instance-- the overall community composition is stabilized over time, becoming less subject to fluctuations [109]. Thus, antibiotic resistance plays a strong role in regulating microbial interactions and shaping microbial communities, possibly representing a prominent factor in the ecological regulation of the gut microbiota.
Antibiotic Resistance Genes are Spread via Horizontal Gene Transfer (HGT)
Bacteria can exchange ARGs via horizontal gene transfer (HGT). Specifically, HGT includes: i) transformation, the acquisition of DNA fragments from the environment; ii) conjugation, the delivery of genetic material from one cell to another through a pilus; iii) transduction, an exchange mediated by phages [97, 110]. All three mechanisms have been implicated in transfer of antibiotic resistance genes (Figure 3) [97, 110].
Generally speaking, HGT can take place among commensals, environmental bacteria or, importantly, between the two. For instance, a porphyranase necessary to process specific seaweed carbohydrates originally carried by a marine Bacteroidetes, Zobellia galactanivorans, was also found in commensal bacteria from Japanese, but not North American subjects [111]. As Japanese diets are highly enriched in seaweed containing porphyran, this work strongly implicated horizontal gene transfer between environmental and gut commensal bacterial species.
HGT of antibiotic resistance genes occurs at high rates among intestinal bacteria. In fact, clindamycin resistance transfer factor gene (BtgA) has been identified as carrying the highest number of SNPs within 200 human gut metagenomes, and other conjugal transfer factors have followed in this ranking [112].
Interestingly, all three mechanisms of HGT seem to be upregulated in bacteria upon antibiotic treatment (Figure 3).
Conjugation
Ciprofloxacin induces DNA damage-related SOS stress responses in E. coli, leading to a 100-fold enhancement in conjugative transfer of SXT, a genetic element conferring resistance to several antibiotics. Notably, in recent decades, the SXT element has emerged, presenting itself quite commonly in V. cholerae isolates from patients, potentially a result of antibiotic-induced resistance transfer [113].
Transduction
The phage metagenome (or “phageome”) recovered from the intestinal content of mice treated with ciprofloxacin or ampicillin is highly enriched in ARGs [114]. Furthermore, bacteriophages recovered from antibiotic-treated mice have been shown to be significantly more capable of transferring resistance to cultured microbiota ex vivo [114]. Phage genes conferring bacterial survival advantages under stress conditions (e.g. DNA repair enzymes), or an increase in fitness (e.g. carbohydrate degradation) were also enriched upon antibiotic treatment, suggesting that the ‘phageome’ can represent a reservoir for bacterial adaptation genes to be spread in times of need.
Transformation
Antibiotics and drugs interfering with DNA replication induce competence in bacteria, allowing acquisition of ARGs and other genetic material from the environment [115]. Treatment of S. pneumoniae with ciprofloxacin or HPUra (an inhibitor of DNA replication) has been shown to stall the progression of the DNA replication fork without inhibiting initiation of DNA replication [115]. This has been reported to result in increased copy numbers and, consequently, enhanced transcription of genes neighboring the origin of replication. Genes encoding for competence functions are located in this region, and consequently, they are upregulated in the presence of these drugs, promoting a state of competence.
Thus, ARGs spread among bacteria through conjugation, transduction and transformation, and this process is promoted by antibiotic exposure. As the human intestine is densely colonized with an amazing variety of microbes, antibiotic therapies are likely to promote diffusion of resistance genes within the microbiota, an aspect of antibiotic therapy that should not be neglected.
Moving Beyond Antibiotics
Considering the important roles of the microbiota in regulating host physiology and the multiple drawbacks of antibiotic use discussed above, finding alternative or complementary strategies to fight infections is imperative. Different promising approaches have been proposed to tackle this problem (Figure 4).
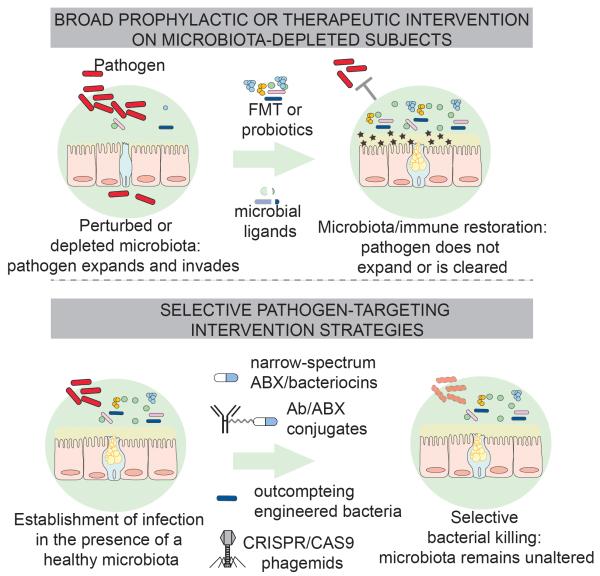
Figure 4. Novel Approaches to Substitute or Complement Antibiotic Therapies.
Antibiotic treatment depletes commensal communities in the gut, decreases mucus layer thickness and expression of AMPs, and predisposes to infection (upper panel). Transfer of microbiota by fecal transplantation can restore a healthy microbiota, mucus production, antimicrobial peptide secretion, and provide colonization resistance against pathogens, that can no longer expand or are cleared. Transfer of selected bacterial communities, as shown in mouse models, can achieve the same effect. Similarly, administration of microbial ligands, here depicted as fragments of bacteria, can restore basal production of mucus and antimicrobial peptides following antibiotic treatment. Lower panel: strategies to selectively deplete pathogens without perturbing the microbiota. All the approaches illustrated have proven to be successful in mouse models, providing high levels of protection and leaving the composition of the surrounding communities, unaltered. Consequently, colonization resistance mechanisms can be potentially preserved.
First and foremost, reforming or establishing a set of complementary public health measures can greatly diminish the need for antibiotic use. As pointed out by Laxminarayan [116], improving sanitation, expanding the use of vaccines, and strengthening hospital infection control have proven extremely effective tools in reducing the needs for antibiotic therapies. Moreover, reducing the unnecessary use of these molecules in farming practices must also be considered (Box 4).
Colonization resistance through microbiota transfer can substitute for antibiotics by restoring commensal communities (Figure 4). Fecal microbiota transplantation (FMT) has been shown to be more effective than conventional antibiotic therapy in the treatment of patients with recurrent C. difficile infection [53, 117]. Bacterial strains [54, 118, 119] and metabolites [54] associated with protection from C. difficile have now been identified, paving the way for eventual replacement of fecal material with selected probiotic strains or effector molecules. VRE colonizes the intestine following antibiotic treatment, prior to spreading to the bloodstream, and thus, represents a major threat for hospitalized patients [83]. In one study, transplantation of fecal microbiota containing Barnesiella species effectively cleared intestinal VRE from colonized mice, a promising finding that requires further investigation [120]. Enterococci carrying the conjugative plasmid pPD1, encoding for bacteriocin bac-21, were shown to outcompete VRE from mice colonized with the pathogen [56]. However, in this model, transfer of the plasmid to commensal bacteria was also observed, raising the possibility of pPD1 conjugation to VRE itself, and therefore, suggesting that caution should be taken with the clinical application of this approach.
Bacterial ligand administration following microbiota depletion can protect mice from infection . Systemic administration of TLR-5 agonist flagellin, or oral administration of TLR-4 agonist LPS, were shown to reinstate resistance to VRE or C. difficile in mice previously treated with antibiotics, through restoration of RegIII-γ production [18-20]. Moreover, R848, a synthetic agonist of TLR-7/8, orally delivered to mice, was reported to protect from VRE colonization [121]. Notably, R848 is already used in the clinics to treat papillomavirus infections, although it is topically administered. Thus, controlled administration of microbial ligands represents an important means by which to restore basal innate immune status and protection in hospitalized subjects undergoing antibiotic treatment.
Computational platforms and high-throughput screenings are currently being exploited in the quest for novel antimicrobial molecules. Screening of 2000 bacterial genomes from the human microbiome project have identified biosynthetic gene clusters (BGCs) encoding thiopeptides, abundantly present at multiple anatomical locations [122]. Lactocillin, a thiopeptide encoded by one such cluster (i.e. bgc66 from L. gasseri), has been produced and characterized, showing strong inhibitory activity against common pathogens such as S. aureus and G. vaginalis, but not against commensals [122]. Moreover, a high throughput screening of small molecules recently identified an inhibitor of riboflavin synthesis, called “ribocil”, that targets a regulatory non-coding region (i.e. riboswitch) in the mRNA of a synthase involved in the pathway, ribB [123]. Ribocil binding inhibits translation of ribB transcript in E. coli, inducing cell death by riboflavin starvation. Accordingly, ribocil administration, although only at high concentrations, significantly reduced bacterial burden in a mouse model of systemic infection with E. coli, suggesting that novel molecular targets may have a promising potential for drug development [123].
Recognition of the marked effect of antibiotics on microbiota composition has led to the search for a set of more narrow-spectrum bactericidal compounds. For instance, Thuricin-CD, a bacteriocin produced by Bacillus thuringiensis, has been shown to be as effective as vancomycin or metronidazole against C. difficile without impacting microbiota composition in a fecal-culture system that models human colon [124]. Along the same lines, the bacteriocin Avidocin-CDs, engineered to selectively target the clinically relevant C. difficile strain BI/NAP1/027, has been reported to promote pathogen clearance in mice upon oral administration. Furthermore, Avidocin-CDs, unlike antibiotics, was not found to interfere with the ability of the microbiota to provide colonization resistance [125].
Exploiting the principle of selective targeting, a recent study cleverly conjugated an analogue of rifampicin (i.e. rifalogue) and an S. aureus-specific antibody in order to target intracellular MRSA [126]. Indeed, intracellular MRSA represent an important pathogen reservoir that is shielded from antibiotic action. Mice challenged with infected cells rather than bacterial particles, were shown to present higher bacterial burdens despite vancomycin treatment [126]. In this study, rifalogue was conjugated to an S. aureus-specific antibody via a cleavable bridge so that opsonized MRSA, once phagocytosed, could be effectively killed by the protease-liberated antibiotic. Importantly, the Ab-rifalogue complex was more effective than uncoupled antibiotics in combating infection in an in vivo mouse model. This approach may be in principle, suitable for many intracellular pathogens.
Finally, a strategy to eliminate selected bacterial targets exploiting the CRISPRCAS9 system was recently proposed [127, 128]. CRISPR-CAS9 is a bacterial immune system that can be easily engineered to cleave DNA sequences of interest. Phagemids (i.e. plasmids carrying the information to package phage particles) bearing CRISPR-CAS9 components were generated to cleave antibiotic resistance genes on chromosomes or plasmids of specific pathogens, such as E. coli and S. aureus [127, 128]. Phage-mediated delivery of this genetic material resulted in efficient killing of the bacteria in both cases. Importantly, no resistance developed against the phagemids, and the sensitivity of the system allowed discrimination of single polymorphic nucleotides. Accordingly, within a consortium of 2-3 bacterial isolates of the same strain, the CRISPR-CAS9 phagemids killed only bacteria carrying the target gene, sparing surrounding microbes in in vivo models, i.e. upon infection of moth larvae with E. coli and of mouse skin with S. aureus [127, 128]. Thus, this system has allowed selective killing of pathogens, even antibiotic-resistant ones, without affecting the surrounding microbiota.
Concluding Remarks
Recent evidence demonstrates the fundamental role of the gut microbiota in directing host physiology. In particular, development of a fully functional immune system requires key induction and maintenance signals from the commensal community, many of which are still likely to be discovered (see Outstanding Questions). Antibiotics, to which we are increasingly exposed, disrupt the equilibrium among commensal populations, and lead to a decreased or altered communication between the human intestinal flora and the underlying mucosa. The effects of such perturbations are long-lasting and can have multiple consequences, including increased susceptibility to infections, the potential to develop allergies, a predisposition to develop metabolic syndrome, the decreased efficacy outcomes of pharmacologic therapies (Box 3), as well as the induction and spread of antibiotic resistance (Box 4, Box 5). Unnecessary antibiotic use should therefore be avoided to prevent any potential adverse consequence on the host, in addition to eliminating the spread of antibiotic resistance. Indeed, new strategies are needed to substitute or complement the use of antibiotic treatments.
Outstanding Questions.
What other physiological processes are influenced by the gut microbiota?
Research is showing that virtually no cell type in the body is oblivious to the presence of commensals. Even the physiology of organs such as the brain or the lungs is impacted by the composition of the gut microbiota, and can be altered upon dysbiosis. Unraveling the interaction network between host and intestinal commensals in different organs could provide novel targets for therapeutic intervention in many disease scenarios.
Can we cure disease by reshaping our commensal communities?
Selected taxa within the microbiota are being associated with different diseases such as atherosclerosis and cancer, and the mechanisms are starting to be elucidated. This paves the way for a true revolution in medicine, where precise antimicrobial therapy against specific intestinal microbes could prevent or treat disease.
What other infectious diseases can we treat by exploiting colonization resistance?
C. difficile infection is now being treated with fecal microbiota transplantation. The procedure has been successfully applied also in the context of VRE infection in animal models, both as preventive and therapeutic strategies. However, transfer of fecal material or selected bacterial communities could treat many other intestinal infections in principle. We predict that in the next few years, novel candidate consortia will be proposed to handle a plethora of infectious threats.
What are the factors that promote or restrict the introduction of health-promoting symbiotic bacteria into the community structure of a host’s microbiota?
The metabolic and structural requirements of specific commensal bacterial species are as yet, largely unknown. Understanding what microbial networks (i.e. other bacterial species) or molecular environments promote or inhibit the beneficial function of selected commensal strains would facilitate the design of more effective therapies based on the administration of such probiotics.
Box 5. Clinician’s Corner.
The normal gut microbiota benefits its host in a number of important ways; it helps with breakdown and absorption of nutrients, prevents infections directly by resisting expansion of potential pathogens, and enhances both innate and adaptive immune system function.
Disrupted gut microbiota states are associated with a variety of diseases, including infections (both inside and outside the gut), autoimmune diseases such as inflammatory bowel disease and diabetes, allergic disease, obesity, and atherosclerosis.
Many antibiotics that are currently used adversely affect beneficial members of the gut microbiota, which puts patients at risk for various disease states. These include infections arising due to increasingly prevalent colonization with antibiotic-resistant pathogens such as VRE, MRSA, and multidrug resistant gram negative Enterobacteriaceae. Healthcare providers could treat their patients more optimally by understanding and recognizing the full impact of these drugs on a healthy gut microbiota.
Potential strategies to prevent the untoward effects of antibiotics on the microbiota include: avoidance of unnecessary use through antibiotic stewardship, development and use of more targeted, narrow-spectrum antibiotics, and replacement of depleted microbes with fecal microbiota transplantation (FMT).
In the future, it may be possible to manipulate the gut microbiome for specific purposes such as prevention of atherosclerosis or optimization of responses to cancer treatment. This could be accomplished in the clinical setting through interventions such as specialized diets, administration of specific bacteria consortia, and tailored antibiotic therapy.
Here, we have highlighted strategic approaches that currently lie at various states of clinical development, namely, the exploitation of colonization resistance through the use of FMT or probiotics, the restoration of appropriate immune responses through delivery of bacterial ligands, as well as the design of narrow-spectrum or highly specific antimicrobial compounds. A rational use of highly specific antibacterial molecules would be intended to selectively deplete detrimental bacterial strains within the microbiota, particularly those associated with inflammation, atherogenesis, carcinogenesis, and drug inactivation. Indeed, it will be exciting to follow how the future application of such strategies unfolds in the clinic.
Trends Box.
The gut microbiota contains trillions of bacteria belonging to hundreds, possibly thousands, of species and is critical for optimal maintenance of host physiological processes.
The microbiota protects against infections and other pathologies by directly inhibiting invading microbes or by orchestrating appropriate immune responses; conversely, metabolites produced by some gut commensals can promote a variety of diseases such as atherosclerosis or cancer.
Antibiotics alter the microbiota composition, resulting in an increased risk of disease, secondary infections, allergy, and obesity. In addition, they promote the spread of drug-resistant pathogens, making the search for alternative clinical approaches imperative.
Novel strategies are being developed to substitute or complement antibiotic therapies, attempting to either selectively target pathogens without perturbing the microbiota, and/or, to re-establish commensal communities along with the protective and beneficial effects they confer to the host.
Acknowledgments
We thank members of the Pamer laboratory, particularly Sejal Morjaria and Sohn G. Kim, for critical reading of the manuscript and valuable suggestions. SB is supported by the Swiss National Science Foundation Early Postdoc Mobility Fellowship. YT is supported by the National Institutes of Health (grant 1K23 AI095398-01 to Y.T.), the Lucille Castori Center for Microbes, Inflammation, and Cancer, and the Tow Foundation. EGP has received funding from NIH Grants RO1 AI095706, AI042135, UO1 AI124275 and PO1 CA023766 and from the Leonard Tow Foundation.
Glossary
- Anti-Microbial Peptides (AMP)
small peptides with bactericidal activity, mainly positively charged, produced by microorganisms and host myeloid and epithelial cells
- Bacteriocins
toxins, largely proteins, secreted by bacteria to kill other bacteria
- β-lactams
antibiotics containing a β-lactam ring in their molecular structure; this class includes penicillins, cephalosporins and carbapenems
- B1 cells
subset of B cells activated by innate sensor triggeringthat produce the vast majority of natural IgM against common microbial structures(secreted independently of antigenic exposure)
- Cathelicidins
heterogeneous family of antimicrobial peptides, including LL-37 in humans and CRAMP in mice, produced mainly by myeloid and epithelial cells
- Colonization resistance
protection against pathogens exerted by commensal bacteria
- Conventionalization
transfer of microbiota into a mouse; often performed by co-housing or gavage (intra-gastric inoculation) of fecal material, to equalize microbial populations in the gut of different mice
- Dendritic Cells (DCs)
myeloid cells specialized in sampling the outer environment through endocytosis, and which initiate immune responses upon antigen presentation to other immune cells
- Dysbiosis
imbalance (alteration in composition) within a microbiota
- DSS-induced colitis
mouse model of colitis promoted by administration in drinking water of dextran sulfate sodium (DSS), which damages the intestinal epithelium and promotes inflammation
- Enterocytes
epithelial cells constituting the intestinal epithelium, characterized by the presence of apical microvilli that enhance their adsorbing functions.
- Germ-free (GF) mice
mice born and raised in isolators in the absence of any detectable microorganism inside or outside their body
- Gnotobiotic mice
born aseptically, bred in laboratories and bearing known strains of microorganisms
- Inflammasome
oligomeric protein complex that drives caspase1-mediated maturation of IL-1β and IL-18 upon sensing of danger-associated signals, thus promoting inflammation
- Innate Lymphoid Cells
cells of lymphoid lineage that lack antigen-specific receptors and rapidly respond to infection or inflammation by producing cytokines. The gut is enriched in a subset defined ILC3, that produces mainly IL-22 and IL-17
- Lamina propria
the connective tissue layer underlying the epithelium in the intestine, highly vascularized and rich in immune cells
- Mesenteric Lymph Nodes (MLNs)
lymph nodes draining the majority of the gastro-intestinal tract
- Microbiome
the collection of genomes within a microbiota, i.e. the pool of genes represented in a bacterial community
- Microbiota
a collection of microbes that inhabit a given environment. In this review, the term is used to specifically indicate bacterial communities residing in the intestine
- NLRPs (including NLRP6)
Nod-like receptor (NLR) protein family that has a role in sensing stress- or pathogen-associated signals and participate in the formation of the inflammasome
- NOD (Non-obese diabetic) mice
mouse model for type I diabetes. NOD mice spontaneously develop insulin-dependent diabetes as a consequence of leukocyte-mediated invasion and destruction of pancreatic β-islets
- Paneth cells
epithelial cells of the small intestine located at the bottom of the crypts of Lieberkühn and specialized in the production of anti-microbial molecules
- Regulatory T cells (Treg)
FoxP3-dependent IL10-producing CD4+ T cells, responsible for dampening the immune response and maintaining tolerance
- Short Chain Fatty Acids (SCFAs)
molecules such as acetate, propionate and butyrate, produced by bacteria through fermentation of fibers in the large intestine, influencing many host physiological processes
- Shotgun sequencing
sequencing approach based on the generation of a library of short sequences from a complex sample (often total DNA or RNA extracted from bacterial communities). the obtained reads can be computationally assembled to reconstruct the entire sequences of the original nucleic acids
- T helper 1 (Th1)
T-bet+ IFNγ-producing CD4+ T cells, involved in the clearance of intracellular pathogens
- T helper 17 (Th17)
RORγt+ IL17-producing CD4+ T cells, specialized in responses to extracellular bacteria and fungi
- T helper 2 (Th2)
GATA3+ IL4/5/13-producing CD4+ T cells, involved in the response against helminths and key players in allergic reactions
- Toll-Like Receptors (TLRs)
dimeric receptors mainly expressed in cells of the innate immune system, recognizing conserved microbial structures and activating immune responses
- Unadjuvanted vaccines
vaccines devoid of adjuvants, i.e. molecules that enhance vaccine immunogenicity by promoting inflammation or slower release of the antigen
- Vertical acquisition
acquisition of maternal microbes in newborns through processes such as delivery (birth canal microbes) and breastfeeding (milk microbes)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annual review of microbiology. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 3.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nature immunology. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nature reviews. Immunology. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyal MS, et al. Feeding the brain and nurturing the mind: Linking nutrition and the gut microbiota to brain development. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:14105–14112. doi: 10.1073/pnas.1511465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsland BJ, Salami O. Microbiome influences on allergy in mice and humans. Current opinion in immunology. 2015;36:94–100. doi: 10.1016/j.coi.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 7.Caballero S, Pamer EG. Microbiota-Mediated Inflammation and Antimicrobial Defense in the Intestine. Annual review of immunology. 2015 doi: 10.1146/annurev-immunol-032713-120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO reports. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 10.Kelly CJ, et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell host & microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinhardt C, et al. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483:627–631. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luna RA, Foster JA. Gut brain axis: diet microbiota interactions and implications for modulation of anxiety and depression. Curr Opin Biotechnol. 2015;32:35–41. doi: 10.1016/j.copbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Mayer EA, et al. Gut/brain axis and the microbiota. The Journal of clinical investigation. 2015;125:926–938. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mu C, et al. Gut Microbiota: The Brain Peacekeeper. Frontiers in microbiology. 2016;7:345. doi: 10.3389/fmicb.2016.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins SM, et al. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 16.Rakoff-Nahoum S, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Cash HL, et al. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinnebrew MA, et al. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. The Journal of infectious diseases. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinnebrew MA, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature medicine. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshmukh HS, et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nature medicine. 2014;20:524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill DA, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nature medicine. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganal SC, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 28.Mazmanian SK, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell host & microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atarashi K, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–812. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 33.Sano T, et al. An IL-23R/IL-22 Circuit Regulates Epithelial Serum Amyloid A to Promote Local Effector Th17 Responses. Cell. 2015;163:381–393. doi: 10.1016/j.cell.2015.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atarashi K, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mortha A, et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 38.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 40.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun J, et al. Pancreatic beta-Cells Limit Autoimmune Diabetes via an Immunoregulatory Antimicrobial Peptide Expressed under the Influence of the Gut Microbiota. Immunity. 2015;43:304–317. doi: 10.1016/j.immuni.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Markle JG, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 43.Fox HS. Androgen treatment prevents diabetes in nonobese diabetic mice. The Journal of experimental medicine. 1992;175:1409–1412. doi: 10.1084/jem.175.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roden AC, et al. Augmentation of T Cell Levels and Responses Induced by Androgen Deprivation. The Journal of Immunology. 2004;173:6098–6108. doi: 10.4049/jimmunol.173.10.6098. [DOI] [PubMed] [Google Scholar]
- 45.Kissick HT, et al. Androgens alter T-cell immunity by inhibiting T-helper 1 differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9887–9892. doi: 10.1073/pnas.1402468111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature medicine. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 47.Proietti M, et al. ATP-gated ionotropic P2X7 receptor controls follicular T helper cell numbers in Peyer's patches to promote host-microbiota mutualism. Immunity. 2014;41:789–801. doi: 10.1016/j.immuni.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Oh JZ, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wlodarska M, et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infection and immunity. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wlodarska M, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan D, et al. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nature medicine. 2015;21:808–814. doi: 10.1038/nm.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 53.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. The New England journal of medicine. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 54.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsiao A, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kommineni S, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suez J, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature. 2014;514:181–186. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 59.Zeevi D, et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 61.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 62.Forslund K, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lewis JD, et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn's Disease. Cell host & microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 65.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshimoto S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 67.Belcheva A, et al. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 68.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature medicine. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. The New England journal of medicine. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z, et al. Non-lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis. Cell. 2015;163:1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Haiser HJ, et al. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winter SE, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seo SU, et al. Distinct Commensals Induce Interleukin-1beta via NLRP3 Inflammasome in Inflammatory Monocytes to Promote Intestinal Inflammation in Response to Injury. Immunity. 2015;42:744–755. doi: 10.1016/j.immuni.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ayres JS, et al. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nature medicine. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pacheco AR, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fonseca DM, et al. Microbiota-Dependent Sequelae of Acute Infection Compromise Tissue-Specific Immunity. Cell. 2015;163:354–366. doi: 10.1016/j.cell.2015.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horai R, et al. Microbiota-Dependent Activation of an Autoreactive T Cell Receptor Provokes Autoimmunity in an Immunologically Privileged Site. Immunity. 2015;43:343–353. doi: 10.1016/j.immuni.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jernberg C, et al. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. The ISME journal. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 82.Buffie CG, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infection and immunity. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. The Journal of clinical investigation. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lewis BB, et al. Loss of Microbiota-Mediated Colonization Resistance to Clostridium difficile Infection With Oral Vancomycin Compared With Metronidazole. The Journal of infectious diseases. 2015;212:1656–1665. doi: 10.1093/infdis/jiv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dominguez-Bello MG, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nature medicine. 2016;22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kozyrskyj AL, et al. Increased risk of childhood asthma from antibiotic use in early life. Chest. 2007;131:1753–1759. doi: 10.1378/chest.06-3008. [DOI] [PubMed] [Google Scholar]
- 88.Risnes KR, et al. Antibiotic exposure by 6 months and asthma and allergy at 6 years: Findings in a cohort of 1,401 US children. American journal of epidemiology. 2011;173:310–318. doi: 10.1093/aje/kwq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korpela K, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nature communications. 2016;7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Russell SL, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO reports. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cox LM, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Levy SB, et al. High frequency of antimicrobial resistance in human fecal flora. Antimicrobial agents and chemotherapy. 1988;32:1801–1806. doi: 10.1128/aac.32.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sommer MO, et al. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science. 2009;325:1128–1131. doi: 10.1126/science.1176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Santiago-Rodriguez TM, et al. Gut Microbiome of an 11th Century A.D. Pre-Columbian Andean Mummy. PloS one. 2015;10:e0138135. doi: 10.1371/journal.pone.0138135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.D'Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 96.Bhullar K, et al. Antibiotic resistance is prevalent in an isolated cave microbiome. PloS one. 2012;7:e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Modi SR, et al. Antibiotics and the gut microbiota. The Journal of clinical investigation. 2014;124:4212–4218. doi: 10.1172/JCI72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oldenburg M, et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science. 2012;337:1111–1115. doi: 10.1126/science.1220363. [DOI] [PubMed] [Google Scholar]
- 99.Cullen TW, et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koch G, et al. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell. 2014;158:1060–1071. doi: 10.1016/j.cell.2014.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Looft T, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu Y, et al. The abundance of antibiotic resistance genes in human guts has correlation to the consumption of antibiotics in animal. Gut microbes. 2014;5:245–249. doi: 10.4161/gmic.27916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Field W, Hershberg R. Alarmingly High Segregation Frequencies of Quinolone Resistance Alleles within Human and Animal Microbiomes Are Not Explained by Direct Clinical Antibiotic Exposure. Genome Biol Evol. 2015;7:1743–1757. doi: 10.1093/gbe/evv102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moore AM, et al. Gut resistome development in healthy twin pairs in the first year of life. Microbiome. 2015;3:27. doi: 10.1186/s40168-015-0090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu N, et al. DNA microarray analysis reveals that antibiotic resistance-gene diversity in human gut microbiota is age related. Scientific reports. 2014;4:4302. doi: 10.1038/srep04302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nature genetics. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee HH, et al. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kelsic ED, et al. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature. 2015;521:516–519. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Costello EK, et al. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hehemann JH, et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 112.Schloissnig S, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Beaber JW, et al. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 114.Modi SR, et al. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Slager J, et al. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell. 2014;157:395–406. doi: 10.1016/j.cell.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 116.Laxminarayan R. Antibiotic effectiveness: balancing conservation against innovation. Science. 2014;345:1299–1301. doi: 10.1126/science.1254163. [DOI] [PubMed] [Google Scholar]
- 117.Kelly CR, et al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol. 2014;109:1065–1071. doi: 10.1038/ajg.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lawley TD, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS pathogens. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reeves AE, et al. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infection and immunity. 2012;80:3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ubeda C, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infection and immunity. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Abt MC, et al. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Science translational medicine. 2016;8:327ra325. doi: 10.1126/scitranslmed.aad6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Donia MS, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Howe JA, et al. Selective small-molecule inhibition of an RNA structural element. Nature. 2015;526:672–677. doi: 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- 124.Rea MC, et al. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4639–4644. doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gebhart D, et al. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio. 2015;6 doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lehar SM, et al. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature. 2015 doi: 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- 127.Bikard D, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nature biotechnology. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Citorik RJ, et al. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nature biotechnology. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS microbiology reviews. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ley RE, et al. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nguyen TL, et al. How informative is the mouse for human gut microbiota research? Disease models & mechanisms. 2015;8:1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xiao L, et al. A catalog of the mouse gut metagenome. Nature biotechnology. 2015;33:1103–1108. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 133.Seedorf H, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Blanton LV, et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351 doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kau AL, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Science translational medicine. 2015;7:276ra224. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schluter J, Foster KR. The evolution of mutualism in gut microbiota via host epithelial selection. PLoS biology. 2012;10:e1001424. doi: 10.1371/journal.pbio.1001424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pickard JM, et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature. 2014;514:638–641. doi: 10.1038/nature13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hand TW, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Johansson ME, et al. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell host & microbe. 2015;18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Diehl GE, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–120. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hepworth MR, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hepworth MR, et al. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science. 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fanning S, et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vetizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dubin K, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nature communications. 2016;7:10391. doi: 10.1038/ncomms10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chang Q, et al. Antibiotics in agriculture and the risk to human health: how worried should we be? Evolutionary applications. 2015;8:240–247. doi: 10.1111/eva.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Meek RW, et al. Nonmedical Uses of Antibiotics: Time to Restrict Their Use? PLoS biology. 2015;13:e1002266. doi: 10.1371/journal.pbio.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kennedy D. Time to deal with antibiotics. Science. 2013;342:777. doi: 10.1126/science.1248056. [DOI] [PubMed] [Google Scholar]