Abstract
G-proteins and their cognate G-protein coupled receptors (GPCRs) function as critical signal transduction molecules that regulate cell survival, proliferation, motility and differentiation. The aberrant expression and/or function of these molecules have been linked to the growth, progression and metastasis of various cancers. As such, the analysis of mutations in the genes encoding GPCRs, G-proteins and their downstream targets provides important clues regarding how these signaling cascades contribute to malignancy. Recent genome-wide sequencing efforts have unveiled the presence of frequent mutations in GNA13, the gene encoding the G-protein Gα13, in Burkitt’s lymphoma and Diffuse Large B cell lymphoma (DLBCL). We found that mutations in the downstream target of Gα13, RhoA, are also present in Burkitt’s lymphoma and DLBCL. By multiple complementary approaches, we now show that that these cancer-specific GNA13 and RHOA mutations are inhibitory in nature, and that the expression of wild type Gα13 in B cell lymphoma cells with mutant GNA13 has limited impact in vitro but results in a remarkable growth inhibition in vivo. Thus, although Gα13 and RhoA activity has previously been linked to cellular transformation and metastatic potential of epithelial cancers, our findings support a tumor suppressive role for Gα13 and RhoA in Burkitt’s lymphoma and DLBCL.
Keywords: G-protein, GPCR, RhoA, tumor suppressor, B cell lymphoma
Introduction
G-protein coupled receptors (GPCRs) represent the largest class of cell surface receptors and when activated by extracellular ligands, they transmit signals involved in cell growth, proliferation, survival, differentiation and motility (1). GPCRs initiate signal transduction cascades by coupling to and activating heterotrimeric G-proteins. Depending on the coupling specificity, a given GPCR may couple to one or more different Gα proteins, which are grouped into four main families: Gα12 (including Gα13), Gαs, Gαi, and Gαq (including Gα11). Each family of G-proteins activates distinct sets of second messenger and kinase signaling cascades (1).
Heterotrimeric G-proteins are major cellular signaling hubs, and they are activated upon binding to GPCRs or non-receptor guanine nucleotide exchange factors (GEFs) (2). Due to their important roles in regulating cell survival, proliferation and movement, it is not surprising that tumors often harbor mutations in or exhibit aberrant expression of G-proteins and their GEFs (3). For example, activating mutations in GNA11 and GNAQ, encoding Gα11 and Gαq respectively, were recently discovered to be the key oncogenic drivers in uveal melanoma (4–6). Gαs is the most frequently mutated G-protein in human cancers and activating mutations in the gene, GNAS, have been found in a variety of neoplasms including pituitary, thyroid, pancreatic, biliary tract, colon and small intestine and a variety of other tumors (3). Furthermore, constitutively active mutants of genes encoding Gαi, Gαo, Gαq Gα12, and Gα13 were found to induce cellular transformation in experimental systems (reviewed in (1, 7)).
Despite the transforming capacity of constitutive Gα12 and Gα13 activity in experimental systems and numerous implications of this G-protein family and downstream targets in cancer metastasis (8–13), activating mutations in the GNA13 and GNA12 genes in patient tumor samples have not been described. However, recent large-scale sequencing efforts have revealed the presence of GNA13 mutations in Burkitt’s lymphoma and Diffuse Large B cell Lymphoma (DLBCL) (14, 15). Interestingly, recent studies in mouse models demonstrated that conditional B cell deficiency in Gα13 or the Gα13 –coupled sphingosine 1 phosphate receptor 2 (S1P2) result in DLBCL-like phenotypes (16, 17). Based on the analysis of deposited sequencing data from tumors in the Catalog of Somatic Mutations in Cancer (COSMIC), mutations in GNA13 in human Burkitt’s lymphoma and DLBCL are highly statistically significant over background cancer mutation rate, with p-value and q-value scores of 0 (3), suggesting these mutations are likely not random. However, unlike the activating GTPase domain mutations found in other G-proteins in cancers, including Gαq and Gαs, the mutations in GNA13 are distributed throughout the gene. Furthermore, we identified additional mutations in the gene of the major downstream effector of Gα13 signaling, RhoA.
In this study, we characterize the mutations identified in GNA13 and RHOA in Burkitt’s lymphoma and DLBCL tumor samples to determine how these mutations affect protein function and signaling capacity. We also evaluated the effects of mutations and wild type (WT) Gα13 expression on tumor growth and progression in xenograft models. Overall, our results support a tumor suppressive role for the Gα13/RhoA axis in Burkitt’s lymphoma and DLBCL. Our data also extend recent findings supporting the presence of disruptive RHOA mutations in peripheral T cell lymphomas, suggesting that disruption of RhoA function may have a broad impact in multiple haematological malignancies (18–22).
Results
Mutations in GNA13 and RHOA are frequent in Burkitt’s Lymphoma and DLBCL tumors
Data from genome-wide sequencing analyses collected through the Catalog of Somatic Mutations in Cancer (COSMIC v72) database reveal the presence of GNA13 mutations in nearly 2% of all haematopoietic and lymphoid malignancies (Figure 1A). Previous statistical analyses of these mutations indicated p-value and q-value (for false discovery rate) scores approaching 0, suggesting they are unlikely to be random, but rather could have important driver mutation functions (3). Of the haematopoietic and lymphoid malignancies evaluated in COSMIC, most of the GNA13 mutations are present in B cell lymphomas, primarily Diffuse Large B cell lymphoma (DLBCL) and Burkitt’s Lymphoma, for which mutations are harbored in approximately 10% of patient tumor samples (Figure 1A). Mutations in GNA13 found in Burkitt’s lymphoma and DLBCL appeared likely to result in loss of function because nearly 22% (5/23) of the DLBCL mutations (17% of overall in both lymphomas) were nonsense, resulting in a premature STOP codon and all other mutations were non-synonymous (Figure 1A). When mapped onto the crystal structure of GNA13, the mutations clustered into multiple regions including the binding interface for the Gβ/γ subunits, the helical domain, conformational switches 1 and 2 (important for nucleotide binding as well as interaction with regulators and effectors), conformational switch 3, the GPCR interface, and directly in the nucleotide pocket (Figures 1A, Supplemental Table 1). An interesting in-frame deletion was observed in the loop connecting the GTPase and the helical domain that can dramatically impair the relative mobility of the two domains. Mutations are therefore not localized into one region and so they could exert a broad influence on the protein function or stability.
Figure 1.
Sequence and structure localization of mutations in GNA13 and RHOA that are observed in Burkitt’s Lymphoma and DLBCL tumors. A) Table of the number and percentage of GNA13 mutations in haematopoietic malignancies overall and in Burkitt’s Lymphoma and DLBCL based on data from COSMIC v72 (top). Linear diagram of mutations along the GNA13 gene and the functional and structural domains of the protein Gα13 (bottom). B) Table of the number and percentage of RHOA mutations in haematopoietic malignancies overall and in Burkitt’s Lymphoma and DLBCL based on data from COSMIC v72 (top). Linear diagram of mutations along the RHOA gene and the functional and structural domains of the protein and the ribbon diagram of RhoA crystal structure (bottom). Structural and functional domains of both proteins are color-coded and indicated on separate lines above the linear diagram and mapped onto the 3D structures. The mutated residue positions are shown as stars on over the corresponding residues of the linear diagrams and as spheres in the 3D structure representation. The nucleotide is shown as yellow skin.
RhoA is the main downstream effector of Gα13 signaling (23–25), therefore we searched the COSMIC database to determine if there were mutations in RHOA in Burkitt’s Lymphoma and DLBCL tumors as well. Although less frequent than the GNA13 mutations, non-redundant RHOA mutations were observed in nearly 6% of Burkitt’s Lymphoma and almost 1% of DLBCL tumors (Figure 1B), as these RHOA mutations were present in separate tumor samples from those with GNA13 mutations. Taken together, the Gα13/RhoA axis is disrupted in more than 10% of these B cell lymphomas. Interestingly, there was a recurrent R5Q mutation, observed in 7/16 RHOA mutations identified in these tumors. Structural analysis of RhoA suggests that R5 is important for GEF binding. Several of the other mutations in RHOA also localized to GEF binding residues, indicating that these mutations could influence RhoA activation due to changes in RhoA-GEF interactions (Figure 1B, Supplemental Table 2).
Characterization of GNA13 mutations indicates loss of function
In order to determine the consequences of the GNA13 mutations on Gα13 protein function, we prepared and characterized 9 of the mutations from the COSMIC data repository, representing a variety of the structural and functional domains. Plasmids encoding empty vector, Gα13 WT, Gα13 mutants based on Burkitt’s lymphoma and DLBCL tumor samples and an established constitutively active Q226L mutant were transfected into HEK293 cells and Gα13 protein expression was assessed by western blot (Figure 2A). Although most of the Gα13 mutants were expressed at similar levels to WT, there were a few including L197Q and F245S that had reduced protein expression. Because the Gα13 antibody recognizes an epitope in the C-terminus of Gα13, disrupted antibody recognition is unlikely to fully explain the relatively low protein expression observed in the L197Q and F245S mutants. Rather, it could be that these mutants affect protein stability, thus reducing expression.
Figure 2.
GNA13 mutations in Burkitt’s lymphoma and DLBCL tumor samples result in loss of downstream transcriptional activity based on SRF-RE luciferase assay. A) Representative western blot for Gα13 expression of cells transfected with Gα13 WT, mutants based on COSMIC data, constitutively active Q226L mutant, or vector control. Antibody detection of α-tubulin was used as a loading control (top). Densitometry quantification of Gα13 protein expression of the mutants relative to WT (bottom) as an average of three independent western blots. Error bars indicate standard deviation. B) SRF-RE luciferase assay of spontaneous activity of cells transfected with Gα13 WT, mutants, constitutively active Q226L mutant, or vector control. Error bars indicate standard deviation. C) SRF-RE luciferase activity of cells expressing SyR-Gi transfected with WT or mutant Gα13-i5 chimeras, in the presence (+) and absence (−) of CNO stimulation. Error bars indicate standard deviation.
To evaluate the functional consequences of these mutants, we tested their activity in a serum response factor response element (SRF-RE) luciferase assay, which is used to selectively monitor gene transcription downstream of Gα13-RhoA signaling. The SRF-RE luciferase assay demonstrated that all 9 Gα13 mutants had significantly reduced basal transcriptional activity relative to WT and constitutively active (Q226L) Gα13 (Figure 2B). Due to promiscuity of most Gα13-coupled GPCRs, which often also couple to other G proteins and/or have ligands that target other GPCRs, a synthetic biology approach was employed to determine the effects of the Gα13 mutants on ligand-activated Gα13 signaling events. The synthetic GPCR (SyR), also known as DREADD (26), is activated by an otherwise biologically inert ligand, clozapine N-oxide (CNO). Because no Gα13-SyR is currently available, an SyR that couples to Gαi was used in combination with a chimeric G-protein that has Gαi-coupling specificity (based on the last 5 amino acids at the C-terminus) but retains Gα13 signaling response. Additionally, in order to ensure that the responses observed were due to Gα13 activity and not influenced by potential Gαi signaling, a C-I mutation was made in the last 5 amino acids of the Gα13-i5 chimeras to make them pertussis toxin (PTX)-insensitive and the assay was performed in the presence of PTX. The SRF-RE assay demonstrated that some of the Gα13 mutants were completely dead whereas others still retained some ligand-activated response (Figure 2C). Nevertheless, overall SRF-RE activity was lower in all the mutants compared with WT (and basal activity was again lower in all the mutants). A recently developed TGF-α shedding assay was used as another measurement of Gα13 activity (27). Gα13 and Gαq activity are known to induce the shedding of tethered TGF-α, and so we could monitor the release of AP-TGF-α into media as a measurement of Gα13 signaling (Figure 3A). Similar to the SRF-RE results, the spontaneous (or basal) activities of all the Gα13 mutants were significantly lower than WT or the Q226L constitutively active mutant (Figure 3B, Supplemental Table 3), whereas a few of the mutants still retained some ligand-induced signaling response as determined using the synthetic receptor system, SyR-Gi with the Gα13-i5 chimeras, (Figure 3C, Supplemental Table 3) and using another receptor that couples to Gα13, the dopamine D2 receptor (Supplemental Figure 1, Supplemental Table 3).
Figure 3.
GNA13 mutations in Burkitt’s lymphoma and DLBCL tumor samples result in loss of function based on a TGFα shedding assay. A) Schematic representation of the AP-TGFα shedding assay used to monitor activity of the Gα13 mutants. B) AP-TGFα shedding assay for spontaneous/basal activity of Gα13 WT and mutants. Error indicates standard deviation. C) AP-TGFα shedding assay dose response curves of CNO stimulation of cells expressing SyR-Gi transfected with WT or mutant Gα13-i5 chimeras.
GNA13 and RHOA mutations impair RhoA activity
RhoA is the main downstream effector of Gα13 signaling, thus we sought to analyze the effects of the GNA13 mutations on RhoA activity and to determine the effect of the recurrent RhoA R5Q mutation. We monitored active GTP-bound RhoA using a pull-down assay for three of the Gα13 mutants and controls. In line with the previous experiments, the Gα13 mutants had considerably reduced levels of active RhoA compared to WT Gα13, constitutively active Gα13 Q226L and the thrombin-stimulated positive control (Figure 4A). Next, we evaluated the activity of the R5Q RhoA mutant that had been identified in several DLBCL and Burkitt’s lymphoma tumors. The R5Q mutant exhibited reduced activity compared with WT RhoA and the constitutively active Q63L RhoA mutant in both an active RhoA pull down assay (Figure 4B) and an SRF-RE luciferase assay (Figure 4C). However, the RhoA R5Q mutant appeared to retain a little activity compared with the dominant negative T19N RhoA mutant (Figure 4C). Formation of actin stress fibers is often used as another indication of RhoA activity, so we performed a stress fiber formation assay using LifeActin-GFP in MDCK cells. Consistent with the SRF and RhoA pull down results, we observed significant actin stress fiber formation in WT and Q63L constitutively active RhoA-transfected MDCK cells, but not with the R5Q mutant or the dominant negative T19N mutant (Figure 4D). Overall, these data indicate that the GNA13 and RHOA mutations observed in DLBCL and Burkitt’s lymphoma tumors result in loss of function of RhoA activity and downstream signaling events.
Figure 4.
Burkitt’s lymphoma and DLBCL GNA13 and RHOA mutations impair RhoA activity. A) GST-Rhotekin-RBD pull down assay to monitor RhoA activity in HEK293 cells transfected with vector control (V), WT, constitutively active Q226L or mutant Gα13 plasmids. Thrombin (Th) stimulation was used as a positive control. Input lysates were probed for Gα13 and for total RhoA and α-tubulin as loading controls. B) GST-Rhotekin-RBD pull down assay to monitor RhoA activity in HEK293 cells transfected with vector control (V) or RhoA WT, constitutively active Q63L, dominant negative T19N or R5Q mutant plasmids. C) SRF-RE luciferase assay detecting spontaneous activity of cells transfected with vector control, RhoA WT, constitutively active Q63L, dominant negative T19N and the R5Q mutant plasmids. Error bars indicate standard deviation. D) Immunofluorescence images of actin stress fibers in MDCK cells co-transfected with LifeActin-GFP and vector control or RhoA WT, constitutively active Q63L, dominant negative T19N or R5Q mutant plasmids.
Restoration of Gα13 WT suppresses tumor growth
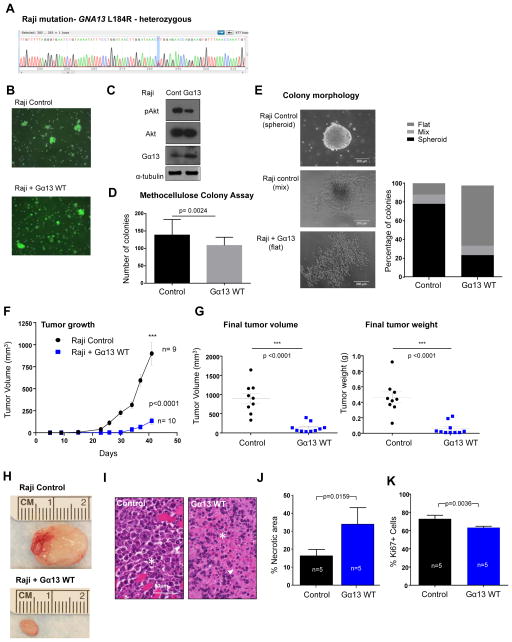
The COSMIC sequencing data indicated the presence of a GNA13 mutation at L184R in the frequently used Raji Burkitt’s lymphoma cell line. Therefore, in order to determine if mutation and thus loss of Gα13 activity provides the cells with a growth or survival advantage, we expressed Gα13 WT in the cells. First we verified the COSMIC sequencing data in the Raji cell line and confirmed the presence of a heterozygous L184R mutation (Figure 5A). Then, we transduced Raji cells using MSCV puro IRES GFP retrovirus with Gα13 WT or vector control. After cell sorting, we were able to achieve a highly GFP-enriched population of cells and verified the expression of Gα13 (Figure 5B). B cell lines are notorious for their difficulties in terms of feasibility of genetic manipulations, which prevented us from deploying similar or complementary strategies to express Gα13 WT in additional relevant cellular systems. Regarding Gα13 function, it was previously reported that Gα13 activity has suppressive effects on Akt phosphorylation (17). Thus, we measured basal Akt phosphorylation at S473 and found that Gα13 WT expression in the Raji cells reduced phosphorylated Akt (Figure 5C). Using in vitro viability and proliferation assays, we did not observe any differences between the control and Gα13 WT-expressing Raji cells (data not shown). In a methocellulose clonogenic assay, WT Gα13–expressing cells showed only a slight reduction in the number of colonies; however, the difference in cell morphology was striking (Figures 5D–E). While the majority of the control Raji cells formed spheroid colonies in the methocellulose, the Raji cells expressing Gα13 WT tended to exhibit a more flattened morphology (Figure 5E). Next we tested the Raji control cells and Raji Gα13 WT-transduced cells in a tumor xenograft model using NSG (NOD SCID gamma) mice. In line with the role of a tumor suppressor, the restoration of Gα13 WT to the Raji cells resulted in significantly delayed and impaired tumor growth and smaller tumor masses and volumes (Figures 5F–H). Histological evaluation of the cells indicated that the Raji cells with Gα13 WT expressed were considerably smaller and more necrotic than the control Raji cells (Figures 5I–J). Additionally, the Raji cells expressing Gα13 WT were significantly less proliferative, based on Ki67 staining and analysis of tumor sections (Figure 5K). These data indicate that restoration and/or overexpression of Gα13 WT influences the survival and/or proliferation of the Raji cells within the tumor microenvironment, which may be the mechanism by which it suppresses tumor growth in vivo.
Figure 5.
Gα13 WT expression has tumor suppressive effects on Raji Burkitt’s Lymphoma xenograft tumor growth. A) Sequence chromatogram of Raji cell DNA validating the presence of a heterozygous T→G mutation resulting in the L184R mutation. B) Immunofluorescence imaging for GFP expression of Raji cells transduced with MSCV puro IRES GFP (PIG) vector control or expressing Gα13 WT. C) Western blot of Raji cells transduced with MSCV puro IRES GFP (PIG) vector control or expressing Gα13 WT. Lysates were probed for Gα13 and pAkt S473 expression and for total Akt and α-tubulin as loading controls. D) Number of colonies formed in a methocellulose clonogenic assay of Raji control and Gα13 WT-expressing cells. Data represents the averages of three independent experiments performed in duplicate or triplicate. Error indicates standard deviation and a two-tailed t-test was used to determine statistical significance. E) Percentage of colonies exhibiting spheroid, flat or mixed morphologies (representative images on right) in Raji control and Gα13 WT-expressing cells. F) Tumor volume measurements (error bars represent standard error of the mean (SEM)) of Raji control (n=9) and Gα13 WT-expressing (G13 WT, n=10) tumor xenografts over time, starting with injections of the tumor cells at Day 0. G) Graphs of the mean (error bars represent SEM) of final tumor masses (left) and final tumor volumes (right) of Raji control and Gα13 WT (G13 WT) tumors. H) Representative image of Raji control and Gα13 WT tumors. A two-tailed t-test was used to determine statistical significance. I) H&E staining of tissue sections from Raji control and Gα13 WT tumors. H&E shows viable round atypical cells infiltrating skeletal muscle, aberrant mitotic figures (arrowhead) an apoptotic cells (star) in the Raji control image. H&E shows the Raji Gα13 WT tumor is highly necrotic with numerous ghost cells, eosinophilic cell bodies lacking nuclear structure (star), and chromatin dust (arrowhead) observed. J) Quantification of necrotic area in Raji control (n=5) and Raji Gα13 WT (n=5) tumors analyzed. Statistics were based on a Mann Whitney test. K) Quantification of Ki67 immunohistochemistry staining in Raji control (n=5) and Raji Gα13 WT (n=5) tumors. Statistics were based on a t-test.
Discussion
Burkitt’s lymphoma and DLBCL are aggressive and prevalent B cell malignancies (28). Although some genetic alterations have been identified and associated with malignancy of these B cell lymphomas, much is still unknown about the majority of the genetic mutations and how these alterations may affect the development and progression of disease (14, 29). Previous genome wide sequencing studies unveiled frequent mutations in GNA13, the gene encoding the G-protein, Gα13 (14, 15), and these mutations were found to be highly statistically significant over background cancer mutation rates (3). Although activity of Gα13 has previously been linked to cellular transformation in fibroblasts as well as tumor cell invasion in several cancers including breast cancer and prostate cancer (7, 30–32), our characterization of the GNA13 mutations present in human Burkitt’s lymphoma and DLBCL tumors indicates that these mutations rather result in loss of function and/or reduced protein stability. Furthermore, we demonstrated that expression of wild-type Gα13 to Burkitt’s lymphoma cells with mutated GNA13 results in reduced tumor growth in xenograft models and altered cell morphology and growth in a methocellulose colony assay.
Recent elegant genetic mouse models support our loss-of-function characterization of GNA13 mutations present in DLBCL and Burkitt’s lymphoma, as mice deficient in lymphoid lineage for Gα13 or the Gα13-coupled receptor, sphingosine 1 phosphate receptor 2 (S1P2), exhibited an increase in total T cell and B cell numbers and resulted in an outgrowth of cells classified as diffuse large B cell lymphoma (16, 17, 33). Specifically, these studies identified a migration defect and an increase in phosphorylated Akt in germinal center (GC) B cells caused by S1P2 or Gα12/Gα13 deficiency. Thus, signaling through the Gα12/Gα13 axis via GPCRs, including S1P2 and the recently characterized P2Ry8, is proposed to help confine B cells to the GC and limit B cell expansion (17). Loss of this axis would therefore allow cells to escape the GC and populate lymph nodes as is characteristic of diffuse large B cell lymphomas. However, in addition to the migration defects observed in these previous studies, our data indicate a direct role for Gα13 in growth suppression of B cells in vivo and suggest Gα13 may influence their survival, and/or differentiation state.
Previous sequencing of non-Hodgkin’s lymphomas reported frequent GNA13 mutations that were enriched in germinal center B cell (GCB) type DLBCL (34). Of interest, DLBCL have been recently classified in multiple distinct groups based on their histological and biological characteristics and morphological features (35). Thus, based on our findings it is possible that DLBCL harboring RHOA or GNA13 mutations may exhibit particular characteristics. Indeed, further characterization of RHOA and GNA13 mutations in relation to subgroup, stage and patient survival is warranted once more information becomes available.
To evaluate the mechanisms by which the GNA13 mutations may alter signaling that could affect the growth and morphology of the B cells, we first investigated possible effects on the major downstream effector, RhoA. The constitutive/basal activity of RhoA was found to be significantly reduced in all 9 of the Gα13 mutants tested compared with WT Gα13. Although a few of the mutants still had some ligand-induced signaling responses, the overall levels of activity were reduced compared with WT. Furthermore, these ligand-induced signaling assays were performed with receptor overexpression systems, so it is possible that these ligand-responses may not be as pronounced in more physiological systems.
Because the Gα13 mutants critically impacted RhoA activity, we investigated whether independent mutations in RHOA were present in DLBCL and Burkitt’s lymphoma tumors by searching the COSMIC database. Indeed, mutations in RHOA were identified in several patient tumors that were independent from those with GNA13 mutations. Intriguingly, a recurrent glutamine substitution at R5 of RhoA was observed in several patient tumors, aligned with the recent description of RHOA mutations, including a recurrent R5Q mutation, in 8.5% of pediatric Burkitt’s Lymphoma cases studied (36), Correlating with the loss of function data observed with the Gα13 mutations, our data indicate that the activity of R5Q is significantly impaired compared with WT RhoA in active RhoA pull down, SRF-RE luciferase and actin stress fiber formation assays. Based on structural modeling and known interactions, R5 appears to be directly involved in the interaction with RhoGEFs (particularly RhoGEFs 11, 12, and DBS). Although no mutations were found in the RhoGEFs most frequently activated by Gα13, p115 (ARHGEF1), LARG (ARHGEF12), or PDZ RhoGEF (ARHGEF11), this is not surprising due to the redundancy of the RhoGEFs and the opportunities for compensation (37, 38). Interestingly, inactivating or dominant negative mutations in RhoA were recently identified in 50–70% of peripheral T cell lymphomas (18–22), suggesting that the Gα13/RhoA pathway may have tumor suppressive functions in other haematological malignancies as well.
Although Gα13 and RhoA have been associated with cellular transformation and characterized as growth promoting in fibroblasts and some epithelial cancer models, this axis appears to have the opposite effect in B cell lymphomas. This sort of duality in function based on cellular context is not unheard of and has been observed with other key pathways, including JNK and transforming growth factor beta signaling pathways (39, 40). Although the precise mechanism by which Gα13/RhoA axis may inhibit cell growth in B cells requires further exploration, the reduction in phosphorylated Akt (Ser473) associated with Gα13 activity could in part affect the growth and survival of B cells (16, 17). Additionally, we observed distinct morphological changes in the cells with mutant Gα13 compared with WT-Gα13 both in the methocelluolose clonogenic assay and in the histology analysis from tumor xenografts, suggesting other factors may play a role. Nevertheless, the reasons why Gα13 and RhoA may exhibit pro-tumorigenic effects in some cancers while having tumor suppressive functions in other cancers requires further investigation. In this regard, once better tools become available for manipulation of B cells and B cell lines, it will be possible to determine how disruption of GNA13 or RHOA in normal human B cells and other Burkitt’s lymphoma and DLBCL lines influences their growth and morphology, thereby helping to elucidate fully the repertoire of tumor suppressive signals induced by Gα13/RhoA in B cells.
Characterization of tumor mutations provides valuable information for how to precisely identify causes of disease and best treatment options. Our data suggest that the mutations identified in the Gα13 signaling pathway in Burkitt’s lymphoma and DLBCL tumors result in loss of function, and that restoration and/or activation of this signaling pathway may help reduce tumor growth and progression. Overall, the characterization of these mutations may enable more precise treatment in the >10% of patients with mutations in GNA13 and RHOA in DLBCL and Burkitt’s lymphoma.
Materials and Methods
Identification of interaction interfaces for mapping and structural characterization of mutations
Structures of RhoA protein and its complexes were retrieved from the PDB. Per-residue contact strengths of RhoA with each of the available co-crystallized binding partners were measured as described in (41, 42) for residue side-chains only and averaged over multiple structures of homologous complexes. Based on this analysis, residues were classified as a part of GAP interface, GEF interface, or nucleotide binding pocket. Conformational switches I and II were found to be extensively involved in binding of both GAPs and GEFs. The analysis for Gα13 was conducted similarly; however, because there are no structures of trimeric α/β/γ complexes for Gα13 and because the N-terminal helix is absent from all available structures, this part of the protein was modeled and Gβ/γ interface was mapped by homology with bovine Gαt (transducin, PDB 1got), rat Gαi1 (PDB 1gp2), and mouse Gαq (PDB 3ah8).
DNA constructs and site directed mutagenesis
MSCV puro IRES GFP (PIG) and pBABE puro retroviral vectors were obtained from Addgene (43, 44). The SyR-Gi plasmid (also known as Gi-DREADD) was originally provided by Dr. Bryan Roth and was subcloned into the pBABE retroviral vector. Gα13 and N-terminally tagged AU5-RhoA were cloned into our in house pCEFL vector and mutations as indicated were prepared by site directed mutagenesis using the QuikChange Lightning Kit (Agilent, Santa Clara, CA). Gα13-i5 chimeras were prepared by PCR substitution of the last 5 amino acids of Gα13 with the Gαi1 sequence including a C-I mutation to make the plasmid pertussis toxin insensitive (the last 5 amino acids were as follows: DIGLF). Sequences were confirmed by DNA sequence analysis (NIDCR shared resource facility). A plasmid for alkaline phosphatase-fused transforming growth factor-α (AP-TGFα) (45) was kindly provided by Prof. Shigeki Higashiyama, Ehime University, Japan. A human dopamine D2 receptor (D2R, long isoform) was inserted into the pCAGGS expression plasmid as previously described (27).
Genomic DNA isolation and sequencing
Raji cells were obtained from ATCC (ATCC CCL-86). Genomic DNA was isolated from 5×106 Raji cells using the QiaAmp DNA isolation kit (Qiagen) and PCR was performed on the genomic DNA in the region surrounding the 184 amino acid position using AccuPrime Pfx Supermix (Life Technologies, Grand Island, NY). PCR primer details are included in supplemental information.
Retrovirus production and infection
Retrovirus from MSCV PIG empty vector and expressing Gα13 WT was produced by transfection of HEK293t/17 cells in a poly-lysine coated 6-well plate with 1 ug of MSCV plasmid DNA, 0.67 μg of gag/pol and 0.33 μg of VSV-G using Turbofect transfection reagent (Life Technologies). Virus for pBABE SyR-Gi was similarly prepared by transfecting 1 μg of pBABE SyR-Gi, 0.67 μg of gag/pol and 0.33 μg of VSV-G. Viral supernatants were collected at 48 h and 72 h post-transfection and filtered with 0.45 μm PVDF-filters onto target cells. For HEK293 cells transduced with pBABE SyR-Gi, 6 μg/ml of polybrene was added with the viral supernatants; cells were then selected with 1 μg/ml puromycin (Life Technologies) to generate a stable line. For transduction of Raji cells with MSCV virus, Raji cells were seeded onto retronectin-coated plates (Takara, Madison, WI) and centrifuged with viral supernatants at 2000 rpm for 2 hr and then incubated overnight at 37°C/5% CO2. MSCV-transduced Raji cells were allowed to recover before FACS sorting for GFP expression using a BD FACSAria III Cell Sorter (NIDCR Shared Resource Facility). Raji cells were cultured in RPMI media (Sigma, St. Louis, MO) supplemented with 10% FBS.
SRF-RE luciferase assay
A serum response factor response element (SRF-RE) luciferase assay was performed by seeding HEK293 cells in a poly-lysine coated 24-well plate and culturing in DMEM supplemented with 10% FBS for 24 h. Cells were then co-transfected with 100 ng Gα13 plasmid DNA or control, 50 ng pSRF-RE-firefly luciferase reporter DNA, and 20 ng pRL-renilla luciferase using lipofectamine 2000 (Life Technologies) transfection reagent. For stimulated SRF response, HEK293 cells stably transduced with SyR-Gi were similarly transfected. The day after transfection, cells were serum starved overnight in the presence of 50 ng/ml pertussis toxin (List Biological Laboratories, Campbell, CA) and then stimulated for 6 h with CNO prior to harvesting the cells. For all SRF-luciferase assays, cells were lysed and luciferase activity was determined using Dual-Glo Luciferase Assay Kit (Promega, Madison, WI). Chemiluminescence was measured using a BioTek Synergy Neo plate reader and the SRF-RE activation was calculated as the ratio of firefly to renilla luciferase levels. The assays were performed three times in duplicates and results are presented as the mean +/− standard deviation.
Rho-GTP pull down assay and western blot
Antibodies to RhoA (catalog # 2117), pAkt (Ser473) (catalog #4060), Akt (catalog #9272), and α-tubulin (catalog #3873) were obtained from Cell Signaling Technology. Gα13 antibody was obtained from Santa Cruz (catalog # sc-26788). Cells for western blot lysates were lysed on ice in RIPA buffer (Sigma) containing protease and phosphatase inhibitors (Thermo Scientific, Pittsburgh PA) and clarified by centrifugation at 13,000 rpm for 10 min at 4°C. The relative Gα13 protein expression levels were quantified by densitometry using ImageJ and normalized to α-tubulin; data represents an average of three independent western blots (mean +/− standard deviation). GST-tagged Rhotekin RBD beads were purchased from Millipore for pull down assays to detect active RhoA. HEK293 cells were seeded in 6 cm plates and transfected with lipofectamine 2000 for 48 h with 4 μg of plasmid DNAs. Cells were serum starved overnight and harvested for pull downs with 350 μl of recommended lysis buffer. Lysates were clarified and 50 μl of lysate was saved for input and the remaining lysate was rotated in tubes with the GST-tagged Rhotekin RBD beads. A three minute thrombin (Sigma) stimulation (1 U/mL) was used as a positive control. Lysates were resolved on SDS-PAGE gels, transferred onto PVDF membranes (Millipore, Billerica, MA), probed with appropriate antibodies and developed with ECL and film (GE healthcare, Piscataway, NJ) or scanned on an Odyssey imager.
Tumor xenograft model and Tissue Analysis
Ten-week old female NOD-scid IL2Rgammanull (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice were obtained from the Jackson Laboratory and randomly assigned for injection with tumor cells. Raji cells transduced with MSCV PIG control or MSCV PIG Gα13 WT were resuspended in phosphate buffered saline (PBS) and 5×106 cells in a 200 μl volume were injected into the right and left flanks of five mice for each group (ten tumors in total for each group. One injection was excluded from the Raji control group due to an injection error). Tumors were monitored two to three times a week and mice were sacrificed all together once the largest tumors reached approximately 1 cm3 volume. Tumor measurements were performed by a blinded individual and the measurements were recorded by an un-blinded person. An initial pilot study was performed with 4 tumors in each group, which was used to determine the appropriate sample size. This study was then expanded and repeated two more times; data trends were the same in all three experiments. Necropsies were performed and tumors were weighed, measured and imaged and tumor sections were cut and fixed in z-fix and embedded in paraffin for histology and immunohistochemistry (IHC) analysis as previously described (46) and elaborated in supplemental information. Individual data points are shown in the scatter dot plots to indicate variance in the data and indicates the mean +/− standard error of the mean. These animal studies were carried out according to National Institutes of Health (NIH) approved protocols (ASP # 13-695) in compliance with the NIH Guide for the Care and Use of Laboratory Mice.
TGFα shedding assay
TGFα shedding assay was performed as described previously (27) and as detailed in supplemental information. Briefly, HEK293A cells were seeded in 12-well plates and transfected using lipofectamine 2000 (Life Technologies) with a combination of the following plasmids: AP-TGFα-encoding plasmid, a GPCR-encoding plasmid and a Gα-encoding plasmid. After 24 h, cells were harvested with a trypsin-EDTA solution, washed with D-PBS, suspended in Hank’s balanced salt solution containing 5 mM HEPES (pH 7.4), and seeded in a 96-well plate and incubated for 30 min. The seeded cells were treated with various concentrations of a GPCR ligand (10 μl per well) and incubated for 1 h. Conditioned media (CM, 80 μl/well) was transferred into a blank 96-well plate. Alkaline phosphatase (AP) solution was added to both the CM plate and the cell plate (80 μl/well). Absorbance at 405 nm of the two plates was measured before and after 1 h incubation at room temperature, using a SpectraMax 340PC384 microplate reader (Molecular Devices, Sunnyvale, CA). TGFα shedding data are shown as the mean +/− standard deviation.
Spontaneous activity of Gα13 was measured by a previously described method with a slight modification (47). Briefly, HEK293A cells were seeded in 96-well plates in Opti-MEM (80 μl/well), transfected with lipofectamine 2000 reagent (0.1 μl/well), the AP-TGFα-encoding plasmids (20 ng/well), Gα13-encoding plasmid (5 to 20 ng/well) and the pCEFL vector plasmid, which was used to adjust total volume of transfected plasmid. After adding the transfection solution, the cells were incubated for 24 h and then AP-TGFα release was measured as described above.
Actin stress fiber formation and confocal imaging
Madin-Darby Canine Kidney (MDCK) cells were seeded onto poly-lysine coated glass cover slips (Fisher) in a 6-well plate and transfected with 1 ug LifeActin GFP and 1 ug of pCEFL RhoA plasmid or vector control using lipofectamine 2000. Cells for immunofluorescence (IF) imaging were washed with PBS and fixed with 2% formaldehyde – PBS solution for 12 min at room temperature. Fixed cells were washed twice with PBS and then mounted onto glass slides (Thermo Scientific/Fisher) with Fluorosave (Millipore/Calbiochem) mounting solution. Confocal images were collected on a Zeiss LSM-700 laser scanning microscope with a 40× oil immersion lens using 488 nm excitation for the LifeActin GFP.
Methocellulose clonogenic assay
Methocult™ (H4434) was purchased from Stem Cell Technologies and a clonogenic assay was performed by seeding 2000 Raji cells in 1 mL of Methocult™ in triplicate in a 6 well plate. Colonies were allowed to grow for 14 days and then number of colonies per well and morphology of the colonies were analyzed. Results are shown as the mean +/− standard deviation. Statistics were calculated using GraphPad Prism 6 using a paired t-test.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Dental and Craniofacial Research intramural program at NIH. We thank Maria S. Degese for her help with immunohistochemistry. A.I. was funded by PRESTO from JST. J.A. was funded by CREST from JST. We thank Miho Morikawa for technical assistance with the TGFα shedding assay. RAD and MSL are supported by the Division of Intramural Research, NIAID, NIH. IK is supported by NIH grants R01 GM071872 and R01 AI118985.
Footnotes
Conflict of Interest
Authors have no conflicts of interest to declare.
References
- 1.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nature reviews Cancer. 2007;7(2):79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Marcos M, Ghosh P, Farquhar MG. GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. The Journal of biological chemistry. 2015;290(11):6697–704. doi: 10.1074/jbc.R114.613414. Epub 2015/01/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Hayre M, Vazquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, et al. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nature reviews Cancer. 2013;13(6):412–24. doi: 10.1038/nrc3521. Epub 2013/05/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer cell. 2014;25(6):831–45. doi: 10.1016/j.ccr.2014.04.016. Epub 2014/06/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457(7229):599–602. doi: 10.1038/nature07586. Epub 2008/12/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–9. doi: 10.1056/NEJMoa1000584. Epub 2010/11/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhanasekaran N, Heasley LE, Johnson GL. G protein-coupled receptor systems involved in cell growth and oncogenesis. Endocr Rev. 1995;16(3):259–70. doi: 10.1210/edrv-16-3-259. [DOI] [PubMed] [Google Scholar]
- 8.Kelly P, Moeller BJ, Juneja J, Booden MA, Der CJ, Daaka Y, et al. The G12 family of heterotrimeric G proteins promotes breast cancer invasion and metastasis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(21):8173–8. doi: 10.1073/pnas.0510254103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly P, Stemmle LN, Madden JF, Fields TA, Daaka Y, Casey PJ. A role for the G12 family of heterotrimeric G proteins in prostate cancer invasion. The Journal of biological chemistry. 2006;281(36):26483–90. doi: 10.1074/jbc.M604376200. [DOI] [PubMed] [Google Scholar]
- 10.Gutkind JS, Coso OA, Xu N. G12 and G13 α subunits of heterotrimeric G proteins: A novel family of oncogenes. In: MSA, editor. G Proteins, Receptors, and Disease. Totowa, NJ: Humana Press; 1998. pp. 101–17. [Google Scholar]
- 11.Juneja J, Casey PJ. Role of G12 proteins in oncogenesis and metastasis. British journal of pharmacology. 2009;158(1):32–40. doi: 10.1111/j.1476-5381.2009.00180.x. Epub 2009/05/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu N, Bradley L, Ambdukar I, Gutkind JS. A mutant alpha subunit of G12 potentiates the eicosanoid pathway and is highly oncogenic in NIH 3T3 cells. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(14):6741–5. doi: 10.1073/pnas.90.14.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu N, Voyno-Yasenetskaya T, Gutkind JS. Potent transforming activity of the G13 alpha subunit defines a novel family of oncogenes. Biochemical and biophysical research communications. 1994;201(2):603–9. doi: 10.1006/bbrc.1994.1744. [DOI] [PubMed] [Google Scholar]
- 14.Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, et al. The genetic landscape of mutations in Burkitt lymphoma. Nature genetics. 2012;44(12):1321–5. doi: 10.1038/ng.2468. Epub 2012/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(10):3879–84. doi: 10.1073/pnas.1121343109. Epub 2012/02/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green JA, Suzuki K, Cho B, Willison LD, Palmer D, Allen CD, et al. The sphingosine 1-phosphate receptor S1P(2) maintains the homeostasis of germinal center B cells and promotes niche confinement. Nat Immunol. 2011;12(7):672–80. doi: 10.1038/ni.2047. Epub 2011/06/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muppidi JR, Schmitz R, Green JA, Xiao W, Larsen AB, Braun SE, et al. Loss of signalling via Galpha13 in germinal centre B-cell-derived lymphoma. Nature. 2014;516(7530):254–8. doi: 10.1038/nature13765. Epub 2014/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nature genetics. 2014;46(4):371–5. doi: 10.1038/ng.2916. Epub 2014/03/04. [DOI] [PubMed] [Google Scholar]
- 19.Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nature genetics. 2014;46(2):171–5. doi: 10.1038/ng.2872. Epub 2014/01/15. [DOI] [PubMed] [Google Scholar]
- 20.Cools J. RHOA mutations in peripheral T cell lymphoma. Nature genetics. 2014;46(4):320–1. doi: 10.1038/ng.2937. Epub 2014/03/29. [DOI] [PubMed] [Google Scholar]
- 21.Manso R, Sanchez-Beato M, Monsalvo S, Gomez S, Cereceda L, Llamas P, et al. The RHOA G17V gene mutation occurs frequently in peripheral T-cell lymphoma and is associated with a characteristic molecular signature. Blood. 2014;123(18):2893–4. doi: 10.1182/blood-2014-02-555946. Epub 2014/05/03. [DOI] [PubMed] [Google Scholar]
- 22.Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nature genetics. 2014;46(2):166–70. doi: 10.1038/ng.2873. Epub 2014/01/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fromm C, Coso OA, Montaner S, Xu N, Gutkind JS. The small GTP-binding protein Rho links G protein-coupled receptors and Galpha12 to the serum response element and to cellular transformation. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(19):10098–103. doi: 10.1073/pnas.94.19.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart MJ, Jiang X, Kozasa T, Roscoe W, Singer WD, Gilman AG, et al. Direct Stimulation of the Guanine Nucleotide Exchange Activity of p115 RhoGEF by Galpha13. Science. 1998;280(5372):2112–4. doi: 10.1126/science.280.5372.2112. [DOI] [PubMed] [Google Scholar]
- 25.Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AG, et al. p115 RhoGEF, a GTPase Activating Protein for Galpha12 and Galpha13. Science. 1998;280(5372):2109–11. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- 26.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(12):5163–8. doi: 10.1073/pnas.0700293104. Epub 2007/03/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue A, Ishiguro J, Kitamura H, Arima N, Okutani M, Shuto A, et al. TGFalpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nature methods. 2012;9(10):1021–9. doi: 10.1038/nmeth.2172. Epub 2012/09/18. [DOI] [PubMed] [Google Scholar]
- 28.Jaffe ES, Pittaluga S. Aggressive B-cell lymphomas: a review of new and old entities in the WHO classification. Hematology / the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2011;2011:506–14. doi: 10.1182/asheducation-2011.1.506. Epub 2011/12/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roschewski M, Staudt LM, Wilson WH. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nature reviews Clinical oncology. 2014;11(1):12–23. doi: 10.1038/nrclinonc.2013.197. Epub 2013/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasheed SA, Teo CR, Beillard EJ, Voorhoeve PM, Casey PJ. MicroRNA-182 and microRNA-200a control G-protein subunit alpha-13 (GNA13) expression and cell invasion synergistically in prostate cancer cells. The Journal of biological chemistry. 2013;288(11):7986–95. doi: 10.1074/jbc.M112.437749. Epub 2013/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly P, Casey PJ, Meigs TE. Biologic functions of the G12 subfamily of heterotrimeric g proteins: growth, migration, and metastasis. Biochemistry. 2007;46(23):6677–87. doi: 10.1021/bi700235f. Epub 2007/05/17. [DOI] [PubMed] [Google Scholar]
- 32.Yagi H, Tan W, Dillenburg-Pilla P, Armando S, Amornphimoltham P, Simaan M, et al. A synthetic biology approach reveals a CXCR4-G13-Rho signaling axis driving transendothelial migration of metastatic breast cancer cells. Science signaling. 2011;4(191):ra60. doi: 10.1126/scisignal.2002221. Epub 2011/09/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cattoretti G, Mandelbaum J, Lee N, Chaves AH, Mahler AM, Chadburn A, et al. Targeted disruption of the S1P2 sphingosine 1-phosphate receptor gene leads to diffuse large B-cell lymphoma formation. Cancer research. 2009;69(22):8686–92. doi: 10.1158/0008-5472.CAN-09-1110. Epub 2009/11/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. Epub 2011/07/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menon MP, Pittaluga S, Jaffe ES. The histological and biological spectrum of diffuse large B-cell lymphoma in the World Health Organization classification. Cancer J. 2012;18(5):411–20. doi: 10.1097/PPO.0b013e31826aee97. Epub 2012/09/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohde M, Richter J, Schlesner M, Betts MJ, Claviez A, Bonn BR, et al. Recurrent RHOA mutations in pediatric Burkitt lymphoma treated according to the NHL-BFM protocols. Genes, chromosomes & cancer. 2014;53(11):911–6. doi: 10.1002/gcc.22202. Epub 2014/07/22. [DOI] [PubMed] [Google Scholar]
- 37.Fukuhara S, Chikumi H, Gutkind JS. RGS-containing RhoGEFs: the missing link between transforming G proteins and Rho? Oncogene. 2001;20(13):1661–8. doi: 10.1038/sj.onc.1204182. [DOI] [PubMed] [Google Scholar]
- 38.Mikelis CM, Palmby TR, Simaan M, Li W, Szabo R, Lyons R, et al. PDZ-RhoGEF and LARG are essential for embryonic development and provide a link between thrombin and LPA receptors and Rho activation. The Journal of biological chemistry. 2013;288(17):12232–43. doi: 10.1074/jbc.M112.428599. Epub 2013/03/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Lin A. Role of JNK activation in apoptosis: a double-edged sword. Cell research. 2005;15(1):36–42. doi: 10.1038/sj.cr.7290262. Epub 2005/02/03. [DOI] [PubMed] [Google Scholar]
- 40.Bachman KE, Park BH. Duel nature of TGF-beta signaling: tumor suppressor vs. tumor promoter. Current opinion in oncology. 2005;17(1):49–54. doi: 10.1097/01.cco.0000143682.45316.ae. Epub 2004/12/21. [DOI] [PubMed] [Google Scholar]
- 41.Qin L, Kufareva I, Holden LG, Wang C, Zheng Y, Zhao C, et al. Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015;347(6226):1117–22. doi: 10.1126/science.1261064. Epub 2015/01/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kufareva I, Rueda M, Katritch V, Stevens RC, Abagyan R. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure. 2011;19(8):1108–26. doi: 10.1016/j.str.2011.05.012. Epub 2011/08/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–84. doi: 10.1016/j.cell.2009.06.016. Epub 2009/08/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic acids research. 1990;18(12):3587–96. doi: 10.1093/nar/18.12.3587. Epub 1990/06/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tokumaru S, Higashiyama S, Endo T, Nakagawa T, Miyagawa JI, Yamamori K, et al. Ectodomain shedding of epidermal growth factor receptor ligands is required for keratinocyte migration in cutaneous wound healing. The Journal of cell biology. 2000;151(2):209–20. doi: 10.1083/jcb.151.2.209. Epub 2000/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Martin D, Molinolo AA, Patel V, Iglesias-Bartolome R, Degese MS, et al. mTOR co-targeting in cetuximab resistance in head and neck cancers harboring PIK3CA and RAS mutations. Journal of the National Cancer Institute. 2014;106(9) doi: 10.1093/jnci/dju215. Epub 2014/08/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue A, Arima N, Ishiguro J, Prestwich GD, Arai H, Aoki J. LPA-producing enzyme PA-PLA(1)alpha regulates hair follicle development by modulating EGFR signalling. The EMBO journal. 2011;30(20):4248–60. doi: 10.1038/emboj.2011.296. Epub 2011/08/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.