Abstract
Purpose
To identify deleterious mutations in the latent transforming growth factor-β–binding protein 2 (LTBP2) gene in sporadic patients with primary congenital glaucoma (PCG) from a Han Chinese population, which had been excluded for mutations in the CYP1B1 gene.
Methods
In this retrospective case–control study, 36 coding exons and adjacent exon–intron boundaries of LTBP2 were amplified with PCR and screened for mutations with Sanger sequencing in DNA samples of 214 sporadic patients with PCG. Sequence variants identified in the patients with PCG were subsequently screened in 100 unaffected control subjects and the unaffected parents of the patients with PCG who had sequence changes in LTBP2.
Results
Eight heterozygous single nucleotide polymorphisms (SNPs) in coding regions of LTBP2 were identified in the patients with PCG. Four of these SNPs were missense changes that resulted in the replacement of amino acids (rs2304707, rs116914994, rs45468895, and rs763035721), two of which (rs2304707 and rs116914994) were also present in the control subjects. No significant differences in the frequencies of the missense SNPs were found between the patients with PCG and the controls. The two missense SNPs, rs45468895 and rs763035721, which were each found in one patient also existed in their unaffected parents, suggesting that these two SNPs were not segregated in these families and are unlikely to be a disease-causative variant. In addition, four synonymous SNPs were detected in the patients with PCG (rs61738025, rs862031, rs199805158, and rs12586758).
Conclusions
The results showed that no deleterious mutations were found in coding regions of LTBP2 in patients with PCG, suggesting that it is not a causal gene for PCG in the Han Chinese population.
Introduction
Primary congenital glaucoma (PCG) is a rare but severe form of glaucoma that can lead to blindness from the infantile period. The characteristic findings of PCG include the classic triad of presenting symptoms in the newborn: epiphora, photophobia, and blepharospasm [1]. Elevated intraocular pressure (IOP) causes optic atrophy in the late stage. The incidence of PCG varies from country to country depending on ethnicity. The highest incidence (1 in 1,250) occurs in the Slovakian Romani population with higher rates of consanguinity, while lower incidence (1 in 18,500 to 1 in 30,000) is observed in Western populations [2,3]. The prevalence in the Han Chinese population is approximately 1 in 5,000 to 1 in 25,000 (1 in 10,000 on average) [4]. Several genetic loci have been linked to PCG in family linkage studies, including GLC3A (Gene ID: 1545, OMIM #: 601771; 2p22-p21) [5], GLC3B (Gene ID: 2728, OMIM #: 600975; 1p36.2-p36.1) [6], GLC3C (gene ID: 399565, OMIM #: 613085; l4q24.2-q24.3) [7], and GLC3D (Gene ID: 4053, OMIM #: 602091; 14q24) [8,9]. Two genes have been identified from these loci: CYP1B1, coding for cytochrome P450, family 1, subfamily B, polypeptide 1 at GLC3A [10], and LTBP2, coding for latent transforming growth factor-β-binding protein 2 at GLC3D [8,9].
Previous studies showed that mutations in CYP1B1 are the predominant cause of PCG [11,12]. Populations with a higher mutation frequency of CYP1B1 in patients with PCG often possess a higher incidence of PCG. For instance, Arab and Romani populations have the highest incidence of PCG, corresponding to the highest mutation frequency of CYP1B1 in patients with PCG (90% to 100%) [13]. However, in our previous study, only 17.2% of patients with PCG had CYP1B1 mutations in a Han Chinese population [14], corresponding to the lower incidence of PCG in the Han Chinese population [4]. The low frequencies of CYP1B1 mutations in patients with PCG from some populations [14,15] indicate that PCG is genetically heterogeneous, and other genes may contribute to the disease.
The GLC3C locus spans a region of around 5.77 Mb that contains more than 40 genes, and it has been confirmed and further mapped within a genetic distance of 0.006 cM [16]. LTBP2 is around 1.3 Mb proximal to the GLC3C locus [8,9]. Ocular expression studies have determined the presence of LTBP2 in the trabecular meshwork and ciliary processes [17]. Recently, LTBP2 has been verified as essential for the development of ciliary zonule microfibrils [18]. Several studies have shown that LTBP2 mutations are associated with secondary glaucoma [19-22]. Null mutations have been found in patients with PCG from Pakistani, Iranian, and Slovakian Romani consanguineous families [8,9,23]. However, the exact role of LTBP2 in the development of PCG remains unclear. Several studies did not discover any deleterious mutations in LTBP2 in sporadic PCG North Indian, British, and American cases [24-26]. Intriguingly, a possible role of LTBP2 has been suggested in the etiology of primary open angle glaucoma (POAG), primary angle closure glaucoma (PACG), and secondary glaucoma, such as megalocornea, spherophakia, ectopia lentis, Weill-Marchesani syndrome, and pseudoexfoliation syndrome [19-22,27,28]. In the present study, we screened for mutations in the LTBP2 gene in a large sample of Chinese patients with PCG, aiming to evaluate the role of LTBP2 in Chinese patients with PCG.
Methods
Study subjects
A total of 214 unrelated patients diagnosed with PCG but without mutations in the CYP1B1 gene were included in this study [14,29]. Briefly, we amplified the coding regions (exons 2 and 3) and the promoter region of CYP1B1 with PCR and screened for CYP1B1 mutations with direct sequencing with forward and reverse primers [14,29]. PCR was performed in a 25 µl mixture containing genomic DNA (50 ng), forward and reverse primers (0.5 µmol/l), magnesium chloride (1.5 mmol/l), deoxyribonucleotide triphosphate (dNTP; 0.2 µmol/ml) and Taq polymerase (1 U; Takara Bio Inc, Shiga, Japan), as per the following procedure: initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 94 °C for 45 s, annealing at a primer-specific temperature for 45 s, extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. We excluded patients who carried CYP1B1 mutations from the present study. Among these sporadic cases, 152 trios that consisted of one affected proband and two unaffected parents had been used in our previous study of GLC3C with the transmission disequilibrium test (TDT) [16]. As the other 62 sporadic cases had only one parent available for genotyping, they were excluded from TDT in our previous study [16]. The diagnosis of PCG was based on the following criteria [29]: 1) Age of diagnosis was less than 3 years old, 2) the cornea was enlarged so that the parameter was bigger than 11 mm or edema with or without Haab striae, 3) IOP was greater than 21 mmHg, and 4) the cup to disk ratio was enlarged or dissymmetric between two eyes. Patients with other ocular or systemic anomalies, such as aniridia, anterior segment dysgenesis, congenital hereditary endothelial dystrophy, neurofibromatosis, and Sturge–Weber syndrome, were excluded.
All patients underwent comprehensive examinations of the anterior segments with slit-lamp and surgical microscope, the structure of anterior chamber angles with gonioscope, and the ocular fundus with a direct ophthalmoscope. IOP was measured with Tono- PEN® (Reichert, Depew, NY) before or during surgery under general anesthesia. Corneal opacity was scored based on the microscope examination, and a higher corneal opacity score indicated a more opaque cornea [29]. The more severe eye was recorded and analyzed when cases were bilateral present.
One hundred ethnically matched unrelated individuals without any ocular or systemic disorders were enrolled as controls. The unaffected parents of the patients with PCG who had sequence changes in LTBP2 were also included in the study. All the controls and healthy parents were older than 18 years and had normal IOP, open angles on gonioscopy, and normal optic nerves on ocular fundus examination. Peripheral blood samples were collected from patients and controls by venipuncture in EDTA vacutainers and stored in −20 °C until further use.
All study subjects were Han Chinese who live in Mainland China. This study adhered to the ARVO statement on human subjects. This study was approved by the Ethics Committee of the Eye and ENT hospital, Fudan University. Informed written consent was obtained from all study subjects after explanation of the nature and possible consequences of the study, in accordance with the tenets of the Declaration of Helsinki.
Screening for LTBP2 mutations with Sanger sequencing
DNA extraction and Sanger sequencing were performed for all 214 patients with PCG using standard methods [30]. The primers were designed to include the intron–exon boundaries of LTBP2 using the software Primer Premier 5.0 (Table 1). Standard PCR conditions were run to amplify all exons and exon–intron boundaries of LTBP2. PCR was performed in a 25 µl mixture containing genomic DNA (50 ng), forward and reverse primers (0.5 µmol/l), magnesium chloride (1.5 mmol/l), deoxyribonucleotide triphosphate (dNTP; 0.2 µmol/ml) and Taq polymerase (1 U; Takara Bio Inc, Shiga, Japan), as per the following procedure: initial denaturation at 95 °C for 5 min, 35 cycles of denaturation at 94 °C for 45 s, annealing at a primer-specific temperature for 45 s, extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. All PCR products were analyzed on 2% agarose gel and stained with ethidium-bromide (EB, 10 mg/ml). Then the right size products were purified with spin-columns technology (QIAQuick PCR Purification Kit; Qiagen, Valencia, CA). Using ABI BigDye chemistry (Applied Biosystems Inc., Foster City, CA), Sanger sequencing was performed on the PCR amplicons and was processed through an automated ABI 3730 XL Sequencer (Applied Biosystems). The sequences were scrutinized for variations alongside reference sequences from the University of California, Santa Cruz Genome Browser website using Sequencher 5.0 software (Gene Codes, Ann Arbor, MI). PCR and Sanger sequencing were subsequently performed on 100 unaffected control samples with identified sequence variants. The unaffected parents of the patients with PCG who had sequence changes in LTBP2 were also screened for the specific SNPs using Sanger sequencing. The PCR products were first sequenced with forward primers. Further sequencing with reverse primers was performed to confirm the identified variants.
Table 1. PCR Primers used for amplification of LTBP2 gene.
| Primer name | Forward Primer | Reverse Primer | Product size (bp) |
|---|---|---|---|
|
Exon1 |
GGGAGAGATGAAAGCAGGCAAGG |
AGGTGGAGGCAGCCAGCAGA |
350 |
|
Exon2 |
CGTTCAAGTCATCACACTCATTCACA |
AAGGGCGAGGGTCCCAGCATT |
666 |
|
Exon3 |
TGTGATAGTTTATGACGGAGTTGTTAGGG |
GCATTGAGGCAGAGCGGGAG |
456 |
|
Exon3–5 |
CTCTGGCTTGGTGTGTCTTTCCC |
CAATGCCCCACCCTTTCCTG |
1110 |
|
Exon5–6 |
GTGGATGTCGTGGTCAGGGAGGT |
GCTAGTCTTGAGAAGGAAATGATGGAG |
982 |
|
Exon7 |
TGGGACAGAAAAGGTGGAGGC |
GTGACACTGAAAGAAGGGGAGAGA |
289 |
|
Exon8 |
CGTCCAGGCTCCTTCCACAG |
TCTTGAGATGATGATTAGGCCACTT |
559 |
|
Exon9 |
GGACACGCTCTCTGTAAACCTCTGA |
TGCCACTCCTCTCCTTTTTTCTTCT |
100 |
|
Exon10–11 |
CCCCTGGGCGAGGAGTTGAA |
GGCAGTGGAGGAAAAGGAGTG |
1027 |
|
Exon12 |
CATCTGTGGAAAGTGCTCTGCTC |
TCCCTGACTCTCCTATGGGCTACT |
329 |
|
Exon13–14 |
CCTCACATCACTCCTCGCTCCC |
GCTGCCATCGGCTGTCTTCC |
578 |
|
Exon15 |
AGAGGGGGCATCCCATTTGTATC |
AGGAGGAAAAGCCAGTCCCAGAG |
354 |
|
Exon15–16 |
GGGGGCATCCCATTTGTATCCT |
CCTATTCACACTGACCCAACACATCC |
789 |
|
Exon17 |
CATTCACCCAGTACCCGTTCTCAC |
GCCCACCGTCTTGGACTTCTCTC |
488 |
|
Exon18 |
ACAACAGCCTCCAAGCCACCTCC |
GTCCATCCCTCTCCCCAACCAAA |
344 |
|
Exon19 |
GCAACAGGGCGGAGGTTACAG |
GCAGACCCGAAGGGGATATAGTG |
788 |
|
Exon20 |
ATTCTGCGGCACGGTGTTTG |
ACTGGGCTGACTTTATGGCTTCC |
409 |
|
Exon21 |
ACTGCTTGGACCTTCTGCTTCTTC |
GGGGGATGCCCTACTATGACCT |
465 |
|
Exon22 |
TTCCCCCTTCTGGGCTTGG |
GGCTTCACCCTTCGTTCTTGG |
445 |
|
Exon23 |
TCTGATTTATGTGCTAAGTGGTGC |
GCTTCTGTTACTGTGTTGAGATGCT |
654 |
|
Exon24 |
TCCGTGAGGTGGTTTGAGGC |
GGGCAGAGGCAGGTAAGAAGAGT |
645 |
|
Exon25 |
ATGGAAGAGGAAGTGGGTGGGG |
ATGGGGCAGCGAGTGTGAGAA |
710 |
|
Exon26 |
AAGCCAACAGCCAGAGGACAAAC |
TGGGGAGTCACGAGTCTCATCAAG |
657 |
|
Exon27 |
TCCAGGTTGAATAGCCGACAC |
AAACTGAGGCACAGGGAGAGG |
422 |
|
Exon28 |
GGCTGACTTGATTTCCCACCCTA |
TTTGTCTTTCTGTGCCTGTCTTATTTC |
697 |
|
Exon29 |
GCTGGAACAGGAGGTAAGGAGATAA |
TGGCAGAATGTGGGGAGTGAG |
626 |
|
Exon30 |
AACAAAGAGGACAGAGAGGAGGAGAAG |
TCAGAGGGTTGGAAATGAGGGTG |
443 |
|
Exon31 |
TCCACCCAACTGGCACTCCTC |
CCAAGTCTGGCTTCCGCATCT |
783 |
|
Exon32 |
AGAGAAACAGGCGGCAGAACC |
AAAATACCAACACTCAGCCATAGGA |
897 |
|
Exon33 |
ATGATAGATTGGGCAGAAATAGAGC |
GTAGGGAGTAGGTACAGGACTTGGTT |
846 |
|
Exon34 |
ATGCCAGGGAAAGCCAAGGA |
TGTGGGACGTGGTGGTGTGAG |
680 |
|
Exon35 |
TGGGAGTAGGGTGGGGAATGG |
AGAAGGTGGGACTGTCGGAGGG |
225 |
| Exon36 | GCCTGATGTCACGGTGTCTTCC | GCTCCCTACAGCACTCTCCTTTG | 281 |
Statistical analysis
Fisher’s exact test was used to compare the allele frequencies of the LTBP2 variants between patients with PCG and controls. A Bonferroni corrected p value of 0.0125 was considered statistically significant.
Results
Clinical features of the patients
Some clinical features of the patients with PCG in this study are shown in Figure 1. At the preoperative examination, the median IOP was 36 mmHg, with an average corneal diameter of 13.5 mm. The median age of diagnosis was 4.5 months, ranging from birth to 3 years old. The median corneal opacity score was 2, indicating mild corneal haze with visible iris. Figure 2 shows the classic clinical features seen in the left eye of a patient with PCG.
Figure 1.

Clinical features of Chinese patients with primary congenital glaucoma.
Figure 2.
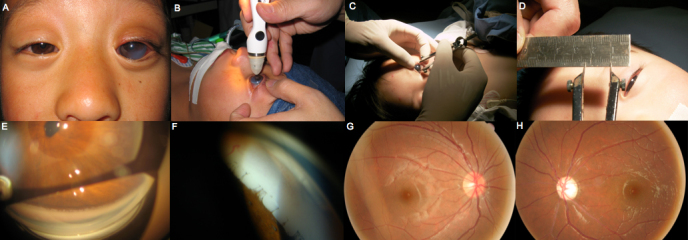
Classic features of Chinese patients with primary congenital glaucoma. A: The left eye is obviously bigger than the right eye and mild corneal haze with visible iris. B: Intraocular pressure (IOP) was measured with Tono-PEN®. C,D: The horizontal diameter of the cornea is about 13 mm measured with bisecting compasses. E,F: The structure of the anterior chamber angle on gonioscopy; absence of angle recess; peripheral iris hypoplasia; tenting of the peripheral iris pigment epithelium. G,H: The fundi photos show a dissymmetric cup disc ratio.
Screening for LTBP2 mutations
Eight single nucleotide polymorphisms (SNPs) were identified in the coding regions of the LTBP2 gene in the patients with PCG. All these variants were inherited in a heterozygous manner. Four missense SNPs resulted in the replacement of amino acids (rs2304707, rs116914994, rs45468895, and rs763035721, Table 2). Two of the SNPs, rs2304707 and rs116914994, which changed amino acids p.Pro319Gln and p.Arg1429Gln, respectively, were also present in the control subjects. No significant differences in the frequencies of these missense SNPs were found between the patients with PCG and the controls. The other two missense SNPs, rs45468895 and rs763035721, which changed amino acids p.Ala1204Val and p.Glu1794Lys, respectively, were each found in one patient with PCG. Their unaffected parents also had the same sequence changes, suggesting that these two SNPs were not segregated in these families and, thus, were unlikely to be a disease-causative variant. In addition, four synonymous SNPs were detected in the patients with PCG (rs61738025, rs862031, rs199805158, and rs12586758).
Table 2. LTBP2 polymorphisms identified from patients with primary congenital glaucoma.
| SNP ID | Exon | Sequence change | Amino acid change | Minor allele frequency (%) |
P value | |
|---|---|---|---|---|---|---|
| Affected | Unaffected | |||||
| rs2304707 | 4 | c.956C>A | p. Pro319Gln | 35/370 (9.5) | 7/90 (7.8) | 0.62 |
| rs45468895 | 24 | c.3611C>T | p. Ala1204Val | 1/428 (0.2) | 0/200 (0.0) | 0.49 |
| rs116914994 | 29 | c.4286G>A | p. Arg1429Gln | 22/428 (5.1) | 16/200 (8.0) | 0.16 |
| rs763035721 | 36 | c.5380G>A | p. Glu1794Lys | 1/428 (0.2) | 0/200 (0.0) | 0.49 |
Fisher’s exact test was used to compare the allele frequencies of the LTBP2 variants between PCG patients and controls. A Bonferroni corrected p value of 0.0125 was considered significant.
Discussion
Null mutations in LTBP2 were initially found to cause PCG in four consanguineous families from Pakistan and in patients of Romani ethnicity [8]. Subsequently, mutations in LTBP2 were identified in patients with PCG from several other populations, such as Iranian and Slovakian Romani families (Table 3) [9,23]. However, several other studies did not discover deleterious mutations in LTBP2 in sporadic North Indian, British, and American patients with PCG [24-26]. In the present study, we screened 214 sporadic patients with PCG from the Han Chinese population for mutations in the LTBP2 gene. We did not find disease-causative mutations in LTBP2 in the patients with PCG, which is consistent with previous studies [24-26]. None of the previously reported pathogenic mutations was detected in our study (Table 3) [8,9,23]. In addition, the SNP rs2304707 identified in this study was also reported as a polymorphism in consanguineous Iranian families with PCG and a Turkish GLC3C-linked family with PCG [9,25]. Another SNP rs862031 has been reported in patients in the United States [26]. None of the other SNPs identified in the present study was reported in patients with PCG in other populations [8,9,23].
Table 3. LTBP2 mutations/ polymorphisms previously reported in patients with primary congenital glaucoma from other populations.
| Exon | Sequence change | Amino acid change | SNP ID | Population | References |
|---|---|---|---|---|---|
| 1 | c.331C>T | p.Gln111X p. Pro319Gln | rs121918356 * | Pakistan/ Gypsy * | 8 * |
| 1 | c.412delG | p.Ala138ProfsX278 | N.A. | Pakistan/ Gypsy | 8, 23 |
| 4 | c.895C>T | p.Arg299X | rs121918355 | Pakistan, Roma / Gypsy | 8 |
| 4 | C>A | p.Pro319Gln | rs2304707 | American | 26 |
| 6 | c.1243–1256 del | p.Glu415ArgfsX596 | N.A. | Pakistan/ Gypsy | 8 |
| 6 | c.1287G>A | p.Leu429Leu | rs61738025 | Iranian | 9, 26 |
| Intron 6–7 | N.A. | g.75070493 | rs3742793 | North Indian | 24 |
| 7 | c.1415delC | p.Ser472fsX3 | N.A. | Iranian | 9 |
| 7 | G>T | p.Ser518Ile | N.A. | American | 26 |
| 7 | C>T | p.Arg548X | N.A. | American | 26 |
| 7 | C>T | p.Asn560Asn | N.A. | American | 26 |
| 14 | T>C | p.Thr802Thr | rs699374 | American | 26 |
| 15 | C>T | p.Thr834Thr | rs862031 | American | 26 |
| 17 | C>T | p.Arg905Arg | rs7145480 | American | 26 |
| 19 | c.2966C>G | p.Pro989Arg | rs76172717 | Iranian | 9 |
| 20 | C>T | p.Ser1031Ser | rs45473602 | American | 26 |
| 22 | G>A | p.Thr1111Thr | rs61729544 | American | 26 |
| 26 | C>T | p.Arg1284Arg | rs61736977 | American | 26 |
| 26 | G>A | p.Pro1297Pro | rs61738013 | American | 26 |
| 29 | G>A | p.Ala1421Ala | rs61738017 | American | 26 |
| 33 | c.4808G>A | p.Arg1603His | rs75200417 | Iranian | 9 |
| 36 | c.5376delC | p.Tyr1793fsX55 | N.A. | Iranian | 9 |
N.A., not avaiblable.
In addition to PCG, LTBP2 has been related to the development of secondary glaucoma, such as megalocornea, spherophakia, ectopia lentis, Weill-Marchesani syndrome, and pseudoexfoliation syndrome [19,21,22,27]. This can be well explained by recent findings in Ltbp2–/– mice [18]. The mice survived to adulthood but developed lens luxation caused by compromised ciliary zonule formation. The suppression of LTBP2 expression in cultured human ciliary epithelial cells by siRNA disrupted the formation of the microfibril meshwork by the cells [18]. Supplementation of recombinant LTBP2 in culture medium not only rescued the microfibril meshwork formation in LTBP2-suppressed ciliary epithelial cells but also restored unfragmented and bundled ciliary zonules in Ltbp2 –/– mouse eyes under organ culture [18]. The LTBP proteins are important components of the extracellular matrix (ECM) that interact with fibrillin microfibrils and have several different roles in microfibril biology. There are four LTBP isoforms in the human genome (LTBP-1, -2, -3, and -4). LTBP-2 is expressed abundantly in elastic tissues and unique in the family as it is the only one that does not bind to latent transforming growth factor-beta (TGF-β) [31]. Several reports indicate that LTBP-2 has a role in the elastic fiber assembly process, especially fibrillin-1 and -2 [18,32]. Whether increased IOP is a primary symptom or a secondary effect of another abnormality caused by LTBP2 mutations in childhood glaucoma remains controversial.
In summary, we screened for mutations in the LTBP2 gene in a large sample of Chinese patients with PCG. We did not detect deleterious mutations in LTBP2, suggesting that this gene is not a causal gene for PCG in the Han Chinese population.
Acknowledgments
The samples used for the analyses described in this manuscript were obtained from the EENT Biobank. We would like to thank all the participants for their valuable contribution to this research. This work was supported in part by the Funds for International Cooperation and Exchange of the National Natural Science Foundation of China (81020108017; Xinghuai Sun) and National Natural Science Foundation of China (81000401, 81570887; Xueli Chen, Yuhong Chen).
References
- 1.Ophthalmology AAo. Childhood Glaucoma. Basic and Clinical Science Course. Vol 2008–2009. USA: Lifelong Education for the Ophthalmologist; 2008. p. 157. [Google Scholar]
- 2.Dimasi DP, Hewitt AW, Straga T, Pater J, MacKinnon JR, Elder JE, Casey T, Mackey DA, Craig JE. Prevalence of CYP1B1 mutations in Australian patients with primary congenital glaucoma. Clin Genet. 2007;72:255–60. doi: 10.1111/j.1399-0004.2007.00864.x. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17718864&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 3.Abu-Amero KK, Edward DP. Primary Congenital Glaucoma. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K, editors. GeneReviews(R). Seattle (WA)1993. [PubMed] [Google Scholar]
- 4.Kaur K, Mandal AK, Chakrabarti S. Primary Congenital Glaucoma and the Involvement of CYP1B1. Middle East Afr J Ophthalmol. 2011;18:7–16. doi: 10.4103/0974-9233.75878. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21572728&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarfarazi M, Akarsu AN, Hossain A, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS. Assignment of a locus (GLC3A) for primary congenital glaucoma (Buphthalmos) to 2p21 and evidence for genetic heterogeneity. Genomics. 1995;30:171–7. doi: 10.1006/geno.1995.9888. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8586416&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 6.Akarsu AN, Turacli ME, Aktan SG, Barsoum-Homsy M, Chevrette L, Sayli BS, Sarfarazi M. A second locus (GLC3B) for primary congenital glaucoma (Buphthalmos) maps to the 1p36 region. Hum Mol Genet. 1996;5:1199–203. doi: 10.1093/hmg/5.8.1199. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8842741&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 7.Stoilov IR, Sarfarazi M. The third genetic locus (GLC3C) for primary congenital glaucoma (PCG) maps to chromosome l4q24.3.: The Association for Research in Vision and Ophthalmology (ARVO) Annual Meeting; 2002. [Google Scholar]
- 8.Ali M, McKibbin M, Booth A, Parry DA, Jain P, Riazuddin SA, Hejtmancik JF, Khan SN, Firasat S, Shires M, Gilmour DF, Towns K, Murphy AL, Azmanov D, Tournev I, Cherninkova S, Jafri H, Raashid Y, Toomes C, Craig J, Mackey DA, Kalaydjieva L, Riazuddin S, Inglehearn CF. Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet. 2009;84:664–71. doi: 10.1016/j.ajhg.2009.03.017. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19361779&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Narooie-Nejad M, Paylakhi SH, Shojaee S, Fazlali Z, Rezaei Kanavi M, Nilforushan N, Yazdani S, Babrzadeh F, Suri F, Ronaghi M, Elahi E, Paisan-Ruiz C. Loss of function mutations in the gene encoding latent transforming growth factor beta binding protein 2, LTBP2, cause primary congenital glaucoma. Hum Mol Genet. 2009;18:3969–77. doi: 10.1093/hmg/ddp338. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=19656777&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 10.Stoilov I, Akarsu AN, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet. 1997;6:641–7. doi: 10.1093/hmg/6.4.641. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=9097971&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 11.Bejjani BA, Lewis RA, Tomey KF, Anderson KL, Dueker DK, Jabak M, Astle WF, Otterud B, Leppert M, Lupski JR. Mutations in CYP1B1, the gene for cytochrome P4501B1, are the predominant cause of primary congenital glaucoma in Saudi Arabia. Am J Hum Genet. 1998;62:325–33. doi: 10.1086/301725. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=9463332&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plasilova M, Stoilov I, Sarfarazi M, Kadasi L, Ferakova E, Ferak V. Identification of a single ancestral CYP1B1 mutation in Slovak Gypsies (Roms) affected with primary congenital glaucoma. J Med Genet. 1999;36:290–4. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10227395&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 13.Abu-Amero KK, Osman EA, Mousa A, Wheeler J, Whigham B, Allingham RR, Hauser MA, Al-Obeidan SA. Screening of CYP1B1 and LTBP2 genes in Saudi families with primary congenital glaucoma: genotype-phenotype correlation. Mol Vis. 2011;17:2911–9. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22128238&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Jiang D, Yu L, Katz B, Zhang K, Wan B, Sun X. CYP1B1 and MYOC mutations in 116 Chinese patients with primary congenital glaucoma. Arch Ophthalmol. 2008;126:1443–7. doi: 10.1001/archopht.126.10.1443. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=18852424&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 15.Kakiuchi T, Isashiki Y, Nakao K, Sonoda S, Kimura K, Ohba N. A novel truncating mutation of cytochrome P4501B1 (CYP1B1) gene in primary infantile glaucoma. Am J Ophthalmol. 1999;128:370–2. doi: 10.1016/s0002-9394(99)00143-9. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10511040&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Chen Y, Wang L, Jiang D, Wang W, Xia M, Yu L, Sun X. Confirmation and further mapping of the GLC3C locus in primary congenital glaucoma. Front Biosci (Landmark Ed) 2011;16:2052–9. doi: 10.2741/3838. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21622161&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 17.Schlotzer-Schrehardt U, Zenkel M, Kuchle M, Sakai LY, Naumann GO. Role of transforming growth factor-beta1 and its latent form binding protein in pseudoexfoliation syndrome. Exp Eye Res. 2001;73:765–80. doi: 10.1006/exer.2001.1084. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11846508&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 18.Inoue T, Ohbayashi T, Fujikawa Y, Yoshida H, Akama TO, Noda K, Horiguchi M, Kameyama K, Hata Y, Takahashi K, Kusumoto K, Nakamura T. Latent TGF-beta binding protein-2 is essential for the development of ciliary zonule microfibrils. Hum Mol Genet. 2014;23:5672–82. doi: 10.1093/hmg/ddu283. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24908666&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan AO, Aldahmesh MA, Alkuraya FS. Congenital megalocornea with zonular weakness and childhood lens-related secondary glaucoma - a distinct phenotype caused by recessive LTBP2 mutations. Mol Vis. 2011;17:2570–9. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22025892&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 20.Haji-Seyed-Javadi R, Jelodari-Mamaghani S, Paylakhi SH, Yazdani S, Nilforushan N, Fan JB, Klotzle B, Mahmoudi MJ, Ebrahimian MJ, Chelich N, Taghiabadi E, Kamyab K, Boileau C, Paisan-Ruiz C, Ronaghi M, Elahi E. LTBP2 mutations cause Weill-Marchesani and Weill-Marchesani-like syndrome and affect disruptions in the extracellular matrix. Hum Mutat. 2012;33:1182–7. doi: 10.1002/humu.22105. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22539340&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 21.Kumar A, Duvvari MR, Prabhakaran VC, Shetty JS, Murthy GJ, Blanton SH. A homozygous mutation in LTBP2 causes isolated microspherophakia. Hum Genet. 2010;128:365–71. doi: 10.1007/s00439-010-0858-8. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20617341&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 22.Desir J, Sznajer Y, Depasse F, Roulez F, Schrooyen M, Meire F, Abramowicz M. LTBP2 null mutations in an autosomal recessive ocular syndrome with megalocornea, spherophakia, and secondary glaucoma. Eur J Hum Genet. 2010;18:761–7. doi: 10.1038/ejhg.2010.11. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=20179738&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azmanov DN, Dimitrova S, Florez L, Cherninkova S, Draganov D, Morar B, Saat R, Juan M, Arostegui JI, Ganguly S, Soodyall H, Chakrabarti S, Padh H, Lopez-Nevot MA, Chernodrinska V, Anguelov B, Majumder P, Angelova L, Kaneva R, Mackey DA, Tournev I, Kalaydjieva L. LTBP2 and CYP1B1 mutations and associated ocular phenotypes in the Roma/Gypsy founder population. Eur J Hum Genet. 2011;19:326–33. doi: 10.1038/ejhg.2010.181. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=21081970&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohanty K, Tanwar M, Dada R, Dada T. Screening of the LTBP2 gene in a north Indian population with primary congenital glaucoma. Mol Vis. 2013;19:78–84. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23378721&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 25.Sharafieh R, Child AH, Khaw PT, Fleck B, Sarfarazi M. LTBP2 gene analysis in the GLC3C-linked family and 94 CYP1B1-negative cases with primary congenital glaucoma. Ophthalmic Genet. 2013;34:14–20. doi: 10.3109/13816810.2012.716486. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=22924778&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 26.Lim SH, Tran-Viet KN, Yanovitch TL, Freedman SF, Klemm T, Call W, Powell C, Ravichandran A, Metlapally R, Nading EB, Rozen S, Young TL. CYP1B1, MYOC, and LTBP2 mutations in primary congenital glaucoma patients in the United States. Am J Ophthalmol. 2013;155:508–17. doi: 10.1016/j.ajo.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jelodari-Mamaghani S, Haji-Seyed-Javadi R, Suri F, Nilforushan N, Yazdani S, Kamyab K, Elahi E. Contribution of the latent transforming growth factor-beta binding protein 2 gene to etiology of primary open angle glaucoma and pseudoexfoliation syndrome. Mol Vis. 2013;19:333–47. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=23401661&dopt=Abstract [PMC free article] [PubMed] [Google Scholar]
- 28.Safari I, Akbarian S, Yazdani S, Elahi E. A Possible Role for LTBP2 in the Etiology of Primary Angle Closure Glaucoma. J Ophthalmic Vis Res. 2015;10:123–9. doi: 10.4103/2008-322X.163783. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=26425313&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Chen Y, Wang L, Jiang D, Wang W, Xia M, Yu L, Sun X. CYP1B1 genotype influences the phenotype in primary congenital glaucoma and surgical treatment. Br J Ophthalmol. 2014;98:246–51. doi: 10.1136/bjophthalmol-2013-303821. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=24227805&dopt=Abstract [DOI] [PubMed] [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–7. doi: 10.1073/pnas.74.12.5463. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=271968&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saharinen J, Keski-Oja J. Specific sequence motif of 8-Cys repeats of TGF-beta binding proteins, LTBPs, creates a hydrophobic interaction surface for binding of small latent TGF-beta. Mol Biol Cell. 2000;11:2691–704. doi: 10.1091/mbc.11.8.2691. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=10930463&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirai M, Horiguchi M, Ohbayashi T, Kita T, Chien KR, Nakamura T. Latent TGF-beta-binding protein 2 binds to DANCE/fibulin-5 and regulates elastic fiber assembly. EMBO J. 2007;26:3283–95. doi: 10.1038/sj.emboj.7601768. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=17581631&dopt=Abstract [DOI] [PMC free article] [PubMed] [Google Scholar]


