Abstract
To explore the phosphoproteome profiles during Xenopus egg activation by Ca2+-stimulation, an automated phosphopeptide purification system involving a titania column was improved by introducing 4-step elution with phosphate buffers. The number of detected phosphopeptides in the tryptic digest of a Xenopus egg cytosol fraction on mass spectrometry (MS) was increased 1.5-fold and the percentage of multiply phosphorylated peptides increased from 17 to 24% with introduction of the 4-step elution method. Phosphopeptides were purified by the improved method from tryptic digests of cytosol fractions of Xenopus eggs without and with a Ca2+-stimulus, and then, analysed by MS. One thousand three hundred and seventy-five and 994 phosphopeptides were reproducibly detected on duplicate MS, respectively. They included 818 and 437 phosphopeptides specific to each digest, respectively. A method involving isobaric tags for relative and absolute quantitation (iTRAQ) was also applied to compare the phosphorylation levels in Xenopus eggs without and with a Ca2+-stimulus, the ratios for 112 phosphopeptides in tryptic digests of these egg cytosol fractions being obtained. It was suggested from all the results that the phosphorylation sites and levels change during Xenopus egg activation for many known and unknown sites on structural proteins, signalling related proteins, cell cycle-related proteins and others.
Keywords: egg, phosphoproteome, proteome, titania, Xenopus
Xenopus eggs have been used in studies on egg maturation, egg activation and other mechanisms, which facilitated the progress of developmental biology (1, 2). It has been shown that signalling pathways including protein phosphorylation and dephosphorylation participate greatly in these regulation mechanisms (1, 3, 4). To clarify the complicated regulation mechanism of egg activation, comprehensive analysis of protein phosphorylation during egg activation is a powerful means. The cell cycle of Xenopus eggs is arrested at the second meiotic metaphase. So, Xenopus eggs without Ca2+-stimulation show meiotic metaphase properties. When eggs are activated by Ca2+-stimulation to mimic fertilization, they show synthetic phase properties. So, we can analyse the changes of protein phosphorylation during activation of Xenopus eggs by comparison of the phosphorylation in Ca2+-unstimulated and Ca2+-stimulated eggs (5). In this study, therefore, we obtained phosphoproteome profiles of Xenopus egg cytosol fractions without and with Ca2+-stimulation as a basis for understanding the egg activation mechanism.
To obtain high quality phosphoproteome data, i) determination of a large number of phosphorylation sites, ii) reproducibility of phosphopeptide determination and iii) getting less-biased data sets should be considered (6, 7). A key step to achieve these purposes is the phosphopeptide purification step. For such purification, immobilized metal ion affinity chromatography (8), titania (9) and others (10) have been applied. Titania columns are superior to immobilized metal ion affinity chromatography columns in the phosphopeptide binding capacity and the recovery of phosphopeptides (11). An on-line phosphoproteomics system based on a titania column and nano-flow liquid chromatography-linear ion trap/Orbitrap tandem MS (LC MS/MS) has been reported (12). We also developed a fully automated phosphopeptide purification system based on a titania column, the purified phosphopeptides being detected by matrix-assisted laser desorption ionization-time of flight MS (11). The system gave a highly purified phosphopeptide fraction in a high yield (11). Although the system gave highly reproducible results, the phosphopeptides of which the sequences could be determined were limited (11).
In this study, we improved the peptide elution method for the phosphopeptide purification system by increasing the number of steps with phosphate buffers from one to four, and the sequences of the purified peptides were determined by LC MS/MS. The phosphopeptides that could be detected on LC MS/MS increased 1.5 times with the change of the elution method from one to four steps. The percentage of multiply phosphorylated peptides increased with the 4-steps elution. The method was applied to analysis of the Xenopus egg phosphoproteome without and with Ca2+-stimulation, and many phosphopeptides including multiply phosphorylated ones could be reproducibly detected. The detected phosphorylation sites included many specific ones as to egg stages. Phosphoproteome data useful for studies on the egg activation mechanism was obtained.
Materials and Methods
Buffers for egg cytosol preparation
Extraction buffer: 50 mM HEPES-KOH (pH 7.7), 250 mM sucrose, 50 mM KCl and 2.5 mM MgCl2; buffer M: 20 mM HEPES-KOH (pH 7.5), 60 mM β-glycerophosphate, 20 mM EGTA and 15 mM MgCl2; MMR: 25 mM HEPES-KOH (pH 7.8), 100 mM NaCl, 2 mM KCl, 1 mM MgSO4, 2 mM CaCl2 and 0.1 mM EGTA and 1/5 MMR: MMR diluted 5-times with 100 mM NaCl.
Solvents for chromatography
Sol.A1, 0.1% trifluoroacetic acid (TFA); Sol.A2, 750 mM TFA in 80% acetonitrile; Sol.A3, 1 M NaCl in 0.1% TFA; Sol.A4, 2-propanol; Sol.B1, 0.1% TFA; Sol.B2, 0.1% TFA in acetonitrile; Sol.B3, 0.5 M sodium phosphate buffer (pH 8.0) and Sol.B4, 20 mM sodium phosphate buffer (pH 2.3) in 3% formic acid.
Preparation of a cytosol fraction of Xenopus eggs without a Ca2+-stimulus
Xenopus laevis eggs were collected, dejelled and then lysed to prepare a cytosol fraction essentially as described previously (13), as follows. Eggs were dejelled with 2% cysteine-NaOH (pH 8.0) at 23°C. After washing three times with 100 mM NaCl and twice with buffer M at 23°C, the eggs were washed twice with cold buffer M containing 100 mM NaCl and 250 mM sucrose at 4°C. Then the eggs were supplemented with 1 mM phenylmethylsulfonylfluoride and packed into tubes by brief centrifugation at 45 g for 10 min. Excess buffer above the packed eggs was removed and the eggs were then crushed by centrifugation at 15,000 g for 10 min. The supernatant between the lipid cap and pellet, i.e. crude extract, was collected and further centrifuged at 200,000 g for 4 h in an RP55S rotor (Hitachi, Tokyo). The supernatant, i.e. cytosol fraction, was stored at −80°C until use.
Preparation of a cytosol fraction of Xenopus eggs with a Ca2+-stimulus
X.laevis eggs were collected, dejelled and then lysed to prepare a cytosol fraction essentially as described previously (13), as follows. Eggs were dejelled with 2% cysteine-NaOH (pH 8.0) at 23°C. After washing three times with 1/5 MMR and twice with MMR, the eggs were treated with 1 µM (final concentration) A23187 calcium ionophore in MMR at 23°C for 5 min. Then, the eggs were washed three times with 1/5 MMR and left at 23°C for 15 min. Thus, treated eggs were washed three times with cold extraction buffer. The extraction buffer was supplemented with 2 mM 2-mercaptoethanol and 1 mM phenylmethylsulfonylfluoride immediately before use. Eggs were packed into tubes by brief centrifugation at 45 g for 10 min. Excess buffer above the packed eggs was removed and the eggs were then crushed by centrifugation at 15,000 g for 10 min at 4°C. The crude extract, i.e. the supernatant between the lipid cap and pellet, was collected and mixed with 10 µg/ml cytochalasin B. The crude extract was further treated in the same way as for that ‘without a Ca2+-stimulus’ to obtain a cytosol fraction with a Ca2+-stimulus.
A typical phosphopeptide purification
Tryptic digestion of Xenopus egg cytosol protein (4 mg) was carried out in 2 M urea as described previously (11). TFA was added to the digest to lower the pH to 2.3 and then the digest was centrifuged at 10,000 g for 10 min to remove insoluble peptides. A fully automated phosphopeptide purification system (11) was used to purify phosphopeptides, as shown in Fig. 1 and as follows. A soluble peptide mixture was directly applied at 1 ml/min to an Oligo R3 column (4.6 mm I.D. × 100 mm, 30 µm; Applied Biosystems), which was washed with Sol.A1 at 2 ml/min for 30 min for clarification through removal of salts and urea, and then the bound peptides were eluted with Sol.A2 and passed through a titania column (4.0 mm I.D. × 10 mm, 5 µm, Titansphare TiO; GL Science) at 1 ml/min for 3 min and then 0.1 ml/min for 15 min. The titania column was exhaustively washed with Sol.A2, Sol.A3 and Sol.A4 at 0.5 ml/min for 5–7 min each. The washing was further repeated for one cycle. Then, with the 1-step method, phosphopeptides on the titania column were isocratically eluted with Sol.B3 at 0.5 ml/min for 20 min. Whereas, with the 4-step method, bound phosphopeptides were eluted stepwise with mixtures made from Sol.B1, Sol.B3 and Sol.B4 at 0.5 ml/min; Step 1: 5 mM phosphate and pH 2.3 for 5 min; Step 2: 20 mM phosphate and pH 2.4 for 20 min; Step 3: 50 mM phosphate and pH 2.6 for 20 min and Step 4: 500 mM phosphate and pH 8.0 for 20 min. This was followed by elution with Sol.B4 at 0.5 ml/min for 5 min to rapidly lower the pH to ensure the complete binding of peptides to the octadecylsilica (ODS) column (4.6 mm I.D. × 100 mm, 3 µm, Capcell Pak MG2; Shiseido, Tokyo) (11). After washing the column with Sol.B1 at 1 ml/min for 23 min, the phosphopeptides were eluted with 80% Sol.B2 at 1 ml/min for 15 min, with monitoring as to the absorbance at 210 nm, and collected for the first 10 min. The collected samples were dried in vacuo for MS. The 4-step elution from an automated phosphopeptide purification system described earlier was performed using the program presented in a Supplementary Table SI.
Fig. 1.
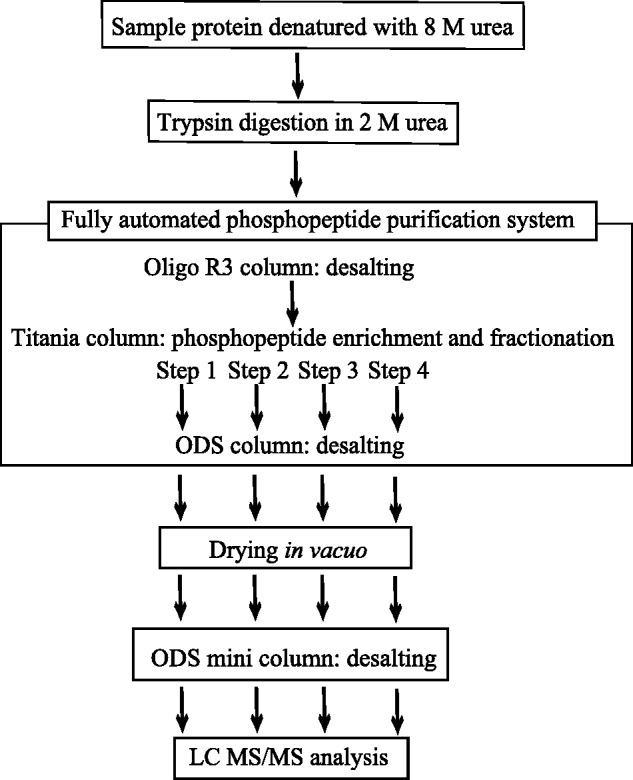
Phosphoproteome analysis by 4-step elution from a fully automated phosphopeptide purification system. A sample protein denatured with 8 M urea was diluted 4 times to reduce the urea concentration and then digested with trypsin. After acidification and removal of insoluble peptide by centrifugation, the thus prepared peptide mixture was directly applied on a fully automated phosphopeptide purification system and eluted in 4-steps. After drying and desalting, the thus purified phosphopeptides were analysed by LC MS/MS.
Nano-flow liquid chromatography-linear ion trap/orbitrap tandem MS
After drying in vacuo, the purified phosphopeptides were dissolved in a small volume of Sol.A1, desalted through a mini ODS column (2.0 mm I.D. × 10 mm, 5 µm, Capcell C18 MG; Shiseido, Tokyo) and then concentrated by solvent evaporation. Subsequently, the phosphopeptides were dissolved in 2% acetonitrile containing 0.1% formic acid and 25 µg/ml EDTA (free acid) to increase the rate of identification of phosphopeptides (14). The phosphopeptides derived from 0.8 mg starting protein were analysed with a nano-flow liquid chromatography-linear ion trap/Orbitrap tandem mass spectrometer (LC MS/MS; LTQ Orbitrap XL; Thermo Fisher Scientific Inc.) equipped with an HPLC nanospray chip integrating a nano LC trap column (0.5 mm I.D. × 1 mm, 3 µm; KYA Technologies Corp.), a nano LC capillary column (75 µm I.D. × 100 mm, 3 µm; KYA Technologies Corp.) and a spray needle. Mobile Phase A comprised 2% acetonitrile containing 0.1% formic acid, and mobile Phase B 98% acetonitrile containing 0.1% formic acid. A 120 min linear gradient from 4 to 33% B followed by a 5 min gradient from 33 to 100% B at a flow rate of 300 nl/min were applied to separate phosphopeptides. The mass spectrometer was operated in the positive ion mode over the 350−2000 mass-to-charge ratio (m/z) range in the data-dependent mode to select the five most intense precursor ions for acquisition of MS/MS spectra. MS detector: Orbitrap; MS/MS detector: linear ion trap; ion transfer capillary: 200°C; electrospray voltage: 2.2 kV; charge states: 2+ and 3+; target value (MS): 2.00 e+06 and target value (MS/MS): 1.00 e+05.
Peptide identification
All MS/MS spectra were searched against the X.laevis subsection of the National Center for Biotechnology Information (NCBI) database using Proteome Discoverer (version 1.1, Thermo Fisher Scientific Inc.). Identification of peptides was performed with the following parameters: enzyme: trypsin; missed cleavages: 2; absolute Xcorr threshold: 0.4; MS tolerance: 10 ppm; MS/MS tolerance: 0.8 Da and dynamic modification: Gln > pyro-Glu (N-term), propionamide (C), oxidation (H, M, W) and phosphorylation (S, T, Y). In addition, confidence values were determined by a target-decoy search strategy to identify phosphopeptides (<0.05 false discovery rate).
Pathway analysis
Phosphoproteins deduced from phosphopeptides determined reproducibly in duplicate analyses of Xenopus egg cytosol digests were selected and pathway analysis was performed with Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) (15) against pathways registered in the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/).
Labelling with iTRAQ reagents
Phosphopeptide samples were purified from tryptic digests of cytosol fractions of Xenopus eggs without and with a Ca2+-stimulus by the one step method (derived from 4 mg protein each), and then labelled separately with a vial of iTRAQ isobaric reagent according to the protocol of ABSciex (114 and 117, respectively; ABSciex, Tokyo). They were then incubated for 1 h and mixed. Ten volumes of Cation Exchange Buffer-Load were added to the mixed sample and then the pH was lowered to 2.5 with 10% formic acid. The sample was injected into a cation-exchange cartridge, washed with 1 ml of the Cation Exchange Buffer-Load and then eluted with Cation Exchange Buffer-Elute. The labelled phosphopeptide fraction was desalted through a mini ODS column (2.0 mm I.D. × 10 mm, 5 µm, Capcell C18 MG; Shiseido), followed by concentration in vacuo.
Determination of relative amounts of phosphopeptides in cytosol digests of Xenopus eggs without and with a Ca2+-stimulus
An iTRAQ-labelled phosphopeptides sample was dissolved in 2% acetonitrile containing 0.1% formic acid and then analysed by LC MS/MS. The procedures and conditions for analysis were the same as those for ‘LC MS/MS’ and ‘Peptide identification’ except for the following. The instrument was operated in the parallel mode, allowing accurate mass measurement of precursors in the iontrap concurrent with the acquisition of data-dependent collision-induced dissociation MS/MS data in the ion trap and high-energy collision-induced dissociation MS/MS data in the Orbitrap for the three most intense precursor ions. Peptide precursor ions were selected with an isolation window of 2 Da and a target value of 1 × 105 at the linear trap quadrupole, and one of 3 Da and a target value of 1 × 106 at the Orbitrap. All MS/MS spectra were searched against the X.laevis subsection of the NCBI database using Proteome Discoverer (version 1.3, Thermo Fisher Scientific Inc.). Dynamic modification: iTRAQ 4plex (Y) and static modification: iTRAQ 4plex (N-term) and iTRAQ 4plex (K) were added to parameters for peptide identification. The ratios of the heights of the 117 (m/z)/114 (m/z) reporter fragment peaks obtained for Ca2+-non-stimulated (114) and stimulated (117) Xenopus egg cytosol proteins were used as indices for changes in the phosphorylation level.
Results
Improved phosphopeptide identification by 4-step elution from a titania column
Stepwise elution from a titania column was performed for a fully automated phosphopeptide purification system (11) to improve phosphopeptide identification. The phosphate concentration of the eluent was increased in 4 steps, as shown in Table I, Condition A, and then the phosphopeptides in each fraction derived from 1.2 mg of starting protein were analysed separately by LC MS/MS (Table I). It was shown that the number of phosphopeptides specifically detected in each step fraction is less than the half of the number of whole phosphopeptides identified in each step fraction (Table I, Condition A). Therefore, a further increase in the number of steps to increase the number of identified phosphopeptides seemed to be not so effective. It was also shown that more than 70% of the phosphopeptides were eluted from the titania column in the first and second steps, i.e. with lower than 50 mM phosphate buffer. Therefore, we tried 4-step elution, Condition B, i.e. 5, 20, 50 and 500 mM phosphate buffer elutions. Phosphopeptides were well dispersed throughout the steps under these conditions (Table I, Condition B). Then, we compared 1- and 4-step elution under Condition B. Phosphopeptides derived from 0.4 mg starting protein and purified under each condition were analysed by LC MS/MS (Table II). The purities of phosphopeptides obtained on 4- and 1-step elution were more than 95%. The number of identified phosphopeptides increased 1.5 times on 4-step elution (Table IIA). In particular, peptides phosphorylated at multiple sites increased 2.2 times (Table IIA). These multiply phosphorylated peptides were tightly bound to the titania column and thus were eluted with higher concentration phosphate buffers (Table IIB). Then, we applied the 4-step elution method to Xenopus egg phosphoproteome analysis.
Table I.
Fractionation of phosphopeptides by stepwise elution from a titania columna
| Elution step | Condition A |
Condition B |
||
|---|---|---|---|---|
| Phosphate concentration (mM) and pH | Number of phosphopeptides detected | Phosphate concentration (mM) and pH | Number of phosphopeptides detected | |
| 1 | 5 (pH 2.3) | 686 (218)b | 5 (pH 2.3) | 638 (201)b |
| 2 | 50 (pH 2.6) | 785 (274)b | 20 (pH 2.4) | 689 (171)b |
| 3 | 200 (pH 3.5) | 395 (112)b | 50 (pH 2.6) | 439 (117)b |
| 4 | 500 (pH 8.0) | 179 (13)b | 500 (pH 8.0) | 444 (133)b |
| Total | 1,161 | 1,174 | ||
aA tryptic digest of a cytosol fraction of Xenopus eggs without a Ca2+-stimulus (4 mg) was applied to an automated phosphopeptide purification system. Phosphopeptides bound on a titania column were eluted stepwise with phosphate buffers, Conditions A or B. The obtained fractions were dried in vacuo and then desalted with ODS tips. The thus clarified phophopeptides derived from 1.2 mg of starting protein were dissolved in 0.1% formic acid-2% acetonitrile and then analysed by LC MS/MS.
bNumbers of phosphopeptides specifically detected in the fraction.
Table II.
Purification of phosphopeptides by 1-step and 4-step elution from a titania columna
| (A) | ||
|---|---|---|
| Kind of phosphopeptide | Number of phosphopeptides detected |
|
| 1-step method | 4-step method | |
| Monophospho-peptide | 383 (83) | 535 (76) |
| Diphospho-peptide | 72 (16) | 130 (18) |
| Triphospho-peptide | 5 (1) | 37 (5) |
| Tetraphospho-peptide | 0 (0) | 4 (1) |
| Total | 460 (100) | 706 (100) |
| (B) | |
|---|---|
| Elution step | Distribution of multiply phosphorylated peptides (%) |
| 1 | 5 |
| 2 | 17 |
| 3 | 36 |
| 4 | 42 |
| Total | 100 |
aA tryptic digest of a cytosol fraction of Xenopus eggs without Ca2+-stimulation (4 mg) was applied to an automated phosphopeptide purification system. Phosphopeptides bound on a titania column were eluted with 1-step or 4-steps under Condition B in Table I with phosphate buffers. The obtained fractions were concentrated in vacuo and then desalted with a small ODS column. The thus clarified phophopeptides derived from 0.4 mg starting protein were dissolved in 0.1% formic acid-2% acetonitrile and then analysed by LC MS/MS. (A) Phosphopeptides detected with the 1-step and 4-step methods were compared. Numbers in parentheses indicate percentages. (B) Distribution of multiply phosphorylated peptides between elution steps with the 4-step method.
Phosphoproteome analysis of Xenopus eggs without and with a Ca2+-stimulus
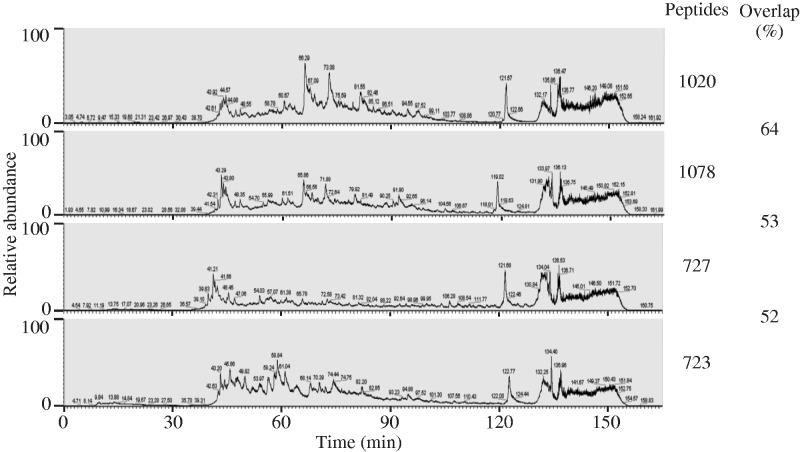
To determine the phosphoproteome profiles of Xenopus eggs without and with a Ca2+-stimulus, phosphopeptides were eluted by the 4-step method from an automated phosphopeptide purification system. Four phosphopeptide fractions derived from cytosol proteins (0.8 mg) of Xenopus eggs without a stimulus were analysed by LC MS/MS. A set of total ion chromatograms obtained on LC MS of these phosphopeptide fractions is shown in Fig. 2. In the first to fourth step fractions, 1,020, 1,078, 727 and 723 phosphopeptides were detected, and some of these phosphopeptides overlapped with each other (Fig. 2). The numbers of unique phosphopeptides determined for a digest of Xenopus eggs without a Ca2+-stimulus were 2,224 and 1,940 in duplicate analyses. Of these phosphopeptides, 1,375 were reproducibly detected in the duplicate analyses (Table III). These phosphopeptides reproducibly determined in the duplicate analyses are listed in a Supplementary Table SII. Phosphopeptides derived from 0.8 mg cytosol protein of Xenopus eggs with a Ca2+-stimulus were also analysed twice by LC MS/MS, 1,341 and 1,504 non-redundant phosphopeptides being detected. Of these phosphopeprides, 994 were detected reproducibly in duplicate analyses (Table III). The latter phosphopeptides are also listed in a Supplementary Table SIII. Among these reproducibly detected phosphopeptides, 818 and 437 were specifically detected in cytosol fractions of Xenopus eggs without and with a Ca2+-stimulus, respectively, and 557 were commonly detected in both fractions (Table III). Proteins containing a site(s) specifically phosphorylated in Xenopus eggs without or with a Ca2+-stimulus are listed in a Supplementary Table SIV.
Fig. 2.
Phosphopeptide detection in a tryptic digest of a cytosol fraction of Xenopus eggs without a Ca2+-stimulus. Phosphopeptides derived from 4 mg cytosolic protein of Xenopus eggs without a Ca2+-stimulus were eluted from the automated phosphopeptide purification system by the 4-step elution method with phosphate buffers, see Condition B in Table I. The four obtained fractions were separately concentrated in vacuo and then desalted with an ODS mini-column. The thus purified phosphopeptide fractions were dissolved in 0.1% formic acid-2% acetonitrile containing 25 µg/ml EDTA, and aliquots corresponding to 0.8 mg starting protein of each fraction were analysed by LC MS/MS. Total ion chromatograms obtained on LC MS of these phosphopeptide fractions eluted from the system in the first to fourth steps are shown from the top to the bottom. The number of phosphopeptides detected in each fraction is indicated on the right of the chromatogram. The percentage of the same phosphopeptide detected in the next fraction is shown as ‘Overlap (%)’ in the figure.
Table III.
Phosphopeptides detected in tryptic digests of cytosol fractions of Xenopus eggs without and with a Ca2+-stimulusa
aCytosol fractions (4 mg protein) of Xenopus eggs without and with a Ca2+-stimulus were digested with trypsin, and the phosphopeptides in each digest were purified by the 4-step elution method as in Fig. 2. The thus purified phosphopeptides derived from 0.8 mg protein were analysed twice for each sample by LC MS/MS as in Fig. 2. Phosphopeptides detected reproducibly in duplicate analyses of each fraction were counted.
bPhosphopeptides specifically detected in the cytosol fraction of eggs without or with a Ca2+-stimulus.
cPhosphopeptides detected in both cytosol fractions.
Phosphopeptides detected reproducibly in tryptic digests of cytosol fractions of Xenopus eggs without and with a Ca2+-stimulus, 1,375 and 994, amounted to 356 and 295 proteins. Pathway analysis of these proteins was performed with DAVID against pathways registered in the KEGG, 9 and 2 pathways being assigned, respectively (Table IV). ‘Progesterone-mediated oocyte maturation’ and ‘mammalian target of rapamycin (mTOR) signalling pathway’ were common for both cytosol fractions, and others were specific to the cytosol of eggs without a Ca2+-stimulus.
Table IV.
Pathways including phosphoproteins detected in cytosol fractions of Xenopus eggs without and with a Ca2+-stimulusa
| Pathway | Phosphoproteins included in the pathway | P-value* | |
|---|---|---|---|
| Without Ca2+-stimulus | |||
| Progesterone-mediated oocyte maturation | 9 | 2.04E-03 | |
| mTOR signalling pathway | 6 | 2.16E-03 | |
| Focal adhesion | 11 | 4.07E-03 | |
| DNA replication | 6 | 5.71E-03 | |
| ErbB signalling pathway | 7 | 8.50E-03 | |
| Mismatch repair | 4 | 2.09E-02 | |
| Homologous recombination | 4 | 2.37E-02 | |
| Base excision repair | 4 | 2.99E-02 | |
| Pyrimidine metabolism | 6 | 4.85E-02 | |
| With Ca2+-stimulus | |||
| mTOR signalling pathway | 8 | 1.99E-05 | |
| Progesterone-mediated oocyte maturation | 10 | 2.93E-04 | |
aThe results of pathway analysis of 356 and 295 phosphoproteins, to which 1,375 and 994 phosphopeptides found in digests of cytosol fractions of Xenopus eggs without and with a Ca2+-stimulus are assigned, were summarized.
*P-value < 0.05.
iTRAQ analysis of protein phosphorylation in Xenopus eggs without and with a Ca2+-stimulus
Phosphopeptide fractions purified by the 1-step elution method with an automated phosphopeptide purification system from the tryptic digests of cytosol fractions of Xenopus eggs without and with a Ca2+-stimulus were labelled separately with 114 and 117 iTRAQ reagents and then mixed. After desalting, the labelled peptides were analysed by LC MS/MS. The 117/114 ratios of the unique 112 phosphopeptides were determined and are listed in a Supplementary Table SV. The distribution of the 117/114 ratios is shown as a histogram in Fig. 3. The median of the ratios was 0.758. The top ten phosphopeptides having small 117/114 ratios (highly down-regulated) and those having large ratios (highly up-regulated) were selected, and are shown in Table V. In this analysis, a protein related to DNA replication: RNASEH2B and a protein related to pyrimidine metabolism: CTBS1-B were shown to be highly down- and up-regulated, respectively. Pathways containing these proteins were also detected on pathway analysis based on phosphopeptides derived from a cytosol digest of Xenopus eggs without a Ca2+-stimulus (Table IV).
Fig. 3.
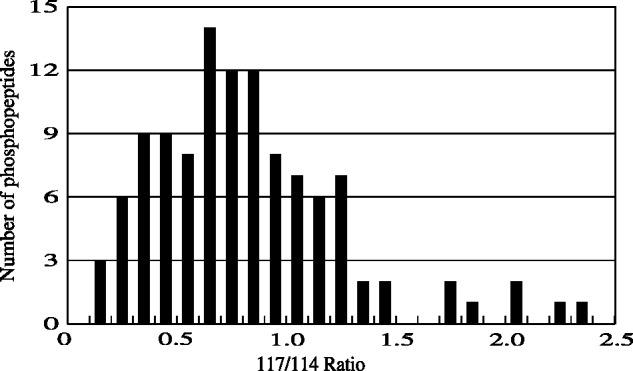
A histogram of the ratios of iTRAQ-labels of phosphopeptides derived from cytosolic proteins of Xenopus eggs without and with a Ca2+-stimulus. Phosphopeptides in tryptic digests of cytosol fractions (3 mg protein) of Xenopus eggs without and with a Ca2+-stimulus were purified with an automated phosphopeptide purification system with 1-step elution method, and then modified with 114- and 117-iTRAQ, respectively. The thus labelled phosphopeptides were mixed, desalted and dissolved in the EDTA-containing solvent. The labelled phosphopeptides derived from 1.6 mg protein were analysed three times by LC MS/MS, and the 117/114 ratios were determined. The histogram was made using 112 phosphopeptides reproducibly detected three times.
Table V.
Ratios of iTRAQ-labelled phosphopeptides purified from tryptic digests of the cytosol fractions of Xenopus eggs without and with a Ca2+-stimulusa
| No. | Protein Accession | Symbol | Peptide sequenceb | Sitec | Name | Ratio 1 | Ratio 2 | Ratio 3 | Average | SD | Pathway/Function |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Highly down-regulated phospho-sites | |||||||||||
| 1 | gi:68534867 | MGC84800 | YRsDNEAFEEK | S12 | MGC84800 protein | 0.119 | 0.125 | 0.114 | 0.119 | 0.006 | vitamin B6 methabolism |
| 2 | gi:147905882 | LOC100049128 | SYPFDGVsPLQLK | S12 | hypothetical protein LOC100049128 | 0.139 | 0.179 | 0.173 | 0.164 | 0.023 | |
| 3 | gi:28422169 | - | TIGGGDDsFNTFFSETGAGK | S48 | Tuba6-prov protein | 0.140 | 0.166 | 0.270 | 0.192 | 0.074 | gap junction |
| 4 | gi:108935850 | MCM2 | GLLYDsDEEEEDRPAR | S123 | DNA replication-licensing factor mcm2 | 0.209 | 0.214 | 0.213 | 0.212 | 0.003 | gap junction, cell cycle |
| 5 | gi:82209768 | DBNL-A | LRsPFLQK | S281 | drebrin-like protein A | 0.152 | 0.236 | 0.267 | 0.219 | 0.064 | |
| 6 | gi:148226825 | ABCF1 | sFFEELSADNK | S37 | ATP-binding cassette, sub-family F, member 1 | 0.231 | 0.205 | 0.244 | 0.227 | 0.022 | |
| 7 | gi:147900770 | RNASEH2B | RVsDAPVEADEDYTK | S250 | ribonuclease H2 subunit B | 0.212 | 0.262 | 0.234 | 0.236 | 0.027 | DNA replication |
| 8 | gi:117949815 | MAPK1 | VADPDHDHTGFLtEYVATR | T188 | MAPK 1 | 0.252 | 0.252 | 0.279 | 0.261 | 0.017 | MAPK signalling |
| 9 | gi:148227966 | SIVA1 | SYPFDSVsPLQLK | S12 | SIVA1, apoptosis-inducing factor | 0.062 | 0.500 | 0.230 | 0.264 | 0.237 | |
| 10 | gi:148227449 | LDLRAP1-B | AFDVLNAsDNHIEDLFR | S229 | low-density lipoprotein receptor adapter protein 1-B | 0.302 | 0.353 | 0.271 | 0.309 | 0.045 | endocytosis |
| Highly up-regulated phospho-sites | |||||||||||
| 1 | gi:148229878 | PNPO | YRsDNESFEEK | S12 | pyridoxamine 5'-phosphate oxidase | 1.386 | 1.309 | 1.366 | 1.354 | 0.043 | vitamin B6 metabolism |
| 2 | gi:57032988 | NVSTFPDEPTsPLQEK | S58 | importin subunit alpha | 1.440 | 1.239 | 1.621 | 1.433 | 0.205 | protein transport | |
| 3 | gi:148226825 | ABCF1 | LSVQAsDEEPDEAPAPK | S105 | ATP-binding cassette, sub-family F, member 1 | 1.391 | 1.486 | 1.470 | 1.449 | 0.055 | |
| 4 | gi:148222844 | TFDP1 | VFVDQNLsPGK | S23 | transcription factor Dp-1 | 1.294 | 1.900 | 2.019 | 1.738 | 0.417 | cell cycle, TGF-beta signalling |
| 5 | gi:82176639 | CTPS1-B | SENSsPDAEIAELK | S575 | CTPS 1-B | 2.158 | 1.844 | 1.345 | 1.783 | 0.440 | pyrimidine metabolism |
| 6 | gi:115528686 | LOC100158391 | QSSSVEQPASPsQSPK | S20 | LOC100158391 protein | 2.220 | 1.457 | 1.968 | 1.881 | 0.416 | |
| 7 | gi:148234990 | USP8 | HPFAAGKEPsEPK | S529 | ubiquitin carboxyl-terminal hydrolase | 2.611 | 1.859 | 1.676 | 2.049 | 0.531 | ubiquitin conjugation |
| 8 | gi:1168995 | CFL1-B | HQLsPEEAK | S25 | cofilin-1-B | 2.077 | 2.046 | 2.059 | 2.061 | 0.017 | regulation of actin cytoskeleton |
| 9 | gi:586046 | STMN1-A | RAsGQAFELILSPPSMDAAPDLSITsPK | S16, S39 | stathmin-1-A | 1.830 | 2.781 | 2.148 | 2.253 | 0.519 | MAPK signalling |
| 10 | gi:66269747 | NAP1L1-A | VEEEDIsGDLK | S143 | nucleosome assembly protein 1 | 2.686 | 1.972 | 2.263 | 2.307 | 0.385 | nucleosome assembly |
aPhosphopeptides in tryptic digests of cytosol fractions of Xenopus eggs without and with a Ca2+-stimulus (3 mg protein each) were purified with an automated phosphopeptide purification system with the 1-step elution method, and then modified with 114- and 117-iTRAQ, respectively. The labelled phosphopeptides were mixed, desalted and then dissolved in an EDTA-containing solvent. Aliquots of labelled phosphopeptides derived from 1.6 mg protein were analysed three times by LC MS/MS, and then the 117/114 ratios were determined. Phosphopeptides of which the 117/114 ratio was reproducibly determined in triplicate analyses numbered 112 (Supplementary Table SV). Among these phosphopeptides, 10 highly down-regulated and 10 highly up-regulated phosphosites are included.
bLower-case letters indicate phosphorylated amino acid residues.
cPhosphorylated sites are shown as residue numbers from the amino-terminal methionine of the original protein.
Discussion
A 4-step elution method for a fully automated phosphopeptide purification system
A stepwise pH-gradient elution method involving a titania column has been reported, and it has been shown that the method is superior to the 1-step elution method for the determination of multiply phosphorylated peptides (16). A pH gradient of between 9.2 and 11.3 is applied for this method. This pH range was difficult to apply to our automated system because when such high pH eluents are directly applied online to an ODS column for desalting, complete binding of phosphopeptides to the column could be difficult (11). Kyono et al. (17) applied successive elution with a 5% ammonium hydroxide solution, a 5% piperidine solution and a 5% pyrrolidine solution in series for phosphopeptide enrichment with titania, obtaining a 1.6-fold increase in the identified phosphopeptide number in comparison with the result obtained with 1-step elution. These elution conditions were also difficult to apply to our automated system because i) many kinds of solvents are necessary and ii) strong alkaline eluents are not suitable for subsequent online desalting with an ODS column (11). In this study, we chose 4-step elution with phosphate buffers made by mixing two kinds of phosphate buffers, of which the pHs were lower than 8, and a TFA solution, the phosphate buffer concentrations thereby being optimized (Table I). Then, a 1.5-fold increase in the number of phosphopeptides detected in comparison with the result obtained with the 1-step elution method was obtained with the fully automated system (Table IIA). Multiply phosphorylated peptides detected increased from 17 to 24% of whole phosphopeptides with the 4-step elution (Table IIA). Multiply phosphorylated peptides were more abundant in the third and fourth step fractions than the first and second ones (Table IIB). So, it was suggested that suppression of ionization of multiply phosphorylated peptides by mono-phosphorylated peptides on MS was reduced in the samples prepared by the 4-step elution method. In addition to the enhanced resolution of mono-phosphorylated peptides, the stepwise elution method was especially useful for identification of multiply phosphorylated sites that may be linked to the receptor signalling process because kinases involved in the process often require multi-site phosphorylation for their activation (18, 19). A fully automated phosphopeptide purification system involving optimized 4-step elution from a titania column is more useful than the original 1-step method for less-biased phosphoproteome analysis.
Validation of phosphorylation sites
To verify the phosphorylation sites determined on MS in this study, sites detected in cytosol proteins in Xenopus eggs without a Ca2+-stimulus were compared with the corresponding phosphorylation sites detected using site-specific methods in human proteins listed in the PhosphoSitePlus database (http://www.phosphosite.org) (20). As summarized in Table VI, many phosphorylation sites determined in Xenopus proteins on MS in this study corresponded to verified phosphorylation sites in human proteins. These results show that the phosphorylation sites detected with our MS method reflect protein phosphorylation in Xenopus eggs. McGivern et al. (4) identified 350 phosphopeptides derived from 224 proteins in a whole extract of Xenopus eggs without a Ca2+-stimulus. We detected 1,375 phosphopeptides derived from 356 proteins in a cytosol fraction of Xenopus eggs without a Ca2+-stimulus reproducibly in duplicate analyses (Supplementary Table SII). Though some phosphopeptides in our and McGivern’s studies overlapped, most were unique in each study. Some of the differences in determined phosphorylation sites between our and McGivern’s studies should be due to the differences in the samples, i.e. a cytosol fraction versus whole eggs, and the phosphopeptide purification method, i.e. titania chromatography versus immobilized metal ion affinity chromatography. So, the list of phosphopeptides derived from Xenopus egg proteins was increased by these studies. These data should be useful for Xenopus egg studies.
Table VI.
Validation of phosphorylation sites on proteins in Xenopus eggs without a Ca2+-stimulusa
| No. |
Xenopus protein |
Human protein |
|||||
|---|---|---|---|---|---|---|---|
| Accession | Name | Site | Accession | Site | SSb | MSc | |
| 1 | gi108935850 | MCM2 | S123 | gi41019490 | S139 | 4 | 370 |
| 2 | gi117949815 | MAPK 1 | T188 | gi119554 | T185 | 974 | 2447 |
| 3 | gi117949815 | MAPK 1 | T193 | gi119554 | T190 | 1 | 25 |
| 4 | gi117949815 | MAPK 1 | Y200 | gi119554 | Y187 | 994 | 4041 |
| 5 | gi117949815 | MAPK 1 | T184 | gi119554 | T181 | 4 | 41 |
| 6 | gi125693 | MAPK-activated protein kinase 1 | S380 | gi20178306 | S380 | 46 | 90 |
| 7 | gi125959 | Lamin-B3 | S389 | gi23503078 | S385 | 1 | 36 |
| 8 | gi125959 | Lamin-B3 | S391 | gi23503078 | S387 | 1 | 29 |
| 9 | gi127855 | N-CAM-1-A | S768 | gi205830665 | S784 | 2 | 19 |
| 10 | gi147900510 | heat shock protein 90 kDa alpha | S260 | gi92090606 | S263 | 4 | 217 |
| 11 | gi147902292 | TTK protein kinase | S847 | gi160112977 | S824 | 1 | 1 |
| 12 | gi147905302 | splicing factor 1 | S72 | gi42544125 | S82 | 1 | 57 |
| 13 | gi148226260 | 4EBP 2 | T42 | gi34921510 | T46 | 1 | 86 |
| 14 | gi148226260 | 4EBP 2 | T33 | gi34921510 | T37 | 1 | 62 |
| 15 | gi148228921 | ribosomal protein S6 kinase, 90 kDa, polypeptide 3 | T362 | gi20178306 | T359 | 35 | 17 |
| 16 | gi148228921 | ribosomal protein S6 kinase, 90 kDa, polypeptide 3 | S366 | gi20178306 | S363 | 36 | 25 |
| 17 | gi148230727 | cyclin-dependent kinase 11B | T744 | gi34978359 | T751 | 1 | 37 |
| 18 | gi148230727 | cyclin-dependent kinase 11B | S754 | gi34978359 | S752 | 1 | 36 |
| 19 | gi148237217 | heterogeneous nuclear ribonucleoprotein H2 (H') | S103 | gi2500576 | S104 | 2 | 165 |
| 20 | gi232034 | Elongation factor 1-beta | S108 | gi119163 | S106 | 3 | 483 |
| 21 | gi232034 | Elongation factor 1-beta | S114 | gi119163 | S112 | 1 | 11 |
| 22 | gi28385985 | Dnm1l | S578 | gi125987821 | S616 | 11 | 145 |
| 23 | gi284521569 | 3-phosphoinositide dependent protein kinase-1 | S192 | gi17380162 | S241 | 43 | 293 |
| 24 | gi54873686 | Hsp90 beta | S226 | gi17865718 | S226 | 3 | 148 |
| 25 | gi54873686 | Hsp90 beta | S254 | gi17865718 | S255 | 4 | 481 |
| 26 | gi82176749 | NDRG2 | S342 | gi20141615 | S350 | 1 | 285 |
| 27 | gi82176749 | NDRG2 | T340 | gi20141615 | T348 | 2 | 192 |
| 28 | gi82180448 | CTPS 1-A | S571 | gi20981706 | S571 | 1 | 24 |
| 29 | gi82180448 | CTPS 1-A | S574 | gi20981706 | S574 | 1 | 31 |
| 30 | gi82180448 | CTPS 1-A | S575 | gi20981706 | S575 | 2 | 49 |
| 31 | gi82184926 | UBX domain-containing protein 1-A | S204 | gi30923268 | S199 | 1 | 9 |
| 32 | gi82184926 | UBX domain-containing protein 1-A | S205 | gi30923268 | S200 | 2 | 10 |
| 33 | gi82197988 | Septin-2B | S217 | gi2500769 | S218 | 3 | 620 |
| 34 | gi82209768 | Drebrin-like protein A | S281 | gi51316115 | S283 | 1 | 75 |
| 35 | gi82236803 | Protein kinase Akt-2-B | S132 | gi1170703 | S128 | 1 | 0 |
aPhosphorylation sites detected in this study on proteins of a cytosol fraction of Xenopus eggs without a Ca2+-stimulus were compared with the corresponding phosphorylation sites detected using site-specific methods in human proteins listed in the PhosphoSitePlus database: http://www.phosphosite.org (20).
bNumber of records in which this modification site was detected using site-specific methods. The method included amino acid sequencing, site-directed mutagenesis, modification site-specific antibodies, specific MS strategies, etc.
cNumber of records in which this modification site was assigned using only proteomic discovery-mode MS.
Proteins phosphorylated at multi-sites
When phosphopeptides derived from 0.8 mg protein were subjected to analysis by LC MS/MS, 1,375 phosphopeptides were reproducibly detected on duplicate MS in the digest of the cytosol fraction of eggs without a Ca2+-stimulus, i.e. eggs in the second meiotic metaphase (Supplementary Table SII). These phosphopeptides included 391 multiply phosphorylated peptides (28%) derived from 80 proteins (Supplementary Table SVI). It is known that proteins containing closely related multi-phosphorylation sites are important in signalling pathways and frequently act as nodes at which two or more signalling pathways are integrated or crossed (21). For example in the case of connexin, it is phosphorylated by mitogen-activated protein kinase (MAPK) at S255, S279 and 282 (22), and the phosphorylation causes the down-regulation of gap junctional communication (23). During mitosis, connexin is phosphorylated by cdc2 kinase in the overlapped region, i.e. S255 and S262, and the phosphorylation seems to correlate with gap junction internalization (24, 25). In this case, the MAPK and cdc2 kinase signalling pathways cross through phosphorylation of the region. Then, we searched the literature on the functions of 80 proteins containing multiply phosphorylated peptides derived from the cytosol fraction of Xenopus eggs without a Ca2+-stimulus. The functions of 53 out of 80 proteins are known (Supplementary Table SVI). These 53 proteins were classified according to their known functions. ‘Proteins related to signalling’ numbered 15 and formed the largest class, followed by ‘proteins related to the cell cycle’: 11, ‘proteins related to transcription’: 10, ‘proteins related to transport’: 6, ‘structural proteins’: 5, ‘proteins related to replication, repair and recombination of DNA’: 4 and ‘proteins related to ribosome assembly’: 1 (Supplementary Table SVI). Many multiply phosphorylated peptides derived from eukaryotic translation initiation factor 4E binding protein 2 (4EBP2) were included (Supplementary Table SVI, gi148226260). These peptides showed that the protein was phosphorylated at two residues in the first cluster: S24, Y30, T32, T33, T37, S40, T41, T42 and T46 and two residues in the second cluster: T66 and T60/S61. The positions of these residues are numbered from the N-terminal methionine of the protein. It is known that the phosphorylation sites of mammalian 4EBP2 are very similar with those of 4EBP1. 4EBP1 has a very similar amino acid sequence to that of 4EBP2, and most Ser, Thr and Tyr residues are conserved in the two proteins (26). Furthermore, it is known that these two mammalian proteins regulate the assembly of initiation complexes required for cap-dependent translation (27). In human 4EBP1, T37, T46, S65 and T70, which correspond to T33, T41, S61 and T66 in Xenopus 4EBP2, are phosphorylated by mTOR, which regulates the binding of 4EBP1 and eukaryotic translation initiation factor 4E (4E) (28). Human 4EBP1 is also phosphorylated at T70 and S65, which correspond to T66 and S61 in Xenopus 4EBP2, by cdc2 and other kinases, respectively, in the mitotic phase, and the phosphorylation affects the interaction of 4EBP1 with 4E (29). It is also known in human 4EBP1 that phosphorylation of S65, T70 and T37/46, which correspond to S61, T66 and T33/42 in Xenopus 4EBP2, is regulated through the MAPK and mTOR signalling pathways (30). These results suggest that the MAPK and mTOR signalling pathways are integrated into the first multi-phosphorylation cluster but that the cdc2 and mTOR signalling pathways are integrated into the second multi-phosphorylation cluster. In the case of 4EBP2 in Xenopus eggs, we showed that the protein is phosphorylated at T66, and also possibly at S61 and T33/42, corresponding to those of human 4EBP1, as shown earlier. These results suggested that 4EBP2 in Xenopus egg acts in a similar manner to human 4EBP1 to integrate signalling pathways: MAPK, mTOR and cdc2, at these multi-phosphorylation clusters. In some cases of function-unknown proteins containing multiply phosphorylated peptides, i.e. LOC733387 protein (gi213625101), MGC80197 protein (gi47938703) and MGC79091 protein (gi49114782), these phosphorylation sites are conserved in human homologues. Therefore, these proteins in Xenopus and man may be regulated by phosphorylation through these sites.
Differences in phosphoproteome of eggs without and with a Ca2+-stimulus
Proteins phosphorylated specifically in Xenopus eggs without or with a Ca2+-stimulus included many kinds of structural proteins, signalling related proteins, cell cycle-related proteins, etc., and function-unknown proteins (Supplementary Table SIV). For more precise comparison, differences in phosphorylation of cytosol proteins of Xenopus egg without and with a Ca2+-stimulus were analysed by iTRAQ (Supplementary Tables SV and Table V). The phosphorylation ratio was decreased for many proteins and increased for some proteins. The median of the ratios was 0.758. In both lists, i.e. Supplementary Tables SIV and SV, many kinds of protein kinases and protein phosphatases related to signalling are included. Then, phosphorylated protein kinases and protein phosphatases detected specifically in eggs without or with a stimulus (Table VII), or of which the phosphorylation levels were determined by iTRAQ (Table VIII) were listed. It was shown that the phosphorylation levels of MAPK1 at T188 and Y190 were lower in eggs with a Ca2+-stimulus (Table VIII). This kinase is known to be involved in the MAPK cascade, and phosphorylation of the human enzyme at corresponding sites causes activation of the signalling cascade (31). It is also known that MAPK1 is phosphorylated and active in unfertilized eggs, and dephosphorylated and inactivated through calcium influx after fertilization, and thus the MAPK pathway is inactivated (32). Our earlier observations were consistent with those for well-known MAPK pathway regulation through phosphorylation and dephosphorylation in eggs.
Table VII.
Protein kinases and phosphatases containing a site(s) phosphorylated specifically in Xenopus eggs without or with a Ca2+-stimulusa
|
Xenopus protein |
Human orthologue protein |
||||
|---|---|---|---|---|---|
| No. | Accession | Name | Sequence and siteb | Accession | Site |
| Protein kinases and phosphatases phosphorylated specifically in Xenopus egges without Ca2+-stimulus | |||||
| 1 | gi147905280 | maternal embryonic leucine zipper kinase | KPIGTGEEFANVIsPERR, S498; SVELDLNQAHIDsAQK, S517 | gi7661974 | S498, E517 |
| 2 | gi148233350 | protein phosphatase 1, regulatory (inhibitor) Subunit 13-like | FPDDVSsPR, S230 | gi 92090607 | |
| 3 | gi148235999 | serine/threonine-protein Phosphatase 4 regulatory Subunit 3 | DPSVDItQDLVDESEEER, T110 or DPSVDITQDLVDEsEEER, S117 | gi39930397 | T110 or S117 |
| 4 | gi160420263 | p21-activated Kinase 2 | GSEPSTAAtDDDDFDDDKAPPPAIAPRPEHTK, T163 | gi143811432 | T169 |
| 5 | gi39573638 | serine/threonine protein kinase ARAF | GAsVSEYPYAEGK, S12 | gi4502193 | |
| 6 | gi4033698 | dual specificity MAPK kinase 1 | KPtPIQLNPNPEGTAVNGTPTAETNLEALQK, T7; KPTPIQLNPNPEGtAVNGTPTAETNLEALQK, T18; KPTPIQLNPNPEGTAVNGtPTAETNLEALQK, T23; KPTPIQLNPNPEGTAVNGTPtAETNLEALQK, T25; or KPTPIQLNPNPEGTAVNGTPTAEtNLEALQK, T28 | gi400274 | T7, S18, T23, S25, or T28 |
| 7 | gi5733091 | MAPK activator XMEK3 | EAFDQPQVsSPTPPR, S26; EAFDQPQVSsPTPPR, S27; or EAFDQPQVSSPtPPR, T29 | gi15080540 | S26, S27, or T28 |
| 8 | gi82182282 | Serine/threonine-protein Phosphatase 4 regulatory Subunit 2-B | SLSDSAVFDDGSQATtPK, T225 | gi158564082 | S226 |
| Protein kinases and phosphatase phosphorylated specifically in Xenopus egges with Ca2+-stimulus | |||||
| 9 | gi11527160 | calcium/calmodulin-dependent protein kinase II variable region of gamma X isoform | GAILTTMLVsR, S28 | gi325197141 | S311 |
| QtSAPVVAATSAANLVEQAAK, T37 or QTsAPVVAATSAANLVEQAAK, S38 | S320 or S321 | ||||
| 10 | gi148236179 | MAPK 12 | HTDsEMTGYVVTR, S180; HTDSEMtGYVVTR, T183; or HTDSEMTGyVVTR, Y185 | gi2851522 | S180, T183, or Y185 |
| ItGTPTQDFVQK, T242; ITGtPTQDFVQK, T244; or ITGTPtQDFVQK, T246 | T242, T244, or P246 | ||||
| 11 | gi82225833 | serine/threonine-protein kinase ULK3 | SESLGQEVLSEsVR, S460 | gi259016166 | S464 |
aProtein kinases and phosphatases containing the phosphopeptides detected specifically in the tryptic digest of the cytosol fraction of Xenopus eggs without or with a Ca2+-stimulus in Table III and Supplementary Table SIV were selected. Phosphopeptides detected reproducibly in duplicate analyses on LC MS/MS were counted.
bLower-case letters indicate phosphorylated amino acid residues.
Table VIII.
Ratios of iTRAQ-labelled phosphopeptides derived from protein kinases and protein phosphatases in Xenopus eggs without and with a Ca2+-stimulusa
|
Xenopus protein |
Human orthologue protein |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. | Accession | Name | Sequence and siteb | 117/114 Ratio |
Accession | Site | ||||
| 1 | 2 | 3 | Average | SD | ||||||
| 1 | gi117949815 | MAPK 1 | VADPDHDHTGFLtEYVATR, T188 | 0.252 | 0.252 | 0.279 | 0.261 | 0.017 | gi66932916 | T185 |
| 2 | gi117949815 | MAPK 1 | VADPDHDHTGFLtEyVATR, T188, Y190 | 0.448 | 0.565 | 0.328 | 0.447 | 0.127 | gi66932916 | T185, Y187 |
| 3 | gi82209768 | ribosomal protein S6 Kinase 2 alpha | GFsFVAPALVEEDAK, S380 | 0.431 | 0.489 | 0.671 | 0.530 | 0.134 | gi292457 | S380 |
| 4 | gi148228921 | ribosomal protein S6 kinase, 90 kDa, polypeptide 3 | DsPGIPAsANAHQLFR, S366, S372 | 0.531 | 0.589 | 0.662 | 0.594 | 0.070 | gi1730070 | S369, S375 |
| 5 | gi147902292 | TTK protein kinase | ILGQLIGLNsPNSISR, S844 | 0.789 | 0.701 | 0.613 | 0.701 | 0.094 | gi23308722 | S821 |
| 6 | gi125694 | ribosomal protein S6 Kinase 2 beta | GFsFVAPVLVEEDAK, S380 | 0.917 | 0.646 | 1.364 | 0.976 | 0.389 | gi15929013 | S380 |
| 7 | gi148228921 | ribosomal protein S6 kinase, 90 kDa, polypeptide 3 | DsPGIPASANAHQLFR, S366 | 0.948 | 1.154 | 1.215 | 1.106 | 0.150 | gi1730070 | S369 |
| 8 | gi148225112 | casein kinase 1, epsilon | ISAsQAsVPFDHLGK, S405, S408 | 1.672 | 0.984 | 1.178 | 1.278 | 0.380 | gi23199991 | S405, S408 |
aPhosphopeptides in tryptic digests of cytosol fractions (3 mg protein) of Xenopus eggs without and with a Ca2+-stimulus were purified with an automated phosphopeptide purification system with the 1-step method, and then modified with 114- and 117-iTRAQ, respectively. The labelled phosphopeptides were mixed, desalted and then dissolved in a EDTA-containing solvent. The labelled phosphopeptides derived from 1.6 mg protein were analysed three times by LC MS/MS, and then the 117/114 ratios were determined. The 117/114 ratios of phosphopeptides derived from protein kinases and protein phosphatases, and reproducibly quantified three times are listed.
bLower-case letters indicate phosphorylated amino acid residues.
It is known that casein kinase I epsilon regulates developmental and oncogenic processes as a positive regulator of the Wnt signalling pathway (33). The enzymatic activity of casein kinase I epsilon is known to be suppressed by autophosphorylation (34). We observed that the phosphorylation levels of the Xenopus enzyme at S405 and S408, which correspond to human enzyme autophosphorylation sites S405 and S408, were elevated in eggs with a Ca2+-stimulus (Table VIII). These results suggest that casein kinase I epsilon is active in Xenopus eggs without a Ca2+-stimulus and suppressed by Ca2+-stimulation through autophosphorylation.
It is also know that the Sonic hedgehog signalling pathway is important in embryogenesis (35). In this pathway, human serine/threonine-protein kinase ULK3 is autophosphorylated at several serine and threonine residues including S464, and thereby is activated (36). The activity is crucial for its function in the pathway (37). We observed the phosphorylation of Xenopus egg ULK3 at S460, which corresponds to S464 of human ULK3, specifically in Ca2+-stimulated eggs (Table VII). This suggests that the Sonic hedgehog signalling pathway is activated by Ca2+-stimulation in Xenopus eggs.
We detected phosphorylation of the Xenopus calcium/calmodulin-dependent protein kinase II variable region of the gamma X isoform, partial (gi11527160) at S28 and T37/S38 specifically in eggs with a Ca2+-stimulus (Table VII). S28 is included in the common region, while T37 and S38 are involved in an alternative splicing region of the enzyme. It is known in mouse eggs that calcium/calmodulin-dependent protein kinase II gamma is activated by calcium/calmodulin through autophosphorylation at T287 and controls egg activation by regulating cell cycle resumption (38). Phosphorylation at T37 or S38 of the enzyme in Xenopus eggs may be involved in the egg activation in an isoform-specific manner, though these are different from the earlier autophosphorylation residues.
Phosphorylation of Xenopus serine/threonine-protein Phosphatase 4 regulatory Subunit 2-B at T225 was observed specifically in eggs without a Ca2+-stimulus (Table VII). Serine/threonine-protein Phosphatase 4 regulatory Subunit 2 is localized to the centrosome and its complex with the protein phosphatase catalytic subunit is known to be essential for maturation of the centrosome, though the regulation mechanism is not known (39). So, further study on the phosphorylation of the subunit in Xenopus eggs may provide some clues elucidating the mechanism.
It is also known that p21-activated Kinase 2 is phosphorylated and inactivated during the process of Xenopus egg maturation (40). When eggs are stimulated with Ca2+ ions, p21-activated kinase is dephosphorylated and activated (40). We observed phosphorylation of Xenopus p21-activated Kinase 2 at T163 specifically in eggs without a Ca2+-stimulus but not in eggs with a Ca2+-stimulus (Table VII). It would be of interest to determine whether the phosphorylation is involved in the regulation of the kinase activity or not.
As seen earlier, our phosphoproteome data reflected the changes during egg activation. Therefore, the data should contribute to future studies on the Xenopus egg activation mechanism and also on mammalian egg activation for which available materials for studies are limited.
Supplementary Data
Supplementary Data are available at JB Online.
Acknowledgements
The authors thank Dr Kentaro Kaneko, Graduate School of Science and Technology, Niigata University, for his excellent help in the LC MS/MS analyses.
Funding
This study was supported by a Grant-in-Aid for Scientific Research (Exploratory Research 19651085 to T. H.) from the Japan Society for the Promotion of Science and a Grant for Research for Promoting Technological Seeds (04-022 to T. H.) from the Japan Science and Technology Agency.
Conflict of Interest
None declared.
Glossary
Abbreviations
- 4EBP
eukaryotic translation initiation factor 4E binding protein
- iTRAQ
isobaric tags for relative and absolute quantitation
- LC MS/MS
nano-flow liquid chromatography-linear ion trap/Orbitrap tandem mass spectrometry
- MAPK
mitogen-activated protein kinase
- MS
mass spectrometry
- mTOR
mammalian target of rapamycin
- m/z
mass-to-charge ratio
- ODS
octadecylsilica
- TFA
trifluoroacetic acid
References
- 1.Hormanseder E., Tischer T., Mayer T.U. (2013) Modulation of cell cycle control during oocyte-to-embryo transitions. EMBO J. 32, 2191–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radford H.E., Meijer H.A., de Moor C.H. (2008) Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim. Biophys. Acta 1779, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krauchunas A.R., Wolfner M.F. (2013) Molecular changes during egg activation. Curr. Top. Dev. Biol. 102, 267–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGivern J.V., Swaney D.L., Coon J.J., Sheets M.D. (2009) Toward defining the phosphoproteome of Xenopus laevis embryos. Dev. Dyn. 238, 1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tunquist B. J., Maller J. L. (2003) Under arrest: cytostatic factor (CSF)-mediated metaphase arrest in vertebrate eggs. Genes Dev. 17, 683–710 [DOI] [PubMed] [Google Scholar]
- 6.Tsai C.F., Hsu C.C., Hung J.N., Wang Y.T., Choong W.K., Zeng M.Y., Lin P.Y., Hong R.W., Sung T.Y., Chen Y.J. (2014) Sequential phosphoproteomic enrichment through complementary metal-directed immobilized metal ion affinity chromatography. Anal. Chem. 86, 685–693 [DOI] [PubMed] [Google Scholar]
- 7.Tape C.J., Worboys J.D., Sinclair J., Gourlay R., Vogt J., McMahon K.M., Trost M., Lauffenburger D.A., Lamont D.J., Jorgensen C. (2014) Reproducible automated phosphopeptide enrichment using magnetic TiO2 and Ti-IMAC. Anal. Chem. 86, 10296–10302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruhler A., Olsen J.V., Mohammed S., Mortensen P., Faergeman N.J., Mann M., Jensen O.N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. (2007) Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics 6, 1103–1109 [DOI] [PubMed] [Google Scholar]
- 10.Thingholm T.E., Jensen O.N., Larsen M.R. (2009) Analytical strategies for phosphoproteomics. Proteomics 9, 1451–1468 [DOI] [PubMed] [Google Scholar]
- 11.Iwase Y., Honma S., Matsuzaki M., Miyakawa Y., Kanno T., Ishii K., Furuichi N., Furukawa K., Horigome T. (2010) A fully automated phosphopeptide purification system for large-scale phosphoproteome analysis. J. Biochem. 147, 689–696 [DOI] [PubMed] [Google Scholar]
- 12.Pinkse M.W.H., Mohammed S., Gouw J.W., van Breukelen B., Vos H.R., Heck A.J.R. (2008) Highly robust, automated, and sensitive online TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J. Proteome Res. 7, 687–697 [DOI] [PubMed] [Google Scholar]
- 13.Takano M., Takeuchi M., Ito H., Furukawa K., Sugimoto K., Omata S., Horigome T. (2002) The binding of lamin B receptor to chromatin is regulated by phosphorylation in the RS region. Eur. J. Biochem. 269, 943–953 [DOI] [PubMed] [Google Scholar]
- 14.Nakamura T., Myint K. T., Oda Y. (2010) Ethylenediaminetetraacetic acid increases identification rate of phosphoproteomics in real biological samples. J. Proteome Res. 9, 1385–1391 [DOI] [PubMed] [Google Scholar]
- 15.Huang D.W., Sherman B.T., Lempicki R.A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park S.S., Maudsley S. (2011) Discontinuous pH gradient-mediated separation of TiO2-enriched phosphopeptides. Anal. Biochem. 409, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kyono Y., Sugiyama N., Imami K., Tomita M., Ishihama Y. (2008) Successive and selective release of phosphorylated peptides captured by hydroxy acid-modified metal oxide chromatography. J. Proteome Res. 7, 4585–4593 [DOI] [PubMed] [Google Scholar]
- 18.Maudsley S., Davidson L., Pawson A.J., Chan R., Lopez de Maturana R., Millar R.P. (2004). Gonadotropin-releasing hormone (GnRH) antagonists promote proapoptotic signaling in peripheral reproductive tumor cells by activating a Gαi-coupling state of the type I GnRH receptor. Cancer Res. 64, 7533–7544 [DOI] [PubMed] [Google Scholar]
- 19.Kim E.K., Choi E.J. (2010) Pathological roles of MAPK signaling pathways in human diseases. Biochim. Biophys. Acta 1802, 396–405 [DOI] [PubMed] [Google Scholar]
- 20.Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C., Li Y., Semenov M., Han C., Baeg G.H., Tan Y., Zhang Z., Lin X., He X. (2002) Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 22.Warn-Cramer B.J., Lampe P.D., Kurata W.E., Kanemitsu M.Y., Loo L.W.M., Eckhart W., Lau A.F. (1996) Characterization of the mitogen-activated protein kinase phosphorylation sites on the connexin-43 gap junction protein. J. Biol. Chem. 271, 3779–3786 [DOI] [PubMed] [Google Scholar]
- 23.Warn-Cramer B.J., Cottrell G.T., Burt J.M., Lau A.F. (1998) Regulation of connexin-43 gap junctional intercellular communication by mitogen-activated protein kinase J . Biol. Chem. 273, 9188–9196 [DOI] [PubMed] [Google Scholar]
- 24.Kanemitsu M.Y., Jiang W., Eckhart W. (1998) Cdc2-mediated phosphorylation of the gap junction protein, connexin43, during mitosis. Cell Growth Differ. 9, 13–21 [PubMed] [Google Scholar]
- 25.Lampe P.D., Kurata W.E., Warn-Cramer B.J., Lau A.F. (1998) Formation of a distinct connexin43 phosphoisoform in mitotic cells is dependent upon p34cdc2 kinase. J. Cell Sci. 111, 833–841 [DOI] [PubMed] [Google Scholar]
- 26.Bidinosti M., Ran I., Sanchez-Carbente M.R., Martineau Y., Gingras A.C., Gkogkas C., Raught B., Bramham C., Sossin W.S., Costa-Mattioli M., DesGroseillers L., Lacaille J.C., Sonenberg N. (2010) Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol. Cell 37, 797–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferguson G., Mothe-Satney I., Lawrence J.C., Jr (2003) Ser-64 and Ser-111 in PHAS-I are dispensable for insulin-stimulated dissociation from eIF4E. J. Biol. Chem. 278, 47459–47465 [DOI] [PubMed] [Google Scholar]
- 28.Mothe-Satney I., Brunn G.J., McMahon L.P., Capaldo C.T., Abraham R.T., Lawrence J.C., Jr (2000) Mammalian target of rapamycin-dependent phosphorylation of PHAS-I in four (S/T)P sites detected by phospho-specific antibodies. J. Biol. Chem. 275, 33836–33843 [DOI] [PubMed] [Google Scholar]
- 29.Heesom K.J., Gampel A., Mellor H., Denton R.M. (2001) Cell cycle-dependent phosphorylation of the translational repressor eIF-4E binding protein-1 (4E-BP1). Curr. Biol. 11, 1374–1379 [DOI] [PubMed] [Google Scholar]
- 30.Herbert T.P., Tee A.R., Proud C.G. (2002) The extracellular signal-regulated kinase pathway regulates the phosphorylation of 4E-BP1 at multiple sites. J. Biol. Chem. 277, 11591–11596 [DOI] [PubMed] [Google Scholar]
- 31.Fan H.Y., Sun Q.Y. (2004) Involvement of mitogen-activated protein kinase cascade during oocyte maturation and fertilization in mammals. Biol. Reprod. 70, 535–547 [DOI] [PubMed] [Google Scholar]
- 32.Kumano M., Carroll D.J., Denu J.M., Foltz K.R. (2001) Calcium-mediated inactivation of the MAP kinase pathway in sea urchin eggs at fertilization. Dev. Biol. 236, 244–257 [DOI] [PubMed] [Google Scholar]
- 33.Sakanaka C., Leong P., Xu L., Harrison S.D., Williams L.T. (1999) Casein kinase Iε in the Wnt pathway: regulation of β-catenin function. Proc. Natl. Acad. Sci. USA 96, 12548–12552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gietzen K.F., Virshup D.M. (1999) Identification of inhibitory autophosphorylation sites in casein kinase Iϵ. J. Biol. Chem. 274, 32063–32070 [DOI] [PubMed] [Google Scholar]
- 35.Simpson F., Kerr M.C., Wicking C. (2009) Trafficking, development and hedgehog. Mech. Dev. 126, 279–288 [DOI] [PubMed] [Google Scholar]
- 36.Maloverjan A., Piirsoo M., Kasak L., Peil L., Osterlund T., Kogerman P. (2010) Dual function of UNC-51-like kinase 3 (Ulk3) in the Sonic hedgehog signaling pathway. J Biol. Chem. 285, 30079–30090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maloverjan A., Piirsoo M., Michelson P., Kogerman P., Osterlund T. (2010) Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway. Exp. Cell Res. 316, 627–637 [DOI] [PubMed] [Google Scholar]
- 38.Backs J., Stein P., Backs T., Duncan F.E., Grueter C.E., McAnally J., Qi X., Schultz R.M., Olson E.N. (2010) The γ isoform of CaM kinase II controls mouse egg activation by regulating cell cycle resumption. Proc. Natl. Acad. Sci. USA 107, 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cohen P.T.W., Philp A., Vazquez-Martin C. (2005) Protein phosphatase 4 – from obscurity to vital functions. FEBS Lett. 579, 3278–3286 [DOI] [PubMed] [Google Scholar]
- 40.Cau J., Faure S., Vigneron S., Labbe J.C., Delsert C., Morin N. (2000) Regulation of Xenopus p21-activated kinase (X-PAK2) by cdc42 and maturation-promoting factor controls Xenopus oocyte maturation. J. Biol. Chem. 275, 2367–2375 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.