Abstract
Tumour suppressor p53, which is encoded by the TP53 gene, is widely known to play an important role in response to DNA damage and various stresses. It has recently been reported that p53 regulates glucose metabolism and that an increase in p53 protein level is induced after serum deprivation or treatments with a natural compound, trans-Resveratrol (Rsv). In this study, we constructed a Luciferase expression vector, pGL4-TP53-551, containing 551 bp of the 5’-upstream region of the human TP53 gene, which was then transfected into HeLa S3 cells. A Luciferase assay showed that Rsv treatment increased the promoter activity of the TP53 gene in comparison to that of PIF1. Detailed deletion and mutation analyses revealed that Nkx-2.5 and E2F-binding elements are required in addition to duplicated GGAA (TTCC), for the regulation of TP53 promoter activity. In this study, it is suggested that the transient induction of TP53 gene expression by Rsv treatment might be partly involved in its anti-aging effect through maintenance of chromosomal DNAs.
Keywords: E2F, ETS, Nkx-2.5, Resveratrol, TP53
Tumour suppressor protein p53, which is encoded by the TP53 gene, is known as a DNA damage or stress responding transcription factor that binds to the consensus sequence, 5’-(A/G)(A/G)(A/G)C(A/T)(A/T) G(C/T)(C/T)(C/T)-3’(1). Genetic mutations on the TP53 gene have been very frequently identified in a variety of tumour cells (2, 3). Thus, p53 is regularly referred to as a ‘guardian of the genome’. Significant biological functions of the p53 protein include induction of cell cycle regulatory factor-encoding genes, regulation of cellular senescence, apoptosis and autophagy (4). With regard to cancer generation, activation of p53 is thought to play an important role in inducing senescence to prevent aberrant fusions or breakages within telomere-shortened chromosomes (5). Furthermore, it has been suggested that cross-talk between telomeres and mitochondria plays a role in the regulation of aging (6, 7). Moreover, p53 is known to accumulate in the cytoplasm and mitochondria in response to various stresses, suggesting that it also regulates mitochondrial functions, including glucose metabolism (8, 9).
Previous studies have shown that expression of the TP53 gene is induced after deprivation of serum from the culture medium of granulosa and HepG2 cells (10, 11). Moreover, it has been reported that glucose deprivation from culture medium induces TP53 gene expression in U2OS cells (12). These results suggest that reduced nutrient or energy stress may induce TP53 gene expression. We have reported that the promoter activities of the genes encoding telomere maintenance-associated factors, including WRN and shelterin proteins, are induced after treatment with caloric restriction (CR) mimetic compounds, such as 2-deoxy-D-glucose (2DG) and trans-Resveratrol (Rsv) (13, 14). We thus hypothesized that serum deprivation or CR mimetic compounds induce the promoter activity of the TP53 gene. To assess the possibility, a Luciferase (Luc) expression plasmid containing 551 bp of the 5’-upstream region of human TP53 was constructed and used for a transfection assay. The Luc reporter assay revealed that the 551-bp region responds to both serum deprivation and Rsv treatment in HeLa S3 cells. A natural polyphenolic CR mimetic compound, Rsv is known to stimulate NAD+-dependent deacetylase sirtuin and elongate lifespan of model animals (15–19). A comparison of the Rsv-inducible human WRN and TERT promoter regions showed that the Sp1/GC-box is common to both (20). However, canonical GC-box sequences are not found in the 551 bp of the human TP53 promoter region.
In this study, we found that deletion of the duplicated GGAA (TTCC) or c-Ets binding element drastically diminished promoter activity. Deletion analyses showed that the GGAA (TTCC) motifs and the Nkx-2.5 element both play essential roles in the regulation of TP53 promoter activity in HeLa S3 cells. In addition, mutation of the E2F-motif apparently reduced the response to Rsv, suggesting that proteins binding to the E2F-motif play an important role in the control of TP53 gene expression in response to Rsv treatment.
Materials and Methods
Materials
Rsv was purchased from Cayman Chemical (Ann Arbor, MI) (21).
Cells and cell culture
Human cervical carcinoma (HeLa S3) cells (13) were grown in Dulbecco’s modified Eagle’s (DME) medium (WAKO Pure Chemical, Tokyo, Japan), supplemented with 10% fetal bovine serum (Biosera, East Sussex, UK) and penicillin-streptomycin at 37°C in a humidified atmosphere with 5% CO2.
Construction of luciferase (Luc) reporter plasmids
The Luc reporter plasmid pGL4-TP53-551 carrying 551 bp of the 5’-flanking region of the human TP53 gene was constructed by a similar procedure to that previously described (13, 14). Similarly, other Luc reporter plasmids were constructed by ligating a polymerase chain reaction (PCR)-amplified DNA fragment into the KpnI/XhoI site of pGL4.10[luc2] (Promega, Madison, WI). The sense and anti-sense primers used for the amplification of the DNA fragments are shown in Table I. The shaded nucleotides (Table I) indicate the mutations that disrupt the c-ETS, Nkx2.5 and E2F binding elements.
Table I.
Primer pairs used for amplifying 5’-upstream regions of the human TP53 gene
| Luc plasmid | Primer | Sequence (5’ to 3’) |
|---|---|---|
| pGL4-TP53-551 | hTP53-4710 | TCGGTACCTCATAAGGCTTACGTTTCCA |
| AhTP53-4160 | ATCTCGAGACGGTGGCTCTAGACTTTTGA | |
| pGL4-TP53-Δ1 | hTP53-4710 | TCGGTACCTCATAAGGCTTACGTTTCCA |
| Ahp53-4734 | ATCTCGAGGCTCCTGGCACAAAGCTGG | |
| pGL4-TP53-Δ2 | hp53-5025 | TCGGTACCTGATGAGAAGAAAGGATCCAG |
| AhTP53-4160 | ATCTCGAGACGGTGGCTCTAGACTTTTGA | |
| pGL4-TP53-Δ3 | hTP53-4710 | TCGGTACCTCATAAGGCTTACGTTTCCA |
| AhTP53-4210 | ATCTCGAGCACATGGGAGGGGAAAACCCC | |
| pGL4-TP53-Δ12 | hp53-5025 | TCGGTACCTGATGAGAAGAAAGGATCCAG |
| Ahp53-4734 | ATCTCGAGGCTCCTGGCACAAAGCTGG | |
| pGL4-TP53-ΔA | hp53-4488 | TCGGTACCCTTCATATTTGACACAATGCAG |
| AhTP53-4160 | ATCTCGAGACGGTGGCTCTAGACTTTTGA | |
| pGL4-TP53-ΔB | hp53-4426 | TCGGTACCAGCTCTGGCTTGCAGAATTTTC |
| AhTP53-4160 | ATCTCGAGACGGTGGCTCTAGACTTTTGA | |
| pGL4-TP53-ΔC | hp53-4349 | TCGGTACCCTCCTCCCCAACTCCATTTC |
| AhTP53-4160 | ATCTCGAGACGGTGGCTCTAGACTTTTGA | |
| pGL4-TP53-ΔD | hp53-4285 | TCGGTACCATGGCGACTGTCCAGCTTTGTG |
| AhTP53-4160 | ATCTCGAGACGGTGGCTCTAGACTTTTGA | |
| pGL4-TP53-ΔE | hp53-4225 | TCGGTACCCCTCCCATGTGCTCAAGACTGG |
| AhTP53-4160 | ATCTCGAGACGGTGGCTCTAGACTTTTGA | |
| pGL4-TP53-ΔF | hp53-4333 | TCGGTACCATTTCCTTTGCTTCCTCCGGC |
| AhTP53-4160 | ATCTCGAGACGGTGGCTCTAGACTTTTGA | |
| pGL4-TP53-ΔG | hp53-4333 | TCGGTACCATTTCCTTTGCTTCCTCCGGC |
| AhTP53-4210 | ATCTCGAGCACATGGGAGGGGAAAACCCC | |
| pGL4-TP53-ΔH | hp53-4295 | TCGGTACCTTACTTGTCATGGCGACTGTCC |
| AhTP53-4160 | ATCTCGAGACGGTGGCTCTAGACTTTTGA | |
| pGL4-TP53-ΔI | hp53-4295 | TCGGTACCTTACTTGTCATGGCGACTGTCC |
| AhTP53-4210 | ATCTCGAGCACATGGGAGGGGAAAACCCC | |
| pGL4-TP53-WT | hp53-4333 | TCGGTACCATTTCCTTTGCTTCCTCCGGC |
| AhTP53-4193 | ATCTCGAGCTTTTAGCGCCAGTCTTGAGCA | |
| pGL4-TP53-m1 | hp53-4333#1 | TCGGTACCATTTGGTTTGCTTCCTCCGGCA |
| AhTP53-4193 | ATCTCGAGCTTTTAGCGCCAGTCTTGAGCA | |
| pGL4-TP53-m3 | hp53-4333 | TCGGTACCATTTCCTTTGCTTCCTCCGGC |
| AhTP53-4193#1 | ATCTCGAGCTTTTAAAACCAGTCTTGAGCA | |
| pGL4-TP53-m13 | hp53-4333#1 | TCGGTACCATTTGGTTTGCTTCCTCCGGCA |
| AhTP53-4193#1 | ATCTCGAGCTTTTAAAACCAGTCTTGAGCA | |
| pGL4-TP53-mEts, pGL4-TP53-mEtNk | EtsM1 | TCCATTTGGTTTGCTTCCTCCGGCAGGCGG |
| EtsM2 | AGCAAACCAAATGGAGTTGGGGAGGAGGGT | |
| pGL4-TP53-mNkx | NkxM1 | TTGCCCGAACTTGTCATGGCGACTGTCCAG |
| NkxM2 | GACAAGTTCGGGCAAGTAATCCGCCTGCCG | |
| pGL4-TP53-mE2F | E2FM3 | AAGACTGGTTTTAAAAGTTTTGAGCTTCTC |
| E2FM4 | TTTAAAACCAGTCTTGAGCACATGGGAGGG |
Luc reporter plasmids, pGL4-TP53-mEts, pGL4-TP53-mNkx and pGL4-TP53-mE2F were made according to a procedure (22) with slight modifications. Briefly, PCR was performed with appropriate sense and antisense primers (Table I) and pGL4-TP53-551 as a template. The DNAs were denatured at 65°C for 20 min and gradually cooled down to 25°C for 20 min, then kept at 25°C for further 20 min. The double-stranded DNA products, which were treated with T4 DNA polymerase (Toyobo) and digested with KpnI and XhoI, were introduced into the MSC of the pGL4.10[luc2] vector. Nucleotide sequences were confirmed by DNA sequencing service (FASMAC, Greiner Japan Inc., Atsugi, Japan) with primers Rv (TAGCAAAATAGGCTGTCCCC) and GL (CTTTATGTTTTTGGCGTCTTCC).
Transient transfection and Luc assay
Plasmid DNAs were transfected into HeLa S3 cells by the DEAE-dextran method in 96-well plates (23, 24). After 24 h of transfection, the culture medium was changed to DME without serum or with 10% fetal calf serum (FCS) containing Rsv (0–40 µM). After a further 24 h of incubation, cells were collected and lysed with 100 µl of 1 x cell culture lysis reagent, containing 25 mM Tris-phospate (pH 7.8), 2 mM DTT, 2 mM 1,2-diaminocyclohexane-N,N,N’,N’,-tetraacetic acid, 10% glycerol and 1% Triton X-100, then mixed and centrifuged at 12,000 × g for 5 s. The supernatant was stored at −80°C. The Luc assay was performed with a Luciferase assay system (Promega) and relative Luc activities were calculated as described previously (14, 21, 24).
Western blot analysis
Western blot analysis was carried out as previously described (13, 21), with antibodies against p53 (BIOSS, Woburn, MA), and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA) followed by the addition of horseradish peroxidase-conjugated secondary antibody (Calbiochem, Darmstadt, Germany). Signal intensities were quantified with a ChemiDoc and ImageLab System (BioRad, Berkeley, CA).
Reverse transcriptase PCR
Reverse transcriptase PCR (RT-PCR) was carried out as described previously (13, 21). First-strand cDNAs were synthesized with ReverTra Ace (Toyobo, Tokyo, Japan), random primers (Takara) and total RNAs extracted from HeLa S3 cells. The sequences (from 5’ to 3’) of primer pairs used to amplify human TP53, GAPDH and β-actin cDNAs were Shp53-265; CTGCCCTCAACAAGATGTTTTG and Ahp53-436; CTATCTGAGCAGCGCTCATGG, hGAPDH556; TGCACCACCAACTGCTTAGC and hGAPDH642; GGCATGGACTGTGGTCATGAG and hbactS541; TGACGGGGTCACCCACACTGTGCCCATC and hbactA1201; CTAGAAGCATTTGCGGTGGACGATGGAG.
Conditions for the PCR were as follows: 94°C for 15 s, 55°C for 20 s and 72°C for 10 s, with 28 (TP53), 19 (GAPDH) and 20 (β-actin) cycles. PCR was performed with BIOTAQ DNA polymerase (BIOLINE, London, UK), and the PCR products were electrophoresed on 5% acrylamide gels and stained with ethidium bromide.
Quantitative real-time PCR
Real-time PCR analysis was carried out using the Mx3000P Real-Time qPCR System (Stratagene, La Jolla, CA) as described previously (13, 21). For PCR amplification, cDNAs were amplified using Thunderbird Real-time PCR Master Mix (Toyobo) and 0.3 µM of each primer pair. The primer pairs for amplifying human TP53 and GAPDH transcripts were Shp53-265/Ahp53-436 and hGAPDH556/hGAPDH642, respectively. Amplification was carried out initially for 1 min at 95°C, followed by 40 cycles (95°C 15 s and 58°C 30 s). Quantitative PCR analysis for each sample was carried out in triplicates. Relative gene expression values were obtained by normalizing CT (threshold cycle) values of target genes in comparison with CT values of the GAPDH gene using the ΔΔCT method.
Results
Effects of serum deprivation on TP53 gene expression and its protein amount in HeLa S3 cells
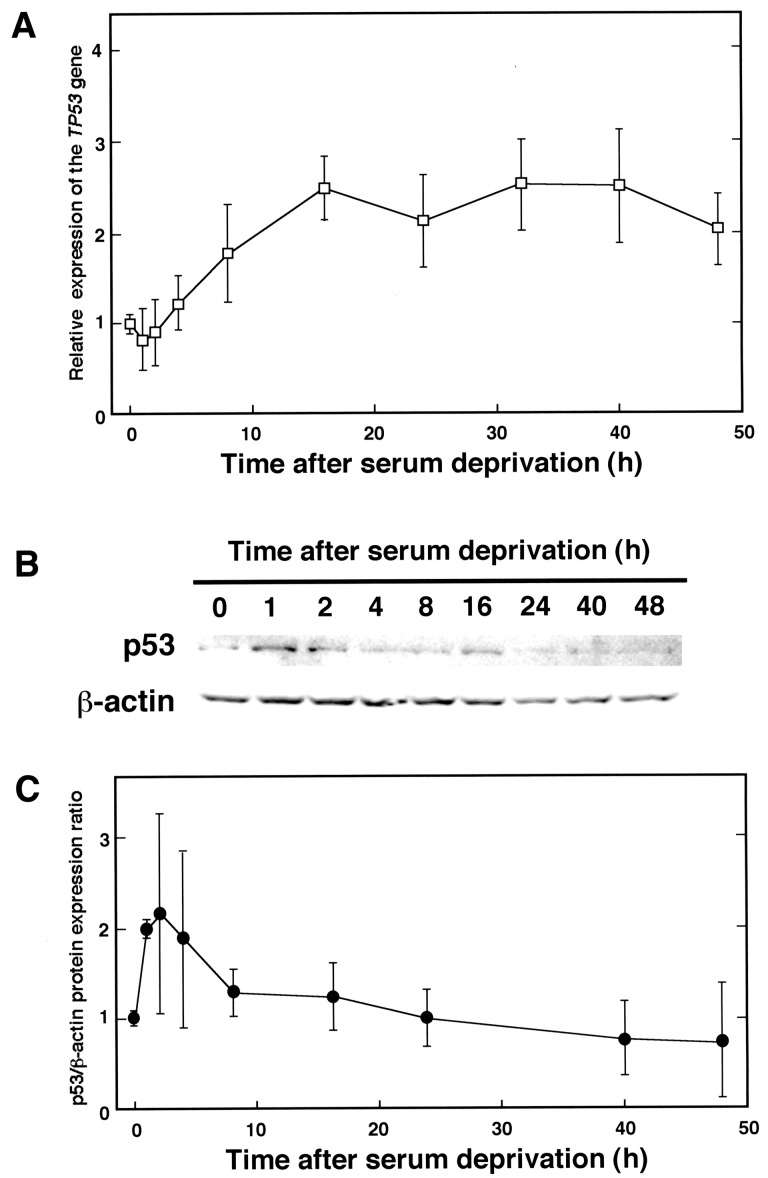
To examine whether human TP53 gene expression is affected by serum deprivation, total RNAs were extracted from cells after changing culture medium to serum-free DME (Fig. 1A). The relative gene expression of TP53 compared with that of GAPDH began to increase slightly from 4 h after withdrawal of serum and then reached a plateau level. Western blot analysis showed that after serum deprivation, the amount of p53 protein reached a peak at 1–4 h and then decreased gradually (Fig. 1B and C). This decrease in protein amount is not only caused by the lowered mRNA level but also by degradation of the p53 protein, non-coding regulatory RNAs or another regulatory mechanism.
Fig. 1.
Changes in TP53 gene expression and p53 protein level after serum deprivation. (A) HeLa S3 cells were cultured in Dulbecco’s modified Eagle’s medium without FCS for 0, 1, 2, 4, 8, 16, 24, 32, 40 and 48 h (lanes 1–10, respectively), then harvested. Total RNAs were extracted from cells, and synthesized cDNAs were subjected to quantitative real-time PCR with appropriate primer pairs to amplify TP53 and GAPDH cDNAs. Results show relative TP53/GAPDH gene expression ratios that were compared with that of untreated cells (0 h). Three independent experiments were done, and results show means ± SD. (B) Similarly treated HeLa S3 cells as in (A) were collected after 0, 1, 2, 4, 8, 16, 24, 40 and 48 h of serum withdrawal. Proteins extracted from cells were separated by a 15% SDS-PAGE, and Western blotting was performed with primary antibodies against p53 and β-actin (upper and lower panels, respectively). (C) Each band was quantified and the results are shown by relative p53/β-actin protein expression ratio. Results are shown as means ± SD from at least three independent experiments.
Upregulation of TP53 gene expression and p53 protein amount in HeLa S3 cells by CRmimetic compounds
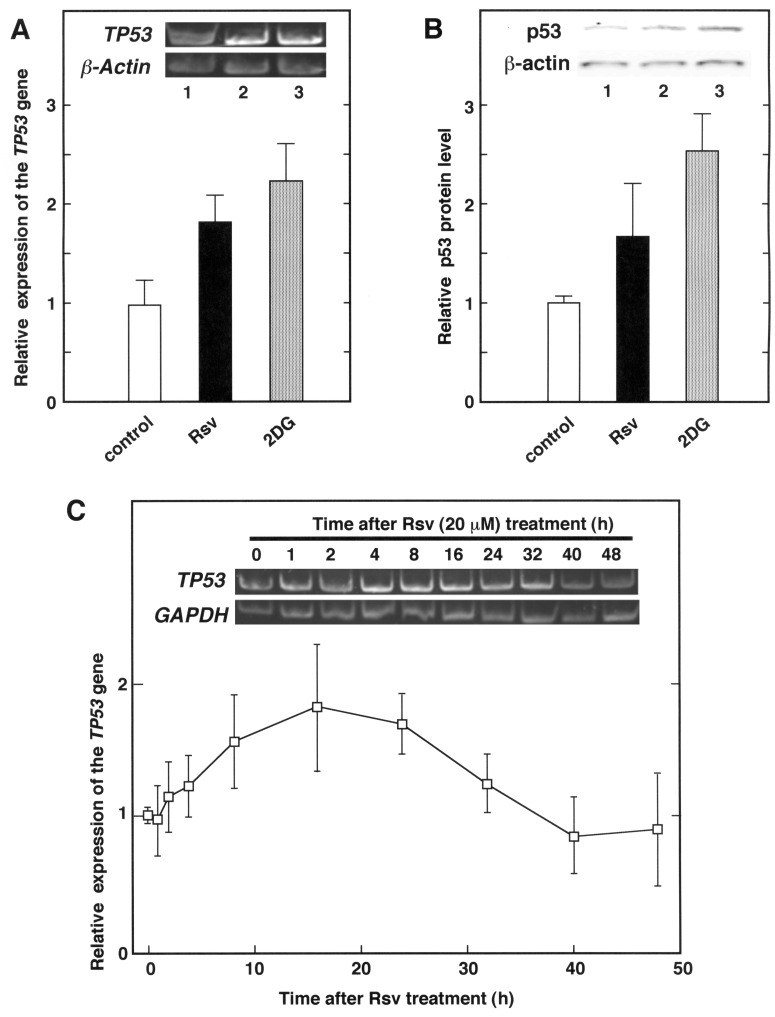
The above results suggested that the TP53 gene expression is transiently up-regulated by nutrient stress to HeLa S3 cells. To examine whether the expression of the TP53 gene and its translated protein product p53 is induced by treatment with CR mimetic compounds, total RNAs and protein extracts from 2DG or Rsv-treated HeLa S3 cells were analysed by RT-PCR and Western blotting, respectively (Fig. 2).
Fig. 2.
Effects of CR mimetic compounds on TP53 gene expression and p53 protein level in HeLa S3 cells. (A) The culture medium of HeLa S3 cells was changed to Dulbecco’s modified Eagle’s (DME) medium containing 10% FCS with 20 µM of Rsv or 8 mM of 2DG (lanes 2 and 3, respectively). Total RNAs were extracted from cells, and synthesized cDNAs were subjected to PCR with appropriate primer pairs to amplify TP53 (upper panel) and β-actin (lower panel) cDNA. The histograms show relative TP53/β-actin gene expression ratio. Results are shown as means ± SD from three independent experiments. (B) HeLa S3 cells similarly treated to (A) were collected, then extracted proteins were separated by a 15% SDS-PAGE, and Western blotting was performed with primary antibodies against p53 and β-actin (upper and lower panels, respectively). Each band was quantified and results show relative p53/β-actin protein expression ratio. Results are shown as means ± SD from three independent experiments. (C) The culture medium of HeLa S3 cells was changed to DME containing 10% FCS with 20 µM of Rsv and harvested after 0, 1, 2, 4, 8, 16, 24, 32, 40 and 48 h. Total RNAs were extracted from cells and synthesized cDNAs were subjected to PCR with primer pairs to amplify TP53 (upper panel) and GAPDH (lower panel) cDNA. Real-time quantitative RT-PCR was carried out to analyse TP53 and GAPDH gene expression in HeLa S3 cells after 20 µM of Rsv treatment for 0 to 48 h. The results show relative TP53/GAPDH gene expression ratio compared with that of Rsv non-treated cells. Results are shown as means ± SD from at least three independent experiments.
As shown in Fig. 2A, the amount of TP53 transcripts in the cells treated with Rsv (20 µM) and 2DG (8 mM) for 24 h increased to approximately 2-fold that of the control cells. The Western blot analyses indicated, as expected, that the relative level of the p53/β-actin increased after treatment with these two CR mimetic drugs (Fig. 2B). Next, HeLa S3 cells were collected at different times after Rsv (20 µM) treatment, and total RNAs were extracted. As shown in Fig. 2C, the relative expression of the TP53 gene reached its peak level in comparison to the GAPDH gene (approximately 3-fold) at 16 h after the Rsv treatment (Fig. 2C).
Isolation and characterization of the 5’-flanking region of the human TP53 gene
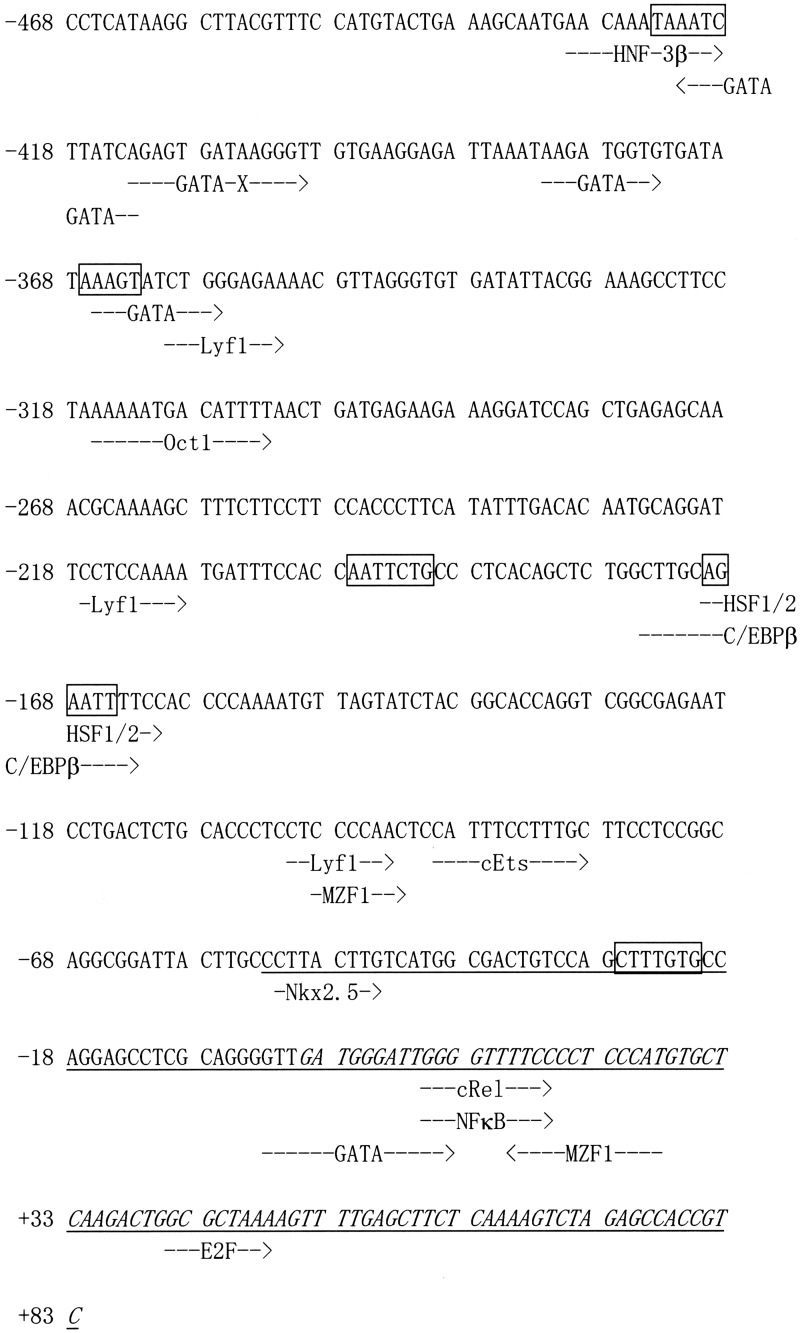
To examine whether the induction of the TP53 transcripts by Rsv occurs with the activation of the promoter, we isolated 551 bp of the 5’-upstream region of the TP53 gene by PCR. Sequence analysis revealed that the pGL4-TP53-551 contains a nucleotide identical to NCBI Sequence ID, NC_018928.2 (nucleotides from 7600471 to 7599921), and that it covers the sequence of the most-upstream 5’- end of the cDNA (Sequence ID; NM_001276760.1, NM_001276761.1, NM_001276696.1, NM_001276695.1 and NM_001126118.1 for variant 1, 2, 3, 4 and 8 of the TP53 mRNA, respectively; GENE ID, 7157 TP53). Interestingly, this 551-bp region also contains a 5’-upstream end of variant 3 of the WRAP53 mRNA (Accession No.NM_001143991.1; GENE ID, 55135 WRAP53) in a reverse orientation to that of the TP53 gene. The transcription start site (TSS) was tentatively set as +1 at the most-upstream 5’ of the TP53 transcripts shown in the database. The TF-SEARCH program (http://www.cbrc.jp/research/db/TFSEARCH.html) suggested that the characteristic recognition sequences of several known transcription factors were found in the 551-bp region (Fig. 3). Although no obvious sequences similar to the TATA or CCAAT boxes were found, putative binding sites for HNF-3β (−430 to −419), GATA-x (−422 to −413, −413 to −400), GATA-1/2/3 (−382 to −374, −3 to +12), GATA-1/2 (−366 to −357), Oct-1 (−316 to −303), HSF-1/2 (−170 to −161), C/EBPβ (−175 to –163), c-Ets (−90 to −79), Nkx-2.5 (−52 to −45), c-Rel/NF-κB (+10 to +19) and E2F (+41 to +48) are contained in the 551-bp region (Fig. 3).
Fig. 3.
Nucleotide sequence of the 5’-flanking region of the human TP53 gene. The nucleotide sequence of the 551-bp fragment that was obtained from PCR is shown. The most-upstream 5’-end of the TP53 cDNA (NM_001276760.1, NM_001276761.1, NM_001276696.1, NM_001276695.1 and NM_001126118.1) is designated nucleotide +1. Putative transcription factor-binding sites (TF-SEARCH score > 87) are indicated by dotted arrows. Boxes represent CdxA sequences. Italic and underlined characters represent the cDNA sequences that overlap the human TP53 (Gene ID: 7157) and WRAP53 (GENE ID: 55135) genes, respectively.
Effect of Rsv on the TP53 promoter activity
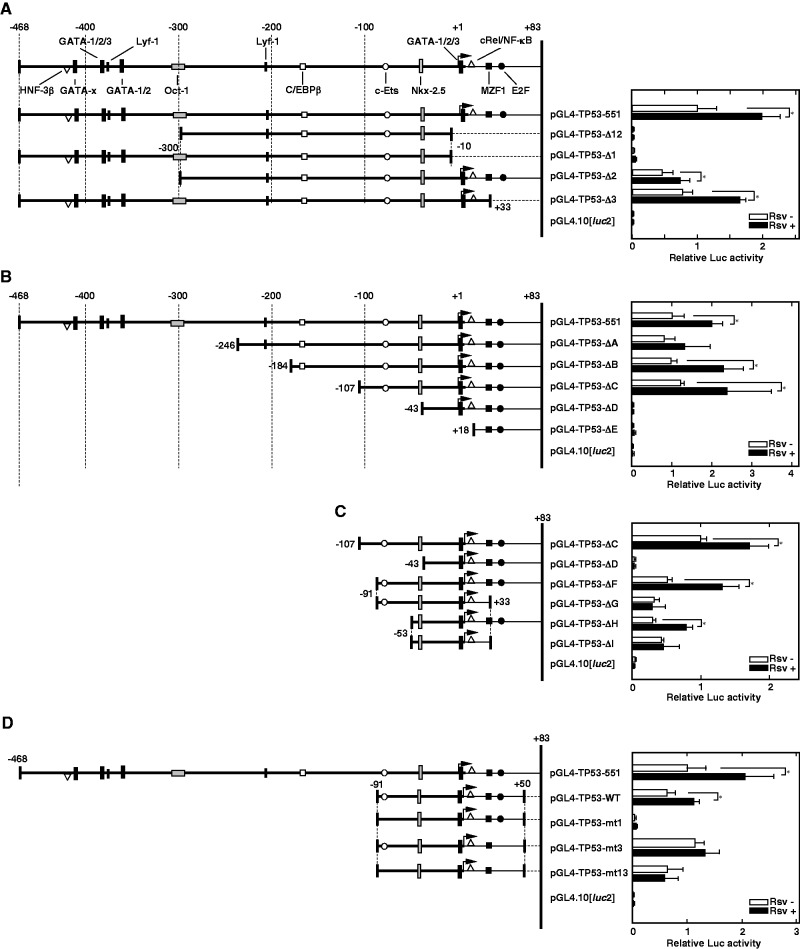
To examine whether the human TP53 promoter is affected by the natural compound Rsv, Luc reporter plasmid pGL4-TP53-551 and its derivative deletion constructs (Fig. 4A–C, left panel) were transiently transfected into HeLa S3 cells. First, the Luc activities of pGL4-TP53-551 and pGL4-TP53-Δ1, Δ2, Δ3 and Δ12-transfected cells were compared to the Luc activity of the pGL4-PIF1-transfected cells (14, 21). As shown in Fig. 4A, the relative Luc activity of the pGL4-TP53-551-transfected cells increased after the addition of Rsv (20 µM) to the cell culture. This induction by Rsv was also observed in pGL4-TP53-Δ2- and Δ3-transfected cells but not in the pGL4-TP53-Δ1- and Δ12-transfected cells. Luc activities of pGL4-TP53-Δ1- and Δ12-transfected cells were almost background level, probably due to their lack of TSSs. The results suggested that the sequence of the 91-nucleotides from −9 to +83 is primarily required for the TP53 promoter activity.
Fig. 4.
Effect of Rsv on human TP53 promoter activity. (A–D) (Left panel) The 5’-flanking region of the human TP53 gene, which has been ligated upstream of the Luciferase gene of the pGL4.10[luc2], is shown. The 5’-end of the cDNA is designated +1. Putative transcription factor binding elements that were found from the TF-SEARCH program (score > 87) are schematically shown. The sequence of the TP53 promoter contained in each deletion construct is schematically indicated. (A) Luc reporter plasmids were transiently transfected into HeLa S3 cells and treated with (closed bars) or without (open bars) Rsv (20 µM) for 24 h, then they were collected after further 24 h incubation. Luc activities were normalized to that of the pGL4-PIF1-transfected cells. Histograms show relative Luc activities of deletion construct transfected cells comparing with that of the pGL4-TP53-551-transfected cells without Rsv treatment. (B–D) Similar experiments as described under (A) were carried out with pGL4-TP53-551, -ΔA, -ΔB, -ΔC, -ΔD, -ΔE, -ΔF, -ΔG, -ΔH, -ΔI, -WT, -mt1, -mt3, -mt13 and pGL4.10[luc2]. Results are shown as means ± SD from five independent experiments. Statistical analysis was performed with the Student’s t test and asterisks indicate values of P < 0.05.
We made further serial deletions from the 5’-upstream by PCR, and these reporter plasmids were transiently transfected into HeLa S3 cells. As shown in Fig. 4B, the deletion of the −105 to −44 region completely abolished the promoter activity, suggesting that the 61 bp, which contains the putative Nkx-2.5 binding sequence, 5’-CTTACTTG-3’, is an essential region for the TP53 promoter function. To narrow the core promoter sequence, additional deletions were made by PCR with the 188-bp fragment of the pGL4-TP53-ΔC (Fig. 4C). Although, Luc activity from cells that were transfected with pGL4-TP53-ΔH was comparatively lower than that of the pGL4-TP53-ΔC-transfected cells (Fig. 4C, right panel), it showed a positive response to Rsv. The result suggests that the region from −53 to +83 contains sets of essential transcription elements, including the Nkx-2.5 and the E2F motifs, respond to Rsv in HeLa S3 cells. Moreover, deletion of 50 bp (+34 to +83) from ΔH abolished the response to Rsv (Fig. 4C, compare ΔI and ΔG, with ΔH and ΔF, respectively). Complete loss of promoter activity was observed by a deletion from nucleotide −53 to −44 (Fig. 4C, compare ΔD with ΔH). These results suggest that Nkx-2.5 element from nucleotide −52 to −45 is essentially required for the TP53 promoter activity and that putative E2F binding sequence (+41 to +48) plays a role in the response to Rsv. While it remains to be shown whether TP53 gene expression is controlled during cardiogenesis, the homeobox protein Nkx-2.5 has been shown to regulate the differentiation of myocardial lineage or cardiogenesis (25, 26).
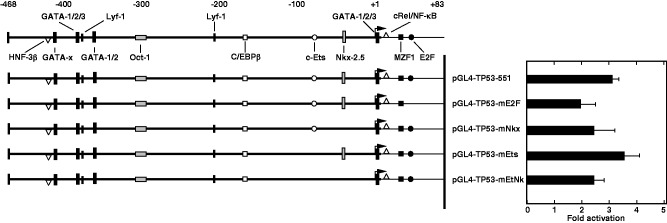
Above results showed that the region from +34 to +83, which contains a putative E2F binding sequence, responds positively to Rsv in HeLa S3 cells. To examine the Rsv-responding elements in detail, point mutations were introduced in the pGL4-TP53-WT plasmid. As shown in Fig. 4D, the mutation on the c-Ets element greatly reduced basal promoter activity with its response to Rsv (pGL4-TP53-mt1). Although the mutation on the E2F element does not affect the basal promoter activity, the response to Rsv was abolished (pGL4-TP53-mt3). Moreover, cells that were transfected with pGL4-TP53-m13, which carries mutations on both c-ETS and E2F elements, showed an apparent promoter activity, but they do not show response to Rsv. Moreover, we performed transfection experiment with plasmids carrying mutations on these elements in the pGL4-TP53-551 plasmid (Fig. 5). The result indicated that point mutations on Nkx-2.5 and E2F binding element reduced the response to Rsv. Introduction of mutations on the c-Ets element did not alter the response to Rsv, suggesting that other responsive element(s) in the region from −468 to −97 is also contributing for the response. Taken together, these results suggest that the TP53 promoter is regulated by c-Ets, Nkx-2.5 and E2F binding elements under the control of the other sequence(s) to respond to Rsv in HeLa S3 cells.
Fig. 5.
Mutation analysis on c-Ets, Nkx-2.5 and E2F binding elements in the 551-bp human TP53 promoter region. Similar experiment as described under the legend to Fig. 4 was carried out with pGL4-TP53-551, pGL4-TP53-mE2F, pGL4-TP53-mNkx, pGL4-TP53-mEts and pGL4-TP53-mEtNk. Histograms show fold induction of Luc activities by Rsv (20 µM) for 24 h treatment. Results are shown as means ± SD from four independent experiments.
Discussion
In this study, we showed that both serum withdrawal and the addition of CR mimetic drugs up-regulate TP53 gene expression in HeLa S3 cells. Moreover, deletion and mutation analyses of the 5’-upstream region of the TP53 gene revealed that c-Ets, Nkx-2.5 and E2F elements are essential for the activation of the promoter in response to Rsv.
Previous studies indicated that various transcription factors, including NF-κB, CREB, c-Myc and c-Ets, are involved in the regulation of TP53 gene expression (12, 27, 28). A palindromic c-Ets element, which is located in very close proximity to the TSS of the TP53 gene (29), has been shown to play an essential role in controlling its promoter function (30). We have reported that the duplicated GGAA (TTCC) motif is frequently found in the promoter regions of the DNA repair factor-encoding human genes (23), including PARP and PARG, which encode poly(ADP-ribose) metabolism regulating enzymes (31, 32). A microarray study of HIV-1 infection in human primary CD4+ T cells revealed interferon-dependent up-regulation of TP53 gene expression (33). However, the proteins that bind to the duplicated GGAA motifs to regulate the TP53 promoter have not been correctly shown. The opposite strand nucleotide sequences from −92 to −79 and −83 to −70 are 5’-GCAAAGGAAATGGA-3’ and 5’-CCGGAGGAAGCAAA-3’, respectively (Fig. 3). Therefore, Ets family class III proteins, such as Spi1, SpiB and SpiC, which recognize the sequence 5’-(A/T/C)(G/A)(A/G/C)GGAA(G/C)(T/C)N-3’ (34), may be candidate binding factors. We have proposed that tandem repeated or duplicated GGAA-motifs serve as a fine-tuning system to respond to a variety of stresses that induce apoptosis and DNA damage/interferon responses (35–37). Thus, in this context, the duplicated GGAA motif in the region from −92 to −70 might be a competitive target for various transcription factors, including the Ets family proteins. It should be noted that the duplicated GGAA (TTCC) plays important roles in the regulation of human RB1 and mouse Rb1 promoter (23, 38). Of note, it has been experimentally shown that GABP α/β protein targets the GGAA motif in the mouse Rb1 promoter region (38).
The response to Rsv was diminished only when the E2F element was eliminated or mutated (Fig. 4), suggesting that the putative E2F binding sequence 5’-TTTAGCGC-3’ (inverted nucleotide sequence from +41 to +48), which is located downstream of the TSS (Fig. 3), is the cis-element that is primarily required to respond to Rsv. Although the TF-SEARCH program did not indicate a tandem repeated element, the nucleotide sequence from +51 to +58 is 5’-TTTTGAGC-3’. Thus, the 18-bp nucleotide from + 41 to +58 consists of a palindromic tandem inverted repeat of the E2F element. It should be noted that the tandem E2F consensus sites are located in the promoter region of the human p107 gene, which encodes Rb-related protein (39). A variety of biological functions, including DNA replication, DNA repair, DNA-damage check point and G1-S transcriptional activation, are regulated by the E2F family proteins, which are encoded by eight distinct genes (E2F1 to E2F8) (40, 41). Given that the E2F family includes both positive and negative transcription regulators (41), the tandem repeated E2F element could serve as a fine-tuning modulator for TP53 transcription in response to cell cycle progression or DNA-damage-inducing signals in a similar mechanism, which is regulated by the duplicated GGAA motifs (35–37). E2F family proteins are known as regulators for p53, and E2F1-p53 cooperation in particular has a significant role in the regulation of apoptosis (40, 41). Recent studies have suggested that p53 up-regulates the transcription of the E2F7 gene, which in turn causes cellular senescence linking the Rb and p53 pathways (42, 43). It was shown that E2F1 binds to E2F element of the E2F7 promoter to activate its transcription (44). Moreover, overexpression of NF-Y up-regulates E2F1, which leads to an increase in the amount and the activity of p53 in mouse embryonic fibroblasts and human cells (45). These lines of evidences imply that Rsv-induced expression of the TP53 gene is controlled by the E2F family proteins. Importantly, duplicated GGAA (TTCC) motifs are present near the TSS of the human RB1 gene (23). Moreover, the RB protein has been suggested to affect and modulate the transcription from E2F target gene promoters (46). Taken together, these findings suggest that the Rsv-induced activation of the TP53 promoter might be associated with interaction between RB and E2F. Therefore, the effect of Rsv to induce cell cycle arrest (47, 48) could be partly explained by the activation of E2Fs that affect G1-S cell cycle progression.
Rsv activates p53 to induce apoptosis and up-regulates p53 protein level in mouse epidermal cells and bovine pulmonary artery endothelial cells (49, 50). It was reported that Rsv induces the expression of the ASPP1 (PPP1R13B) gene, which encodes the apoptosis stimulation protein of p53, via the induction of E2F1 in breast cancer cells (51). The observation suggests a possible mechanism through which Rsv effectively evokes apoptosis by the up-regulation of TP53 and ASPP1 in an E2F1-dependent manner. We have reported that Rsv induces SIRT1 promoter activity in HeLa S3 cells (52, 53). It is noteworthy that the human SIRT1 promoter is activated by serum deprivation in a p53-dependent manner (54). This suggests that the transient induction of the SIRT1 gene occurs as a result of the effect of p53 after Rsv treatment. It is widely known that p53 controls the expression of genes that encode DNA-damage response factors and cell cycle/apoptosis regulating factors (1, 55). Moreover, in the presence and even absence of stress, p53 protein localizes in the mitochondria as well as the nuclei (9, 56), suggesting that it does not only control transcription in nuclei, but also that it affects metabolic reactions in mitochondria (9). It is noteworthy that p53 regulates mitochondrial respiration, inducing expression of the SCO2 gene, which encodes the synthesis of cytochrome c oxidase 2 (57). The p53 is also reported to affect metabolic stress and aging (58). Furthermore, p53 regulates malic enzymes in human and mouse cells to induce cellular senescence without causing apoptosis (59). The functions of p53 as a key regulator for glycolysis/oxidative phosphorylation balance in a cell have been reviewed (60).
A recent study showed that telomere dysfunction induces the expression of the Tp53 gene to suppress the function of Pgc-1α (7), implying that telomeres exert signals to mitochondria. Greater damage to chromosomes will affect mitochondrial proteins, including Bcl-2, Bcl-XL, BAX and cytochrome c, to induce cell death (61). Our previous study showed that the promoter activities of TERT, WRN and several shelterin-encoding genes are induced by Rsv in HeLa S3 cells (14, 21). Besides GC-boxes and Sp1 binding elements, the duplicated GGAA (TTCC) motif is a common sequence in the 5’-upstream regions of the TERT and WRN genes (13, 33). Furthermore, recent studies in cancer genomes revealed that mutations on the GGAA (TTCC) motifs or the creation of Ets binding elements in the TERT promoter are frequently found in human melanoma (62, 63). Thus, co-induction of the TP53 and TERT genes through the duplicated GGAA motifs may account for keeping telomeres to their appropriate length. Variety of clinical trials showed that health-promoting responses, including reduction in generation of reactive oxygen species (ROS) and induction of insulin sensitivity, are caused by Rsv (64). Further investigations are required to elucidate the mechanisms by which Rsv-induced signals regulate the cellular senescence, lifespan and longevity of organisms.
Acknowledgements
The authors are grateful to S. Larsen for critical reading of the manuscript and discussion.
Funding
This work was supported in part by a Grant-in Aid from the Ministry of Education, Culture, Sports, Science and Technology Japan (No. 24510270) and a Research Fellowship from the Research Center for RNA Science, RIST, Tokyo University of Science.
Conflict of Interest
None declared.
Glossary
Abbreviations
- 2DG
2-deoxy-D-glucose
- CR
caloric restriction
- DME medium
Dulbecco’s modified Eagle’s (DME) medium
- FCS
fetal calf serum
- Luc
Luciferase
- PCR
polymerase chain reaction
- Rsv
trans-Resveratrol
- RT-PCR
reverse transcriptase-polymerase chain reaction
- TSS
transcription start site
References
- 1.Menendez D., Inga A., Resnick M.A. (2009) The expanding universe of p53 targets. Nat. Rev. Cancer 9, 724–737 [DOI] [PubMed] [Google Scholar]
- 2.Brosh R., Rotter V. (2009) When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer 9, 701–713 [DOI] [PubMed] [Google Scholar]
- 3.Vousden K.H., Lane D. (2007) p53 in health and disease. Nat. Rev. Mol. Cell Biol. 8, 275–283 [DOI] [PubMed] [Google Scholar]
- 4.Zilfou J.T., Lowe S.W. (2009) Tumor suppressive functions of p53 in Cold Spring Harbor Perspectives in Biology (Levine A.J., Lane D., eds.) pp. 1–12, Cold Spring Harbor Laboratory Press, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong K.K., Sharpless N.E., DePinho R.A. (2011) Chapter 4, Telomeres, telomerase, and Cancer in Cancer, Principles & Practice of Oncology—Primer of the Molecular Biology of Cancer (DeVita V.T., Lawrence T.S., Rosenberg S.A., eds.) pp. 58–69, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 6.Sahin E., DePinho R.A. (2012) Axis of aging: telomerase, p53 and mitochondria. Nat. Rev. Mol. Cell Biol. 13, 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahin E., Colla S., Liesa M., Moslehi J., Müller F.L., Guo M., Cooper M., Kotton D., Fabian A.J., Walkey C., Maser R.S., Tonon G., Foerster F., Xiong R., Wang Y.A., Shukla S.A., Jaskelioff M., Martin E.S., Heffernan T.P., Protopopov A., Ivanova E., Mahoney J.E., Kost-Alimova M., Perry S.R., Bronson R., Liao R., Mulligan R., Shirihai O.S., Chin L., DePinho R.A. (2011) Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung E.C., Vousden K.H. (2010) The role of p53 in glucose metabolism. Curr. Opin. Cell Biol. 22, 186–191 [DOI] [PubMed] [Google Scholar]
- 9.Green D.R., Kroemer G. (2009) Cytoplasmic functions of the tumor suppressor p53. Nature 458, 1127–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng X., Maruo T., Matsuo H., Takekida S., Deguchi J. (1998) Serum deprivation-induced apoptosis in cultured porcine granulosa cells is characterized by increased expression of p53 protein, Fas antigen and Fas ligand and by decreased expression of PCNA. Endocr. J. 45, 247–253 [DOI] [PubMed] [Google Scholar]
- 11.Zhuge J., Cederbaum A.I. (2006) Serum deprivation-induced HepG2 cell death is potentiated by CYP2E1. Free Radic. Biol. Med. 40, 63–74 [DOI] [PubMed] [Google Scholar]
- 12.Okoshi R., Ando K., Suenaga Y., Sang M., Kubo N., Kizaki H., Nakagawara A., Ozaki T. (2009) Transcriptional regulation of tumor suppressor p53 by cAMP-responsive element-binding protein/AMP-activated protein kinase complex in response to glucose deprivation. Genes Cells 14, 1429–1440 [DOI] [PubMed] [Google Scholar]
- 13.Zhou B., Ikejima T., Watanabe T., Iwakoshi K., Idei Y., Tanuma S., Uchiumi F. (2009) The effect of 2-deoxy-D-glucose on Werner syndrome RecQ helicase gene. FEBS Lett. 583, 1331–1336 [DOI] [PubMed] [Google Scholar]
- 14.Uchiumi F., Oyama T., Ozaki K., Tanuma S. (2011) Characterization of 5’-flanking regions of various human telomere maintenance factor-encoding genes in DNA Repair (Kruman I., ed.) pp. 585–596, InTech - Open Access Publisher, Rijeka, Croatia [Google Scholar]
- 15.Stefani M., Markus M.A., Lin R.C., Pinese M., Dawes I.W., Morris B.J. (2007) The effect of resveratrol on a cell model of human aging. Ann. NY Acad. Sci. 1114, 407–418 [DOI] [PubMed] [Google Scholar]
- 16.Kaeberlein M. (2010) Resveratrol and rapamycin: are they anti-aging drugs? Bioessays 32, 96–99 [DOI] [PubMed] [Google Scholar]
- 17.Howitz K.T., Bitterman K.J., Cohen H.Y., Lamming D.W., Lavu S., Wood J.G., Zipkin R.E., Chung P., Kisielewski A., Zhang L.L., Scherer B., Sinclair D.A. (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196 [DOI] [PubMed] [Google Scholar]
- 18.Wood J.G., Rogina B., Lavul S., Howitz K., Helfand S.L., Tatar M., Sinclair D. (2004) Sirtuin activators mimic caloric restriction and delay aging in metazoans. Nature 430, 686–689 [DOI] [PubMed] [Google Scholar]
- 19.Viswanathan M., Kim S.K., Berdichevsky A., Guarente L.A. (2005) A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev. Cell 9, 605–615 [DOI] [PubMed] [Google Scholar]
- 20.Uchiumi F., Higami Y., Tanuma S. (2010) Regulations of telomerase activity and WRN gene expression in Telomerase: Composition, Functions and Clinical Implications (Gagnon A.N., ed.) pp. 95–103, Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 21.Uchiumi F., Watanabe T., Hasegawa S., Hoshi T., Higami Y., Tanuma S. (2011) The effect of resveratrol on the Werner Syndrome RecQ helicase gene and telomerase activity. Curr. Aging Sci. 4, 1–7 [DOI] [PubMed] [Google Scholar]
- 22.Lee J., Lee H.J., Shin M.K., Ryu W.S. (2004) Versatile PCR-mediated insertion mutagenesis. Biotechniques 36, 398–400 [DOI] [PubMed] [Google Scholar]
- 23.Uchiumi F., Watanabe T., Tanuma S. (2010) Characterization of various promoter regions of the human DNA helicase-encoding genes and identification of duplicated ets (GGAA) motifs as an essential transcription regulatory element. Exp. Cell Res. 316, 1523–1534 [DOI] [PubMed] [Google Scholar]
- 24.Uchiumi F., Larsen S., Tanuma S. (2014) Application of DEAE-dextran to an efficient gene transfer system in Dextran: Chemical Structure, Application and Potential Side Effects (Figgs G. P., ed.) pp. 143–156. Nova Science Publishers, Hauppauge, NY [Google Scholar]
- 25.Lints T.J., Parsons L.M., Hartley L., Lyons I., Harvey R.P. (1993) Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119, 419–431 [DOI] [PubMed] [Google Scholar]
- 26.Pucéat M. (2005) Rb and LEK1: a “pas de deux” in cardiogenesis. Cell Cycle 4, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 27.Evan G.I., Wyllie A.H., Gilbert C.S., Littlewood T.D., Land H., Brooks M., Waters C.M., Penn L.Z., Hancock D.C. (1992) Induction of apoptosis in fibroblasts by c-myc protein. Cell 69, 119–128 [DOI] [PubMed] [Google Scholar]
- 28.Hellin A.C., Calmant P., Gielen J., Bours V., Merville M.P. (1998) Nuclear factor—kappaB-dependent regulation of p53 gene expression induced by daunomycin genotoxic drug. Oncogene 16, 1187–1195 [DOI] [PubMed] [Google Scholar]
- 29.Sementchenko V.I., Watson D.K. (2000) Ets target genes: past, present and future. Oncogene 19, 6533–6548 [DOI] [PubMed] [Google Scholar]
- 30.Baillat D., Laitem C., Leprivier G., Margerin C., Aumercier M. (2009) Ets-1 binds cooperatively to the palindromic Ets-binding sites in the p53 promoter. Biochem. Biophys. Res. Commun. 378, 213–217 [DOI] [PubMed] [Google Scholar]
- 31.Uchiumi F., Sakakibara G., Sato J., Tanuma S. (2008) Characterization of the promoter region of the human PARG gene and its response to PU.1 during differentiation of HL-60 cells. Genes Cells 13, 1229–1248 [DOI] [PubMed] [Google Scholar]
- 32.Uchiumi F., Watanabe T., Ohta R., Abe H., Tanuma S. (2013) PARP1 gene expression is downregulated by knockdown of PARG gene. Oncol. Rep. 29, 1683–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imbeault M., Ouellet M., Tremblay M.J. (2009) Microarray study reveals that HIV-1 induces rapid type-I interferon-dependent p53 mRNA up-regulation in human primary CD4+ T cells. Retrovirol. 6, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei G.H., Badis G., Berger M.F., Kivioja T., Palin K., Enge M., Bonke M., Jolma A., Varjosalo M., Gehrke A.R., Yan J., Talukder S., Turunen M., Taipale M., Stunnenberg H.G., Ukkonen E., Hughes T.R., Bulyk M.L., Taipale J. (2010) Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uchiumi F., Miyazaki S., Tanuma S. (2011) The possible functions of duplicated ets (GGAA) motifs located near transcription start sites of various human genes. Cell. Mol. Life Sci. 68, 2039–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uchiumi F., Larsen S., Masumi A., Tanuma S. (2013) The putative implications of duplicated GGAA-motifs located in the human interferon regulated genes (ISGs) in Genomics I-Humans, Animals and Plants (iConcept ed.) pp. 87–105, iConcept Press Ltd., Hong Kong [Google Scholar]
- 37.Uchiumi F., Larsen S., Tanuma S. (2015) Transcriptional regulation of the human genes that encode DNA repair- and mitochondrial function-associated proteins in DNA Repair (Chen C., ed.) pp. 129–167, InTech - Open Access Publisher, Rijeka, Croatia [Google Scholar]
- 38.Deléhouzée S., Yoshioka T., Sawa C., Sawada J., Ito T., Omori M., Wada T., Yamaguchi Y., Kabe Y., Handa H. (2005) GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells 10, 717–731 [DOI] [PubMed] [Google Scholar]
- 39.Zhu I., Xie E., Chang L.S. (1995) Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins. Mol. Cell Biol. 15, 3552–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polager S., Ginsberg D. (2009) p53 and E2f: partners in life and death. Nat. Rev. Cancer 9, 738–748 [DOI] [PubMed] [Google Scholar]
- 41.Bertoli C., Skotheim J.M., de Bruin R.A.M. (2013) Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Res. 14, 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carvajal L.A., Hamard P.J., Tonnessen C., Manfredi J.J. (2012) E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev. 26, 1533–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aksoy O., Chicas A., Zeng T., Zhao Z., McCurrach M., Wang X., Lowe S.W. (2012) The atypical E2F family member E2F7 couples the p53 and Rb pathways during cellular senescence. Genes Dev. 26, 1546–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Stefano L., Jensen M.R., Helin K. (2003) E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22, 6289–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gurtner A., Fuschi P., Martelli F., Manni I., Artuso S., Simonte G., Ambrosino V., Antonini A., Folgiero V., Falcioni R., Sacchi A., Piaggio G. (2010) Transcription factor NF-Y induces apoptosis in cells expressing wild-type p53 through E2F1 upregulation and p53 activation. Cancer Res. 70, 9711–9720 [DOI] [PubMed] [Google Scholar]
- 46.Dick F.A., Rubin S.M. (2013) Molecular mechanisms underlying RB protein function. Nat. Rev. Mol. Cell. Biol. 14, 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casanova F., Quarti J., da Costa D.C., Ramos C.A., da Silva J.L., Fialho E. (2012) Resveratrol chemosensitizes breast cancer cells to meiphalan by cell cycle arrest. J. Cell Biochem. 113, 2586–2596 [DOI] [PubMed] [Google Scholar]
- 48.Quoc Trung L., Espinoza J.L., Takami A., Nakao S. (2013) Resveratrol induces cell cycle arrest and apoptosis in malignant NK cells via JAK2/STAT3 pathway inhibition. PLoS One 8, e55183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang C., Ma W., Goranson A., Dong Z. (1999) Resveratrol suppresses cell transformation and induces apoptosis through a p53-dependent pathway. Carcinogenesis 20, 237–242 [DOI] [PubMed] [Google Scholar]
- 50.Hsieh T.C., Juan G., Darzynkiewicz Z., Wu J. M. (1999) Resveratrol increases nitric oxide synthase, induces accumulation of p53 and p21WAF1/CIP1, and suppresses cultured bovine pulmonary artery endothelial cell proliferation by perturbing progression through S and G2. Cancer Res. 59, 2596–2601 [PubMed] [Google Scholar]
- 51.Shi Y., Yang S., Troup S., Lu X., Callaghan S., Park D.S., Xing Y., Yang X. (2011) Resveratrol induces apoptosis in breast cancer cells by E2F1-mediated up-regulation of ASPP1. Oncol. Rep. 25, 1713–1719 [DOI] [PubMed] [Google Scholar]
- 52.Uchiumi F., Tachibana H., Larsen S., Tanuma S.I. (2013) Effect of lignin glycosides extracted from pine cones on the human SIRT1 promoter. Pharm. Anal. Acta. 4, 266 [Google Scholar]
- 53.Uchiumi F., Tachibana H., Abe H., Yoshimori A., Kamiya T., Fujikawa M., Larsen S., Honma A., Ebizuka S., Tanuma S. (2012) Effects of thujaplicins on the promoter activities of the human SIRT1 and telomere maintenance factor encoding genes. Pharm. Anal. Acta. 3, 159 [Google Scholar]
- 54.Shang L., Zhou H., Xia Y., Wang H., Gao G., Chen B., Liu Q., Shao C., Gong Y. (2009) Serum withdrawal up-regulates human SIRT1 gene expression in a p53-dependent manner. J. Cell. Mol. Med. 13, 4176–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gurkar A., Chu K., Lee S.W. (2013) Chapter 12, p53 in DNA Damage, Repair and Cancer Therapeutics in DNA Repair and Cancer—From Bench to Clinic. (Madhusudan S., Wilson D.M., III, eds.) pp. 372–425, CRC press, Taylor and Francis Group, Boca Raton, FL [Google Scholar]
- 56.Ferecatu I., Bergeaud M., Rodríguez-Enfedaque A., Le Floch N., Oliver L., Rincheval V., Renaud F., Vallette F.M., Mignotte B., Vayssière J.L. (2009) Mitochondrial localization of the low level p53 protein in proliferative cells. Biochem. Biophys. Res. Commun. 387, 772–777 [DOI] [PubMed] [Google Scholar]
- 57.Matoba S., Kang J.G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P. M. (2006) p53 regulates mitochondrial respiration. Science 312, 1650–1653 [DOI] [PubMed] [Google Scholar]
- 58.Vousden K.H., Ryan K.M. (2009) p53 and metabolism. Nat. Rev. Cancer 9, 691–700 [DOI] [PubMed] [Google Scholar]
- 59.Jiang P., Du W., Mancuso A., Wellen K.E., Yang X. (2013) Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature 493, 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kruiswijk F., Labuschagne C.F., Vousden K.H. (2015) p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393–400 [DOI] [PubMed] [Google Scholar]
- 61.Tait S.W., Green D.R. (2010) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621–632 [DOI] [PubMed] [Google Scholar]
- 62.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. (2013) Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horn S., Figl A., Rachakonda P.S., Fischer C., Sucker A., Gast A., Kadel S., Moll I., Nagore E., Hemminki K., Schadendorf D., Kumar R. (2013) TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 [DOI] [PubMed] [Google Scholar]
- 64.Vang O. (2013) What is new for resveratrol? Is a new set of recommendations necessary? Ann. NY Acad. Sci. 1290, 1–11 [DOI] [PubMed] [Google Scholar]