Abstract
Somatic embryogenesis (SE) is one of the methods with the highest potential for the vegetative propagation of commercially important coniferous species. However, many conifers, including Scots pine (Pinus sylvestris L.), are recalcitrant to SE and a better understanding of the mechanisms behind the SE process is needed. In Scots pine SE cultures, embryo production is commonly induced by the removal of auxin, addition of abscisic acid (ABA) and the desiccation of cell masses by polyethylene glycol (PEG). In the present study, we focus on the possible link between the induction of somatic embryo formation and cellular stress responses such as hydrogen peroxide protection, DNA repair, changes in polyamine (PA) metabolism and autophagy. Cellular PA contents and the expression of the PA metabolism genes arginine decarboxylase (ADC), spermidine synthase (SPDS), thermospermine synthase (ACL5) and diamine oxidase (DAO) were analyzed, as well as the expression of catalase (CAT), DNA repair genes (RAD51, KU80) and autophagy-related genes (ATG5, ATG8) throughout the induction of somatic embryo formation in Scots pine SE cultures. Among the embryo-producing SE lines, the expression of ADC, SPDS, ACL5, DAO, CAT, RAD51, KU80 and ATG8 showed consistent profiles. Furthermore, the overall low expression of the stress-related genes suggests that cells in those SE lines were not stressed but recognized the ABA + PEG treatment as a signal to trigger the embryogenic pathway. In those SE lines that were unable to produce embryos, cells seemed to experience the ABA + PEG treatment mostly as osmotic stress and activated a wide range of stress defense mechanisms. Altogether, our results suggest that the direction to the embryogenic pathway is connected with cellular stress responses in Scots pine SE cultures. Thus, the manipulation of stress response pathways may provide a way to enhance somatic embryo production in recalcitrant Scots pine SE lines.
Keywords: autophagy, catalase, DNA repair, drought stress, Pinus, polyethylene glycol, ROS protection
Introduction
Somatic embryogenesis (SE) is a process in which somatic cells are triggered toward the embryogenic pathway by exogenous signals under in vitro conditions (Fehér 2005). In conifers, SE provides a favorable model for studying factors that affect embryo development because the development of somatic embryos resembles that of zygotic embryogenesis (von Arnold et al. 2002). Furthermore, SE is currently one of the methods with the highest potential for the vegetative propagation of many commercially important coniferous species (Lelu-Walter et al. 2013). The ability to produce somatic embryos depends on plant species, genotype, developmental stage and growth conditions (Fehér 2005), as well as the tissue used for the initiation of embryogenic cultures (Stasolla et al. 2002). Despite the fact that somatic embryo formation has been achieved for a variety of angiosperm and gymnosperm species, many coniferous species are recalcitrant to SE (Bonga et al. 2010). For Scots pine (Pinus sylvestris L.), the number of successful SE initiations as well as the yields of somatic embryos are low and SE lines tend to lose their embryogenic potential over time during in vitro cultivation (Keinonen-Mettälä et al. 1996, Häggman et al. 1999, Lelu et al. 1999, Niskanen et al. 2004, Park et al. 2006).
Immature zygotic embryos surrounded by megagametophytes are the most responsive explants for the initiation of Scots pine SE cultures (Häggman et al. 1999, Lelu et al. 1999, Niskanen et al. 2004), whose development through the SE process encompasses four distinct stages: initiation, proliferation, maturation and germination, followed by acclimatization to ex vitro conditions (see Figure S1 available as Supplementary Data at Tree Physiology Online). Transition between the developmental stages is induced by manipulation of the culture medium composition, including the concentration of growth regulators (Häggman et al. 2006). The maturation stage is a crucial step for SE because it is during this stage that the embryogenic cell masses are induced to produce somatic embryos (Stasolla et al. 2002). In Scots pine SE cultures, embryo production is induced by the removal of auxin, addition of abscisic acid (ABA) and the subsequent desiccation of the embryogenic cell masses by an osmoticum, most commonly polyethylene glycol (PEG) (Stasolla et al. 2002). The use of PEG not only triggers the production of somatic embryos (von Arnold et al. 2002) but also arouses a state of osmotic stress by restricting water uptake (Attree and Fowke 1993). Polyethylene glycol treatments may cause osmotic stress-related morphological changes such as cellular shrinkage and decrease cell viability in Scots pine proembryogenic cell cultures (Muilu-Mäkelä et al. 2015).
Stress conditions alter cell metabolism. Numerous abiotic stresses may cause oxidative stress via the accumulation of reactive oxygen species (ROS) in cells. Reactive oxygen species, which include hydroxyl radicals, alkoxy radicals and hydrogen peroxide (H2O2) among numerous other species, are very reactive, can damage cellular structures and ultimately lead to cell death (Gill and Tuteja 2010). Hydrogen peroxide, one of the most stable forms of ROS, is a weak agent, which can directly oxidize proteins or produce toxic hydroxyl radicals (Mhamdi et al. 2012). To defend their cells against ROS damage, plants have utilized both enzymatic and nonenzymatic antioxidant agents for the removal of these substances (Gill and Tuteja 2010). While plants contain several types of H2O2-metabolizing proteins, catalases (CATs, H2O2 oxidoreductase, EC 1.11.1.6.) are highly active peroxisomal enzymes, which convert H2O2 into water and molecular oxygen probably to minimize its accumulation and further conversion to hydroxyl radicals (Mhamdi et al. 2012). In Scots pine zygotic embryos, CAT expresses strongly throughout the seed development and also in mature seeds (Vuosku et al. 2015).
Reactive oxygen species can damage DNA, which if unrepaired, may block critical cellular processes and eventually lead to cell death. DNA double-strand breaks (DSBs) present a particularly severe form of damage whose rapid repair is critical for the survival of the whole organism. Thus, plants use two systems for DSB repair: homologous recombination (HR) and non-homologous end-joining (NHEJ) (Kimura and Sakaguchi 2006). Homologous recombination utilizes a homologous sequence from a sister chromatid or a homologous chromosome as a template for the synthesis of new DNA, with an important role played by Rad51 protein in all of the three stages of the process (Krejci et al. 2012). Non-homologous end-joining is initiated by Ku70/Ku80 heterodimer and repairs DSBs without homology, and thus is more prone to errors (Mladenov and Iliakis 2011). During the Scots pine zygotic embryogenesis, both HR and NHEJ are used for DNA repair (Vuosku et al. 2009). Generally, NHEJ is a more common mechanism for DSB repair in plants (Kimura and Sakaguchi 2006) and, furthermore, Arabidopsis (Arabidopsis thaliana L.) with a nonfunctional Rad51 gene exhibits normal vegetative development (Li et al. 2004).
The accumulation of nonenzymatic antioxidant components such as polyamines (PAs) may play a role in the protection of cells against ROS damages. In plants, PAs are connected to abiotic stress tolerance (reviewed by Liu et al. 2007, Groppa and Benavides 2008, Alcázar et al. 2010). In addition, PAs are essential for both zygotic embryogenesis and SE of Pinus sp. (Amarasinghe et al. 1996, Minocha et al. 1999, Vuosku et al. 2006, Noceda et al. 2009). Polyamines are low molecular weight aliphatic cations, which may provide protection through several mechanisms such as the direct scavenging of reactive agents, by inducing conformational changes in DNA, by physically blocking the interaction between DNA and harmful agents or by a combination of the above-mentioned mechanisms (Basu et al. 1987, Feuerstein et al. 1990, Ha et al. 1998, Fujisawa and Kadoma 2005). The most common PAs in plant tissues are putrescine (Put), spermidine (Spd) and spermine (Spm), which are found in cells as free amines or amide conjugates (Kaur-Sawhney et al. 2003). The isomer of Spm, thermospermine (tSpm), has also been found to be a common PA in plants (Takano et al. 2012). Cellular PA levels are regulated via biosynthesis, degradation and transport pathways (Kusano et al. 2008; see Figure S2 available as Supplementary Data at Tree Physiology Online).
Autophagy as an intracellular catabolic process is upregulated in response to stress conditions (Han et al. 2011). In autophagy, cytoplasmic components are degraded in the vacuole to provide raw materials and energy, and to eliminate components that are toxic or damaged (Yoshimoto 2012). Two types of autophagy are well known in plants: microautophagy and macroautophagy. Macroautophagy, the more extensively studied of the two pathways, involves a double-membrane vesicle, the autophagosome, whose outer membrane merges with the tonoplast and releases the inner membrane and its content to the vacuolar lumen to be degraded (Yoshimoto 2012). The autophagy-related ATG5 gene involved in the macroautophagic machinery has previously been shown to be essential for the zygotic embryogenesis of Scots pine (Vuosku et al. 2015) and for the SE of Norway spruce (Picea abies L.) (Minina et al. 2013). ATG8, another gene involved in macroautophagy, is a ubiquitin-like protein, which is required for the formation of the autophagosome (Nakatogawa et al. 2007). Plants possess nine different ATG8 genes designated ATG8a to ATG8i (Yoshimoto et al. 2004). In wheat (Triticum dicoccoides), ATG8 has been found to be a positive regulator in osmotic and drought stress responses (Kuzuoglu-Ozturk et al. 2012).
Here, we study how the induction of somatic embryo formation affects H2O2 protection, DNA repair, PA metabolism and autophagy, i.e., the mechanisms related to oxidative stress defense in Scots pine embryogenic cell cultures. We show that the direction of cells to the embryogenic pathway is tightly connected with cellular stress responses. In the embryo-producing SE lines, both stress defense and PA metabolism-related genes follow consistent expression profiles that differ considerably from the expression profiles of these genes in the SE lines that were unable to produce embryos.
Materials and methods
Establishment of embryogenic tissue
Immature seed cones were collected from two open-pollinated elite Scots pine mother trees K818 and K884 growing in Punkaharju, Finland (61°48′N; 29°17′E), during one growing season as described in Vuosku et al. (2009). The collection was repeated four times in July throughout the period of embryo development. The cones were collected on 5 July, 12 July, 19 July and 26 July, and each time, 50 SE initiations were made from both mother trees. Thus, the total number of initiations was 400. Immature seeds were excised from the cones and sterilized with PPM™ solution (Preservative for Plant Tissue Culture, Plant Cell Technology, Washington, DC, USA). The seed coat was removed and embryogenic cultures were initiated from immature zygotic embryos with suspensor tissues surrounded by the megagametophyte. The initiation and proliferation of the embryogenic cell masses were carried out as described in Sarjala et al. (1997) on a basal Douglas Fir cotyledon revised (DCR) medium at room temperature in the dark (Gupta and Durzan 1985; modified by Becwar et al. 1990). In the present study, however, DCR medium was gelled by 2.5 g l−1 Phytagel™ (Sigma-Aldrich Co., St Louis, MO, USA).
Induction and maturation of somatic embryos
The SE cultures were evaluated after 2 months of proliferative growth and eight vigorous SE lines (biological replicates) were selected for the induction of somatic embryo development and maturation. Four SE lines (K818-1, -2, -3 and -4) originating from the mother tree K818 and four SE lines (K884-5, -6, -7 and -8) originating from K884 were used in the experiments. In addition to these young SE lines, two old SE lines, K818-9 and K884-10, which had been established 2 years earlier and kept on routine subculturing since then, were included in the study. After proliferation, the embryogenic cell masses were cut into 2 g pieces and transferred onto growth-regulator-free DCR0 medium. Ten plates containing seven cell mass pieces were set up from each SE line and one plate per SE line was used on every sampling date. After 2 weeks on DCR0 medium, the first sampling was performed, and the embryogenic cell masses were transferred onto DCR medium with 90 μM ABA, 7% (w/v) PEG 4000 and 5 g l−1 Phytagel™ for 2 weeks. Thereafter, the second sampling was performed and the cell masses were subsequently transferred onto DCR medium with ABA but without PEG for 2 weeks. After the third sampling, the cell masses were transferred back onto DCR0 medium with 2.5 g l−1 Phytagel™ and observed for embryo production for the next 8 weeks (see Figure S3 available as Supplementary Data at Tree Physiology Online). The cell mass samples were frozen in liquid nitrogen and stored at −80 °C until use.
Analysis of PAs by high-performance liquid chromatography
For the PA (Put, Spd and Spm/tSpm) analyses, samples of 250–300 mg fresh weight (FW) from embryogenic cell masses were homogenized in liquid nitrogen and extracted in 5% (w/v) perchloric acid. The crude extract was used to determine free PAs. Perchloric acid-soluble conjugated PAs were determined from hydrolyzed supernatant. The samples were dansylated and analyzed with high-performance liquid chromatography as described in detail in Flores and Galston (1982), Smith and Davies (1985) and Sarjala and Kaunisto (1993). This method does not distinguish between Spm and potential tSpm.
RNA isolation, reverse transcription and cDNA cloning
Total RNA from the embryogenic cell masses (300 mg) was extracted for the gene expression analyses using the automatic magnetic-based KingFisher™ mL method (Thermo Fisher Scientific, Waltham, MA, USA) with the MagExtractor® total RNA purification kit (Toyobo, Osaka, Japan) according to the manufacturer’s instructions. The samples were treated with RNase-free DNase (Thermo Fisher Scientific) and purified with the NucleoSpin® RNA Clean-Up kit (Macherey-Nagel, Oensingen, Switzerland). RNA yields were measured three times using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific), and 200 ng of each RNA sample was subsequently used for the cDNA synthesis by the SuperScript® VILO™ cDNA Synthesis kit (Thermo Fisher Scientific). No intact RNA could be extracted from the SE lines K818-1 and K884-7, and thus they were removed from the analyses.
The fragment of the putative Scots pine ATG8 gene was amplified by standard polymerase chain reaction (PCR) using cDNA as a template, gene-specific primers and DyNAzyme™ EXT polymerase (Thermo Fisher Scientific). The fragment with the appropriate length was gel purified by Montage DNA Gel Extraction Kit (Merck Millipore, Darmstadt, Germany), cloned by TOPO TA Cloning Kit (Thermo Fisher Scientific) and sequenced by an Applied Biosystems 3730 DNA analyzer.
Quantitative real-time PCR
The real-time reverse-transcription PCR analysis (Q-RT-PCR) was used for the quantification of the expression of the Scots pine PA metabolism genes arginine decarboxylase (ADC; AF306451), spermidine synthase (SPDS; HM236827), tSpm synthase (ACL5; HM236828) and diamine oxidase (DAO; HM236829), CAT (EU513163), DNA repair genes RAD51 (JN566226) and KU80 (JN566225), as well as the autophagy-related genes ATG5 (KM046993) and ATG8 (KP864676). The alignments of the amino acid sequences of the Scots pine putative genes ADC, SPDS, ACL5, DAO, CAT, RAD51, KU80, ATG5 and ATG8 with the corresponding amino acid sequences of Arabidopsis are presented in Data S1 available as Supplementary Data at Tree Physiology Online. No spermine synthase (SPMS) genes have been found from gymnosperms so far (Minguet et al. 2008). The ACT primers, which have been designed against Pinus contorta Dougl. ex Loud. actin gene (M36171) containing an intron in the sequence between the primers, were used for revealing possible genomic DNA contamination in the cDNA samples. The functioning of the ACT primers has been previously shown in Jaakola et al. (2004). In our preceding study, we used both absolute and relative Q-RT-PCR analyses for studying gene expressions during the Scots pine zygotic embryogenesis and found the results mainly consistent with each other (Vuosku et al. 2015), although it has been previously reported that it is challenging to find suitable endogenous reference genes for the pine embryogenesis (Gonçalves et al. 2005). In the present study, the absolute Q-RT-PCR was chosen, because it allows the precise quantification of the target mRNA (reverse transcribed to cDNA) based on a standard curve constructed in the same quantification assay. The standard curves were generated using serial 10-fold dilutions of synthesized RNA molecules to control variability during both RT and PCR steps of Q-RT-PCR runs. The DNA templates from which the RNA molecules could be transcribed were amplified by basic PCR procedure using gene-specific primers (see Table S1 available as Supplementary Data at Tree Physiology Online). The upstream primers contained T7 promoter sequence (TAATACGACTCACTATAGGG) and the downstream primers contained poly(T) tail at their 5′ end. The DNA molecules were subsequently used as templates for in vitro transcription by T7 RNA polymerase. The numbers of standard RNA molecules added to the reverse-transcription reactions were calculated using the molecular weights of the oligonucleotides and Avogadro’s constant (6.022 × 1023 mol−1).
The PCR amplification conditions were optimized for the LightCycler® 480 instrument (Roche Diagnostics, Espoo, Finland), and the subsequent PCR runs showed a single PCR product during melting curve and electrophoretic analysis. The real-time PCR amplifications were performed using LightCycler® 480 SYBR Green I Master mix (Roche Molecular Biochemicals, Mannheim, Germany), 2 μM gene-specific primers (see Table S2 available as Supplementary Data at Tree Physiology Online) and 2 μl cDNA (1 : 10 dilution) in the reaction volume of 20 μl. The real-time PCR amplification was initiated by incubation at 95 °C for 10 min, followed by 45 cycles of 10 s at 95 °C, 10 s at 58 °C and 10 s at 72 °C.
Phylogenetic analyses
For studying the evolution of plant ATG5 and ATG8 sequences, phylogenetic analyses were performed using the neighbor-joining algorithm (Tamura et al. 2011). Nucleotide sequences for the analyses were obtained by BLAST searches against the Scots pine ATG5 and ATG8 sequences in the NCBI databases (http://www.ncbi.nlm.nih.gov). The phylogenetic analyses were conducted in MEGA 5.10 and the bootstrap method (Felsenstein 1985) with 1000 replicates was used to evaluate the confidence of the reconstructed trees. Bootstrap values between 70 and 100% have been suggested to indicate significant support for a branch (Soltis and Soltis 2003).
Statistical analyses
The significance of differences in the PA contents between the SE lines (biological replicates) originated from the different mother trees as well as between the sampling dates was analyzed with the nonparametric Mann–Whitney U test, whereas the significance of differences in the number of mRNA transcripts was analyzed by one-way analysis of variance. Statistical analyses included the young SE lines excluding K818-1 and K884-7, and were conducted using R software package (v. 3.0.2) (Ihaka and Gentleman 1996).
Results
Efficiency of Scots pine SE
From the original 400 Scots pine SE initiations, 11 (i.e., 2.75%) started to produce embryogenic cell mass. During the maturation stage, two SE lines, K818-4 and K884-8, produced somatic embryos. Thus, considering the original number of SE initiations, the efficiency of SE was 0.5%.
Polyamine metabolism during the induction of somatic embryo development
All SE lines contained Put, Spd and Spm/tSpm. Considering both free and conjugated PAs, Put was the most abundant PA, followed by Spd and Spm/tSpm (Figure 1). The concentrations of free PAs varied between the SE lines, but no differences between the SE lines originating from the different mother trees nor between the sampling days were observed. In the beginning of the experiment, the concentration of free Spd was slightly higher in the embryo-producing cell lines compared with the SE lines that were not able to produce embryos. Also, the decrease of all free PA concentrations during the ABA + PEG treatment was typical for the embryo-producing SE lines. The concentration of free Put and Spd continued to decrease during ABA treatment in K818-4 but not in K884-8. In the two old SE lines, the concentrations of free PAs were generally higher than in the young SE lines.
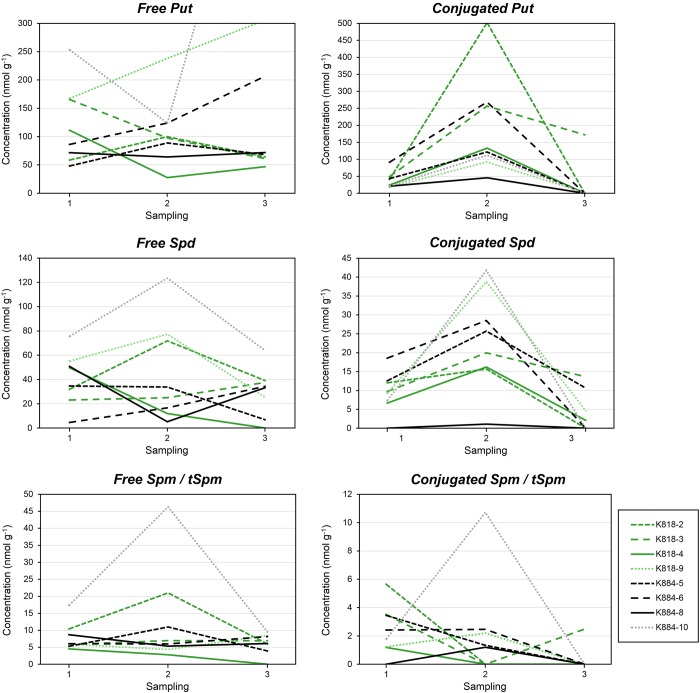
Figure 1.
Free and soluble conjugated PA concentrations (nmol g−1 FW) in Scots pine embryogenic cell masses representing different cell lines during the induction of SE. The SE lines originated from the mother trees K818 (gray lines (green lines online)) and K884 (black lines). From the young SE lines, two (solid lines) out of six produced embryos and four did not (dashed lines). The old SE lines are marked with small dash (with lighter colors online). Every observation point represents an arithmetic mean from two PA analyses.
The concentrations of conjugated Put and Spd increased during the ABA + PEG treatment and decreased during the ABA treatment in all SE lines. The differences were significant between the first and second sampling days (P = 0.009) and between second and third sampling days (P = 0.016) in conjugated Put but not in Spd. The concentration of conjugated Spm/tSpm showed remarkable variation between the SE lines. In the old SE lines, the concentration of conjugated Spd was higher compared with the young SE lines as well as the concentration of conjugated Spm/tSpm in the old SE line K884-10. Mother tree had no effect on the concentrations of conjugated PAs.
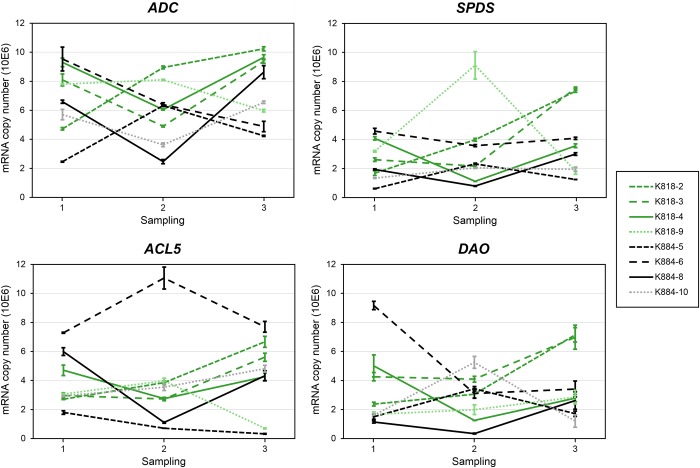
Generally, the expression of the PA genes ADC, SPDS, ACL5 and DAO varied between the SE lines and no difference between the SE lines originating from the different mother trees was observed. However, in the embryo-producing SE lines, all four PA genes showed similar expression profiles: their expression decreased during the ABA + PEG treatment and increased during the ABA treatment (Figure 2) when tissue differentiation was expected to proceed. In the old SE lines, the expression of the PA biosynthetic genes was not higher than in the young SE lines despite the high PA concentrations in the old SE lines.
Figure 2.
Expression of PA metabolism genes ADC, SPDS, ACL5 and DAO (mRNA copy number) in Scots pine embryogenic cell masses representing different cell lines during the induction of SE. The SE lines originated from the mother trees K818 (gray lines (green lines online)) and K884 (black lines). From the young SE lines, two (solid lines) out of six produced embryos and four did not (dashed lines). The old SE lines are marked with small dash (with lighter colors online). Every observation point represents an arithmetic mean ± standard error from three Q-RT-PCR analyses.
Effects of the induction of somatic embryo development on H2O2 protection, DNA repair and autophagy-related gene expression
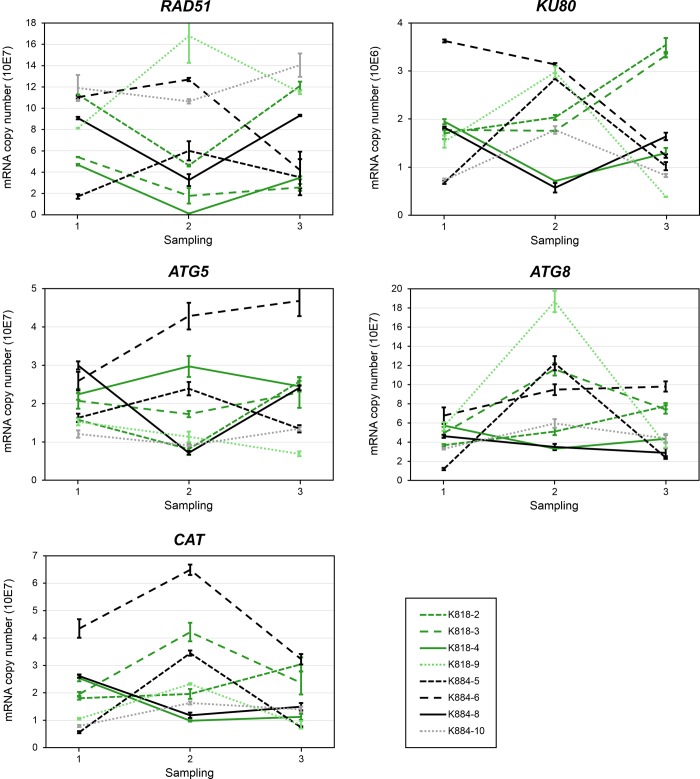
The expression profile of CAT in the embryo-producing SE lines was similar to that of the PA genes: decreasing during the ABA + PEG treatment and increasing during the ABA treatment. In all the other cell lines, except K818-2, the expression of CAT was opposite to that of the embryo-producing cell lines, increasing during the ABA + PEG treatment and decreasing during the ABA treatment. Mother tree had no effect on the expression of CAT (Figure 3).
Figure 3.
Expression of stress-related genes RAD51, KU80, ATG5, ATG8 and CAT (mRNA copy number) in Scots pine embryogenic cell masses representing different cell lines during the induction of SE. The SE lines originated from the mother trees K818 (gray lines (green lines online)) and K884 (black lines). From the young SE lines, two (solid lines) out of six produced embryos and four did not (dashed lines). The old SE lines (K818-9 and K884-10) are marked with small dash (with lighter colors online). Every observation point represents an arithmetic mean ± standard error from three Q-RT-PCR analyses.
Also, the expression of DNA repair genes RAD51 and KU80 decreased during the ABA + PEG treatment and increased during the ABA treatment in the embryo-producing SE lines. In the SE lines that did not produce embryos, there was considerable variation in the expression of both genes, and no difference between the SE lines originating from the different mother trees was observed (Figure 3).
In the expression of ATG5, there was considerable variation among all the SE lines, and no similar expression pattern was observed between the embryo-producing SE lines. In K818-4, ATG5 expression increased during the ABA + PEG treatment and decreased during the ABA treatment, whereas in K884-8, the expression of ATG5 was the opposite, decreasing during the ABA + PEG treatment and increasing during the ABA treatment (Figure 3). The ATG5-sequence-based phylogeny (see Figure S4 available as Supplementary Data at Tree Physiology Online) was consistent with the current view on the evolution of green plants in which morphologically simple photosynthetic forms, such as unicellular green algae, gave rise to multicellular forms and, further, morphologically simple plants, such as bryophytes, were followed by more complex flowering forms (Qiu and Palmer 1999).
In the expression of ATG8, there was considerable variation between the SE lines that did not produce embryos. In the embryo-producing SE lines, the expression of ATG8 was notably low and slightly decreasing during the ABA + PEG treatment. During the ABA treatment, however, the expression of ATG8 slightly increased in K818-4 but continued to decrease in K884-8. Mother tree had no effect on the expression of ATG5 or ATG8 (Figure 3). The Scots pine ATG8 gene, sequenced in the present study, was submitted to GenBank with the accession number KP864676. In the phylogenetic analysis, Scots pine ATG8 shows close relationship with the ATG8f genes of dicot angiosperms (see Figure S5 available as Supplementary Data at Tree Physiology Online).
Discussion
The efficiency of Scots pine SE is dependent on the mother tree (Lelu et al. 1999, Niskanen et al. 2004, Park et al. 2006, Lelu-Walter et al. 2008, Aronen et al. 2009). Niskanen et al. (2004) found that the effect of parents’ genotypes varies during the SE process and the maternal effect is considerable at the initiation stage and at the somatic embryo maturation stage. Even though selected mother trees, which produced somatic embryos in previous studies (Häggman et al. 1999, Niemi et al. 2002, Niemi and Häggman 2002, Aronen et al. 2009, Latutrie and Aronen 2013), were used in the present study, the proportion of successfully proliferating cellular masses as well as the proportion of embryo-producing lines remained low. Previously, variable initiation success percentages such as 0.2–9% (Keinonen-Mettälä et al. 1996), 0.2–4% (Häggman et al. 1999), 1–22.5% (Lelu et al. 1999), 1–42% (Niskanen et al. 2004), 2.5–19.7% (Park et al. 2006), 3–25% (Lelu-Walter et al. 2008) and 3–30% (Aronen et al. 2009) have been reported for Scots pine embryogenic cultures. In the present study, more cell lines were eliminated both at the initiation stage and the maintenance culture stage than at the maturation stage, which is consistent with the results previously reported by Niskanen et al. (2004).
Polyamines play an important role in both zygotic embryogenesis and SE in conifers (Minocha et al. 1993, 1999, 2004, Vuosku et al. 2006), and PA contents vary in embryogenic cells during the different stages of the SE process (Minocha et al. 2004, Vuosku et al. 2012). In the present study, the induction of Scots pine somatic embryo development was connected by the specific expression profiles of the PA metabolism-related genes. The Scots pine embryo-producing SE lines were characterized by similar expression in the PA genes, but the same kind of consistency was not observed in any of the SE lines that did not produce embryos. However, the PA gene expression profiles did not fully correspond with the PA concentrations, pointing out the multiple levels in the regulation of PA metabolism (Kusano et al. 2008, Perez-Leal and Merali 2012).
Previously, free Spd has been shown to be the most common PA at the maturation stage of somatic embryos of Scots pine (Niemi et al. 2002) and other pine species (Minocha et al. 1999), as well as in developing zygotic embryos of Scots pine (Vuosku et al. 2006). In the present study, the concentration of free Spd was highest in the SE lines that were able to produce embryos in the beginning of the maturation phase, suggesting the importance of free Spd in the embryo production. However, Put was the most abundant PA both in the embryo-producing SE lines and in the SE lines that did not produce embryos. That was unexpected because high Put concentrations characterize the exponential growth phase in Scots pine embryogenic cultures (Vuosku et al. 2012), and furthermore, high Put concentrations have been associated with the inability to induce somatic embryo production in Austrian pine (Pinus nigra Arn. ssp. Austriaca) (Noceda et al. 2009).
The concentrations of free and conjugated Spm/tSpm were low throughout the entire early maturation phase, suggesting that Spm/tSpm plays a minor role in the induction and maturation of somatic embryos. Spm/tSpm seems not to be essential for plant embryogenesis because the loss-of-function mutants of the SPMS and ACL5 genes of Arabidopsis also showed normal embryo development (Imai et al. 2004, Kakehi et al. 2008). In adult Arabidopsis plants, Spm deficiency had no effect on the phenotype but caused hypersensitivity to drought and salt stress (Yamaguchi et al. 2006, 2007), whereas loss-of-function mutations of the AtACL5 gene resulted in a severely dwarfed phenotype (Hanzawa et al. 2000). The role of auxin as the main regulator of vascular differentiation is well documented (Miyashima et al. 2013), and recent findings on xylem differentiation have proposed a model of complex functional interaction between auxin, tSpm and HD—ZIP III genes (Milhinhos et al. 2013, Baima et al. 2014). In SE of pine species, including Scots pine, it is common that the proliferation type of growth will continue if auxin has not been removed at least for one passage before transferring cell masses to maturation medium (first with ABA and PEG and thereafter with ABA only). However, as demonstrated in Arabidopsis embryos (Liu et al. 1993), it is obvious that auxin plays a role also in the Scots pine SE. Taken together, the observations in the previous studies and the low Spm/tSpm concentrations found in the present study reveal that the tSpm-related regulation mechanisms controlling auxin signaling during xylem differentiation may not operate during early embryo formation.
In Scots pine SE cultures, the prolonged maintenance culture stage seems to cause permanent changes to the PA metabolism that may partly be a reason for the loss of embryogenic potential. Free PA concentrations were generally higher in the old SE lines than in the young ones, which suggests that the old lines ignored the signals triggering the cells to the embryogenic path and stayed at the proliferative stage. Previously, we found that the high concentration of free Put is typical for the Scots pine embryogenic cultures during exponential growth phase (Vuosku et al. 2012). Also, in red spruce (Picea rubens Sarg.) and Norway spruce embryogenic cultures, PA concentrations are higher at the proliferative stage than during the induction of somatic embryos (Minocha et al. 1993).
Catalases are considered as a sink for H2O2 (Mhamdi et al. 2012) which, besides causing oxidative stress, is an important signaling molecule in stress responses (Veal and Day 2011) as well as in the development of somatic embryos of conifers (Zhang et al. 2010). In Japanese larch (Larix leptolepis) SE, the amount of H2O2 increased dramatically after transfer onto ABA + PEG medium while the expression of CAT remained low. Later, however, when the amount of H2O2 increased again, it was followed by an increase in the expression of CAT as well (Zhang et al. 2010). In conifer seeds, both metabolic activity and moisture content fluctuate drastically during zygotic embryogenesis because the maturation drying is an essential part of the developmental process (Kapik et al. 1995, Carrier et al. 1999, Silveira et al. 2004, Vuosku et al. 2006). Thus, the sources of ROS production, connected to basic cellular and specific seed developmental processes, also vary (Bailly et al. 2008). In our previous study, we found that during the Scots pine seed development, high amounts of H2O2 are generated in the seed coat, flight wing, nucellar cap, nucellar layers and the megaspore membranes, whereas H2O2 could not be localized in the embryo or megagametophyte tissues (Vuosku et al. 2015). At the early embryogeny of a developing Scots pine seed, strong CAT expression was detected in the cells of the dominant embryo and in the megagametophyte cells. CAT expression, however, faded out in the megagametophyte when the embryogenesis proceeded to the late developmental stage/maturation drying but increased again during the imbibition phase of the seed germination (Vuosku et al. 2015). In liquid cultures of Scots pine embryogenic cells, CAT expression increased during proliferative growth (Vuosku et al. 2012) but CAT was not upregulated under PEG-induced stress (Muilu-Mäkelä et al. 2015). As a whole, the findings from Scots pine zygotic embryogenesis and SE suggest that CAT protection against H2O2 damage is more connected to active metabolism than to dehydration-related oxidative stress. In the present study, CAT expression decreased during the ABA + PEG treatment in the embryo-producing SE lines and increased in the SE lines that were unable to produce embryos. The results suggest that the SE lines that were unable to produce embryos experienced the PEG treatment mostly as drought stress and responded uncharacteristically by increasing the CAT expression in order to remove H2O2. Alternatively, intense CAT expression in those SE lines may be associated with active metabolism during cell proliferation and, thus, with the inability to suppress proliferative growth and direction to the embryo-producing pathway.
The expression of RAD51 and KU80 revealed that the Scots pine embryogenic cells use both HR and NHEJ to repair DSBs. The use of the HR pathway may result from the fact that the SE lines were derived from immature embryos in which the preventing of DNA mutations is especially important. In somatic cells, DSBs are usually repaired via the NHEJ pathway in plants (Waterworth et al. 2011). In the embryo-producing SE lines, the expression profiles of both RAD51 and KU80 suggest that the DNA damages accumulate during the ABA + PEG treatment and the repairing processes start later, during the ABA treatment.
Many of the SE lines that were unable to produce embryos showed increased ATG8 expression during the ABA + PEG treatment, indicating the activation of autophagy as a stress response. In the embryo-producing SE lines, ATG8, instead, was expressed slightly throughout the induction phase and showed even a decreasing trend during the ABA + PEG treatment. Unlike all the other genes, ATG5 was expressed differently in the embryo-producing SE lines. This is consistent with our previous results showing that the gene expression level regulation of ATG5 is associated with developmental cell death rather than with osmotic stress caused by maturation drying in the Scots pine embryogenesis (Vuosku et al. 2015).
All in all, our findings provide a new viewpoint on the factors affecting the induction of somatic embryo development in coniferous embryogenic cultures. The Scots pine SE lines producing somatic embryos responded at the gene expression level to the ABA + PEG treatment similarly to each other but very differently compared with the SE lines unable to produce embryos. The triggering of the embryo-producing pathway was connected to the specific changes in PA metabolism as well as to the moderate stress responses. That is to say, the SE lines experienced the ABA + PEG treatment as a signal to induce embryo production. Instead, the SE lines that did not produce embryos seemed to initiate stress defense by removing H2O2, repairing DNA and destroying cellular components by autophagy. The results suggest that the function of CAT may be critical to embryo production, although further protein level studies are needed to confirm the possible role of CAT. We propose that the manipulation of stress response pathways may provide a new way to enhance somatic embryo production in recalcitrant Scots pine lines.
Supplementary data
Supplementary data for this article are available at Tree Physiology online.
Conflict of interest
None declared.
Funding
The research was funded by the Academy of Finland (Project 121994 to T.S.) and Thule Institute at the University of Oulu (to H.H.). The research was conducted as a part of the Bioeconomy Research Community of the University of Oulu.
Supplementary Material
Acknowledgments
We are grateful to the personnel of the Natural Resources Institute Finland for conducting the cone collections and Ms Eeva Pihlajaviita for her skillful technical help. Our sincere thanks also to Ms Taina Uusitalo at the Genetics and Physiology Department in the University of Oulu for her skillful help in the laboratory.
References
- Alcázar R, Altabella T, Marco F, Bortolotti C, Reymond M, Koncz C, Carrasco P, Tiburcio AF (2010) Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231:1237–1249. doi:10.1007/s00425-010-1130-0 [DOI] [PubMed] [Google Scholar]
- Amarasinghe V, Dhami R, Carlson JE (1996) Polyamine biosynthesis during somatic embryogenesis in interior spruce (Picea glauca × Picea engelmannii complex). Plant Cell Rep 15:495–499. doi:10.1007/BF00232981 [DOI] [PubMed] [Google Scholar]
- Aronen T, Pehkonen T, Ryynänen L (2009) Enhancement of somatic embryogenesis from immature zygotic embryos of Pinus sylvestris. Scand J For Res 24:372–383. doi:10.1080/02827580903228862 [Google Scholar]
- Attree SM, Fowke LC (1993) Embryogeny of gymnosperms: advances in synthetic seed technology of conifers. Plant Cell Tissue Organ Cult 35:1–35. doi:10.1007/BF00043936 [Google Scholar]
- Bailly C, El-Maarouf-Bouteau H, Corbineau F (2008) From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C R Biol 331:806–814. doi:10.1016/j.crvi.2008.07.022 [DOI] [PubMed] [Google Scholar]
- Baima S, Forte V, Possenti M, Peñalosa A, Leoni G, Salvi S, Barbara Felici B, Ruberti I, Morelli G (2014) Negative feedback regulation of auxin signaling by ATHB8/ACL5-BUD2 transcription module. Mol Plant 7:1006–1025. doi:10.1093/mp/ssu051 [DOI] [PubMed] [Google Scholar]
- Basu HS, Shafer RH, Marton LJ (1987) A stopped-flow H-D exchange kinetic study of spermine-polynucleotide interactions. Nucleic Acids Res 15:5873–5886. doi:10.1093/nar/15.14.5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becwar MR, Nagmani R, Wann SR (1990) Initiation of embryogenic cultures and somatic embryo development in loblolly pine (Pinus taeda). Can J For Res 20:810–817. doi:10.1139/x90-107 [Google Scholar]
- Bonga JM, Klimaszewska KK, von Aderkas P (2010) Recalcitrance in clonal propagation, in particular of conifers. Plant Cell Tissue Organ Cult 100:241–254. doi:10.1007/s11240-009-9647-2 [Google Scholar]
- Carrier DJ, Kendall EJ, Bock CA, Cunningham JE, Dunstan DI (1999) Water content, lipid deposition, and (+)-abscisic acid content in developing white spruce seeds. J Exp Bot 50:1359–1364. doi:10.1093/jxb/50.337.1359 [Google Scholar]
- Fehér A. (2005) Why somatic plant cells start to form embryos? Plant Cell Monogr 2:85–101. doi:10.1007/7089_019 [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi:10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Feuerstein BG, Pattabiraman N, Marton LJ (1990) Molecular mechanics of the interactions of spermine with DNA: DNA bending as a result of ligand binding. Nucleic Acids Res 18:1271–1282. doi:10.1093/nar/18.5.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores HE, Galston AW (1982) Analysis of polyamines in higher plants by high performance liquid chromatography. Plant Physiol 69:701–706. doi:10.1104/pp.69.3.701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa S, Kadoma Y (2005) Kinetic evaluation of polyamines as radical scavengers. Anticancer Res 25:965–969. [PubMed] [Google Scholar]
- Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930. doi:10.1016/j.plaphy.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Gonçalves S, Cairney J, Maroco J, Oliveira MM, Miguel C (2005) Evaluation of control transcripts in real-time RT-PCR expression analysis during maritime pine embryogenesis. Planta 222:556–563. doi:10.1007/s00425-005-1562-0 [DOI] [PubMed] [Google Scholar]
- Groppa MD, Benavides MP (2008) Polyamines and abiotic stress: recent advances. Amino Acids 34:35–45. doi:10.1007/s00726-007-0501-8 [DOI] [PubMed] [Google Scholar]
- Gupta PK, Durzan DJ (1985) Shoot multiplication from mature trees of Douglas-fir (Pseudotsuga menziesii) and Sugar pine (Pinus lambertiana). Plant Cell Rep 4:177–179. doi:10.1007/BF00269282 [DOI] [PubMed] [Google Scholar]
- Ha HC, Sirisoma NS, Kuppusamy P, Zweier JL, Woster PM, Casero RA Jr (1998) The natural polyamine spermine functions directly as a free radical scavenger. Proc Natl Acad Sci USA 95:11140–11145. doi:10.1073/pnas.95.19.11140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häggman H, Jokela A, Krajnakova J, Kauppi A, Niemi K, Aronen T (1999) Somatic embryogenesis of Scots pine: cold treatment and characteristics of explants affecting induction. J Exp Bot 50:1769–1778. doi:10.1093/jxb/50.341.1769 [Google Scholar]
- Häggman H, Vuosku J, Sarjala T, Jokela A, Niemi K (2006) Somatic embryogenesis of pine species: from functional genomics to plantation forestry. In: Mujib A, Samaj J (eds) Somatic embryogenesis. In Series: Plant Cell Monographs, Volume 2 Springer, New York, pp 119–140. [Google Scholar]
- Han S, Yu B, Wang Y, Liu Y (2011) Role of plant autophagy in stress response. Protein Cell 2:784–791. doi:10.1007/s13238-011-1104-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y, Takahashi T, Michael AJ, Burtin D, Long D, Pineiro M, Coupland G, Komeda Y (2000) ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J 19:4248–4256. doi:10.1093/emboj/19.16.4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman RR (1996) A language for data analysis and graphics. J Comput Graph Stat 5:299–314. [Google Scholar]
- Imai A, Akiyama T, Kato T, Sato S, Tabata S, Yamamoto KT, Takahashi T (2004) Spermine is not essential for survival of Arabidopsis. FEBS Lett 556:148–152. doi:10.1016/S0014-5793(03)01395-4 [DOI] [PubMed] [Google Scholar]
- Jaakola L, Pirttilä AM, Vuosku J, Hohtola A (2004) Method based on electrophoresis and gel extraction for obtaining genomic DNA-free cDNA without DNase treatment. Biotechniques 37:744–748. [DOI] [PubMed] [Google Scholar]
- Kakehi J, Kuwashiro Y, Niitsu M, Takahashi T (2008) Thermospermine is required for stem elongation in Arabidopsis thaliana. Plant Cell Physiol 49:1342–1349. doi:10.1093/pcp/pcn109 [DOI] [PubMed] [Google Scholar]
- Kapik RH, Dinus RJ, Dean JFD (1995) Abscisic acid and zygotic embryogenesis in Pinus taeda. Tree Physiol 15:485–490. doi:10.1093/treephys/15.7-8.485 [DOI] [PubMed] [Google Scholar]
- Kaur-Sawhney R, Tiburcio AF, Altabella T, Galston AW (2003) Polyamines in plants: an overview. J Cell Mol Biol 2:1–12. [Google Scholar]
- Keinonen-Mettälä K, Jalonen P, Eurola P, von Arnold S, von Weissenberg K (1996) Somatic embryogenesis of Pinus sylvestris. Scand J For Res 11:242–250. doi:10.1080/02827589609382933 [Google Scholar]
- Kimura S, Sakaguchi K (2006) DNA repair in plants. Chem Rev 106:753–766. doi:10.1021/cr040482n [DOI] [PubMed] [Google Scholar]
- Krejci L, Altmannova V, Spirek M, Zhao X (2012) Homologous recombination and its regulation. Nucleic Acids Res 40:5795–5818. doi:10.1093/nar/gks270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano T, Berberich T, Tateda C, Takahashi Y (2008) Polyamines: essential factors for growth and survival. Planta 228:367–381. doi:10.1007/s00425-008-0772-7 [DOI] [PubMed] [Google Scholar]
- Kuzuoglu-Ozturk D, Cebeci O, Bala Y, Akpinar A, Mitou G, Korkmaz G, Gozuacik D, Budak H (2012) Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta 236:1081–1092. doi:10.1007/s00425-012-1657-3 [DOI] [PubMed] [Google Scholar]
- Latutrie M, Aronen T (2013) Long-term cryopreservation of embryogenic Pinus sylvestris cultures. Scand J For Res 28:103–109. doi:10.1080/02827581.2012.701325 [Google Scholar]
- Lelu M-A, Bastien C, Drugeault A, Gouez M-L, Klimaszewska K (1999) Somatic embryogenesis and plantlet development in Pinus sylvestris and Pinus pinaster on medium with and without growth regulators. Physiol Plant 105:719–728. doi:10.1034/j.1399-3054.1999.105417.x [Google Scholar]
- Lelu-Walter M-A, Bernier-Cardou M, Klimaszewska K (2008) Clonal plant production from self- and cross-pollinated seed families of Pinus sylvestris (L.) through somatic embryogenesis. Plant Cell Tissue Organ Cult 92:31–45. doi:10.1007/s11240-007-9300-x [Google Scholar]
- Lelu-Walter M-A, Thompson D, Harvengt L, Sanchez L, Toribio M, Pâgues LE (2013) Somatic embryogenesis in forestry with a focus on Europe: state-of-the-art, benefits, challenges and future direction. Tree Genet Genomes 9:883–899. doi:10.1007/s11295-013-0620-1 [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101:10596–10601. doi:10.1073/pnas.0404110101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CM, Xu ZH, Chua NH (1993) Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 5:621–630. doi:10.1105/tpc.5.6.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-H, Kitashiba H, Wang J, Ban Y, Moriguchi T (2007) Polyamines and their ability to provide environmental stress tolerance to plants. Plant Biotechnol 24:117–126. doi:10.5511/plantbiotechnology.24.117 [Google Scholar]
- Mhamdi A, Noctor G, Baker A (2012) Plant catalases: peroxisomal redox guardians. Arch Biochem Biophys 525:181–194. doi:10.1016/j.abb.2012.04.015 [DOI] [PubMed] [Google Scholar]
- Milhinhos A, Prestele J, Bollhöner B et al. (2013) Thermospermine levels are controlled by an auxin-dependent feedback loop mechanism in Populus xylem. Plant J 75:685–698. doi:10.1111/tpj.12231 [DOI] [PubMed] [Google Scholar]
- Minguet EG, Vera-Sirera F, Marina A, Carbonell J, Blazquez MA (2008) Evolutionary diversification in polyamine biosynthesis. Mol Biol Evol 25:2119–2128. doi:10.1093/molbev/msn161 [DOI] [PubMed] [Google Scholar]
- Minina EA, Filonova LH, Fukada K et al. (2013) Autophagy and metacaspase determine the mode of cell death in plants. J Cell Biol 203:917–927. doi:10.1083/jcb.201307082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocha R, Kvaalen H, Minocha SC, Long S (1993) Polyamines in embryogenic cultures of Norway spruce (Picea abies) and red spruce (Picea rubens). Tree Physiol 13:365–377. doi:10.1093/treephys/13.4.365 [DOI] [PubMed] [Google Scholar]
- Minocha R, Smith DR, Reeves C, Steele KD, Minocha SC (1999) Polyamine levels during the development of zygotic and somatic embryos of Pinus radiata. Physiol Plant 105:155–164. doi:10.1034/j.1399-3054.1999.105123.x [Google Scholar]
- Minocha R, Minocha SC, Long S (2004) Polyamines and their biosynthetic enzymes during somatic embryo development in red spruce (Picea rubens sarg.). In Vitro Cell Dev Biol Plant 40:572–580. doi:10.1079/IVP2004569 [Google Scholar]
- Miyashima S, Sebastian J, Lee JY, Helariutta Y (2013) Stem cell function during plant vascular development. EMBO J 32:178–193. doi:10.1038/emboj.2012.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E, Iliakis G (2011) Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res 711:61–72. doi:10.1016/j.mrfmmm.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Muilu-Mäkelä R, Vuosku J, Hamberg L, Latva-Mäenpää H, Häggman H, Sarjala T (2015) Osmotic stress affects polyamine homeostasis and phenolic content in proembryogenic liquid cell cultures of Scots pine. Plant Cell Tissue Organ Cult 122:709–726. doi:10.1007/s11240-015-0805-4 [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130:165–178. doi:10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Niemi K, Häggman H (2002) Pisolithus tinctorius promotes germination and forms mycorrhizal structures in Scots pine somatic embryos in vitro. Mycorrhiza 12:263–267. doi:10.1007/s00572-002-0181-x [DOI] [PubMed] [Google Scholar]
- Niemi K, Sarjala T, Chen X, Häggman H (2002) Spermidine and methylglyoxal bis(guanylhydrazone) affect maturation and endogenous polyamine content of Scots pine embryogenic cultures. J Plant Physiol 159:1155–1158. doi:10.1078/0176-1617-00634 [Google Scholar]
- Niskanen A-M, Lu J, Seitz S, Keinonen K, von Weissenberg K, Pappinen A (2004) Effect of parent genotype on somatic embryogenesis in Scots pine (Pinus sylvestris). Tree Physiol 24:1259–1265. doi:10.1093/treephys/24.11.1259 [DOI] [PubMed] [Google Scholar]
- Noceda C, Salaj T, Pérez M, Viejo M, Cañal MJ, Salaj J, Rodriguez R (2009) DNA demethylation and decrease on free polyamines is associated with the embryogenic capacity of Pinus nigra Arn. cell culture. Trees 23:1285–1293. doi:10.1007/s00468-009-0370-8 [Google Scholar]
- Park YS, Lelu-Walter M-A, Harvengt L, Trontin JF, MacEacheron I, Klimaszewska K, Bonga JM (2006) Initiation of somatic embryogenesis in Pinus banksiana, P. strobus, P. pinaster and P. sylvestris at three laboratories in Canada and France. Plant Cell Tissue Organ Cult 86:87–101. doi:10.1007/s11240-006-9101-7 [Google Scholar]
- Perez-Leal O, Merali S (2012) Regulation of polyamine metabolism by translational control. Amino Acids 42:611–617. doi:10.1007/s00726-011-1036-6 [DOI] [PubMed] [Google Scholar]
- Qiu YL, Palmer JD (1999) Phylogeny of early land plants: insights from genes and genomes. Trends Plant Sci 4:26–30. doi:10.1016/S1360-1385(98)01361-2 [DOI] [PubMed] [Google Scholar]
- Sarjala T, Kaunisto S (1993) Needle polyamine concentrations and potassium nutrition in scots pine. Tree Physiol 13:87–96. doi:10.1093/treephys/13.1.87 [DOI] [PubMed] [Google Scholar]
- Sarjala T, Häggman H, Aronen T (1997) Effect of exogenous polyamines and inhibitors of polyamine biosynthesis on growth and free polyamine contents of embryogenic Scots pine callus. J Plant Physiol 150:597–602. doi:10.1016/S0176-1617(97)80325-2 [Google Scholar]
- Silveira V, Balbuena TS, Santa-Catarina C, Floh EIS, Guerra MP, Handro W (2004) Biochemical changes during seed development in Pinus taeda L. Plant Growth Regul 44:147–156. doi:10.1023/B:GROW.0000049410.63154.ed [Google Scholar]
- Smith MA, Davies PJ (1985) Separation and quantification of polyamines in plant tissue by high performance liquid chromatography of their dansyl derivatives. Plant Physiol 78:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS (2003) The role of phylogenetics in comparative genetics. Plant Physiol 132:1790–1800. doi:10.1104/pp.103.022509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasolla C, Kong L, Yeung EC, Thorpe AT (2002) Maturation of somatic embryos in conifers: morphogenesis, physiology, biochemistry, and molecular biology. In Vitro Cell Dev Biol Plant 38:93–105. doi:10.1079/IVP2001262 [Google Scholar]
- Takano A, Kakechi J-I, Takahashi T (2012) Thermospermine is not a minor polyamine in the plant kingdom. Plant Cell Physiol 53:606–616. doi:10.1093/pcp/pcs019 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal E, Day A (2011) Hydrogen peroxide as a signaling molecule. Antioxid Redox Signal 15:147–151. doi:10.1089/ars.2011.3968 [DOI] [PubMed] [Google Scholar]
- von Arnold S, Sabala I, Bozhkov P, Dyachok J, Filonova L (2002) Developmental pathways of somatic embryogenesis. Plant Cell Tissue Organ Cult 69:233–249. doi:10.1023/A:1015673200621 [Google Scholar]
- Vuosku J, Jokela A, Läärä E, Sääskilahti M, Muilu R, Sutela S, Altabella T, Sarjala T, Häggman H (2006) Consistency of polyamine profiles and expression of arginine decarboxylase in mitosis during zygotic embryogenesis of Scots pine. Plant Physiol 142:1027–1038. doi:10.1104/pp.106.083030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuosku J, Sarjala T, Jokela A, Sutela S, Sääskilahti M, Suorsa M, Läärä E, Häggman H (2009) One tissue, two fates: different roles of megagametophyte cells during Scots pine embryogenesis. J Exp Bot 60:1375–1386. doi:10.1093/jxb/erp020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuosku J, Suorsa M, Ruottinen M, Sutela S, Muilu-Mäkelä R, Julkunen-Tiitto R, Sarjala T, Neubauer P, Häggman H (2012) Polyamine metabolism during exponential growth transition in Scots pine embryogenic cell culture. Tree Physiol 32:1274–1287. doi:10.1093/treephys/tps088 [DOI] [PubMed] [Google Scholar]
- Vuosku J, Sutela S, Kestilä J, Jokela A, Sarjala T, Häggman H (2015) Expression of catalase and retinoblastoma-related protein genes associates with cell death processes in Scots pine zygotic embryogenesis. BMC Plant Biol 15:88 doi:10.1186/s12870-015-0462-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, Drury GE, Bray CM, West CE (2011) Repairing breaks in the plant genome: the importance of keeping it together. New Phytol 192:805–822. doi:10.1111/j.1469-8137.2011.03926.x [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Takahashi Y, Berberich T, Imai A, Miyazaki A, Takahashi T, Michael A, Kusano T (2006) The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett 580:6783–6788. doi:10.1016/j.febslet.2006.10.078 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Takahashi Y, Berberich T, Imai A, Takahashi T, Michael AJ, Kusano T (2007) A protective role for the polyamine spermine against drought stress in Arabidopsis. Biochem Biophys Res Commun 352:486–490. doi:10.1016/j.bbrc.2006.11.041 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K. (2012) Beginning to understand autophagy, an intracellular self-degradation system in plants. Plant Cell Physiol 53:1355–1365. doi:10.1093/pcp/pcs099 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16:2967–2983. doi:10.1105/tpc.104.025395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S-G, Han S-Y, Yang W-H, Wei H-L, Zhang M, Qi L-W (2010) Changes in H2O2 content and antioxidant enzyme gene expression during the somatic embryogenesis of Larix leptolepis. Plant Cell Tissue Organ Cult 100:21–29. doi:10.1007/s11240-009-9612-0 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.