Abstract
Objective
Abnormal expression of microRNA-215 has been identified in a variety of solid cancers. However, little is known about the expression pattern of microRNA-215 in acute myeloid leukemia. This study was to investigate the status of microRNA-215 expression and further analyze its clinical significance in acute myeloid leukemia.
Methods
Real-time quantitative polymerase chain reaction assay was performed to evaluate the expression level of microRNA-215 in 113 patients with acute myeloid leukemia. Besides, the relationship between microRNA-215 levels and clinical and pathological factors was explored.
Results
Compared with the healthy individuals, microRNA-215 expression in acute myeloid leukemia patients was significantly down-regulated (P= 0.001). MicroRNA-215 low-expressed patients had higher white blood cells than microRNA-215 high-expressed patients (P= 0.014). The incidence of FLT3/ITD mutation in the patients with low microRNA-215 expression was significantly higher than those with high microRNA-215 expression (P= 0.025). MicroRNA-215 low-expressed patients had significantly shorter overall survival than microRNA-215 high-expressed patients in both non-M3 acute myeloid leukemia patients and cytogenetically normal patients (P= 0.017 and P= 0.044, respectively). Meanwhile, multivariate analysis confirmed the adverse prognostic value of microRNA-215 expression in acute myeloid leukemia patients with non-M3 subtypes.
Conclusions
Our study demonstrates that reduced microRNA-215 expression is a common event and is associated with poor clinical outcome in acute myeloid leukemia.
Keywords: MiR-215, acute myeloid leukemia, prognosis, real-time quantitative PCR
Introduction
Acute myeloid leukemia (AML), a common type of hematopoietic malignant disease, is characterized by increased self-renewal, differentiation arrest and malignant proliferation of leukemia progenitor cells, which ultimately interferes with normal production of blood cells (1). Both genetic abnormalities and epigenetic alterations play crucial roles in the above-mentioned pathogenetic process (2). Moreover, it is proven that microRNAs (miRNAs), as a new kind of participators of epigenetic regulation besides DNA methylation and histone modifications, also occupy an indispensable position during leukemogenesis and provide useful prognostic information of AML (3,4).
MicroRNAs are a class of small non-coding RNAs that are identified as the critical regulators of gene expression at the post-transcriptional level by binding to the 3′-untranslated region of their target mRNAs and thus generally participate in a variety of biological processes including cell proliferation, differentiation, viability and apoptosis (5,6). Recently, dysregulation of miRNAs has been linked to cancer initiation and progression, which provides a new perspective to understanding of the process of carcinogenesis (7,8). Furthermore, accumulating studies have also identified that aberrant expression of unique miRNAs in pathological processes is closely associated with the diagnosis as well as prognosis of hematological malignancies (9–11).
MicroRNA-215 (miR-215), identified from the location on chromosome 1q41, has been proven to be a p53-inducible miRNA with the capability of enhancing p21 levels and mediating cell cycle arrest (12–14). Previous studies showed that miR-215 could act as a tumor suppressor gene, and down-regulation of miR-215 was identified in several cancers, such as myeloma (14), nephroblastoma (15), esophageal adenocarcinoma (16), breast cancer (BC) (17) and colon cancer (18,19). However, the status of miR-215 expression and its prognostic value remain unclear in AML. Thus, our study was aimed to investigate the expression pattern and analyze its clinical significance in the patients with AML.
Patients and methods
Patients and samples
The current investigation was approved by the Ethics Committee and Institutional Review Board of the Affiliated People' Hospital of Jiangsu University, China. After written informed consents were signed, a total of 138 bone marrow samples were collected from 25 healthy people who were the hematopoietic stem cell donors and 113 de novo AML patients from January 2008 to August 2015. Based on French–American–British (FAB) and World Health Organization (WHO) criteria combined to immunophenotyping and cytogenetic analysis, the diagnosis and classification of AML patients were established (20–23), including the cases with low-percentage blasts (<20%) in bone marrow with the detection of cytogenetic aberrations, such as t(15;17) (q22;q12). The relevant clinical and laboratory features of the patients are presented in Table 1.
Table 1.
Correlation between miR-215 expression and patients' parameters
| Patients' parameters | Low (n= 47) | High (n= 66) | P |
|---|---|---|---|
| Sex, male/female | 32/15 | 35/31 | 0.124 |
| Median age, years (range) | 57 (20–93) | 57.5 (21–87) | 0.641 |
| Median WBC, ×109/l (range) | 18.4 (0.3–197.7) | 5.9 (0.5–528.0) | 0.014 |
| Median hemoglobin, g/l (range) | 76 (40–138) | 74 (32–133) | 0.108 |
| Median platelets, ×109/l (range) | 28.5 (3–399) | 40 (6–447) | 0.058 |
| BM blasts, % (range) | 45.5 (1–94.5) | 45.8 (3–97.5) | 0.622 |
| FAB | 0.989 | ||
| M0 | 0 | 1 | |
| M1 | 4 | 4 | |
| M2 | 23 | 32 | |
| M3 | 7 | 10 | |
| M4 | 10 | 12 | |
| M5 | 3 | 6 | |
| M6 | 0 | 1 | |
| WHO | 1.000 | ||
| AML with t(8;21) | 7 | 9 | |
| APL with t(15;17) | 7 | 10 | |
| AML with 11q23 translocation | 0 | 1 | |
| AML without maturation | 4 | 4 | |
| AML with maturation | 16 | 23 | |
| Acute myelomonocytic leukemia | 10 | 13 | |
| Acute monoblastic and monocytic leukemia | 3 | 4 | |
| Acute erythroid leukemia | 0 | 1 | |
| Karyotype classification | 0.667 | ||
| Favorable | 14 | 19 | |
| Intermediate | 28 | 34 | |
| Poor | 4 | 10 | |
| No data | 1 | 3 | |
| Karyotype | 0.913 | ||
| Normal | 21 | 28 | |
| t(8;21) | 7 | 9 | |
| t(15;17) | 7 | 10 | |
| 11q23 | 0 | 1 | |
| Complex | 4 | 9 | |
| Others | 7 | 6 | |
| No data | 1 | 3 | |
| Gene mutation | |||
| C/EBPA (+/−) | 3/42 | 12/53 | 0.094 |
| NPM1 (+/−) | 6/39 | 5/60 | 0.352 |
| FLT3/ITD (+/−) | 11/34 | 5/60 | 0.025 |
| C-KIT (+/−) | 3/42 | 0/65 | 0.066 |
| NRAS or KRAS (+/−) | 2/40 | 6/55 | 0.467 |
| IDH1/2 (+/−) | 4/38 | 2/59 | 0.222 |
| DNMT3A (+/−) | 6/36 | 3/58 | 0.154 |
| U2AF1 (+/−) | 1/41 | 4/57 | 0.646 |
| CR(+/−) | 17/28 | 31/32 | 0.326 |
WBC, white blood cells; FAB, French–American–British classification; AML, acute myeloid leukemia; CR, complete remission.
Treatment protocol for AML patients was described previously (24). For non-M3 patients, one or two courses with standard of cytarabine (100 mg/m2) plus daunorubicin (45 mg/m2) 7 + 3 induction therapy were given. Patients who achieved complete remission (CR) were given subsequent high- or medium-dose cytarabine-based chemotherapy treatment for consolidation. For patients older than 65 years, CHG protocol (cytarabine 10 mg/m2 q12 h for 14 days, homoharringtonine 1 mg daily for 14 days and G-CSF 200 g/m2 for 14 days) was administered. For the patients with acute promyelocytic leukemia (APL), induction therapy consisted of oral all-trans retinoic acid (ATRA) 45 mg/m2 per day until morphologic CR and intravenous daunorubicin 45 mg/m2 for 3 days and cytarabine 100 mg/m2 for 7 days. Patients in CR received three monthly consolidation courses consist of daunorubicin (45 mg/m2 for 3 days) and cytarabine (100 mg/m2 for 7 days) as the first, followed by mitoxantrone (8 mg/m2 per day for 3 days) and cytarabine (100 mg/m2 for 7 days) as the second, and the homoharringtonine (2 mg/m2 daily for 7 days) and cytarabine (100 mg/m2 for 7 days) at the last. Patients who were negative for PML/RARA transcript at the end of consolidation were started on maintenance therapy with oral mercaptopurine (50 mg/m2 per day), oral methotrexate (15 mg/m2 per week) and oral ATRA (45 mg/m2 per day for 15 days every 3 months) over 2 years.
RNA isolation and reverse transcription
The mirVana miRNA isolation kit (Ambion, Austin, TX, USA) was used to extract the total RNA. Reverse transcription was performed to synthesize cDNA using MiScript Reverse Transcription Kit (Qiagen, catalog no. 218061). The operations mentioned above were conducted in accordance with the manufacturer's protocols.
MiR-215 level detection
The primers of miR-215 transcript used for real-time quantitative polymerase chain reaction (RQ-PCR) were 5′-GCATGACCTATGAATTGACAGAC-3′ and the manufacturer-provided miScript Universal primer (Qiagen, catalog no. 218061). RQ-PCR was performed using miScript SYBR green PCR kit (Qiagen, catalog no. 218073) in an ABI 7300 Thermo cycler (Applied Biosystems, Foster City, CA, USA). The cycling conditions of the reactions are as follows: 94°C for 15 min for initial denaturation, followed by 40 cycles at 94°C for 15 s for denaturation, 55°C for 30 s for annealing and 70°C for 30 s for extension. Relative expression levels were determined by using the 2−ΔΔCt method from the relevant signals. U6 small nuclear RNA was selected as the endogenous normalizer.
Gene mutation detection
The detections of IDH1/2, DNMT3A, NRAS or KRAS, NPM1, C-KIT and U2AF1 mutations were reported previously (25–28). All samples determined positive by high-resolution melting analysis (HRMA) were further confirmed by direct DNA sequencing. FLT3/ITD and CCAAT enhancer binding protein alpha (C/EBPA) mutations were detected using DNA sequencing (29,30).
Statistical analyses
The statistical analyses in this study were performed using Statistical Program for Social Sciences (SPSS) software, version 20.0 (SPSS, Chicago, IL). Mann–Whitney's U-test and Pearson's chi-square analysis or Fisher's exact test were used to compare the difference of continuous variables and categorical variables between the groups, respectively. Overall survival (OS), defined as the time between the initial diagnosis and death or the last follow-up, was compared to show any significant associations between miR-215 expression and the survival of the AML patients according to Kaplan–Meier method and a Cox proportional hazards model was performed further to determine the impact of miR-215 expression. A two-tailed P-value <0.05 was considered to indicate a statistically significant result.
Results
Down-regulation of miR-215 in AML patients
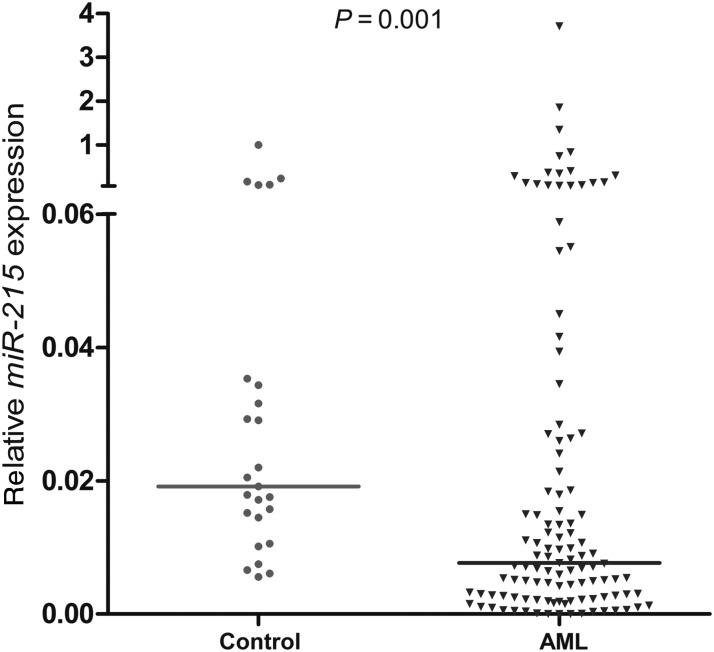
The median level of miR-215 transcript in controls was 0.019 with a range from 0.006 to 1.000. Compared with controls, we demonstrated a significantly decreased expression of miR-215 in AML patients (range 0.000–3.713, median 0.008) (P= 0.001) (Fig. 1).
Figure 1.
Relative expression levels of miR-215 expression in acute myeloid leukemia (AML) patients and controls. The level of miR-215 was significantly lower in AML patients than in healthy controls (P= 0.001).
Correlation between miR-215 expression and clinical characteristics in AML
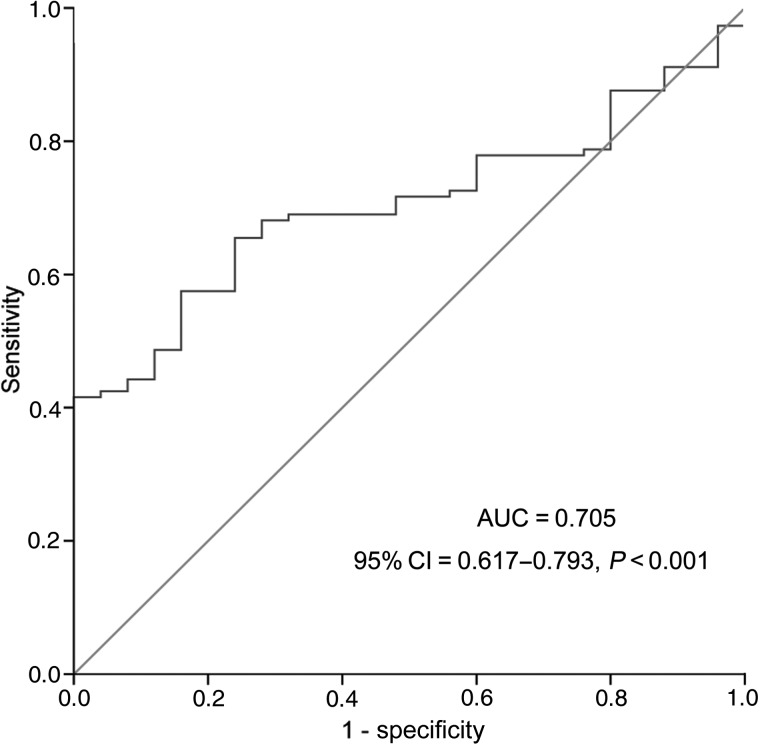
To explore the clinical relevance of miR-215 expression in AML, receiver operating characteristic curve (ROC) analysis was performed to divide the whole patients into two groups (low miR-215 expression and high miR-215 expression) on the basis of the level of miR-215 expression. ROC analysis showed that at the cut-off value of 0.0054 of miR-215 expression level, the sensitivity and the specificity were 42% and 100%, respectively (Fig. 2). The area under the curve (AUC) was 0.705 (95% confidence interval= 0.617–0.793, P< 0.001). The clinical and laboratory features in AML patients at time of diagnosis with and without miR-215 low expression are presented in Table 1. As shown in Table 1, lower levels of miR-215 were associated with a higher white blood cell (WBC) counts (P= 0.014). There was no significant association of the miR-215 expression level between the two groups in other clinical features including sex, age, hemoglobin (HB) counts, platelets (PLT) counts, FAB or WHO classifications and cytogenetic abnormalities (P> 0.05).
Figure 2.
Receiver operating characteristic curve analysis using miR-215 for discriminating AML patients from normal controls.
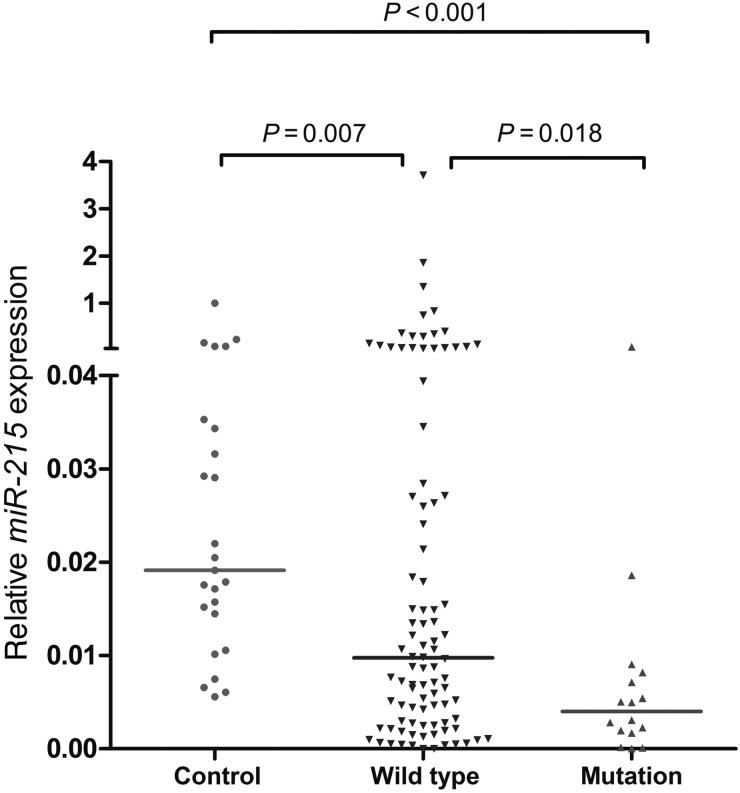
Among the 10 gene mutations, no one except for FLT3/ITD mutation was validated to have a difference between miR-215 low-expressed and high-expressed patients (Table 1). AML patients with low miR-215 expression had significantly higher incidence of FLT3/ITD mutation as compared with those with high miR-215 expression (P= 0.025). Additionally, the relative miR-215 expression was compared in three groups (FLT3/ITD-positive AML, FLT3/ITD-negative AML and controls). Analysis of miR-215 expression by RQ-PCR in AML patients compared with controls validated that miR-215 to be significantly down-expressed in the FLT3/ITD-positive AML patients in comparison to both FLT3/ITD-negative AML patients and controls (P= 0.018 and P< 0.001, respectively) (Fig. 3). Due to the largest number of patients with cytogenetically normal AML (CN-AML), the correlation between gene mutations and miR-215 expression was further analyzed. However, differences could not be found in the distribution of gene mutations between patients with and without miR-215 low expression.
Figure 3.
Relative expression levels of miR-215 in FLT3-ITD-positive AML patients (FLT3/ITD mutation), FLT3-ITD-negative AML patients (FLT3/ITD wild-type) and controls. MiR-215 was significantly down-expressed in the AML patients with FLT3/ITD mutation in comparison to both AML patients with FLT3/ITD wild-type and healthy controls (P= 0.018 and P< 0.001, respectively).
Correlation between miR-215 expression and survival in AML
A total of 108 newly diagnosed patients had the follow-up data. In the whole AML patients, the rate of CR after induction therapy in the miR-215 low-expressed group (37.8%, 17/45) was similar to the miR-215 high-expressed group (49.2%, 31/63) (P= 0.326). However, among non-M3 patients, the cases with low miR-215 expression tended to have lower CR rate than those with high miR-215 expression (28.2 versus 47.3%, P= 0.086). There was no significant difference in CR rate between the two groups among CN-AML patients (P= 0.555).
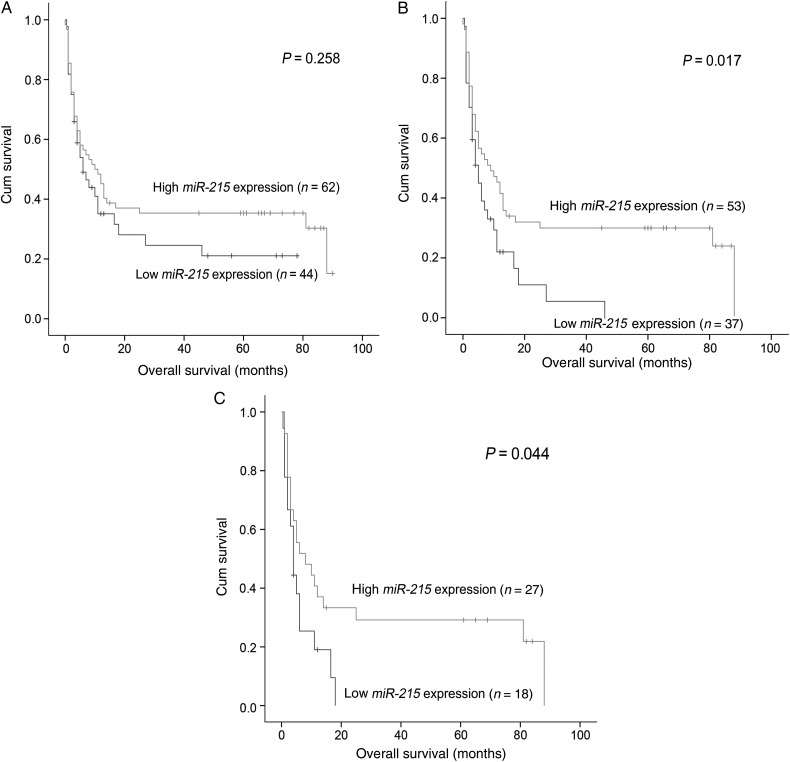
Survival data were obtained for 106 AML patients with median follow-up time of 8 months ranging from 1 to 90 months. No significant difference in OS was observed between the groups with low and high miR-215 expression in the whole cohort of AML patients (P= 0.258) (Fig. 4A). However, the non-M3 AML patients with low miR-215 expression had shorter OS (median 4 months) than those with high miR-215 expression (median 9 months) (P= 0.017) (Fig. 4B). Likewise, among CN-AML patients, the low miR-215 expression group also had a shorter OS (P= 0.044) (Fig. 4C). Moreover, multivariate analysis identified that low miR-215 expression was an independent prognostic factor besides age, karyotype classifications and U2AF1 mutation among non-M3 AML patients (Table 2). Among CN-AML patients, multivariate analysis including the same variables except for karyotype classification and U2AF1 mutation confirmed that low miR-215 expression tended to predict the adverse prognosis independently (Table 3).
Figure 4.
The impact of miR-215 expression on overall survival of AML patients. (A) All patients; (B) non-M3 patients and (C) cytogenetically normal AML (CN-AML) patients.
Table 2.
Multivariate analyses of prognostic factors for overall survival (OS) in 96 non-M3 AML patients
| Hazard ratio (95% CI) | P value | |
|---|---|---|
| Age (>60/≤60 years) | 1.988 (1.191–3.320) | 0.009 |
| WBC (≥30/<30 ×109/l) | 1.044 (0.604–1.804) | 0.877 |
| Karyotype classification (poor/intermediate/favorable) | 2.716 (1.462–5.045) | 0.002 |
| miR-215 expression (low/high) | 2.309 (1.350–3.953) | 0.002 |
| U2AF1 mutation (+/−) | 4.261 (1.578–11.506) | 0.004 |
Table 3.
Multivariate analyses of prognostic factors for OS in 49 cytogenetically normal AML patients
| Hazard ratio (95% CI) | P value | |
|---|---|---|
| Age (>60/≤60 years) | 2.012 (0.984–4.117) | 0.056 |
| WBC (≥30/<30 ×109/l) | 1.290 (0.649–2.564) | 0.467 |
| miR-215 expression (low/high) | 1.949 (0.951–3.984) | 0.068 |
Discussion
Many indicators have been involved in the diagnosis, prognosis and treatment of AML by far, such as karyotypes, mutations in FLT3, C-KIT, NPM1 and C/EBPA genes (23,31). However, the numbers of adults with normal cytogenetical AML are >40%, and the frequencies of gene mutations are relatively low in AML actually (<30%) (32). For most AML patients, especially for those who failed to be characterized by relevant cytogenetic or molecular changes, difficulty still exists to estimate their survival and prognosis. Accordingly, it is quite necessary for finding more valuable biomarkers to improve our understanding of the biology of leukemia and identify those who tend to have adverse prognosis as well as optimize treatment strategies in CN-AML patients. Nowadays, considerable progress has been made in identifying, characterizing and applying new molecular markers (33,34), including miRNAs that were known to be dysregulated during oncogenesis. Due to their advantages of easy detection and less degradation, a growing number of miRNAs has been reported to be biomarkers involved in tumor progression, diagnosis, prognosis and so on (35–37). Here, we focused on miR-215, whose overexpression could induce cell cycle arrest, cell detachment and apoptosis in a partially but not completely p53-dependent pathway (13). MiR-215 was also identified as a putative anti-oncogenic miRNA through modulating a number of genes: up-regulated miR-215 causes a decrease in clonogenicity and mediates the repression of cell cycle and stemness genes downstream of CDX1 by targeting BMI1 in colon cancer (18,38), decreased cell migration and invasion in renal cell carcinoma (RCC) by targeting SIP1/ZEB2 and MMP3 (39); increased apoptosis by targeting XIAP in non-small cell lung cancer (40). However, up-regulation of miR-215 can also be observed in several studies such as gastric cancer, cervical cancer and hepatocellular carcinoma (41–43). These data may reflect that miR-215 played distinct roles in many types of cancers with particular tissue origin. Moreover, lots of reports have found that the expression pattern of miR-215 is associated with the clinical outcome of several cancers (44). Karaayvaz et al. found that low expression of miR-215 was correlated with poor prognosis in colon cancer (19). Khella et al. demonstrated that decreased miR-215 expression was a negative prognostic biomarker in RCC (39). Zhou et al. verified that the down-regulation of miR-215 was an unfavorable prognostic factor in BC (17). Prognostic significance of miR-215 expression has been revealed in many solid tumors, which suggests that it could serve as a potential role of prognostic biomarker and thereby create a more predictable future when refers to the clinical treatment of patients.
To our best knowledge, this is the first time that reports the clinical significance of miR-215 expression in AML. Our study presented that down-regulation of miR-215 expression was a common event in AML. In this study, miR-215 low expression was validated to be associated with shorter OS in non-M3 AML patients and CN-AML patients and was proven to be an adverse prognostic factor in non-M3 AML. We also confirmed in multivariate analysis that miR-215 can serve as a significant and independent predictor of AML. Taken together, these results indicated that miR-215 functioned as a tumor suppressor in AML as well and miR-215 might be an important modulator involved in AML development. Obviously, our results deserve further studies to be confirmed before miR-215 expression can be used routinely as a potential biomarker for risk stratification in AML.
Previous studies have reported that FLT3/ITD mutation, known as a poor prognostic factor in leukemia, was associated with higher peripheral WBC counts and a higher bone marrow blast percentage (45). Furthermore, several miRNAs have already been reported to be associated with FLT3/ITD. For example, expression of miR-155 was overexpressed and involved in the pathogenesis of AML with FLT3/ITD mutation (11,46). Additionally, miR-16 repression could participate in the process of up-regulating Pim-1 oncogene, a regulator of FLT3/ITD signaling in FLT3/ITD expressing cells (47). Our study showed that miR-215 was down-regulated in AML patients with FLT3/ITD mutation and the patients with low miR-215 expression had a high incidence of FLT3/ITD mutation. The exact relationship between down-regulation of miR-215 and FLT3/ITD mutation need further experimental studies.
However, there still exits some limitations such as the small number of patient cases, potential non-applicability of the used cut-off value and some missing data for gene mutations in this study. Of course, more samples with the follow-up data are surely worthy to be employed to take insight into the more accurate clinical significance of miR-215 in AML.
In summary, down-regulated miR-215 expression is a common event in de novo AML patients and might act as an independent risk factor for prognosis in non-M3 AML.
Funding
This work was supported by National Natural Science foundation of China [81270630, 81172592], Six Major Talent Summit Project in Jiangsu Province (WSN-112), Science and Technology Special Project in Clinical Medicine of Jiangsu Province [BL2012056], 333 Project of Jiangsu Province [BRA2013136], Medical Key Talent Project of Zhenjiang and Social Development Foundation of Zhenjiang [SH2014044,SH2014086].
Conflict of interest statement
None declared.
References
- 1.Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer 2003;3:89–101. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer 2010;10:23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jongen-Lavrencic M, Sun SM, Dijkstra M et al. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood 2008;111:5078–85. [DOI] [PubMed] [Google Scholar]
- 4.Dixon-McIver A, East P, Mein CA et al. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One 2008;3:e2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R, Su B. Small but influential: the role of microRNAs on gene regulatory network and 3′UTR evolution. J Genet Genomics 2009;36:1–6. [DOI] [PubMed] [Google Scholar]
- 6.Sotiropoulou G, Pampalakis G, Lianidou E et al. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA 2009;15:1443–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu W, Sun M, Zou GM et al. MicroRNA and cancer: current status and prospective. Int J Cancer 2007;120:953–60. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Getz G, Miska EA et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834–8. [DOI] [PubMed] [Google Scholar]
- 9.Marcucci G, Radmacher MD, Maharry K et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008;358:1919–28. [DOI] [PubMed] [Google Scholar]
- 10.Li Q, Liu L, Li W. Identification of circulating microRNAs as biomarkers in diagnosis of hematologic cancers: a meta-analysis. Tumor Biol 2014;35:10467–78. [DOI] [PubMed] [Google Scholar]
- 11.Garzon R, Volinia S, Liu CG et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008;111:3183–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georges SA, Biery MC, Kim SY et al. Coordinated regulation of cell cycle transcripts by p53-inducible microRNAs, miR-192 and miR-215. Cancer Res 2008;68:10105–12. [DOI] [PubMed] [Google Scholar]
- 13.Braun CJ, Zhang X, Savelyeva I et al. P53-Responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 2008;68:10094–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichiorri F, Suh SS, Rocci A et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010;18:367–81. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Senanayake U, Das S, Vesely P et al. miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated and their common target ACVR2B is strongly expressed in renal childhood neoplasms. Carcinogenesis 2012;33:1014–21. [DOI] [PubMed] [Google Scholar]
- 16.Wijnhoven BP, Hussey DJ, Watson DI et al. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg 2010;97:853–61. [DOI] [PubMed] [Google Scholar]
- 17.Zhou SW, Su BB, Zhou Y et al. Aberrant miR-215 expression is associated with clinical outcome in breast cancer patients. Med Oncol 2014;31:259. [DOI] [PubMed] [Google Scholar]
- 18.Faltejskova P, Svoboda M, Srutova K et al. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med 2012;16:2655–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karaayvaz M, Pal T, Song B et al. Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer 2011;10:340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett JM, Catovsky D, Daniel MT et al. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French–American–British Cooperative Group. Ann Intern Med 1985;103:620–5. [DOI] [PubMed] [Google Scholar]
- 21.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood 2002;100:2292–302. [DOI] [PubMed] [Google Scholar]
- 22.Lo CF, Foa R. Diagnostic and prognostic advances in the immunophenotypic and genetic characterization of acute leukaemia. Eur J Haematol 1995;55:1–9. [DOI] [PubMed] [Google Scholar]
- 23.Slovak ML, Kopecky KJ, Cassileth PA et al. Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: a Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood 2000;96:4075–83. [PubMed] [Google Scholar]
- 24.Li Y, Lin J, Yang J et al. Overexpressed let-7a-3 is associated with poor outcome in acute myeloid leukemia. Leuk Res 2013;37:1642–7. [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Yao DM, Qian J et al. Recurrent DNMT3A R882 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One 2011;6:e26906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Yao DM, Qian J et al. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol 2012;91:519–25. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Qian J, Sun A et al. RAS mutation analysis in a large cohort of Chinese patients with acute myeloid leukemia. Clin Biochem 2013;46:579–83. [DOI] [PubMed] [Google Scholar]
- 28.Qian J, Yao DM, Lin J et al. U2AF1 mutations in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. PLoS One 2012;7:e45760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin LI, Chen CY, Lin DT et al. Characterization of CEBPA mutations in acute myeloid leukemia: most patients with CEBPA mutations have biallelic mutations and show a distinct immunophenotype of the leukemic cells. Clin Cancer Res 2005;11:1372–9. [DOI] [PubMed] [Google Scholar]
- 30.Qian J, Lin J, Qian W et al. Overexpression of miR-378 is frequent and may affect treatment outcomes in patients with acute myeloid leukemia. Leuk Res 2013;37:765–8. [DOI] [PubMed] [Google Scholar]
- 31.Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol 2013;88:318–27. [DOI] [PubMed] [Google Scholar]
- 32.Rowley JD. Chromosomal translocations: revisited yet again. Blood 2008;112:2183–9. [DOI] [PubMed] [Google Scholar]
- 33.Masetti R, Togni M, Astolfi A et al. DHH-RHEBL1 fusion transcript: a novel recurrent feature in the new landscape of pediatric CBFA2T3-GLIS2-positive acute myeloid leukemia. Oncotarget 2013;4:1712–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kralik JM, Kranewitter W, Boesmueller H et al. Characterization of a newly identified ETV6-NTRK3 fusion transcript in acute myeloid leukemia. Diagn Pathol 2011;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–66. [DOI] [PubMed] [Google Scholar]
- 36.Sun X, Li Y, Yu J et al. miR-128 modulates chemosensitivity and invasion of prostate cancer cells through targeting ZEB1. Jpn J Clin Oncol 2015;45:474–82. [DOI] [PubMed] [Google Scholar]
- 37.Xue Q, Sun K, Deng HJ et al. MicroRNA-338–3p inhibits colorectal carcinoma cell invasion and migration by targeting smoothened. Jpn J Clin Oncol 2014;44:13–21. [DOI] [PubMed] [Google Scholar]
- 38.Jones MF, Hara T, Francis P et al. The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc Natl Acad Sci USA 2015;112:E1550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khella HW, Bakhet M, Allo G et al. miR-192, miR-194 and miR-215: a convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis 2013;34:2231–9. [DOI] [PubMed] [Google Scholar]
- 40.Ye M, Zhang J, Zhang J et al. Curcumin promotes apoptosis by activating the p53-miR-192–5p/215-XIAP pathway in non-small cell lung cancer. Cancer Lett 2015;357:196–205. [DOI] [PubMed] [Google Scholar]
- 41.Deng Y, Huang Z, Xu Y et al. MiR-215 modulates gastric cancer cell proliferation by targeting RB1. Cancer Lett 2014;342:27–35. [DOI] [PubMed] [Google Scholar]
- 42.Liang H, Li Y, Luo RY et al. MicroRNA-215 is a potential prognostic marker for cervical cancer. J Huazhong Univ Sci Technolog Med Sci 2014;34:207–12. [DOI] [PubMed] [Google Scholar]
- 43.Zhang ZQ, Meng H, Wang N et al. Serum microRNA 143 and microRNA 215 as potential biomarkers for the diagnosis of chronic hepatitis and hepatocellular carcinoma. Diagn Pathol 2014;9:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol 2012;59:467–74. [PubMed] [Google Scholar]
- 45.Heinrich MC. Targeting FLT3 kinase in acute myelogenous leukemia: progress, perils, and prospects. Mini Rev Med Chem 2004;4:255–71. [DOI] [PubMed] [Google Scholar]
- 46.Faraoni I, Laterza S, Ardiri D et al. MiR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: correlation with NPM1 and FLT3 mutation status. J Hematol Oncol 2012;5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim KT, Carroll AP, Mashkani B et al. MicroRNA-16 is down-regulated in mutated FLT3 expressing murine myeloid FDC-P1 cells and interacts with Pim-1. PLoS One 2012;7:e44546. [DOI] [PMC free article] [PubMed] [Google Scholar]