Abstract
The recent advance in identifying risk genes has provided an unprecedented opportunity for developing animal models for psychiatric disease research with the goal of attaining translational utility to ultimately develop novel treatments. However, at this early stage, successful translation has yet to be achieved. Here, we review recent advances in modeling psychiatric disease, discuss utility and limitations of animal models, and emphasize the importance of shifting from behavioral analysis to identifying neurophysiological defects, which are likely more conserved across species and thus increase translatability. Looking forward, we envision that preclinical research will align with clinical research to build a common framework of comparable neurobiological abnormalities and form subgroups of patients based on similar pathophysiology. Experimental neuroscience can then use animal models to discover mechanisms underlying distinct abnormalities and develop strategies for effective treatments.
Introduction
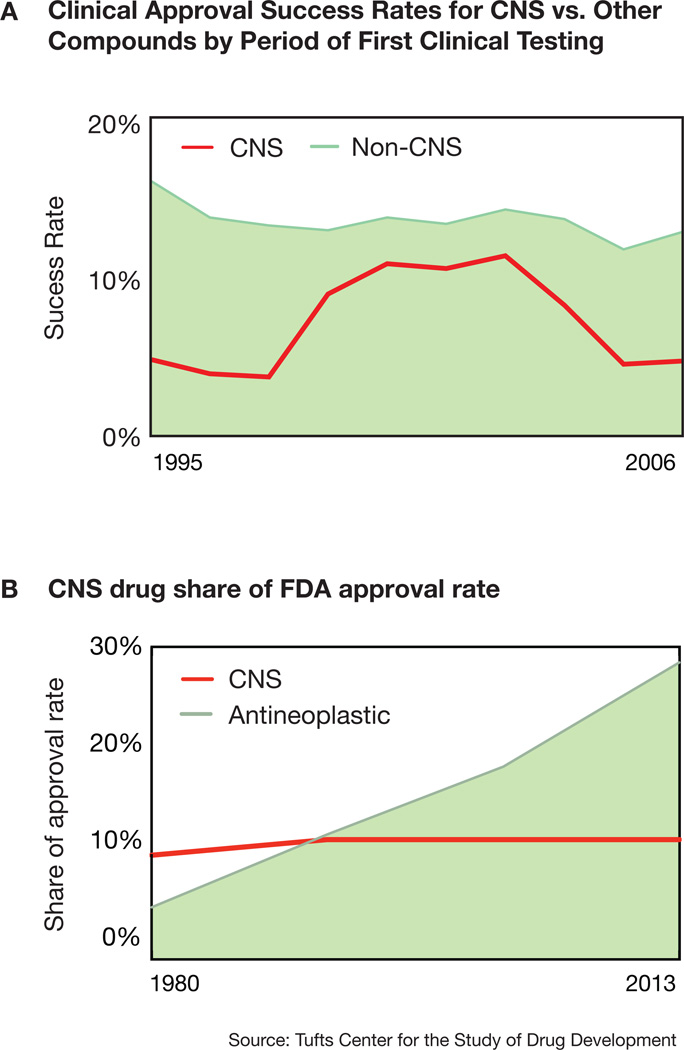
According to the latest NIMH estimates from 2006, more than three times as many people in the United States paid expenses for care related to psychiatric disorders as compared to those requiring cancer treatments (cost statistics on http://nimh.nih.gov). Strikingly, most of the drug treatments these patients receive were discovered serendipitously decades ago and are often unspecific and ineffective. The current outlook for developing novel compounds is also bleak given consistently lower clinical approval success rates of CNS compounds compared to their non-CNS counterparts (Figure 1), and the lack of mechanistic understanding that indicates no clear path to success. Consequently, pharmaceutical companies drastically reduced R&D expenditures into psychiatric disorders. This is in stark contrast to a comparatively growing number of treatments that are being developed and approved in the US for other non-CNS diseases with increasingly understood pathophysiological mechanisms, such as neoplastic diseases (Figure 1).
Figure 1. Clinical approval of CNS-drugs.
A) The clinical approval success rates for CNS drugs fall far below drugs for non-CNS disorders between 1995 and 2006. Except for a period of increased approvals of so-called me-too-drugs, the approval rates were consistently low with about 5 in a 100 compounds receiving approval.
B) In contrast to compounds for CNS disorders, the share of FDA approval rate for antineoplastic drugs increased substantially between 1995 and 2006, since therapy evolved from unspecific cytotoxic compounds to highly cancer-specific compounds. Source: Tufts Center for the Study of Drug Development92.
To set the stage for a similar development in psychiatric disease research, we must gain a better understanding of the pathophysiology of these disorders, including improving our understanding of the heterogeneity of the disorders. First, we must advance our knowledge regarding disease etiology. Since most psychiatric disorders are highly heritable, identifying genetic factors conveying risk is a crucial step. Current large-scale genetic studies are already taking this step by discovering numerous risk genes for various psychiatric diseases1–5. Second, due to the lack of access to diseased brain tissues, we must use model systems to investigate neurophysiological abnormalities that may be caused by genetic variants and mutations. While no model systems will ever perfectly phenocopy human disease, we can use cellular models for the interrogation of conserved molecular pathways or animal models to dissect complex neural circuit defects that may underlie particular phenotypic abnormalities found in patients. Third, beyond the assessment of observable signs in affected individuals, we need to identify clusters of affected individuals with similar neurophysiological abnormalities that have been molecularly understood and targeted for treatment development in model systems. Eventually, we will be able to give these more homogenous clusters of patients interventions developed for their specific pathophysiological mechanism (personalized medicine).
The past decade has seen a large increase in the number of rodent models generated for mechanistic research and treatment development. However, many of the early studies using these models have focused on behavioral characterizations. Only recently, animal model studies are starting to reveal mutation-specific neural circuit defects that might be relevant to disease pathology (see review in6,7). The lack of deep understanding of disease-relevant cellular and circuit mechanisms is a bottleneck for successful translation in psychiatry research.
In this perspective, we discuss the utility and limitations of animal models. We emphasize the importance of using animal models that are based on disease etiology, the difficulties and approaches in modeling polygenic disorders, the necessity to shift emphasis from behavioral studies to neurophysiological characterization with a focus on translatable molecular and neural circuit mechanisms that are evolutionary conserved. Finally, we envision an integrated path forward that may enable us to better translate preclinical findings into effective treatments for psychiatric diseases.
Animal models and disease etiology
During the last decade, a host of animal models for psychiatric disease research have been developed. Generally, neuroscientists evaluate these models in terms of construct validity, face validity and predictive validity8,9. Construct validity in the context of animal models for psychiatric disease research refers to the degree to which the model is based on disease etiology, such as environmental or genetic risk conditions for developing the disease. An animal model’s face validity is its phenotypic resemblance to the human disease, while its predictive validity describes the similarities in treatment response between the animal model and human patients.
Construct validity is impossible to achieve in an animal model if we do not understand the etiology. Little is known about etiology of psychiatric disease and no biomarkers are available except for a few syndromic disorders such as Rett syndrome and Fragile X syndrome where genetic lesions are known and used for diagnosis. Thus, clinicians assess patients based on phenotypic presentation and observable signs. For example, using guidelines of the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5), a patient with impaired reciprocal social interactions combined with restricted, repetitive behaviors manifesting in early childhood would be diagnosed with autism spectrum disorder. Such heavily phenotype-based diagnoses in psychiatric diseases have inspired similar phenotypic assessment in animal models, particularly mice. For instance, comparison of the inbred mouse strain BTBR T+tf/J with the inbred mouse strain C57/BL6 reveals that these mice show relatively less reciprocal social interaction, more grooming and different ultrasonic vocalizations10. Thus the BTBR mice have been proposed to be used as an autism model with high face validity and are used for ongoing preclinical drug testing11,12.
Similarly, it has been suggested that hyper-locomotion induced by Amphetamine, a psychostimulant that causes psychosis-like episodes in humans, is a predictively valid model for schizophrenia, given that treatment with approved antipsychotic drugs normalizes locomotion in these mice13. This hypothesis is supported by findings showing that schizophrenia is associated with increased amphetamine-induced synaptic dopamine concentrations14 and genetic association of the dopamine receptor DRD2 with schizophrenia4. At the same time, animal models centered on face and predictive validity have significant drawbacks. First, behaviors considerably diverged during the 80 million years since the last common ancestor of humans and mice, and should only be interpreted as a correlate of neural circuit function, and ideally only in models based on an evolutionarily proximal species. Second, approaches focused on predictive validity with the ultimate goal of discovering new therapeutic targets for the development of new compounds are inherently flawed, since they are biased towards the same molecular pathways that have been targeted in the past to limited success. Third, making inferences regarding human pathophysiology from these models is substantially complicated by the fact that they do not reflect the etiology of ASD or schizophrenia in human patients on the basis of current knowledge. An alternative approach is to deemphasize face and predictive validity and only consider them in models with high construct validity15.
How can we generate animal models that do not merely mirror phenotypic presentation, but are built on disease etiology? High heritability indicates that genetics is among the most important contributors to the development of psychiatric disease16,17.
In fact, it is widely assumed that genetic and environmental factors interact and converge on the same molecular pathways and, in combination, exert either protective or adverse effects17. In other words, the consequence of an environmental experience such as stress depends on the presence or absence of certain genetic variants within an individual. To date, reported environmental influences on psychiatric disease include (but are not limited to) maternal stress or infection during pregnancy, birth complications, infections, stressful life events, and drug abuse. While the field of modeling gene and environment interactions in animals is still in its infancy and facing challenges related to uncertainties regarding nature and quantitative parameters of environmental factors, several studies have already shown promising results and excellent reviews on this exciting topic can be found elsewhere17–19. In the future, it may be crucial to explore and define standardized paradigms mimicking the exact time course and nature of gene-environment interplay in humans to determine to what extent, and how such interplay might affect developmental trajectories and neurophysiology.
The most significant advances in understanding etiology of psychiatric disorders in the past decade have come from genetic studies. Recent genome-wide association studies, copy number variation studies and whole exome sequencing studies have identified a large number of genetic risk factors for the development of psychiatric disease20 and references therein). Here, we outline how such genetic findings provide neuroscientists with the unprecedented opportunity to generate animal models that are similar in etiology to human disease and hence may prove to be more valid tools for the dissection of disease-associated pathways and circuits.
Utility and limitations of current animal models
A range of model organisms from fruit flies to zebrafish to mice15,21,22 has been successfully used to investigate gene to phenotype relationships and discover relevant molecular mechanisms underlying disease. Because the etiology and clinical expression of psychiatric disease is complex and related to the unique biology of humans, genetic findings will by no means enable researchers to generate animal models that recapitulate all phenotypic features of any one DSM-defined disorder. However, simpler units of intermediate disease phenotypes associated with genetic variants, called endophenotypes, are amenable to interrogation23,24. Compared to a disorder classified by a combination of complex symptoms, these quantifiable endophenotypes are thought to arise from the interaction of fewer gene products and can be neurophysiological, biochemical, neuroanatomical or behavioral. Using this approach, one hope in the field is deconstructing complex traits into fewer, distinct cellular and circuit mechanisms and subsequently reconstruct a general neurobiological logic, which will help explain and better predict consequences of similar genetic variants, acting singularly or in combination with non-genetic factors.
Both common and rare genetic variants have been used to model gene-endophenotype relationships in animals. Here we define common variants as polymorphisms that occur in more than 5% of the human population. In animal models, such variants have been used to generate useful models that display endophenotypes, some of which have also been observed in humans25–27. The advantage of this approach is that investigators can study the relevance and endophenotypic impact of polymorphisms of interest in fairly large cohorts of human subjects and animal models in parallel using comparable experimental paradigms in both species25. While this approach has yet to be taken one step further to the demonstration of causality through circuit manipulation, the adoption of robust methodology and focus on comparable endophenotypes hold great promise for translation.
Modeling neural circuit abnormalities in autism spectrum disorder
Currently, most animal disease modeling studies focus on highly penetrant rare mutations. Specifically, here we highlight genetic mouse models for interrogation of neural circuit abnormalities in autism spectrum disorders, a psychiatric disease where the discovery of highly penetrant variants in patients has enabled the study of monogenic models. Many such genetic models have been developed in the past decade, a few of which are shown in Table 1.
Table 1.
Recently developed human genetics-based animal models point towards synaptic mechanisms
| Genetic mouse model | Cellular and neurophysiological abnormality | Behavioral abnormality |
|---|---|---|
| Shank2 knockout96,97 | Reduced hippocampal glutamatergic neurotransmission, reduced spine density, increased glutamate receptor expression96 or reduced NMDAR function97 |
Excessive grooming, increased social interaction, abnormal vocalizations |
| Shank3 knockout-Ankyrin repeat98–100 Shank3 knockout-PDZ domain29 Shank3 knockout-Exon 21101 Shank3 overexpression102 |
Impaired hippocampal synaptic transmission and LTP, reduced PSD proteins, reduced activity-dependent AMAPR98–100 Reduced cortico-striatal neurotransmission, reduced PSD and PSD proteins, reduced spine density, increased dendritic length, striatal hypertrophy29 Hippocampal synaptic defects, increased mGluR5 in PSD101. Abnormal EEG, decreased mIPSC frequency and increased sEPSC in the hippocampus, increased spine density, increased excitatory synaptic markers and reduced inhibitory markers102. |
Abnormal social behaviors, communication repetitive behaviors and learning Excessive grooming, impaired social increased anxiety29 Spatial learning and memory defects, deficits, hypersensitivity to heat, Increased locomotor activity, hypersensitive amphetamine, abnormal circadian |
| CATNAP2 knockout31 | Abnormal EEG, cortical neuronal migration abnormalities, reduced cortical neuronal synchrony, reduced number of interneurons in striatum and cortex |
Excessive grooming, epileptic behavior, abnormal vocalizations |
| Neuroligin-3 knockout and R451C knock-in33 | Increased inhibitory synaptic transmission in somatosensory cortex, increased expression of inhibitory neuron markers in hippocampus and somatosensory cortex |
Impaired social interaction, enhanced |
| SynGAP1 knockout of one allele32 | Elevated excitatory synaptic transmission during development, premature spine maturation, abnormal dendritic spine size and shape, abnormal E/I balance in the hippocampus |
Seizures, learning deficit, hyperactivity |
| MeCP2 knockout103; MeCP2 microglia rescue34; MeCP2 astrocyte rescue104 |
Reduced neuronal cell size, reduced number of dendritic branches, microglia phagocytic activity |
Decreased body weight, decreased shortened lifespan |
| Neurexin-1α knockout30 | Reduced excitatory synaptic transmission in the hippocampus | Decreased prepulse-inhibition, impaired nest-building, improved |
These monogenic models have collectively revealed several cellular and neurophysiological abnormalities that may be related to autism pathology encompassing synaptic dysfunction and abnormal dendritic spine morphology, excitation-inhibition imbalance, and glia cell dysfunction. First, excitatory synaptic dysfunction in the hippocampus or the striatum is a consistent defect found in mutant mice lacking synaptic organizing proteins such as neurexin-1α, Shank2- or Shank328–30. In addition, both Shank2- and Shank3-deficient mice display altered expression of synaptic proteins, reduced density and abnormal shape of dendritic spines. Second, excitation-inhibition imbalance is implicated in a range of monogenic ASD models including homozygous deletion of CNTNAP2 in mice, a gene important for clustering potassium channels and neurodevelopment; mice heterozygous for SynGAP, a gene involved in dendritic spine development; and mice with the ASD-associated R451C missense mutation in synaptic organizing gene Neuroligin-3.31–33. Third, studies in MeCP2, a gene transcription regulator, knockout mice suggest that distinct cellular entities, particularly astroglia34 and microglia (35; but also see36) may play a significant role in the neurobiology of Rett syndrome.
Among a host of behavioral abnormalities, several of these studies reported abnormal social interactions and repetitive self-grooming in the mice. When investigators interpret these findings, a prevailing idea is that these behaviors are comparable to DSM-defined symptoms of ASD and causatively related to the cellular and neural circuit abnormalities observed. Consequently, a common view is that approaches correcting some circuit defects or the abnormal behaviors in animal models could directly translate to treatments in affected individuals.
However, this interpretation is problematic for at least three reasons. First, although the cellular and neurophysiological defects reported are likely to be relevant to the disorder and should thus be studied further, they might neither be necessary nor sufficient to cause a particular abnormal behavior. Thus, it is crucial to establish a causal relationship between an abnormal behavior and a specific cellular or neural circuit defect, the latter of which ideally can also be identified in human patients. For example, decades of clinical research indicated that cortico-striatal-thalamo-cortical circuits are abnormally active in patients with obsessive-compulsive disorder37,38, but only the experimental optogenetic perturbation in rodent models demonstrated definitively that abnormal activity of this circuit drives compulsive behaviors39,40.
Second, while exome sequencing analysis of de novo mutations suggest that most mutations associated with ASD are missense and different mutations in the same gene can either be loss- or gain-of-function41, animal model studies generally tend to adopt knockout approaches when attempting to understand disease relevance of the gene. The limitations of this approach are exemplified by studies showing that mice deficient for Neuroligin-3 and mice carrying the ASD-associated R451C knock-in mutation display different phenotypic and neurobiological abnormalities. Unlike Neuroligin-3 deficient mice, mice harboring R451C human mutation express residual amounts of mutant protein that may result in an unexpected gain-of-function33,42. Thus, inferences made from an animal model carrying a certain mutation may only be valid for that specific mutation or a similar set of mutations. A possible strategy is to exclusively model patient-specific mutations in animals using knock-in approaches. Admittedly, studying the entirety of mutations found in affected individuals is currently unpractical even in rodents, yet screening for loss- or gain-of-function in cell culture and in simple animal models like the zebrafish may help group mutations by their possible mechanism of action.
The third challenge to linking mutations with circuit dysfunctions causing behaviors is that mouse and human behaviors as well as the circuits underlying these behaviors are not always directly comparable. When attempting to align clinical and preclinical findings, investigators should bear two things in mind. One, DSM-5 and other symptom classifications are designed to provide a common framework for clinical purposes in the absence of mechanistic understanding and disease-relevant biomarkers. This clinical framework cannot be used to directly guide the common preclinical framework, which primarily concerns neurobiological measures and molecular signatures. Thus, a specific mouse behavior should not be regarded as an equivalent to a human symptom, but interpreted as a readout that can be used to study the underlying neurobiological defects. In addition, despite the fact that mice and humans have homologous brain regions and complex behavioral repertoires, there may be significant divergence of neural circuitry, or even repurposing of existing circuits, caused by the unique evolutionary pressures on mice and humans. Specifically, unlike the similar evolutionary history and role of cortico-striatal-thalamo-cortical circuitry in repetitive behavior in humans, non-human primates and rodents37–40,43,44, neural circuits that underlie rodent social behavior and primate social behavior may be vastly different, given their different evolutionary histories45.
In contrast to these behavioral differences and the function of evolutionarily more recent neural circuits which may diverge widely between rodents and primates, synaptic genes and their cellular functions are largely conserved throughout vertebrate and invertebrate evolution46. Thus, shifting emphasis from behavioral resemblance to studying evolutionarily conserved circuits and neurophysiological correlates of disease-specific mutations may significantly increase the translatability of preclinical studies. Among many models that have been developed recently, the monogenic models listed in Table 1 revealed several cellular and circuit abnormalities that are implicated in pathophysiology of psychiatric disease. In addition, various abnormal behaviors have been demonstrated in these mice. It should be noted that the cellular and molecular abnormalities are relevant to the mutation, but not necessarily causative for the abnormal behavior observed.
Tackling polygenicity
The aforementioned monogenic models have significantly enhanced our understanding of underlying molecular, cellular and circuitry defects caused by these particular mutations. That being said, a major limitation with this approach is that we can only generate monogenic models for highly penetrant variants (e.g. Shank3, MeCP2). Based on current knowledge, such highly penetrant variants may account for only 5–15% of cases in ASD, whereas the majority of patients with psychiatric disease may harbor many variants of small effect size that cumulatively confer genetic risk to disease41,47–49. In addition, the majority of variants may localize to non-coding regions, which are difficult to study due to limited understanding of the function of these regions and poor sequence conservation between species. For schizophrenia, the generation of animal models that reflect disease etiology is impeded by the lack of highly penetrant variants that are replicated across more than just a few families47. In a few cases, such as deletions of Neurexin-1, a gene important for synapse development, mutations are highly penetrant and found in many patients, but clinically they manifest as many different disorders50,51. Thus, until further genetic and neurobiological studies reveal more information about these complex diseases, relatively simple genetic animal models are not within reach and alternative approaches are required to elucidate the fundamental biology of polygenic diseases.
iPSCs and polygeneticity
Currently, a promising approach to understanding polygenicity is to use patient-derived induced pluripotent stem cells (iPSC) - the strength of this approach is that these cells have the same complex genetics as the affected indiviudal, which is crucial for dissecting the pathophysiology of polygenic diseases. While most initial human iPSC studies focused on monogenetic disorders52–55, a few new studies are starting to reveal interesting cellular phenotypes in iPSC-derived neurons from patients of unknown genetic causes56,57. However, despite great promises, there are currently still several obstacles hampering significant advances in modeling psychiatric diseases using these cells. Briefly, the main hurdles when using neurons differentiated from human iPSC are line to line variability due to culture conditions and genetic backgrounds; the neurons differentiated from iPSCs are less mature than adult human neurons with few spines, a subcellular compartment strongly implicated in pathophysiology of psychiatric disorders; lack of robust ways to differentiate the vast variety of neuronal cell types; and neurons differentiated from iPSC do not form complex neural circuits, which substantially constrains the complexity of neurophysiological endophenotypes that can be studied and related to relevant behavioral endophenotypes.
To overcome these hurdles, more sophisticated approaches for using iPSC-derived neurons have been explored. One of them is the development of organoids for studying local circuits58,59. These organoids contain cortical layer-like structures and multiple neuronal cell types60 and have been used to study neural developmental defects in ASD61. Another approach is to generate chimeric models by injecting fluorescently labeled human iPSC into the developing brain of rodents or primates. If the fluorescently labeled iPSCs becomes an integral part of the developing and migrating neuronal progenitor pool, iPSC-derived neurons may become incorporated into functional circuits62,63. In an experimental setting, fundamental biological processes can then be investigated in patient iPSC and control iPSC in the same animal with different fluorescent labeling, which intriguingly may also solve in vitro differentiation issues and the problem of neuronal maturation including dendritic spine formation. The developing brain, being the natural environment for differentiating neurons, endogenously provides all the factors necessary for differentiation and the physiological formation of mature spines. Although the study of iPSC-derived neurons is still in early stages, further developing these approaches could be critical for studying polygenic disorders.
GWASs and polygenecity
In addition to patient iPS cells, we might also use GWAS data to gain insights on polygenicity of psychiatric disorders. GWAS studies reveal common genomic loci and alleles that convey increased risks for the disease rather than identifying large effect rare mutations20. Risk alleles and other genetic or non-genetic (such as environmental) factors may converge on altering functions of the same disease-relevant pathway and circuit and thus collectively push it towards pathological states. Therefore, highly significant GWAS results allow us, with confidence, to identify genes that function in disease-relevant pathways and circuits. With the increasing numbers of risk alleles identified by GWAS and large numbers of rare variants discovered in whole exome and whole genome sequencing, it is conceivable that systematic analysis of risk gene functions with cell type-specificity in simpler model systems may allow us to subgroup risk genes into converging pathways based on their effects on cellular and circuits function. These pathway-specific gene list can in turn help bioinformatic analysis of large scale genetic data to identify potential polygenic combinations and guide experimental validation.
Primate models for studying higher brain function and dysfunction
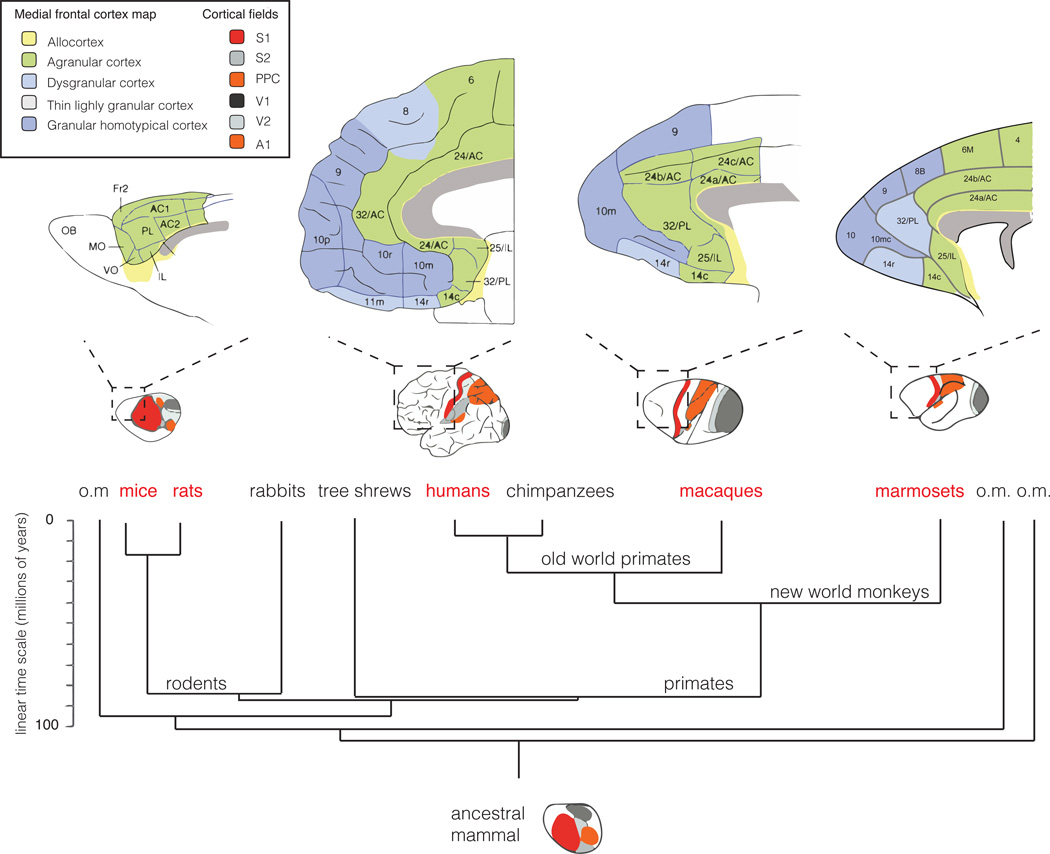
The ability to genetically modify the mouse genome has revolutionized biomedical research including neuroscience. Mouse and rat models have been and will likely continue to be the main mammalian models for studying brain function and dysfunction. However, mice and humans are separated by 80 million years of evolution, which led to significant divergence in the structure and function of the brain, such as in the prefrontal cortex, which is one of the largest and most developed portions of the human brain whereas rodents have only a rudimentary prefrontal cortex and lack some of the counterparts of the primate prefrontal cortical regions (Figure 2). Thus mice do not exhibit the same complexity in cognitive functions that are mediated by these regions in primates on the basis of current knowledge64,65. There are also many unique functional circuits and related behaviors that are frequently affected in human psychiatric disorders and almost impossible to study in rodents such as face recognition, eye gazing and vocalization, all of which play important roles in social cognition and social communication66–68.These differences have led to the exploration of primate models for research of higher brain function and psychiatric disorders69.
Figure 2. The change in cortical fields and medial frontal cortex architecture since the last common ancestor of rodents and humans.
Bottom: The common ancestor of mice, monkeys, and humans is likely to have displayed extended somatosensory areas, but small parietal fields. On a gross scale, similar organization can be found in the rodent brain, whereas humans and monkeys, most probably driven by their visual specialization, have profoundly expanded parietal fields and reduced somatosensory areas. Considering brain architecture revealed by histological staining, the absence of a well-developed granular homotypical cortex in rodents is striking. Although rodents may possess functionally analogous regions, distinct cell-type composition and computations in these regions, which are implicated in psychiatric disease in patients, may be unique to primates. Illustrations are schematic and not drawn to scale. A1: primary auditory cortex, AC1 and AC2: anterior cingulate cortex area 1 and 2, Fr2: frontal area 2, IL: infralimbic cortex, MO: medial orbital cortex, OB: olfactory bulb, PL: prelimbic cortex, PPC: posterior parietal cortex, S1 and 2: primary and secondary somatosensory cortex, V1 and V2: primary and secondary visual cortex, VO: ventro orbital cortex. Numbers correspond to Brodmann areas. Adapted and modified from93–95.
Although potential advantages of primate models will be discussed below, as for any other animal model in biomedical research, it remains imperative to address any scientific questions in the phylogenetically lowest adequate species possible and to minimize adverse experience of the animal. With regard to the use of primate models, there are additional ethical concerns, and these experiments should only be carried out if deemed absolutely necessary.
One issue relevant to the use of primate models more generally, even beyond neuroscience research, has been that until recently, precise genetic manipulations in mammals have been limited to rodents. The development of highly efficient CRISPR genome-editing technology has made it feasible to directly manipulate the genome in zygotes70–72, thus expanding genetic manipulations to many species including primates73. A second issue is reproduction: although macaque monkeys are the most commonly used non-human primates in neuroscience research, they are less desirable as a routine genetic model due to their long generation time and slow reproduction. Macaque monkeys live up to 30 and 40 years in captivity, reach sexual maturity at the age of 3–4 years and give birth once a year to a single offspring. Thus, establishing a sizable transgenic colony means years of waiting.
Among primates, an attractive species to generate genetic models for the investigation of psychiatric disorders is the common marmoset74. The common marmoset is a small (300–400g) New World monkey that is not evolutionarily as close to humans as the Old World monkeys such as macaques. However, from the practical point of generating transgenic models, the marmoset has several advantages75. First and most importantly, marmosets reach sexual maturity around 12–15 months and thus breeding is much faster than with macaques. Furthermore, marmosets give birth twice a year, usually with non-identical twins from each birth. This rapid reproduction cycle is an advantage for generating transgenic animals. Similarly, the relatively rapid maturation of marmosets compared to macaques is an advantage for longitudinal studies of postnatal development and for studying late onset brain disorders. With regard to behavioral characteristics, marmosets are highly social, with strong family structures and complex vocal communication, and thus could be a promising model for studying social cognition and communication, behaviors affected in ASD and schizophrenia. Like macaques but unlike rodents, marmosets have a well-developed prefrontal cortex, a region critical for the cognitive functions that are impaired in many psychiatric disorders65.
However, primate models do have their limitations. Although primates are evolutionarily closer to humans than rodents, they are by no means perfect models for the human brain and human behavior. Most notably, primates do not speak but communicate in vocal calls that are rudimentary compared with human language. Second, while primates have a well-developed prefrontal cortex, transcriptomic analysis has revealed significant differences in cortical gene transcript expression patterns and complexity between primates and humans76. Thus, primates may be better models for human brains only in some aspects. At this early stage, neither failed nor successful translational attempts based on primate models have been made, which would be necessary to assess the value of primate models for human disease. Third, unlike for research in rodent models, extensive tools for detailed circuit interrogation and functional manipulation are not yet readily available. Fourth, studying genetic variants that may be related to more variable, subtle changes and thus require larger cohorts to warrant robust hypothesis testing is unpractical in primates. These scientific limitations, together with the ethical considerations mentioned above and the high cost of maintaining primate colonies, call for caution and clear reasoning regarding scientific necessity when considering primate models.
The pursuit of translatability: aligning preclinical to clinical research
The shortcomings associated with the assessment of animal models based on face, construct and predictive validity in the past as outlined above and the low translatability of findings from animal models to human patients indicate that the validity evaluation and study of animal models should fundamentally change64. Specifically, NIMH recently launched the Research Domain Criteria (RDoC) initiative that spearheads the initiative to extend clinical research beyond the mere assessment of subjective symptoms and observable signs to an assessment that is also based on objective genetic and neurobiological measures65. We suggest that preclinical animal research should follow this example by de-emphasizing face and predictive validity and focusing on neurobiological validity, encompassing neuropathology, molecular pathways, and cellular and circuit mechanisms66. Admittedly, assessing neurobiological validity in an animal model is still difficult due to the lack of knowledge regarding the precise nature of neurophysiological abnormalities in human patients. However, it is worth noting that with advanced neuroimaging, electroencephalography (EEG) and magnetencephalography (MEG) recording, and transcranial magnetic stimulation (TMS) approaches, ongoing clinical research has great potential to build a framework of disease correlates in the near future; and we already know several neurophysiological abnormalities in patients, which can guide mechanistic studies in animal models and be considered for validity assessment of an animal model (reverse translation).
This is exemplified by schizophrenia research, where reported neurophysiological alterations in human patients include abnormalities in local gamma and sleep spindle activity67–70, long-range functional connectivity71,72, impaired long-range structural connectivity (73 and references therein), and morphological changes, such as decreased dendritic spine density and cortical thinning74–76.
How are these neurophysiological phenomena studied in human patients? Functionally, local oscillatory cortical activity arises from synchronous activation of large neuronal ensembles. This is measured with extra-cranial EEG, electrocorticography (ECoG) or MEG as steady-state evoked potentials (SSEP) or as strength of gamma-oscillation in testing paradigms engaging the auditory or visual sensory systems69,70,77,78. Similarly, one can use covariance analysis of the fMRI BOLD response from two regions (functional magnetic resonance imaging, blood oxygen level dependent) to study long-range functional connectivity of neuronal ensembles in distant brain regions.71,72. The morphological framework for such oscillations is laid by the hard-wiring and structural connectivity of brain regions. Here, diffusion tensor imaging (DTI) allows the visualization and quantitative assessment of long-range projections on the macroscopic scale. Lastly, both dendritic spine counting upon Golgi-impregnation in postmortem tissues and structural MRI in patients can be carried out to study structural alterations such as atrophy or hypertrophy of certain regions.
Can preclinical research move towards employing the same or equivalent approaches in animal models to study neurobiological correlates? Although such studies outlined above are still scarce in animal models, various examples are listed in table 2, together with a non-exhaustive summary of both widely and rarely adopted useful methodologies for the elucidation of functional and morphological neurophysiological disease correlates. Using and developing more such methods to better align preclinical research with clinical work requires basic scientists to closely collaborate with clinical scientists to learn disease relevant knowledge.
Table 2.
Preclinical and clinical research converges through use of comparable approaches in conserved domains.
| Neurophysiological domain |
Technology | Examples for findings in human patients |
Examples for application in animal models |
Highlights |
|---|---|---|---|---|
| Functional connectivity: Local synchrony |
Wireless Electroencephalography (EEG), Electrocorticography (ECoG), Magnetencephalography (MEG) |
Reduced gamma power and abnormal sleep spindles in schizophrenia67–70 |
Auditory processing deficits in 15q13.3 deletion heterozygous mice 80 and abnormal spike discharges after seizure onset in CNTNAP2 mutant mice31 |
Longitudinal study Possible in freely |
| Functional connectivity: Long-range synchrony |
Functional Magnet Resonance Imaging (fMRI), Positron Emission Tomography (PET), paired electrophysiology in two regions |
Abnormal functional connectivity in schizophrenia71,72 |
Impaired fronto-parietal synchrony in 22q11.2 deletion schizophrenia model79 |
fMRI: Repeated non imaging/longitudinal unbiased whole-brain equivalent readout |
| Structural connectivity | Diffusion Tensor Imaging (DTI), CLARITY, electron microscopy |
Abnormal connectivity in schizophrenia 73 |
No defects in NLGN-3 knockin mice with DTI studies105 |
DTI: Longitudinal CLARITY: probing native three-dimensional |
| Anatomy | Three-dimensional magnet resonance imaging (MRI) Golgi-impregnation |
Increased caudate volume in ASD patients81 Reduced spine density in schizophrenia |
Increased caudate volume and reduced spine number in Shank3- deficient mice29 |
MRI: Longitudinal Equivalent readout models |
| Gene and protein expression, transcriptional dynamics, single cell transcriptome |
In situ hybridization (ISH), Immunohistochemistry, mass spectrometry, bilsulfite sequencing, ChIP, isoform-specific RNA seq |
Reduced parvalbumin expression in prefrontal cortex in schizophrenia post- mortem tissue106, Mid-fetal transcriptional networks in ASD87 |
Abnormal synaptic protein expression in Shank2- and Shank3- deficient mice29,96 Reduced parvalbumin-positive interneurons numbers in CNTNAP2 mutant mice31 |
High sensitivity Potential to identify developmental periods Identification of converging pathways |
| Neuronal transmission and synaptic plasticity |
Ex vivo and in vivo electrophysiology (sharp electrode, stereotrodes, tetrodes, large electrode arrays) |
Impaired LTP and LTD in schizophrenia107,108 |
Abnormal synaptic transmission in Shank2- and Shank2-deficient mice 28,29 |
Unprecedented temporal robustness and molecular with pharmacology |
This non-exhaustive summary lists various useful techniques that help reveal neurobiological abnormalities, which are largely conserved and therefore likely comparable between human patients and animal models. Note that invasive preclinical approaches have the potential to identify molecular signatures and pathways that may serve as treatment targets, whereas classical clinically used approaches allow for the longitudinal study of treatment effects while also ensuring better translatability.
Regarding the investigation of functional properties, two studies are of particular interest. Using paired recording of spikes and field potentials with multiple recording electrodes in a model for deletion of schizophrenia-associated 22q11.2 region, one study revealed impaired fronto-temporal synchrony as a neurophysiological abnormality79, while the other study adopted ECoG measurements in mice lacking the schizophrenia-associated 15q13.3 homologue to reveal impairments of SSEP and a reduction of evoked gamma power80. In terms of structural changes that correlate with disease, a study investigating gross anatomy of the brain found an increased caudate volume in a mouse model for ASD29, which is consistent with findings in human patients81. Macroscopic structural mapping of fiber tracts using diffusion tensor imaging (DTI) is not yet routinely performed in common laboratory animals. With improved resolution down to 50 µm3 for ex vivo and 200 µm3 for in vivo interrogations, this technique can be used in animal models such as the common marmoset82. In addition, recently developed tissue-clearing techniques such as Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/Immunostaining/In situ hybridization-compatible Tissue-hYdrogel, (CLARITY) for mesoscale optical investigation, which renders the brain transparent in its native three-dimensional state, can be used to reveal morphological abnormalities in animal models83. Together with classical electrophysiological and molecular interrogation of disease-relevant brain circuits in model organisms chosen based evolutionary conservation of such circuits, such as basal ganglia and amygdala for studying compulsions and innate fear in rodents, respectively7,84–86, these approaches help elucidate neurophysiological disease correlates that are likely comparable between humans and animal models.
With these approaches, it is conceivable that investigators will be able to make more valid inferences about human pathophysiology and better predictions of treatment response in patients, but which other opportunities can be explored to improve our understanding of pathophysiology? Reviewing the recently developed animal models, arguably one of the most striking disconnects is the fact that interrogation of neurobiological mechanisms is largely conducted in adult animals, while a substantial portion of psychiatric disease is developmental. Thus, instead of revealing developmental neurobiological abnormalities that are potentially causal to the abnormal neurobiology observed in the adult and still malleable to interventions, current studies focus on studying potentially less malleable consequences of such developmental abnormalities at the adult stage. Specifically, it is conceivable that molecular signatures during development characterize fundamental neurobiological wiring and circuit maturation steps with different potentials regarding reversibility, such that some circuit abnormalities are reversible in adulthood, while others may only be sensitive to treatment in early development32. Regarding future preclinical research, one critical advantage of the technologies outlined above is that they permit longitudinal studies on functional and morphological abnormalities along development with potential to identify critical plasticity periods or neurophysiological and molecular signatures of prodromal stages87. In an experimental setting, animal models offer the unique opportunity to invasively probe early interventions targeted at linked candidate pathways or circuits by utilizing drug treatments or deep brain stimulation and transcranial magnetic stimulation, respectively88, while also being able to study these effects on defined neurobiological abnormalities at various stages along development.
Despite the notion that focusing on neurophysiological defects is critical for better translatability of studies using animal models, studying behavior will not become irrelevant, since behavior is an important organism-level readout of circuit dysfunction and correction. Rather, bearing in mind the limitation that a change in animal behavior upon experimental treatment is by itself insufficient as readout for successful treatment response, future preclinical work may use it as one of many readouts for the correction of the mechanistically understood neurobiological abnormalities.
The path forward: convergent science
Attempts to develop effective treatments for psychiatric diseases with the help of animal models were largely unsuccessful in the past decades. In brief, the main reasons for this disconnect are that heterogeneous patient populations were treated as homogeneous units, the fundamental underlying biological mechanisms were and mostly remain unknown, and the readouts used for treatment response in preclinical research were poor predictors of treatment response in the clinical setting.
Psychiatric diseases stand in contrast to other diseases, such as cancer, where preclinical research significantly advanced our understanding of the underlying mechanisms and revealed many robust biomarkers. For some types of cancer, mechanistic understanding and the availability of robust biomarkers facilitated the evolution of therapy from unspecific cytotoxic drugs to the highly effective compounds that target cancer subtype-specific pathways and thus personalized medicine89,90.
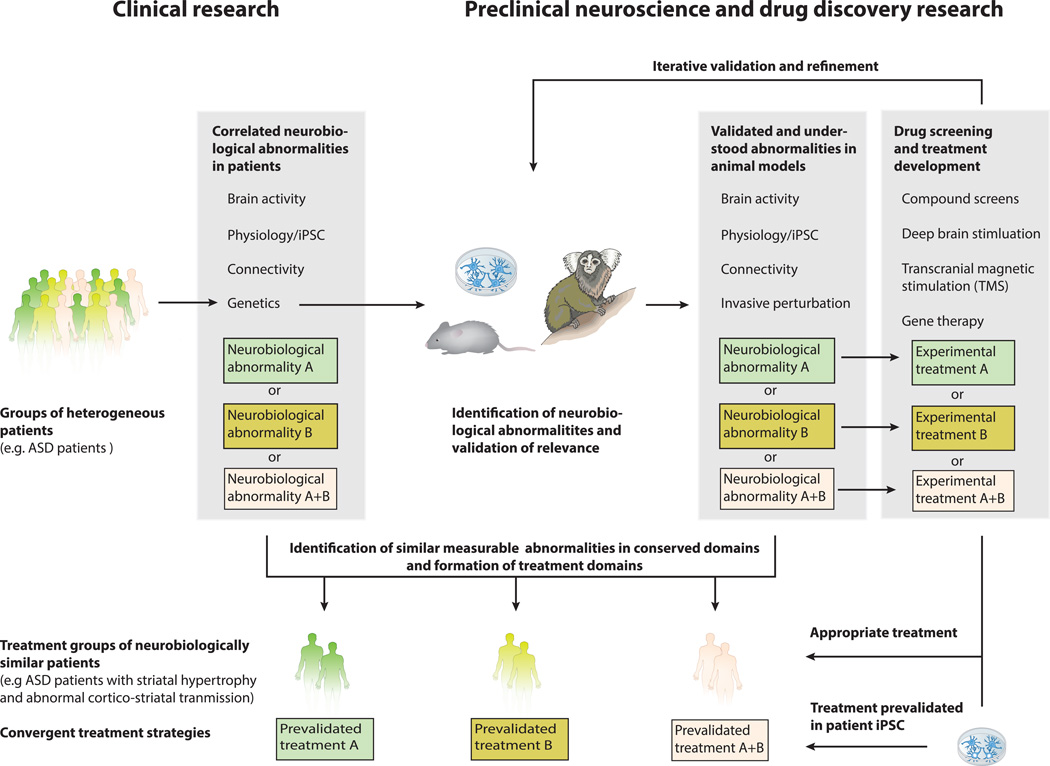
To introduce a similar process to psychiatric disease research, we envision a path forward that leads to the convergence of clinical research, preclinical neuroscience, and drug discovery (Figure 3). This path comprises of four key steps. First, driven by the new NIMH Research Domain Criteria (RDoC) program, groups of highly heterogeneous patients diagnosed with a given disorder are deconstructed, parsed, and categorized into more homogeneous clusters on the basis of genetics, observable signs and neurobiological abnormalities revealed by imaging and neurophysiology65,91. Second, mirroring the principles of RDoC, preclinical neuroscience uses genetic-based animal models and patient iPSC models to identify, understand and validate the relevance and underlying mechanisms of neurobiological abnormalities found in patients. Third, preclinical researchers develop treatment strategies targeting distinct/relevant neurobiological abnormalities with specific compounds and other interventions, such as deep brain stimulation or transcranial magnetic stimulation, while utilizing animal and cellular models to iteratively validate and refine these treatment strategies. In this step, it is critical to identify comparable abnormalities in conserved domains between humans and animal models. Finally, treatment domains and more homogeneous clusters of patients are formed based on shared measureable abnormalities and biomarkers that are conserved and comparable between animal/cellular models and human patients. In clinical trials, these treatment groups receive the appropriate treatment developed based on shared mechanisms, some of which could potentially have been pre-validated in patient iPSC-derived neurons and thus significantly increasing the chances of treatment success.
Figure 3. The path forward: convergence of clinical and preclinical research.
In this hypothetic example of the way forward in treatment development, a heterogeneous group of ASD patients is sub-grouped on the basis of genetics and comprehensive Research Domain Criteria (RDoC). The genetic information obtained in this process also informs preclinical neuroscience and enables the generation of animal models that are similar to the patient in their biological basis. Mirroring the patient RDoC, the same neurophysiological abnormalities are identified, understood and tested in terms of their relevance to disease. Shown schematically here, testing causation through invasive perturbation in animal models mirrors the importance where correlative genetic observation stands in human patients. Next, these valid models are used for invasive and iterative treatment development. Finally, homogeneous treatment domains are formed based on comparable abnormalities in conserved domains and patients within these clusters receive the appropriate treatment for their specific defect, e.g. patient with neurophysiological abnormality A receives the treatment that has been developed for the corresponding defect in animal models. It is also conceivable that another group of patients displays neurobiological abnormalities A and B and thus receives a combinatorial treatment of A and B.
Similar to cancer research, this convergent approach will help close the gap between clinical and preclinical research, establish a fundamental understanding of pathophysiology, and bring more precise and effective treatments to patients. Although this will unlikely result in a singular treatment strategy for all patients with the same DSM diagnosis, several specific mechanism-based treatments can be used in combination, potentially even cutting across similar disorders, to match the patients’ specific needs.
Acknowledgments
We thank James Hawrot, Patricia Monteiro and Charles Jennings for their contribution through valuable discussion and critical reading of the manuscript. T.K. is supported by the Henry E. Singleton fellowship. G.F. is supported by the National Institute of Mental Health (5R01MH097104), the Poitras Center for Affective Disorders Research at MIT, Stanley Center for Psychiatric Research at Broad Institute of MIT and Harvard, Nancy Lurie Marks Family Foundation, Simons Foundation Autism Research Initiative (SFARI) and Simons Center for the Social Brain at MIT.
References
- 1.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–808. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang K, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maloney SE, Rieger MA, Dougherty JD. Identifying essential cell types and circuits in autism spectrum disorders. International review of neurobiology. 2013;113:61. doi: 10.1016/B978-0-12-418700-9.00003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monteiro P, Feng G. Learning from Animal Models of Obsessive-Compulsive Disorder. Biological Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willner P. Validation criteria for animal models of human mental disorders: learned helplessness as a paradigm case. Progress in neuro-psychopharmacology and biological psychiatry. 1986;10:677–690. doi: 10.1016/0278-5846(86)90051-5. [DOI] [PubMed] [Google Scholar]
- 9.McKinney WT, Bunney WE. Animal model of depression: I. Review of evidence: implications for research. Archives of general psychiatry. 1969;21:240–248. doi: 10.1001/archpsyc.1969.01740200112015. [DOI] [PubMed] [Google Scholar]
- 10.McFarlane HG, et al. Autism-like behavioral phenotypes in BTBR T+tf/J mice. Genes, brain, and behavior. 2008;7:152–163. doi: 10.1111/j.1601-183X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- 11.Silverman J, Oliver C, Karras M, Gastrell P, Crawley J. AMPAKINE enhancement of social interaction in the BTBR mouse model of autism. Neuropharmacology. 2013;64:268–282. doi: 10.1016/j.neuropharm.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silverman JL, Tolu SS, Barkan CL, Crawley JN. Repetitive self-grooming behavior in the BTBR mouse model of autism is blocked by the mGluR5 antagonist MPEP. Neuropsychopharmacology. 2010;35:976–989. doi: 10.1038/npp.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lourenço DSA, et al. Effect of riluzole on MK-801 and amphetamine-induced hyperlocomotion. Neuropsychobiology. 2002;48:27–30. doi: 10.1159/000071825. [DOI] [PubMed] [Google Scholar]
- 14.Breier A, et al. Schizophrenia is associated with elevated amphetamine-induced synaptic dopamine concentrations: Evidence from a novel positron emission tomography method. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2569–2574. doi: 10.1073/pnas.94.6.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13:1161–1169. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardno AG, et al. Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Archives of general psychiatry. 1999;56:162–168. doi: 10.1001/archpsyc.56.2.162. [DOI] [PubMed] [Google Scholar]
- 17.Caspi A, Moffitt TE. Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci. 2006;7:583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- 18.Kannan G, Sawa A, Pletnikov MV. Mouse models of gene-environment interactions in schizophrenia. Neurobiology of disease. 2013;57:5–11. doi: 10.1016/j.nbd.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klengel T, Binder Elisabeth B. Epigenetics of Stress-Related Psychiatric Disorders and Gene × Environment Interactions. Neuron. 86:1343–1357. doi: 10.1016/j.neuron.2015.05.036. [DOI] [PubMed] [Google Scholar]
- 20.McCarroll SA, Feng G, Hyman SE. Genome-scale neurogenetics: methodology and meaning. Nature neuroscience. 2014;17:756–763. doi: 10.1038/nn.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haesemeyer M, Schier AF. The study of psychiatric disease genes and drugs in zebrafish. Current Opinion in Neurobiology. 2015;30:122–130. doi: 10.1016/j.conb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zweier C, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. The American Journal of Human Genetics. 2009;85:655–666. doi: 10.1016/j.ajhg.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. The American journal of psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 24.Gould TD, Gottesman II. Psychiatric endophenotypes and the development of valid animal models. Genes, brain, and behavior. 2006;5:113–119. doi: 10.1111/j.1601-183X.2005.00186.x. [DOI] [PubMed] [Google Scholar]
- 25.Dincheva I, et al. FAAH genetic variation enhances fronto-amygdala function in mouse and human. Nature communications. 2015;6 doi: 10.1038/ncomms7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mague SD, et al. Mouse model of OPRM1 (A118G) polymorphism has sex-specific effects on drug-mediated behavior. Proceedings of the National Academy of Sciences. 2009;106:10847–10852. doi: 10.1073/pnas.0901800106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z-Y, et al. Genetic Variant BDNF (Val66Met) Polymorphism Alters Anxiety-Related Behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Böckers TM, et al. Synaptic Scaffolding Proteins in Rat Brain ANKYRIN REPEATS OF THE MULTIDOMAIN Shank PROTEIN FAMILY INTERACT WITH THE CYTOSKELETAL PROTEIN α-FODRIN. Journal of Biological Chemistry. 2001;276:40104–40112. doi: 10.1074/jbc.M102454200. [DOI] [PubMed] [Google Scholar]
- 29.Peca J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peñagarikano O, et al. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clement JP, et al. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 2012;151:709–723. doi: 10.1016/j.cell.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabuchi K, et al. A Neuroligin-3 Mutation Implicated in Autism Increases Inhibitory Synaptic Transmission in Mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derecki NC, et al. Wild-type microglia arrest pathology in a mouse model of Rett syndrome. Nature. 2012;484:105–109. doi: 10.1038/nature10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lioy DT, et al. A role for glia in the progression of Rett/'s syndrome. Nature. 2011;475:497–500. doi: 10.1038/nature10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, et al. Wild-type microglia do not reverse pathology in mouse models of Rett syndrome. Nature. 2015;521:E1–E4. doi: 10.1038/nature14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxena S, Brody AL, Schwartz JM, Baxter LR. Neuroimaging and frontal-subcortical circuitry in obsessive-compulsive disorder. The British journal of psychiatry. Supplement. 1998:26–37. [PubMed] [Google Scholar]
- 38.Saxena S, Rauch SL. FUNCTIONAL NEUROIMAGING AND THE NEUROANATOMY OF OBSESSIVE-COMPULSIVE DISORDER. Psychiatric Clinics of North America. 2000;23:563–586. doi: 10.1016/s0193-953x(05)70181-7. [DOI] [PubMed] [Google Scholar]
- 39.Burguière E, Monteiro P, Feng G, Graybiel AM. Optogenetic Stimulation of Lateral Orbitofronto-Striatal Pathway Suppresses Compulsive Behaviors. Science. 2013;340:1243–1246. doi: 10.1126/science.1232380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmari SE, et al. Repeated Cortico-Striatal Stimulation Generates Persistent OCD-Like Behavior. Science. 2013;340:1234–1239. doi: 10.1126/science.1234733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nature neuroscience. 2014;17:764–772. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rothwell PE, et al. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158:198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grabli D, et al. Behavioural disorders induced by external globus pallidus dysfunction in primates: I. Behavioural study. Brain. 2004;127:2039–2054. doi: 10.1093/brain/awh220. [DOI] [PubMed] [Google Scholar]
- 44.Reiner A, Medina L, Veenman CL. Structural and functional evolution of the basal ganglia in vertebrates. Brain Research Reviews. 1998;28:235–285. doi: 10.1016/s0165-0173(98)00016-2. [DOI] [PubMed] [Google Scholar]
- 45.Shultz S, Opie C, Atkinson QD. Stepwise evolution of stable sociality in primates. Nature. 2011;479:219–222. doi: 10.1038/nature10601. [DOI] [PubMed] [Google Scholar]
- 46.Bayés À, et al. Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PloS one. 2012;7:e46683. doi: 10.1371/journal.pone.0046683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neale BM, Sklar P. Genetic analysis of schizophrenia and bipolar disorder reveals polygenicity but also suggests new directions for molecular interrogation. Current opinion in neurobiology. 2015;30:131–138. doi: 10.1016/j.conb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 48.Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Current Opinion in Genetics & Development. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Geschwind DH. Genetics of autism spectrum disorders. Trends in Cognitive Sciences. 2011;15:409–416. doi: 10.1016/j.tics.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rujescu D, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Human molecular genetics. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ching MS, et al. Deletions of NRXN1 (neurexin - 1) predispose to a wide spectrum of developmental disorders. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2010;153:937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paşca SP, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nature medicine. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shcheglovitov A, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hook V, et al. Human iPSC neurons display activity-dependent neurotransmitter secretion: Aberrant catecholamine levels in schizophrenia neurons. Stem cell reports. 2014;3:531–538. doi: 10.1016/j.stemcr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eiraku M, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 59.Sato T, et al. Single Lgr5 stem cells build crypt villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 60.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mariani J, et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kriks S, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson/'s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Espuny-Camacho I, et al. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 64.Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotech. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 65.Insel T, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. The American journal of psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 66.Belzung C, Lemoine M. Criteria of validity for animal models of psychiatric disorders: focus on anxiety disorders and depression. Biology of mood & anxiety disorders. 2011;1:9. doi: 10.1186/2045-5380-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferrarelli F, et al. Thalamic dysfunction in schizophrenia suggested by whole-night deficits in slow and fast spindles. Am J Psychiatry. 2010;167:1339–1348. doi: 10.1176/appi.ajp.2010.09121731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wamsley EJ, et al. Reduced sleep spindles and spindle coherence in schizophrenia: mechanisms of impaired memory consolidation? Biological psychiatry. 2012;71:154–161. doi: 10.1016/j.biopsych.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwon JS, et al. Gamma Frequency–Range Abnormalities to Auditory Stimulation in Schizophrenia. Archives of general psychiatry. 1999;56:1001–1005. doi: 10.1001/archpsyc.56.11.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teale P, et al. Cortical source estimates of gamma band amplitude and phase are different in schizophrenia. NeuroImage. 2008;42:1481–1489. doi: 10.1016/j.neuroimage.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang X, et al. Altered default mode and fronto-parietal network subsystems in patients with schizophrenia and their unaffected siblings. Brain Res. 2014;1562:87–99. doi: 10.1016/j.brainres.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 72.Chai XJ, et al. Abnormal medial prefrontal cortex resting-state connectivity in bipolar disorder and schizophrenia. Neuropsychopharmacology. 2011;36:2009–2017. doi: 10.1038/npp.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kubicki M, et al. A review of diffusion tensor imaging studies in schizophrenia. J Psychiatr Res. 2007;41:15–30. doi: 10.1016/j.jpsychires.2005.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuperberg GR, et al. Regionally localized thinning of the cerebral cortex in schizophrenia. Archives of general psychiatry. 2003;60:878–888. doi: 10.1001/archpsyc.60.9.878. [DOI] [PubMed] [Google Scholar]
- 75.Shenton ME, et al. Abnormalities of the Left Temporal Lobe and Thought Disorder in Schizophrenia. New England Journal of Medicine. 1992;327:604–612. doi: 10.1056/NEJM199208273270905. [DOI] [PubMed] [Google Scholar]
- 76.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Archives of general psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 77.Spencer KM, et al. Neural synchrony indexes disordered perception and cognition in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17288–17293. doi: 10.1073/pnas.0406074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sutter EE. The brain response interface: communication through visually-induced electrical brain responses. J. Microcomput. Appl. 1992;15:31–45. [Google Scholar]
- 79.Sigurdsson T, Stark KL, Karayiorgou M, Gogos JA, Gordon JA. Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature. 2010;464:763–767. doi: 10.1038/nature08855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fejgin K, et al. A mouse model that recapitulates cardinal features of the 15q13.3 microdeletion syndrome including schizophrenia- and epilepsy-related alterations. Biological psychiatry. 2014;76:128–137. doi: 10.1016/j.biopsych.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 81.Hollander E, et al. Striatal volume on magnetic resonance imaging and repetitive behaviors in autism. Biological psychiatry. 2005;58:226–232. doi: 10.1016/j.biopsych.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 82.Okano H, Mitra P. Brain-mapping projects using the common marmoset. Neuroscience Research. 2015;93:3–7. doi: 10.1016/j.neures.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 83.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Janak PH, Tye KM. From circuits to behaviour in the amygdala. Nature. 2015;517:284–292. doi: 10.1038/nature14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stephenson-Jones M, Samuelsson E, Ericsson J, Robertson B, Grillner S. Evolutionary Conservation of the Basal Ganglia as a Common Vertebrate Mechanism for Action Selection. Current Biology. 2011;21:1081–1091. doi: 10.1016/j.cub.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 86.Graybiel AM. The basal ganglia. Current Biology. 2000;10:R509–R511. doi: 10.1016/s0960-9822(00)00593-5. [DOI] [PubMed] [Google Scholar]
- 87.Willsey AJ, et al. Coexpression Networks Implicate Human Midfetal Deep Cortical Projection Neurons in the Pathogenesis of Autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen R, Romero G, Christiansen MG, Mohr A, Anikeeva P. Wireless magnetothermal deep brain stimulation. Science. 2015;347:1477–1480. doi: 10.1126/science.1261821. [DOI] [PubMed] [Google Scholar]
- 89.de Bono JS, Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467:543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 90.Collins FS, Varmus H. A New Initiative on Precision Medicine. New England Journal of Medicine. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Insel TR, Cuthbert BN. Brain disorders? Precisely. Science. 2015;348:499–500. doi: 10.1126/science.aab2358. [DOI] [PubMed] [Google Scholar]
- 92.Tufts, C.f.t.S.o.D.D. CNS drugs take longer to develop, have lower success rates, than other drugs. CSDD Impact Report. 2014;16 [Google Scholar]
- 93.Wise SP. Forward frontal fields: phylogeny and fundamental function. Trends in neurosciences. 2008;31:599–608. doi: 10.1016/j.tins.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cooke D, Goldring A, Recanzone GH, Krubitzer . The Evolution of Parietal Areas Associated with Visuomanual Behavior: From Grasping to Tool Use. In: Chalupa L, Werner J, editors. The New Visual Neurosciences. Cambridge, MA: MIT Press; 2014. pp. 1049–1063. [Google Scholar]
- 95.Burman KJ, Rosa MG. Architectural subdivisions of medial and orbital frontal cortices in the marmoset monkey (Callithrix jacchus) Journal of Comparative Neurology. 2009;514:11–29. doi: 10.1002/cne.21976. [DOI] [PubMed] [Google Scholar]
- 96.Schmeisser MJ, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- 97.Won H, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- 98.Bozdagi O, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15–15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang X, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Human molecular genetics. 2011:ddr212. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang M, et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. The Journal of Neuroscience. 2012;32:6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kouser M, et al. Loss of predominant shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. The Journal of Neuroscience. 2013;33:18448–18468. doi: 10.1523/JNEUROSCI.3017-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han K, et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature. 2013;503:72–77. doi: 10.1038/nature12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nature genetics. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 104.Lioy DT, et al. A role for glia in the progression of Rett/'s syndrome. Nature. 2011;475:497–500. doi: 10.1038/nature10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kumar M, et al. High Resolution Magnetic Resonance Imaging for Characterization of the Neuroligin-3 Knock-in Mouse Model Associated with Autism Spectrum Disorder. PloS one. 2014;9:e109872. doi: 10.1371/journal.pone.0109872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hashimoto T, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Frantseva MV, et al. Evidence for impaired long-term potentiation in schizophrenia and its relationship to motor skill learning. Cerebral cortex (New York, N.Y. : 1991) 2008;18:990–996. doi: 10.1093/cercor/bhm151. [DOI] [PubMed] [Google Scholar]
- 108.Hasan A, et al. Impaired long-term depression in schizophrenia: a cathodal tDCS pilot study. Brain stimulation. 2012;5:475–483. doi: 10.1016/j.brs.2011.08.004. [DOI] [PubMed] [Google Scholar]