Abstract
Members of the G protein coupled receptor (GPCR) family play key roles in many physiological functions and have been extensively exploited pharmacologically to treat diseases. Individual GPCRs exert diverse and distinct effects on cellular physiology and transduce signals by activating heterotrimeric G proteins. Mammalian genomes encode 16 different G protein alpha subunits, and each one of them has distinct properties. Here, we developed a single-platform, optical strategy for the direct monitoring of G protein activation in live cells, and using it we profiled the activities of individual GPCRs across a range of different G proteins, simultaneously quantifying both magnitude of their signaling and activation rates. We report that GPCRs engage multiple G proteins with varying efficacy and kinetics, generating fingerprint-like profiles that define individual receptors. We found that different classes of GPCR ligands, including full and partial agonists, allosteric modulators, and antagonists distinctly affected these fingerprints to functionally bias GPCR signaling. Finally, we showed that intracellular signaling modulators further altered the G protein–coupling profiles of GPCRs, which suggests that their differential expression may alter signaling outcomes in a cell-specific manner. . These observations suggest that the diversity of the effects of GPCRs on cellular physiology may be determined by their differential engagement of multiple G proteins with varying signal magnitudes and activation kinetics, properties that may be exploited pharmacologically.
Introduction
Signaling through G protein–coupled receptors (GPCRs) controls a vast number of physiological processes, ranging from the action of hormones and neurotransmitters to cell migration and differentiation (1). The disruption of GPCR signaling frequently contributes to various pathophysiological conditions, including cancer, neurological disorders, and metabolic syndromes (2–5). As such, GPCRs are among the most successful and tractable drug targets, and they account for about 30 to 40% of the medications currently on the market (6, 7). Despite their importance, there are substantial challenges in understanding the mechanisms of GPCR signaling, as well as the actions of drugs on these receptors. Perhaps one of the biggest unresolved questions is to understand how GPCRs receive, encode, and convert diverse extracellular cues into a precise set of signaling reactions that change cellular responses in a characteristic fashion. There are more than 800 GPCRs encoded in mammalian genomes and there is likely an even greater number of stimuli that they respond to. However, the activation of an individual receptor generates a distinct message that the cells can distinguish from others.
In the canonical model, GPCR signaling is initiated when a ligand-bound receptor activates heterotrimeric G proteins on the inner leaflet of the plasma membrane by catalyzing the exchange of GDP for GTP on the G protein α subunit (Gα), causing it to release the Gβγ subunits (which form a single unit). Both GTP-bound Gα and free Gβγ subunits transduce the signal by engaging intracellular effector molecules until the GTP is hydrolyzed and the subunits re-associate (8). In addition to activating G proteins, GPCRs can also engage β-arrestin scaffolds that can transmit a signal independently of G proteins (9). This signaling model was substantially revised to account for the discovery that GPCRs exhibit functional selectivity, which manifests in the activation of different pathways depending on the nature of the ligand, the interactions that receptors are engaged in, or both (10, 11). It is thought that this signaling flexibility is determined by the ability of GPCRs to adopt various conformational states that translate into differential interactions with molecules downstream of the receptors that transduce signals (12).
One of the best examples of the functional selectivity of GPCRs is the differential engagement of G proteins versus β-arrestins in a ligand-directed fashion (11). Whereas G protein vs. β-arrestin selectivity provides an important insight into the mechanisms that generate signaling diversity, our understanding of the whole spectrum of the functional selectivity of GPCRs is still in its infancy and many rules and mechanisms have yet to be determined. Defining the functional selectivity of GPCRs will help to explain the unique “code conversion” process for individual receptors supporting their distinct effects on cellular physiology. Furthermore, there is a growing appreciation that this selectivity could be exploited pharmacologically by designing biased, small-molecule agonists and modulators to extend the precision of therapeutic interventions (13, 14).
All known GPCRs share the ability to activate G proteins, and this step is likely the largest source of functional selectivity (15). Mammalian genomes contain 16 different genes that encode Gα subunits, which serve as direct targets of the guanine nucleotide exchange factor (GEF) activity of GPCRs, and an equally diverse repertoire of Gβγ isoforms that facilitate Gα activation (15, 16). Whereas different Gβγ subunits are thought to be functionally interchangeable (17), Gα subunits display distinct and nonredundant properties, regulating various effectors and thus consequently defining a host of cellular responses (1, 18, 19). It is common to downplay the actual diversity of Gα subunits, grouping all GPCRs into four large, functional classes according to the family of Gα subunits that they are assumed to activate most effectively; that is, Gi/o, Gq/11, Gs, and G12/13. However, rapidly growing evidence indicates that many GPCRs can couple to multiple G proteins across classes and that their ability to activate individual members of even the same class can vary substantially (20–23). This evidence, combined with reports that the coupling of GPCRs to G proteins is influenced by the ligands (21, 24–27) and possibly by the interacting partners of GPCRs (28), paints a far more complex picture in which GPCRs likely use a multivalent and flexible code based on their G protein coupling selectivity. Although there have been several insightful reports characterizing the G protein coupling profiles of GPCRs (29–32), most attempts were limited by the narrow range of the G proteins that were used or by indirect evaluations, which has left the functional selectivity of most GPCRs across their various G protein targets virtually unexplored.
Here, we revealed the complex profiles of functional bias with which GPCRs activate G proteins. We applied a quantitative and systematic interrogation of GPCR activity across an exhaustive spectrum of G proteins in live cells to generate comprehensive substrate preference maps for representative receptors. Determining both the kinetics and extent of G protein activation enabled us to independently sample the catalytic activities of GPCRs and their signaling efficacy toward various target G proteins. We report that various GPCRs have distinct profiles of G protein engagement, both in terms of signaling kinetics and the extent of activation, , effectively generating characteristic signatures, or “fingerprints.” We demonstrated that these GPCR fingerprints exhibited clear functional bias and were differentially affected by receptor ligands and intracellular signaling modulators. These findings open up the exploration of the selective effects produced by GPCR activation and may help in understanding how the diversity of signaling by GPCRs is generated.
Results
Comprehensive real-time monitoring of GPCR activity toward multiple G proteins in live cells
Few, if any, individual GPCRs have been evaluated for their ability to activate an exhaustive set of possible G protein targets with quantitative discrimination. We reasoned that because the GPCR-catalyzed dissociation or re-arrangement of G protein heterotrimers, which then leads to the exposure of their effector-binding surfaces, is a key event in the activation of all G proteins, a suitable approach would have to rely on monitoring transitions in the heterotrimer. Among several previously proposed strategies (33–35), we favored monitoring Gβγ release (36) because of its universality and the ability to avoid the need to modify the GPCRs or Gα subunits, which may substantially impede their physiological states and make quantitative comparisons between individual pairs of GPCRs and G proteins difficult, if not impossible. This strategy monitors changes in agonist-induced interactions of Venus-tagged Gβγ with the reporter, an effector-like peptide derived from G protein Receptor Kinase 3 (GRK3) tagged with Renilla luciferase (Rluc) by bioluminescence resonance energy transfer (BRET). Although measuring Gβγ release with this approach has been useful for assaying select G proteins (37, 38), the rather weak and noisy nature of BRET signals has prevented the detection of low-efficiency reactions while limiting the throughput, the temporal resolution, and the quantitative accuracy needed for comparative profiling of GPCR activities on many G proteins.
To make such analysis possible, we employed a NanoBRET strategy, by fusing the engineered blue-shifted bright luciferase from Oplophorus gracilirostris (NLuc) (39) to the lipid-modified reporter peptide GRK3ct (masGRK3ct-NLuc), and studying its interactions with Venus-tagged Gβ1γ2 (Fig. 1A). The introduction of NLuc into HEK293T/17 cells markedly increased the luminescence signal and improved the variability and reproducibility of the agonist-induced signal (Fig. 1, B and C, and fig. S1, A to E), making it possible to reliably record kinetics with millisecond resolution (fig. S1, F and G). The improvement in the signal to noise ratio was so pronounced that we could even resolve signals from previously intractable G proteins and GPCRs; for example, signaling through G12/13 proteins (fig. S1H) (40). Finally, we benchmarked the NanoBRET sensor approach against the electrophysiological detection of G protein activation by monitoring the activity of a G protein–coupled inwardly-rectifying K+ (GIRK) channel (fig. S1, I to K), an ion channel that is directly gated by Gβγ subunits and is ubiquitously used to assess GPCR signaling in native cells and reconstituted systems (41). When constructs were ectopically expressed in human embryonic kidney (HEK) 293T/17 cells, we obtained very similar kinetics for the GPCR-driven responses between NanoBRET biosensors and the patch clamp recordings (fig. S1, I to K). Indeed, the activation rates that we observed were very similar to those of GPCR-stimulated GIRKs in native cells (42, 43), suggesting that the conditions of this assay closely match the in vivo setting. This finding further demonstrates the ability of the system to resolve the fast, physiologically relevant kinetics of GPCR signaling.
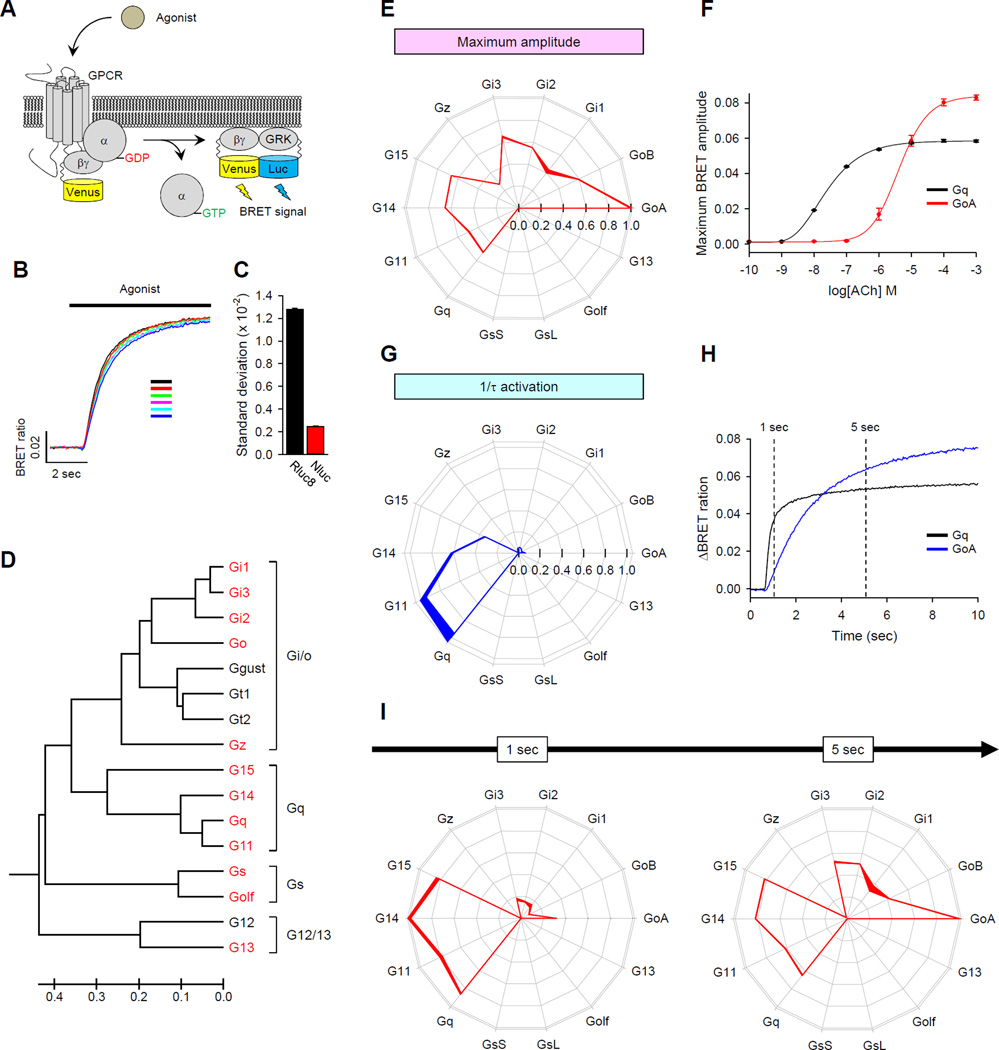
Fig. 1. Fingerprinting GPCR activity by measuring signaling efficacy and kinetics across a set of G proteins.
(A) Schematic representation of the BRET assay. Activation of a GPCR by agonist leads to the dissociation of inactive heterotrimeric G proteins into active GTP-bound Gα and Venus-Gβγ subunits. The free Venus-Gβγ then interacts with the Gβγ effector mimetic masGRK3ct-Nluc to produce the BRET signal. (B) Representative response profile showing the BRET signal generated by the D2 dopamine receptor in the presence of Gαo. Dopamine (100 µM) was applied to the cells and six independent reactions were conducted in parallel. (C) Quantification of response variability between the different indicated sensors. (D) Repertoire of mammalian Gα subunits. G proteins marked in red were successfully reconstituted in the NanoBRET system. Scale bar below represents relative evolutionary distance. (E to I) Fingerprinting responses of the M3 muscarinic acetylcholine receptor (M3R) to the physiological ligand acetylcholine (ACh). (E) Quantification of the maximal response amplitudes generated by M3R. The maximum amplitudes from the 14 different G proteins were normalized to the largest value to obtain comparative agonist efficacy and were plotted at corresponding vertices in the wheel diagram. The thickness of the lines connecting each data point represents the SEM of four experiments performed in parallel. (F) Dose-response curve relationships of two representative signaling pathways in response to Ach. Data are means ± SEM of four experiments. (G) Quantification of the G protein activation rates catalyzed by the M3R. Activation rate constants from 14 different G proteins were normalized to the response that produced the maximum value and are plotted for each of the G proteins tested. Data are means ± SEM of four experiments. (H) Comparison of the time-courses of activation of GoA and Gq. Each trace represents the mean of the responses measured in eight wells. (I) Maximal response amplitudes recorded at different times [1 and 5 s marked in (H)] after agonist application. Data are means ± SEM of four experiments.
We extended this approach to reconstitute signaling to monitor the activity of 14 different G proteins, thereby covering essentially the entire repertoire of mammalian Gα subunits with the exception of the sensory G proteins of the Gt subfamily (gustducin and the rod and cone transducins), which have very restricted expression profiles, and G12, which we were unable to reconstitute (Fig. 1D and figs. S2 to S4). The following optimization steps were applied to reconstitute these systems. First, we found that Gα15, Gα14, and Gαolf required co-expression with the molecular chaperones Ric-8A or Ric-8B (44, 45) for the formation of functional G protein complexes (fig. S2). Second, we optimized the stoichiometry of Gα and Venus-Gβγ by titrating the amount of Gα subunits against a constant amount of Venus-Gβγ for all pairs, and then choosing the transfection condition that resulted in the lowest basal BRET ratio and the highest maximum amplitude for each G protein (fig. S3). Finally, for each of the G protein and GPCR combinations, we ensured the specificity of the change in BRET signal by the application of appropriate receptor antagonists, which effectively reversed the signal (fig. S4).
Under these optimized conditions, the localization of Venus-Gβγ on the plasma membrane was dependent upon its co-expression with the Gα subunits, which ensured that there were stoichiometric ratios of subunits in G protein heterotrimers that could be activated by GPCRs at the plasma membrane (fig. S5). Consistent with this observation, we detected no change in the BRET signal upon applying agonist to cells that did not express exogenous Gα subunits, indicating that the signal in our system was specifically driven by the Gα subunits that were expressed (fig. S6). Furthermore, the abundance of Venus-Gβγ in cells transfected with plasmids encoding different Gα subunits was similar to the amount of endogenous Gβγ present in the cells (fig. S7), suggesting that our experimental conditions were close to being physiological. Furthermore, with all of the GPCR-Gα combinations that we tested, the application of a saturating concentration of agonist did not produce a saturated BRET response, as determined by referencing the agonist-induced signals against signals in the presence or absence of Gα (fig. S8), indicating that relative differences in the strengths of signals generated by different combinations of receptor and Gα subunit could be evaluated in this system. Thus, this assay enables the direct comparison of the efficacy and activation kinetics of most of the G proteins in a live cell.
Independent assessment of GPCR functional activity by measuring the efficacy and kinetics of activation for individual G proteins
For our initial experiments, we chose the well-studied M3 muscarinic acetylcholine receptor (M3R) because of its reported diversity of G protein activation and its substantial clinical relevance (46). Measurement of the maximal BRET response amplitudes, which were stimulated by a physiological agonist for this receptor, acetylcholine (ACh), revealed a distinct profile of its G protein selectivity (Fig. 1E). The use of a saturating concentration of agonist to achieve total receptor occupancy enabled us to approximate the efficacy of the ligand-GPCR pair on each of the G proteins by measuring the amplitude of the maximal response. Under these conditions, the M3R coupled to 10 of the 14 G proteins tested, including all members of the Gi/o and Gq families, but it had no activity towards Gs or G12/13 proteins.
We unexpectedly found that M3R activated Gi/o proteins to a greater extent than it activated Gq proteins, despite it being generally considered a Gq-specific GPCR. Indeed, of all of the G proteins tested, GoA was activated by M3R to the greatest extent (Fig. 1E). To obtain further insights into this observation, we performed classical dose-response studies to compare the dependence of the M3R-mediated activation of GoA and Gq on the concentration of acetylcholine (Fig. 1F). As expected, acetylcholine was more potent at activating Gq (~190 fold) than it was at activating GoA (EC50 = 2.4 ± 0.1 × 10−8 M and 4.5 ± 0.9 × 10−6 M, respectively). However, the efficacy of GoA activation by the M3R exceeded that of Gq activation by ~30% (BRET maximum amplitude = 0.082 ± 0.014 vs 0.058 ± 0.006). These findings suggest that the efficacy and potency of M3R–generated responses may be differentially determined by the identity of the G protein to which the receptor couples.
Considering that GPCRs serve as GEFs for heterotrimeric G proteins and thus act to accelerate their activation, we next assessed the kinetics of the response onset as a more direct proxy of the catalytic activity of the receptor. We quantified the activation rate constant 1/τ (s−1) across different G proteins by fitting traces with a single exponent function (Fig. 1G). In this kinetic domain, the M3R had the largest effect on the activation of the Gq family members Gq and G11, which were activated the fastest (5.9 ± 0.3 s−1 and 5.7 ± 0.2 s−1, for Gq and G11, respectively), which was followed by G14 and G15 (3.6 ± 0.1 s−1 and 2.0 ± 0.1 s−1, respectively). Activation of Gi/o family members was markedly slower (GoA: 0.38 ± 0.01 s−1; GoB: 0.18 ± 0.01 s−1; Gi1: 0.26 ± 0.01 s−1; Gi2: 0.37 ± 0.01 s−1; Gi3: 0.25 ± 0.01 s−1; Gz: 0.024 ± 0.001 s−1). Note that the difference in the kinetics of activation of GoA and Gq by the M3R was consistent with the corresponding difference in their potencies, which suggests that potency differences are underpinned by differences in the catalytic GEF activity of GPCRs towards target G proteins (Fig. 1F and 1H). By contrast, the efficacies measured in our assay likely reflect the extent of heterotrimer dissociation (Fig. 1F and 1H).
The observation that the kinetics of G protein activation do not necessarily correlate with the maximal amplitude of activation emphasized the importance of determining both parameters when examining the G protein selectivity of GPCRs. For example, the M3R produces different fingerprints depending on the reaction time (Fig. 1I). Assessing the activity during the initial phase (~1 s) showed that M3R primarily activated Gq, G11, G14, and G15, whereas the activation of Gi/o became more prominent as the system reached steady state upon prolonged stimulation with agonist (Fig. 1I). We therefore conclude that kinetic considerations together with steady-state measurements of receptor efficacies form a characteristic GPCR fingerprint that is sufficient for the comprehensive interpretation of the functional activity and selectivity of the GPCR.
Distinct G protein activation fingerprints of GPCRs
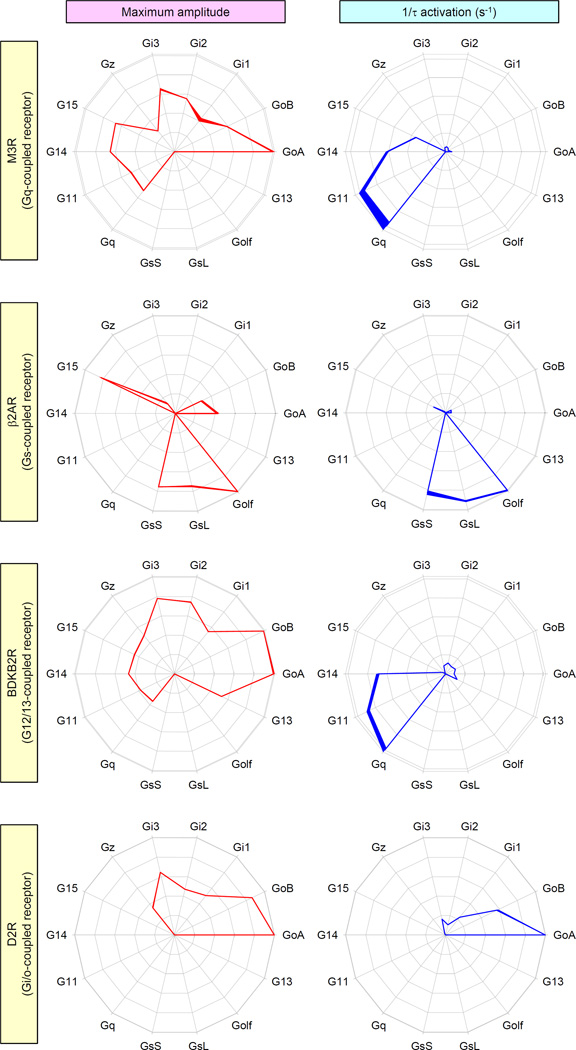
To better understand the scope of differences in the coupling of GPCRs to their various target G proteins, we evaluated the fingerprints of representative receptors, including the dopamine D2 receptor (D2R), the β2 adrenergic receptor (β2AR), and the bradykinin B2 receptor (BDKB2R) which are classically defined as coupled to Gi, Gs, and G12/13 family members, respectively, and compared them to the fingerprint of the Gq-coupled M3R. For each chosen receptor, we measured both the maximal response amplitude that was induced by a saturating concentration of an endogenous agonist and the activation kinetics across the entire panel of G proteins. The data revealed distinct coupling profiles for each of the tested receptors (Fig. 2).
Fig. 2. Characteristic profiles of G protein activation distinguish various GPCRs from each other.
Several GPCRs that belong to different subfamilies were examined for the specificity of their G protein coupling by measuring two parameters: maximum amplitude of the BRET signal (red) and activation rates (blue). Cells expressing M3R, β2AR, the bradykinin B2 receptor (BDKB2R), or the dopamine D2 receptor (D2R) were activated by saturating concentrations (100 µM) of their respective endogenous agonists: acetylcholine, adrenaline, bradykinin, and dopamine. The data reflecting maximum BRET amplitude and activation rate are plotted as relative activity values after normalization against the G protein species that exhibited maximal activity. Data are means ± SEM of four to six experiments.
Similar to the M3R, the pattern of G proteins mobilized by β2AR was complex and covered three classes and seven types of G proteins. We found that in addition to coupling to the Gs isoforms and Golf, the β2AR exhibited substantial coupling to G15, Go, and Gz; however, we were unable to find any evidence of the coupling of the β2AR to Gi1, Gi2, or Gi3. In terms of activation kinetics, the β2AR activated Gs and Golf proteins more quickly than others, which again revealed the dissociation of the maximal extent of activation from the timing of activation across its target G proteins. Yet another pattern was exhibited by the BDKB2R, which efficiently coupled to all members of the Gi/o, Gq, and G12/13 families, but not the Gs family, with characteristic differences between individual G proteins (Fig. 2). Again, this receptor exhibited disproportionately faster activation of Gq and G11, with less rapid activity towards G14, and markedly slower activation of other G protein types. Consistent with expectations, we found that D2R coupled exclusively to members of the inhibitory Gi/o family, eliciting the greatest response through the Go isoforms, which were followed by Gi1, Gi2, Gi3, and Gz. In this case, the activation kinetics matched this pattern, suggesting that, in contrast to other tested receptors, the extent and timing of G protein activation by the D2R is proportionately scaled across its G protein targets. Overall, our analysis of several receptors revealed diverse profiles of their G protein selectivity with distinct characteristics in terms of efficacy and timing of their G protein activation.. .
Shaping GPCR fingerprints by intracellular regulators of G protein signaling
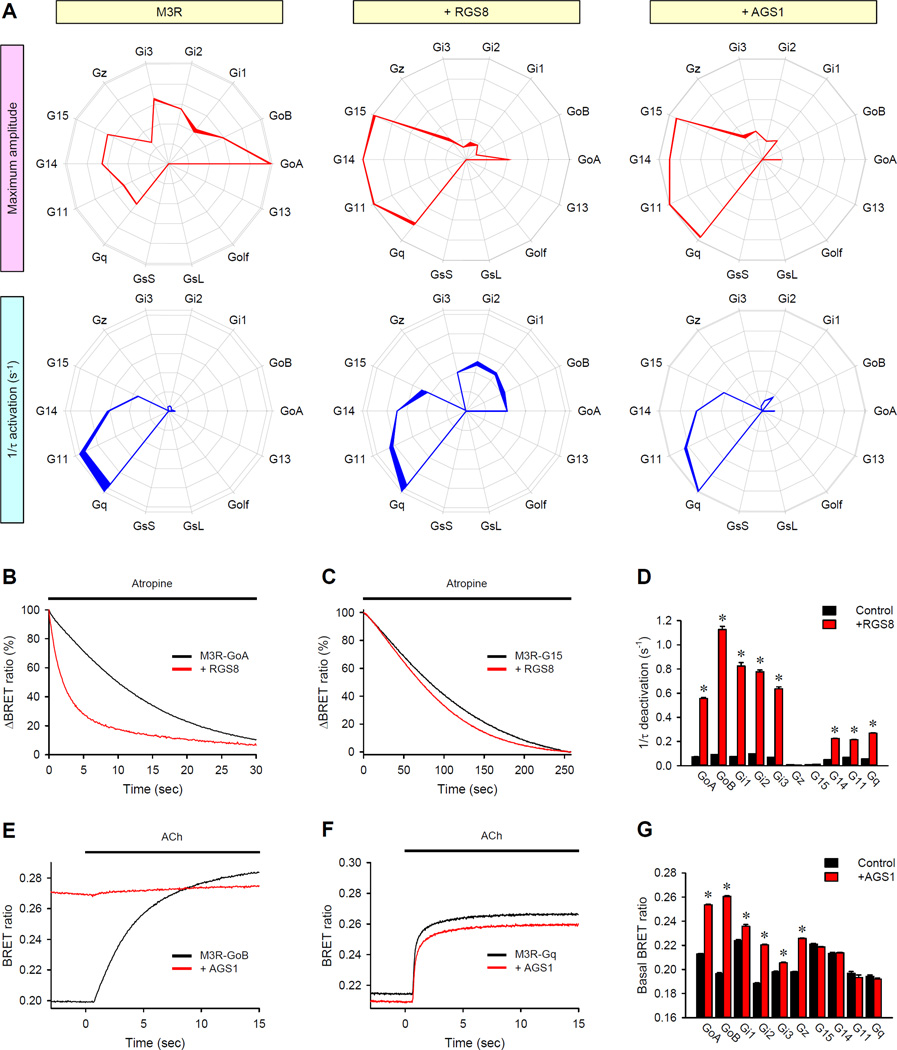
In native cells, G protein signaling is substantially shaped by various regulators that directly or indirectly influence the strength, kinetics, or both of the GPCR responses (28). Because these signaling regulators exhibit tissue- and cell–specific patterns of expression, we hypothesized that they may contribute to establishing selective GPCR signaling patterns. To determine whether these factors markedly influenced GPCR fingerprints, we focused on two families: regulators of G protein signaling (RGS) proteins (47) and activators of G protein signaling (AGS) proteins (48). Representative members of these families, RGS8 (49) and AGS1 (50), are well-characterized to serve as GTPase-activating proteins (GAPs) and GEFs, respectively.
Introduction of RGS8 into the HEK293T/17 cells substantially changed the profile of M3R signaling (Fig. 3A). RGS8 suppressed the BRET signal generated specifically by all Gi/o family members, with the exception of Gz, relative to the signals of other G proteins. At the same time, RGS8 increased the activation speed of the response through the Gi and Go proteins, consistent with the known behavior of RGS proteins (49, 51). This RGS protein selectively muted M3R signaling through Gi/o family members while making the responses faster. To investigate the mechanistic underpinning of the effects produced by RGS8, we examined the deactivation phase of the BRET response after the addition of the muscarinic antagonist atropine. Analysis of the reaction time course indicated that RGS8 substantially accelerated the deactivation of Go, but had no effect on the deactivation kinetics of G15 (Fig. 3, B and C). Complete profiling of the deactivation kinetics of M3R in the presence and absence of RGS8 further revealed asymmetric effects of this protein on the termination of signaling through different G protein substrates (Fig. 3D and fig. S9). These data suggest that RGS8 exerts changes in the fingerprint of the M3R by selectively accelerating G protein deactivation.
Fig. 3. Major classes of intracellular G protein regulators have distinct effects on GPCR fingerprints.
(A) The G protein coupling profiles of the M3 receptor were examined in cells in the absence of regulatory molecules (left), in the presence of RGS8 (middle), or in the presence of AGS1 (right). Data are means ± SEM of four experiments. (B and C) Effect of RGS8 on the deactivation rates of Go and G15. Cells were pretreated with 100 µM acetylcholine (ACh) for 35 s and then were treated with 1 mM muscarinic antogonist atropine. Traces correspond to the deactivation phase of the responses of GoA (B) and G15 (C) in the absence and presence of RGS8, and are the average of 12 experiments, normalized to the response at the time of atropine application. (D) The deactivation rate constants in the absence (black) or presence (red) of RGS8 were measured for all responding G proteins. Data are means ± SEM of four experiments. (E and F) Effect of AGS1 on the activation of GoB and Gq. Cells were cotransfected with plasmids encoding M3R (E and F) and either GoB (E) or Gq (F) with (red) or without (black) plasmid encoding AGS1. BRET signals before (basal) and after the application of Ach were recorded. Each trace is an average of six replicates. (G) Quantification of changes in the basal BRET ratio for the indicated G proteins measured in the absence (black) or presence (red) of AGS1. Data are means ± SEM of six experiments. The unpaired t-test was used to test for statistically significant differences between nuntransfected cells and RGS8-expressing (D) or AGS1-expressing cells (G). *P < 0.001.
In addition to their function as receptor-independent GEFs, several AGS proteins are also capable of modulating GPCR-stimulated responses in vivo (52). Hence, we tested their effect on GPCR fingerprints, focusing on a representative member of the family, AGS1. AGS1 had a very similar effect on the M3R fingerprint in respect to the effects on the amplitude as that of RGS8, but did not change the G protein profile of the M3R in terms of the activation kinetics (Fig. 3A). However, investigation of the effect of AGS1 on the time-course of the BRET response revealed a different mechanism. Consistent with its activity as a GEF, AGS1 increased the basal BRET ratio for the Go, but not Gq proteins (Fig. 3, E and F). The degree of this effect varied across other G protein types, with the most prominent influence of AGS1 being on the Gi/o proteins and the least effect (or no effect) being on the Gq proteins (Fig. 3G). Thus, the ability of AGS1 to change the M3R fingerprint likely originates from its competition with the receptor for binding to some G proteins, because the activation of G proteins by AGS1 makes them unavailable for coupling to M3R. Together, these data suggest that intracellular regulatory molecules change the hierarchical order of G protein coupling through distinct molecular mechanisms, and that the differential abundances of these molecules in various cells likely shapes the specificity of the G protein coupling of GPCRs.
Biasing the G protein activation profiles of muscarinic receptors by synthetic ligands
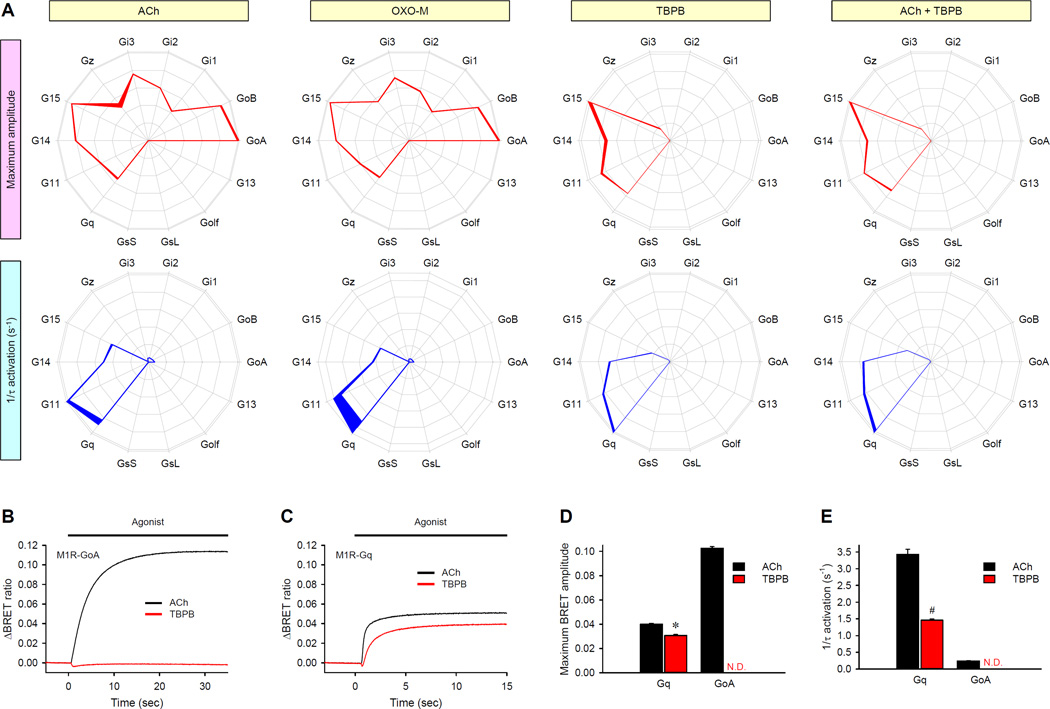
Ligand-directed signaling, in which the relative engagement of downstream pathways is dependent on the nature of the agonist, is becoming an accepted mode of GPCR function (10, 53). We therefore investigated whether this signaling bias could be initiated, at least in part, at the level of differential G protein engagement. To address this question, we first used our fingerprinting approach to profile the activity of the M1 muscarinic acetylcholine receptor (M1R), which has become a prominent target for the treatment of Alzheimer’s disease and schizophrenia and has spurred the development of well-characterized ligands with distinct properties (54, 55). We compared the action of the endogenous physiological ligand ACh to an orthosteric agonist Oxotremorine-M (OXO-M) (56) and a highly-selective M1R bitopic agonist 1-(1′-(2-methylbenzyl)-1,4′-bipiperidin-4-yl)-1H-benzo[d]imidazol-2(3H)-one (TBPB), which bind to topographically distinct sites, the orthosteric and the allosteric sites, respectively (Fig. 4) (57). The results showed that ACh and OXO-M produced virtually indistinguishable profiles in respect to the effects on both the amplitude and kinetics of the response (Fig. 4A). However, the G protein activation profile stimulated by TBPB was markedly different (Fig. 4A). TBPB completely failed to support the activation of Go, Gi1, Gi2, or Gi3 (Fig. 4, A and B); however, TBPB still efficiently activated Gq, G11, G14, and G15 proteins (Fig. 4, A and C). At the same time, signaling through Gz remained the same as with ACh and OXO-M. Another selective effect of TBPB was a decrease in the rate of G15 activation in comparison to that of G14, without changing the ratio of their maximal activation. Furthermore, the effects of TBPB persisted even when it was co-applied with Ach, because it was capable of over-writing the Ach-dependent fingerprint of M1R (Fig. 4A), suggesting that the bias in G protein usage caused by TBPB is likely to contribute to its unique pharmacological profile in vivo. TBPB similarly inhibited the OXO-M–mediated coupling of M1R to GoA, but not Gq (fig. S10, A and B).
Fig. 4. Synthetic GPCR ligands can bias the G protein coupling profiles of GPCRs.
(A) Four different agonist application conditions (yellow boxes) were examined for their effects on the G protein fingerprints of the M1R using two parameters: maximum amplitude (red) and activation rates (blue). Saturating concentrations (100 µM) of ACh, OXO-M, TBPB, or ACh and TBPB were applied to the M1R-expressing cells. Data are means ± SEM of six experiments. (B and C) Individual comparison of the activation of GoA (B) and Gq (C) by ACh (black) or TBPB (red). Each trace represents the mean of 12 replicates. (D) Direct comparison of the effects of the indicated agonists on amplitudes of the responses of GoA and Gq to M1R. (E) Direct comparison of the effects of the indicated agonists on the activation kinetics of GoA and Gq by M1R. Data are means ± SEM of six replicates. *P < 0.001 by paired t test. N.D., not detected.
To investigate the mechanism of the TBPB-directed bias, we applied ACh and TBPB to the same cells, which were reconstituted with either GoA or Gq (Fig. 4, B to E). When performing the analysis in parallel, we found that both agonists elicited responses with similar amplitudes in the Gq-based system (Fig. 4, C and D). However, the activation kinetics of Gq was markedly slower in response to TBPB than in response to ACh (Fig. 4, C and E). Despite the greater efficacy of activation of GoA relative to that of Gq, its onset kinetics were markedly slower than those for Gq, when ACh was used (Fig. 4, D and E). Accordingly, the slower G protein activation velocity by TBPB-stimulated M1R was insufficient to sustain the activation of GoA (Fig. 4B).
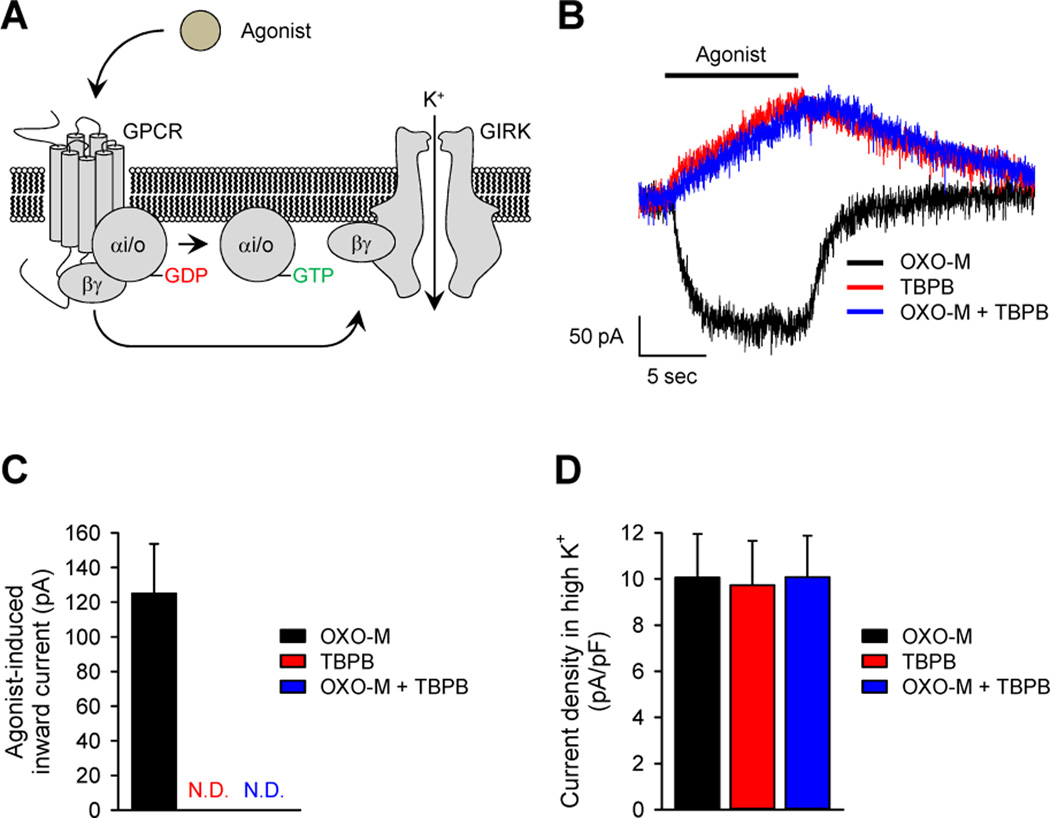
We further examined the relevance of ligand-directed differential G protein activation to endogenous signaling with cultured hippocampal neurons that express M1R at high abundance (58, 59). In this system, we monitored the opening of GIRK channels (Fig. 5A), which are selectively sensitive to the activation of Gi/o proteins (60). Consistent with its ability to activate Gi/o proteins, application of the M1R agonist OXO-M induced a substantial amount of inward GIRK current (Fig. 5, B and C). Removal of the agonist resulted in the return of the current flow back to baseline because of channel closure. The kinetics of the activation and inactivation of the response closely matched the expected behavior of GIRK channels in these neurons, which is observed upon activation of various Gi/o-coupled GPCRs (61).
Fig. 5. Ligand-dependent coupling of muscarinic receptors to GIRK channels in native hippocampal neurons.
(A) Schematic representation of the activation of GIRK channels by GPCRs. The binding of agonist to a Gi/o-coupled GPCR leads to an interaction between Gβγ and the GIRK channel, which evokes an inward-rectifying K+ current. (B) Representative traces of GIRK currents in hippocampal neurons evoked by a saturating concentration (100 µM) of the indicated agonists. (C) Maximal current amplitudes of GIRK responses elicited by agonist were measured 10 s after agonist application. The application of TBPB either in the absence or presence of OXO-M did not evoke any inward current. (D) Current densities in the presence of a high concentration of K+ were measured to assess ligand-independent ion flow through inward-rectifying potassium channels. The amount of current was recorded before the application of each indicated agonist. All electrophysiological data were recorded from a total of seven neurons. Data are means ± SEM.
In contrast, the application of TBPB, which did not activate Gi/o class proteins in the BRET fingerprinting assay, failed to produce GIRK-mediated inward currents (Fig. 5, B and C). Furthermore, and consistent with the BRET experimental data (fig. S10, C and D), TBPB also inhibited GIRK currents elicited by OXO-M when both ligands were co-applied to the neurons. These effects were specific to the application of agonist, and no change in channel gating was evident in response to the application of medium containing a high potassium concentration, which nonspecifically increased the ion flow from inward rectifier K+ channels (Fig. 5D). Thus, the analysis of agonist-driven G protein bias by BRET fingerprinting can be extrapolated to understand the behavior of an endogenous system. At a mechanistic level, these data support the notion that binding of ligand to the allosteric pocket of the M1R is capable of inducing a distinct conformational change in the GPCR (12), which likely generates distinct conformational changes in the cytoplasmic region of the receptor, thus altering its G protein specificity. The data further suggest that the ability of GPCR ligands to bias the selection of G proteins may be related to the ability of these ligands to induce conformational states in the receptor that affect the catalytic efficiency of the receptors towards G proteins, rather than modulating the activation extent of individual G proteins.
Ligand-biased G protein agonism in the opioid receptor family
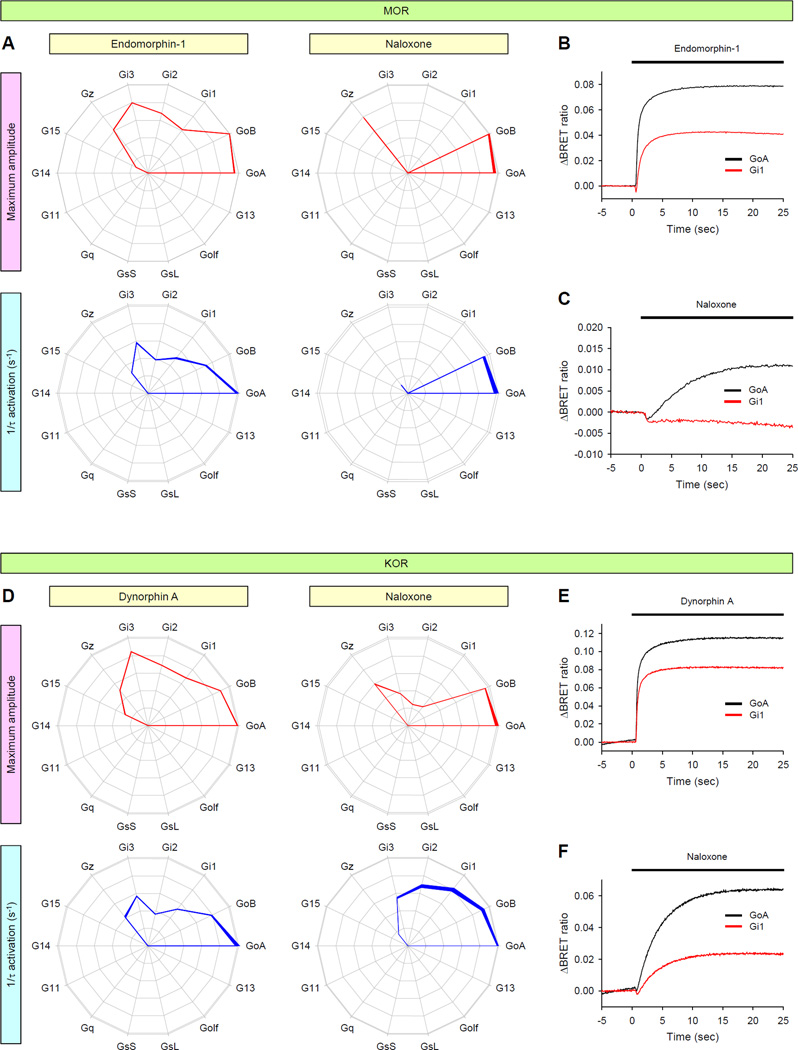
We further asked whether another functional class of ligands, antagonists, might also exhibit bias when sampled across the entire gamut of possible G protein targets, as opposed to simply being inert competitors of agonists. For these studies, we evaluated the µ-, δ-, and κ-opioid receptors (MOR, DOR, and KOR, respectively), which play essential roles in nociception and have resulted in the availability of a diverse pharmacology, including naloxone, which is commonly used to reverse opioid overdose, but is also used for the treatment of addiction and for chronic pain management in combination with partial agonists (62, 63). Naloxone binds to all of the opioid receptors with high affinity, and it is commonly described as competitive antagonist (64). To see whether naloxone acted as a true antagonist across multiple G protein species, we compared its effects with those of physiologically relevant endogenous agonists on the three opioid receptors (Fig. 6 and fig. S11). Stimulation of MOR, DOR, and KOR with their respective endogenous agonists, endomorphin-1, enkephalin, and dynorphin A, produced similar fingerprints showing differential activation of Go, Gi1 to Gi3, Gz, and G15 proteins. As expected for a pure antagonist, naloxone did not stimulate DOR to activate any G proteins (fig. S11). In contrast, we detected weak, but consistent, signals from multiple G proteins when naloxone was applied to cells expressing MOR or KOR (Fig. 6). Furthermore, we saw evidence of differential G protein engagement by the two receptors, which suggests that naloxone exerts functionally biased effects. When exposed to MOR, naloxone activated both Go and Gz, but completely failed to engage Gi1, Gi2, Gi3, or G15 (Fig. 6, A to C). However, when exposed to KOR, naloxone induced the activation of all of the Gi/o proteins, but not G15. Naloxone also changed the relative activation rates of Gα subunits and produced activation rate fingerprints that are different from the dynorphin A-induced fingerprint (Fig. 6, D to F), which suggests that naloxone is not just a weak agonist, but that it also induces KOR to adopt a conformation that differs from that of the dynorphin A–bound KOR.
Fig. 6. GPCR fingerprinting reveals the selective activation of G proteins by opioid receptors in response to a classical antagonist.
(A to F) Endogenous agonists (endomorphin-1 or dynorphin A) and a classical antagonist (naloxone) were examined for their effects on the G protein coupling specificities of MOR (A to C) and KOR (D to F) using two parameters: maximum amplitude (red) and activation rates (blue). Saturating concentrations (100 µM) of the indicated ligands were applied. Data are means ± SEM of six to twelve experiments. (B, C, E and F) Direct comparison of the activation of GoA (black) and Gi1 (red) by MOR (B and C) and KOR (E and F) in response to endomorphin-1 (B), dynorphin A (E), or naloxone (C and F). Each trace represents an average of six replicates.
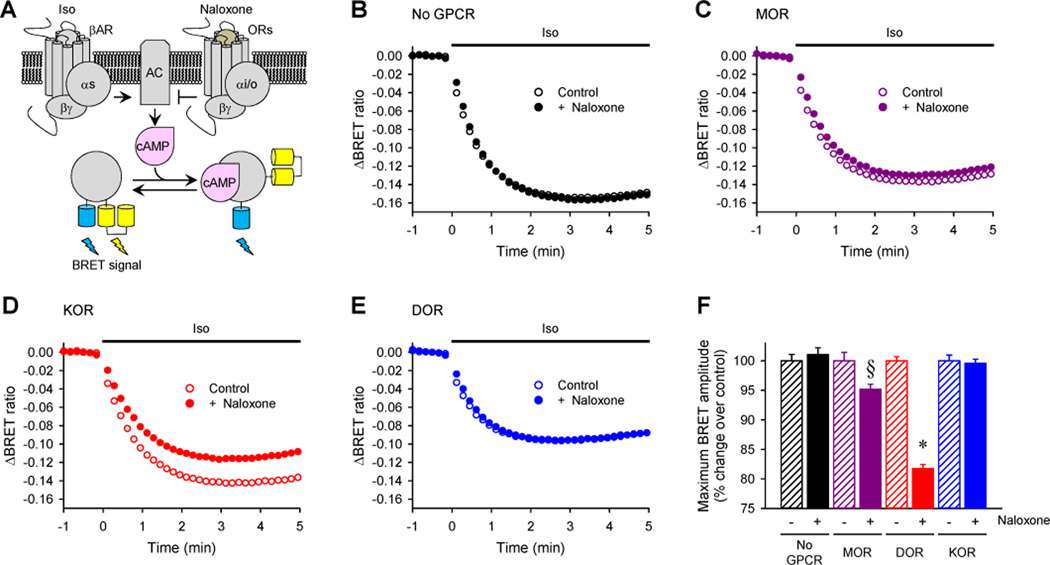
To determine the relevance of the varying fingerprints produced by naloxone to the ability of the opioid receptors to modulate endogenous intracellular signaling pathways, we examined the effects of naloxone on intracellular cyclic adenosine monophosphate (cAMP) concentrations. We used an EPAC-based BRET sensor to study the real-time kinetics of changes in cAMP production in HEK293T/17 cells in response to the stimulation of the β2AR by its agonist isoproterenol (ISO). In this paradigm, the activity of the Gi/o-coupled opioid receptors was assessed by their ability to suppress ISO-mediated increases in cAMP concentration (Fig. 7A). First, we established that , pretreatment of the cells not expressing exogenous opioid receptors with naloxone did not change the time-course or extent of the change in the BRET signal caused by ISO (Fig. 7B). In contrast, pretreatment of MOR- or KOR-expressing cells with naloxone inhibited the ISO-induced generation of cAMP, whereas no such effect was evident in DOR-expressing cells (Fig 7, C to F). These observations are consistent with the fingerprinting data, which suggested that naloxone stimulated the activation of Gi/o proteins through MOR and KOR, and that KOR exhibited stronger coupling to Gi proteins when compared to MOR (Fig. 6). Thus, the traditional opioid receptor antagonist naloxone appears to be a biased agonist that differentially affects signaling across different opioid receptor types. Together, these observations highlight the value of examining the entire spectrum of G protein selectivity with respect to kinetics and signaling extent for revealing previously uncharacterized actions of these ligands.
Fig. 7. The biased G protein coupling specificities of opioid receptor subtypes in response to naloxone results in differential modulation of cAMP production.
(A) Schematic representation of the assay paradigm. Transfected cells expressing opioid receptors were pre-incubated with naloxone before the β2AR agonist ISO was applied to stimulate cAMP production. The kinetics of the amplitude of the cAMP signal were determined in real time with a BRET-based cAMP sensor that exhibits a decreased BRET signal upon cAMP binding. (B to E) Effect of naloxone on ISO-stimulated cAMP production in HEK 293T/17 cells expressing no opioid receptor (B), MOR (C), KOR (D), or DOR (E). The cells were cotransfected with plasmids encoding the indicated opioid receptors together with Nluc-Epac-VV. Before the activation of endogenous βARs with ISO, transfected cells were incubated with (closed circle) or without (open circles) 100 µM naloxone for 5 min. The cells were then treated with 1 µM isopreterenol at time zero. Each trace represents the mean of 12 replicates. (F) Quantification of changes in maximal BRET amplitudes induced by naloxone for each of the opioid receptors. §P < 0.05 and *P < 0.0001 by paired t-test.
Discussion
Here, we revealed the complexity of the mechanisms by which GPCRs convert extracellular signals into G protein activation. signal conversion process at the GPCRs. We demonstrated that these receptors have distinct patterns of activity and differentially engage multiple G proteins with characteristic signatures, which we call fingerprints. Indeed, individual receptors could be distinguished by their distinct profile of G protein activation, which we separately evaluated in terms of both signaling efficacy and kinetics. Furthermore, these GPCR fingerprints showed distinct functional bias in terms of G protein engagement depending on the chemical identity of the ligand activating the receptor and on the intracellular environment.
Characterizing GPCR actions through the direct monitoring of both activation kinetics and response magnitude toward multiple G protein targets
We obtained mechanistic insight into selectivity of GPCRs by quantifying the kinetics of G protein activation. GPCRs catalyze the conversion of the inactive G protein heterotrimer into active Gα-GTP and Gβγ species. Therefore, measuring the rate of this reaction represents perhaps the most direct proxy of the catalytic activity of GPCRs. We found that the activation rates of different G proteins by the same GPCR varied substantially and that the relative speeds with which various G proteins were activated contributed to the characteristic signature of a GPCR. Thus, in our studies, the catalytic efficiency of a GPCR was defined by measuring its activation rate constants 1/τ (s−1) for different G proteins, which were used to determine the relative selectivity of the GPCR.
Because individual G protein subunits have distinct functional characteristics, directly monitoring their activation offers clear mechanistic and prognostic advantages over examining changes in downstream signaling reactions, which are pervasively used to assess GPCR function (65). Although the activation rates serve as a proxy for the GEF activity of GPCRs, maximal BRET amplitudes are influenced by the propensity of different heterotrimers to liberate Gβγ subunits (which depends on the affinity between Gα and Gβγ), which substantially determines the efficacy of the measured responses (66). Furthermore, we observed that G protein activation rates did not necessarily correlate with the maximal extent of their engagement by a given GPCR. Thus, a receptor can catalyze a very fast response of limited extent through one type of G protein while at the same time supporting a slow, but large, response through another. This property may vary with the identity of the receptor and the ligand that activates it, generating distinct profiles both in terms of the kinetics and magnitude of G protein activaiton, a distinction that is lost when the reaction is monitored by measuring end-points of downstream signals.
Another limitation associated with the use of the downstream signaling measurements to assess GPCR function is the inability to compare the efficacies of responses mediated by different G proteins. In the context of the classical receptor occupancy model, efficacy is considered an intrinsic property of the ligand and receptor pair, and it is customarily assumed to be the same for all responses elicited by the pair (67). Therefore, for practical purposes, the efficacies of GPCR ligands are frequently assessed indirectly by their ability to affect downstream signaling reactions, for example cAMP accumulation, mitogen-activated protein kinase activation, Ca2+ mobilization, or gene expression regulation, or by the binding of non-hydrolyzable GTPγS to G proteins with little regard for G protein heterogeneity. Our observations reveal that all tested GPCRs couple to multiple G proteins with different efficacies, suggesting that this parameter might be substantially influenced by the intrinsic properties of the individual G protein subunits that couple to a particular GPCR. This finding calls for the consideration of the identity of the target G proteins when determining the efficacies of GPCR ligands.
Pharmacological implications of exhaustive G protein profiling
We propose that the characterization of GPCR activity in respect to both kinetics and extent of G protein activation has the following implications. First, it will aid in the process of distinguishing between and predicting the signaling consequences of phasic and tonic neurotransmission mediated by different receptors. Indeed, the timing of signal transmission events is broadly modulated under various physiological and pathological conditions (2, 68). Our analysis showed that even when the signal transmission was mediated by the same receptor, it could lead to the engagement of different G proteins depending on the duration of action of the neurotransmitter or hormone. Thus, GPCRs have an intrinsic ability to dynamically adjust response properties over time, engaging different sets of G proteins in response to acute vs chronic stimulation situations.
Second, it seems important to consider the signaling bias among the entire range of G proteins in drug development campaigns. Although the duration of action of small-molecule drugs cannot be easily controlled on the timescale of receptor activation, knowing the assortment of G proteins that they can activate might be helpful in designing agonists with targeted properties. For example, we found that the M1R allosteric agonist TBPB elicited different profiles of G protein activation than those of the endogenous agonist acetylcholine or the orthosteric agonist OXO-M. Although the full range of the behavioral effects of TBPB remains to be established, it clearly has antipsychotic-like activity and shows efficacy in decreasing Aβ production (57), which suggests that these beneficial effects likely arise from the distinct and selective set of G proteins that TBPB enables M1R to activate. Similarly, the highly selective agonistic properties of naloxone toward opioid receptors may underlie the success of its use as an adjuvant for weak opioid agonists for the management of pain and dependence (62, 63), and this can likely be exploited further. Thus, it is conceivable that optimal therapeutic efficacy could be achieved through the selective activation of only a subset of the G proteins within the repertoire of a GPCR, possibly circumventing adverse side effects.
G protein coupling diversity and GPCR signaling in native environment
Traditionally, GPCRs have been thought to establish their signaling specificity by coupling to a single class of G proteins; however, there is growing evidence that several GPCRs activate multiple G proteins in native cells. One of the best-documented cases is provided by the β2AR. In cardiac myocytes, β2AR couples to both Gs and Gi proteins to regulate the contraction rate and stimulate a pro-survival response, respectively (69, 70). This dual coupling of the β2AR appears to be also preserved in macrophages (71). Another, perhaps more extreme, example is provided by the thyrotropin receptor, which couples to members of all four G protein subfamilies in native thyroid cells (72). Although direct evidence is limited, it seems that many GPCRs may couple to multiple G proteins both in vivo and in vitro (73). Thus, the quantitative strategy for the exhaustive analysis of G protein coupling profiles of GPCRs in the model environment that we have introduced in this study will likely be useful for understanding the spectrum of GPCR responses by extrapolating these observations to the native environment.
It is very likely that, in native cells under physiologically relevant conditions, a range of factors influence the responses of GPCRs. First, in many cells, several G proteins are present at the same time and thus are potentially competing for activation by the receptor. Second, native cells usually have varying abundances of different G proteins, which further amplify responses through some G protein species, but dampen responses through others. In extreme cases, some G proteins might be present at negligible amounts, even if they couple with the highest efficiency to a particular GPCR. In addition, the repertoire of Gα subunits may also vary throughout development, which could contribute to changes in the nature of the responses to stimulation of a GPCR (74, 75). Third, a host of differentially abundant intracellular factors, such as RGS, AGS, and Ric8 proteins, can further shape GPCR fingerprints. These considerations suggest that the GPCR fingerprints in native cells are likely more complex than anything that could be modeled in our reconstituted systems. Nevertheless, our approach enables us to begin the elucidation of this complexity by surveying the intrinsic properties of receptors in terms of their G protein coupling characteristics in an environment largely devoid of complicating factors. Essentially, we are proposing a means of assaying the innate target preferences of individual GPCRs in a model environment. In other words, our overexpression system identifies those G proteins that could be activated by a given GPCR, but this coupling may not necessarily happen in native cells. Making use of these profiles to understand the signaling reactions stimulated by GPCRs in native cells will ultimately require knowing the full repertoire of G protein subunits and their regulators that is available in each individual cell of interest, as well as their relative abundances. Together, gene expression information obtained through single-cell genomic approaches and intrinsic GPCR coupling profiles should be sufficient to predict the consequences of GPCR activation. We think it is likely that the next generation of drug development efforts for GPCRs will need to take this information into account and consider the desired shape of the GPCR responses in the context of the specificity of the receptors and the particular molecular landscape of the therapeutically relevant cells.
Conclusion
In summary, we demonstrated the existence of another dimension of functional bias by GPCRs: the differential engagement of multiple target G proteins, which was revealed by quantitative analysis of the extent of G protein mobilization and their activation kinetics by the receptors. We hope that profiling GPCRs for their biased coupling to G proteins with our exhaustive fingerprinting technology will prove useful for understanding the physiological functions of GPCRs and the diversity of their cellular effects, as well as for drug discovery.
Material and Methods
Genetic constructs for reporters, receptors, and G proteins
pcDNA3.1+ plasmids encoding the M3 muscarinic acetylcholine receptor, the β2AR, the bradykinin B2 receptor, the δ-opioid receptor (DOR), and the κ-opioid receptor (KOR) were purchased from the Missouri S&T cDNA Resource Center. cDNA encoding the M1 muscarinic acetylcholine receptor was purchased from OriGene. Plasmid encoding the Flag-tagged, long isoform of the D2 dopamine receptor was a gift from Dr. Abraham Kovoor (University of Rhode Island). Plasmids encoding GABAB R1 and GABAB R2 were provided by Dr. Kevin Wickman (University of Minnesota). Plasmid encoding the Flag-tagged µ-opioid receptor (MOR) was a gift from Dr. Pin-Yee Law (University of Minnesota). pcDNA3.1+ plasmids encoding GαoB, Gαz, Gα11, Gα14, Gα15, Gαs long isoform (GαsL), Gαolf, and Gα13 were purchased from the Missouri S&T cDNA Resource Center. pCMV5 plasmids encoding GαoA, Gαi1, Gαi2, Gαi3, Gαq, and Gαs short isoform (GαsS) were gifts from Dr. Hiroshi Itoh (Nara Institute of Science and Technology, Japan). Plasmids encoding masGRK3ct-Rluc8, Venus 156-239-Gβ1, and Venus 1–155-Gγ2 were gifts from Dr. Nevin Lambert (Georgia Regents University) (36). Flag-tagged Ric-8A in pcDNA3 and Flag-tagged Ric-8B in pcDNA3.1 were gifts from Dr. Jean-Pierre Montmayeur (CNRS, France) (76) and Dr. Bettina Malnic (Universidade de São Paulo, Brazil) (77), respectively. pcDNA3.1+ plasmids encoding AGS1 and triple HA-tagged RGS8 were purchased from the Missouri S&T cDNA resource Center. The masGRK3ct-Nluc construct contained cDNA encoding amino acid residues 495 to 688 of bovine GRK3 (NP_776925), which was preceded by a myristic acid attachment peptide (mas; MGSSKSKTSNS). The stop codon of GRK3 was replaced with a GGGS linker (78), which was followed by the NanoLuc (Nluc). Nluc-Epac-VV constructs were generated by replacing mTurquoise of the FRET-based cAMP sensor (78) with Nluc.
Cell culture and transfection
HEK 293T/17 cells were chosen because of their high transfection efficiency (79). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), MEM non-essential amino acids (Life Technologies), 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 µg/ml) at 37°C in a humidified incubator containing 5% CO2. Culture dishes (6-cm) were coated by incubation for 10 min at 37°C with 2.5 ml of Matrigel solution [approximately 10 µg/ml of growth factor-reduced Matrigel (BD Biosciences) in culture medium]. For transfections, cells were seeded in the 6-cm dishes containing the Matrigel solution at a density of 4 × 106 cells/dish. We found that Matrigel decreased the toxicity of the DNA-transfection reagent complex without affecting transfection efficiency. Four hours later, the cells were transfected with the appropriate expression constructs (total of 7.5 µg DNA per dish) with the reagents PLUS (7.5 µl/dish) and Lipofectamine LTX (12 µl/dish). The cells were transfected with the Venus 156-239-Gβ (0.42 µg), Venus 1-155-Gγ2 (0.42 µg), and masGRK3ct-Rluc8 (0.42 µg) or masGRK3ct-Nluc (0.42 µg) constructs in addition to the different amounts of constructs for the GPCR and Gα of interest. According to our observation (fig. S2) that Gα15, Gα14, and Gαolf required co-expression with molecular chaperones to generate functional G protein complexes, we cotransfected cells with the following combinations of constructs: Gα15 with Ric-8A, Gα14 with Ric-8A, and Gαolf with Ric-8B. We used 0.42 µg of each Ric-8 construct per transfection. Cells were cotransfected with a pcDNA3.1-based construct encoding the catalytic subunit of pertussis toxin (PTX-S1) (0.42 µg) and constructs encoding Gαz, Gα15, Gα14, Gα11, Gαq, GαsS, GαsL, Gαolf, or Gα13 to ensure that the small BRET signals were not contaminated by the possible recruitment of endogenous Gαi/o proteins. The empty vector pcDNA3.1 was used to normalize the amount of DNA in each transfection.
Primary hippocampal cultures
Primary cultures of hippocampal neurons were prepared as described previously with minor modifications (42). All experiments with mice were performed in accordance with NIH guidelines and were approved by the IACUC protocol at the Scripps Research Institute. Neonatal mice (days 0 to 2 after birth) were used as a tissue source. Neurons were incubated at 37°C, 5% CO2, and half of the Neurobasal A/B27 based culture medium was replaced with fresh medium every 3 to 4 days. Neurons were cultured for 10 to 18 days before being used for experiments.
Electrophysiological recordings of GIRK channel activity
For whole-cell recordings, primary hippocampal neurons and transfected HEK 293T/17 cells on coverslips were transferred to a chamber containing a low-K+ bath solution [145 mM NaCl, 4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5.5 mM D-glucose, 5 mM HEPES/NaOH (pH 7.4)]. Borosilicate patch pipettes (3 to 5 MΩ) were filled with recording solution [130 mM KCl, 10 mM NaCl, 1 mM EGTA/KOH (pH 7.2), 0.5 mM MgCl2, 10 mM HEPES/KOH (pH 7.2), 2 mM Na2ATP, 5 mM phosphocreatine, 0.3 mM GTP]. Agonist-induced currents were measured at room temperature with a high-K+ bath solution [120 mM NaCl, 25 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5.5 mM D-glucose, 5 mM HEPES/NaOH (pH 7.4)]. The high-K+ bath solution was applied with or without 100 µM agonist directly to the cells with a SF-77B rapid perfusion system (Warner Instruments, Inc.). Current responses to the application of agonist were measured at a holding potential of −80 mV. Membrane potentials and whole-cell currents were recorded with hardware (Axopatch-700B amplifier, Digidata 1440A) and software (pCLAMP v. 10.3) obtained from Molecular Devices. All currents were low-pass filtered at 2 kHz, sampled at 5 kHz, and stored on computer hard disk for subsequent analysis with Clampfit v. 10.3 software. Only those experiments in which access resistances were stable and low (<20 MΩ) were included in the analysis.
BRET experiments
Agonist-dependent cellular measurements of BRET between Venus-Gβ1γ2 and masGRK3ct-Rluc8 or masGRK3ct-Nluc were performed as described previously (80) to examine the activation of G protein signaling in live cells. Sixteen to twenty-four hours after transfection, HEK 293T/17 cells were washed once with phosphate-buffered saline (PBS) containing 5 mM EDTA (EDTA/PBS), and were detached by incubation in EDTA/PBS at room temperature for 5 min. Cells were harvested by centrifugation at 500g for 5 min and were resuspended in BRET buffer (PBS containing 0.5 mM MgCl2 and 0.1% glucose). Approximately 50,000 to 100,000 cells per well were distributed in 96-well flat-bottomed white microplates (Greiner Bio-One). The Rluc substrate, ViviRen (Promega), was dissolved in ethanol at a final concentration of 20 mM and stored at −20°C. ViviRen was dissolved in BRET buffer immediately before use and added to the cell suspension at a final concentration of 20 µM. The Nluc substrate furimazine was purchased from Promega and used according to the manufacturer’s instructions. BRET measurements were made with a micro plate reader (POLARstar Omega; BMG Labtech) equipped with two emission photomultiplier tubes, which enabled the detection of two emissions simultaneously with the highest possible resolution of 20 ms per data point. All measurements were performed at room temperature. The BRET signal was determined by calculating the ratio of the light emitted by Venus-Gβ1γ2 (535 nm with a 30-nm band path width) to the light emitted by masGRK3ct-Rluc8 or masGRK3ct-Nluc (475 nm with a 30-nm band path width). The average baseline value (basal BRET ratio) recorded before stimulation of cells with agonist was subtracted from the experimental BRET signal values to obtain the ΔBRET ratio.
Fluorescence and luminescence measurements
For luminescence measurements, 25 µl of the cell suspension (containing 50,000 to 100,000 cells) was added to each well of a white, opaque-bottom, 96-well plate. Then, 25 µl of the appropriate 2X luciferase substrate was applied (ViviRen for Rluc8; and furimazine for Nluc), incubated for 3 min to enable the luminescence to peak, and total luminescence was measured with no emission filter. For fluorescence measurements, 100 µl of the cell suspension was added to each well of a black, opaque-bottom, 96-well plate. Untransfected cells (mock) were used as a blank control. Fluorescence was measured by exciting the cells at 480 nm and recording the emission at 530 nm.
Generation of a phylogenetic tree of Gα subunits
The protein sequence divergence of human Gα subunits was demonstrated by phylogenetic analysis. Human Gα subunit protein sequences were aligned and a phylogram was generated with MEGA 6 using default parameters (81). The GenBank accession numbers of the Gα subunits are as follows: Gαo, AF493894; Gαi1, NM_002069; Gαi2, AF493906; Gαi3, M27543; Gαt1, NM_000172; Gαt2, AF493909; Gαgust, NM_001102386; Gαz, J03260; Gαq, U40038; Gα11, AF493900; Gα14, NM_004297; Gα15, AF493904; Gαs, X04409; Gαolf, AF493893; Gα12, NM_007353; Gα13, NM_006572.
Fluorescence confocal microscopy
HEK 293T/17 cells were seeded onto Matrigel-coated coverslips and transfected as described earlier. After 16 to 24 hours, the cells were fixed with 4% paraformaldehyde in PBS for 20 min. The coverslips were washed once with PBS and mounted on glass slides with Fluoromount (Sigma). Microscopy was performed with a TCS SP8 confocal microscope (Leica).
Western blotting
For each sample, approximately 5 × 106 cells were lysed in 500 µl of sample buffer [125 mM Tris-HCl (pH 6.8), 4 M urea, 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.16 mg/ml bromophenol blue]. Western blotting analysis of proteins was performed after samples were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to membranes. Blots were blocked with 5% skim milk in PBS containing 0.1% Tween 20 (PBST) for 30 min at room temperature, which was followed by a 90-min incubation with specific antibodies diluted in PBST containing 1% skim milk. Blots were washed in PBST and incubated for 45 min with a 1:10,000 dilution of secondary antibodies conjugated with horseradish peroxidase in PBST containing 1% skim milk. Proteins were visualized on x-ray film by SuperSignal West Femto substrate (Pierce).
Data analysis
The rate constants (1/τ) of the activation and deactivation phases were obtained by fitting a single exponential curve to the traces with Clampfit Ver. 10.3 software (Molecular Devices). Statistical analysis was performed with GraphPad Prism software Ver. 4.02.
[Below I’ve pasted the sections of the materials and methods from the supplementary file. Please incorporate them appropriately into the main Materials and Methods above]
Supplementary Material
Acknowledgments
We thank Dr. Abraham Kovoor for the gift of the D2 receptor expression, Dr. Hiroshi Itoh for supplying Gα expression constructs, Dr. Nevin Lambert for sharing GRK3CT and Gβγ-Venus constructs, Dr. Jean-Pierre Montmayeur for supplying Ric8A and Dr. Bettina Malnic for providing Ric8B constructs.
Funding: This work was supported by NIH grants EY018139, DA026405, and DA036596 to K.A.M.
Footnotes
Author contributions: I.M. participated in project design, performed experiments and data analysis, interpreted the data, and drafted and revised the manuscript; G.M.K. performed experiments and data analysis; O.O. performed electrophysiological recordings of GIRK channel activity; C.D.J. and K.X. generated expression plasmids; and K.A.M. was responsible for project design, data interpretation, and manuscript writing.
Competing interests: The authors declare that they have no competing interests.
Fig. S1. Characterization of the performance of the Nluc-based BRET assay.
Fig. S2. Effects of Ric-8A and Ric-8B on the expression of Gαsubunits and their responsiveness to agonist.
Fig. S3. Optimization of the stoichiometry of Gαand Venus-Gβγ.
Fig. S4. Traces showing real-time activity measurements of 14 different G proteins.
Fig. S5. The plasma membrane localization of Venus-Gβγ is dependent on co-expression with Gα subunits to ensure 1:1 complex stoichiometry.
Fig. S6. Exogenous GPCR and Gα stimulate BRET responses in the assay.
Fig. S7. The abundances of heterotrimers of Gα/ and enus-Gβγ in cells transiently transfected with plasmids encoding 14 different Gα subunits are similar.
Fig. S8. GPCR responses are within the dynamic range of the assay.
Fig. S9. Fingerprinting of the deactivation phase.
Fig. S10. Direct comparison of the agonist-induced coupling of M1R to different G proteins.
Fig. S11. Characterization of DOR fingerprints.
References and Notes
- 1.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiological reviews. 2005;85:1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 2.O’Hayre M, Vazquez-Prado J, Kufareva I, Stawiski EW, Handel TM, Seshagiri S, Gutkind JS. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nature reviews. Cancer. 2013;13:412–424. doi: 10.1038/nrc3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catapano LA, Manji HK. G protein-coupled receptors in major psychiatric disorders. Biochim Biophys Acta. 2007;1768:976–993. doi: 10.1016/j.bbamem.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vassart G, Costagliola S. G protein-coupled receptors: mutations and endocrine diseases. Nature reviews. Endocrinology. 2011;7:362–372. doi: 10.1038/nrendo.2011.20. [DOI] [PubMed] [Google Scholar]
- 5.Blad CC, Tang C, Offermanns S. G protein-coupled receptors for energy metabolites as new therapeutic targets. Nat Rev Drug Discov. 2012;11:603–619. doi: 10.1038/nrd3777. [DOI] [PubMed] [Google Scholar]
- 6.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 7.Rask-Andersen M, Almen MS, Schioth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10:579–590. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- 8.Neer EJ. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 9.Shukla AK, Xiao K, Lefkowitz RJ. Emerging paradigms of beta-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem Sci. 2011;36:457–469. doi: 10.1016/j.tibs.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenakin T. Functional selectivity and biased receptor signaling. J Pharmacol Exp Ther. 2011;336:296–302. doi: 10.1124/jpet.110.173948. [DOI] [PubMed] [Google Scholar]
- 11.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 14.Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends in molecular medicine. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermans E. Biochemical and pharmacological control of the multiplicity of coupling at G-protein-coupled receptors. Pharmacol Ther. 2003;99:25–44. doi: 10.1016/s0163-7258(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 16.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 17.Smrcka AV. G protein betagamma subunits: central mediators of G protein-coupled receptor signaling. Cell Mol Life Sci. 2008;65:2191–2214. doi: 10.1007/s00018-008-8006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbard KB, Hepler JR. Cell signalling diversity of the Gqalpha family of heterotrimeric G proteins. Cell Signal. 2006;18:135–150. doi: 10.1016/j.cellsig.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends in pharmacological sciences. 2001;22:368–376. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 20.Inoue A, Ishiguro J, Kitamura H, Arima N, Okutani M, Shuto A, Higashiyama S, Ohwada T, Arai H, Makide K, Aoki J. TGFalpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nat Methods. 2012;9:1021–1029. doi: 10.1038/nmeth.2172. [DOI] [PubMed] [Google Scholar]
- 21.Sauliere A, Bellot M, Paris H, Denis C, Finana F, Hansen JT, Altie MF, Seguelas MH, Pathak A, Hansen JL, Senard JM, Gales C. Deciphering biased-agonism complexity reveals a new active AT1 receptor entity. Nat Chem Biol. 2012;8:622–630. doi: 10.1038/nchembio.961. [DOI] [PubMed] [Google Scholar]
- 22.Michal P, El-Fakahany EE, Dolezal V. Muscarinic M2 receptors directly activate Gq/11 and Gs G-proteins. J Pharmacol Exp Ther. 2007;320:607–614. doi: 10.1124/jpet.106.114314. [DOI] [PubMed] [Google Scholar]
- 23.Kandola MK, Sykes L, Lee YS, Johnson MR, Hanyaloglu AC, Bennett PR. EP2 receptor activates dual G protein signaling pathways that mediate contrasting proinflammatory and relaxatory responses in term pregnant human myometrium. Endocrinology. 2014;155:605–617. doi: 10.1210/en.2013-1761. [DOI] [PubMed] [Google Scholar]
- 24.Thomas RL, Mistry R, Langmead CJ, Wood MD, Challiss RA. G protein coupling and signaling pathway activation by m1 muscarinic acetylcholine receptor orthosteric and allosteric agonists. J Pharmacol Exp Ther. 2008;327:365–374. doi: 10.1124/jpet.108.141788. [DOI] [PubMed] [Google Scholar]
- 25.Griffin MT, Figueroa KW, Liller S, Ehlert FJ. Estimation of agonist activity at G protein-coupled receptors: analysis of M2 muscarinic receptor signaling through Gi/o,Gs, and G15. J Pharmacol Exp Ther. 2007;321:1193–1207. doi: 10.1124/jpet.107.120857. [DOI] [PubMed] [Google Scholar]
- 26.Buch TR, Heling D, Damm E, Gudermann T, Breit A. Pertussis toxin-sensitive signaling of melanocortin-4 receptors in hypothalamic GT1-7 cells defines agouti-related protein as a biased agonist. J Biol Chem. 2009;284:26411–26420. doi: 10.1074/jbc.M109.039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart GD, Sexton PM, Christopoulos A. Detection of novel functional selectivity at M3 muscarinic acetylcholine receptors using a Saccharomyces cerevisiae platform. ACS Chem Biol. 2010;5:365–375. doi: 10.1021/cb900276p. [DOI] [PubMed] [Google Scholar]
- 28.Ritter SL, Hall RA. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat Rev Mol Cell Biol. 2009;10:819–830. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wenzel-Seifert K, Seifert R. Molecular analysis of beta(2)-adrenoceptor coupling to G(s)-, G(i)-, and G(q)-proteins. Mol Pharmacol. 2000;58:954–966. doi: 10.1124/mol.58.5.954. [DOI] [PubMed] [Google Scholar]
- 30.Daaka Y, Luttrell LM, Lefkowitz RJ. Switching of the coupling of the beta2-adrenergic receptor to different G proteins by protein kinase A. Nature. 1997;390:88–91. doi: 10.1038/36362. [DOI] [PubMed] [Google Scholar]
- 31.Xiao RP, Avdonin P, Zhou YY, Cheng H, Akhter SA, Eschenhagen T, Lefkowitz RJ, Koch WJ, Lakatta EG. Coupling of beta2-adrenoceptor to Gi proteins and its physiological relevance in murine cardiac myocytes. Circ Res. 1999;84:43–52. doi: 10.1161/01.res.84.1.43. [DOI] [PubMed] [Google Scholar]
- 32.Fribourg M, Moreno JL, Holloway T, Provasi D, Baki L, Mahajan R, Park G, Adney SK, Hatcher C, Eltit JM, Ruta JD, Albizu L, Li Z, Umali A, Shim J, Fabiato A, MacKerell AD, Jr, Brezina V, Sealfon SC, Filizola M, Gonzalez-Maeso J, Logothetis DE. Decoding the signaling of a GPCR heteromeric complex reveals a unifying mechanism of action of antipsychotic drugs. Cell. 2011;147:1011–1023. doi: 10.1016/j.cell.2011.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohse MJ, Nuber S, Hoffmann C. Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacol Rev. 2012;64:299–336. doi: 10.1124/pr.110.004309. [DOI] [PubMed] [Google Scholar]
- 34.Gales C, Rebois RV, Hogue M, Trieu P, Breit A, Hebert TE, Bouvier M. Real-time monitoring of receptor and G-protein interactions in living cells. Nat Methods. 2005;2:177–184. doi: 10.1038/nmeth743. [DOI] [PubMed] [Google Scholar]
- 35.Lazar J, Bondar A, Timr S, Firestein SJ. Two-photon polarization microscopy reveals protein structure and function. Nat Methods. 2011;8:684–690. doi: 10.1038/nmeth.1643. [DOI] [PubMed] [Google Scholar]
- 36.Hollins B, Kuravi S, Digby GJ, Lambert NA. The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell Signal. 2009;21:1015–1021. doi: 10.1016/j.cellsig.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuchs T, Saunders-Pullman R, Masuho I, Luciano MS, Raymond D, Factor S, Lang AE, Liang TW, Trosch RM, White S, Ainehsazan E, Herve D, Sharma N, Ehrlich ME, Martemyanov KA, Bressman SB, Ozelius LJ. Mutations in GNAL cause primary torsion dystonia. Nat Genet. 2013;45:88–92. doi: 10.1038/ng.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masuho I, Xie K, Martemyanov KA. Macromolecular Composition Dictates Receptor and G Protein Selectivity of Regulator of G Protein Signaling (RGS) 7 and 9-2 Protein Complexes in Living Cells. J Biol Chem. 2013;288:25129–25142. doi: 10.1074/jbc.M113.462283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riobo NA, Manning DR. Receptors coupled to heterotrimeric G proteins of the G12 family. Trends in pharmacological sciences. 2005;26:146–154. doi: 10.1016/j.tips.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Luscher C, Slesinger PA. Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat Rev Neurosci. 2010;11:301–315. doi: 10.1038/nrn2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ostrovskaya O, Xie K, Masuho I, Fajardo-Serrano A, Lujan R, Wickman K, Martemyanov KA. RGS7/Gbeta5/R7BP complex regulates synaptic plasticity and memory by modulating hippocampal GABABR-GIRK signaling. eLife. 2014;3:e02053. doi: 10.7554/eLife.02053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Posokhova E, Wydeven N, Allen KL, Wickman K, Martemyanov KA. RGS6/Gbeta5 complex accelerates IKACh gating kinetics in atrial myocytes and modulates parasympathetic regulation of heart rate. Circ Res. 2010;107:1350–1354. doi: 10.1161/CIRCRESAHA.110.224212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gabay M, Pinter ME, Wright FA, Chan P, Murphy AJ, Valenzuela DM, Yancopoulos GD, Tall GG. Ric-8 proteins are molecular chaperones that direct nascent G protein alpha subunit membrane association. Sci Signal. 2011;4:ra79. doi: 10.1126/scisignal.2002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan P, Thomas CJ, Sprang SR, Tall GG. Molecular chaperoning function of Ric-8 is to fold nascent heterotrimeric G protein alpha subunits. Proc Natl Acad Sci U S A. 2013;110:3794–3799. doi: 10.1073/pnas.1220943110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wess J, Eglen RM, Gautam D. Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nat Rev Drug Discov. 2007;6:721–733. doi: 10.1038/nrd2379. [DOI] [PubMed] [Google Scholar]
- 47.Hollinger S, Hepler JR. Cellular regulation of RGS proteins: Modulators and integrators of G protein signaling. Pharmacol Rev. 2002;54:527–559. doi: 10.1124/pr.54.3.527. [DOI] [PubMed] [Google Scholar]
- 48.Blumer JB, Cismowski MJ, Sato M, Lanier SM. AGS proteins: receptor-independent activators of G-protein signaling. Trends in pharmacological sciences. 2005;26:470–476. doi: 10.1016/j.tips.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Saitoh O, Kubo Y, Miyatani Y, Asano T, Nakata H. RGS8 accelerates G-protein-mediated modulation of K+ currents. Nature. 1997;390:525–529. doi: 10.1038/37385. [DOI] [PubMed] [Google Scholar]
- 50.Cismowski MJ, Ma C, Ribas C, Xie X, Spruyt M, Lizano JS, Lanier SM, Duzic E. Activation of heterotrimeric G-protein signaling by a ras-related protein. Implications for signal integration. J Biol Chem. 2000;275:23421–23424. doi: 10.1074/jbc.C000322200. [DOI] [PubMed] [Google Scholar]
- 51.Lambert NA, Johnston CA, Cappell SD, Kuravi S, Kimple AJ, Willard FS, Siderovski DP. Regulators of G-protein signaling accelerate GPCR signaling kinetics and govern sensitivity solely by accelerating GTPase activity. Proc Natl Acad Sci U S A. 2010;107:7066–7071. doi: 10.1073/pnas.0912934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tse MK, Wong YH. Neuronal functions of activators of G protein signaling. Neuro-Signals. 2013;21:259–271. doi: 10.1159/000337263. [DOI] [PubMed] [Google Scholar]
- 53.Maudsley S, Martin B, Luttrell LM. The origins of diversity and specificity in g protein-coupled receptor signaling. J Pharmacol Exp Ther. 2005;314:485–494. doi: 10.1124/jpet.105.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melancon BJ, Tarr JC, Panarese JD, Wood MR, Lindsley CW. Allosteric modulation of the M1 muscarinic acetylcholine receptor: improving cognition and a potential treatment for schizophrenia and Alzheimer’s disease. Drug discovery today. 2013;18:1185–1199. doi: 10.1016/j.drudis.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conn PJ, Jones CK, Lindsley CW. Subtype-selective allosteric modulators of muscarinic receptors for the treatment of CNS disorders. Trends in pharmacological sciences. 2009;30:148–155. doi: 10.1016/j.tips.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang XP, Nagy PI, Williams FE, Peseckis SM, Messer WS., Jr Roles of threonine 192 and asparagine 382 in agonist and antagonist interactions with M1 muscarinic receptors. Br J Pharmacol. 1999;126:735–745. doi: 10.1038/sj.bjp.0702301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones CK, Brady AE, Davis AA, Xiang Z, Bubser M, Tantawy MN, Kane AS, Bridges TM, Kennedy JP, Bradley SR, Peterson TE, Ansari MS, Baldwin RM, Kessler RM, Deutch AY, Lah JJ, Levey AI, Lindsley CW, Conn PJ. Novel selective allosteric activator of the M1 muscarinic acetylcholine receptor regulates amyloid processing and produces antipsychotic-like activity in rats. J Neurosci. 2008;28:10422–10433. doi: 10.1523/JNEUROSCI.1850-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dasari S, Gulledge AT. M1 and M4 receptors modulate hippocampal pyramidal neurons. J Neurophysiol. 2011;105:779–792. doi: 10.1152/jn.00686.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fisahn A, Yamada M, Duttaroy A, Gan JW, Deng CX, McBain CJ, Wess J. Muscarinic induction of hippocampal gamma oscillations requires coupling of the M1 receptor to two mixed cation currents. Neuron. 2002;33:615–624. doi: 10.1016/s0896-6273(02)00587-1. [DOI] [PubMed] [Google Scholar]
- 60.Wang W, Whorton MR, MacKinnon R. Quantitative analysis of mammalian GIRK2 channel regulation by G proteins, the signaling lipid PIP2 and Na+ in a reconstituted system. eLife. 2014;3:e03671. doi: 10.7554/eLife.03671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sodickson DL, Bean BP. Neurotransmitter activation of inwardly rectifying potassium current in dissociated hippocampal CA3 neurons: interactions among multiple receptors. J Neurosci. 1998;18:8153–8162. doi: 10.1523/JNEUROSCI.18-20-08153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mauger S, Fraser R, Gill K. Utilizing buprenorphine-naloxone to treat illicit and prescription-opioid dependence. Neuropsychiatric disease and treatment. 2014;10:587–598. doi: 10.2147/NDT.S39692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen KY, Chen L, Mao J. Buprenorphine-naloxone therapy in pain management. Anesthesiology. 2014;120:1262–1274. doi: 10.1097/ALN.0000000000000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raynor K, Kong H, Chen Y, Yasuda K, Yu L, Bell GI, Reisine T. Pharmacological characterization of the cloned kappa-, delta-, and mu-opioid receptors. Mol Pharmacol. 1994;45:330–334. [PubMed] [Google Scholar]
- 65.Thomsen W, Frazer J, Unett D. Functional assays for screening GPCR targets. Current opinion in biotechnology. 2005;16:655–665. doi: 10.1016/j.copbio.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 66.Digby GJ, Sethi PR, Lambert NA. Differential dissociation of G protein heterotrimers. J Physiol. 2008;586:3325–3335. doi: 10.1113/jphysiol.2008.153965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kenakin T. Drug efficacy at G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2002;42:349–379. doi: 10.1146/annurev.pharmtox.42.091401.113012. [DOI] [PubMed] [Google Scholar]
- 68.Nishiguchi KM, Sandberg MA, Kooijman AC, Martemyanov KA, Pott JW, Hagstrom SA, Arshavsky VY, Berson EL, Dryja TP. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. 2004;427:75–78. doi: 10.1038/nature02170. [DOI] [PubMed] [Google Scholar]
- 69.Chesley A, Lundberg MS, Asai T, Xiao RP, Ohtani S, Lakatta EG, Crow MT. The beta(2)-adrenergic receptor delivers an antiapoptotic signal to cardiac myocytes through G(i)-dependent coupling to phosphatidylinositol 3’-kinase. Circ Res. 2000;87:1172–1179. doi: 10.1161/01.res.87.12.1172. [DOI] [PubMed] [Google Scholar]
- 70.Xiao RP. Beta-adrenergic signaling in the heart: dual coupling of the beta2-adrenergic receptor to G(s) and G(i) proteins. Sci STKE. 2001;2001:re15. doi: 10.1126/stke.2001.104.re15. [DOI] [PubMed] [Google Scholar]
- 71.Magocsi M, Vizi ES, Selmeczy Z, Brozik A, Szelenyi J. Multiple G-protein-coupling specificity of beta-adrenoceptor in macrophages. Immunology. 2007;122:503–513. doi: 10.1111/j.1365-2567.2007.02658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Laugwitz KL, Allgeier A, Offermanns S, Spicher K, Van Sande J, Dumont JE, Schultz G. The human thyrotropin receptor: a heptahelical receptor capable of stimulating members of all four G protein families. Proc Natl Acad Sci U S A. 1996;93:116–120. doi: 10.1073/pnas.93.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Offermanns S. G-proteins as transducers in transmembrane signalling. Prog Biophys Mol Biol. 2003;83:101–130. doi: 10.1016/s0079-6107(03)00052-x. [DOI] [PubMed] [Google Scholar]
- 74.Foley JF, Singh SP, Cantu M, Chen L, Zhang HH, Farber JM. Differentiation of human T cells alters their repertoire of G protein alpha-subunits. J Biol Chem. 2010;285:35537–35550. doi: 10.1074/jbc.M110.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rius RA, Mollner S, Pfeuffer T, Loh YP. Developmental changes in Gs and G(olf) proteins and adenylyl cyclases in mouse brain membranes. Brain research. 1994;643:50–58. doi: 10.1016/0006-8993(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 76.Fenech C, Patrikainen L, Kerr DS, Grall S, Liu Z, Laugerette F, Malnic B, Montmayeur JP. Ric-8A, a Galpha protein guanine nucleotide exchange factor potentiates taste receptor signaling. Front Cell Neurosci. 2009;3:11. doi: 10.3389/neuro.03.011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Von Dannecker LE, Mercadante AF, Malnic B. Ric-8B promotes functional expression of odorant receptors. Proc Natl Acad Sci U S A. 2006;103:9310–9314. doi: 10.1073/pnas.0600697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Klarenbeek JB, Goedhart J, Hink MA, Gadella TW, Jalink K. A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PLoS One. 2011;6:e19170. doi: 10.1371/journal.pone.0019170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masuho I, Martemyanov KA, Lambert NA. Monitoring G Protein Activation in Cells with BRET. Methods Mol Biol. 2015;1335:107–113. doi: 10.1007/978-1-4939-2914-6_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.