Abstract
Periaxin is expressed in mammalian Schwann cells and lens fiber cells, and has been identified in a screen for cytoskeleton-associated proteins. Charcot–Marie–Tooth 4F is caused by losses or mutations of the periaxin gene. The periaxin gene encodes two protein isoforms, namely, L-periaxin and S-periaxin. S-periaxin contains 147 amino acid residues and has an N-terminal PDZ domain. In this paper, S-periaxin was reported to be homodimerized through the formation of intermolecular disulfide bonds with its Cys88 and Cys139 residues under mild oxidation conditions. The covalent dimer of S-periaxin was also observed by western blot analysis and bimolecular fluorescence complementation analyses. S-periaxin dimerization formation could be regulated by cellular redox fluctuations. These results offer a possible mechanism to the formation of periaxin complexes, improvement of complex stability, and establishment of a link between the extracellular matrix and the cytoskeleton.
Keywords: S-periaxin, intermolecular disulfide bond, dimer, cysteine, bimolecular fluorescence complementation analyses
Introduction
Periaxin was initially identified from myelinating Schwann cells in the mammal peripheral nervous system (PNS), and it plays an essential role in stabilizing the Schwann cell–axon unit through membrane–protein interactions [1,2]. Periaxin protein constitutes 16% of the protein content of the PNS [3]. Periaxin gene deletions or mutations cause demyelinating peripheral neuropathies, such as Charcot–Marie–Tooth disease 4F, peroneal muscular atrophy, hereditary motor and sensory neuropathy [1,4], and recessive Dejerine–Sottas neuropathy [5]. In lens fiber cells, periaxin interacts with ezrin, periplakin, and desmoyokin, which constitutes a macromolecular cytoskeletal complex of ezrin–periplakin–periaxin–desmoyokin (EPPD) [6]. The effect of assembling is found to be involved in the maturation, packing, and membrane organization of lens fiber cells [6].
Periaxin has two isoforms: L-periaxin (147 kD and 1461 amino acid residues) and S-periaxin (16 kD and 147 amino acid residues), which is a truncated isoform [7]. Both proteins have an N-terminal PDZ (PSD-95/Discs Large/ZO-1) protein-binding domain (Fig. 1A) [7,8]. Aside from the PDZ domain, L-periaxin can be further divided into a basic nuclear localization signal region, a long-repeat domain, and a C-terminal acidic domain [9,10]. It has been found that periaxin can dimerize with its PDZ domain [11]. In the absence of the PDZ domain, ΔPDZ-Prx mice are barely functional [12]. This phenomenon may be attributed to the distinctive clustering of the periaxin complex in the Schwann cell plasma membrane [7,11,13].
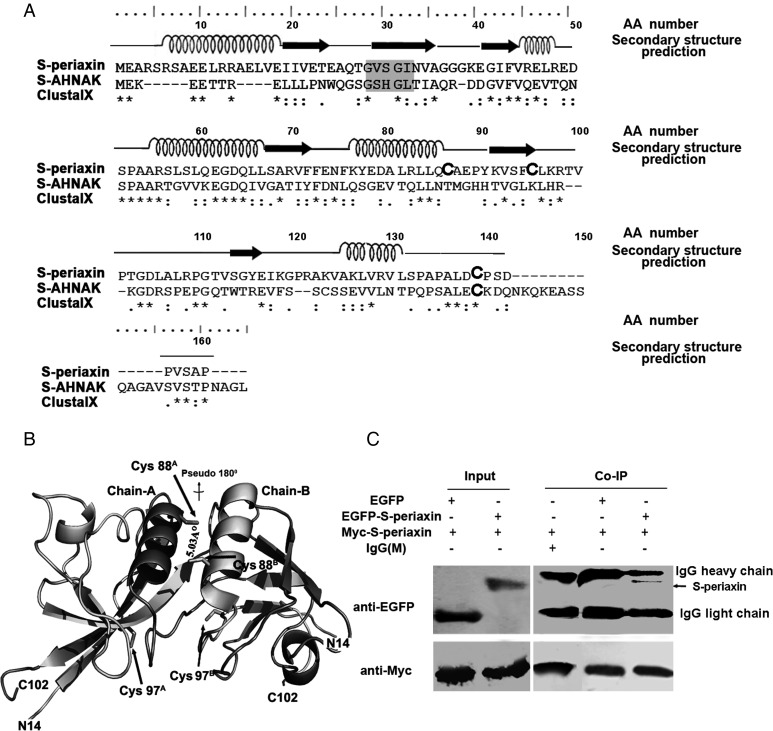
Figure 1.
Protein conservation and homodimerization of full-length S-periaxin (A) Amino acid sequence alignment of S-periaxin (NP_066007.1) and AHNAK isoform 2 (NP_076965.2) with AA number corresponding to the tops of written numerically, and secondary structure prediction of S-periaxin based on the PHD program. (B) The periaxin–PDZ dimer (PDB: 4CMV) is shown in a cartoon model representation by Pymol, with each fragment of secondary structure colored individually. The dimeric 2-fold axis is indicated in deep color (Chain A) and light color (Chain B), and the Cys88 and Cys97 residues are easily identifiable. (C) Verification of the S-periaxin dimerization by co-immunoprecipitation (Co-IP) assay. HeLa cells were transfected with the indicated plasmids for 48 h. Lysates were subjected to Co-IP with IgG or anti-Myc antibody, and then analyzed by western blot analysis with anti-EGFP and anti-Myc antibodies.
The PDZ domain consists of an ∼90-amino acid protein-binding motif that interacts with the cytoplasmic tail of plasma membrane proteins or with the cortical cytoskeleton, which is involved in the assembly of macromolecular signaling complexes [8,14]. The PDZ domain of periaxin is poorly conserved, and the highest sequence identity is found in the N-terminal PDZ-like domain of giant AHNAK nucleoprotein 2 (Fig. 1A) [14]. The PDZ domain of periaxin or AHNAK includes a unique subfamily that may link the extracellular matrix to the cytoskeleton network [15]. The dimerization structures of periaxin–PDZ domain have been presented (PDB: 4CMV), showing that an intertwined, domain-swapped dimer exhibits a head-to-tail antiparallel orientation (Fig. 1B) [14].
The function of the PDZ domain includes the recognition of internal peptide motifs, hetero- and/or homodimerization, as well as the interactions with membrane phospholipids [16,17]. The PDZ domain of periaxin also guides its translocation from the nucleus to the cytoplasm [7]. A nuclear export signal (NES) is identified specifically from amino acid 73–86 of the L-periaxin–PDZ domain, and the nuclear export activity of L-periaxin is found to be inhibited by NES mutation or by LMB treatment [18]. L-periaxin and S-periaxin have the same PDZ domain (Fig. 1A), but S-periaxin is restricted only to the cytoplasm or nucleus of Schwann cells. This restriction can be attributed to protein domain simplification and functional uniqueness [15], and the function of S-periaxin is likely to regulate an mRNA splicing [15].
To date, most functions of S-periaxin are unknown. However, S-periaxin is likely to participate in large molecular complexes through the interactions of its PDZ domain, which is directly involved in biological functions [18]. In the present study, we found that S-periaxin can form dimers or oligomers under non-reducing conditions, and cysteine residues 88 and 139 may be involved in the intermolecular disulfide bond formation in vivo and in vitro.
Materials and Methods
Plasmid construction
The DNA sequence encoding S-periaxin was amplified by PCR by using the following primers: forward primer (5′GAGGAATTCATGGAGGCCAGGAGCCGGAGTGCCGAGGAGCTGAGGCGGGCGGAG3′) and reverse primer (5′ CGAGTCGACTCACGGCGCAGAGACCGGATCGCTGGGGCAGTCCAGGGC3′). The template is a cDNA library from rat RSC96 Schwann cells (Type Culture Collection of the Chinese Academy of Sciences, Shanghai, China). The resultant PCR product digested with EcoRI and SalI (Thermo Scientific, Waltham, USA) was inserted into vector pET-M-3C, in which thrombin protease cleavage sites replaced by human rhinovirus 3c protease cleavage sites,generated from pET15b (Novagen, Madison, USA) to obtain the recombinant plasmid pET-M-3C-S-periaxin. The 441 bp open reading frame was verified by DNA sequence analysis (HuaDa, Beijing, China). The mutants (C88/G, C97/G, C139/G, C88,97/G, C88,139/G, C97,139/G, and C88,97,139/G) were generated by Easy Mutagenesis System (TransGen Biotech, Beijing, China). Furthermore, S-periaxin or its mutants were also cloned into mammalian pCMV-Tag-3B (Myc tag) and pEGFP (EGFP tag) vector (Invitrogen).
Protein expression and purification
The production of recombinant S-periaxin or its mutants was expressed in Escherichia coli BL21 with 0.3 mM IPTG. The cells were harvested and resuspended in lysis buffer (20 mM Tris–HCl, 500 mM NaCl, 5 mM imidazole, pH 7.9). The constructs were lysed by sonication, and the debris was removed by centrifugation at 17,000 g at 4°C for 30 min. The supernatant was applied to a nickel-nitrilotriacetic acid (Ni-NTA) affinity column (GE Healthcare, Bethesda, USA). Recombinant protein was eluted with elution buffer (20 mM Tris–HCl, 500 mM NaCl, 500 mM imidazole, pH 7.9). After concentration by ultrafiltration, the protein was loaded onto Sephacryl S-200 gel filtration chromatography with a buffer containing 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 2 mM DTT, and 15 mM β-mercaptoethanol. The purified protein was collected, concentrated, and then stored at 4°C.
Cell culture, transfection and western blot analysis
HeLa cells were maintained at 37°C with 5% CO2 in RPMI-1640 containing 10% fetal bovine serum. Cells were seeded onto six-well plates and cultured overnight prior to transfections. At 60%–70% confluence, the cells were transfected individually with Myc-tagged wild-type (WT) S-periaxin or its mutants by using TurboFect transfection reagent (Thermo Scientific) according to the manufacturer's instructions. Approximately 48 h post-transfection, HeLa cells were harvested, and the free sulfhydryl groups of proteins were blocked by incubation with 30 mM N-ethylmaleimide (NEM) on ice for 30 min [19]. Then cells were lysed in 400 μl of lysis buffer (20 mM Tris–HCl, 150 mM NaCl, 1% Triton X-100, 1 mM PMSF, sodium pyrophosphate, β-glycerophosphate, EDTA, and leupeptin). The supernatant of cell lysates was separated by 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) without a reducing agent. S-periaxin was transferred onto a PVDF membrane (Millipore, Billerica, USA). The membrane was incubated with rabbit anti-Myc antibody (TransGen Biotech) as the primary antibody, followed by incubation with horseradish peroxidase (HRP)-labeled secondary antibody (TransGen Biotech). Finally, the immunoblots were visualized using an enhanced chemiluminescence reaction kit (Engreen Biosystem, Beijing, China).
Co-IP
HeLa cells were transiently transfected with pCMV-tag3B-S-periaxin/pEGFP- S-periaxin or pCMV-tag3B-S-periaxin/pEGFP. At 48 h post-transfection, whole cell lysates were harvested in 400 μl of lysis buffer, kept on ice for 15 min and centrifuged at 4°C. The supernatant was collected and incubated with 2 μl of anti-Myc antibodies or 2 μl of mouse IgG as the control and allowed to rotate overnight at 4°C. Protein A+G agarose (Beyotime, Shanghai, China) was added and incubated for 4 h, followed by washing with lysis buffer. The samples were separated by 12% SDS-PAGE and analyzed by western blot analysis with the corresponding antibodies. The antibodies used were rabbit anti-Myc antibody and mouse anti-EGFP antibody (TransGen Biotech).
BiFC plasmid construction and imaging of mCherry fluorescence in E. coli
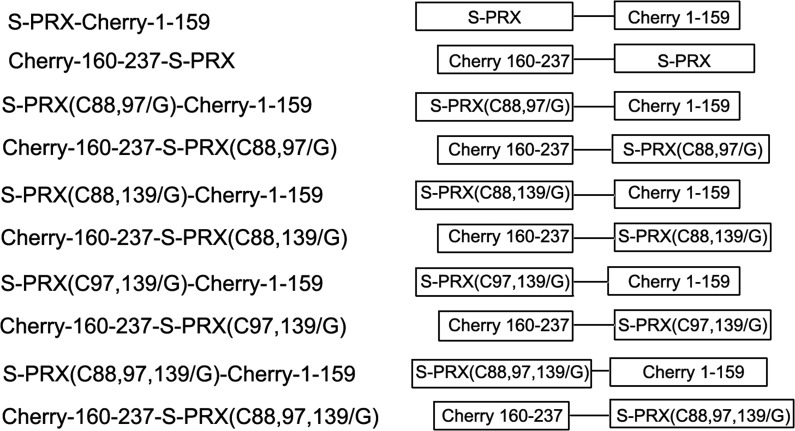
Bimolecular fluorescence complementation (BiFC) plasmids are shown in Fig. 2. The mCherry proteins were split at positions between amino acids 159 and 160 [20]. The N- and C-terminal coding regions of mCherry were amplified by PCR from plasmid pmCherry-C1 (Invitrogen). The gene fragment of mCherry1-159 was cut by SalI and HindIII (Thermo Scientific) and then inserted into the same pQE-30 sites to generate pCherry1-159. The gene fragment of mCherry160-237 was digested by BamHI and EcoRI (Thermo Scientific), and then inserted into the corresponding sites of pET-28a to generate pCherry160-237. The DNA sequence of S-periaxin (WT and its mutants) was inserted into pCherry1-159 or pCherry160-237 to construct pCherry1-159-S-periaxin (WT and its mutants) and pCherry160-237-S-periaxin (WT and its mutants). All sequences were verified by DNA sequencing.
Figure 2.
Schematic presentation of BiFC fusion proteins used in this study mCherry fragment (aa 1–159) was fused to the C terminus of S-periaxin (WT or its mutants) proteins. mcherry fragment (aa 160–237) was fused to the N terminus of S-periaxin (WT or its mutants) proteins.
Escherichia coli cultures that express various fusion proteins of pCherry1-159 or pCherry160-237 (Fig. 2) were maintained on Luria-Bertani medium at 37°C, induced with 0.3 mM IPTG, and grown for an additional 3 h. The bacterial cells were collected and resuspended in 100 µl of phosphate saline buffer [21]. The fluorescence of the E. coli cells was visualized using the Delta Vision Microscopy Imaging Systems (GE Healthcare) with the TRITC channel.
Results
Homodimerization of full-length S-periaxin in transfected HeLa cells
Periaxin isoform 1 and 2 (NP_066007.1 and NP_870998.2) were identified from Homo sapiens through an NCBI database search. The homology of periaxin to other proteins is very low, and the only conserved domain is the PDZ domain of N-terminal [14]. Sequence alignment revealed an overall sequence identity of 30% between S-periaxin (NP_066007.1) and AHNAK isoform 2 (NP_076965.2) at the amino acid level (Fig. 1A). Furthermore, the crystal structures of the PDZ homology domain of periaxin (PDB: 4CMV; 14–104 aa) were verified [14]. We paid special attention to the structure model. As shown in Fig. 1B, the core of the PDZ dimer is formed by a 6-stranded antiparallel β sheet by chain A and B, and the cores of α2 helices (Phe75–Tyr90) from the two chains of PDZ dimer structure are placed side by side [14]. An extended conformation in the N-terminal (Met1–Arg13) and C-terminal (Gly105–Pro147) of S-periaxin is modeled based on SAXS data [14], thereby suggesting that S-periaxin is elongated and the moiety points of carboxyl terminus remain exposure the most.
L-periaxin dimerization had been shown by yeast two-hybrid experiments, pull-downs from nerve lysates, and CO-IP from mammalian cells that are over-expressing L-periaxin constructs [11]. To confirm the dimerization form of S-periaxin, HeLa cells were transiently transfected with pCMV-tag3B-S-periaxin/pEGFP-S-periaxin. Myc-tagged S-periaxin was shown to co-precipitate with EGFP-tagged S-periaxin. No specific binding was observed with the EGFP control or the irrelevant mouse IgG. These results indicate that S-periaxin forms homodimers in vivo (Fig. 1C).
Oligomerization of S-periaxin without reducing reagents in vitro
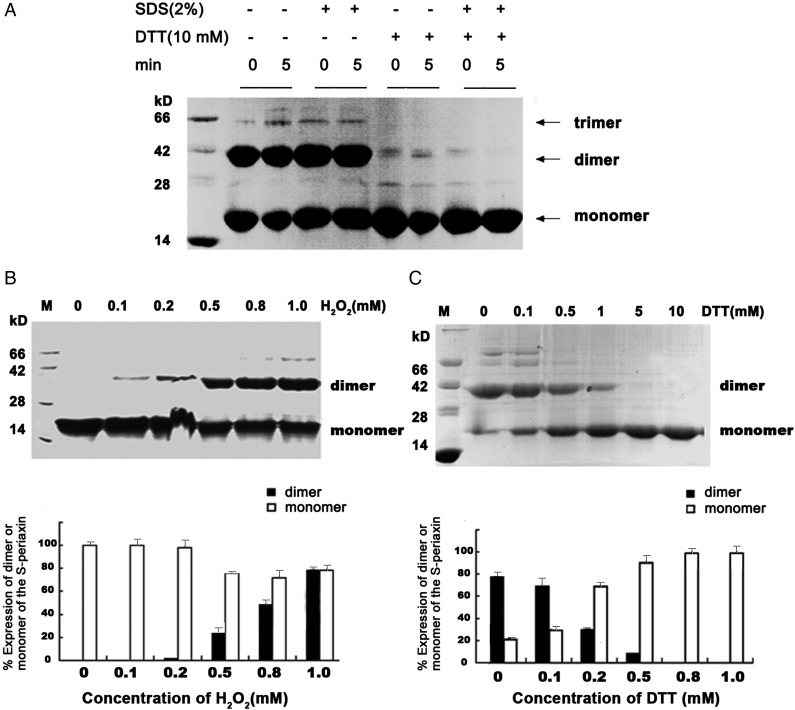
The recombinant His-S-periaxin was expressed in BL21 cells and its molecular weight was estimated to be ∼15 kD (Fig. 3A). Without any reducing reagent in the loading buffer, different oligomers of S-periaxin were detected, which suggests that S-periaxin may oligomerize under oxidation (Fig. 3A, lanes 2–5). Approximately 10 mM DTT was added to Laemmli loading sample buffer to reduce the tetrameric and trimeric protein bands. A few S-periaxin dimers were detected after incubation for at least 5 min (Fig. 3A, lanes 6–9).
Figure 3.
S-periaxin forms dimers through disulfide bond (A) His-S-periaxin was boiled for 0 or 5 min without (lanes 2–5) or with 10 mM DTT (lanes 6–9) or 2% SDS (lanes 4–5 and 8–9) in loading buffer. Proteins were visualized by Coomassie staining analysis. (B) SDS-PAGE analysis of the oxidation of S-periaxin with H2O2. (C) SDS-PAGE analysis of the redox of S-periaxin with DTT. Quantitative assay of the dimeric S-periaxin with H2O2 or with DTT were shown.
The recombinant S-periaxin in 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 2 mM DTT, and 15 mM β-mercaptoethanol was desalted by using a P10 column and quickly transferred to a buffer that contains 20 mM Tris–HCl (pH 8.0) and 100 mM NaCl. Then it was treated with different concentrations of H2O2 (0–1 mM) for 30 min at room temperature and analyzed by non-reducing SDS-PAGE. H2O2 treatment induced the formation of an S-periaxin dimer in a dose-dependent manner (Fig. 3B). The S-periaxin dimer in 20 mM Tris–HCl (pH 8.0) and 100 mM NaCl can be fully converted back to the monomer with the addition of 5 mM DTT (Fig. 3C). These findings indicate that the intermolecular disulfide bond formed by oxidation is involved in the oligomerization of S-periaxin.
Mediation of S-periaxin dimerization via intermolecular disulfide bonds
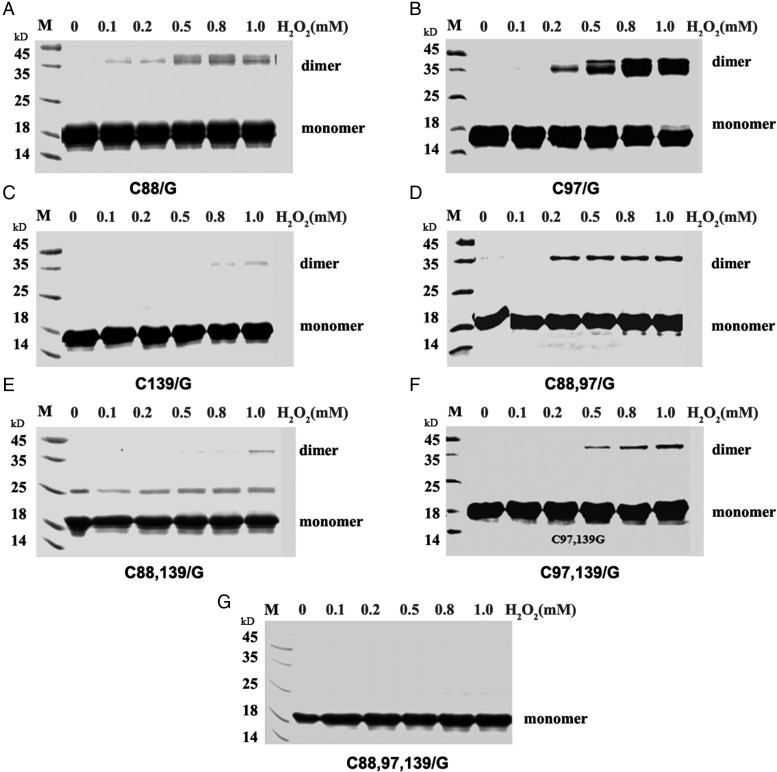
To investigate the role of the three Cys (C88, C97, and C139) in the formation of intermolecular disulfide bond, S-periaxin and its mutants (C88/G, C97/G, C139/G C88,97/G, C88,139/G, C97,139/G, and C88,97,139/G) were subject to oxidation with H2O2 in vitro. In single mutations, only Cys88 and Cys139 showed an obvious weak tendency to form an H2O2-induced dimer, unlike WT (S-periaxin-Cherry1-159/Cherry160-237-S-periaxin) and C97/G (Fig. 4A–C). Mutation of C88,139/G in S-periaxin also had a greater tendency to reduce dimer formation than C88,97/G, and C97,139/G (Fig. 4D–F). When all Cys were mutated to Gly, C88,97,139/G only showed a monomer band (Fig. 4G). These results indicate that Cys of S-periaxin are involved in dimerization through the formation of intermolecular disulfide bonds. Cys88 and Cys139 undergo oxidation in S-periaxin, while Cys97 was relatively inert to oxidation.
Figure 4.
Cys 88 and Cys 139 of S-periaxin are more susceptible to oxidation SDS-PAGE analysis of the oxidation of the S-periaxin mutants with H2O2. (A) S-periaxin-C88/G, (B) C97/G, (C) C139/G, (D) C88,97/G, (E) C88,139/G, (F) C97,139/G and (G) C88,97,139/G were incubated with indicated concentrations of H2O2 for 30 min at room temperature before SDS-PAGE.
Dimer formation of S-periaxin through intermolecular disulfide formation in vivo
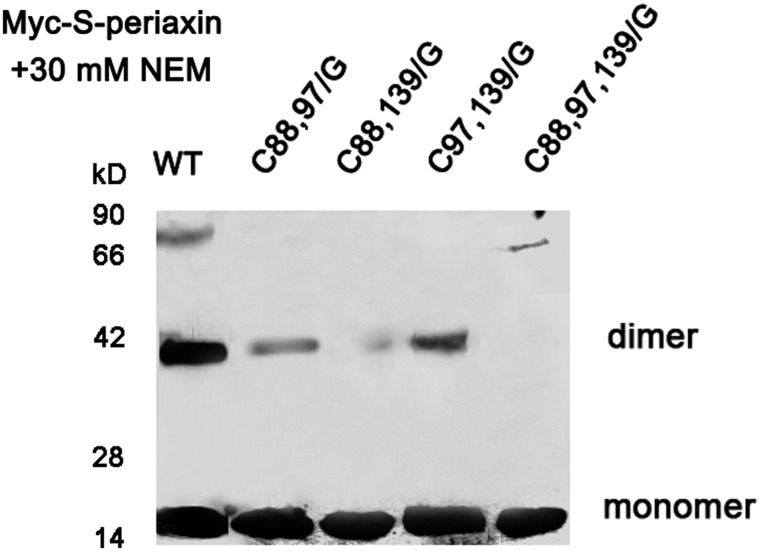
To determine whether S-periaxin can form a dimer in vivo, Myc-tagged S-periaxin (WT, C88,97/G, C88,139/G, C97,139/G, or C88,97,139/G) was transfected into HeLa cells. Before lysis, these cells were collected and incubated with 30 mM NEM to block reactive sulfhydryl groups [19]. The cell lysates were loaded onto non-reducing SDS-PAGE. The dimer of S-periaxin and its mutants was detected by western blot analysis using anti-Myc antibody. Interestingly, on non-reducing SDS-PAGE, both monomer and dimer were detected in WT Myc-S-periaxin and in three mutants (C88,97/G, C88,139/G, and C97,139/G), while only monomer was detected in the C88,97,139/G mutant (Fig. 5), suggesting that Cys residues are involved in the assembling of S-periaxin. However, the intermolecular disulfide bond is not the only important factor involved in the oligomerization of the molecule.
Figure 5.
Dimer conformation of S-periaxin requires Cys88 and Cys139 HeLa cells were transfected with Myc-S-periaxin (WT, C88,97/G, C88,139/G, C97,139/G, and C88,97,139/G). Whole cell extracts were separated under non-reducing conditions and detected by western blot analysis using anti-Myc antibody.
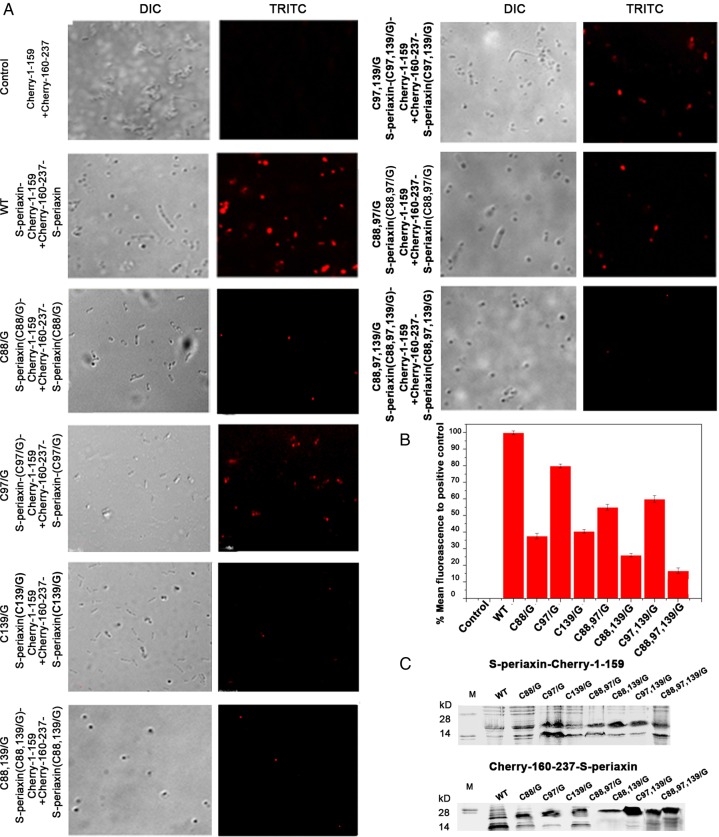
Subsequently, BiFC analysis was performed to obtain the intermolecular disulfide bond information of S-periaxin in E. coli BL21 cells. S-periaxin and its mutant fusion with mCherry fragments were transformed into BL21, and all the fusion proteins were expressed in supernatant fluid (Fig. 6C). The emission of mCherry-RFP fluorescence by the various strains was monitored [22,23]. Co-inoculation of pCherry-1-159-S-periaxin/pCherry-160-237-S-periaxin constructs resulted in strong red fluorescent signals located in small aggregations all over the cytoplasm, thereby indicating the self-interaction of S-periaxin. S-periaxin can form dimers in living cells (Fig. 6A). Red fluorescence was found in little aggregates from Cys mutation C97/G, C88,97/G and C97,139/G. However, the oligomerization of C88/G, C139/G, and C88,97,139/G was relatively lower than that of S-periaxin (Fig. 6B). In contrast, the mutations of S-periaxin did not completely abolish BiFC fluorescence, which means that other factors may also contribute to S-periaxin dimerization, among which the spatial structure of the PDZ domain is one such factor [16].
Figure 6.
Visualization of S-periaxin interactions using the mCherry-based BiFC system (A) E. coli BL-21 cells were transformed with 9 BiFC fusion constructs (WT and mutants); (B) the emission of mCherry fluorescence was monitored; (C) 12% SDS-PAGE analysis of the co-expressed cherry-1-159 and 160-237 fusion proteins in E. coli BL21 cells. WT and mutants of S-periaxin protein were loaded in each lane. M, molecular markers were showed in the left lane.
Discussion
L-periaxin and S-periaxin have the same N-terminal PDZ domain. However, L-periaxin predominantly localizes in the nucleus (adaxonal membrane) during axon ensheathment and early myelination, and then localizes to the abaxonal Schwann cell membrane (opposite the basal lamina) that is undergoing myelination [1]. This process is unlikely for S-periaxin, which distributes in the cytoplasm (occasionally in the nucleus) [15].
The PDZ domain is a protein–protein interacting module that plays an important role in the organization of signaling complexes [15]. For example, PICK1 (protein interacting with C-kinase 1) PDZ was reported to be redox-regulated by the formation of a reversible intermolecular disulfide bond that engages Cys44 in the β2–β3 loop under mild oxidative conditions [24]. The most common target for cellular oxidants is Cys residues in proteins. Upon exposure to oxidants such as H2O2, free sulfhydryl groups in the Cys residues can be oxidized into sulfenic, sulfinic, or sulfonic acids. H2O2 treatment induced the formation of intermolecular disulfide bonds between S-periaxin in vitro. In this report, we demonstrated that Cys88 and Cys139 in S-periaxin play a role in oxidation-induced disulfide bond formation, while Cys97 is relatively inert to oxidation.
Furthermore, from the PDZ crystal structure of periaxin (Fig. 1B), we found that the α2 helices (Phe75–Tyr90) from the two chains of the periaxin dimer structure are side by side, and the distance between Cys88A and Cys88B is ∼5.03 Å, which meets the standard of a disulfide bond. In comparison, the distance between Cys97A and Cys97B or Cys88A–Cys97B is more than 10 Å, suggesting that disulfide bonds cannot be formed between these sites. The conformation of the C-terminal (including that of Cys139) exhibits extended structures. Although the homology of S-periaxin to other proteins is low, the C-terminal extensions gradually increase the possibility of oligomer formation through the Cys139–Cys139 intermolecular disulfide bond.
Various techniques, including yeast two-hybrid screening, Co-IP, glutathione S-transferase pull-down, and the recently developed tandem affinity purification assays, have been used to detect and study protein–protein interactions [11,25,26]. BiFC, which was developed by Hu et al. [27], detects weak or otherwise transient protein–protein interactions in vivo. BiFC is based on the principle that the N- and C-terminal fragments of fluorescent proteins and its derivatives alone do not fluoresce. However, if they are fused to interacting proteins, the two non-functional halves can be brought into close proximity and form a functional fluorophore as a result of specific protein interactions [28,29]. In this study, we performed BiFC analysis to obtain the intermolecular disulfide bond information of S-periaxin in bacterial cells. Results showed that S-periaxin (WT and its mutants) could form inter-subunit disulfide bonds in vivo. However, inter-disulfide bond formation was not the main reason for the dimerization or oligomerization of S-periaxin.
Other factors may also be involved in the oligomerization of S-periaxin in the cytoplasm. The mutation of the GLGF motif of the L-periaxin–PDZ domain into GLAA attenuated homodimerization (Fig. 1A), suggesting that the canonical peptide binding groove of the periaxin–PDZ domain may participate in homodimerization [11]. To date, the function of S-periaxin has not yet been revealed. The dimerization of S-periaxin may provide a clue to the hypothesis that S-periaxin may interact with L-periaxin by PDZ dimerization to regulate the function of L-periaxin in Schwann cells [15]. These questions are critical to future studies.
S-periaxin was found to be homodimerized through the formation of intermolecular disulfide bonds between its Cys88 and Cys139 residues under mild oxidative conditions. Cys97 residue is relatively inert to oxidation. We propose that the molecular conformation of S-periaxin could be regulated by cellular redox fluctuations under physiological and pathophysiological conditions.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (No. 31170748).
References
- 1.Scherer SS, Xu YT, Bannerman PG, Sherman DL, Brophy PJ. Periaxin expression in myelinating Schwann cells: modulation by axon-glial interactions and polarized localization during development. Development 1995, 121: 4265–4273. [DOI] [PubMed] [Google Scholar]
- 2.Williams AC, Brophy PJ. The function of the Periaxin gene during nerve repair in a model of CMT4F. J Anat 2002, 200: 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patzig J, Jahn O, Tenzer S, Wichert SP, de Monasterio-Schrader P, Rosfa S, Kuharev J et al. Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J Neurosci 2011, 31: 16369–16386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guilbot A, Williams A, Ravise N, Verny C, Brice A, Sherman DL, Brophy PJ et al. A mutation in periaxin is responsible for CMT4F, an autosomal recessive form of Charcot-Marie-Tooth disease. Hum Mol Genet 2001, 10: 415–421. [DOI] [PubMed] [Google Scholar]
- 5.Boerkoel CF, Takashima H, Stankiewicz P, Garcia CA, Leber SM, Rhee-Morris L, Lupski JR. Periaxin mutations cause recessive Dejerine-Sottas neuropathy. Am J Hum Genet 2001, 68: 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maddala R, Skiba NP, Lalane RR, Sherman DL, Brophy PJ, Rao PV. Periaxin is required for hexagonal geometry and membrane organization of mature lens fibers. Dev Biol 2011, 357: 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dytrych L, Sherman DL, Gillespie CS, Brophy PJ. Two PDZ domain proteins encoded by the murine periaxin gene are the result of alternative intron retention and are differentially targeted in Schwann cells. J Biol Chem 1998, 273: 5794–5800. [DOI] [PubMed] [Google Scholar]
- 8.Khan Z, Lafon M. PDZ domain-mediated protein interactions: therapeutic targets in neurological disorders. Curr Med Chem 2014, 21: 2632–2641. [DOI] [PubMed] [Google Scholar]
- 9.Sherman DL, Brophy PJ. A tripartite nuclear localization signal in the PDZ-domain protein L-periaxin. J Biol Chem 2000, 275: 4537–4540. [DOI] [PubMed] [Google Scholar]
- 10.Gillespie CS, Lee M, Fantes JF, Brophy PJ. The gene encoding the Schwann cell protein periaxin localizes on mouse chromosome 7 (Prx). Genomics 1997, 41: 297–298. [DOI] [PubMed] [Google Scholar]
- 11.Sherman DL, Fabrizi C, Gillespie CS, Brophy PJ. Specific disruption of a Schwann cell dystrophin-related protein complex in a demyelinating neuropathy. Neuron 2001, 30: 677–687. [DOI] [PubMed] [Google Scholar]
- 12.Wu LM, Williams A, Delaney A, Sherman DL, Brophy PJ. Increasing internodal distance in myelinated nerves accelerates nerve conduction to a flat maximum. Curr Biol 2012, 22: 1957–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Court FA, Brophy PJ, Ribchester RR. Remodeling of motor nerve terminals in demyelinating axons of periaxin-null mice. Glia 2008, 56: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han H, Kursula P. Periaxin and AHNAK nucleoprotein 2 form intertwined homodimers through domain swapping. J Biol Chem 2014, 289: 14121–14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Morree A, Droog M, Grand ML, Bisschop IJ, Impagliazzo A, Frants RR, Klooster R et al. Self-regulated alternative splicing at the AHNAK locus. FASEB J 2012, 26: 93–103. [DOI] [PubMed] [Google Scholar]
- 16.Ernst A, Appleton BA, Ivarsson Y, Zhang Y, Gfeller D, Wiesmann C, Sidhu SS. A structural portrait of the PDZ domain family. J Mol Biol 2014, 426: 3509–3519. [DOI] [PubMed] [Google Scholar]
- 17.Grillo-Bosch D, Choquet D, Sainlos M. Inhibition of PDZ domain-mediated interactions. Drug Discov Today Technol 2013, 10: e531–e540. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, Zhang L, Yang T. Nuclear export of L-periaxin, mediated by its nuclear export signal in the PDZ domain. PLoS One 2014, 9: e91953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zha XM, Wang R, Collier DM, Snyder PM, Wemmie JA, Welsh MJ. Oxidant regulated inter-subunit disulfide bond formation between ASIC1a subunits. Proc Natl Acad Sci USA 2009, 106: 3573–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan JY, Cui ZQ, Wei HP, Zhang ZP, Zhou YF, Wang YP, Zhang XE. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein-protein interactions in living cells. Biochem Biophys Res Commun 2008, 367: 47–53. [DOI] [PubMed] [Google Scholar]
- 21.Tsuchisaka A, Theologis A. Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc Natl Acad Sci USA 2004, 101: 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zilian E, Maiss E. An optimized mRFP-based bimolecular fluorescence complementation system for the detection of protein-protein interactions in planta. J Virol Methods 2011, 174: 158–165. [DOI] [PubMed] [Google Scholar]
- 23.Zilian E, Maiss E. Detection of plum pox potyviral protein-protein interactions in planta using an optimized mRFP-based bimolecular fluorescence complementation system. J Gen Virol 2011, 92: 2711–2223. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Yu J, Jia Y, Pan L, Shen C, Xia J, Zhang M. Redox-regulated lipid membrane binding of the PICK1 PDZ domain. Biochemistry 2010, 49: 4432–4439. [DOI] [PubMed] [Google Scholar]
- 25.Ma H, McLean JR, Chao LF, Mana-Capelli S, Paramasivam M, Hagstrom KA, Gould KL et al. A highly efficient multifunctional tandem affinity purification approach applicable to diverse organisms. Mol Cell Proteomics 2012, 11: 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Laval SH, van Remoortere A, Baudier J, Benaud C, Anderson LV, Straub V et al. AHNAK, a novel component of the dysferlin protein complex, redistributes to the cytoplasm with dysferlin during skeletal muscle regeneration. FASEB J 2007, 21: 732–742. [DOI] [PubMed] [Google Scholar]
- 27.Hu CD, Chineno Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 2002, 9: 789–798. [DOI] [PubMed] [Google Scholar]
- 28.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys 2008, 37: 465–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerppola TK. Bimolecular fluorescence complementation: visualization of molecular interactions in living cells. Methods Cell Biol 2008, 85: 431–470. [DOI] [PMC free article] [PubMed] [Google Scholar]