Abstract
The effect of apocynin on the activity of arylamine N-acetyltransferases (NATs) in excised liver samples was examined using eighteen Sprague-Dawley rats. Three groups of six animals each were fed a normal diet alone or a treatment of 50 or 100 mg/kg/day of apocynin via gavages for eight (8) weeks. Chronic in vivo administration of apocynin led to significant (p < 0.001) reduction of in vitro liver NAT activity up to 93% as compared with untreated rats (18.80 ± 2.10 μmols p-anisidine/min/μg liver protein). In vitro exposure of untreated liver homogenates to apocynin led to a dose-dependent inhibition of NAT activity with IC50 = 0.69 ± 0.02 mM. In silico modelling of apocynin tautomers and radical species into human NAT crystal structures supported the hypothesis that thiol functionalities in NAT enzymes may be crucial in apocynin binding. The involvement of human NAT enzymes in different pathological conditions, such as cancer, has encouraged the research for selective NAT inhibitors in both humans and animal models with possible chemopreventive properties.
Apocynin, 4-hydroxyl-3-methoxyacetophenone or acetovanillone, was originally isolated from the roots of Apocynum cannabinum L. (Apocynacaeae) and also found in Picrorhiza kurroa Royle ex Benth. (Scrophulariaceae); this natural product has been traditionally used in the Aryurvedic medicine for a range of illnesses including heart and lung diseases1.
 |
In recent years, apocynin has gained significance as a potential prodrug for multiple diseases, although its complete mechanism of action is not fully elucidated1. Apocynin is considered to act as an antioxidant because it prevents the activity of NAD(P)H oxidase enzyme from generating reactive oxygen species (ROS), such as superoxide anion (O2−)2, and thus may be useful in the treatment of a variety of illnesses which are triggered or exacerbated by an elevated inflammatory response.
Initially, apocynin was found to inhibit NAD(P)H oxidase in neutrophiles3, wherein its inhibitory effect appeared to be closely mediated by myeloperoxidase (MPO) enzyme4. The reaction of peroxidases generates radical species of apocynin, which subsequently form dimers5; these dimers are capable of oxidizing essential cysteine thiol groups within the sub-units of NAD(P)H oxidase6, thereby inhibiting the formation of the complex and its catalytic activity7. Nevertheless, some controversy does exist around the exclusive antioxidant role of apocynin for ROS formation processes and unfavourable pro-oxidant effects8. Apocynin widely seems to be useful in vitro and in vivo models for lipid peroxidation9, atherosclerosis10, kidney injuries11, and ischemia12. Moreover, apocynin shows low cytotoxicity13, and has chemopreventive properties14.
Many natural organic compounds are known to display a valuable potential for cancer-prevention in chemically induced carcinogenesis models15,16,17. Tumour-preventive strategies may include the use of phytochemicals for either their antioxidant properties which allow the modulation of the intracellular redox status finally leading to the apoptosis of tumour cells, or their inhibitory potency towards some metabolic pathways which activate procarcinogens18.
Whilst apocynin has been shown to be an inhibitor of certain isoforms of cytochrome P450 (CYP) enzymes19,20, its impact on other drug metabolizing enzymes has not been reported to date. Therefore, we investigated the effects of apocynin on the activity of arylamine N-acetyltransferase (NAT), which is an enzyme involved in the activation and phase-II metabolism of different carcinogenic xenobiotics via N-,O-, or N,O-acetylation21.
NAT gene is found in a variety of prokaryotic and eukaryotic species21. Chromosome 8 from the human genome contains two polymorphic NAT loci, (HUMAN)NAT1 and (HUMAN)NAT2, which encode for two functionally distinct enzymes and have long been associated with pharmacogenetics21. Also a non-functional NAT pseudogene (NATP) is encountered on the same chromosome21.
Rodents possess three NAT loci in their genome and their corresponding enzyme products have conventionally been used as animal models to study human NATs22. In particular, (RAT)NAT2 enzyme shows a high functional homology with (HUMAN)NAT1, whereas active (RAT)NAT1 and (RAT)NAT3 have a partial analogy in relation to (HUMAN)NAT2 and NATP respectively, with rodent NAT3 having a lower cytosolic production and very little catalytic activity (Table 1, Supplementary Figure S1)22,23. Moreover, the arrangement of the three rat NAT loci on chromosome 16 appeared to be highly similar to that of the three mouse NATs on chromosome 822; also, the mouse NAT enzymes (namely (MOUSE)NAT1, (MOUSE)NAT2 and (MOUSE)NAT3) were shown to N-acetylate similar probe substrates with an analogous catalytic efficiency as compared to (RAT)NAT1, (RAT)NAT2, and (RAT)NAT3 respectively (Table 1, Supplementary Figure S1)22,24,25,26,27.
Table 1. Percentage Of Primary Sequence Identity of Human, Mouse, and Rat Nat Proteins against (Human)NAT1 Isoform and Probe NAT Substrate Specificities.
| (HUMAN)NAT1 | – | |||||||
| (HUMAN)NAT2 | 81 | – | ||||||
| (MOUSE)NAT1 | 75 | 72 | – | |||||
| (MOUSE)NAT2 | 82 | 75 | 82 | – | ||||
| (MOUSE)NAT3 | 68 | 66 | 68 | 73 | – | |||
| (RAT)NAT1 | 76 | 75 | 92 | 83 | 69 | – | ||
| (RAT)NAT2 | 81 | 74 | 81 | 95 | 73 | 83 | – | |
| (RAT)NAT3 | 69 | 67 | 75 | 73 | 88 | 71 | 72 | – |
| (HUMAN) NAT1 | (HUMAN) NAT2 | (MOUSE) NAT1 | (MOUSE) NAT2 | (MOUSE) NAT3 | (RAT) NAT1 | (RAT) NAT2 | (RAT) NAT3 | |
| isoenzyme-selective substratea | pABglub | isoniazid | isoniazid | pABglub | – | isoniazid | pABglub | – |
Since extensive studies on human NAT genotype and phenotype have revealed a close relationship between NATs and different pathological conditions, such as cancer, looking for selective inhibitors of NAT enzymes in both humans and animal models has constituted a valuable tool to develop possible chemopreventive agents21. In particular, (HUMAN)NAT1 is up-regulated in several cancer types, thereby leading to cancer cell growth and resistance to chemotherapy, and has emerged as a viable candidate for drug development, which should lead to small molecule inhibitors for preclinical and clinical evaluation28. A few examples of phytochemical molecules were previously screened as possible inhibitors of human NATs from human liver29.
We report here the first study showing the inhibitory potency of apocynin against NAT activity in rat liver both ex vivo, following treatment with apocynin, and in vitro.
Results and Discussion
Inhibition of NAT activity in rat livers following treatment with apocynin
In this report we investigated the effects of apocynin on liver NAT activity in rats using both a system strategy which measures NAT enzymatic activity in livers excised from rats treated with apocynin and a molecular approach directly on native NAT activity in rat liver.
Previous investigations reported that rats fed 120 mg apocynin/kg showed 80% recovery of un-metabolized apocynin in the urine30 and a diet of apocynin 50–100 mg/kg had a low physiological impact on rats without toxic side effects30,31,32. Also, no gender-specific differences in rat NAT expression have been observed in the liver33. Thus, we initially investigated the impact of apocynin as un-metabolized natural biotic on liver NAT activity in rats fed normal diet or 50–100 mg apocynin/kg body for eight weeks. The activity of NAT enzymes in liver homogenates was measured using p-anisidine (pANS) which was previously shown to be a good arylamine substrate catalyzed by all active human and mouse NAT isoforms27.
In particular, liver NAT activity in rats fed 50 or 100 mg apocynin/kg (1.44 ± 0.35 and 4.51 ± 0.83 μmols/min/μg liver protein ex vivo respectively) appeared to be significantly reduced (p < 0.001) as compared to liver NAT activity in untreated rats (18.80 ± 2.21 μmols/min/μg liver protein) (Fig. 1). However, no statistically significant (p > 0.5) difference was observed between the two NAT activity measures following either of the two treatments with apocynin. Using a range of lower in vivo doses of apocynin would help better establish in vivo dose-response relationship in future pharmacokinetic studies. As far as we are aware, no report of the impact of a per os treatment with apocynin on liver NAT activity in rats without apparent undesired systemic side effects has been published previously.
Figure 1. Impact of a diet containing apocynin on the activity of rat liver NATs.
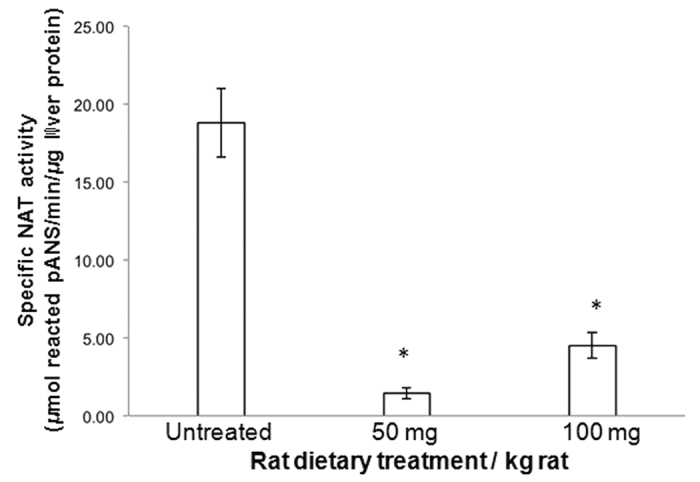
The NAT activity of the liver S9 fractions (3–8 μg/μl protein) obtained from rats fed a diet free of apocynin (untreated rats) or varying amounts of apocynin (treated rats) was determined ex vivo using pANS and AcCoA, as described in methods. The average value of NAT activity for the untreated group was compared with each NAT activity value for the treated groups, and statistical significance at p < 0.001 is indicated by an asterisk (*). Assays were conducted in triplicate and all values are expressed as mean ± standard deviation values.
In order to determine whether apocynin has also inhibitory effects against native NAT enzymes in normal rat liver, we focused on analyzing the molecular impact of apocynin on the S9 fractions prepared from liver samples of untreated rats.
Initially, apocynin or acetyl Coenzyme A (AcCoA) were allowed to preincubate with S9 liver fractions from untreated rat for either 5 or 15 minutes before starting the catalytic reaction with the other reagents. In enzymatic assays with AcCoA and pANS substrates, NAT activity was significantly reduced (p < 0.05) upon immediate addition of apocynin (assay B, Fig. 2); however, a comparable decrease of NAT activity occurred when apocynin was allowed to preincubate 5 or 15 minutes with liver homogenates prior to the addition of the two NAT substrates (assays C-D, Fig. 2). The effects of a time-dependent incubation of liver homogenates with apocynin on NAT activity apparently are not statistically dissimilar.
Figure 2. Impact of varying preincubation times of liver homogenates with apocynin and AcCoA on the inhibition of rat liver NAT activity.
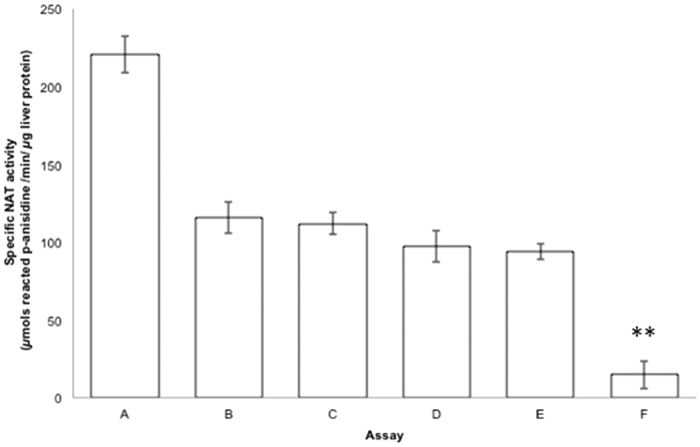
NAT activity in the S9 fractions (9.60 μg protein) from untreated rats was determined in vitro as described in methods (A). The inhibitory potency of apocynin (0.49 mM) was tested at different assay conditions: in assays (B–D) apocynin was preincubated with S9 fractions for 0, 5′ or 15′ respectively; in assays E and F, S9 fractions were preincubated with AcCoA for 5′ or 15′ respectively. Appropriate enzymatic control assays were performed for each condition. Assays (B–F) were all statistically different from assay A (p < 0.05); ** denotes statistically significant difference (p < 0.001) between assays E and F.
Overall, these initial results suggested a close interaction between apocynin and at least a NAT isoenzyme within rat liver homogenates, which generated inhibition of NAT activity. Additionally, apocynin showed a clear dose-dependent inhibitory potency against liver NAT activity in vitro with an IC50 value of 0.69 ± 0.02 mM (Fig. 3). IC50 values are known to be dependent on the amount of reactive enzyme in the assay34; therefore, the micromolar order of magnitude of this IC50 might be related to the use of liver cell lysates as source of impure NAT enzymes, instead of pure recombinant proteins. Moreover, the IC50 value obtained appeared to be of particular interest in comparison to previous investigations where other mammalian liver-specific enzymes, such as CYP1A235 and CYP1B136, were reported to be inhibited fairly weakly by apocynin in lysates from engineered prokaryotic cells over-expressing CYP enzymes37.
Figure 3. In vitro inhibition of NAT activity from rat liver samples by apocynin.
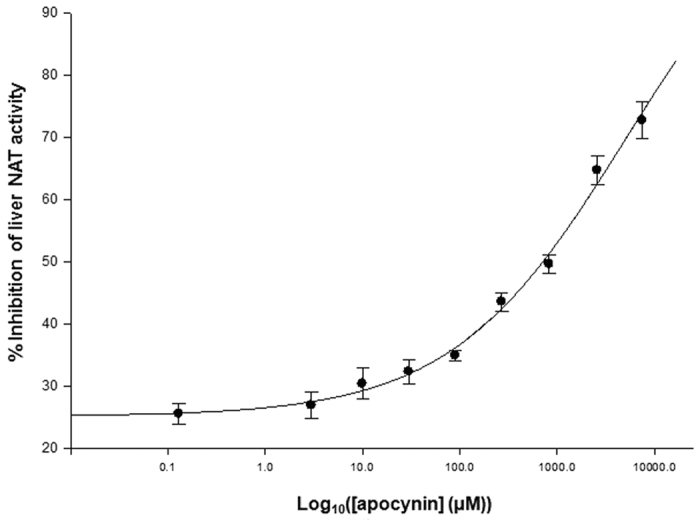
The impact of varying concentrations of apocynin (0 and 10 mM) on the activity of rat liver NAT from S9 fractions (9.60 μg protein) of untreated rat livers was investigated in-vitro, as described in methods. Assays were conducted in triplicate and all values are expressed as mean ± standard deviation values.
On the other hand, when AcCoA was preincubated with S9 fractions for 5 minutes, N-acetylation of pANS was equally lowered in the presence of apocynin (assay E, Fig. 2) as compared to assays B, C and D; this similarity looked consistent with the AcCoA preincubation times used in other enzymatic protocols using dimethylaminobenzaldehyde (DMAB) with cell lysates38. However, the NAT activity was doubly reduced (p < 0.001) when AcCoA was allowed to interact with S9 fractions for 15 minutes prior to the addition of apocynin and pANS (assay F, Fig. 2). This additional inhibition are likely to be related to the longer preincubation times of liver homogenates with AcCoA, since S-acetylated NAT intermediates can be spontaneously hydrolyzed in enzyme assays39, and the Coenzyme A (CoA) species formed upon AcCoA depletion can cause further inhibition of NAT activity39,40.
Enzymatic assays measuring CoA formation in NAT reaction with the colorimetric Ellman’s reagent (5,5′-dithiobis-(2-nitrobenzoic acid), DTNB) were also attempted to ascertain the extent of apocynin inhibition of NAT activity in assay F and also investigate whether apocynin can be a possible substrate of NAT enzymes41; however, the background noise in all spectrophotometric assay controls was very high due to the free thiol species in cell homogenates reacting with DTNB, and valuable measurements were not allowed.
Hypothetical modes of NAT enzyme inhibition by apocynin
Amongst the studies investigating apocynin as functional enzyme inhibitor, apocynin, its radical metabolites and other polyphenolic compounds were proposed to inhibit the activity of NADPH oxidase by interacting with selective cysteine thiol groups of cytosolic sub-units thereby preventing complex formation. Two possible mechanisms were therefore suggested: a Michael conjugation between essential cysteine thiolates and the electrophilic quinone derivatives of phenols42,43, or oxidation of specific cysteine thiol groups by the phenolic radical species generated by peroxidases7,44,45.
In the family of mammalian NATs, cysteine residues have been well characterized in both NAT catalysis mechanism and protein structure. In particular, NAT reaction with AcCoA and arylamine substrates has been well established to follow a bi-bi ping-pong mechanism. In the first step, AcCoA binds to eukaryotic NAT protein to form a NAT-AcCoA complex, then the Cys68 thiolate within the active site is acetylated via nucleophilic addition to AcCoA, and finally CoA is delivered as first product. In the second phase, the incoming arylamine binds to the acetylated-NAT intermediate, the acetyl group is transferred to the amine substrate, and an N-acetylated product is finally delivered (Supplementary Figure S2)46,47. Structural investigations on Syrian hamster NAT2 helped determine this catalytic mechanism, and also highlighted the singular reactivity of the nucleophilic Cys68 thiolate towards electrophiles, such as iodo- or bromo-acetamide, in comparison with the other cysteine residues39. Additionally, the position and the structural functionality of the other cysteines in human NAT isoenzymes were determined via crystallography and helped visualize key intramolecular interactions in protein folding48.
Considering the tautomeric species of apocynin (1) (Fig. 4a), the two possible modes of NADPH oxidase inhibition by apocynin and the role of cysteine residues in NATs, a couple of hypotheses can be proposed concerning the inhibition of NAT enzymes by apocynin in liver cell homogenates: the nucleophilic Cys68 thiolate could react with the apocynin quinonemethide tautomer (2) via Michael addition (Fig. 4b), possibly generating a covalent inactive adduct; alternatively, the other cysteine thiols (3 to 4) in rat NATs could be oxidized by radical apocynin metabolites (3), probably disrupting correct NAT folding (Fig. 4c). Also, apocynin can be hypothesized to act as a non-covalent competitive inhibitor ligand of rat NAT enzymes for its similarity in steric size or chemical shape to known arylamine NAT substrates, such as 4-aminoveratrole27.
Figure 4. Hypothetical mechanisms of NAT inhibition by apocynin.
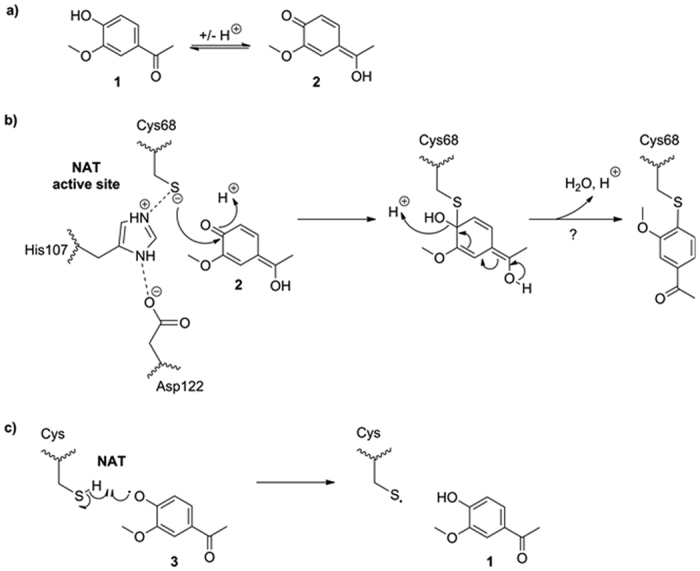
(a) Tautomers of apocynin (1). (b) Possible Michael addition between Cys68 thiolate and apocynin quinonemethide tautomer (2) within NAT active site. Cys68, His107 and Asp122 constitute the catalytic triad in mammalian NAT enzymes46,47. (c) Possible interaction between exposed thiols groups of NATs and radical apocynin metabolites (3).
Structural simulations and biochemical validation of preliminary hypotheses
Modelling 1 and 2 or visualizing the position of oxidisable cysteines within rat NAT protein structures would be ideal to examine our hypotheses. To date, only crystal structures of human NAT enzymes are available as mammalian species48; nevertheless, they have constituted highly reliable templates for inhibitor studies involving rodent NATs because of their high percentages of residue identity (>70%) (Table 1, Supplementary Figure S1)27,49,50.
Therefore, modelling 1 and 2 into human NAT structures was attempted to explore possible distinctive intermolecular interactions between the ligand and human NAT enzymes, as models for rodent NATs. Initially, 2 was successfully docked in the crystal structures of human NATs ((HUMAN)NAT1: PDB 2PQT, (HUMAN)NAT2: PDB 2PFR)48, and high reliable results were obtained. In particular, the affinity energies (kcal.mol−1) of all the simulations with (HUMAN)NAT2 structure were lower than those with (HUMAN)NAT1, which would implicate a better affinity of 2 for (HUMAN)NAT2 than the (HUMAN)NAT1 isoform.
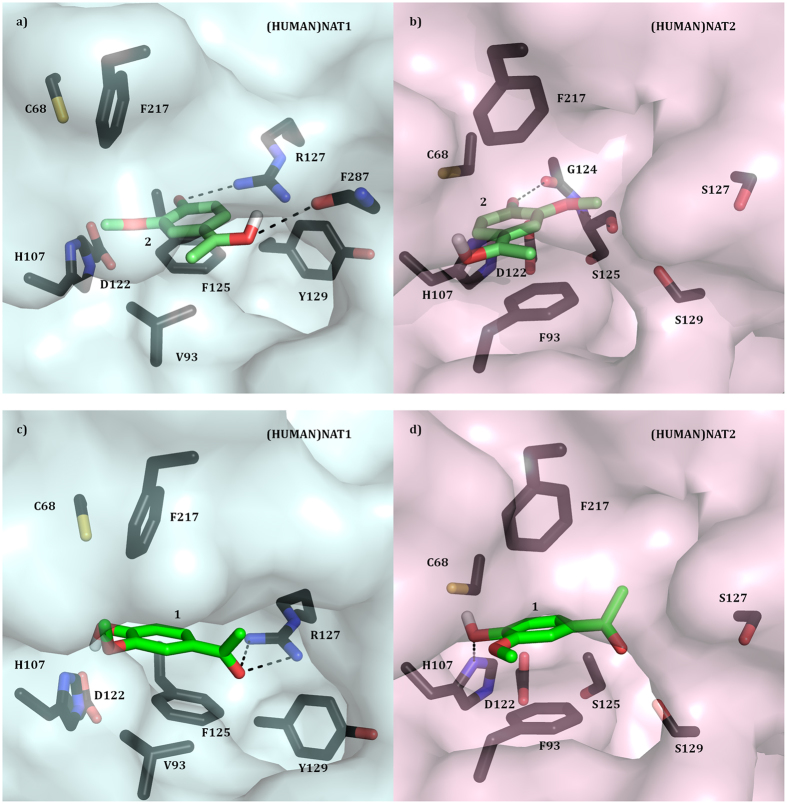
In one of the best simulations obtained, 2 appeared to fit within the active site of both human NATs consistent with our mechanistic hypothesis proposing a Michael addition reaction between the ligand and the enzyme (Figs 4b and 5a,b). Moreover, the compound appeared to be accommodated more internally in (HUMAN)NAT2 active site than that of (HUMAN)NAT1, which seemed coherent with the affinity energy values obtained. In details, the carbonyl carbon of the ligand was shown to point towards the thiolate functionality of C68 (4 Å (HUMAN)NAT2, 5 Å (HUMAN)NAT1), and its planar quinone ring to be sandwiched hydrophobically between the benzyl rings of two opposite phenylalanine residues (F93 in (HUMAN)NAT2, and F125 & F217 in (HUMAN)NAT1, 3–4 Å) (Fig. 5a,b). Also, the polar functionalities of 2 were proposed to interact with some polar groups within the active site via hydrogen bonds: the carbonyl O of the ligand with G124 side chain carbonyl in (HUMAN)NAT2 (3 Å) or R127 arginine guanidine in (HUMAN)NAT1 (3 Å); the methoxy O of 2 with the side chain carbonyl of S125 in (HUMAN)NAT2 (3 Å) or I106 in (HUMAN)NAT1 (3 Å) (Fig. 5a,b).
Figure 5. Substrate binding pockets of (HUMAN)NAT1 and (HUMAN)NAT2 with apocynin tautomers docked.
(a,b) The active site of (HUMAN)NAT1 (a) and (HUMAN)NAT2 (b)48 with docked apocynin quinonemethide tautomer (2) is shown in surface and stick representation respectively. The residues involved in ligand binding, substrate catalysis and substrate selectivity are shown in stick representation and labelled with carbon atoms in dark blue (a) or dark pink (b), nitrogen in blue, oxygen in red, and sulphur in orange. 2 is labelled with carbon atoms in green, oxygen in red, and hydrogen in white. (c,d) The active site of (HUMAN)NAT1 (c) and (HUMAN)NAT2 (d)48 with docked 1 is shown in surface and stick representation respectively. The residues involved in ligand binding, substrate catalysis and substrate selectivity are shown in stick representation and labelled with carbon atoms in dark blue (a) or dark pink (b), nitrogen in blue, oxygen in red, and sulphur in orange. 1 is labelled with carbon atoms in green, oxygen in red, and hydrogen in white.
In order to substantiate this first preliminary hypothesis, dilution experiments were performed using different concentrations of apocynin. Following preincubation of liver cell lysates with apocynin, inhibition of NAT activity by this compound was reversed upon 1000-fold dilution (Fig. 6). This would not support our first mechanistic proposition that apocynin could act as irreversible inhibitor of NAT enzymes. Nevertheless, further experiments with pure recombinant NAT proteins would be helpful.
Figure 6. Reversible inhibition of liver NAT activity by apocynin.
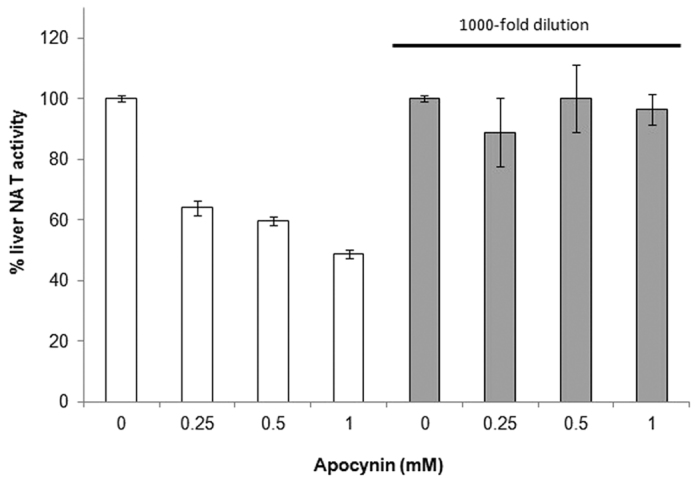
Apocynin inhibition of liver NAT activity appears to be reversible upon dilution. Liver lysates were incubated with varying amounts of apocynin (0–1 mM) in the assay for 30 min. The samples were subsequently diluted 1000 times, and evaluation of residue liver NAT activity was conducted using AcCoA and pANS as substrates, as described in methods. Assays were conducted in triplicates.
To date, panoply of diverse sulphydryl reactive compounds has been used as irreversible inhibitors of mammalian NATs, only to elucidate the involvement of a Cys residue in the catalytic mechanism of NATs39,51,52,53,54,55 or propose hypotheses for the putative endogenous role of human NATs56,57,58. Other recent studies have also shown irreversible inhibition of human NAT isoforms by different heavy metals59,60, and some of the oxidizing products resulting from the oxidation metabolism of aromatic amines61. The eventuality that apocynin could act as covalent inhibitor of NAT enzymes would have offered some advantages in drug design, since specific covalent inhibitors are usually associated with lower doses and a longer duration of action, and avoiding resistance62.
Additionally, reliable docking results were successfully obtained when 1 (which is the most stable tautomer of apocynin in solution) was docked into the active site of both human NATs63, with a general better affinity of the ligand for (HUMAN)NAT2 for the lower affinity energies estimated. In the simulations with both human NATs, the benzene ring of the compound appeared to lay between the benzyl rings of two opposite phenylalanine residues (as shown in previous simulations with 2) via hydrophobic π-π stacking (3 Å), and its hydroxyl tail can form a hydrogen bridge with C68 thiolate (4 Å) (Fig. 5c,d). Distinctively, the hydroxyl functionality of 1 seemed to interact also with H106 side chain imidazole nitrogen via a hydrogen bond in (HUMAN)NAT2 (Fig. 5d), whereas 1 carbonyl functionality was proposed to be locked by R127 side chain guanidine via two hydrogen bridges in (HUMAN)NAT1 (Fig. 5c).
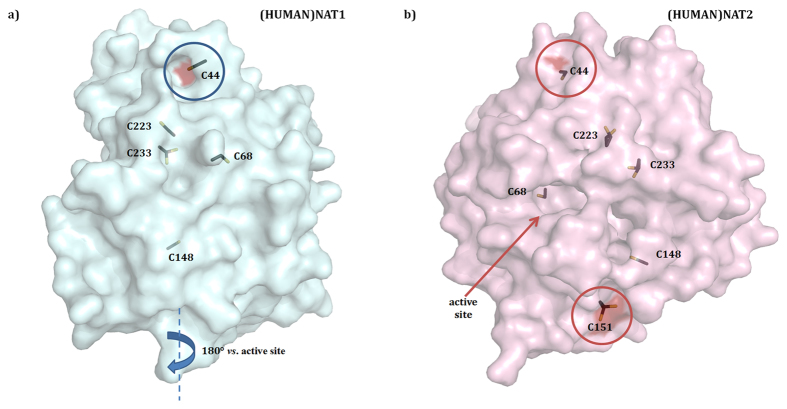
Thirdly, in order to evaluate the alternative hypothesis that the radical species 3 generated by MPO can inhibit NAT enzymes via oxidation of cysteine thiols (Fig. 4c), like it was earlier suggested for the cytosolic components of the NADPH oxidase complex7,44,45, the positions of cysteine residues in human NAT structures were examined, since most of the cysteines in mammalian NAT sequences are identical, except the additional C49 in (RAT)NAT1 (Table 2, Supplementary Figure S1). In both human NAT crystal structures, most of the cysteines are interlocked inside the protein and thus are unlikely to react with 3 for oxidation. Only C44 thiol functionality looks to point outward the protein surface (Fig. 7a,b) and can be available for oxidation by 3: this might modify protein unfolding and subsequently inactivate the enzyme. Indeed, in both human NAT proteins, C44 thiol seems to form a hydrogen bridge with the L40 side chain carbonyl oxygen, which in turn interacts with the side chain polar groups of residues N41 (NH), I42 (NH), H43 (CO) via additional hydrogen bonds. This intricate intramolecular scaffolding appears to be consistent with the hypothesis that C44 plays a key role in correct protein folding, whose alteration upon oxidation can compromise the activity of the enzyme.
Table 2. Comparison of Cysteine Residues in Mouse and Rat NATs versus Human NAT Isoenzymes.
| Residues | ||||||
|---|---|---|---|---|---|---|
| 44 | 68 | 148 | 151 | 223 | 233 | |
| (HUMAN)NAT1 | C | C | C | R | C | C |
| (RAT)NAT2 | C | C | A | R | C | C |
| (MOUSE)NAT2 | C | C | A | R | C | C |
| (MOUSE)NAT1 | C | C | A | L | C | C |
| (RAT)NAT1 | C | C | A | R | C | C |
| (HUMAN)NAT2 | C | C | C | C | C | C |
Figure 7. Position of cysteine residues in human NAT crystal structures.
Overall surface structures of (HUMAN)NAT1 (a) and (HUMAN)NAT2 (b) are shown with cysteine side chains highlighted in stick representation as modelled in the original PDB files48. Sections of protein surface corresponding to outward cysteine thiols are coloured in dark red.
The key residues here highlighted in the formation of the complex human NAT-apocynin are analogously conserved in all rodent NAT sequences. However, additional studies about functional selectivity of rat NATs can be useful to better establish structural analogies between rat and human NAT isoenzymes and build virtual models of rat NAT proteins based on human NAT structures27.
In general, the complex environment of cellular lysates would demand further experiments to depict the molecular mechanism of inhibition of rat NAT activity by apocynin. Also, it is important to ascertain whether apocynin selectively affects NAT functionality in liver against a larger panel of other liver-specific enzymes, as either reactive radical or unmetabolised species, or whether indirectly down-regulates NAT gene expression levels upon oral administration. Also, studies examining NAT and myeloperoxidase activities in liver homogenates can contribute to understand the mode of inhibition of NAT by apocynin.
As (RAT)NAT1 gene is much higher expressed in the liver than (RAT)NAT233and has an expression pattern similar to those of (HUMAN)NAT2 and (MOUSE)NAT1, which give homologous phase-II drug metabolizing enzyme products27,64, it can be suggested that most of the NAT activity encountered in rat liver homogenates can be related to (RAT)NAT1 (Table 1)22. It is therefore crucial to evaluate the selectivity of the inhibitory potency of apocynin against each rat NAT enzyme and establish whether apocynin might be also a potent isoenzyme-specific inhibitor against a panel of other pure mammalian NAT isoenzymes to better characterize its mode of inhibition and unravel its pharmacological potential.
As association between human NATs and different types of cancer has proposed the enzymes as possible pharmacological targets28,65, the evidence of apocynin acting as in vivo inhibitor of rat liver NAT activity might be of particular interest in pharmacology: importantly, apocynin has a good safety profile in all animal30,31,32 and human66,67 experiments reported to date, and the inactivation of NAT enzymes by apocynin can be useful to explore its potential chemopreventive effects towards arylamine carcinogenesis, since no evidence has been reported to date.
In summary, the current study shows for the first time that apocynin, a non-toxic natural organic antioxidant, constitutes a potent inhibitor of NAT activity in rat livers both in vivo and in vitro experiments. Following virtual modelling analyses, thiol functionalities in NATs are hypothesized to play an important role for apocynin binding to the enzyme. Since NAT enzymes in humans have been associated with a wide range of cancers and proposed as potential drug targets38,49,64, looking for possible NAT-selective inhibitors from the available range of non-toxic phytochemicals can constitute a valid approach to develop novel chemopreventive agents.
Methods
General Experimental Procedures
All chemical reagents were of analytical grade. Apocynin (Acetovanillone) was purchased from Sigma-Aldrich, MO, USA; purity >98%. Complete Protease Inhibitor Cocktail tablets, EDTA free, were purchased from Roche Diagnostics, GmbH, Mannheim, Germany. All other chemicals were purchased from Sigma-Aldrich, MO, USA, unless otherwise stated.
Animal Care
The animals used were housed and cared for at the animal house at the University of the West Indies Mona. Experimental procedures on the animals were conducted after receiving ethical approval from UHWI/UWI/FMS ethics committee (protocol number AN 1 11/12). The principles of laboratory animal care were followed in accordance with specific national laws where applicable. Eighteen (18) adult male Sprague-Dawley rats of similar age (20 weeks), weighing 250–300 g, were divided into three (3) groups, with 6 rats per each group, as previously described32. The animals were housed in the animal house, under standard conditions of 12-h light/dark cycle. The group of untreated rats (1) was fed a normal diet consisting of rat chow and tap water ad libitum. Groups 2 and 3 were fed the normal diet together with apocynin 50 mg or 100 mg/kg/day respectively via gavages. The rats were treated for 8 weeks until they were sacrificed. We maintained an aseptic environment throughout the course of study at the best possible conditions.
Liver Homogenate Preparation
4 g of liver tissue were dissected from each adult rat immediately after cervical dislocation and washed quickly in PBS. The tissues were minced and homogenized at 440 rpm (25% w/v) in chilled buffer (10 mM KPB, pH 7.4) containing 0.15 M KCl and 0.1 mM PMSF protease inhibitors (Roche Diagnostics, GmbH, Mannheim, Germany). The suspensions were spun in a refrigerated (4 °C) centrifuge at 9000 g for 20 min. The supernatants from each tube were labelled as S9 fractions and stored in −80 °C for further analysis.
Protein Quantification
The amounts of proteins in all S9 fractions were determined using a modified Lowry assay kit (Thermo Scientific, Rockford IL, USA) in triplicates, as previously described68.
Determination of Arylamine N-Acetyltransferase Activity
NAT activities in crude tissue homogenates were carried out using a modified method with pANS as substrate40. Routinely, reactions were conducted in triplicate at 37 °C in a final volume of 200 μL which contained 100 μL of an appropriately diluted S9 fraction such that NAT activity was within a linear range, with 40 μL of 1 mM pANS in Tris-EDTA buffer (20 mM Tris-HCl, 10 mM NaCl, 1 mM EDTA and pH 7.5) and 40 μL of 1 mM acetyl coenzyme A (AcCoA) was added to start the reaction. After 30 minutes the reaction was quenched by adding 100 μL of 30% (w/v) cold trichloroacetic acid. The assay mixture was centrifuged to remove possibly precipitated protein and 200 μL of 5% (w/v) DMAB were subsequently added. This allowed the spectrophotometric detection of unacetylated arylamine substrate at 450 nm using a UV-spectrophotometer (μQuant universal microplate spectrophotometer, Bio-Tek Instruments, Winooski, VT, USA). pANS was chosen as arylamine substrate in all assays for its widespread reactivity with all mouse NAT enzymes homologues27. Enzymatic NAT activity values are expressed as μmols of N-acetylated p-ANS/min/μg of liver protein.
Assessment of Inhibition of NAT Activity by Apocynin
For detection of inhibition of NAT activity by apocynin, minor modifications were introduced to the arylamine acetylation assay protocol used40. N-acetylation of pANS by liver homogenates was measured in the presence of apocynin following preincubation of the appropriately diluted S9 fractions (9.60 μg protein) with either apocynin (0.49 mM, 0, 5 or 15 min) or AcCoA (0.20 mM, 5 or 15 min).
Varying concentrations of apocynin (0 and 7.5 mM) were also assayed in vitro against NAT activity from diluted liver S9 fractions (9.60 μg protein) of untreated rats. Percentage inhibition was determined as the ratio of specific activity with the inhibitor related to specific activity without inhibitor. Assays were conducted in triplicate and all values are expressed as mean ± standard deviation values. To assess the nature of binding, liver lysates were incubated with varying concentrations of apocynin (0–1 mM) for 30 min prior to dilution by 1000 fold. Each sample was then assessed for residual NAT activity as earlier described.
Data Analysis
Statistical significance of differences between average values was determined by ANOVA, the order of significance was determined using the Newman-Kuel test and the level of significance was set at p = 0.05. IC50 values were determined by plotting % inhibition values against log([apocynin(mM)]) and fitting the data in Sigma Plot software (version 10.0) and in the enzyme kinetics module (version 1.3), using a non-linear regression analysis model.
Virtual Modelling Simulations
Protein sequence alignments and comparisons were performed by using ClustalW2 from the European Bioinformatics Institute69. Apocynin tautomers were drawn in 3D using ChemBio3D Ultra 12.0 and its ground state conformation predicted before it was docked into the catalytic pocket of the human NAT structures ((HUMAN)NAT1: PDB 2PQT, (HUMAN)NAT2: PDB 2PFR)48. Protein and substrate structures were defined as pdbqt files and protein-structure interactions were analyzed using Autodock Vina70. After adding polar hydrogen atoms to the NAT protein and defining the rotatable bonds of the ligand, the active site pocket was defined as the docking site and possible solutions were ranked according to affinity energy (lowest to highest; kcal.mol−1). The docking results were visualized in 3D using PyMOL71.
Additional Information
How to cite this article: Francis, S. et al. Treatment of Rats with Apocynin Has Considerable Inhibitory Effects on Arylamine N-Acetyltransferase Activity in the Liver. Sci. Rep. 6, 26906; doi: 10.1038/srep26906 (2016).
Supplementary Material
Acknowledgments
We are grateful to D. Douglas and A. Baker for technical assistance, Prof. E. Sim, University of Oxford, for constructive comments and input, and Dr. P. Brown, UWI, for kind donation of some research materials and tools. We are also grateful to National Health Fund (Jamaica) and Forest Conservation Fund (Jamaica) for financial support.
Footnotes
Author Contributions S.F. conducted biological and biochemical assays and contributed to writing the manuscript. N.L. conducted in silico modelling and NAT schematics and contributed to writing the manuscript. C.N. and R.D. designed experiments and contributed to writing the manuscript.
References
- Stefanska J. & Pawliczak R. Apocynin: Molecular Aptitudes. Mediators of Inflammation 2008, 106507, 10.1155/2008/106507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa C. A. et al. Antioxidant Treatment With Tempol and Apocynin Prevents Endothelial Dysfunction and Development of Renovascular Hypertension. American Journal of Hypertension 22, 1242–1249, 10.1038/ajh.2009.186 (2009). [DOI] [PubMed] [Google Scholar]
- Simons J. M., Hart B. A., Ching T. R. I. V., Van Dijk H. & Labadie R.P. Metabolic activation of natural phenols into selective oxidative burst agonists by activated human neutrophils. Free Radical Biol. Med. 8, 251–258 (1990). [DOI] [PubMed] [Google Scholar]
- de Almeida A. C., dos Santos Vilela M. M., Condino-Neto A. & Ximenes V. F. The Importance of Myeloperoxidase in Apocynin-Mediated NADPH Oxidase Inhibition. ISRN Inflammation 2012, 7, 10.5402/2012/260453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenes V. F., Kanegae M. P. P., Rissato S. R. & Galhiane M. S. The oxidation of apocynin catalyzed by myeloperoxidase: Proposal for NADPH oxidase inhibition. Archives of Biochemistry and Biophysics 457, 134–141 (2007). [DOI] [PubMed] [Google Scholar]
- Kanegae M. P., da Fonseca L. M., Brunetti I. L., de Oliveira Silva S. & Ximenes V. F. The reactivity of ortho-methoxy-substituted catechol radicals with sulfhydryl groups: contribution for the comprehension of the mechanism of inhibition of NADPH oxidase by apocynin. Biochemical Pharmacology 74, 457–464 (2007). [DOI] [PubMed] [Google Scholar]
- Stolk J., Hiltermann T. J., Dijkman J. H. & Verhoeven A. J. Characteristics of the inhibition of NADPH oxidase activation in neutrophils by apocynin, a methoxy-substituted catechol. American Journal of Respiratory Cell and Molecular Biology 11, 95–102 (1994). [DOI] [PubMed] [Google Scholar]
- Riganti C., Costamagna C., Bosia A. & Ghigo D. The NADPH oxidase inhibitor apocynin (acetovanillone) induces oxidative stress. Toxicology and Applied Pharmacology 212, 179–187 (2006). [DOI] [PubMed] [Google Scholar]
- Wang Q. et al. Apocynin protects against global cerebral ischemia–reperfusion-induced oxidative stress and injury in the gerbil hippocampus. Brain Research 1090, 182–189 (2006). [DOI] [PubMed] [Google Scholar]
- Yu J., Weïwer M., Linhardt R. J. & Dordick J. S. The role of the methoxyphenol apocynin, a vascular NADPH oxidase inhibitor, as a chemopreventative agent in the potential treatment of cardiovascular diseases. Current Vascular Pharmacology 6, 204–217 (2008). [DOI] [PubMed] [Google Scholar]
- Chirino Y. I., Sánchez-González D. J., Martínez-Martínez C. M., Cruz C. & Pedraza-Chaverri J. Protective effects of apocynin against cisplatin-induced oxidative stress and nephrotoxicity. Toxicology 245, 18–23 (2008). [DOI] [PubMed] [Google Scholar]
- Doddo J. M. & Pearse D. B. Effect of the NADPH oxidase inhibitor apocynin on ischemia-reperfusion lung injury. American Journal of Physiology-Heart and Circulatory Physiology 279, H303–H312 (2000). [DOI] [PubMed] [Google Scholar]
- Pedroso L. S., Fávero G. M., de Camargo L. E. A., Mainardes R. M. & Khalil N. M. Effect of the o-methyl catechols apocynin, curcumin and vanillin on the cytotoxicity activity of tamoxifen. Journal of Enzyme Inhibition and Medicinal Chemistry 28, 734–740 (2013). [DOI] [PubMed] [Google Scholar]
- Suzuki S. et al. Apocynin, an NADPH oxidase inhibitor, suppresses rat prostate carcinogenesis. Cancer Science 104, 1711–1717 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badal S. et al. Cytochrome P450 1 enzyme inhibition and anticancer potential of chromene amides from Amyris plumieri. Fitoterapia 82, 230–236 (2011). [DOI] [PubMed] [Google Scholar]
- Francis S. & Delgoda R. A patent review on the development of human cytochrome P450 inhibitors. Expert opinion on therapeutic patents 24, 699–717 (2014). [DOI] [PubMed] [Google Scholar]
- Francis S., Morris D., Shields M., Jacobs H. & Delgoda R. In Natural Products: Structure, Bioactivity and Applications (ed Ramiro E. Goncalves & Marcos Cunha Pinto ) 35–54 (Nova Publisher, 2012). [Google Scholar]
- Hu R. & Kong A. N. Activation of MAP kinases, apoptosis and nutrigenomics of gene expression elicited by dietary cancer-prevention compounds. Nutrition 20, 83–88 (2004). [DOI] [PubMed] [Google Scholar]
- Pietersma A. et al. Evidence against the involvement of multiple radical generating sites in the expression of the vascular cell adhesion molecule-1. Free radical research 28, 137–150 (1998). [DOI] [PubMed] [Google Scholar]
- Swart P., van der Merwe K. J., Swart A. C., Todres P. C. & Hofmeyr J. Inhibition of cytochrome P-450 (11) beta by some naturally occurring acetophenones and plant extracts from the shrub Salsola tuberculatiformis. Planta medica 59, 139–143 (1993). [DOI] [PubMed] [Google Scholar]
- Sim E., Fakis G., Laurieri N. & Boukouvala S. Arylamine N-acetyltransferases–from drug metabolism and pharmacogenetics to identification of novel targets for pharmacological intervention. Adv Pharmacol 63, 169–205 (2012). [DOI] [PubMed] [Google Scholar]
- Walraven J. M., Doll M. A. & Hein D. W. Identification and characterization of functional rat arylamine N-acetyltransferase 3: comparisons with rat arylamine N-acetyltransferases 1 and 2. J Pharmacol Exp Ther 319, 369–375, 10.1124/jpet.106.108399 (2006). [DOI] [PubMed] [Google Scholar]
- Walraven J. M., Barker D. F., Doll M. A. & Hein D. W. Tissue expression and genomic sequences of rat N-acetyltransferases rNat1, rNat2, rNat3, and Functional characterization of a novel rNat3*2 genetic variant. Toxicol Sci 99, 413–421, 10.1093/toxsci/kfm159 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura A. et al. Mouse N-acetyltransferase type 2, the homologue of human N-acetyltransferase type 1. Biochemical Pharmacology 75, 1550–1560, 10.1016/j.bcp.2007.12.012 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fretland A. J., Doll M. A., Gray K., Feng Y. & Hein D. W. Cloning, sequencing, and recombinant expression of NAT1, NAT2, and NAT3 derived from the C3H/HeJ (rapid) and A/HeJ (slow) acetylator inbred mouse: functional characterization of the activation and deactivation of aromatic amine carcinogens. Toxicology and Applied Pharmacology 142, 360–366, 10.1006/taap.1996.8036 (1997). [DOI] [PubMed] [Google Scholar]
- Boukouvala S., Price N. & Sim E. Identification and functional characterization of novel polymorphisms associated with the genes for arylamine N-acetyltransferases in mice. Pharmacogenetics 12, 385–394 (2002). [DOI] [PubMed] [Google Scholar]
- Laurieri N. et al. Differences between murine arylamine N-acetyltransferase type 1 and human arylamine N-acetyltransferase type 2 defined by substrate specificity and inhibitor binding. BMC Pharmacology and Toxicology 15, 68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher N. J. & Minchin R. F. Arylamine N-acetyltransferase 1: a novel drug target in cancer development. Pharmacol Rev 64, 147–165, 10.1124/pr.110.004275 (2012). [DOI] [PubMed] [Google Scholar]
- Kukongviriyapan V. et al. Inhibitory effects of polyphenolic compounds on human arylamine N-acetyltransferase 1 and 2. Xenobiotica 36, 15–28, 10.1080/00498250500489901 (2006). [DOI] [PubMed] [Google Scholar]
- Daly J. W., Axelrod J. & Witkop B. Dynamic aspects of enzymatic O-methylation and-demethylation of catechols in vitro and in vivo. Journal of Biological Chemistry 235, 1155–1159 (1960). [PubMed] [Google Scholar]
- ‘t Hart B. A., Copray S. & Philippens I. Apocynin, a Low Molecular Oral Treatment for Neurodegenerative Disease. BioMed research international 2014, 10.1155/2014/298020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwokocha C. R. et al. Apocynin ameliorates cadmium-induced hypertension through elevation of endothelium nitric oxide synthase. Cardiovascular Toxicology 13, 357–363 (2013). [DOI] [PubMed] [Google Scholar]
- Barker D. F. et al. Quantitative tissue and gene-specific differences and developmental changes in Nat1, Nat2, and Nat3 mRNA expression in the rat. Drug Metab Dispos 36, 2445–2451, 10.1124/dmd.108.023564 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland R. A. In Evaluation of Enzyme Inhibitors in Drug Discovery 57–121 (John Wiley & Sons, Inc., 2013). [Google Scholar]
- Elbarbry F. A., McNamara P. J. & Alcorn J. Ontogeny of hepatic CYP1A2 and CYP2E1 expression in rat. J Biochem Mol Toxicol 21, 41–50, 10.1002/jbt.20156 (2007). [DOI] [PubMed] [Google Scholar]
- Walker N. J. et al. Rat CYP1B1: an adrenal cytochrome P450 that exhibits sex-dependent expression in livers and kidneys of TCDD-treated animals. Carcinogenesis 16, 1319–1327, 10.1093/carcin/16.6.1319 (1995). [DOI] [PubMed] [Google Scholar]
- Francis S., Nwokocha C. R. & Delgoda R. In In-vitro Bioassays and Chromatographic Analyses Used to Screen Natural Products from Jamaica in the 21st Century (ed Green C. E. ) 49–60 (Research Signpost, 2015). [Google Scholar]
- Laurieri N. et al. Small molecule colorimetric probes for specific detection of human arylamine N-acetyltransferase 1, a potential breast cancer biomarker. J Am Chem Soc 132, 3238–3239, 10.1021/ja909165u (2010). [DOI] [PubMed] [Google Scholar]
- Wang H., Liu L., Hanna P. E. & Wagner C. R. Catalytic mechanism of hamster arylamine N-acetyltransferase 2. Biochemistry 44, 11295–11306, 10.1021/bi047564q (2005). [DOI] [PubMed] [Google Scholar]
- Andres H. H., Klem A. J., Szabo S. M. & Weber W. W. New spectrophotometric and radiochemical assays for acetyl-CoA: arylamine N-acetyltransferase applicable to a variety of arylamines. Analytical biochemistry 145, 367–375 (1985). [DOI] [PubMed] [Google Scholar]
- Brooke E. W. et al. An approach to identifying novel substrates of bacterial arylamine N-acetyltransferases. Bioorganic & Medicinal Chemistry 11, 1227–1234, 10.1016/s0968-0896(02)00642-9 (2003). [DOI] [PubMed] [Google Scholar]
- Hart B. A. et al. Reaction products of 1-naphthol with reactive oxygen species prevent NADPH oxidase activation in activated human neutrophils, but leave phagocytosis intact. Free Radic Biol Med 8, 241–249 (1990). [DOI] [PubMed] [Google Scholar]
- Clark R. A., Volpp B. D., Leidal K. G. & Nauseef W. M. Two cytosolic components of the human neutrophil respiratory burst oxidase translocate to the plasma membrane during cell activation. J Clin Invest 85, 714–721, 10.1172/JCI114496 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-Pale M., Weïwer M., Yu J., Linhardt R. J. & Dordick J. S. Inhibition of human vascular NADPH oxidase by apocynin derived oligophenols. Bioorganic & medicinal chemistry 17, 5146–5152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon H. & Boo Y. C. Laminar shear stress enhances endothelial cell survival through a NADPH oxidase 2-dependent mechanism. Biochemical and Biophysical Research Communications 430, 460–465 (2013). [DOI] [PubMed] [Google Scholar]
- Westwood I. M. et al. Structure and mechanism of arylamine N-acetyltransferases. Curr Top Med Chem 6, 1641–1654 (2006). [DOI] [PubMed] [Google Scholar]
- Delgoda R., Lian L.-Y., Sandy J. & Sim E. NMR investigation of the catalytic mechanism of arylamine N-acetyltransferase from Salmonella typhimurium. Biochimica et Biophysica Acta (BBA)-General Subjects 1620, 8–14, 10.1016/S0304-4165(02)00500-7 (2003). [DOI] [PubMed] [Google Scholar]
- Wu H. et al. Structural basis of substrate-binding specificity of human arylamine N-acetyltransferases. J Biol Chem 282, 30189–30197 (2007). [DOI] [PubMed] [Google Scholar]
- Laurieri N. et al. A novel color change mechanism for breast cancer biomarker detection: naphthoquinones as specific ligands of human arylamine N-acetyltransferase 1. PLoS One 8, e70600, 10.1371/journal.pone.0070600 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egleton J. E. et al. Structure–activity relationships and colorimetric properties of specific probes for the putative cancer biomarker human arylamine N-acetyltransferase 1. Bioorganic & Medicinal Chemistry 22, 3030–3054, 10.1016/j.bmc.2014.03.015 (2014). [DOI] [PubMed] [Google Scholar]
- Wang H., Vath G. M., Gleason K. J., Hanna P. E. & Wagner C. R. Probing the mechanism of hamster arylamine N-acetyltransferase 2 acetylation by active site modification, site-directed mutagenesis, and pre-steady state and steady state kinetic studies. Biochemistry 43, 8234–8246 (2004). [DOI] [PubMed] [Google Scholar]
- Cheon H. G., Boteju L. W. & Hanna P. E. Affinity alkylation of hamster hepatic arylamine N-acetyltransferases: isolation of a modified cysteine residue. Mol Pharmacol 42, 82–93 (1992). [PubMed] [Google Scholar]
- Cheon H. G. & Hanna P. E. Effect of group-selective modification reagents on arylamine N-acetyltransferase activities. Biochem Pharmacol 43, 2255–2268 (1992). [DOI] [PubMed] [Google Scholar]
- Guo Z., Wagner C. R. & Hanna P. E. Mass Spectrometric Investigation of the Mechanism of Inactivation of Hamster Arylamine N-Acetyltransferase 1 by N-Hydroxy-2-Acetylaminofluorene. Chemical Research in Toxicology 17, 275–286, 10.1021/tx030045o (2004). [DOI] [PubMed] [Google Scholar]
- Sticha K. R., Bergstrom C. P., Wagner C. R. & Hanna P. E. Characterization of hamster recombinant monomorphic and polymorphic arylamine N-acetyltransferases: bioactivation and mechanism-based inactivation studies with N-hydroxy-2-acetylaminofluorene. Biochem Pharmacol 56, 47–59 (1998). [DOI] [PubMed] [Google Scholar]
- Voice R. A., Manis M. & Weber W. W. Inhibition of human red blood cell N-acetyltransferase. Drug Metab Dispos 21, 181–183 (1993). [PubMed] [Google Scholar]
- Dairou J., Atmane N., Dupret J. M. & Rodrigues-Lima F. Reversible inhibition of the human xenobiotic-metabolizing enzyme arylamine N-acetyltransferase 1 by S-nitrosothiols. Biochem Biophys Res Commun 307, 1059–1065 (2003). [DOI] [PubMed] [Google Scholar]
- Dairou J., Atmane N., Rodrigues-Lima F. & Dupret J. M. Peroxynitrite irreversibly inactivates the human xenobiotic-metabolizing enzyme arylamine N-acetyltransferase 1 (NAT1) in human breast cancer cells: a cellular and mechanistic study. J Biol Chem 279, 7708–7714 (2004). [DOI] [PubMed] [Google Scholar]
- Deng Z. J. et al. Interaction of human arylamine N-acetyltransferase 1 with different nanomaterials. Drug Metab Dispos 42, 377–383, 10.1124/dmd.113.055988 (2014). [DOI] [PubMed] [Google Scholar]
- Sanfins E. et al. Carbon black nanoparticles impair acetylation of aromatic amine carcinogens through inactivation of arylamine N-acetyltransferase enzymes. ACS Nano 5, 4504–4511, 10.1021/nn103534d (2011). [DOI] [PubMed] [Google Scholar]
- Liu L., Wagner C. R. & Hanna P. E. Human arylamine N-acetyltransferase 1: in vitro and intracellular inactivation by nitrosoarene metabolites of toxic and carcinogenic arylamines. Chemical Research in Toxicology 21, 2005–2016, 10.1021/tx800215h (2008). [DOI] [PubMed] [Google Scholar]
- Singh J., Petter R. C., Baillie T. A. & Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov 10, 307–317, 10.1038/nrd3410 (2011). [DOI] [PubMed] [Google Scholar]
- Turner A. B. Quinone methides. Quarterly Reviews, Chemical Society 18, 347–360, 10.1039/qr9641800347 (1964). [DOI] [Google Scholar]
- Sim E., Abuhammad A. & Ryan A. Arylamine N-acetyltransferases: from drug metabolism and pharmacogenetics to drug discovery. Br J Pharmacol 171, 2705–2725, 10.1111/bph.12598 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein D. W. N-acetyltransferase SNPs: emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Expert Opinion on Drug Metabolism & Toxicology 5, 353–366, 10.1517/17425250902877698 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanska J. et al. Apocynin decreases hydrogen peroxide and nitrate concentrations in exhaled breath in healthy subjects. Pulm Pharmacol Ther 23, 48–54, 10.1016/j.pupt.2009.09.003 (2010). [DOI] [PubMed] [Google Scholar]
- Stefanska J. et al. Apocynin reduces reactive oxygen species concentrations in exhaled breath condensate in asthmatics. Exp Lung Res 38, 90–99, 10.3109/01902148.2011.649823 (2012). [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L. & Randall R. J. Protein measurement with the Folin phenol reagent. J biol Chem 193, 265–275 (1951). [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O. & Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31, 455–461, 10.1002/jcc.21334 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L. PyMOL: An open-source molecular graphics tool. DeLano Scientific, San Carlos, California, USA; http://www.ccp4.ac.uk/newsletters/newsletter40/11_pymol.pdf (2002) (Date of access:15/11/2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.