Abstract
The molecular mechanisms that control male strobilus development in conifers are largely unknown because the developmental stages and related genes have not yet been characterized. The determination of male strobilus developmental stages will contribute to genetic research and reproductive biology in conifers. Our objectives in this study were to determine the developmental stages of male strobili by cytological and transcriptome analysis, and to determine the stages at which aberrant morphology is observed in a male-sterile mutant of Cryptomeria japonica D. Don to better understand the molecular mechanisms that control male strobilus and pollen development. Male strobilus development was observed for 8 months, from initiation to pollen dispersal. A set of 19,209 expressed sequence tags (ESTs) collected from a male reproductive library and a pollen library was used for microarray analysis. We divided male strobilus development into 10 stages by cytological and transcriptome analysis. Eight clusters (7324 ESTs) exhibited major changes in transcriptome profiles during male strobili and pollen development in C. japonica. Two clusters showed a gradual increase and decline in transcript abundance, respectively, while the other six clusters exhibited stage-specific changes. The stages at which the male sterility trait of Sosyun was expressed were identified using information on male strobilus and pollen developmental stages and gene expression profiles. Aberrant morphology was observed cytologically at Stage 6 (microspore stage), and differences in expression patterns compared with wild type were observed at Stage 4 (tetrad stage).
Keywords: cDNA microarray, conifer, Cryptomeria japonica, male reproduction
Introduction
Male strobili are one of the most important organs for conifers, and many studies associated with seed orchard management have reported on pollen development (Owens and Blake 1985, Bonnet-Masimbert and Webber 1995, Owens 2004, 2006, 2008, Fernando 2013). Reproduction in many conifers begins 5–10 years after planting (Williams 2009), and the number of male strobili and female cones fluctuates from year to year. Most conifers rarely bear male strobili or female cones in their juvenile period without artificial treatment to stimulate reproductive onset (Williams 2009). Male strobilus and female cone induction in many Pinaceae has been achieved through application of a gibberellin (GA4 and GA7) mixture in combination with various cultural treatments (Owens and Blake 1985). In addition to GA application, the effects of girdling and root pruning on induction of male strobili and female cones have also been studied (Pharis 1975, Owens and Blake 1985, Bonnet-Masimbert and Webber 1995, Almqvist 2003).
Male strobilus and pollen development in conifer species have been investigated, especially with respect to morphological traits (Hashizume 1962, Williams 2009). In Pinus taeda, the timing and pattern of initiation and differentiation of male strobili have been investigated (Greenwood 1980), and male strobilus development was divided into six stages based only on morphological traits, without observation of cytological traits (Bramlett and Bridgwater 1989, Williams 2009). Harrison and Slee (1992) reported on pollen cone differentiation in Pinus caribaea based on a microscopic study. The reproductive biology, especially with a focus on reproductive cycle and seed production, of western white pine (Pinus monticola Dougl.), lodgepole pine (Pinus contorta Dougl.), and western white larch (Larix occidentalis Nutt) has been reported (Owens 2004, 2006, 2008), and Fernando (2013) has reviewed these studies. Pollen and pollen wall development have also been well studied in conifers (Kurmann 1989, Rowley et al. 2000, Uehara and Sahashi 2000, Fernando et al. 2005, Owens et al. 2008). In addition to these cytological analyses, further studies that consider male strobilus development in connection with the genes involved are necessary to understand male reproduction in conifers.
Conifer species in which reproductive onset can be controlled with easy treatment are desirable for studies on reproductive biology. Male reproductive organs have been well studied in Cryptomeria japonica D. Don, Japanese cedar (Futamura et al. 2008, Moriguchi et al. 2012, Tsubomura et al. 2012, Ujino-Ihara et al. 2012, Kurita et al. 2013), because GA3 treatment has a strong effect on male reproductive onset. GA3 treatment of seedlings, even of 1-year-old individuals, enables male strobilus and female cone set (Hashizume 1959). Male strobili are easily induced by GA3 application in July, as are female cones from the end of July to the end of August (Hashizume 1959). Male strobilus and female cone formation are initiated about 1–2 months after GA3 treatment, and later development is similar to natural development (Hashizume 1962).
Fortunately, male-sterile mutants in C. japonica have been discovered from natural stands, and they have been classified into several types (ms-1, ms-2, ms-3, ms-4) based on cytological observation and results of test crosses (Saito et al. 1998, Hosoo et al. 2005, Ueuma et al. 2009, Miyajima et al. 2010, Miura et al. 2011, Moriguchi et al. 2014). Some male-sterile mutants are due to nuclear mutations controlled by a pair of recessive genes (Taira et al. 1999, Moriguchi et al. 2014). The characterization of male-sterile mutants in C. japonica may lead to understanding of the molecular mechanisms of male strobilus and pollen development.
The molecular mechanisms controlling reproductive onset in conifers are largely unknown, and genes related to male strobilus and pollen development have not yet been characterized, with some exceptions, including MADS-box genes (Katahata et al. 2014), FT (FLOWERING LOCUS T)-like genes (Klintenäs et al. 2012), gibberellin metabolism genes (Niu et al. 2014), and LEAFY (Dornelas and Rodriguez 2005, Shiokawa et al. 2008). In recent years, expressed sequence tags (ESTs) have been obtained on a large scale in C. japonica (Futamura et al. 2008, Mishima et al. 2014, Nose and Watanabe 2014). Expressed sequence tags expressed in male reproductive organs have been obtained from full-length cDNA libraries, and ∼20,000 ESTs are available to the public (Futamura et al. 2008). Transcriptome analysis using these ESTs may allow characterization of each stage in the male strobilus developmental process. In this study, our objectives were (i) the classification of male strobilus developmental stages by cytological and transcriptome analysis, and (ii) the determination of the aberrant stage or stages in the newly discovered male-sterile mutant ‘Sosyun’ to understand genes related to male strobilus and pollen development.
Materials and methods
Plant material and sampling
One wild type clone (WT1, 7-year-old seedling) was used for cytological analysis and another wild type clone (WT2, Usui2, 17-year-old graft) and a male-sterile mutant (MT, Sosyun, 4-year-old cutting) were used for cytological and gene expression analysis. They were planted in Hitachi, Ibaraki, Japan (36°69N, 140°69E; elevation 52 m).
On 4 July 2011, these individuals were sprayed with 100 ppm GA3 (Kyowa-Hakko, Japan) to promote setting of male strobili. Sampling of male strobili began at the end of August, when they started to develop, and continued to the end of the following March, the pollen dispersal season. At each sampling time, twigs of ∼3 cm in length with male strobili were collected from the top of the shoots of each tree. The materials for gene expression analysis were stored at −80 °C, and the materials for cytological analysis were fixed in a solution of formalin : acetic acid : alcohol (FAA, 5 : 5 : 90 by volume) and stored at 4 °C until use.
Cytological analysis
Male strobili of ordinary size were collected from the twigs fixed in FAA and were soaked in water for a week. Since the size of male strobili of C. japonica varied within one cluster, extremely large or small strobili were eliminated by eye. The samples were prepared following the modified method of the Kawamoto system (Kawamoto and Shimizu 2000). The samples were frozen in cooled hexane and freeze-embedded with 4–5% carboxymethyl cellulose in the coolant. A specially prepared adhesive film, Cryofilm (Leica, Tokyo, Japan), was fastened to the cut surface of the samples. Longitudinal sections ∼7 µm thick were cut from the embedded blocks using a Leica CM3050S research cryostat (Leica, Bensheim, Germany). The sections were stained with hematoxylin and eosin for cytological observation. The sections were observed under a Leica Leitz DMR microscope (Leica). Male strobilus developmental stages were defined by observation.
Suppression subtractive hybridization
A third wild type clone (WT3, Ohtawara1, 11-year-old graft) and a male-sterile mutant (MT, Sosyun, 11-year-old graft) were used for suppression subtractive hybridization (SSH) libraries. At the beginning of July 2006, these individuals were sprayed with 100 ppm GA3, and sampling of male strobili began on 19 September and continued to 17 October 2006. At each sampling time, twigs of ∼3 cm in length with male strobili were collected from the top of the shoots of each tree. The materials were stored at −80 °C until use. A PCR-Select cDNA Subtraction kit (Takara Bio Inc., Kusatsu, Japan) was used for SSH. First, we constructed four male strobili cDNA pools (cDNA pools 1, 2, 3 and 4); these were respectively derived from the pollen mother cell (PMC) stage of WT3, the PMC stage of MT, the tetrad stage of WT3 and the tetrad stage of MT. Then we made four SSH libraries in the following way. Library SSH_A was constructed using cDNA pool 1 (tester) and cDNA pool 2 (driver). Library SSH_B was constructed using cDNA pool 3 (tester) and cDNA pool 4 (driver). Library SSH_C was constructed using cDNA pool 2 (tester) and cDNA pool 1 (driver). Library SSH_D was constructed using cDNA pool 4 (tester) and cDNA pool 3 (driver). Double-stranded cDNA was synthesized and digested with restriction enzyme RsaI. Tester cDNAs were divided into two groups, one ligated to specific adaptor 1 in the Takara kit and the other to specific adaptor 2R. Each denatured adaptor-ligated tester mix was separately hybridized with a single-stranded driver cDNA for 8 h at 68 °C to enrich non-pairing sequences unique to the tester cDNA pool. A second hybridization using single-stranded driver cDNA as well as the products of the first hybridization from both tester aliquots (adaptor 1-linked and adaptor 2R-linked molecules) was performed for 16 h at 68 °C. Products from the second hybridization were diluted in 200 µl of dilution buffer of this kit, heated at 68 °C for 7 min, and stored at −20 °C. Cloning and sequencing were carried out according to Kurita et al. (2011). Finally, 278, 184, 281 and 180 ESTs were obtained from the SSH_A, SSH_B, SSH_C, and SSH_D library, respectively. In total, 923 ESTs were submitted to the DNA Data Bank of Japan (HX970307 through HX971791).
Microarray chip design
A microarray chip was designed according to the Roche Nimblegen protocol. A custom microarray comprising a total of 75,160 oligonucleotide probes was designed using the EST sequences in the ForestGEN public database (Forest EST and Genome database, http://forestgen.ffpri.affrc.go.jp) and the ESTs collected from SSH libraries of male strobili (Kurita et al. 2011) and libraries of MT and WT3 male strobili (described above).
We chose 18,080 ESTs from the male reproductive organ library (Futamura et al. 2008) and pollen libraries of the ForestGEN database. The male reproductive organ library was constructed using male strobili collected from August to mid-November (Futamura et al. 2008). This library might contain samples of the early and middle stages of male strobilus development and the pollen library might contain samples of the late stages. From the SSH libraries, 1129 ESTs were selected. Altogether, 3773 probes were based on the ESTs from SSH libraries and 68,061 probes were based on the ESTs from the ForestGEN database. The ESTs were spotted in triplicate or quadruplicate on the array. The array comprised 19,209 individual ESTs. Using the TAIR8 blastX algorithm (http://www.arabidopsis.org/index.jsp), 18,688 ESTs were annotated and 521 ESTs had no hits.
RNA preparation and microarray analysis
Male strobili were collected from each twig and were observed by microscope according to the methods previously described. Then, each sample stage was defined and strobili from each stage used for total RNA isolation. Total RNA was isolated using Plant RNA Isolation Reagent (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. This RNA was reacted with recombinant DNase I (Takara Bio Inc.) and purified using an RNeasy plant mini kit (Qiagen, Hilden, Germany). The quality of RNA was checked using an Agilent 2100 bioanalyzer and RNA 6000 Nano Kit (Agilent Technologies, Tokyo, Japan) before cDNA synthesis. Three biological replicates of each sample were used in the analysis. Double-stranded cDNA was synthesized from 10 µg of total RNA using a SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) and labeled with Cy3 random nonamers with a One-Color DNA Labeling Kit (Roche Nimblegen, Madison, WI, USA). The following steps were carried out with equipment and software from the same manufacturer. The custom 4 × 72K array was incubated at 42 °C for 18 h in a Hybridization System 4 (Roche Nimblegen), and washed at room temperature. The microarray slide was scanned at 2 µm resolution using an MS 200 Microarray Scanner (Roche Nimblegen), generating corresponding 532 nm TIFF images. The data were imported into Nimblescan software (Roche Nimblegen) to quantify the signal intensities of the spots on the image. The microarray design and data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus database (GSE64663).
Data processing and statistical analyses
Normalization of signal intensity was carried out with Nimblescan software. Expressed sequence tags with raw signal intensity <100 in two-thirds of all samples were excluded. The array data were normalized according to the Robust Multichip Average algorithm. The datasets were normalized through two following normalization steps using the Subio platform (Subio Inc., Kagoshima, Japan), log-based transformation of the data, and global normalization (to the 75th percentile). We calculated the average log2 ratios at each time point, and excluded ESTs with expression levels that hardly varied (between −1.0 and 1.0) through all stages. One-way analysis of variance (Benjamini–Hochberg false discovery rate <0.05) was executed to identify ESTs with expression levels that varied for at least one time point. Clustering and categorizing were performed on the Subio platform. We selected the annotated ESTs with low E-values (E-value <1e−5) and categorized them using a database of Clusters of Orthologous Groups (COG) from seven eukaryotic genomes (Tatusov et al. 2003).
Real-time PCR
Primer pairs were designed for each sequence using Primer3 software (Rozen and Skaletsky 1999), and the details of these primers are shown in Table S1 available as Supplementary Data at Tree Physiology Online. For SYBR Green real-time PCR assays, the amplification efficiency of all primer pairs was optimized with genomic DNA from the WT2 clone using the StepOnePlus Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific).
Total RNA (500 ng in a final volume of 20 µl) extracted from male strobili at each stage was reverse-transcribed using the High Capacity RNA-to-cDNA Kit (Applied Biosystems) according to the manufacturer’s protocol. Quantitative real-time PCR was performed on the total RNA samples using Power SYBR Green PCR Master Mix (Applied Biosystems) on the StepOnePlus Real-Time PCR System. PCR mixtures were prepared according to the manufacturer’s instructions and contained 300 nM of both forward and reverse gene-specific primers and 2 µl of the 50-fold diluted reverse transcription reaction (total 1 ng) in a final volume of 20 µl. All reactions were heated to 95 °C for 10 min; this denaturation step was followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The PCR products were subjected to melting curve analysis; the conditions were incubation at 60–95 °C with a temperature increment of 0.3 °C s−1. We used C. japonica ubiquitin as an internal control gene, amplified with specific primers (Nose and Watanabe 2014). Relative expression was measured by the ΔΔCt method (Pfaffl 2001). The experiments were conducted three times. The specificity of each amplification was checked by melting analysis.
Results
Cytological analysis of C. japonica male strobilus development
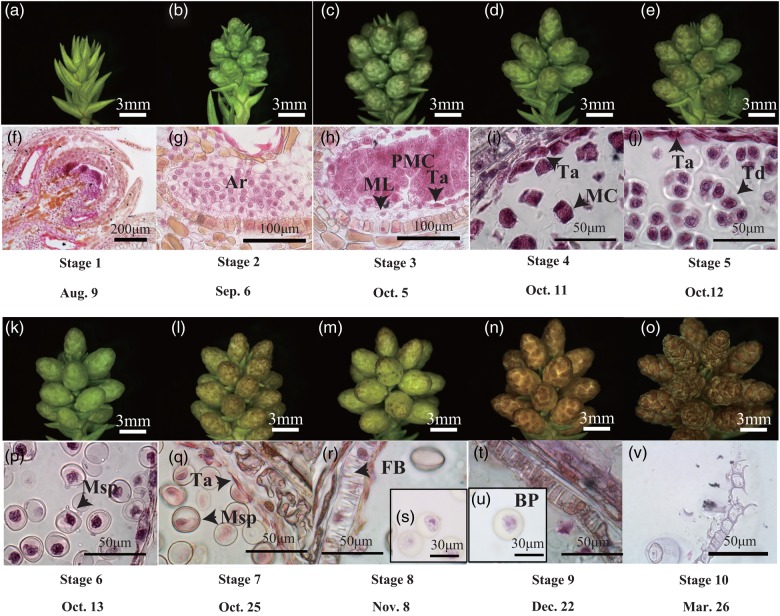
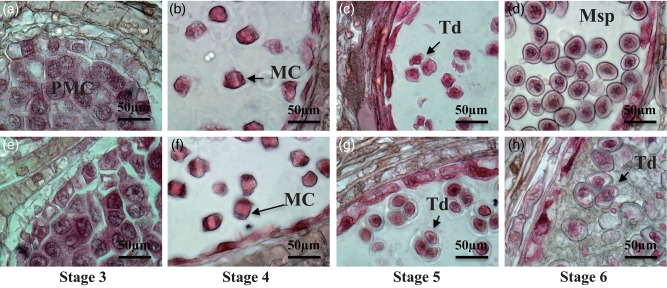
We divided C. japonica male strobilus development into 10 stages by observation of longitudinal sections of WT2 male strobili (Figure 1). Male strobili of C. japonica develop from axils. In late August, the point when the scale primordia were visible in the axils was defined as Stage 1 (Figure 1a and f). At the end of September, following scale formation, the microsporangia were differentiated on the basis of the appearance of scales (Stage 2, Figure 1b and g). Microsporangia wall, middle layer, tapetum and PMCs were recognized from the end of September to the beginning of October (Stage 3, Figure 1c and h). Pollen mother cells entered meiosis (Stage 4, Figure 1d and i), and meiosis was completed and tetrads developed (Stage 5, Figure 1j) in the middle of October. The callose wall surrounding the tetrads degenerated and the microspores were released into the microsporangium (Stage 6, Figure 1k and p). The tapetum degenerated at the end of October (Stage 7, Figure 1l and q), and fibrous bands formed in the microsporangial wall at the beginning of November (Stage 8, Figure 1r and s). Pollen mitotic division occurred and mature pollen grains developed in late December (Stage 9, Figure 1t and u). Dehiscence occurred and pollen grains were released in March (Stage 10, Figure 1v).
Figure 1.
Development of C. japonica male strobili and pollen. (a–e) Longitudinal section of male strobili at Stages 1–5; (f–j) male strobilus at Stages 1–5; (k–o) male strobilus at Stages 6–10; (p–v) male strobilus at Stages 6–10; sections were stained with eosin and hematoxylin. Ar, archesporial cell; BP, bicellular pollen; FB, fibrous band; MC, meiotic cell; ML, middle layer; Msp, microspore; PMC, pollen mother cell; Ta, tapetum; Td, tetrad.
The correspondence between C. japonica male strobilus developmental stages and Arabidopsis anther developmental stages (Sanders et al. 1999) is shown in Table 1. In Arabidopsis thaliana, primary parietal and primary sporogenous layers are derived from archesporial cells and further divisions of each layer generate the secondary parietal layers and sporogenous cells, respectively, at Stage 3. Stage 2 of C. japonica corresponded to Stage 3 of A. thaliana. Microspore mother cells appearing at Stage 5 in A. thaliana corresponded to Stage 3 of C. japonica. Entering meiosis, formation of tetrads and appearance of microspores occurs at Stages 6–8 in A. thaliana, corresponding with C. japonica pollen developmental Stages 4–6, respectively. Stages 7 and 8 in C. japonica, tapetum degeneration to appearance of fibrous bands, corresponded with Stages 10 and 11 of A. thaliana, respectively. Pollen mitotic division occurs at Stage 11 in A. thaliana and at Stage 9 in C. japonica. While the pollen grains of C. japonica have two cells at Stage 9 (Figure 1u), the anther contains tricellular pollen grains at Stages 12 in A. thaliana. Pollen release occurs at Stage 13 in A. thaliana and at Stage 10 in C. japonica. Stages 14 and 15 of A. thaliana are stamen senescence, and therefore these stages were not observed in C. japonica.
Table 1.
Cytological characterization of the 10 developmental stages of male strobili of C. japonica.
| C. japonica male strobilus stages and cytological characterization | Anther stage of Arabidopsis (Sanders et al. 1999) | Microsporangiate strobilus development of P. taeda (Williams 2009) |
|---|---|---|
| 1. The scale primordia are visible in the axils | — | 1 |
| 2. The archesporial cells are differentiated in the base of the scales | 3 | 2 |
| 3. The intact tapetum layer is well defined | 5 | 3 |
| 4. Microspore mother cells enter meiosis | 6 | 3 |
| 5. Meiosis completed; tetrads of micropsores free within each locule | 7 | 3 |
| 6. Callose wall surrounding tetrads degenerates and individual microspores released | 8 | 3 |
| 7. Tapetum degeneration initiated | 10 | 3 |
| 8. ‘Fibrous bands’ appear in endothecium | 11 | 3 |
| 9. Pollen mitotic division occurrs | 11–12 | 3 |
| 10. Dehiscence; pollen release | 13 | 4–6 |
The correspondence between morphological development of C. japonica and P. taeda male strobili (Bramlett and Bridgwater 1989, Williams 2009) is also shown in Table 1. In the P. taeda male strobilus developmental classification, at Stage 1 male strobili are encased in bud scales at tips of vegetative shoots; at Stage 2 the individual male strobilus emerges from its bud scales; at Stage 3 the male strobilus exudes liquid when pressed; at Stage 4 pollen release starts; at Stage 5 pollen release reaches a maximum; and at Stage 6 pollen release is completed (Williams 2009). Stages 1 and 2 of C. japonica corresponded to Stages 1 and 2 of P. taeda, respectively. Stages 3–9 and Stage 10 of C. japonica corresponded to Stage 3 and Stages 4–6 of P. taeda, respectively.
Gene expression analysis
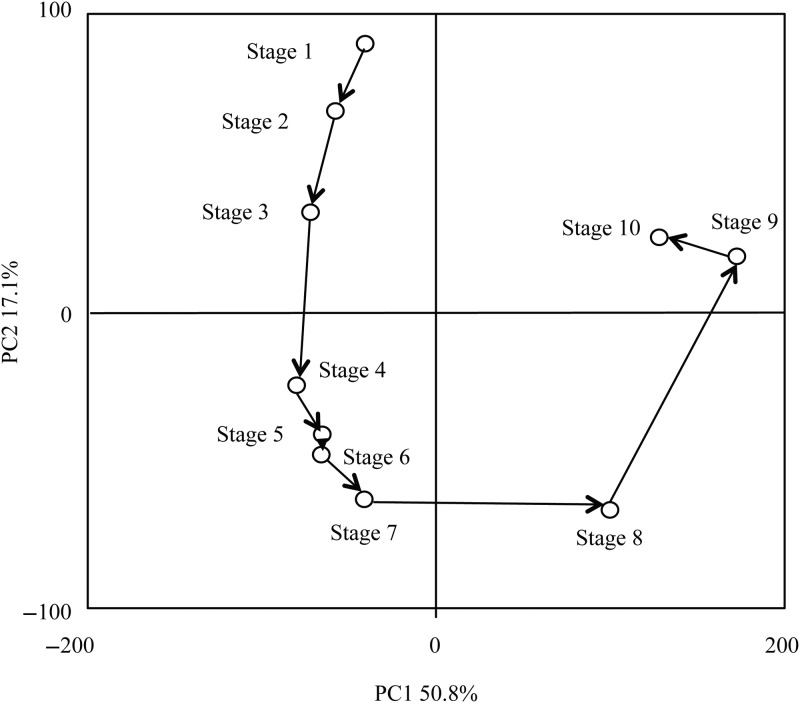
Gene expression patterns during male strobilus development were examined by microarray analysis to identify ESTs that were differentially expressed among the 10 stages. Principal component analysis was performed on the expression patterns of 8079 ESTs. The developmental stages were clustered into four groups, Stages 1–3, Stages 4–7, Stage 8 and Stages 9–10 (Figure 2).
Figure 2.
Principal component analysis of 10 stages in male strobilus development for 8079 ESTs differentially expressed among stages.
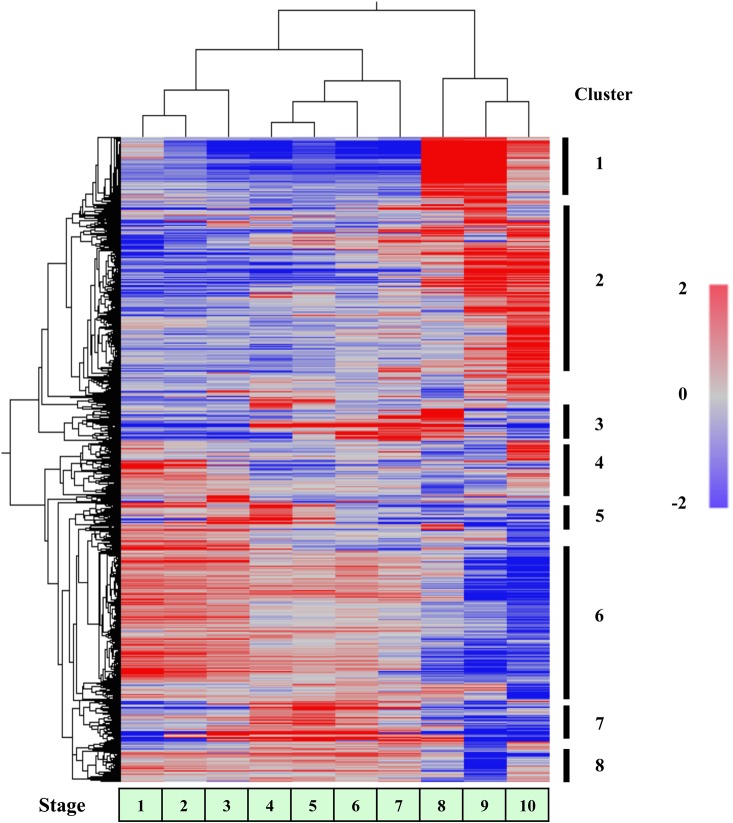
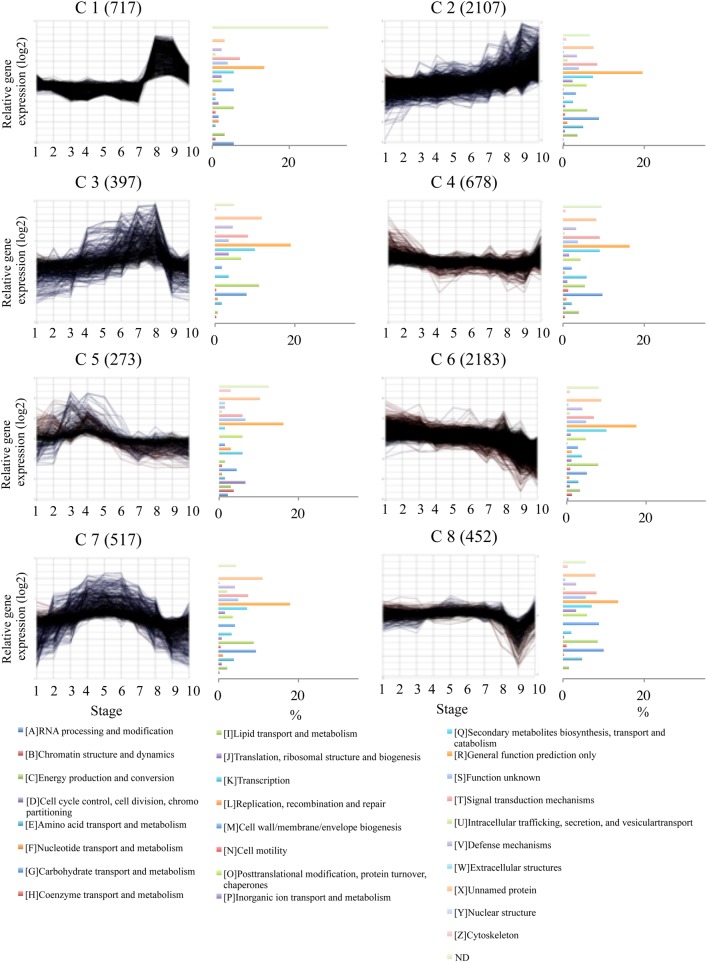
The 8079 ESTs were sorted into several clusters according to their expression profiles during male strobili and pollen development using the Subio platform tree clustering tool (Pearson correlation, Figure 3). Eight major clusters (7324 ESTs) were further examined (Figures 3 and 4). Two clusters, C2 and C6, showed a gradual increase and decline; these included the majority of all profiles (58.5%). Other profiles showed stage-specific expression patterns. Four clusters exhibited a transient increase at multiple stages (C1, C3, C5 and C7). C1 had transient increases in the late developmental stages, 8 and 9. Two clusters (C3 and C7) showed a transient increase at the middle developmental stages, 3–8. C5 exhibited a transient increase at the early developmental stages, 2–4. C8 showed a transient decrease at Stage 9. C4 showed a transient increase in the early and late stages, 1 and 10. Of the total 7324 ESTs, 38.5% (C1 and C2) were expressed only in the late developmental stages, and 46.0% (C3, C5, C6 and C7) were expressed in the early and middle developmental stages.
Figure 3.
Tree clustering analysis of expression of 8079 ESTs in developing WT2 male strobili. Numbered bars on the right indicate distinct clusters of ESTs used for further analysis.
Figure 4.
Category distribution in the eight expression clusters. Clusters were obtained for 7324 EST expression profiles using the Pearson correlation method. Cluster number and number of transcripts belonging to each cluster are labeled on each chart. In the histogram of the functional category distribution, each category is expressed as the percentage of the number of ESTs belonging to that cluster.
A total of 4198 ESTs can be assigned a putative function based on E-value <1e−5, and were classified into 26 functional categories. The ratio of each category to each cluster was calculated (Figure 4). In C1 the ratio of category [A], ‘RNA processing’, was higher than in the other clusters. The ratio of category [I], ‘Lipid transport’, in C3 and the ratio of category [D], ‘Cell cycle control, cell division, chromosome partitioning’, in C5 were high compared with other clusters. The ratio of category [G], ‘Carbohydrate transport and metabolism’, in C7 and C8 was higher than in other clusters.
Expressed sequence tags that changed in expression fourfold or more at a single stage (P < 0.05) compared with the other nine stages were identified. As a result, 15 ESTs at Stage 1, three ESTs at Stage 2, 35 ESTs at Stage 3, 15 ESTs at Stage 4, two ESTs at Stage 5, 19 ESTs at Stage 6, 24 ESTs at Stage 7, 79 ESTs at Stage 8, 59 ESTs at Stage 9, and 222 ESTs at Stage 10 were identified (see Table S2 available as Supplementary Data at Tree Physiology Online). The expression of these ESTs was confirmed by real-time PCR at all stages (Figure 5 and Figure S1 available as Supplementary Data at Tree Physiology Online).
Figure 5.
Real-time PCR analysis of five ESTs showing stage-specific expression. Bar graphs show relative expression from real-time PCR and line graphs show processed signals from microarray analysis. Error bar shows the standard deviation.
Cytological and gene expression analysis of male-sterile mutant male strobili
Male strobili of MT were compared with WT2 during Stages 3–6, the pollen developmental stages (Figure 6). At Stages 3–5, when PMCs developed to tetrads, the MT phenotype was similar to that of WT2. At Stage 6, the degenerating callose wall and pollen release period, an abnormal phenotype was observed in MT microsporangia (Figure 6h). The tetrads of MT were not separated and they had thinner walls than the microspores of WT2. The cytoplasm of tetrads of MT was slightly expanded. The microsporangia of MT were filled with debris and the tetrads were embedded in them. The debris remained until Stage 10, and the microspores did not develop further.
Figure 6.
Male strobili and pollen development of WT2 and MT at Stages 4–6. (a–d) Longitudinal sections of wild type male strobili at Stages 4–6; (e–h) longitudinal sections of mutant type male strobili at Stages 4–6; MC, meiotic cell; Msp, microspore; PMC, pollen mother cell; Td, tetrad.
Similarly, gene expression of MT during Stages 3–6 was analysed by microarray. Principal component analysis and clustering was performed on the expression patterns of the 8079 ESTs differentially expressed among the 10 stages in WT2 (see Figures S2 and S3 available as Supplementary Data at Tree Physiology Online). The changes in gene expression among stages of MT were similar to those of WT2, with slight differences at Stages 4–6 (see Figures S2 and S3 available as Supplementary Data at Tree Physiology Online).
We identified the ESTs for which expression changed at least fourfold (P < 0.05) between MT and WT2 at each developmental stage (Table 2 and Table S3 available as Supplementary Data at Tree Physiology Online). The largest number of upregulated and downregulated ESTs was recognized at Stage 5 (105 and 152 ESTs, respectively), compared with expression in WT2. The number of upregulated or downregulated ESTs at any stage was 188 and 209, respectively, and the total number of ESTs that changed in expression fourfold at any stage was 395. Of 109 ESTs downregulated at Stage 4, 92 were also downregulated at Stage 5, and 70 ESTs were downregulated from Stages 4 to 6. Details on upregulated and downregulated ESTs at Stages 4–6, omitting the ESTs with high E-values, are shown in Tables 3 and 4. Of the 46 downregulated ESTs, 14 were related to terpenoid biosynthesis, four belonged to the CYP450 family and three were disease resistance responsive family proteins and O-methyltransferase. Of the upregulated ESTs, four were identified as a disease resistance family protein.
Table 2.
The number of ESTs at each stage with ≥ fourfold change in expression compared with WT2.
| Stage | Fourfold up in MT | Fourfold down in MT |
|---|---|---|
| 3 (PMC) | 95 | 64 |
| 4 (MEI) | 97 | 109 |
| 5 (TED) | 105 | 152 |
| 6 (MSP) | 103 | 110 |
| 3–6 | 40 | 32 |
| 4–6 | 46 | 70 |
Table 3.
Expressed sequence tags downregulated at Stages 4–6 in MT compared with WT2, omitting the ESTs with high E-values.
| Measurement ID | TAIR | Annotation | E-value |
|---|---|---|---|
| Cj_ES_Contig_0091 | AT4G23340.1 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 2.00e−13 |
| Cj.13983 | AT5G05600.1 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 7.00e−12 |
| Cj.11285 | AT4G05040.4 | Ankyrin repeat family protein | 6.00e−13 |
| Cj.11004 | AT5G24960.1 | Cytochrome P450, family 71, subfamily A, polypeptide 14 | 2.00e−21 |
| Cj_ssh_male_e_026 | AT5G36110.1 | Cytochrome P450, family 716, subfamily A, polypeptide 1 | 3.00e−67 |
| Cj_ssh_male_e_198 | AT5G36110.1 | Cytochrome P450, family 716, subfamily A, polypeptide 1 | 7.00e−29 |
| Cj.26959 | AT2G45580.1 | Cytochrome P450, family 76, subfamily C, polypeptide 3 | 2.00e−30 |
| Cj.21151 | AT2G21100.1 | Disease resistance-responsive (dirigent-like protein) family protein | 6.00e−17 |
| Cj.20835 | AT5G42500.1 | Disease resistance-responsive (dirigent-like protein) family protein | 2.00e−08 |
| Cj.8130 | AT5G42500.1 | Disease resistance-responsive (dirigent-like protein) family protein | 2.00e−19 |
| Cj.17487 | AT5G22500.1 | Fatty acid reductase 1 | 4.00e−26 |
| Cj.20412 | AT1G08510.1 | Fatty acyl-ACP thioesterases B | 2.00e−26 |
| Cj.25774 | AT1G08510.1 | Fatty acyl-ACP thioesterases B | 1.00e−27 |
| Cj.19951 | AT5G51950.2 | Glucose-methanol-choline (GMC) oxidoreductase family protein | 3.00e−37 |
| Cj.11111 | AT4G33790.1 | Jojoba acyl CoA reductase-related male sterility protein | 4.00e−40 |
| Cj.13061 | AT3G53880.1 | NAD(P)-linked oxidoreductase superfamily protein | 1.00e−41 |
| Cj.1269 | AT5G04950.1 | Nicotianamine synthase 1 | 3.00e−23 |
| Cj.12365 | AT1G09240.1 | Nicotianamine synthase 3 | 1.00e−30 |
| Cj.15578 | AT4G35160.1 | O-methyltransferase family protein | 5.00e−14 |
| Cj.6619 | AT4G35160.1 | O-methyltransferase family protein | 3.00e−12 |
| Cj.21447 | AT1G77520.1 | O-methyltransferase family protein | 4.00e−17 |
| Cj.11351 | AT4G24340.1 | Phosphorylase superfamily protein | 2.00e−44 |
| Cj_AE_Contig_0085 | AT5G15780.1 | Pollen Ole e 1 allergen and extensin family protein | 1.00e−09 |
| Cj.4624 | AT5G15780.1 | Pollen Ole e 1 allergen and extensin family protein | 2.00e−10 |
| Cj.2222 | AT5G17170.2 | Rubredoxin family protein | 4.00e−26 |
| Cj.2343 | AT1G21200.1 | Sequence-specific DNA binding transcription factors | 3.00e−23 |
| Cj.2987 | AT4G16740.2 | Terpene synthase 03 | 8.00e−50 |
| Cj.2096 | AT1G70080.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 6.00e−11 |
| Cj.14040 | AT1G79460.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 2.00e−17 |
| Cj_ssh_male_e_239 | AT5G48110.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 7.00e−09 |
| Cj_ssh_male_e_020 | AT4G02780.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 3.00e−16 |
| Cj_ssh_male_e_009 | AT3G29410.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 2.00e−17 |
| Cj_ssh_male_e_187 | AT1G79460.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 5.00e−40 |
| Cj.36 | AT3G25810.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 2.00e−27 |
| Cj.21863 | AT1G33750.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 2.00e−19 |
| Cj.8750 | AT4G02780.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 5.00e−22 |
| Cj.11599 | AT4G02780.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 2.00e−32 |
| Cj.8462 | AT1G79460.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 8.00e−25 |
| Cj.8670 | AT1G79460.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 3.00e−38 |
| Cj.2241 | AT4G02780.1 | Terpenoid cyclases/protein prenyltransferases superfamily protein | 7.00e−18 |
| Cj.16674 | AT1G15780.1 | Unknown protein | 2.00e−20 |
| Cj.25366 | AT2G36835.1 | Unknown protein | 6.00e−18 |
| Cj.21025 | AT1G52825.1 | Unknown protein | 5.00e−21 |
Table 4.
Expressed sequence tags upregulated at Stages 4–6 in MT compared with WT2, omitting the ESTs with high E-values.
| Measurement ID | TAIR | Annotation | E-value |
|---|---|---|---|
| Cj.11098 | AT5G12270.1 | 2-Oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein | 3.00e−21 |
| Cj.21820 | AT1G77380.1 | Amino acid permease 3 | 2.00e−27 |
| Cj.6636 | AT2G41890.1 | Curculin-like (mannose-binding) lectin family protein/PAN domain-containing protein | 2.00e−22 |
| Cj.10292 | AT2G34930.1 | Disease resistance family protein/LRR family protein | 7.00e−26 |
| Cj.12287 | AT2G34930.1 | Disease resistance family protein/LRR family protein | 2.00e−14 |
| Cj.25224 | AT5G45050.2 | Disease resistance protein (TIR-NBS-LRR class) | 5.00e−25 |
| Cj.14439 | AT1G69550.1 | Disease resistance protein (TIR-NBS-LRR class) | 6.00e−19 |
| Cj.10817 | AT5G65350.1 | Histone 3 11 | 3.00e−11 |
| Cj.5057 | AT2G39050.1 | Hydroxyproline-rich glycoprotein family protein | 9.00e−06 |
| Cj.14392 | AT5G55250.2 | IAA carboxylmethyltransferase 1 | 2.00e−12 |
| Cj.3467 | AT1G51980.2 | Insulinase (peptidase family M16) protein | 9.00e−27 |
| Cj.3691 | AT2G37050.3 | Leucine-rich repeat protein kinase family protein | 1.00e−13 |
| Cj.12169 | AT5G64410.1 | Oligopeptide transporter 4 | 2.00e−56 |
| Cj.13917 | AT1G63770.5 | Peptidase M1 family protein | 1.00e−17 |
| Cj.6976 | AT1G74180.1 | Receptor like protein 14 | 8.00e−10 |
| Cj.14309 | AT3G11010.1 | Receptor like protein 34 | 8.00e−23 |
| Cj.19053 | AT3G12000.1 | S-locus related protein SLR1, putative (S1) | 1.00e−28 |
| Cj.13990 | AT5G67360.1 | Subtilase family protein | 1.00e−09 |
| Cj.21905 | AT4G01870.1 | tolB protein-related | 1.00e−23 |
| Cj.13548 | AT5G15390.1 | tRNA/rRNA methyltransferase (SpoU) family protein | 1.00e−33 |
| Cj.26297 | AT4G27570.1 | UDP-glycosyltransferase superfamily protein | 1.00e−12 |
| Cj.9453 | AT5G64190.1 | Unknown protein | 4.00e−32 |
Discussion
Male strobilus development of C. japonica based on cytological and transcriptome analysis
The process of male strobilus development (Hashizume 1962), pollen wall development (Uehara and Sahashi 2000), and histological comparison of fertile and sterile pollen development (Hosoo et al. 2005, Ueuma et al. 2009) have been reported in C. japonica. However, previous reports did not focus on gene expression or molecular analysis. Based on cytological observation and transcriptome analysis, male strobilus development of C. japonica can be divided into 10 stages. It took 8 months from male strobilus initiation to pollen maturation, as previously reported (Hashizume 1962). Identification of the stages by cytological analysis is difficult from Stages 3 to 6, because it takes only a few days to progress through them. In addition, even for the same tree and the same clusters, pollen developmental stages are slightly different among strobili. In comparison with P. taeda male strobilus development (Williams 2009), almost all stages were categorized as Stage 3 (Table 1). Internal cytological observation of the microsporangium is necessary for male strobilus developmental stage classification, in addition to external morphological traits. In Pinus banksiana, the process of sporopollenin formation and microsporangial development has been reported (Dickinson and Bell 1972), and so has pollen cone and pollen development of eastern white pine and lodgepole pine (Owens 2004, 2006). From PMCs to microspore formation, C. japonica and pine are quite similar in time period and morphological traits. Since mature pollen grains have two cells in C. japonica, late pollen development is different from that of pine, which contains four to five cells (Fernando et al. 2010).
In angiosperms, such as A. thaliana and Oryza sativa, developmental stages of anthers and pollen are well characterized by cytological and transcriptome analysis (Regan and Moffatt 1990, Smyth et al. 1990, Sanders et al. 1999, Ikeda et al. 2004, Wang et al. 2005, Li et al. 2006) and are useful for investigation of mutants. Smyth et al. (1990) divided the flower development of A. thaliana into 12 stages based on observation using an electron microscope. Sanders et al. (1999) divided anther development of A. thaliana into 15 stages using a light microscope, and Regan and Moffatt (1990) divided pollen development into 10 stages based on staining and microscopic observation. Ikeda et al. (2004) categorized spikelet development of rice. These developmental stages have been used to determine the stage at which aberrant traits can be observed in mutants. These morphological developmental stages have often been cited by other studies to compare morphological traits or transcriptomes of mutants and wild type to investigate anther developmental genes.
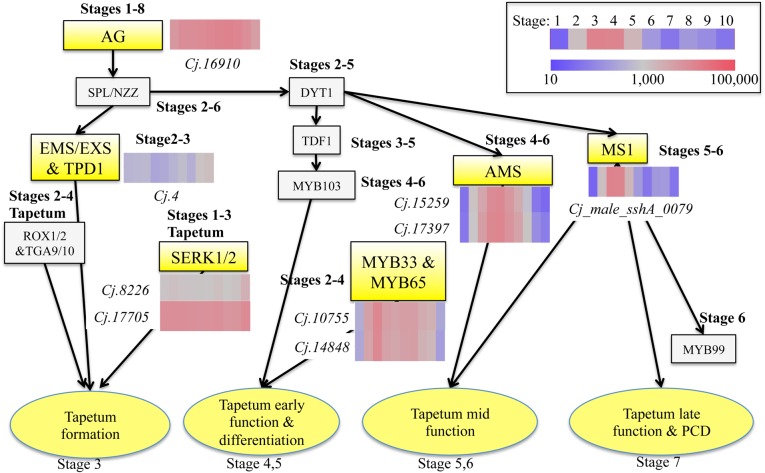
Angiosperms and gymnosperms are quite different in the late stages of pollen development and pollen tube growth (Dickinson and Bell 1972, Fernando et al. 2005). In this study, the process of male strobilus and pollen development of C. japonica, which occurs from Stage 3 (tapetum differentiates) to Stage 6 (microspores are released from tetrads) based on cytological observations, was similar to anther development of A. thaliana. In A. thaliana, genes related to anther development are well characterized and their functions have been analysed (Parish and Li 2010, Huang et al. 2011), while the genes related to male strobilus development in conifers are still unknown. Some genes already characterized in conifers, e.g., A9 (Kurita et al. 2013), LEAFY (Shiokawa et al. 2008) and gibberellin metabolism genes (Niu et al. 2014), are highly conserved among angiosperms. We investigated the expression patterns of C. japonica ESTs homologous to A. thaliana genes involved in anther development (Figure 7). Arabidopsis mutants defective in these genes are male-sterile. AG is a C-function gene required for stamen identity (Bowman et al. 1991, Liu and Fan 2013). EMS/EXS (Canales et al. 2002) and TPD1 (TAPETAL DETERMINANT1) (Yang et al. 2003) are genes encoding leucine-rich repeat receptor-like kinases (LRR-RLKs), and regulate development of archesporial cells from the L2 layer. SERK1/2 (Colcombet et al. 2005) also encodes LRR-RLKs, but SERK1/2 is not regulated by SPL/NZZ (NOZZLE/SPOROCYTELESS) (Liu et al. 2009), as opposed to EMS. MYB33 and MYB65 act redundantly to facilitate tapetal development and may play a role in tapetal starch mobilization in the seed aleurone layer (Millar 2005, Wilson and Zhang 2009). AMS (Sorensen et al. 2003) and MS1 (Wilson et al. 2001) are regulated by DYT1 (DYSFUNCTIONAL TAPETUM1) and are expressed in the tapetum (Wilson and Zhang 2009). MS1 expression starts in the tapetum as the callose wall separating the tetraspores begins to degrade in A. thaliana (Yang et al. 2007). AG-like gene, SERK-like gene, MYB33&65-like genes, AMS-like gene and MS1-like gene were similar to gene expression in addition to sequence across species. It is likely that these genes play a major role for tapetum development also in C. japonica.
Figure 7.
Proposed gene regulation network for tapetum development in Arabidopsis and the expression patterns of candidate ESTs in C. japonica. Stage numbers next to genes are expected expression stages presumed from the comparison between Arabidopsis and C. japonica. This figure is adapted from Huang et al. (2011) and Parish and Li (2010).
Cytological and transcriptome analysis of male-sterile mutant pollen
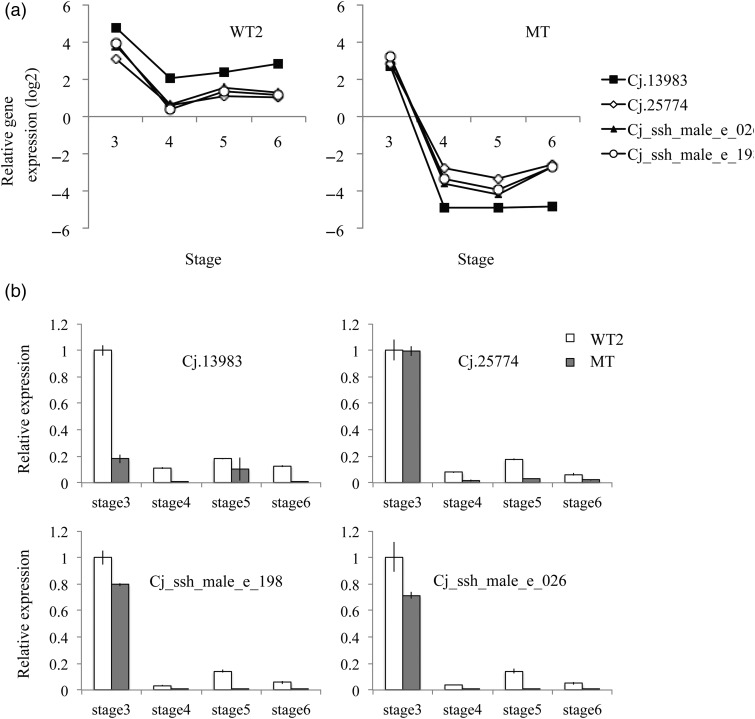
In Sosyun, which is a male-sterile mutant, abnormality of the microsporangium was observed at Stage 6 by cytological analysis. The microsporangium of Sosyun filled with debris, presumably secreted from the tapetum. The abundant debris is regarded as the cause of pollen grains not being released from the microsporangium. Saito et al. (1998) reported that microspores gradually enlarged after individual microspores separated from tetrads in the male sterile mutant ‘Toyama MS’, classified as ms-1. In ‘Shindai1’, classified as ms-2, and ‘Shindai5’, classified as ms-3, microsporogenesis proceeds normally but the microspores clump by the time of pollen release (Yoshii and Taira 2007). In four clones of male-sterile mutant lines (Fukushima-1, Fukushima-2, Shindai-11 and Shindai-12) with abnormal tetrads, the release of Ubisch bodies from the tapetum is not observed after callose dissolution, and the amount of translucent amorphous substances gradually increases (Miura et al. 2011). Sosyun is considered to be a similar type to male-sterile mutants reported in Miura et al. (2011) because of the presence of debris in the microsporangium. The gene expression pattern of Sosyun differed from WT2 at Stages 4–6. Therefore, the difference in its gene expression pattern at Stage 6 appears to be correlated with the abundant debris in the microsporangium. Some ESTs were downregulated at Stage 4 in both WT2 and Sosyun (Figure 8). However, from Stages 4 to 6, they were not expressed at all in Sosyun, while they were slightly expressed in WT2. The protein annotations of these ESTs are 2-oxoglutarate and Fe (II)-dependent oxygenase superfamily protein, CYP450 family and fatty acyl-ACP thioesterases B (Table 3). There are no reports of these genes being directly related to male strobilus development, but they are likely related to pollen sac development.
Figure 8.
Four ESTs with expression remarkably downregulated in MT at Stages 4–6. (a) Microarray analysis, (b) real-time PCR analysis. Error bar means standard deviation.
Supplementary data
Supplementary data for this article are available at Tree Physiology Online.
Conflict of interest
None declared.
Funding
This study is a part of the project on ‘Technology development for circulatory food production systems responsive to climate change’ supported by the Ministry of Agriculture, Forestry and Fisheries, Japan. This study was also supported by a Grant-in-Aid from the Forestry Agency, Ministry of Agriculture, Forestry and Fisheries from 2008 to 2013, and by the Japan Society for Promotion of Science, KAKENHI Grant Number 26850103.
Supplementary Material
References
- Almqvist C. (2003) Timing of GA4/7 application and the flowering of Pinus sylvestris grafts in the greenhouse. Tree Physiol 23:413–418. doi:10.1093/treephys/23.6.413 [DOI] [PubMed] [Google Scholar]
- Bonnet-Masimbert M, Webber JE (1995) From flower induction to seed production in forest tree orchards. Tree Physiol 15:419–426. doi:10.1093/treephys/15.7-8.419 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112:1–20. [DOI] [PubMed] [Google Scholar]
- Bramlett DL, Bridgwater FE (1989) Pollen development classification system for loblolly pine. In: Proceedings of the 20th Southern Forest Tree Improvement Conference, June 26–30, 1989 Charleston, South Carolina Southern Forest Experiment Station, USDA Forest Service, Charleston, SC, pp 116–121. [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H (2002) EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr Biol 12: 1718–1727. doi:10.1016/S0960-9822(02)01151-X [DOI] [PubMed] [Google Scholar]
- Colcombet J, Boisson-Dernier A, Ros-Palau R, Vera CE, Schroeder JI (2005) Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17:3350–3361. doi:10.1105/tpc.105.036731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson HG, Bell PR (1972) The rôle of the tapetum in the formation of sporopollenin-containing structures during microsporogenesis in Pinus banksiana. Planta 107:205–215. doi:10.1007/BF00397936 [DOI] [PubMed] [Google Scholar]
- Dornelas MC, Rodriguez A (2005) A FLORICAULA/LEAFY gene homolog is preferentially expressed in developing female cones of the tropical pine Pinus caribaea var. caribaea. Genet Mol Biol 28:299–307. doi:10.1590/S1415-47572005000200021 [Google Scholar]
- Fernando DD. (2013) The pine reproductive process in temperate and tropical regions. New For 43:33–352. [Google Scholar]
- Fernando DD, Lazzaro MD, Owens JN (2005) Growth and development of conifer pollen tubes. Sex Plant Reprod 18:149–162. doi:10.1007/s00497-005-0008-y [Google Scholar]
- Fernando DD, Quinn CR, Brenner ED, Owens JN (2010) Male gametophyte development and evolution in extant gymnosperms. Int J Plant Dev Biol 4(Special Issue 1):47–63. [Google Scholar]
- Futamura N, Totoki Y, Toyoda A et al. (2008) Characterization of expressed sequence tags from a full-length enriched cDNA library of Cryptomeria japonica male strobili. BMC Genomics 9:383 doi:10.1186/1471-2164-9-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood MS. (1980) Reproductive development in loblolly pine: I. The early development of male and female strobili in relation to the long shoot growth behavior. Am J Bot 67:1414–1422. [Google Scholar]
- Harrison DLS, Slee MU (1992) Long shoot terminal bud development and the differentiation of pollen- and seed-cone buds in Pinus caribaea var. hondurensis. Can J For Res 22:1656–1668. doi:10.1139/x92-219 [Google Scholar]
- Hashizume H. (1959) The effect of gibberellin upon flower formation in Cryptomeria japonica. J Jpn For Soc 41:375–381 (in Japanese with English summary). [Google Scholar]
- Hashizume H. (1962) Initiation and development of flower buds in Cryptomeria japonica. J Jpn For Soc 44:312–319 (in Japanese with English summary). [Google Scholar]
- Hosoo Y, Yoshii E, Negishi K, Taira H (2005) A histological comparison of the development of pollen and female gametophytes in fertile and sterile Cryptomeria japonica. Sex Plant Reprod 18:81–89. doi:10.1007/s00497-005-0003-3 [Google Scholar]
- Huang MD, Hsing YIC, Huang AHC (2011) Transcriptomes of the anther sporophyte: availability and uses. Plant Cell Physiol 52:1459–1466. doi:10.1093/pcp/pcr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Sunohara H, Nagato Y (2004) Developmental course of inflorescence and spikelet in rice. Breed Sci 54:147–156. doi:10.1270/jsbbs.54.147 [Google Scholar]
- Katahata SI, Futamura N, Igasaki T, Shinohara K (2014) Functional analysis of SOC1-like and AGL6-like MADS-box genes of the gymnosperm Cryptomeria japonica. Tree Genet Genomes 10:317–327. doi:10.1007/s11295-013-0686-9 [Google Scholar]
- Kawamoto T, Shimizu M (2000) A method for preparing 2- to 50-μm-thick fresh-frozen sections of large samples and undecalcified hard tissues. Histochem Cell Biol 113:331–339. [DOI] [PubMed] [Google Scholar]
- Klintenäs M, Pin PA, Benlloch R, Ingvarsson PK, Nilsson O (2012) Analysis of conifer FLOWERING LOCUS T/TERMINAL FLOWER1-like genes provides evidence for dramatic biochemical evolution in the angiosperm FT lineage. New Phytol 196:1260–1273. doi:10.1111/j.1469-8137.2012.04332.x [DOI] [PubMed] [Google Scholar]
- Kurita M, Taniguchi T, Nakada R, Kondo T, Watanabe A (2011) Spatiotemporal gene expression profiles associated with male strobilus development in Cryptomeria japonica by suppression subtractive hybridization. Breed Sci 61:174–182. doi:10.1270/jsbbs.61.174 [Google Scholar]
- Kurita M, Konagaya K, Watanabe A, Kondo T, Ishii K, Taniguchi T (2013) The promoter of an A9 homolog from the conifer Cryptomeria japonica imparts male strobilus-dominant expression in transgenic trees. Plant Cell Rep 32:319–328. doi:10.1007/s00299-012-1365-2 [DOI] [PubMed] [Google Scholar]
- Kurmann MH. (1989) Pollen wall formation in Abies concolor and a discussion on wall layer homologies. Can J Bot 67:2489–2504. doi:10.1139/b89-319 [Google Scholar]
- Li N, Zhang DS, Liu HS et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18:2999–3014. doi:10.1105/tpc.106.044107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Fan X-D (2013) Tapetum: regulation and role in sporopollenin biosynthesis in Arabidopsis. Plant Mol Biol 83:165–175. doi:10.1007/s11103-013-0085-5 [DOI] [PubMed] [Google Scholar]
- Liu X, Huang J, Parameswaran S et al. (2009) The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol 151:1401–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA. (2005) The Arabidopsis GAMYB-Like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17:705–721. doi:10.1105/tpc.104.027920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima K, Fujiwara T, Iki T, Kuroda K, Yamashita K, Tamura M, Fujisawa Y, Watanabe A (2014) Transcriptome sequencing and profiling of expressed genes in cambial zone and differentiating xylem of Japanese cedar (Cryptomeria japonica). BMC Genomics 15:219 doi:10.1186/1471-2164-15-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Nameta M, Yamamoto T, Igarashi M, Taira H (2011) Mechanisms of male sterility in four Cryptomeria japonica individuals with obvious visible abnormality at the tetrad stage. J Jpn For Soc 93:1–7 (in Japanese with English summary) doi:10.4005/jjfs.93.1 [Google Scholar]
- Miyajima D, Yoshii E, Hosoo Y, Taira H (2010) Cytological and genetic studies on male sterility in Cryptomeria japonica D. Don (Shindai 8). J Jpn For Soc 92:106–109 (in Japanese with English summary) doi:10.4005/jjfs.92.106 [Google Scholar]
- Moriguchi Y, Ujino-Ihara T, Uchiyama K et al. (2012) The construction of a high-density linkage map for identifying SNP markers that are tightly linked to a nuclear-recessive major gene for male sterility in Cryptomeria japonica D. Don. BMC Genomics 13:95 doi:10.1186/1471-2164-13-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi Y, Ueno S, Higuchi Y, Miyajima D, Itoo S, Futamura N, Shinohara K, Tsumura Y (2014) Establishment of a microsatellite panel covering the sugi (Cryptomeria japonica) genome, and its application for localization of a male-sterile gene (ms-2). Mol Breed 33:315–325. doi:10.1007/s11032-013-9951-8 [Google Scholar]
- Niu S, Yuan L, Zhang Y, Chen X, Li W (2014) Isolation and expression profiles of gibberellin metabolism genes in developing male and female cones of Pinus tabuliformis. Funct Integr Genomics 14:697–705. doi:10.1007/s10142-014-0387-y [DOI] [PubMed] [Google Scholar]
- Nose M, Watanabe A (2014) Clock genes and diurnal transcriptome dynamics in summer and winter in the gymnosperm Japanese cedar (Cryptomeria japonica (L.f.) D.Don). BMC Plant Biol 14:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel BSJ (2008) Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol 147:1046–1061. doi:10.1104/pp.108.117457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens JN. (2004) The reproductive biology of western white pine. Extension Note No. 04. Forest Genetics Council of British Columbia, Victoria, BC. [Google Scholar]
- Owens JN. (2006) The reproductive biology of lodgepole pine. Extension Note No. 07. Forest Genetics Council of British Columbia, Victoria, BC. [Google Scholar]
- Owens JN. (2008) The reproductive biology of western larch. Extension Note No. 08. Forest Genetics Council of British Columbia, Victoria, BC. [Google Scholar]
- Owens JN, Blake MD (1985) Forest tree seed production. A review of the literature and recommendations for future research. Information Report PI-X-53 Petawawa National Forestry Institute, Canadian Forest Service, Petawawa, ON, 161 p. [Google Scholar]
- Parish RW, Li SF (2010) Death of a tapetum: a programme of developmental altruism. Plant Sci 178:73–89. doi:10.1016/j.plantsci.2009.11.001 [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45 doi:10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharis RP. (1975) Promotion of flowering in conifers by gibberellins. For Chron 51:244–248. doi:10.5558/tfc51244-6 [Google Scholar]
- Regan S, Moffatt BA (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2:877–889. doi:10.1105/tpc.2.9.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JR, Skvarla JJ, Walles B (2000) Microsporogenesis in Pinus sylvestris L. VIII. Tapetal and late pollen grain development. Plant Syst Evol 225:201–224. doi:10.1007/BF00985468 [Google Scholar]
- Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386. [DOI] [PubMed] [Google Scholar]
- Saito M, Taira H, Furuta Y (1998) Cytological and genetical studies on male sterility in Cryptomeria japonica D. Don. J For Res 3:167–173. doi:10.1007/BF02762139 [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB (1999) Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex Plant Reprod 11:297–322. doi:10.1007/s004970050158 [Google Scholar]
- Shiokawa T, Yamada S, Futamura N et al. (2008) Isolation and functional analysis of the CjNdly gene, a homolog in Cryptomeria japonica of FLORICAULA/LEAFY genes. Tree Physiol 28:21–28. doi:10.1093/treephys/28.1.21 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2:755–767. doi:10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H (2003) The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J 33:413–423. doi:10.1046/j.1365-313X.2003.01644.x [DOI] [PubMed] [Google Scholar]
- Taira H, Saito M, Furuta Y (1999) Inheritance of the trait of male sterility in Cryptomeria japonica. J For Res 4:271–273. doi:10.1007/BF02762782 [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinfomatics 4:41 doi:10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubomura M, Fukatsu E, Nakada R, Fukuda Y (2012) Inheritance of male flower production in Cryptomeria japonica (sugi) estimated from analysis of a diallel mating test. Ann For Sci 69:867–875. doi:10.1007/s13595-012-0223-2 [Google Scholar]
- Uehara K, Sahashi N (2000) Pollen wall development in Cryptomeria japonica (Taxodiaceae). Grana 39:267–274. doi:10.1080/00173130052504298 [Google Scholar]
- Ueuma H, Yoshii E, Hosoo Y, Taira H (2009) Cytological study of a male-sterile Cryptomeria japonica that does not release microspores from tetrads. J For Res 14:123–126. doi:10.1007/s10310-009-0114-z [Google Scholar]
- Ujino-Ihara T, Iwata H, Taguchi Y, Tsumura Y (2012) Identification of QTLs associated with male strobilus abundance in Cryptomeria japonica. Tree Genet Genomes 8:1319–1329. doi:10.1007/s11295-012-0518-3 [Google Scholar]
- Wang Z, Liang Y, Li C et al. (2005) Microarray analysis of gene expression involved in anther development in rice (Oryza sativa L.). Plant Mol Biol 58:721–737. doi:10.1007/s11103-005-8267-4 [DOI] [PubMed] [Google Scholar]
- Williams CG. (2009) Conifer reproductive biology. Springer, New York, 169 p. [Google Scholar]
- Wilson ZA, Zhang DB (2009) From Arabidopsis to rice: pathways in pollen development. J Exp Bot 60:1479–1492. doi:10.1093/jxb/erp095 [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ (2001) The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J 28:27–39. doi:10.1046/j.1365-313X.2001.01125.x [DOI] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA (2007) MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19:3530–3548. doi:10.1105/tpc.107.054981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-L, Xie L-F, Mao H-Z, Puah CS, Yang W-C, Jiang L, Sundaresan V, Ye D (2003) TAPETUM DETERMINANT1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15:2792–2804. doi:10.1105/tpc.016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii E, Taira H (2007) Cytological and genetical studies on male sterile sugi (Cryptomeria japonica D. Don), Shindai 1 and Shindai 5. J Jpn For Soc 89:26–30 (in Japanese with English summary) doi:10.4005/jjfs.89.26 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.