Abstract
Human cancer genome sequencing has recently revealed that genes encoding subunits of SWI/SNF chromatin remodeling complexes are frequently mutated across a wide variety of cancers, and several subunits of the complex have been shown to have bona fide tumor suppressor activity1. However, whether mutations in SWI/SNF subunits result in shared dependencies is unknown. Here we show that EZH2, a catalytic subunit of the Polycomb repressive complex 2 (PRC2), is essential in all tested cancer cell lines and xenografts harboring mutations of the SWI/SNF subunits ARID1A, PBRM1, and SMARCA4, which are several of the most frequently mutated SWI/SNF subunits in human cancer but that co–occurrence of a Ras pathway mutation correlates with abrogation of this dependence. Surprisingly, we demonstrate that SWI/SNF mutant cancer cells are primarily dependent upon a non–catalytic role of EZH2 in stabilization of the PRC2 complex, and only partially dependent on EZH2 histone methyltransferase activity. These results not only reveal a shared dependency of cancers with genetic alterations in SWI/SNF subunits, but also suggest that EZH2 enzymatic inhibitors now in clinical development may not fully suppress the oncogenic activity of EZH2.
SWI/SNF (BRG1 Associated Factors, BAF) complexes contribute to transcriptional regulation and DNA repair by hydrolyzing ATP to remodel chromatin structure. The complexes consist of combinatorial assemblies of approximately 15 subunits, including lineage restricted variant subunits, resulting in a diversity of SWI/SNF complexes that contribute to the control of lineage–specific gene expression2. Cancer genome sequencing studies revealed that at least nine SWI/SNF subunits are recurrently mutated in 20% of all cancers and mouse studies have shown that SWI/SNF subunits are bona fide tumor suppressors3–71,3,8–18. Although recurrent mutation of nine subunits suggests a shared oncogenic mechanism, the tumor spectra associated with each subunit are distinct and diverse phenotypic consequences arise from ablation of SWI/SNF subunit genes in mice.
Genetic studies in Drosophila first identified antagonistic links between Polycomb group genes (PcG) and the SWI/SNF complex19, revealing that mutations in the Swi/Snf complex are capable of suppressing phenotypes associated with PcG mutations and that PcG proteins can block SWI/SNF–mediated nucleosome mobilization20–22. PRC2 consists of four core subunits: EZH2, the catalytic subunit that methylates H3K27 to repress transcription, as well as SUZ12, EED, and RaAp46/48. High levels of EZH2 often correlate with tumor stage and poor prognosis, and ablation of EZH2 can block proliferation and survival in cell lines and mouse models23–25. Consequently, EZH2 is a potential therapeutic target and several inhibitors are in development, including clinical trials26–29. Efforts to therapeutically target EZH2 have generally focused upon inhibition of its histone methyltransferase activity, although it remains unclear whether this is the central mechanism by which EZH2 can promote cancer.
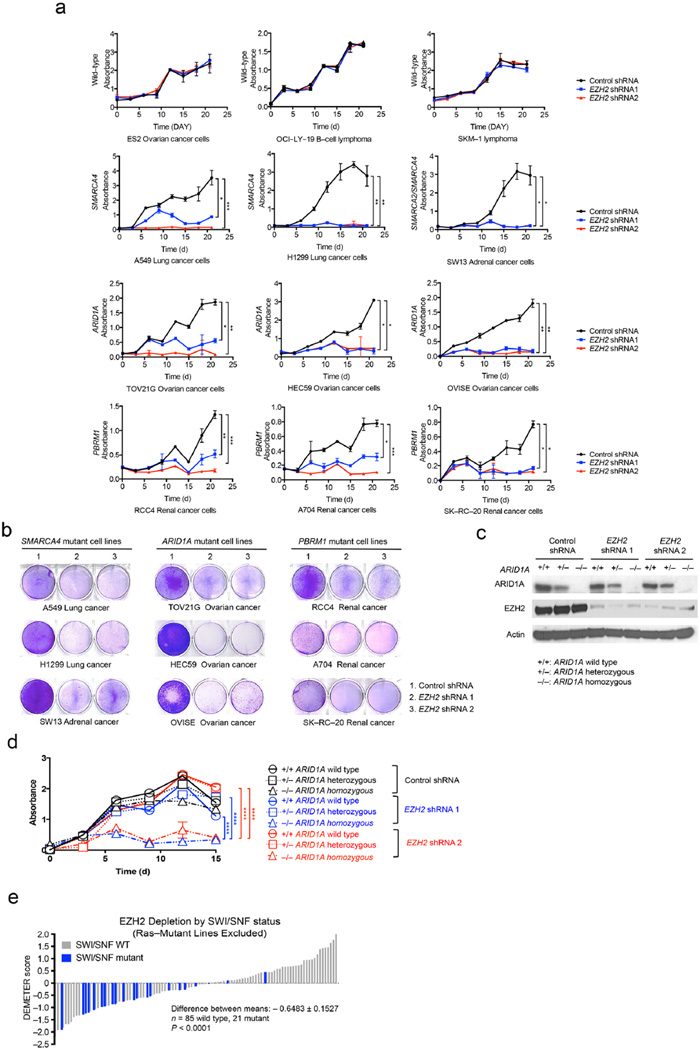
In mammals, we and others have demonstrated an antagonistic relationship between EZH2 and the SMARCB1 (SNF5, INI1, BAF47) subunit of the SWI/SNF complex, which results in genetic dependence upon EZH2 in SMARCB1–mutant cancers26–29. However, whether a similar relationship broadly exists between EZH2 and mutated SWI/SNF subunits is unclear. To evaluate this, we utilized human cancer cell lines mutant for the ARID1A, SMARCA4/BRG1, PBRM1, or SMARCB1/SNF5 subunits of SWI/SNF30–32 and four control lines (ES2, SKM–1, Toledo, and OCI–LY–19) that express both wild–type SWI/SNF complex members and EZH2/PRC2 complex members (Supplementary Fig. 1a). Knockdown of EZH2 reduced H3K27 di– and tri–methylation (Supplementary Fig. 1b). Depletion of EZH2 did not affect control cell lines (Fig. 1a, Supplementary Fig. 1c) but did impair proliferation and colony formation of SWI/SNF–mutant cancer cell lines (Figs. 1a–b and Supplementary Figs. 1d–e). Re–expression of wild–type EZH2 using a construct not recognized by the shRNAs rescued H3K27 tri–methylation, cell proliferation, and colony formation in a dose–dependent manner (Supplementary Fig. 2). To further examine the dependency of SWI/SNF mutant cancers on EZH2, we utilized isogenic knockout of ARID1A in HCT116 cells (wild–type, heterozygous deficient, or homozygous deficient for ARID1A) (Fig. 1c). EZH2 knockdown impaired proliferation and colony formation only in cell lines with homozygous ARID1A inactivation (Fig. 1d and Supplementary Fig. 2b).
Figure 1. SWI/SNF mutant cancer cells require EZH2.
SWI/SNF mutant and wild–type cell lines were transduced with either control or EZH2–targeting shRNAs. Proliferation was assessed by MTT assay. (a) Proliferation curves of SWI/SNF wild–type cell lines (ES2, OCI–LY–19, and SKM–1) and mutant cancer cell lines including SMARCA4 mutant (A549, H1299, and SW13), ARID1A mutant (TOV21G, HEC59, and OVISE), and PBRM1 mutant (RCC4, A704, and SK–RC–20). Error bars indicate means ± s.d. (n = 3). (Ordinary one–way ANOVA, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001) (b) Colony formation assays. Cells transduced with either control or EZH2 shRNAs were seeded at low density in standard 6–well plates. Colonies were visualized by crystal violet staining. The assays are representative of replicates of three independent experiments. (c) Western blot analysis of HCT116 isogenic cells (ARID1A wild-type parental, +/+; ARID1A heterozygous. +/−; or ARID1A homozygous deficient, −/−) after inducing with control or EZH2 shRNA. Actin was used as a loading control. (d). MTT proliferation curves of ARID1A isogenic cell lines. Error bars indicate means ± s.d. (Ordinary one–way ANOVA, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (e) Distribution of EZH2 shRNA dependency based on SWI/SNF mutational status. Each bar represents one cell line; cell lines with homozygous inactivating SWI/SNF mutations are indicated in blue. Lower DEMETER scores indicates more dependency on EZH2. The statistical difference of the EZH2 dependency between SWI/SNF mutant and wild–type cells was calculated using an unpaired, two–sample Welch’s t–test.
To more broadly test the hypothesis that SWI/SNF subunit mutations confer dependency on EZH2, we used data from Project Achilles, genome scale shRNA screens designed to identify essential genes using hundreds of cancer cell lines1,13,33. We first evaluated whether cell lines that contained EZH2 gain–of–function activating mutations were sensitive to PRC2 subunit (EZH2, SUZ12, or EED) loss. While there were only three such lines, all scored as dependent on each PRC2 subunit34,35 (Supplementary Figs. 3a–c). When compared to SWI/SNF wild–type lines, lines that contain biallelic inactivating mutations of a SWI/SNF subunit (here on, termed SWI/SNF mutant) were also dependent on EZH2 (P = 0.0087), EED (P = 0.0023), and SUZ12 (P = 0.0314) (Supplementary Figs. 3d–f). Notably, in each case, however, there were SWI/SNF mutant cell lines that did not exhibit dependency. Since it has been previously reported that loss of PRC2 subunits can potentiate the transforming effect of Ras–pathway mutations36, we asked whether activating Ras pathway mutations (in KRAS, HRAS, NRAS, or BRAF) accounted for SWI/SNF mutant cell lines that did not display PRC2 dependence. Lines with wild–type Ras pathways were in fact more dependent on EZH2 (P = 0.0009), EED (P = 0.0039), and SUZ12 (P < 0.0001) than lines with activating Ras pathway mutations (Supplementary Figs. 3g−i). When we removed lines with Ras mutations from our analysis, SWI/SNF mutant lines displayed a greater dependence on EZH2 (P < 0.0001), EED (P = 0.0005), and SUZ12 (P < 0.0001) (Fig. 1e and Supplementary Figs. 3j–k). Strikingly, almost all of the SWI/SNF mutant cell lines that did not show PRC2 dependence in the initial analysis contained activating Ras pathway mutations (compare Supplementary Figs. 3D–F to Fig. 1E and Supplementary Figs. 3j–k). Taken together, these data indicate that PRC2 is a general dependency of SWI/SNF mutant cell lines, but that the presence of Ras mutation can abrogate this dependence. This suggests that, Ras pathway mutation may reduce dependence upon EZH2 and even perhaps that EZH2 inhibition might enhance proliferation in the setting of Ras mutation.
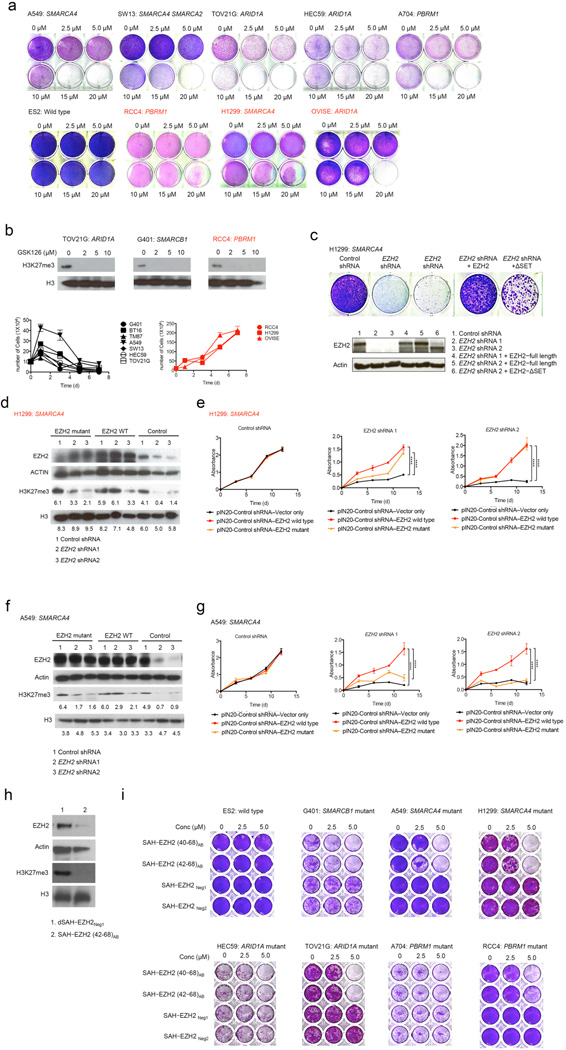
We next investigated the effect of a small–molecule inhibitor of EZH2 on SWI/SNF mutant cancer cell proliferation. GSK126 is an S–adenosyl–methionine–competitive binder of EZH2 that inhibits its histone methyltransferase activity and induces a loss of H3K27 trimethylation without affecting levels of total histone H3 or other PRC2 components27. Positive control SMARCB1 mutant rhabdoid tumor cell lines were most sensitive (Supplementary Fig. 4a), consistent with our prior identification of EZH2 dependence in these cancers. Six of ten cancer cell lines containing SWI/SNF subunit mutations also showed sensitivity beginning at 2.5 µM (Fig. 2a and Supplementary Fig. 4b). To more directly evaluate the effects of SWI/SNF subunit mutation, we generated mouse embryonic fibroblasts (MEFs) conditional for Arid1a. GSK126 efficiently reduced the levels of H3K27 tri–methylation in both cases, but Arid1A depleted MEFs showed higher sensitivity to EZH2 inhibition compared to the wild−type MEFs (Supplementary Figs. 4c–e). We next examined the several SWI/SNF mutant lines that did not respond until a 10–fold increase in concentration up to 20 µM (Figs. 2a−b). Whether the cells were sensitive or resistant to GSK126, H3K27 tri–methylation levels were markedly decreased even at the lowest dose of 2 µM (Fig. 2b). We also tested these cells with additional enzymatic inhibitors (EZ005, GSK343) with similar results (Supplementary Figs. 4f–g).
Figure 2. The catalytic activity is only partially responsible for EZH2 dependence.
(a) Treatment of A549 and SW13 (SMARCA4/SMARCA2 mutant), TOV21G and HEC59 (ARID1A mutant), and A704 (PBRM1 mutant), with GSK126 for 7 d impaired colony formation whereas ES2 (SWI/SNF wild–type), RCC4 (PBRM1 mutant), H1299 (SMARCA4 mutant), and OVISE (ARID1A mutant) cells were relatively resistant. (b) GSK126–sensitive (TOV21G, G401) and resistant (RCC4) cell lines were treated with increasing doses of GSK126 for 7 d. Immunoblots show levels of H3K27 tri–methylation and total H3. Proliferation curves are for cells treated with GSK126 10 µM ± s.d. (n = 3). (c) Effects of EZH2 shRNA knockdown and rescue with either full–length EZH2 or catalytically–dead EZH2–ΔSET (n=3). (d–g) Immunoblot analysis of EZH2 and H3K27me3 expression in GSK126–resistant (H1299) and sensitive (A549) cell lines and MTT proliferation assays before or after replacement with control (vector–only), wild–type, or point mutant EZH2. H3K27 tri–methylation and H3 blots are quantified. Actin and H3 were used as loading controls. (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by 2–way Anova). Error bars indicate means ± s.d. (n = 3). (h) Immunoblot of EZH2 and H3K27 tri–methylation in G401 cells treated with SAH–EZH2 (42–68)AB compared to negative control (dSAH–EZH2Neg1). (i) Colony formation of SWI/SNF mutant and wild–type cells in response to dSAH–EZH2 stapled peptides (n=2).
Given that all SWI/SNF mutant lines were sensitive to knockdown of EZH2 but displayed discrepant responses to the EZH2 enzymatic inhibitor, we sought to further characterize the mechanistic and functional requirements for EZH2. Expression of EZH2–ΔSET in which the methyltransferase domain has been deleted, conferred substantial rescue activity (Fig. 2c), suggesting that loss of the enzymatic function of EZH2 only partially accounted for the effects of EZH2 knockdown. We also tested the effect of a catalytically inactive point mutant EZH2 (F672I/H694A/R732K). Unlike wild–type EZH2, catalytically inactive mutant EZH2 did not rescue the levels of H3K27 tri–methylation (Figs. 2d, 2f and Supplementary Figs. 4h, 4j) but did rescue the effect of EZH2 knockdown in the GSK–resistant cell lines (Figs. 2d–g and Supplementary Figs. 4h–n).
To further investigate the response discrepancies between shRNA mediated knockdown and response to enzymatic inhibitors, we employed the recently described stabilized alpha–helix of EZH2 (SAH–EZH2) stapled peptides block H3K27 tri–methylation in a dose–dependent manner by disrupting formation of the EZH2–EED interaction within PRC237. In contrast to the enzymatic inhibitors, inhibition of PRC2 function via stapled peptide results in degradation of EZH2, thus eliminating not only its enzymatic activity but also any structural contribution. We treated our panel of genetically distinct cancer cell lines with two different SAH–EZH2 stapled peptides (Supplementary Table 1). Treatment with either SAH–EZH2 stapled peptide resulted in decreased levels of both EZH2 and H3K27 tri–methylation and impaired the growth of SWI/SNF mutant cells, while control peptides had no effect (Figs. 2h and 2i). Notably, the RCC4 and H1299 cell lines, which were resistant to the GSK126 enzymatic inhibitor, were sensitive to the SAH–EZH2 peptides (Fig. 3B). These results suggest that the growth of some SWI/SNF mutant cancers is dependent upon EZH2, but independent of its catalytic activity.
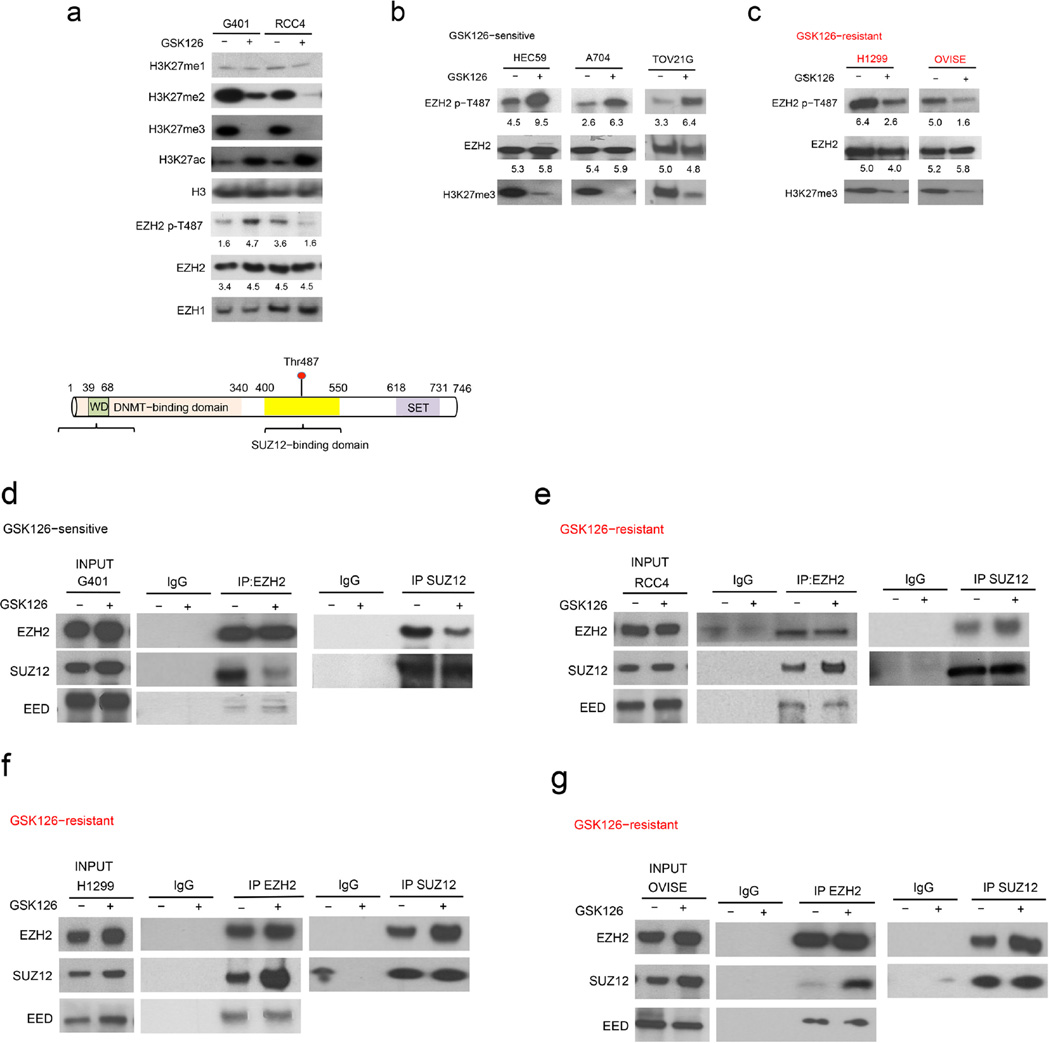
Figure 3. Disruption of PRC2 stability occurs in sensitive cells following enzymatic inhibitor treatment.
(a) Evaluation of H3K27 methylation and acetylation, total EZH1 and EZH2, and EZH2 phosphorylation in GSK126–sensitive (G401) and resistant (RCC4) cell lines following treatment with GSK126 for 7 d. EZH2 p–T487 and EZH2 blots are quantified and the numbers for quantifications are written below each blot. (b) Schematic of the location of Thr487 within EZH2. EED–, SUZ12–, and DNA methyltransferase (DNMT)–binding domains and the SET domain are indicated. (c–f) The integrity of PRC2 complex was evaluated by immunoblot using EZH2 or SUZ12 antibodies in both GSK126–sensitive (G401) and resistant (RCC4, H1299, and OVISE) cell lines. The assays are representative of replicates of three independent experiments.
To search for potential mechanisms underlying these differential responses, we first tested the effect of GSK126 upon H3K27 tri–methylation in both sensitive and resistant cell lines and found that it was equally effective (Fig. 3a). Previous studies have indicated that phosphorylation of EZH2 at Thr487, which is located in the SUZ12 binding domain (Fig. 3a), destabilizes the PRC2 complex37. We therefore evaluated Thr487 phosphorylation in cell lines treated with GSK126. Levels of Thr487 increased in sensitive cell lines (G401, HEC59, and, A704, and TOV21G) upon GSK126 treatment (Fig. 3b and Supplementary Fig. 5a) but were substantially decreased in resistant cell lines (RCC4, OVISE and, H1299) to GSK126 (Fig. 3a and Fig. 3c). Correspondingly, in sensitive cells, the addition of GSK126 destabilized the SUZ12−EZH2 interaction (Fig. 3d) but had the opposite effect in resistant cells (Fig. 3e–g). Other GSK126–sensitive cell lines (HEC59, A704, and TOV21G) also showed increased levels of Thr487 phosphorylation and reduced SUZ12–EZH2 interaction following GSK126 treatment (Supplementary Fig. 5a). Two additional resistant cell lines (H1299 and OVISE) showed substantial reductions in Thr487 phosphorylation and stabilization of SUZ12–EZH2 interactions (Fig. 3c and Figs. 3e–g). Consistent with the previous report5, we found that Thr487 phosphorylated EZH2 did not interact with SUZ12 (Supplementary Fig. 5b). Notably, we found that interaction with EED, which occurs in an amino terminal domain of EZH2 far from Thr487, was minimally affected by GSK126 treatment (Fig. 4d–g). SWI/SNF wild–type control lines (OCI–LY–19 and Toledo), which are not dependent upon EZH2 in any assay, showed no change in the levels of phosphor–EZH2 in response to GSK126 (Supplementary Fig. 5c). Collectively, these results both reveal a shared dependency of SWI/SNF mutant cancers upon PRC2 integrity and further indicate that enzymatic inhibitors of EZH2 may not fully suppress its oncogenic activity unless capable of disrupting the protein interactions of the PRC2 complex.
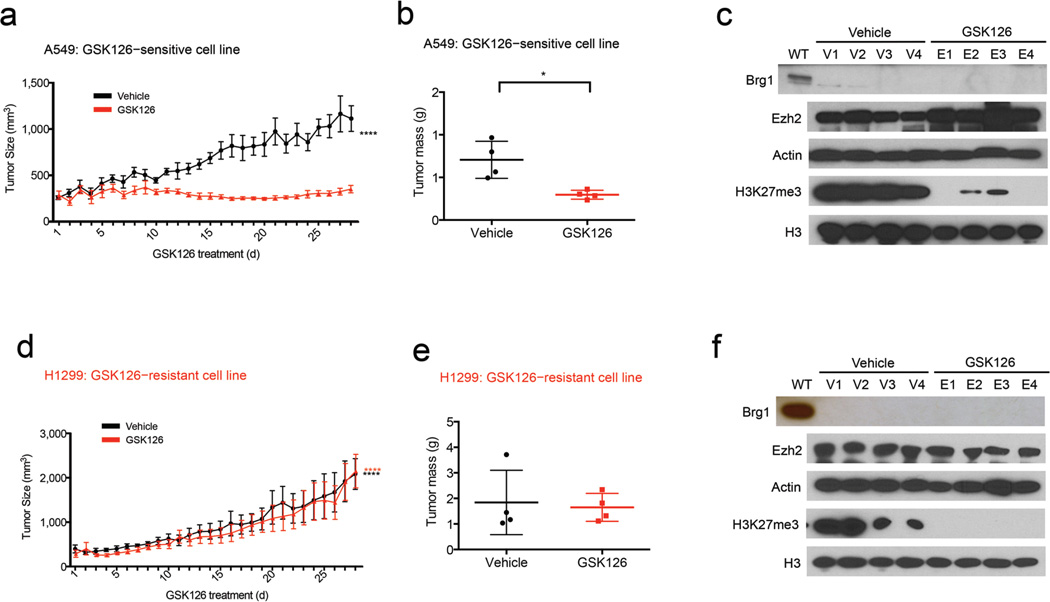
Figure 4.
In vivo inhibition of H3K27me3 via GSK126 caused regression of tumor growth of GSK126–sensitive cancers, but not GSK126–resistant cancers. Recipient mice received xenografts of A549 (a) a GSK126–sensitive line or H1299 (d) a GSK126–resistant cell line and once tumors reached 200 mm3, mice were randomized to treatment with either GSK126 or vehicle control (n = 4), ****P < 0.0001 (ordinary one–way ANOVA). (b and e) Mass of the dissected tumors (n = 4), *P = 0.0237 (unpaired t–test). (c and f) Immunoblot using the indicated antibodies for tumors isolated from mice treated with vehicle control or GSK126; Actin and H3 were used as loading controls.
To determine whether the sensitive and resistant response to EZH2 inhibiting compound could be observed in vivo, we injected a sensitive SWI/SNF mutant cell line, A549, and a resistant SWI/SNF mutant cancer cell line, H1299, into the flanks of recipient mice. Once the tumors reached 200 mm3 volume we randomly assigned mice into two groups (n = 4 mice per group) and began daily treatment with vehicle control (20% Captisol) or GSK126 (150 mg/kg in Captisol) by intraperitoneal injection for 28 d. Consistent with the effects observed in culture, GSK126 reduced the levels of H3K27me3 and blocked the growth of sensitive A549 tumors (Figs. 4a−c and Supplementary Fig. 5d) but had no effect on the growth of the resistant H1299 tumors (Figs. 4d−f and Supplementary Fig. 5e).
We conclude that the dependence upon EZH2 in some SWI/SNF–mutant cancers is largely dictated by a non–enzymatic contribution of EZH2, perhaps to stabilization of PRC2, rather than from the enzymatic activity of EZH2 per se. As EZH2 has been shown to have both PRC2–dependent/enzyme–dependent functions in transcriptional repression as well as PRC2–independent and enzymatic–independent contributions to the recruitment of co–regulators and to transcriptional regulation37, our findings suggest that mutation of SWI/SNF subunits can create dependence upon both functions of EZH2. Collectively, these results reveal a shared dependency of SWI/SNF mutant cancers upon EZH2 function and further indicate that enzymatic inhibitors of EZH2 may not fully suppress its oncogenic activity unless capable of disrupting the protein interactions of the PRC2 complex. Thus, these findings inform the mechanistic requirements of next–generation of EZH2 inhibitors for optimal blockade of pathologic PRC2 activity.
METHODS
Mice
Animals were maintained under conditions approved by the Institutional Animal Care and Use Committee at the Dana–Farber Cancer Institute. All of the procedures were monitored by the technical services of the Dana–Farber Cancer Institute mouse facility and approved by the Institutional Animal Care and Use Committee (IACUC). Crl:NU-Foxn1nu (NU/NU NUDE) mice were used in this study (Charles river, USA). All mice were 4–6 weeks of age female. Mice were kept in a cage (up to five animals in each cage) and fed sterile food and water. Randomization: animals of the same age, sex and genetic background were randomly assigned to treatment groups. Pre–established exclusion criteria were based on IACUC guidelines, and included systemic disease, toxicity, respiratory distress, interference with eating and drinking and substantial weight loss. During the study period most of the animals appeared to be in good health. In all experiments the animals were randomly assigned to the treatment groups.
Cell lines
Control cell lines (SKM–1, ES2, OCI–LY–19, Toledo) and SWI/SNF subunit mutation containing cancer cell lines (ARID1A mutant cancer cell lines, TOV21G, HEC59, OVISE; SMARCA4 mutant cancer cell lines, H1299, A549, SW13; PBRM1 mutant cancer cell lines, RCC4, A704, SK–RC–20) were obtained from American Type Culture Collection (Manassas). HCT116 ARID1A isogenic cell lines are heterozygous or homozygous knockout of ARID1A generated by knocking in premature stop codon (Q456*) obtained from Horizondiscovery Group. Cell lines were grown in recommended media supplemented with FBS and 1% penicillin/streptomycin at 37 °C and 5% CO2. All cell lines tested negative for mycoplasma contamination.
Inhibitor treatment
Cell lines were treated with EZH2 catalytic activity inhibitor, GSK126 (Excess Bio), EZ005 (Courtsey of Jay Bradner’s laboratory, Dana–Farber Cancer Institute), GSK343 (Sigma Aldrich) for 7 d.
Western blotting and antibodies
Cells were treated with DMSO or GSK126 for the indicated period of time and washed two times with 1× PBS. Cellular pellets were washed with buffer A (20 mM HEPES, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothritol, and protease inhibitor cocktail) with 0.2% Triton X–100, and incubated on ice for 5 min. After centrifugation at 600 g, the nuclei were resuspended in buffer A without Triton X–100. Nuclei were then washed with buffer A without Triton X–100. Lysates were resuspended in Buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, and protease inhibitor cocktail) and incubated on ice for 30 m. After centrifugation at 1,700g, at 4 °C, for 5 min, the nuclei were then washed with buffer B. Immunoblotting was performed. Antibodies to EZH2 (D2C9; 1:1,000), SUZ12 (D39F6; 1:1,000), H3 (9715; 1:6,000), Tri–methyl–Histone H3 Lys27 (C36B11; 1:1,000), Acetyl–H3K27 (D5E4; 1:1,000) antibodies were purchased from Cell Signaling. EED (05–1320, clone AA19; 1:1,000) antibody was purchased from Millipore. EZH2 phospho–Thr487 (EPR1410, ab109398; 1:1,000) was purchased from Abcam.
Immunoprecipitation
DMSO or inhibitor treated cells were washed twice with 1X PBS and whole cell lysates were prepared using lysis buffer (20 mM HEPES, pH 7.8, 150 mM NaCl, 10 mM EDTA, 2 mM EGTA, and 2 mM dithiothreitol, and 0.1% Nonidet P–40). The cell lysates were mixed with Dynabeads conjugated with anti–EZH2 antibody (Cell Signaling) and rotated at 4 °C overnight before removal of the supernatant. The resulting samples were analyzed by Western blot analysis using anti–SUZ12 (Cell Signaling) and anti–EED (Milipore) antibodies.
Plasmids
Plasmids for wild–type and SET–domain truncated (SET) EZH2 were kindly provided by Myles Brown (DFCI). Plasmids for null (vector only), wild–type, and point–mutant EZH2 (mutant) were kindly provided by Stuart H. Orkin (DFCI). shRNA against the human EZH2 genes and control shRNAs were cloned into pLKO.1–puro vector (Addgene). To generate lentiviral particles, 293T cells were cotransfected with an envelope plasmid (pVSVG), packaging vector (psPAX2), and shRNA expression vector using Fugene (Promega). The medium containing lentiviral particles was harvested 48–72 h post-transfection and used to transduce cancer cell lines. Cell lines at 24 h post–transduction were selected in the presence of 1 to 2 µg/ml puromycin. EZH2 specific shRNA (shRNA #1; 5’–TATTGCCTTCTCACCAGCTGC–3’, shRNA #2; 5’–CGGAAATCTTAAACCAAGAAT–3’, shRNA #3; 5’–GAAACAGCTGCCTTAGCTTCA–3’)
MTT cell proliferation assay
Cell growth was monitored by absorbance using the MTT assay according to manufacturer’s instructions (Roche). Cells were plated in 24–well plates (1×104 cells/well) and kept under indicated conditions. At the indicated times, MTT reagent (Roche) was added and cells incubated for 4 h at 37 °C then soluble reagent was added and incubated at 37 °C overnight. Cell growth was measured in a microplate reader.
Project Achilles Data Analysis
Project Achilles (www.broadinstitute.org/achilles) has been previously described1. The dataset used in this study is composed of data from cell lines that were screened with a ~ 55K shRNA library, described previously1(and data from additional cell lines that were screened with a ~ 98K shRNAs library (manuscript in preparation). The shRNA–level data was collapsed to gene–level using DEMETER. DEMETER is a computational method that estimates gene-level suppression effects in large–scale RNAi screens from shRNA–level determinations (manuscript in preparation). For each shRNA, the approach models gene-related effects and seed-sequence related effects in each cell line, resulting in a better estimation of gene effects than achieved by existing methods such as ATARiS2. The DEMETER scores for EZH2, EED, and SUZ12 used in this study are reported in Supplementary data. The complete dataset will be published elsewhere. SWI/SNF mutational status was determined from the data reported in Supplemental data. Nonsense and frameshift mutations with allelic frequencies > 0.9 were considered homozygous inactivating mutations. A few cell lines with two inactivating mutations in independent alleles (based on frequency) were included as mutant lines as indicated. Cell lines with other SWI/SNF mutations that were of unclear functional significance (heterozygous, missense, splice site) were removed from the analysis. Ras pathway mutations were annotated based on the data in Oncomap 3.03. EZH2 activating mutations were previously reported4. DEMETER scores were graphed and the difference between means calculated and analyzed with an unpaired, two-sample Welch’s t−test using GraphPad Prism.
In vivo tumour growth in xenograft model
4–6 week old female nude mice were used for xenograft studies. Approximately 1×106 viable A549 and H1299 cells were resuspended in Matrigel (BD Biosciences) and injected subcutaneously into one flank of each mouse. Tumors were allowed to grow until the size reaches ~ 200 mm3. Mice were then randomized into groups that received GSK126 or vehicle: GSK126 150 mg/kg/d intraperitoneally dissolved in 20% Captisol at pH 4.5. The xenografted tumour volume was measured everyday by caliper and determined by direct measuring two dimensions of the tumors and calculated using the formula: (width2 × length)/2. Growth curves were plotted as mean tumor sizes ± standard error of the mean (SEM).
Supplementary Material
Acknowledgments
K.H.K. was supported by an award from National Cancer Center. This work was supported by US National Institutes of Health grants R01CA172152 (C.W.M.R.), R01CA113794 (C.W.M.R.) and U01CA176058 (W.C.H.). W.K. was supported by Claudia Adams Barr grant. T.P.H. was supported by an award from the National Institute of General Medical Sciences (T32GM007753). The Cure AT/RT Now foundation, the Avalanna Fund, the Garrett B. Smith Foundation, Miles for Mary (C.W.M.R.), an LLS SCOR Project Grant (L.D.W.), and the Todd J. Schwartz Memorial Fund (L.D.W.) provided additional support.
Conflict of Interest Statement:
L.D.W. is a scientific advisory board member and consultant for Aileron Therapeutics.
Via the Dana-Farber Cancer Institute Novartis Drug Discovery Program, C.W.M.R. and W.C.H. receive research support and consulting fees from the Novartis Institutes for Biomedical Research.
Footnotes
Author Contributions: K.H.K. and C.W.M.R. designed the study; K.H.K., W.W., and J.R.H. performed the experiments; W.K. provided stapled peptides; T.P.H., V.F., and A.T. analyzed Achilles data; K.H.K. and C.W.M.R. wrote the manuscript with comments from all authors.
REFERENCES
- 1.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 4.Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69:8223–8230. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan B, et al. Roles of deletion of Arid1a, a tumor suppressor, in mouse ovarian tumorigenesis. J Natl Cancer Inst. 2014;106 doi: 10.1093/jnci/dju146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bultman SJ, et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- 7.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- 8.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiegand KC, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 11.Li M, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gui Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sausen M, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shain AH, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A. 2012;109:E252–E259. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Gallo M, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–1315. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennison JA. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 20.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 21.Tamkun JW, et al. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 22.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 23.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 25.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi W, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 28.McCabe MT, et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27) Proc Natl Acad Sci U S A. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 30.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knutson SK, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson BG, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medina PP, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Human mutation. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 34.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao DD, et al. ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome Res. 2013;23:665–678. doi: 10.1101/gr.143586.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Raedt T, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247–251. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- 37.Kim W, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Methods
- 1.Cowley GS, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Scientific data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shao DD, et al. ATARiS: computational quantification of gene suppression phenotypes from multisample RNAi screens. Genome research. 2013;23:665–678. doi: 10.1101/gr.143586.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacConaill LE, et al. Profiling critical cancer gene mutations in clinical tumor samples. PloS one. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nature genetics. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.