Abstract
Na+-activated K+ channels are members of the Slo family of large conductance K+ channels that are widely expressed in the brain, where their opening regulates neuronal excitability. These channels are fascinating for the biological roles they fulfill as well as for their intriguing biophysical properties, including conductance levels ten times most other K+ channels and gating sensitivity to intracellular Na+. Here we present the structure a complete Na+-activated K+ channel, Slo2.2, in the Na+-free state, determined by cryo-electron microscopy at a nominal resolution of 4.5 Å. The channel is composed of a large cytoplasmic gating ring within which resides the Na+-binding site and a transmembrane domain that closely resembles voltage-gated K+ channels. In the structure, the cytoplasmic domain adopts a closed conformation and the ion conduction pore is also closed. The structure provides a first view of a member of the Slo K+ channel family, which reveals features explaining their high conductance and gating mechanism.
Potassium channels control the excitability of electrically active cells by regulating the resting membrane potential in response to a variety of stimuli1. One such stimulus is an increase in intracellular Na+, which occurs following repeated membrane depolarization. Slo2.2, also known as Slack or KCNT1, is opened by increases in intracellular Na+2-8. This channel is widely expressed in the brain and shapes neuronal excitability, especially in neurons that fire action potentials at high frequencies8-12. Mutations in KCNT1, the gene that encodes the Slo2.2 protein, are linked to a variety of intellectual disabilities including malignant migrating partial seizures in infancy13-17, autosomal dominant nocturnal frontal lobe epilepsy18, and Ohtahara syndrome19. Slo2.2 has also been found in other cell types, including nociceptive and sensory neurons, where it is hypothesized to influence pain sensitivity, and in epithelial cells of the thick ascending limb of Henle's loop, where it is involved in ion reabsorption20-23.
Slo2.2 is a member of the Slo family of large conductance K+ channels, which are characterized by a transmembrane domain (TMD) containing six or seven transmembrane helices and a large cytoplasmic domain (CTD) containing two regulator of K+ conductance (RCK) domains. High-resolution structural data do not currently exist for a full-length Slo channel, but structures of isolated CTDs of Slo1, a Ca2+- and voltage-activated channel, were determined in Ca2+-free (closed) and Ca2+-bound (open) conformations24-26. In these structures the CTDs are organized into tetrameric gating rings that expand upon Ca2+ binding25. This expansion appears sufficient to open an inner helix gate in the transmembrane channel25. However, solvent accessibility experiments carried out in Slo1 and in Slo2.1 (a near relative of Slo2.2, also Na+- activated) have led to a hypothesis that these channels never fully close an inner gate, but instead close at the selectivity filter27-29. Thus, the basic question - where is the gate in Slo channels - has remained unanswered.
Cryo-EM Analysis
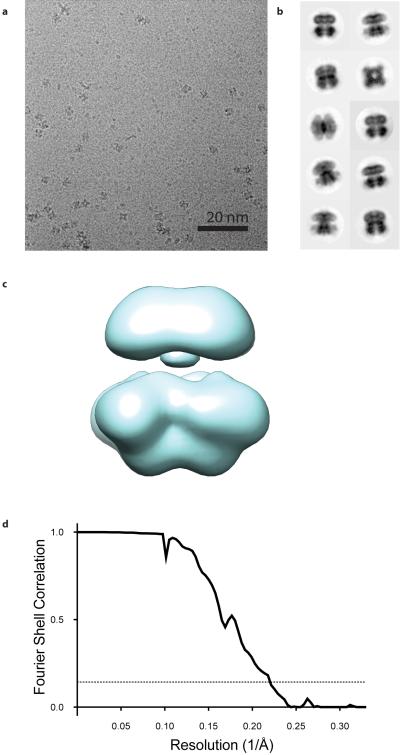
To determine the structure of a full-length Slo2.2 channel a construct encoding the entire Chicken KCNT1 gene (84% sequence identity to human KCNT1; Extended Data Fig. 1) was heterologously expressed in Spodoptera frugiperda cells. Images of frozen-hydrated preparations of detergent- and lipid-solubilized Slo2.2 tetramers in the absence of Na+ were recorded using a direct electron detector (Extended Data Fig. 2a). Using single-particle analysis a cryo-EM density map of a Slo2.2 tetramer was calculated at a nominal resolution of 4.5 Å with C4 symmetry imposed (Extended Data Fig. 2d and Extended Data Table 1). The density map contains two domains into which the tetrameric structures of a Slo1 gating ring24 and the Kv chimera α subunit30 could be manually fitted.
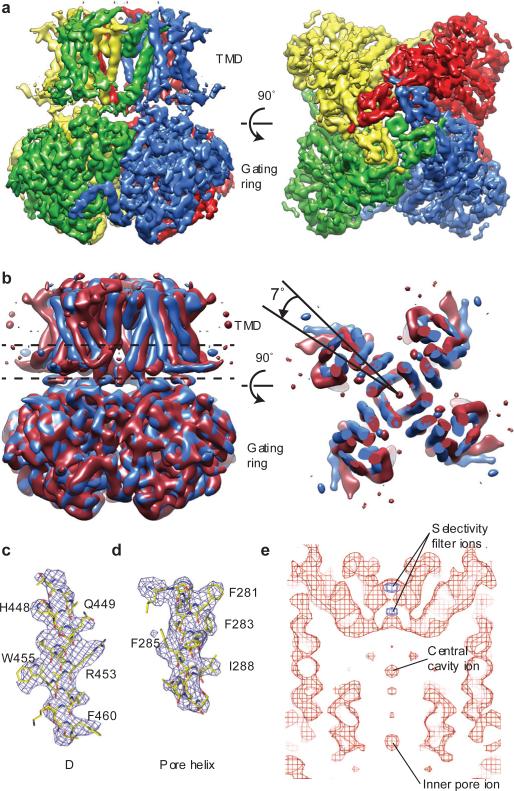
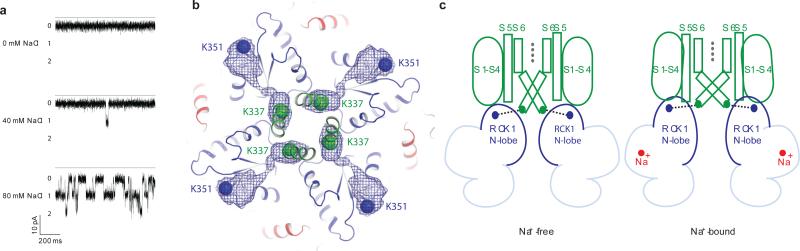
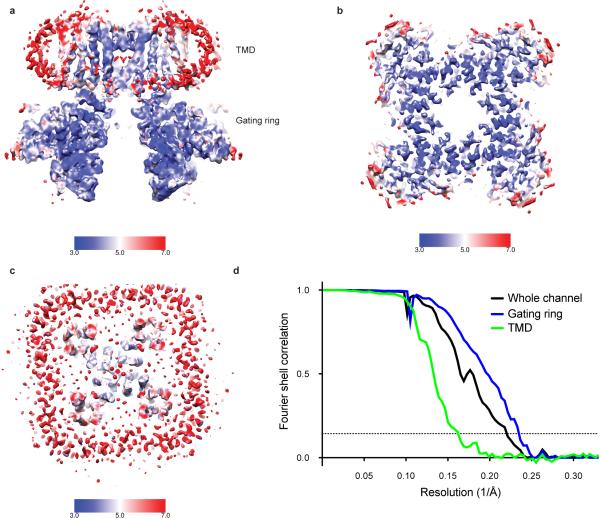
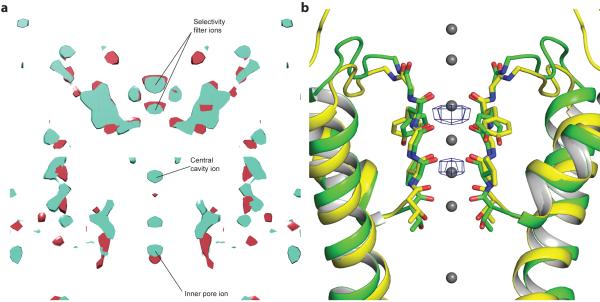
During fitting it became apparent that the density corresponding to the periphery of the TMD was of significantly poorer quality than the density corresponding to the gating ring (Extended Data Fig. 3a). To assess the source of disorder in the TMD, three-dimensional maximum likelihood classification of the particle images was performed, yielding five similar but nonidentical subclasses. These subclasses were related by a rotation of the TMD with respect to the gating ring about the four-fold axis (Fig. 1b). In the two most extreme subclasses the rotation angle is 7°. Consequently, the larger mass of the gating rings resulted in them being well aligned in the reconstruction while the smaller TMD is blurred, especially at the perimeter furthest from the four-fold axis. Separate focused refinements of the TMD and the gating ring using soft masks improved the maps for both domains (Fig. 1a) and provided enough detail to build a model starting with a voltage-dependent K+ channel structure for the TMD and a Slo1 gating ring structure for the gating ring (Fig. 1c - e and Extended Data Fig. 6 and Extended Data Table 1). We note that strong K+ ion density (6.5σ) is present in the selectivity filter and weaker density (4σ) in the central cavity and in the inner pore closer to the cytoplasm (Fig. 1e and Extended Data Fig. 5a). Prior knowledge of the location of K+ ions in the selectivity filter and central cavity of K+ channels from x-ray crystallographic studies supports our assignment of these densities as ions rather than noise along the 4-fold axis (Extended Data Fig. 5b)31.
Figure 1. Cryo-EM structure of chicken Slo2.2.
a, Cryo-EM density map of chicken Slo2.2 following focused refinements of the TMD and the gating ring. The gating ring map is filtered to 4.2 Å and the TMD map is filtered to 4.5 Å. b, Density map of two of the 3D subclasses filtered to 7 Å and aligned by their gating rings. The density slice corresponds to the region of the TMD between the dashed lines. c-d, Fragments of the density map corresponding to helix αC in the gating ring (c) and the pore helix in the TMD (d). The refined model is shown in sticks. Large side chains that were used to register the sequence are highlighted. e, Central section of the density map through the TMD calculated at 4σ (red) and 6.5σ (blue) with densities corresponding to K+ ions highlighted.
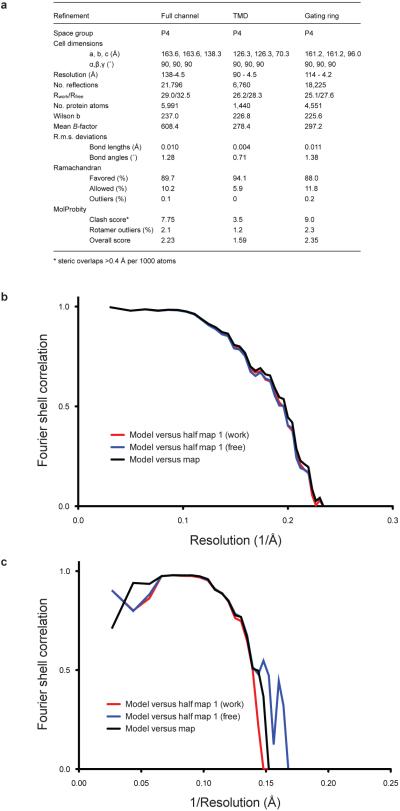
The gating ring and TMD models were built and refined independently against phases and amplitudes calculated from the focused refinement maps using reciprocal space algorithms. The gating ring was refined to a resolution of 4.2 Å using p4 symmetry, the nominal resolution of the map, with Rw ork/Rfree = 25.5/28.0 and good geometry (Extended Data Figs. 3d and 4a,b and Extended Data Table 1). The nominal resolution of the TMD map is 5.2 Å, but local resolution estimates combined with the resolved features of the map indicate that the map, particularly within the pore domain, contains information that extends to higher resolution (Fig. 1 a, c - e, Extended Data Figure 3c, d, Extended Data Table 1). The TMD structure was therefore refined using p4 symmetry to 4.5 Å, the nominal resolution of the full channel map, with Rw ork/Rfree = 25.7/27.1 and good geometry (Extended Data Fig. 4a,c and Extended Data Table 1). A model of the complete channel was generated by manually docking the independently refined TMD and gating ring models into the full channel map. The fit was improved by rigid body refinement of the gating ring and TMD against amplitudes and phases extracted from the full map (Fig. 2b and Extended Data Table 1). The final refined model contains residues 244-337, 351-612, 769-860, 865-1019, 1041-1171. In addition, unregistered polyalanine α-helical models were built for S0 (amphipathic helix similar to those present in Kv channels), S1, the linker between S1 and S2, S2, S3 and S4 in the TMD and αX’, αY’ and αZ’ in the gating ring (Extended Data Fig. 1).
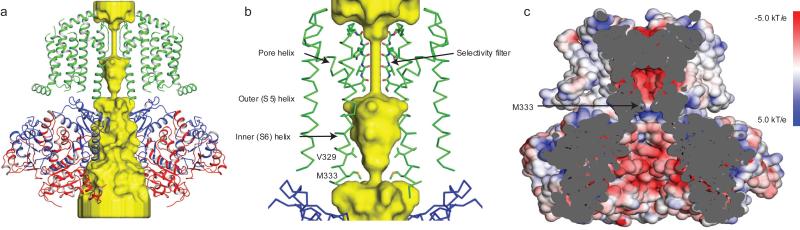
Figure 4. Slo2.2 ion conduction pathway.
a, Surface representation of the Slo2.2 pore and ribbon diagram with front and rear subunits excluded for clarity. The K+- accessible surface was determined using the HOLLOW script (see methods section for details). b, The Slo2.2 constriction site is formed by Met 333 from the inner helix of each of the four subunits. c, Electrostatic surface potential of the Slo2.2 ion conduction pathway colored from red (−5.0 kT/e) to blue (5.0 kT/e).
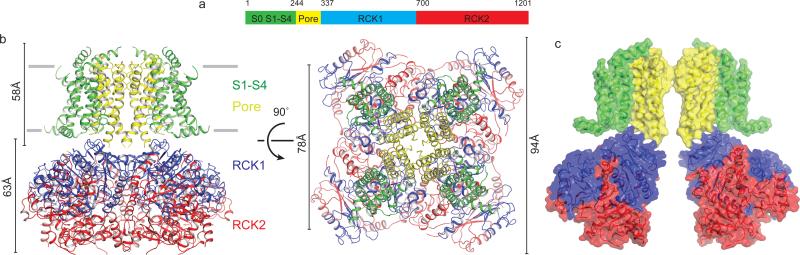
Figure 2. Architecture of Slo2.2.
a, Domain organization of chicken Slo2.2. b, Ribbon diagram of Slo2.2. The S1-S4 domain is colored in green, the pore domain is colored in yellow, the RCK1 domain in blue and the RCK2 domain in red. The approximate width of the membrane is marked by the grey lines. c, Surface depiction of Slo2.2 with front and rear subunits removed for clarity.
Structure of Slo2.2
The tetrameric Slo2.2 channel is 120 Å in length along an axis perpendicular to the membrane plane (Fig. 2b). More than half of this length corresponds to the gating ring, which protrudes into the cytoplasm. Viewed down the 4-fold axis the TMD is approximately 80 Å on square edge and the gating ring is 95 Å (Fig. 2b). Each subunit polypeptide chain builds up the Slo2.2 channel in 3 layers, a TMD layer and two RCK domain layers (RCK1 and RCK2), which form the gating ring (Fig. 2b, c).
The TMD is similar in topology to a voltage-dependent K+ (Kv) channel, to which Slo2.2 is related32. The central ion conduction pore is surrounded in the membrane by voltage sensor-like domains, which we refer to as S1-S4 domains to emphasize their non-voltage-dependent uniqueness (Fig. 2b, c). As in Kv channels, the S1-S4 domains are arranged as appendages that project laterally into the lipid membrane. At a level of greater structural detail we note two important distinctions between the TMDs of Kv1.2 and Slo2.2.
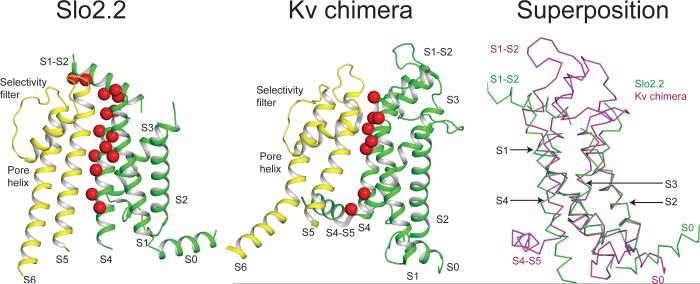
The first distinction is the manner in which the surface of the S1-S4 domain forms its contacts with the pore. In the Kv channel the voltage sensor's interaction with the pore occurs mainly through the extracellular half of S1 and through the S4-S5 linker (Fig. 3). This configuration leaves the charged S4 helix free to move in response to voltage differences across the membrane and exert force on the S4-S5 linker to gate the channel. In Slo2.2 the extracellular half of S1, nearly the entire S4, and a helix from the S1-S2 connecting segment form an extensive contact surface with the pore. The Kv voltage sensor and Slo2.2 S1-S4 domains have essentially the same helical structure, but their different orientations with respect to the pore reconfigure the contact surface (Fig. 3). In Slo2.2 S4 runs antiparallel to and in close contact with S5, as if its function is to help glue the S1-S4 domain to the pore rather than to provide a mobile voltage sensor. This possibility is consistent with the amino acid sequence of Slo2.2, which shows two positive and two negative charged amino acids in or near S4 instead of the excess positive charges observed in Kv channel S4s and in the S4 sequence of Slo1 and Slo333. Further, mutations involving these positive and negative charges in Slo2.2 do not produce large changes in voltage-dependence34. Thus, S1-S4 does not appear to function as a mobile voltage sensor in Slo2.2. We further note a surprising observation, which may be related to the differences in S4: we cannot detect cryo-EM density for an S4-S5 helical linker in Slo2.2. Higher resolution data will be required to tell whether this linker is partially disordered or simply not present as a helix. Its absence in Slo2.2 is conspicuous, and implies that this region serves a different function than in Kv channels.
Figure 3. Interactions between pore and S1-S4 domains.
Ribbon diagram of Slo2.2 (left) S1-S4 domain and Kv chimera (middle) voltage-sensor domain colored in green with the pore domain colored in yellow. Red spheres represent the residues on S1, S4 and the S1-S2 linker that are close enough to interact with the pore domain. Superposition of the S1-S4 domain of Slo2.2 with the voltage-sensor domain of Kv chimera (right) by aligning S1, S2 and S3.
The second distinction is the manner in which the TMD connects to the gating ring. The Kv channel contains a CTD consisting of a T1 domain and beta subunit. These are held at a distance of nearly 15 Å away from the membrane plane and consequently ions can gain access the transmembrane pore through lateral openings between the TMD and CTD, near the membrane surface35. In contrast, in Slo2.2 the TMD and gating ring form a complementary interface near the membrane surface. This is most easily appreciated in a surface representation of Slo2.2 (Fig. 2c). In this picture the gap at the perimeter of the interface would be filled in if a full side-chain model (instead of poly-alanine) of the S1-S4 domain had been built. The relatively snug fit between the TMD and gating ring in Slo2.2 has important implications for ion conduction and gating.
Ion Conduction
In contrast to the Kv channel, the continuous pore across the membrane and through the gating ring makes Slo2.2 more like an inward rectifier K+ channel, which also has an extended ion pathway due to a tightly engaged CTD36,37 (Fig. 4a). In inward rectifiers, however, the extended pore is narrower and contains binding sites for blocking ions, a property that gives rise to inward rectification36,38. In contrast, in Slo2.2 the gating ring creates a massive funnel that begins nearly 40 Å wide and gradually narrows as it approaches the pore inside the TMD. The inner surface of the funnel is highly electronegative owing to the presence of many aspartate and glutamate amino acids (Fig. 4c). This electrostatically negative funnel, by functioning as a cation-attractor, undoubtedly contributes to the unusually high conductance (nearly 200 pS for Slo2.2: about 20 times that of a canonical Kv channel such as Shaker39) of Slo channels (Extended Data Fig. 7a). Indeed, Slo channels are the highest conductance K+ channels known and a recent mutagenesis study with Slo1 showed that the gating ring contributes to the high conductance40.
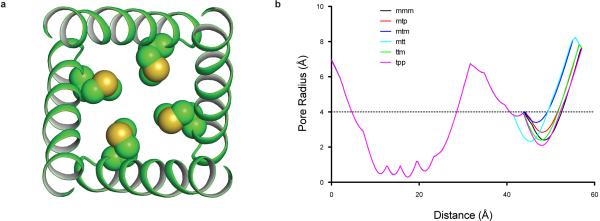
The ion conduction pathway leading from the cytoplasm up to the selectivity filter has a single constriction formed by the crossing of four transmembrane S6 (inner) helices (Fig. 4b). The pore constricts to a minimum radius at Met 333, which points its beta carbon towards the center of the pore. Cryo-EM density for this side chain beyond the beta carbon is not well resolved; however, modeling the six most frequently observed rotamers of Met yields a range of pore diameters – defined by diagonally opposed van der Waals surfaces – ranging from 4 Å to 6 Å (Fig. 4b and Extended Data Fig. 8). A hydrated K+ ion has a diameter closer to 8 Å, therefore, the structure is compatible with a closed conformation, consistent with the Na+-free conditions under which this structure was determined. Adding to the physical barrier that the van der Waals constriction at the level of Met 333 would present to a permeating K+ ion, the local electrostatic environment would raise the barrier further (Fig. 4c). The band of blue color seen in Figure 4c (positive electrostatic potential) coincident with the constriction is due to Arg 335 and Lys 337 on the inner helices and Arg 396 on the surface of the gating ring; these positive-charged amino acids impose a long-range cation-repulsive potential. Thus, the inner helix gate appears closed to K+ diffusion by virtue of its narrow constriction and positive electrostatic potential at the level of the constriction.
Gating inferences from structure
The Slo2.2 gating ring is similar in overall structure to the gating rings in Slo1 and Slo324,25,41. Two non-identical RCK domains on the C terminus of each subunit assemble through a ‘flexible interface’; four RCK domain pairs then assemble with each other through ‘assembly interfaces’ to form the ring (Fig. 5). The assembly interface is essential to the function of Slo1 because it contains the high-affinity Ca2+-binding site known as the ‘Ca2+ bowl’26. Instead of a Ca2+ bowl, Slo2.2 has an extended α-helix, αQ’, which interacts across the assembly interface with an adjacent subunit (Fig. 5). A helix-turn-helix structure (αX’, αY’ and αZ’) is also unique to the assembly interface in Slo2.2. This helical elaboration is provided by a ~110-residue insertion in the RCK2 domain that is conserved and unique to Slo2 channels (Extended Data Fig. 1). Putative Na+-binding site amino acids, identified through mutagenesis, do not fall within the assembly interface42. If this means that Na+ in Slo2.2 activates from a different locus than Ca2+ in Slo1 we should not be too surprised. After all, the prokaryotic K+ channel MthK contains a Ca2+-activated gating ring in which Ca2+ binds near the flexible interface rather than the assembly interface43. Apparently the conformations of RCK-based gating rings can be modulated in a variety of ways.
Figure 5. Slo2.2 gating ring.
Wire diagram of the Slo2.2 gating ring with the boundary of one subunit highlighted (left). The flexible interface that mediates interactions between the RCK1 and RCK2 domains of each subunit and the assembly interface that mediates interactions between adjacent subunits are shown. The assembly interface for Slo2.2 (middle) and Slo1 (right) with one subunit shown as a surface and the second shown as ribbons. The RCK1 domains are colored in blue and the RCK2 domains are colored in red. The Ca2+ ion resolved in the Slo1 Ca2+ bowl is shown as a yellow sphere.
A possible locus for Na+ binding was identified in two studies using mutagenesis42,44. Both studies found that Asp 812 (or the corresponding residue in Slo2.1, Asp 757) reduced Na+ activation. In one study a second residue (His 817 in Slo2.2) was also found to affect Na+ activation, however, position 817 is located 15 Å away from Asp 812 and thus could not possibly form a single binding site along with Asp 81242. Additional studies including a structure of Slo2.2 in the presence of Na+ will be required to better understand Na+ activation in Slo2.2.
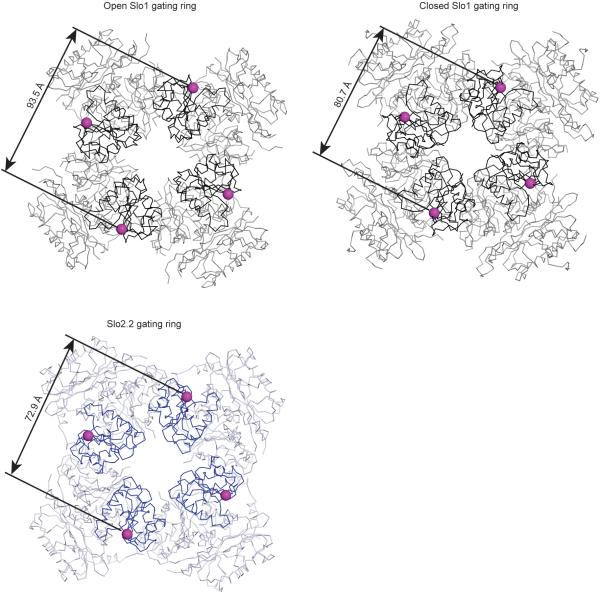
Crystal structures of isolated (from the TMD) Slo1 gating rings in the absence and presence of Ca2+ show that when Ca2+ binds to the Ca2+ bowl the N-lobe of RCK1 undergoes a conformational change24,25. This causes the four N-lobes (one from each subunit) on the surface of the gating ring to expand like the petals of an opening flower (Extended Data Fig. 9). This expansion is illustrated by marking a residue (Lys 343) on the N-lobe that connects to S6 (the inner helix gate) through a linker (Extended Data Fig. 9). The distance between diagonally opposed Lys 343 residues is approximately 81 Å and 94 Å in closed and opened Slo1 gating rings, respectively. The equivalent residue in Slo2.2 (Lys 351) measures 73 Å from its diagonally opposed molecular symmetry mate, and the position of the N-lobe is similar to that in the closed Slo1 gating ring. Thus, the Slo2.2 gating ring is closed, as is the inner helical gate to which it is connected. Cryo-EM density shows a weak connection between Lys 351 on the N-lobe and Lys 337 on S6 (Fig. 6b). This connection supports the idea that expansion of the N-lobes upon Na+ binding to the gating ring is mechanically connected to pore opening through this connecting linker. But the structure raises an additional mechanistic possibility. The interface formed between complementary-shaped surfaces on the TMD and gating ring involves the RCK1 N-lobes. If indeed the N-lobes expand when Na+ binds, as occurs when Ca2+ binds to the gating ring of Slo1, then the expansion would do more than pull on the linkers to S6; it would also presumably exert forces onto the TMD through the protein-protein interface45. Such a mechanism of force transfer across the interface might explain why the S1-S4 domain in Slo2.2 is firmly attached through a large interface with the pore. – so that conformational changes within the gating ring can be efficiently transmitted to the pore via the S1-S4 domain.
Figure 6. Slo2.2 gating.
a, Representative single channel recordings of Slo2.2 in planar lipid bilayers with symmetrical 0 - 80 mM NaCl and 150 mM KCl when held at −80 mV. b, Cryo-EM density connects Lys 337, the C-terminal residue of the TMD, to Lys 351, the N-terminal residue of the gating ring, the Cα positions of which are highlighted by spheres. The density is filtered to 5 Å. c, Model for Slo2.2 channel activation. Na+ binding to the gating ring results in an upward movement of the RCK1 N-lobes, which pull apart the inner S6 helices and push up the cytoplasmic ends of S4 and S5, thereby opening the pore.
In summary, this study visualizes a Na+-activated Slo2.2 K+ channel in its ligand-free, closed conformation. The structure supports three important concepts concerning the function of this class of K+ channels: (i) A closed inner helix gate, analogous to that observed46 or deduced47 in other K+ channels, would prevent K+ conduction through restrictive van der Waal surfaces and an unfavorable electrostatic environment; (ii) Expansion of N-lobes on the gating ring would open the pore through a polypeptide linker and through the protein-protein interface between the gating ring and TMD; (iii) A massive electrostatic funnel facing the cytoplasm helps to explain why Slo channels are the highest conductance K+ channels.
Methods
Expression and Purification
A synthetic gene fragment encoding residues 1 to 1201 of Chicken Slo2.2 (GI:73920088) was purchased from Bio Basic Inc. The resulting fragment was cloned into a modified pFastbac vector (Invitrogen) containing green fluorescent protein and a 1D4 antibody recognition sequence (TETSQVAPA) on the C terminus. Sf9 (Spodoptera frugiperda) cells infected with the baculovirus were cultured at 27°C for 72 hours in supplemented Grace's insect cell medium (Invitrogen). Cells were washed with ice-cold phosphate-buffered saline and extracted for 3 hours at 4°C with buffer containing 50 mM Hepes pH 7.4, 300 mM KCl and 40 mM dodecyl-β-D-maltopyranoside (DDM) in the presence of a protease inhibitor cocktail (2 μg/ml leupeptin, 2 μg/ml aprotinin, 2 μg/ml pepstatin A, 1 mM benzamidine, 100 μg/ml 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, and 100 μM phenylmethane sulphonylfluoride). The insoluble fraction was removed by centrifugation at 35k × g for 45 minutes at 4°C and the remaining soluble fraction was incubated with 1D4-affinity resin pre-equilibrated with 20 mM Hepes pH 7.4, 300 mM KCl and 4 mM DDM. The suspension was mixed for 5 hours at 4°C. Beads were collected on a column by gravity and then washed with 10 column volumes of wash buffer (20 mM Hepes pH 7.4, 300 mM KCl, 4 mM DDM and 0.1 mg/ml 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE): 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) (3:1 w/w)). The protein was digested with PreScission protease (20:1 w/w ratio) on the column overnight at 4°C to remove the affinity tag and then eluted with 2 column volumes of wash buffer. Concentrated protein was further purified by size-exclusion chromatography on a Superose 6 (GE Life Sciences) column in 20 mM Hepes pH 7.4, 300 mM KCl, 1.5 mM DDM and 0.05 mg/ml POPE:POPG (3:1 w/w). Peak fractions were pooled and concentrated to 3-5 mg/ml for cryo-electron microscopy analysis.
Electron microscopy sample preparation and imaging
3.5 μl of purified channel was pipetted onto a glow discharged copper Quantifoil R1.2/1.3 holey carbon grids (Quantifoil). Grids were blotted for 3.5 s at ~85% humidity and flash frozen in liquid nitrogen-cooled liquid ethane using an FEI Vitrobot Mark IV (FEI). Grids were transferred to an FEI Titan Krios electron microscope operating at an acceleration voltage of 300 keV. Images were recorded in an automated fashion on a Gatan K2 Summit (Gatan) detector set to in super-resolution counting mode with a super-resolution pixel size of 0.52 Å using SerialEM48. Inelastically scattered electrons were filtered by a Gatan Imaging Filter with a slit width of 30 eV (Gatan). Images were recorded for 5 s with a sub-frame exposure time of 200 ms and a dose of ~8 electrons per physical pixel (1.04 Å at the image plane) for a total accumulated dose of ~40 electrons per Å2 on the specimen over 25 sub-frames (~1.5 electrons per Å2 per sub-frame).
Image processing and map calculation
Dose-fractionated super-resolution images were 2 × 2 binned (resulting in a pixel size of 1.04 Å) for whole-frame motion correction with motioncorr49. Following motion correction, the sum of the 25 aligned sub-frames in the image stack was 4 × 4 binned (pixel size of 4.16 Å pixel) for manual particle selection. 40,643 particles were manually selected in BOXER50 from 2,243 images and then extracted from the motion-corrected images in RELION51. The effects of the contrast transfer function were estimated by ctffind352 and CTER53. 22,154 particles were subjected to iterative stable alignment and classification54 (ISAC), specifying 200 particles per group and a pixel error threshold of 2; 6 generations yielded a set of 642 stable class averages. The classes were inspected manually and those whose images contained portions of more than one particle or that had other obvious defects were discarded. The remaining 472 classes were used to generate an initial model with the multi_shc command in SPARX55 with and without imposing C4 symmetry. The results were similar for both cases, so the symmetrized version was used as the initial model for 3D refinement (Extended Data Fig. 3c.).
The initial set of 40,643 particles was subjected to 2D classification in RELION from which particles classified into low-abundance classes were removed, resulting in a stack of 35,586 particles. These 35,586 particles were segregated into 6 classes by RELION's 3D classification algorithm using the initial model generated in SPARX. Four of the six classes appeared similar by visual inspection in UCSF Chimera56 and were combined into a stack of 24,231 particles for 3D refinement in RELION. Particle polishing was applied to the particles using RELION 1.3 to correct for individual particle movements and for attenuation of the high-resolution terms due to rapid movement of the initial few frames and radiation damage in the later frames57, yielding a map that achieved a resolution of 4.5 Å as assessed by Fourier shell correlation of independently refined half datasets and using the 0.143 cut-off criterion58. Local resolution estimates calculated using ResMap demonstrated that the periphery of the TMD was poorly ordered compared to the pore and the gating ring59.
3D maximum-likelihood classification of the 24,231 particles was performed using FREALIGN60, starting from the orientation parameters determined by RELION. Five subclasses were specified and the resolution was limited to 8.8 Å for classification and 6.6 Å for refinement to minimize the effects of high-resolution overfitting. Maps of each of the subclasses were calculated at 7 Å and were superimposed based on their gating rings using UCSF Chimera. The orientation of the TMDs varied by up to 7°.
In order to minimize the effects of the rotational variability, soft masks were generated (5 pixel cushion and 5 pixel cosine edge fall-off) for both the gating ring and TMD and applied to the reconstructions during refinement61. The gating ring mask was applied at the end of every round of refinement in RELION, resulting in a map that reached a resolution of 4.2 Å. The variation of the TMD made it difficult to align the entire data set, so instead the two most similar classes from the 3D classification were combined, yielding a subset of 11,303 images. The angular parameters of these images were determined by FREALIGN using an iterative process in which two refinement cycles were run with the mask followed by two without the mask. This iterative procedure of refining with and without mask application was necessary due to the small mass of the TMD relative to the whole channel. The map of the transmembrane region achieved a final resolution of 5.2 Å by FSC.
Model building
The structure of the closed Slo1 gating ring24 (PDB code: 3NAF) was docked into the gating ring map using UCSF Chimera and then manually rebuilt in coot62 to fit the density. For the regions lacking homology with Slo1, secondary structure elements were first built into the density map and then connected with manually built loops. Once the backbone was traced, the sequence was registered using a combination of identification of large side chains and sequence homology with Slo1. The structure of the Kv chimera α subunit30 (PDB code: 2R9R) was docked into the TMD map using UCSF Chimera using the selectivity filter and pore helix as a reference. The positions and tilts of S5 and S6 were adjusted by moving the helices as rigid domains to fit the density and then loops connected them were manually rebuilt. The sequence was registered using the presence of large side chains, the proline kink and sequence homology among K+ channels. The S1-S4 domain was built as 6 polyalanine helices in coot (S0 - 15 residues, S1 - 28 residues, S1-S2 - 17 residues, S2 and S3 - 53 residues, S4 - 26 residues). The helices were assigned based upon homology with Kv chimera and roughly agree in length with those predicted by hydropothy analysis. A model of the full channel was generated by manually docking the independently determined structures for the gating ring and TMD into the map of the full channel.
Reciprocal space refinement
The masked cryo-EM density maps were translated to a new unit cell that extended 10 Å from the model in all directions and then solvent flattened by creating a mask that extends 3 Å away from the model and setting the density outside of the mask to approximate the solvent content of a protein crystal. Reciprocal space structure factors were calculated from the translated and solvent-flattened maps using FFT63. The phases were blurred in a resolution-dependent fashion by setting FOM = FSC and subsequently used to calculate Hendrickson-Lattman coefficients. Coordinate and B-factor refinement of the gating ring and TMD models were performed using Phenix64 using X-ray scattering factors against a maximum likelihood with experimental phase probability distribution target function with 5% of the reflections excluded as a free set to monitor overfitting. Secondary structure, geometric, B-factor and Ramachandran restraints were employed throughout refinement. Rigid body refinement of the full channel model was performed Phenix64 using X-ray scattering factors against a maximum likelihood with experimental phase probability distribution target function with 5% of the reflections excluded as a free set to monitor overfitting.
Overfitting of the models during refinement was also evaluated using a method previously described61. Briefly, all atoms of the refined models were randomly displaced by 0.1 Å and then subjected to one round of coordinate and B-factor refinement against phases and amplitudes calculated from one of the independently determined half-maps. Following this refinement cycle, the Fourier shell correlation was calculated between the resulting model and both of the half maps as well as the full map. Structure figures were prepared with UCSF Chimera56, Pymol (Pymol version 1.7.2 Schrödinger, LLC), Hollow65 and APBS66,67. All structure calculations were performed using software compiled by SBGrid68.
Reconstitution and electrophysiological recordings from planar lipid bilayers
Purified channels were reconstituted into octyl maltoside (Anatrace)-solubilized 3:1 (wt:wt) POPE:POPG lipid vesicles as described69. Detergent was removed by dialysis for 5 days against detergent-free buffer containing 10 mM HEPES-KOH pH 7.5, 450 mM KCl, and 2 mM dithiothreitol at 4°C, with daily buffer exchanges. After 5 days, all residual detergent was removed by incubating the reconstituted channels with Bio-Beads (Bio-Rad) for 2 hr at room temperature. The reconstituted channels were aliquoted and flash frozen into liquid nitrogen prior to storage at −80°C.
Planar lipid bilayer experiments were performed as described previously70,71. Lipids of desired compositions were prepared by dissolving argon-dried lipids in decane to a final concentration of 20 mg/ml. Lipid solutions were painted over a 300 μm hole in a polystyrene partition that separated the two chambers to form the planar lipid bilayer. Reconstituted channels were pipetted onto the chamber side of the bilayer after thinning of a planar lipid bilayer had been detected via monitoring of electrical capacitance. All recordings were performed using the voltage-clamp method in whole-cell mode. Analogue signals were filtered at 1 kHz using a low-pass Bessel filter on an Axopatch 200B amplifier (Molecular Devices) in whole-cell mode and digitized at 10 kHz using a Digidata 1400A analogue-to-digital converter (Molecular Devices). The pClamp software suite (Molecular Devices) was used to control membrane voltage and record current. All electrophysiological recordings were replicated in at least three independent experiments.
Extended Data
Extended Data Figure 1. Sequence alignment of Slo channels.
a, Sequence alignment of chicken Slo2.2 with human Slo2.2 and human Slo2.1. b, Predicted position of transmembrane helices in Slo2.2 S1-S4 domain based upon hydropothy analysis using Jpred 472. c-d, Structure-based sequence alignment of chicken Slo2.2 TMD with rat Kv chimera (c) and chicken Slo2.2 gating ring with human Slo1 gating ring (d). Helices are colored in blue and beta strands are colored in red.
Extended Data Figure 2. Full channel 3D reconstruction of Chicken Slo2.2.
a, Representative micrograph of detergent- and lipid-solubilized Slo2.2 in vitreous ice. b, 10 selected 2D class averages. c, Ab-initio initial model of Slo2.2. d, Fourier shell correlation (FSC) curve of the full channel reconstruction with the nominal resolution estimated to be 4.5 Å based on the FSC = 0.143 criterion.
Extended Data Figure 3. Focused refinement of the gating ring and the TMD.
a, 3D density map of the full channel reconstruction colored according to local resolution. b, 3D density map calculated following focused refinement using a mask to only include the gating ring colored according to local resolution. c, 3D density map calculated following focused refinement using a mask to only include the TMD colored according to local resolution. d, Fourier shell correlation of the full channel reconstruction (4.5 Å), the gating ring-focused refinement reconstruction (4.2 Å) and the TMD-focused refinement reconstruction (5.2 Å).
Extended Data Figure 4. Validation of the Slo2.2 model.
a, Refinement statistics for Slo2.2 full channel, TMD and gating ring models. b-c, FSC curves for cross-validation of the refined gating ring (b) and TMD (c) models. Black curve is the refined model compared to the full data set, red curve is the refined model compared to half map 1 (used during test refinement) and blue curve is the refined model compared to half map 2 (not used during test refinement).
Extended Data Figure 5. K+ ions in Slo2.2.
a, Central section of the density maps of the two independently calculated half maps with densities corresponding to K+ ions highlighted. b, Superposition of the Slo2.2 selectivity filter (green) with KcsA (PDB: 1K4C) selectivity filter (yellow). Density peaks resolved in Slo2.2 selectivity fitler at 6.5σ are shown as blue mesh. K+ ions resolved in KcsA are shown as grey spheres.
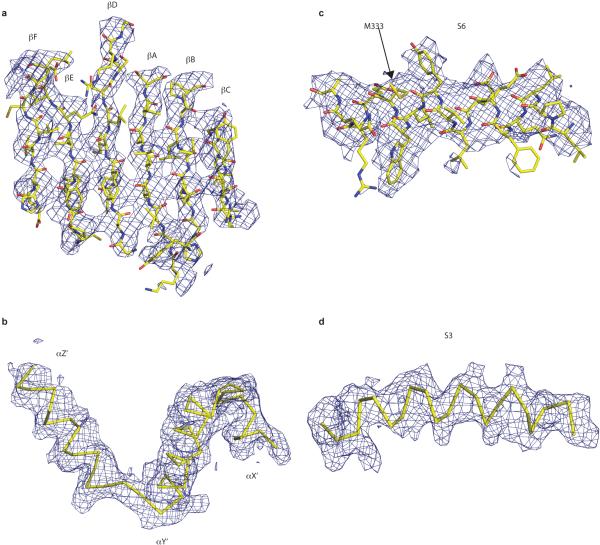
Extended Data Figure 6. Representative segments of the cryo-EM density map.
Selected regions of the gating ring density (a,b) and TMD density (c,d) maps with refined model.
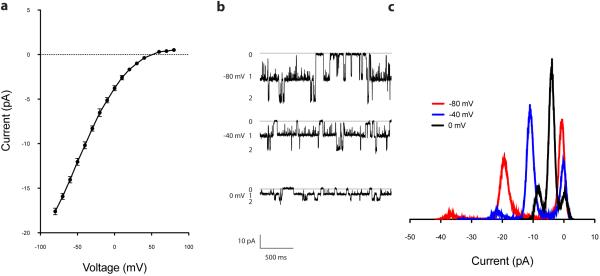
Extended Data Figure 7. Single channel conductance of Slo2.2.
a, Single channel current-voltage relationship (mean ± s.e.m.) for Slo2.2 in planar lipid bilayers. Single channel conductance is ~200 pS. b, Representative recordings of Slo2.2 held at −80, −40 and 0 mV in planar lipid bilayers. Chamber solution contained 135 mM NaCl, 15 mM KCl and cup solution contained 150 mM KCl. c, Histogram of Slo2.2 currents when held at −80, −40 and 0 mV.
Extended Data Figure 8. Inner helix gate.
a, Ribbon diagram of the Slo2.2 pore with Met 333 side chains modeled as spheres. b, Pore radius plot for Slo2.2 with Met 333 modeled as each of the six most frequently observed rotamers.
Extended Data Figure 9. Slo2.2 gating ring is in a closed conformation.
Wire diagram of Slo1 gating ring in the open (top left) and closed (top right) conformations. The mobile RCK1 N-lobe is colored in black and the rest of the gating ring is colored in grey. The N-terminal residue of the gating ring, Lys 343, is shown as a sphere. Wire diagram of the Slo2.2 gating ring (bottom) with the RCK1 N-lobe colored in blue and the rest of the gating ring colored in white. The N-terminal residue of the gating ring, Lys 351, is shown as a sphere.
Extended Data Table 1.
3D reconstructions of chicken Slo2.2 by cryo-electron microscopy
| Refinement | Full channel | TMD | Gating ring |
|---|---|---|---|
| Space group | P4 | P4 | P4 |
| Cell dimensions | |||
| a, b, c (Å) | 163.6, 163.6, 138.3 | 126.3, 126.3, 70.3 | 161.2, 161.2, 96.0 |
| α,β,γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 138-4.5 | 90 - 4.5 | 114 - 4.2 |
| No. reflections | 21,796 | 6,760 | 18,225 |
| Rwork/Rfree | 29.0/32.5 | 26.2/28.3 | 25.1/27.6 |
| No. protein atoms | 5,991 | 1,440 | 4,551 |
| Wilson b | 237.0 | 226.8 | 225.6 |
| Mean B-factor | 608.4 | 278.4 | 297.2 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.010 | 0.004 | 0.011 |
| Bond angles (°) | 1.28 | 0.71 | 1.38 |
| Ramachandran | |||
| Favored (%) | 89.7 | 94.1 | 88.0 |
| Allowed (%) | 10.2 | 5.9 | 11.8 |
| Outliers (%) | 0 1 | 0 | 0.2 |
| MolProbity | |||
| Clash score* | 7.75 | 3.5 | 9.0 |
| Rotamer outliers (%) | 2.1 | 1.2 | 2.3 |
| Overall score | 2.23 | 1.59 | 2.35 |
steric overlaps >0.4 Å per 1000 atoms
Acknowledgements
We thank Zhiheng Yu and Jason de la Cruz at the HHMI Janelia Cryo-EM facility for assistance in data collection, Stephen Harrison and Simon Jenni for assistance with phenix refinement of cryo-EM density maps and members of the MacKinnon laboratory for discussions. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1053575. This work was supported in part by GM43949. R.K.H. is a Howard Hughes Medical Institute postdoctoral fellow of the Helen Hay Whitney Foundation and T.W. and R.M. are investigators of the Howard Hughes Medical Institute.
Footnotes
Author Contributions R.K.H. performed the experiments. P.Y. provided assistance with protein expression and purification. Z.L. aided with sample preparation and data collection. Y.H. provided assistance with protein expression. T.W. aided with initial model generation and map interpretation. R.K.H and R.M. designed the experiments and analyzed results. R.K.H. and R.M. prepared the manuscript with input from all coauthors.
Author Information The 3D cryo-EM density maps of Slo2.2 with low-pass filter and amplitude modification have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-3062 (Slo2.2 whole channel), EMD-3063 (Slo2.2 gating ring) and EMD-3064 (Slo2.2 TMD). Atomic coordinates for the atomic model of full length Slo2.2, Slo2.2 gating ring and Slo2.2 TMD have been deposited in the Protein Data Bank under accession numbers 5A6E, 5A6F and 5A6G, respectively.
The authors declare no competing financial interests.
References
- 1.Hille B. Ionic channels of excitable membranes. 2nd edn Sinauer Associates; 1992. [Google Scholar]
- 2.Bader CR, Bernheim L, Bertrand D. Sodium-activated potassium current in cultured avian neurones. Nature. 1985;317:540–542. doi: 10.1038/317540a0. [DOI] [PubMed] [Google Scholar]
- 3.Dryer SE, Fujii JT, Martin AR. A Na+-activated K+ current in cultured brain stem neurones from chicks. J Physiol. 1989;410:283–296. doi: 10.1113/jphysiol.1989.sp017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haimann C, Bader CR. Sodium-activated potassium channel in avian sensory neurons. Cell Biol Int Rep. 1989;13:1133–1139. doi: 10.1016/0309-1651(89)90027-1. [DOI] [PubMed] [Google Scholar]
- 5.Kameyama M, et al. Intracellular Na+ activates a K+ channel in mammalian cardiac cells. Nature. 1984;309:354–356. doi: 10.1038/309354a0. [DOI] [PubMed] [Google Scholar]
- 6.Schwindt PC, Spain WJ, Crill WE. Long-lasting reduction of excitability by a sodium-dependent potassium current in cat neocortical neurons. J Neurophysiol. 1989;61:233–244. doi: 10.1152/jn.1989.61.2.233. [DOI] [PubMed] [Google Scholar]
- 7.Yan Y, Yang Y, Bian S, Sigworth FJ. Expression, purification and functional reconstitution of slack sodium-activated potassium channels. J Membr Biol. 2012;245:667–674. doi: 10.1007/s00232-012-9425-7. doi:10.1007/s00232-012-9425-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan A, et al. The sodium-activated potassium channel is encoded by a member of the Slo gene family. Neuron. 2003;37:765–773. doi: 10.1016/s0896-6273(03)00096-5. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharjee A, Gan L, Kaczmarek LK. Localization of the Slack potassium channel in the rat central nervous system. J Comp Neurol. 2002;454:241–254. doi: 10.1002/cne.10439. doi:10.1002/cne.10439. [DOI] [PubMed] [Google Scholar]
- 10.Kaczmarek LK, et al. Regulation of the timing of MNTB neurons by short-term and long-term modulation of potassium channels. Hear Res. 2005;206:133–145. doi: 10.1016/j.heares.2004.11.023. doi:10.1016/j.heares.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 11.Wallen P, et al. Sodium-dependent potassium channels of a Slack-like subtype contribute to the slow afterhyperpolarization in lamprey spinal neurons. J Physiol. 2007;585:75–90. doi: 10.1113/jphysiol.2007.138156. doi:10.1113/jphysiol.2007.138156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang B, Desai R, Kaczmarek LK. Slack and Slick K(Na) channels regulate the accuracy of timing of auditory neurons. J Neurosci. 2007;27:2617–2627. doi: 10.1523/JNEUROSCI.5308-06.2007. doi:10.1523/JNEUROSCI.5308-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. doi:10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barcia G, et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat Genet. 2012;44:1255–1259. doi: 10.1038/ng.2441. doi:10.1038/ng.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishii A, et al. A recurrent KCNT1 mutation in two sporadic cases with malignant migrating partial seizures in infancy. Gene. 2013;531:467–471. doi: 10.1016/j.gene.2013.08.096. doi:10.1016/j.gene.2013.08.096. [DOI] [PubMed] [Google Scholar]
- 16.McTague A, et al. Migrating partial seizures of infancy: expansion of the electroclinical, radiological and pathological disease spectrum. Brain. 2013;136:1578–1591. doi: 10.1093/brain/awt073. doi:10.1093/brain/awt073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanderver A, et al. Identification of a novel de novo p.Phe932Ile KCNT1 mutation in a patient with leukoencephalopathy and severe epilepsy. Pediatr Neurol. 2014;50:112–114. doi: 10.1016/j.pediatrneurol.2013.06.024. doi:10.1016/j.pediatrneurol.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heron SE, et al. Missense mutations in the sodium-gated potassium channel gene KCNT1 cause severe autosomal dominant nocturnal frontal lobe epilepsy. Nat Genet. 2012;44:1188–1190. doi: 10.1038/ng.2440. doi:10.1038/ng.2440. [DOI] [PubMed] [Google Scholar]
- 19.Martin HC, et al. Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis. Hum Mol Genet. 2014;23:3200–3211. doi: 10.1093/hmg/ddu030. doi:10.1093/hmg/ddu030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang F, et al. TMEM16C facilitates Na(+)-activated K+ currents in rat sensory neurons and regulates pain processing. Nat Neurosci. 2013;16:1284–1290. doi: 10.1038/nn.3468. doi:10.1038/nn.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu R, et al. Slack channels expressed in sensory neurons control neuropathic pain in mice. J Neurosci. 2015;35:1125–1135. doi: 10.1523/JNEUROSCI.2423-14.2015. doi:10.1523/JNEUROSCI.2423-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu S, Das P, Fadool DA, Kaczmarek LK. The slack sodium-activated potassium channel provides a major outward current in olfactory neurons of Kv1.3−/− super-smeller mice. J Neurophysiol. 2010;103:3311–3319. doi: 10.1152/jn.00607.2009. doi:10.1152/jn.00607.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulais M, Lachheb S, Teulon J. A Na+− and Cl− -activated K+ channel in the thick ascending limb of mouse kidney. J Gen Physiol. 2006;127:205–215. doi: 10.1085/jgp.200509360. doi:10.1085/jgp.200509360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Yang Y, Ye S, Jiang Y. Structure of the gating ring from the human large-conductance Ca(2+)-gated K(+) channel. Nature. 2010;466:393–397. doi: 10.1038/nature09252. doi:10.1038/nature09252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan P, Leonetti MD, Hsiung Y, MacKinnon R. Open structure of the Ca2+ gating ring in the high-conductance Ca2+-activated K+ channel. Nature. 2012;481:94–97. doi: 10.1038/nature10670. doi:10.1038/nature10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan P, Leonetti MD, Pico AR, Hsiung Y, MacKinnon R. Structure of the human BK channel Ca2+-activation apparatus at 3.0 A resolution. Science. 2010;329:182–186. doi: 10.1126/science.1190414. doi:10.1126/science.1190414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garg P, Gardner A, Garg V, Sanguinetti MC. Structural basis of ion permeation gating in Slo2.1 K+ channels. J Gen Physiol. 2013;142:523–542. doi: 10.1085/jgp.201311064. doi:10.1085/jgp.201311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkens CM, Aldrich RW. State-independent block of BK channels by an intracellular quaternary ammonium. J Gen Physiol. 2006;128:347–364. doi: 10.1085/jgp.200609579. doi:10.1085/jgp.200609579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Xia XM, Lingle CJ. Cysteine scanning and modification reveal major differences between BK channels and Kv channels in the inner pore region. Proc Natl Acad Sci U S A. 2011;108:12161–12166. doi: 10.1073/pnas.1104150108. doi:10.1073/pnas.1104150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. doi:10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. doi:10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 32.Joiner WJ, et al. Formation of intermediate-conductance calcium-activated potassium channels by interaction of Slack and Slo subunits. Nat Neurosci. 1998;1:462–469. doi: 10.1038/2176. doi:10.1038/2176. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Zeng X, Lingle CJ. Slo3 K+ channels: voltage and pH dependence of macroscopic currents. J Gen Physiol. 2006;128:317–336. doi: 10.1085/jgp.200609552. doi:10.1085/jgp.200609552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai L, Garg V, Sanguinetti MC. Activation of Slo2.1 channels by niflumic acid. J Gen Physiol. 2010;135:275–295. doi: 10.1085/jgp.200910316. doi:10.1085/jgp.200910316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. doi:10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 36.Tao X, Avalos JL, Chen J, MacKinnon R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science. 2009;326:1668–1674. doi: 10.1126/science.1180310. doi:10.1126/science.1180310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whorton MR, MacKinnon R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 2011;147:199–208. doi: 10.1016/j.cell.2011.07.046. doi:10.1016/j.cell.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo D, Lu Z. Interaction mechanisms between polyamines and IRK1 inward rectifier K+ channels. J Gen Physiol. 2003;122:485–500. doi: 10.1085/jgp.200308890. doi:10.1085/jgp.200308890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heginbotham L, MacKinnon R. Conduction properties of the cloned Shaker K+ channel. Biophys J. 1993;65:2089–2096. doi: 10.1016/S0006-3495(93)81244-X. doi:10.1016/S0006-3495(93)81244-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budelli G, Geng Y, Butler A, Magleby KL, Salkoff L. Properties of Slo1 K+ channels with and without the gating ring. Proc Natl Acad Sci U S A. 2013;110:16657–16662. doi: 10.1073/pnas.1313433110. doi:10.1073/pnas.1313433110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leonetti MD, Yuan P, Hsiung Y, Mackinnon R. Functional and structural analysis of the human SLO3 pH- and voltage-gated K+ channel. Proc Natl Acad Sci U S A. 2012;109:19274–19279. doi: 10.1073/pnas.1215078109. doi:10.1073/pnas.1215078109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Rosenhouse-Dantsker A, Tang QY, Noskov S, Logothetis DE. The RCK2 domain uses a coordination site present in Kir channels to confer sodium sensitivity to Slo2.2 channels. J Neurosci. 2010;30:7554–7562. doi: 10.1523/JNEUROSCI.0525-10.2010. doi:10.1523/JNEUROSCI.0525-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–522. doi: 10.1038/417515a. doi:10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 44.Thomson SJ, Hansen A, Sanguinetti MC. Identification of the Intracellular Na+ Sensor in Slo2.1 Potassium Channels. J Biol Chem. 2015 doi: 10.1074/jbc.M115.653089. doi:10.1074/jbc.M115.653089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang H, et al. Activation of Slo1 BK channels by Mg2+ coordinated between the voltage sensor and RCK1 domains. Nat Struct Mol Biol. 2008;15:1152–1159. doi: 10.1038/nsmb.1507. doi:10.1038/nsmb.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
References for Methods section
- 47.Webster SM, Del Camino D, Dekker JP, Yellen G. Intracellular gate opening in Shaker K+ channels defined by high-affinity metal bridges. Nature. 2004;428:864–868. doi: 10.1038/nature02468. doi:10.1038/nature02468. [DOI] [PubMed] [Google Scholar]
- 48.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. doi:10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. doi:10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. doi:10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 51.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. doi:10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 53.Penczek PA, et al. CTER-rapid estimation of CTF parameters with error assessment. Ultramicroscopy. 2014;140:9–19. doi: 10.1016/j.ultramic.2014.01.009. doi:10.1016/j.ultramic.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. doi:10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. doi:10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 56.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. doi:10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife. 2014;3:e03665. doi: 10.7554/eLife.03665. doi:10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 59.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. doi:10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. doi:10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. doi:10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. doi:10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. doi:10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. doi:10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. doi:10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dolinsky TJ, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. doi:10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. doi:10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morin A, et al. Collaboration gets the most out of software. Elife. 2013;2:e01456. doi: 10.7554/eLife.01456. doi:10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. doi:10.1038/nature05416. [DOI] [PubMed] [Google Scholar]
- 70.Miller C. Ion channel reconstitution. Plenum Press; 1986. [Google Scholar]
- 71.Ruta V, Jiang Y, Lee A, Chen J, MacKinnon R. Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature. 2003;422:180–185. doi: 10.1038/nature01473. doi:10.1038/nature01473. [DOI] [PubMed] [Google Scholar]
- 72.Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv332. doi:10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]