By engineering a mutant mouse strain that preserves the scaffolding function of NLRC4, Dixit et al. show cooperativity between it and another NOD family sensor, NLRP3.
Abstract
NLRC4 and NLRP3, of the NOD-like receptor (NLR) family of intracellular proteins, are expressed in innate immune cells and are thought to nucleate distinct inflammasome complexes that promote caspase-1 activation, secretion of the proinflammatory cytokines IL-1β and IL-18, and a form of cell death termed pyroptosis. We show that NLRP3 associates with NLRC4 in macrophages infected with Salmonella typhimurium or transfected with flagellin. The significance of the interaction between the NLRC4 NACHT domain and NLRP3 was revealed when Nlrc4S533A/S533A bone marrow–derived macrophages (BMDMs) expressing phosphorylation site mutant NLRC4 S533A had only a mild defect in caspase-1 activation when compared with NLRC4-deficient BMDMs. NLRC4 S533A activated caspase-1 by recruiting NLRP3 and its adaptor protein ASC. Thus, Nlrc4S533A/S533A Nlrp3−/− BMDMs more closely resembled Nlrc4−/− BMDMs in their response to S. typhimurium or flagellin. The interplay between NLRP3 and NLRC4 reveals an unexpected overlap between what had been considered distinct inflammasome scaffolds.
NLRC4 and NLRP3 each have a central nucleotide-binding oligomerization NACHT domain and carboxy terminus leucine-rich repeats (LRRs). NLRC4 has an amino terminus caspase-activation and recruitment domain (CARD), whereas NLRP3 has a pyrin domain (Lamkanfi and Dixit, 2009). Analyses of macrophages lacking NLRC4 or NLRP3 suggest that these proteins activate caspase-1 in response to distinct stimuli. For example, NLRC4 is essential for caspase-1 activation in macrophages infected with intracellular pathogens including Salmonella typhimurium (Mariathasan et al., 2004) and Pseudomonas aeruginosa (Sutterwala et al., 2007), whereas NLRP3 is required for caspase-1 activation by diverse agents including ATP, nigericin, and crystalline substances such as monosodium urate (Mariathasan et al., 2006; Martinon et al., 2006).
How NLRP3 senses cellular perturbations remains contentious, but mechanistic details of NLRC4 activation by pathogen-associated molecules are emerging. Bacterial flagellin or components of bacterial type 3 secretion systems, such as PrgJ or needle protein, bind to neuronal apoptosis inhibitory proteins (NAIPs), and this promotes interactions between the NAIPs and NLRC4 (Kofoed and Vance, 2011; Zhao et al., 2011; Yang et al., 2013). The ligand-bound NAIPs appear to drive conformational changes in NLRC4 that expose the NACHT domain for the recruitment and activation of further NLRC4 molecules (Hu et al., 2013, 2015; Diebolder et al., 2015; Zhang et al., 2015). Phosphorylation of NLRC4 Ser533 has also been implicated in NLRC4 activation by S. typhimurium (Qu et al., 2012). Specifically, immortalized NLRC4-deficient myeloid cells reconstituted with wild-type NLRC4 displayed infection-induced caspase-1 activation, whereas cells reconstituted with mutant NLRC4 S533A did not. Unfortunately, the cryo-electron microscopy structures of NAIP-oligomerized NLRC4 are low resolution and provide little insight into how phosphorylation of NLRC4 Ser533 contributes to inflammasome assembly.
We further explored the significance of NLRC4 Ser533 phosphorylation using macrophages from NLRC4 S533A knock-in mice. Our results indicate that this posttranslational modification is required for optimal caspase-1 activation, but it is not essential. NLRC4 S533A, like wild-type NLRC4, can bind to NLRP3 protein that is induced after infection and then NLRP3 engages ASC to activate caspase-1.
RESULTS AND DISCUSSION
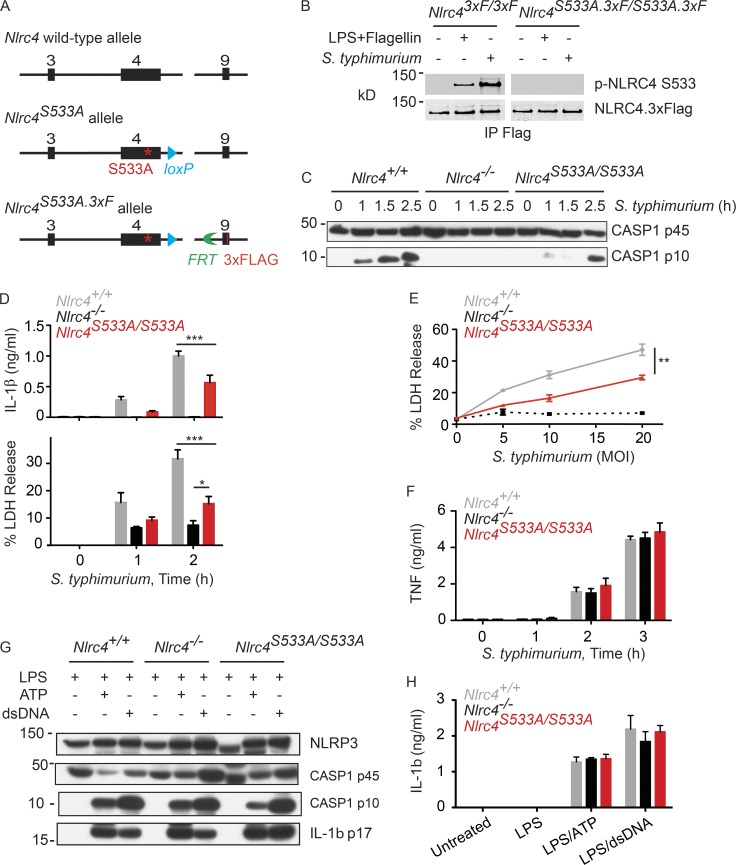
NLRC4 mutation S533A reduces but does not abolish NLRC4 inflammasome activity
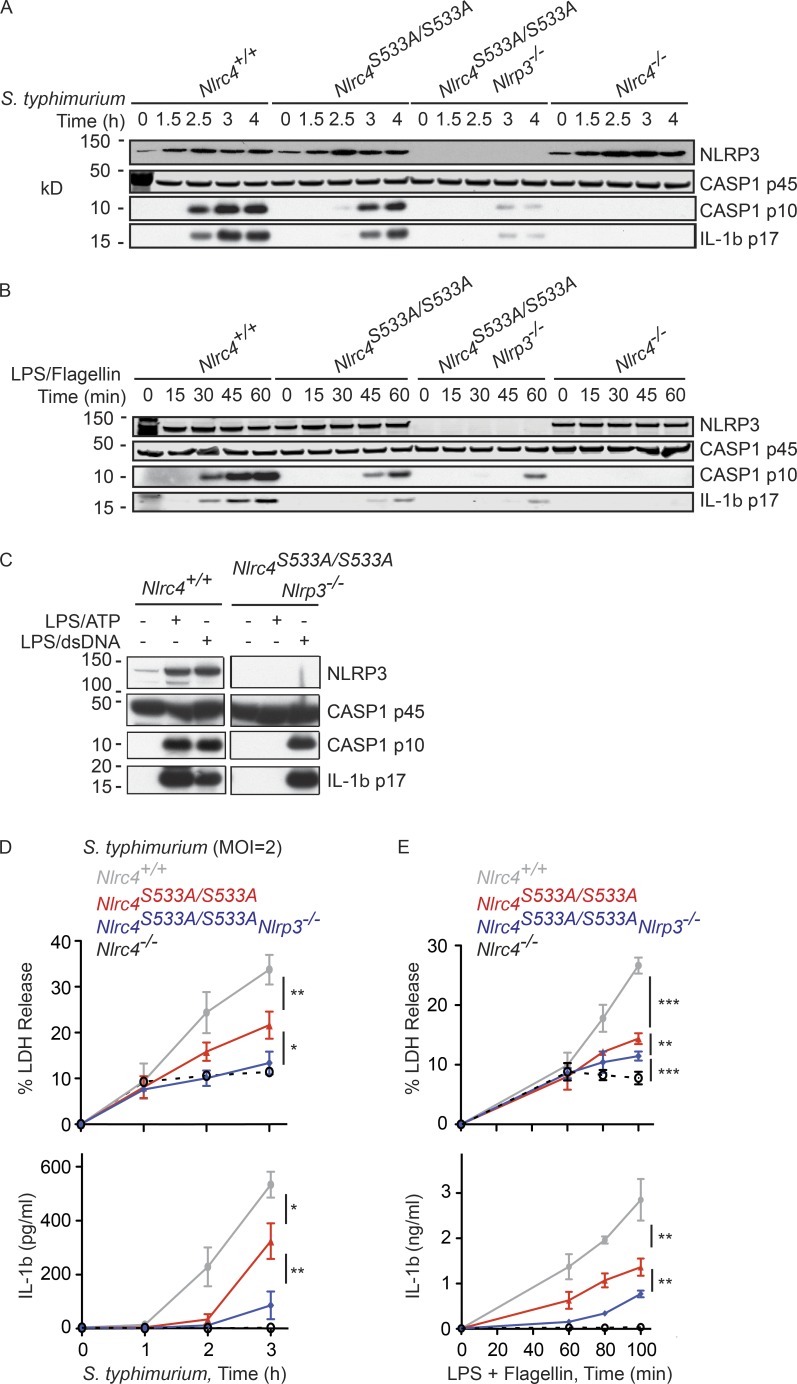
Primary BMDMs infected with S. typhimurium expressing the type 3 secretion system SPI-1 phosphorylate NLRC4 on Ser533 (Qu et al., 2012). To explore the significance of NLRC4 Ser533 phosphorylation, we created Nlrc4S533A/S533A knock-in mice expressing the NLRC4 S533A mutant (Fig. 1 A). A 3xFlag epitope tag was fused to the carboxy terminus in a second round of gene targeting to facilitate NLRC4 protein detection and to allow comparisons to the wild-type NLRC4.3xFlag knock-in mouse (Nlrc43xF/3xF) that we generated previously (Qu et al., 2012). Nlrc4S533A.3xF/S533A.3xF and Nlrc43xF/3xF BMDMs expressed similar amounts of NLRC4 protein but, as expected, Ser533 phosphorylation was detected only in wild-type cells after infection with S. typhimurium, or after stimulation with LPS followed by flagellin transfection (Fig. 1 B). Interestingly, Nlrc4S533A/S533A BMDMs differed from Nlrc4−/− BMDMs in that they produced cleaved, active caspase-1 after infection, albeit ∼1.5 h later than Nlrc4+/+ BMDMs (Fig. 1 C). Consistent with this finding, Nlrc4S533A/S533A BMDMs exhibited delayed caspase-1–dependent release of IL-1β and lactate dehydrogenase (LDH), the latter a measure of cell death (Fig. 1, D and E). Secretion of TNF after infection was not impaired by the NLRC4 S533A mutation, indicating a specific defect in caspase-1 activation (Fig. 1 F). In addition, only NLRC4-dependent caspase-1 activation was compromised because Nlrc4S533A/S533A BMDMs exhibited normal caspase-1 cleavage (Fig. 1 G) and IL-1β secretion (Fig. 1 H) in response to ATP or transfected double-stranded DNA (dsDNA), which stimulate NLRP3 and AIM2 inflammasome activity, respectively (Mariathasan et al., 2006; Jones et al., 2010).
Figure 1.
Attenuated caspase-1 activation, IL-1β secretion, and pyroptosis in Nlrc4S533A/S533A BMDMs infected with S. typhimurium. (A) Strategy for the generation of Nlrc4S533A/S533A and Nlrc4S533A.3xF/S533A.3xF mice. Black boxes indicate exons. (B) Western blots of NLRC4 immunoprecipitated with anti-Flag antibody from BMDMs at 4 h after either infection (multiplicity of infection [MOI] = 10) or flagellin transfection. p-NLRC4, phosphorylated NLRC4. (C–F) BMDMs infected with S. typhimurium. MOI was 10 unless indicated. Western blots represent cells and their culture medium (C). Graphs show IL-1β, LDH, and TNF released into the medium (D–F). (G) Western blots of BMDMs. (H) IL-1β secretion by BMDMs. Graphed data (mean ± SD) are pooled from three independent experiments for a total of two to three mice of each genotype. P-values were determined by the Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
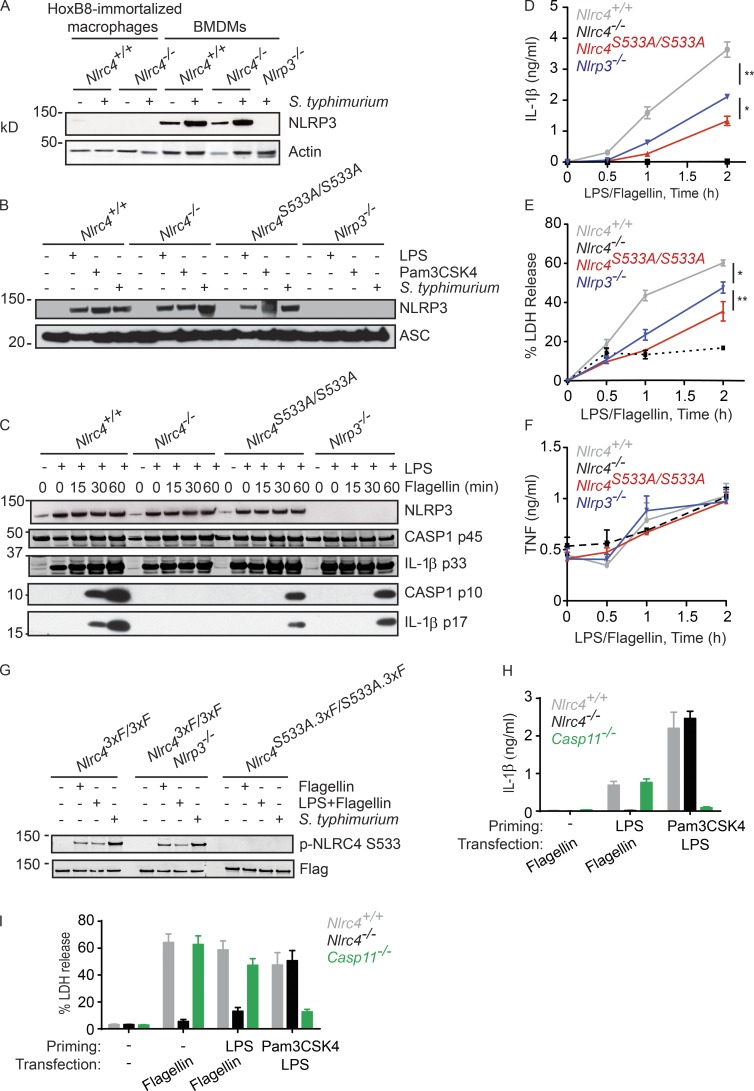
NLRP3 contributes to the NLRC4 inflammasome
The milder inflammasome activation defect of Nlrc4S533A/S533A BMDMs compared with Nlrc4−/− BMDMs prompted us to revisit our earlier experimental system of Nlrc4−/− immortalized macrophage progenitors reconstituted with NLRC4 S533A. Macrophages differentiated from these progenitors failed to activate caspase-1 after infection with S. typhimurium (Qu et al., 2012). These immortalized macrophages induced NLRP3 poorly in response to S. typhimurium when compared with primary BMDMs (Fig. 2 A). Given that both NLRP3 and NLRC4 promoted caspase-1 activation after a long infection with stationary phase S. typhimurium expressing little SPI-1 (Broz et al., 2010a), we wondered if NLRP3 might have an unrecognized role during acute infection of primary BMDMs with SPI-1–expressing bacteria. To focus solely on NLRC4-dependent signaling events, we primed BMDMs with LPS to mimic infection-induced up-regulation of NLRP3 (Fig. 2 B), and then transfected the cells with flagellin from S. typhimurium to stimulate NLRC4 specifically (Franchi et al., 2006; Miao et al., 2006). Nlrc4S533A/S533A and Nlrp3−/− BMDMs processed caspase-1 and pro-IL-1β slower than Nlrc4+/+ BMDMs (Fig. 2 C) and, over the course of an experiment, this translated into reduced IL-1β (Fig. 2 D) and LDH release (Fig. 2 E). Nlrc4−/− BMDMs, as expected, failed to cleave caspase-1 and secrete IL-1β, whereas TNF secretion was equivalent between all the genotypes (Fig. 2 F). Phosphorylation of NLRC4 Ser533 was normal in Nlrp3−/− BMDMs (Fig. 2 G), indicating that NLRP3 contributed to NLRC4-dependent inflammasome signaling by some other mechanism.
Figure 2.
NLRP3, but not caspase-11, contributes to NLRC4-dependent activation of caspase-1. (A and B) Western blots of immortalized macrophages (A) or BMDMs (A, B). Cells were primed for 4 h with LPS or Pam3CSK4, or infected for 2.5 h with S. typhimurium (MOI = 5). (C) Western blots of BMDMs and their culture medium. (D–F) Graphs show IL-1β (D), LDH (E) and TNF (F) released from BMDMs. (G) Western blots of NLRC4 immunoprecipitated with anti-FLAG antibody from BMDMs at 4 h after either infection (MOI = 10) or flagellin transfection. p-NLRC4, phosphorylated NLRC4. (H and I) Graphs show IL-1β (H) and LDH (I) released from BMDMs. Graphed data (mean ± SD) are pooled from three independent experiments for a total of three mice of each genotype. P-values were determined by the Student’s t test. *, P < 0.05; **, P < 0.01.
LPS priming induces expression of caspase-11, as well as NLRP3 (Kayagaki et al., 2013). Unlike NLRP3, however, caspase-11 was dispensable for LPS/flagellin-induced secretion of IL-1β and LDH (Fig. 2, H and I). In control experiments, caspase-11 deficiency impaired IL-1β and LDH release in response to intracellular LPS as previously described (Kayagaki et al., 2013). These data indicate that NLRP3-dependent caspase-1 activation in BMDMs treated with LPS/flagellin was not merely a consequence of residual LPS from the priming step entering the cytoplasm during transfection and engaging caspase-11 (Kayagaki et al., 2011, 2013).
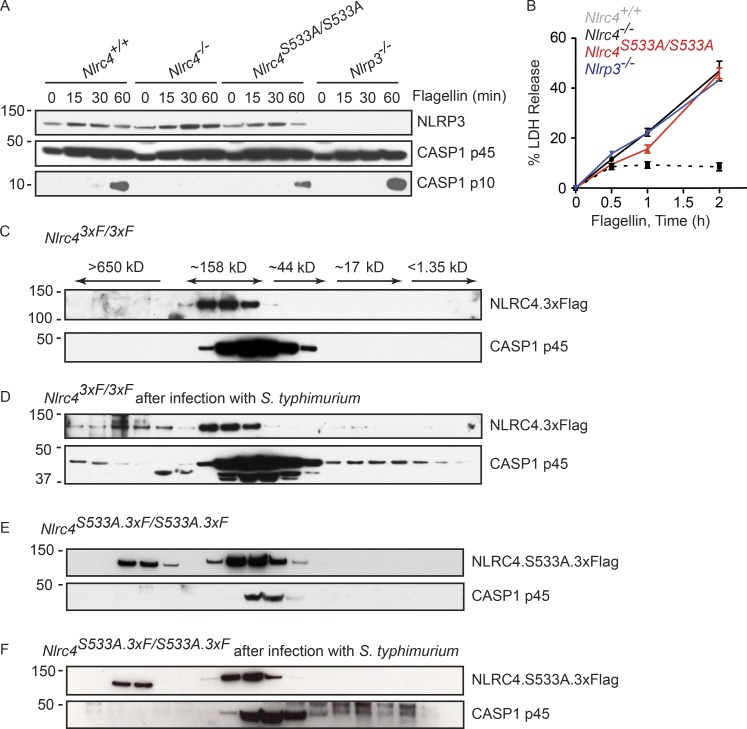
Given that NLRP3 is induced in BMDMs after infection with S. typhimurium or after treatment with TLR agonists (Fig. 2 B), we reasoned that NLRP3 probably contributed little to flagellin-induced inflammasome signaling if BMDMs were not primed with LPS. Indeed, caspase-1 processing in Nlrc4+/+ BMDMs transfected with flagellin without priming was equivalent to that seen in similarly treated Nlrc4S533A/S533A and Nlrp3−/− BMDMs (Fig. 3 A). In addition, LDH release in the Nlrc4+/+, Nlrc4S533A/S533A, and Nlrp3−/− cultures was equivalent at 2 h after transfection with flagellin (Fig. 3 B). Collectively, our data suggest that up-regulation of NLRP3 after LPS stimulation of TLR4 allows NLRP3 to support NLRC4-dependent activation of caspase-1.
Figure 3.
NLRP3 contributes less to NLRC4-dependent inflammasome activation in the absence of priming. (A) Western blots of BMDMs and their culture medium. (B) Graph shows LDH release. (C–F) Western blots of BMDM lysates fractionated by size exclusion chromatography. In D and F, cells were infected for 3 h (MOI = 10). All cultures contained pancaspase inhibitor Z-VAD-FMK to limit pyroptosis. One less fraction was collected in E and F than in C and D. Graphed data (mean ± SD) are pooled from three independent experiments for a total of three mice of each genotype.
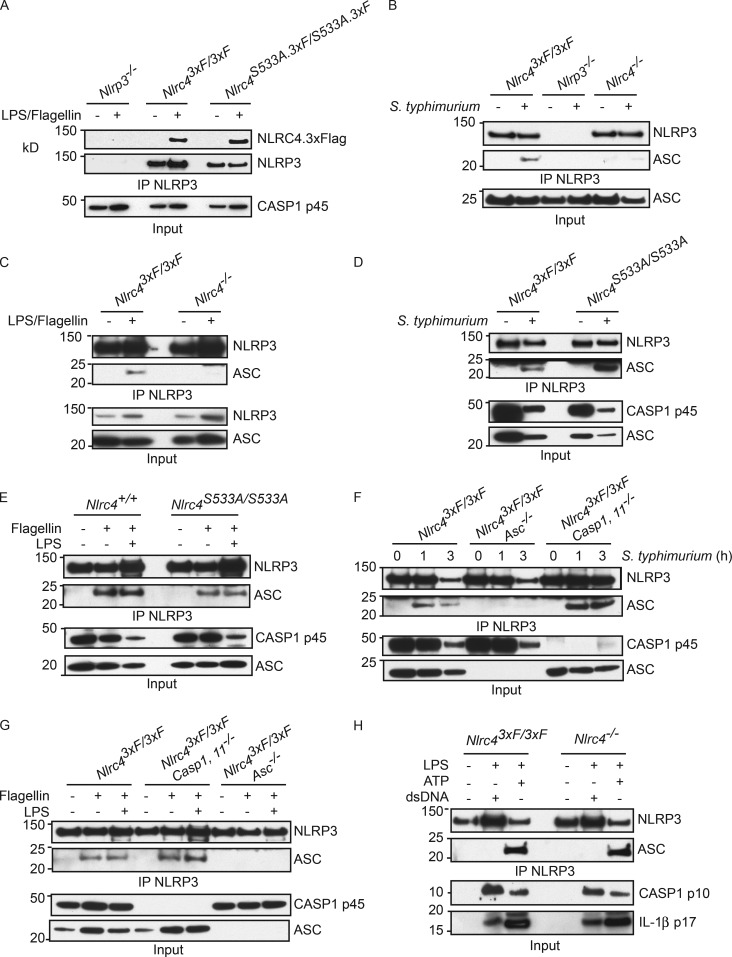
NLRP3 interacts with wild-type NLRC4 and NLRC4 S533A
Confocal and super-resolution microscopy of macrophages infected with S. typhimurium has suggested that NLRC4 and NLRP3 co-localize with ASC and caspase-1 (Man et al., 2014). Size exclusion chromatography of Nlrc43xF/3xF BMDM lysates indicated that a portion of NLRC4 and pro–caspase-1 shifted into macromolecular complexes after S. typhimurium infection (Fig. 3, C and D). We failed to detect NLRP3 in any of the chromatography fractions, probably reflecting that our NLRP3 antibody was not sensitive enough to detect low levels of NLRP3. Equivalent analyses of Nlrc4S533A.3xF/S533A.3xF BMDMs revealed that some NLRC4 S533A was found in macromolecular complexes even in uninfected cells (Fig. 3 E). Of note, however, pro–caspase-1 was absent from macromolecular complexes both before and after infection of Nlrc4S533A.3xF/S533A.3xF BMDMs with S. typhimurium (Fig. 3, E and F). This last result was consistent with a role for NLRC4 S533 phosphorylation in the optimal recruitment of pro–caspase-1 into the NLRC4 inflammasome.
NLRC4.3xFlag and NLRC4.S533A.3xFlag both coimmunoprecipitated with NLRP3 after BMDMs were treated with LPS/flagellin (Fig. 4 A). Therefore, NLRC4 S533 phosphorylation is dispensable for the interaction of NLRP3 with NLRC4. NLRP3 also coprecipitated ASC in an NLRC4-dependent manner (Fig. 4, B and C), but phosphorylation on NLRC4 S533 was not required (Fig. 4, D and E). As expected, caspase-1 was dispensable for the NLRC4-dependent interaction between NLRP3 and ASC (Fig. 4, F and G). In control experiments, LPS-primed BMDMs were transfected with dsDNA or treated with ATP (Fig. 4 H). Consistent with ATP stimulation engaging NLRP3 and ASC independent of NLRC4 (Mariathasan et al., 2004, 2006), NLRP3 associated with ASC after LPS/ATP treatment even in Nlrc4−/− BMDMs. Consistent with dsDNA engaging the AIM2 inflammasome and not the NLRP3 inflammasome (Jones et al., 2010), NLRP3 failed to interact with ASC after LPS/dsDNA treatment (Fig. 4 H).
Figure 4.
Activated NLRC4 recruits NLRP3 and ASC. (A–H) Western blots after immunoprecipitation of NLRP3 from BMDMs stimulated in the presence of Z-VAD-FMK. Cells were harvested at 3 h after flagellin transfection (A, C, E, and G), at 3 h after S. typhimurium infection (MOI = 10; B and D), at 4 h after transfection with dsDNA (H), or 30 min after ATP stimulation (H). Western blots are representative of three independent experiments.
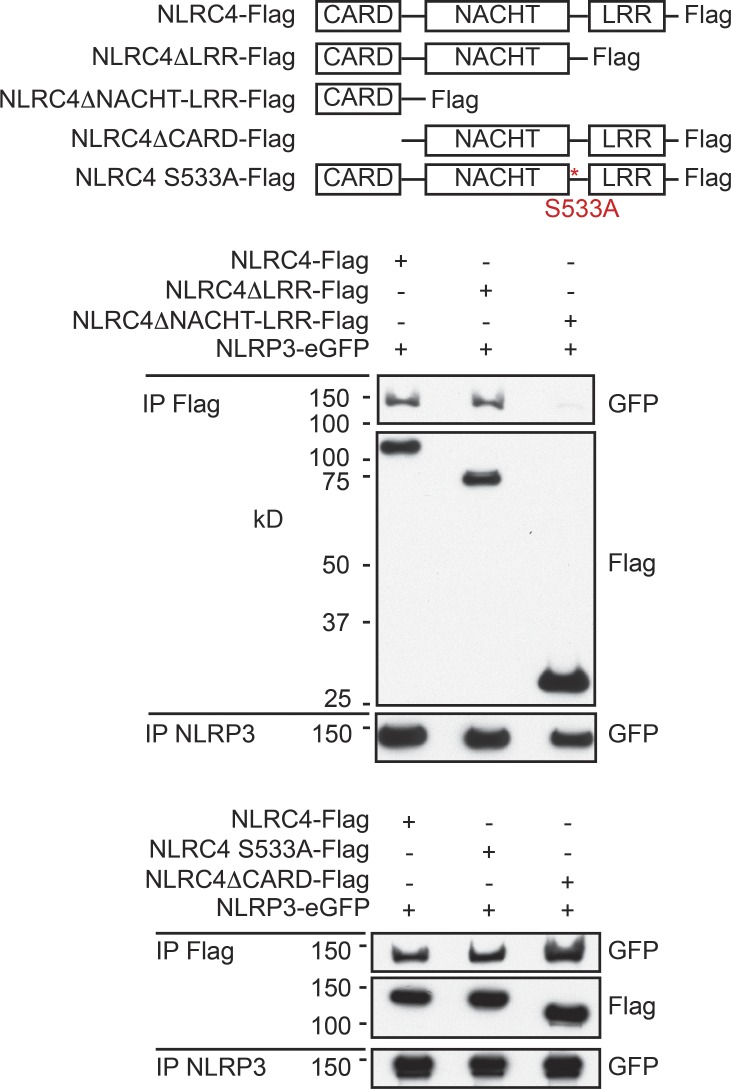
We explored the domains in NLRC4 required for binding to NLRP3 in a 293T overexpression system. The LRRs, CARD, and phosphorylation of S533 in NLRC4 were dispensable for the binding of NLRC4-Flag to NLRP3-eGFP (Fig. 5). The interaction was abolished, however, by loss of both the NACHT domain and LRRs in NLRC4. In light of these results, we hypothesized that caspase-1 activation in Nlrc4S533A/S533A BMDMs infected with S. typhimurium was delayed rather than abolished because the NLRC4 S533A mutant could recruit NLRP3 and ASC. Consistent with such a model, Nlrc4S533A/S533A Nlrp3−/− BMDMs showed a marked reduction in NLRC4-dependent processing of caspase-1 and IL-1β after S. typhimurium infection (Fig. 6 A) or LPS/flagellin treatment (Fig. 6 B) when compared with Nlrc4S533A/S533A BMDMS, although we note that processing was not abolished completely. It is unclear if this residual processing of caspase-1 reflects the involvement of yet a third NLR family member or if the NLRC4 S533A mutant itself retains some limited capacity to recruit pro-caspase-1 directly via its CARD. In control experiments, Nlrc4S533A/S533A Nlrp3−/− BMDMs processed caspase-1 and IL-1β normally after stimulation with LPS/dsDNA (Fig. 6 C). Nlrc4S533A/S533A Nlrp3−/− BMDMs also released less IL-1β and LDH than Nlrc4S533A/S533A BMDMs after infection with S. typhimurium (Fig. 6 D) or treatment with LPS/flagellin (Fig. 6 E). These findings indicate that NLRP3 is in large part responsible for NLRC4-dependent caspase-1 activation in Nlrc4S533A/S533A BMDMs.
Figure 5.
NLRP3 interacts with the NACHT domain in NLRC4. Western blots after immunoprecipitation of NLRC4 or NLRP3 from HEK293T cells transfected with the constructs indicated for 24 h. Western blots are representative of three independent experiments.
Figure 6.
NLRP3 contributes to NLRC4-dependent inflammasome activity. (A–C) Western blots of BMDMs and their culture medium. Cells were infected with S. typhimurium (MOI = 5; A) or stimulated for 4 h with transfected dsDNA or for 30 min with ATP (C). (D and E) Graphs show LDH and IL-1β released by BMDMs infected with S. typhimurium (MOI = 2) (D) or treated with LPS/flagellin (E). Graphed data (mean ± SD) are pooled from three independent experiments for a total of three mice of each genotype. P-values were determined by the Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
A model arises wherein up-regulated NLRP3 is recruited to NLRC4 to amplify caspase-1 activation. We speculate that a NAIP—NLRC4 interaction permits NLRP3 recruitment and then a conformational change in NLRP3 permits its interaction with ASC. Whereas the CARD of NLRC4 can bind directly to the CARD of pro–caspase-1 (Poyet et al., 2001), the pyrin domain of NLRP3 presumably engages the pyrin domain of ASC, and then the CARD in ASC binds to the CARD in pro–caspase-1. Our simplified model does not speak to the role that ASC appears to have in NLRC4 signaling that is independent of NLRP3. For example, NLRP3 is dispensable for the formation of an ASC focus that processes caspase-1, IL-1β, and IL-18 in BMDMs infected with S. typhimurium (Mariathasan et al., 2004, 2006; Broz et al., 2010a;b). In this complex, perhaps ASC is recruited via its CARD when NLRP3 is absent. Notably, NLRC4 deficiency has a much greater impact than ASC deficiency on caspase-1–dependent pyroptosis in BMDMs infected with S. typhimurium (Mariathasan et al., 2004). Some NLRC4 is proposed to activate caspase-1 in distinct complexes lacking ASC without causing caspase-1 autoproteolysis (Broz et al., 2010b).
MATERIALS AND METHODS
Mice
Nlrc4S533A/S533A mice (Genoway) and Nlrc4S533A.3xF/S533A.3xF mice (Taconic) were generated from C57BL/6 embryonic stem cells. For Nlrc4S533A/+ mice, TaqMan primers 5′-CCTACAAGGACTTTCAGTCACCAA-3′ and 5′-CATCTTGCTCAGTGGTATTTCTCAGA-3′ amplified an 83-bp fragment that was detected by a wild-type (AGGCAGGAATCAA) or knock-in (AGGCAGGAAGCCA) probe. For Nlrc4S533A.3xF/+ mice, primers 5′-ACGGGCTGGGAGTTTGA-3′ and 5′-CTTGTAGGTGGTGCGAATCA-3′ amplified a 219-bp wild-type fragment and a 285-bp knock-in fragment. Nlrc43xF/3xF, Casp1,11−/−, Asc−/−, and Nlrp3−/− mice were described previously (Mariathasan et al., 2004, 2006; Kayagaki et al., 2011; Qu et al., 2012). The Genentech animal care and use committee approved all mouse protocols.
Reagents
Antibodies recognized NLRP3 (Cryo-2; Adipogene), FLAG (M2; Sigma-Aldrich), ASC (8E4.1; Genentech), caspase-1 (sc-514; Santa Cruz Biotechnology, Inc.), IL-1β (GTX74034; GeneTex), phospho-NLRC4 S533 (GEN-82 clone 3–3; Genentech), and NLRC4 (hamster monoclonal 1D4 for immunoprecipitation and a rabbit polyclonal for blotting; Genentech). Other reagents included Z-VAD-FMK (MBL), S. typhimurium flagellin (InvivoGen), LPS from Escherichia Coli O111:B4 (Invivogen), Pam3CSK4 (Invivogen), and DOTAP transfection reagent (Roche). LDH release was measured with a Cytotoxicity Detection kitplus (Roche). Secreted IL-1β and TNF were measured with Meso Scale Discovery mouse kits.
Macrophage cultures
Bone marrow cells were cultured for 5–7 d in DMEM containing 20% L cell-conditioned medium, 10% heat-inactivated FBS, 100 U/ml penicillin, 10 mM l-glutamine, and 100 µg/ml streptomycin. BMDMs were replated in 6- or 24-well plates for experiments. BMDMs were primed with 0.5 µg/ml E. coli LPS serotype O1101:B4 for 4 h, and then stimulated with 5 mM ATP (Sigma-Aldrich) for 30 min, DOTAP-transfected flagellin for 4 h, or 1 µg/ml Lipofectamine 2000-transfected pcDNA3.1 (Invitrogen) for 4 h. S. typhimurium was from D. Monack (Stanford University). Bacteria were grown in LB overnight at 37°C and subcultured for 3–4 h before macrophage infection.
Plasmids
Mouse NLRC4 (wild-type or S533A) with a C-terminal FLAG tag was cloned into retroviral vector pMX-GFP (Cell Biolabs). NLRC4ΔNACHT-LRR-FLAG and NLRC4ΔCARD-FLAG were generated in pcDNA3.1(-) (Life Technologies). NLRC4ΔLRR-FLAG and NLRP3-eGFP were generated in pHUSH-GFP (Genentech).
Immunoprecipitation
BMDM soluble lysates were incubated with anti-FLAG agarose or anti-NLRP3 antibody overnight at 4°C. Anti-NLRC4 antibody was added for 2 h at 4°C. NLRP3 or NLRC4 complexes were captured with protein A/G beads. Beads were washed three times with 20 mM Tris pH 7.5, 2.42 mM MgCl2, 92.5 mM NaCl, 0.1% Triton X-100 supplemented with protease and phosphatase inhibitors.
ACKNOWLEDGMENTS
All authors were employees of Genentech. The authors declare no additional conflicts of interest.
Footnotes
Abbreviations used:
- CARD
- caspase activation and recruitment domain
- LDH
- lactate dehydrogenase
- LRR
- leucine-rich repeat
- MOI
- multiplicity of infection
- NAIP
- neuronal apoptosis inhibitory protein
- NLR
- nod-like receptor
References
- Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V.M., and Monack D.M.. 2010a Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207:1745–1755. 10.1084/jem.20100257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., von Moltke J., Jones J.W., Vance R.E., and Monack D.M.. 2010b Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 8:471–483. 10.1016/j.chom.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebolder C.A., Halff E.F., Koster A.J., Huizinga E.G., and Koning R.I.. 2015. Cryoelectron Tomography of the NAIP5/NLRC4 Inflammasome: Implications for NLR Activation. Structure. 23:2349–2357. 10.1016/j.str.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., et al. . 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7:576–582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Hu Z., Yan C., Liu P., Huang Z., Ma R., Zhang C., Wang R., Zhang Y., Martinon F., Miao D., et al. . 2013. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 341:172–175. 10.1126/science.1236381 [DOI] [PubMed] [Google Scholar]
- Hu Z., Zhou Q., Zhang C., Fan S., Cheng W., Zhao Y., Shao F., Wang H.W., Sui S.F., and Chai J.. 2015. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 350:399–404. 10.1126/science.aac5489 [DOI] [PubMed] [Google Scholar]
- Jones J.W., Kayagaki N., Broz P., Henry T., Newton K., O’Rourke K., Chan S., Dong J., Qu Y., Roose-Girma M., et al. . 2010. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. USA. 107:9771–9776. 10.1073/pnas.1003738107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., et al. . 2011. Non-canonical inflammasome activation targets caspase-11. Nature. 479:117–121. 10.1038/nature10558 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Wong M.T., Stowe I.B., Ramani S.R., Gonzalez L.C., Akashi-Takamura S., Miyake K., Zhang J., Lee W.P., Muszyński A., et al. . 2013. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 341:1246–1249. 10.1126/science.1240248 [DOI] [PubMed] [Google Scholar]
- Kofoed E.M., and Vance R.E.. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595. 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M., and Dixit V.M.. 2009. Inflammasomes: guardians of cytosolic sanctity. Immunol. Rev. 227:95–105. 10.1111/j.1600-065X.2008.00730.x [DOI] [PubMed] [Google Scholar]
- Man S.M., Hopkins L.J., Nugent E., Cox S., Glück I.M., Tourlomousis P., Wright J.A., Cicuta P., Monie T.P., and Bryant C.E.. 2014. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. USA. 111:7403–7408. 10.1073/pnas.1402911111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D.M., Vucic D., French D.M., Lee W.P., Roose-Girma M., Erickson S., and Dixit V.M.. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 430:213–218. 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- Mariathasan S., Weiss D.S., Newton K., McBride J., O’Rourke K., Roose-Girma M., Lee W.P., Weinrauch Y., Monack D.M., and Dixit V.M.. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 440:228–232. 10.1038/nature04515 [DOI] [PubMed] [Google Scholar]
- Martinon F., Pétrilli V., Mayor A., Tardivel A., and Tschopp J.. 2006. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 440:237–241. 10.1038/nature04516 [DOI] [PubMed] [Google Scholar]
- Miao E.A., Alpuche-Aranda C.M., Dors M., Clark A.E., Bader M.W., Miller S.I., and Aderem A.. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569–575. 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Poyet J.L., Srinivasula S.M., Tnani M., Razmara M., Fernandes-Alnemri T., and Alnemri E.S.. 2001. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276:28309–28313. 10.1074/jbc.C100250200 [DOI] [PubMed] [Google Scholar]
- Qu Y., Misaghi S., Izrael-Tomasevic A., Newton K., Gilmour L.L., Lamkanfi M., Louie S., Kayagaki N., Liu J., Kömüves L., et al. . 2012. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature. 490:539–542. 10.1038/nature11429 [DOI] [PubMed] [Google Scholar]
- Sutterwala F.S., Mijares L.A., Li L., Ogura Y., Kazmierczak B.I., and Flavell R.A.. 2007. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204:3235–3245. 10.1084/jem.20071239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J., and Shao F.. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA. 110:14408–14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen S., Ruan J., Wu J., Tong A.B., Yin Q., Li Y., David L., Lu A., Wang W.L., et al. . 2015. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 350:404–409. 10.1126/science.aac5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.N., Lu Q., Xu H., Liu L., and Shao F.. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]