de Villartay et al. describe a patient with a DNA repair factor mutation that leads to an increased sensitivity to DNA-damaging agents and, ultimately, to mild bone marrow failure and microcephaly.
Abstract
Inherited bone marrow failure syndromes are human conditions in which one or several cell lineages of the hemopoietic system are affected. They are present at birth or may develop progressively. They are sometimes accompanied by other developmental anomalies. Three main molecular causes have been recognized to result in bone marrow failure syndromes: (1) defects in the Fanconi anemia (FA)/BRCA DNA repair pathway, (2) defects in telomere maintenance, and (3) abnormal ribosome biogenesis. We analyzed a patient with mild bone marrow failure and microcephaly who did not present with the typical FA phenotype. Cells from this patient showed increased sensitivity to ionizing radiations and phleomycin, attesting to a probable DNA double strand break (dsb) repair defect. Linkage analysis and whole exome sequencing revealed a homozygous nonsense mutation in the ERCC6L2 gene. We identified a new ERCC6L2 alternative transcript encoding the DNA repair factor Hebo, which is critical for complementation of the patient’s DNAdsb repair defect. Sequence analysis revealed three structured regions within Hebo: a TUDOR domain, an adenosine triphosphatase domain, and a new domain, HEBO, specifically present in Hebo direct orthologues. Hebo is ubiquitously expressed, localized in the nucleus, and rapidly recruited to DNAdsb’s in an NBS1-dependent manner.
Cells from living organisms are facing DNA damages all through their lives, originating from either internal sources or inflicted by external genotoxics (Sancar et al., 2004). DNA double strand breaks (dsb’s) are considered to be the most toxic DNA lesion, which results in cell death if not properly repaired. Two main DNA repair mechanisms are used to cope with DNAdsb’s. Homologous recombination (HR), considered to be the most faithful process because it uses the homologous sister chromatid as a template, is restricted to the S/G2 phases of the cell cycle. The nonhomologous end-joining (NHEJ) pathway, which does not rely on a DNA template, is considered more error prone. NHEJ can cope with DNAdsb’s happening in all phases of the cell cycle. One prototypical example of a programmed DNA-damaging process in the hemopoietic system is the somatic rearrangement of T cell receptors and immunoglobulin genes during the maturation of immature lymphocytes, V(D)J recombination. The efficient repair of DNAdsb’s introduced during V(D)J recombination is required to maintain genome integrity, thus preventing the development of cancer and other DNA instability disorders (Alt et al., 2013). Defects in critical NHEJ factors are associated with the stalling of V(D)J recombination and the resulting arrest in lymphoid cell development leading to SCID both in human conditions and animal models (de Villartay et al., 2003). In some instances, such as Cernunnos/XRCC4-like factor (Xlf) deficiency, SCID is also associated with extraimmunological manifestations, such as microcephaly (Buck et al., 2006a), owing to the critical role of NHEJ in the central nervous system. DNA damage and repair is also an important aspect of the homeostasis of hemopoietic stem cells (HSCs) in the bone marrow (Walter et al., 2015). Indeed, defects in the NHEJ factors DNA-dependent protein kinase catalytic subunit and Cernunnos/Xlf are accompanied by HSC dysfunction, resulting in progressive bone marrow failure (Zhang et al., 2011; Avagyan et al., 2014).
Inherited bone marrow failure syndrome (IBMFS) is a generic term for clinically heterogeneous syndromes that have in common defects in the development and/or survival of various hemopoietic cell lineages (Dokal and Vulliamy, 2008). IBMFS can affect all types of blood cells, thus resembling aplastic anemia, or be restricted to only a few cell subsets like the specific red cell aplasia characteristic of Diamond-Blackfan anemia. IBMFS can appear very early in childhood or develop more progressively and is often accompanied by a series of other developmental manifestations such as facial features, skeletal anomalies, skin pigmentation, pulmonary diseases, leukoplakia, or nail dystrophy. Fanconi anemia (FA) is the most frequent cause of IBMFS and is characterized by progressive bone marrow failure often accompanied by onset of hematological malignancies (Moldovan and D’Andrea, 2009). Mutations identified in >18 different genes are responsible for FA (FANC-A to FANC-T; Hira et al., 2015; Rickman et al., 2015; Virts et al., 2015; Wang and Smogorzewska, 2015). The various FANC proteins, which are at the crossroad of several DNA repair pathways including HR, nucleotide excision repair, and trans-lesion synthesis, are of critical importance to removing interstrand DNA cross-links during replication. Indeed, cells from FA patients present with increased sensitivity to DNA cross-linking agents such as mitomycin C (MMC) and elevated spontaneous or MMC-induced chromosomal aberrations. Besides impairment of the FA DNA repair pathway, other molecular anomalies can lead to IBMFS. Telomere dysfunctions, which often but not always result in telomere shortening, as observed in dyskeratosis congenita or its more severe form Hoyeraal-Hreidarsson syndrome, are responsible for early and severe IBMFS observed in pediatric patients (Glousker et al., 2015). Lastly, abnormal ribosome biogenesis is a recognized cause of a certain type of IBMFS such as observed in Diamond-Blackfan anemia, Shwachman-Diamond Syndrome, and dyskeratosis congenita (Ruggero and Shimamura, 2014). For most of these human conditions, the extent to which the bone marrow is affected often requires the transplantation of HSCs as the sole curative treatment.
Here, we analyzed a patient, born from consanguineous parents, who presented with a mild IBMFS, not necessitating HSC transplantation, which mostly affects the generation of platelets, and a slight microcephaly. Clinical and biological findings excluded known causes of IBMFS. Instead, we found that the patient’s cells were sensitive to the DNAdsb-damaging agents ionizing radiations (IRs) and phleomycin (phleo) and only weakly sensitive to MMC. Whole exome sequencing (WES) revealed a homozygous nonsense mutation in Hebo, a new DNA repair factor encoded by the ERCC6L2 gene.
RESULTS
A patient with mild IBMFS and microcephaly
A 13-yr-old boy was admitted to the pediatric emergency department for diffuse petechiaes and bruises developing a few days after a febrile episode of unknown etiology. The cell blood count (CBC) revealed profound thrombocytopenia (platelet count of 4 G/liter) associated with mild anemia (hemoglobin level of 90 g/liter), macrocytosis (mean red cell volume of 104 fl), and moderate neutropenia (neutrophil count of 0.7 × 109/liter). Bone marrow smears and biopsies showed reduced cellularity (<15%) without myelodysplastic features, consistent with aplastic anemia. No cytogenetic abnormalities were found.
The patient was the fourth of five children of a healthy consanguineous (first cousins) couple. One of his sisters died at 7 mo of unexplained sudden infant death, and his youngest brother was affected by developmental delay consecutive to perinatal hypoxia. All other children and both parents had a normal CBC. The patient’s past medical history was unremarkable except for learning difficulties and attention deficit disorder. Birth weight (3,070 g) and height (49 cm) were normal and appropriate for gestational age (39 wk), but a mild microcephaly was noted at birth (head circumference of 34 cm). Additional findings included increased hemoglobin F levels (7%) suggesting mild dyserythropoiesis, vascular abnormalities in the right frontal lobe, and bilateral pyeloureteral junction abnormalities. There were no cardiopulmonary defects or mucocutaneous changes. A few weeks later, CBC parameters improved spontaneously and stabilized (Table S1). The patient is now 20 yr old, and CBCs show a hemoglobin level of 120 g/liter, stable macrocytosis (mean red cell volume of 104 fl), reticulocyte count of 75 × 109/liter, moderate neutropenia (neutrophil count of 1.04 × 109/liter), thrombopenia (platelet count of 59 × 109/liter), and a subnormal lymphocyte count (1.47 × 109/liter). Previous immunophenotyping had revealed a slight diminution in T lymphocytes (871/µl), with a significant decrease in naive CD4 T cells (49% CD45RA+/CD4+) as well as a B lymphocyte deficiency (92/µl). Immunoglobulin levels were not affected (14 g/liter IgG, 1.06 g/liter IgA, and 1.17 g/liter IgM). There has been no clonal evolution or progression to myelodysplasia. He achieved normal puberty development but remained <1 SD for weight and head circumference and <0.5 SD for height. We found neither gastrointestinal nor skeletal abnormalities, nor fatty infiltration of the pancreas suggestive of Shwachman-Diamond syndrome. However, the association of the bone marrow failure and the microcephaly/dysmorphy suggested a possible FA in this patient.
Exclusion of classical FA or telomeropathy syndromes
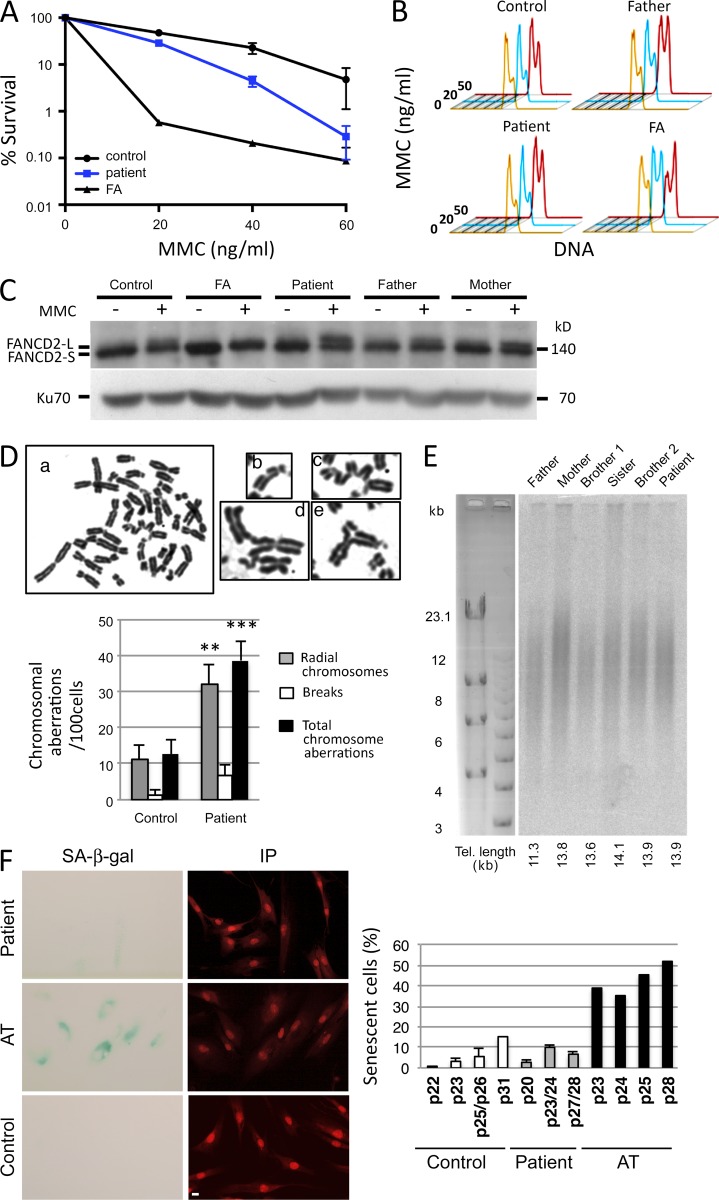
One hallmark of FA-deficient cells is a pronounced sensitivity to the DNA cross-linking agent MMC (Pinto et al., 2009). The sensitivity of the patient’s SV40-transformed fibroblasts to increasing doses of MMC, although significantly higher than that of control cells, was by no means comparable to that of cells from an FA patient (Fig. 1 A). Moreover, an EBV lymphoblastoid cell line from the patient did not demonstrate the typical G2/M accumulation after MMC treatment observed in FA cells and only arrested at the highest dose (50 ng/ml), similarly to lymphoblastoid cells from his father and an unrelated control (Fig. 1 B). Lastly, although a large subset of FA patients with mutations in the FA core proteins unveils an impaired ubiquitination of FANCD2 after MMC treatment (Garcia-Higuera et al., 2001), this was again not the case with our patient’s cells (Fig. 1 C). Altogether, these analyses ruled out a major defect in the Fanconi pathway as the primary cause of the bone marrow failure identified in this patient. Nevertheless, a significant increase in chromosomal aberrations (i.e., chromosomal breakages and radial chromosomes) was found in the patient’s cells upon exposure with MMC (Fig. 1 D), aberrations that are reminiscent of those associated with FA (Auerbach, 2009), which together with the slight increase in MMC sensitivity (Fig. 1 A) raised the possibility of either a hypomorphic FA condition or the existence of a different type of DNA repair defect responsible for the patient’s clinical condition. Telomere dysfunction constitutes another likely cause of early bone marrow failure (Touzot et al., 2012). However, analysis of telomere length in the peripheral blood lymphocytes of the patient, compared with that of three siblings and his parents, did not demonstrate any abnormal shortening of telomeres in these cells with a mean telomere length of ∼11–14 kb in all individuals (Fig. 1 E). In accord with the apparent normal telomere function, primary fibroblasts from the patient did not experience premature replicative senescence as judged by senescence-associated (SA) β-galactosidase (β-gal) staining (Fig. 1 F).
Figure 1.
FA-like phenotype. (A) MMC sensitivity of SV40-transformed fibroblasts from the patient, FA, and control. (B) G2/M arrest in EBV lymphoblastoid B cells from the patient, his father, an unrelated healthy control, and an FA patient. Cells were either untreated (orange) or treated with 20 ng/ml (blue) or 50 ng/ml (red) MMC for 48 h. (C) Ubiquitination of FANCD2 after MMC treatment (150 ng/ml for 48 h) of EBV B cells from the patient, his parents, an unrelated healthy control, and an FA patient. (A–C) The experiment was performed three times. (D) Increase of chromosomal aberrations in cells from the patient after MMC treatment. Representative metaphase from SAS50 fibroblasts treated with MMC (50 ng/ml for 24 h). (a) Examples of chromosomal aberrations found in metaphase from the patient’s fibroblasts. (b) Chromosomal breaks. (c–e) Radial chromosomes. The histogram shows the frequency of chromosomal aberrations (i.e., breaks, radial chromosomes, and total aberrations) found in metaphase spreads from patient fibroblasts compared with normal fibroblasts at similar passage. Data are mean ± SEM. n = 75 and 71 metaphases, respectively. **, P = 0.002; ***, P = 0.0003 for Student’s t tests. This experiment was performed two times. (E) Estimation of telomere (Tel.) length from whole blood cells by terminal restriction fragment analysis. This Southern blot has been performed once. (F) Representative image of the SA–β-gal staining (cyan) in patient fibroblasts and in AT fibroblasts to measure cellular senescence. Nuclei were stained with propidium iodide (IP; red). Quantification of the percentage of SA–β-gal–positive cells versus total cells determined at different cell culture passages (p) in the different fibroblast cultures (control, patient, and AT). The percentage of SA–β-gal–positive cells in patient’s cultures remains at a low level (<11.5% at p28) compared to that of control cultures (15% at passage p31), whereas AT cells, known to present a premature senescent phenotype, exhibited a strong percentage of SA–β-gal–positive cells at earlier passages (39% at p23). This experiment was performed two times. Bar, 10 µm.
Patient cells have defects in DNAdsb repair
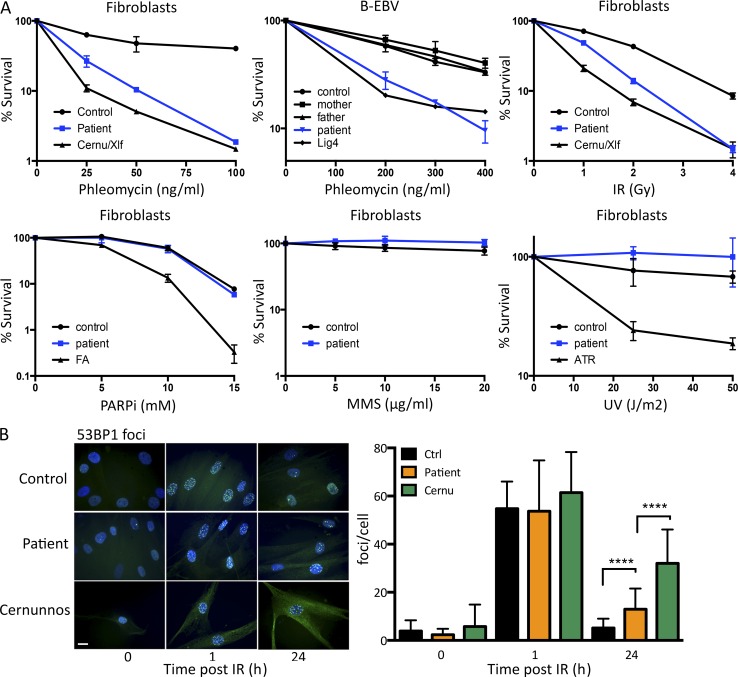
To get further insight into the DNA repair pathway that might be affected in this patient, we analyzed his SV40-transformed fibroblasts and EBV B cells for sensitivity against a panel of genotoxic drugs causing various types of DNA lesions (Fig. 2 A). Although the patient’s fibroblasts were not overtly sensitive to methyl methanesulfonate, UV light, or poly–ADP-ribose polymerase inhibitor, a sharp sensitivity to both phleo and IR was noticed, almost comparable to that observed with NHEJ-deficient Cernunnos/Xlf mutant cells. The sensitivity to phleo was not restricted to fibroblasts, as the EBV B cells demonstrated a sensitivity similar to that of cells from a DNA ligase IV patient, in contrast to the normal sensitivity observed in EBV B cells from the two parents and an unrelated control (Fig. 2 A). Increased sensitivity to both phleo and IR suggested a possible defect in DNAdsb repair, as observed in various NHEJ-deficient conditions. DNAdsb repair capacity of the patient’s primary fibroblasts was analyzed through the kinetics of 53BP1 irradiation-induced foci (IRIF) formation 1 h and 24 h after IR (Fig. 2 B). γ-H2AX or 53BP1 foci were used as surrogate markers of DNAdsbs (Rogakou et al., 1998). Although 53BP1 IRIF, which form upon DNA damages, were no longer present after 24 h in control cells, attesting for the efficient DNA repair process, they persisted in Cernunnos/Xlf mutant cells, attesting for the DNAdsb repair deficiency in these cells. Likewise, a statistically significant retention of 53BP1 IRIF was noted in the patient’s cells, although to a lesser extent as compared with the Cernunnos-deficient situation.
Figure 2.
DNA repair defect in patient’s cells. (A) Survival of SV40-transformed and EBV B cell lines from the patient and appropriate negative controls upon treatment with increasing doses of various genotoxic agents. These experiments were performed between one and four times depending on the drug. PARPi, poly–ADP-ribose polymerase inhibitor. MMS, methyl methanesulfonate. ATR, AT and rad3-related protein. (B) Kinetics of irradiation-induced 53BP1 foci formation in primary fibroblasts from the patient, a healthy control, and a Cernunnos-mutated patient. A representative image of cells treated with 2 Gy IR and quantification of IRIF is shown. Data are mean ± SD. ****, P < 0.0001 for a Mann-Whitney test. This experiment was performed two times. Bar, 10 µm. Ctrl, control. Cernu, Cernunnos.
From this series of analyses in the patient’s cells, we conclude that a general DNA repair defect affecting resolution of DNAdsb’s is the probable cause of the bone marrow failure and other developmental defects presented by this child.
A nonsense mutation in the ERCC6L2 gene
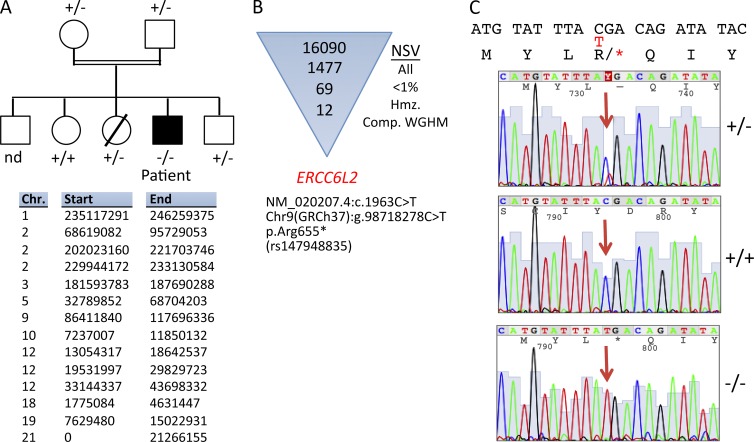
To firmly exclude an FA molecular defect, we performed WES and initially filtered the results on the 18 known FA-causing genes (Wang and Smogorzewska, 2015) and did not identify variants with frequencies <1% in the public and in-house databases for these genes. The patient being born to consanguineous parents, we also undertook whole genome homozygosity mapping (WGHM) from DNA in the family, which highlighted 14 compatible chromosomal regions (Fig. 3 A). These regions were subsequently used to filter nonsynonymous variants (NSVs) identified through WES performed on the patient’s DNA after an initial filtering on homozygous variants with frequencies <1% in single nucleotide polymorphism and in-house databases (Fig. 3 B). A nonsense NSV in the ERCC6L2 gene (Chr9 [GRCh37]:g.98718278C > T, c.1963C > T, and p.Arg655*) was selected as the best candidate given the plausible implication of this factor in the DNA damage response (DDR; Fig. 3 B). Although this variant was already described in a database (Single Nucleotide Polymorphism database accession no. rs147948835), its overall frequency of 1.6 × 10−4 and of 2.6 × 10−4 in the European population (The Exome Aggregation Consortium) was low enough to consider it as a possible pathological NSV. Moreover, this NSV had a very high PHRED score of 49 using combined annotation-dependent depletion (Kircher et al., 2014). Sanger resequencing confirmed the appropriate inheritance of the R655X mutation in the patient’s family with respect to WGHM results (Fig. 3 C). The same mutation was initially described by Tummala et al. (2014).
Figure 3.
Genetic analysis. (A) Pedigree of the family and candidate chromosome (Chr.) regions (GRCh37/hg19) identified by linkage analysis in the patient’s family by WGHM. nd, not determined. (B) Identification of NSV in the ERCC6L2 after filtering of WES NSV to homozygous (hmz.) regions as defined by WGHM and with a frequency <1% in various NSV databases. Comp., compatible. (C) Representative electropherogram of Sanger resequencing covering the three genotypes of the ERCC6L2 mutation identified in the family. The red T indicates the nucleotide change relative to the C above it. The arrows indicate the position of the nucleotide change on the electropherogram. Asterisks indicate stop condons.
Identification of Hebo
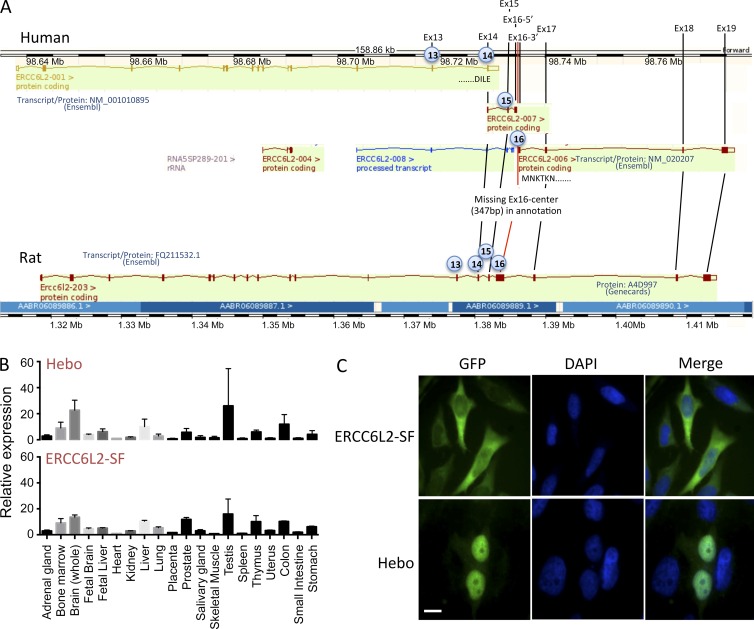
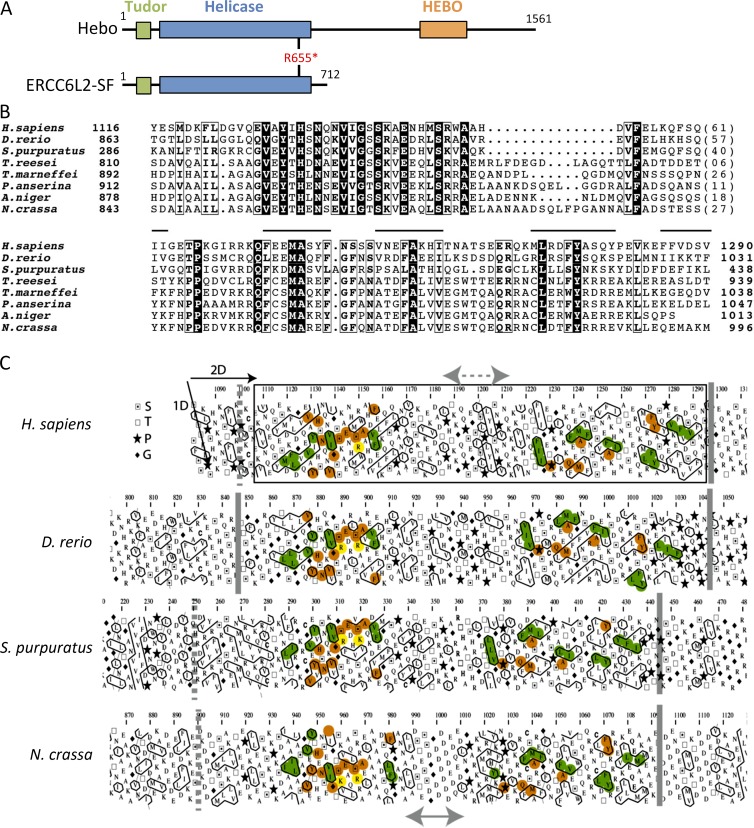
Analyzing the assembly of the ERCC6L2 gene in various species through the Ensembl database (Fig. 4 A), we were puzzled by the difference between the longer transcript annotated in the rat (ENSRNOT00000025965) coding for a 1,533-aa protein and the 712-aa human annotated protein, raising the possibility of an error in the automatic annotation of the human counterpart. Blast analysis of the rat ERCC6L2 open reading frame (orf) over the human genomic region spanning the ERCC6L2 gene established that the first 14 exons correspond to the human ERCC6L2-001 annotated transcript, whereas the additional five exons present in the rat matched short alternative human annotated transcripts (ERCC6L2-006 and ERCC6L2-007) but with a gap of 347 bp within the putative exon 16. Altogether, this suggested the possible expression of a longer ERCC6L2 transcript in humans, comparable with the rat situation but not annotated as such in the Ensembl database. We designed PCR primers in the first exon of transcript ERCC6L2-001 and in the last exon of transcript ERCC6L2-006 to amplify cDNA from human activated peripheral blood leukocytes. This resulted in a 4.7-kb specific band attesting to the existence of an expressed ERCC6L2 long form transcript in humans (unpublished data). Sequencing the PCR product established the 4,686-bp orf encoding a 1,561-aa protein showing 73% identity with the rat ERCC6L2 counterpart. The alternative splicing event between the short and long isoforms intervenes through the usage of a splice donor site within exon 14 after amino acid E711. This ERCC6L2 long isoform is now quoted in GenBank (accession no. NM_020207) after a recent update of human genome sequence annotation but without experimental evidence of its expression. For simplicity, we propose to rename this new isoform of the ERCC6L2 gene Hebo (helicase mutated in bone marrow failure). Hebo differs from the ERCC6L2 short isoform (SF) by the replacement of the last V712 residue with a new 850-aa peptide. Both isoforms include the R655X mutation identified in the patient.
Figure 4.
Identification of Hebo. (A) Schematic representation of the annotated ERCC6L2 gene in human (top) and rat (bottom) as provided by the Ensembl database. Exons 1–14 correspond to the human SF of ERCC6L2. A splice donor site within exon 14 would lead to alternative splicing from transcript 001 to transcript 007, incorporating new exon 15 and part of exon 16. Annotation is interrupted in exon 16 in transcript 007. Transcript 006 would incorporate the end of exon 16 onward to the end of the putative cDNA at exon 19 when compared with the structure of the rat gene. A gap of 347 bp between the two parts of exon 16 present in transcripts 007 and 006, respectively, was filled in through PCR amplification of cDNA. The existence of the ERCC6L2 long form (Hebo) was validated through full-length PCR amplification of cDNA. (B) Real-time quantitative RT-PCR of ERCC6L2-SF and Hebo-specific transcripts in a panel of tissues. This experiment was performed two times. Data are mean ± SEM. (C) Cellular localization of GFP-Hebo and GFP–ERCC6L2-SF in 293T-transfected cells. This experiment was performed three times. Bar, 10 µm.
We next evaluated the pattern of expression of the short and long ERCC6L2 transcripts in various tissues. cDNA from a panel of tissues was analyzed by real-time quantitative PCR using pairs of oligonucleotide primers and probes specific for the ERCC6L2-SF and Hebo isoform. The expression of ERCC6L2 is ubiquitous without evidence for a tissue-specific regulation of the splicing between the two isoforms (Fig. 4 B). Finally, the subcellular localization of ERCC6L2-SF and Hebo was determined after transfection of 293 T cells with ERCC6L2-SF and Hebo fused to GFP. Although ERCC6L2-SF is mostly localized in the cytoplasm, Hebo localization is restricted to the nucleus as expected for a DNA repair factor (Fig. 4 C).
Functional validation of Hebo mutation
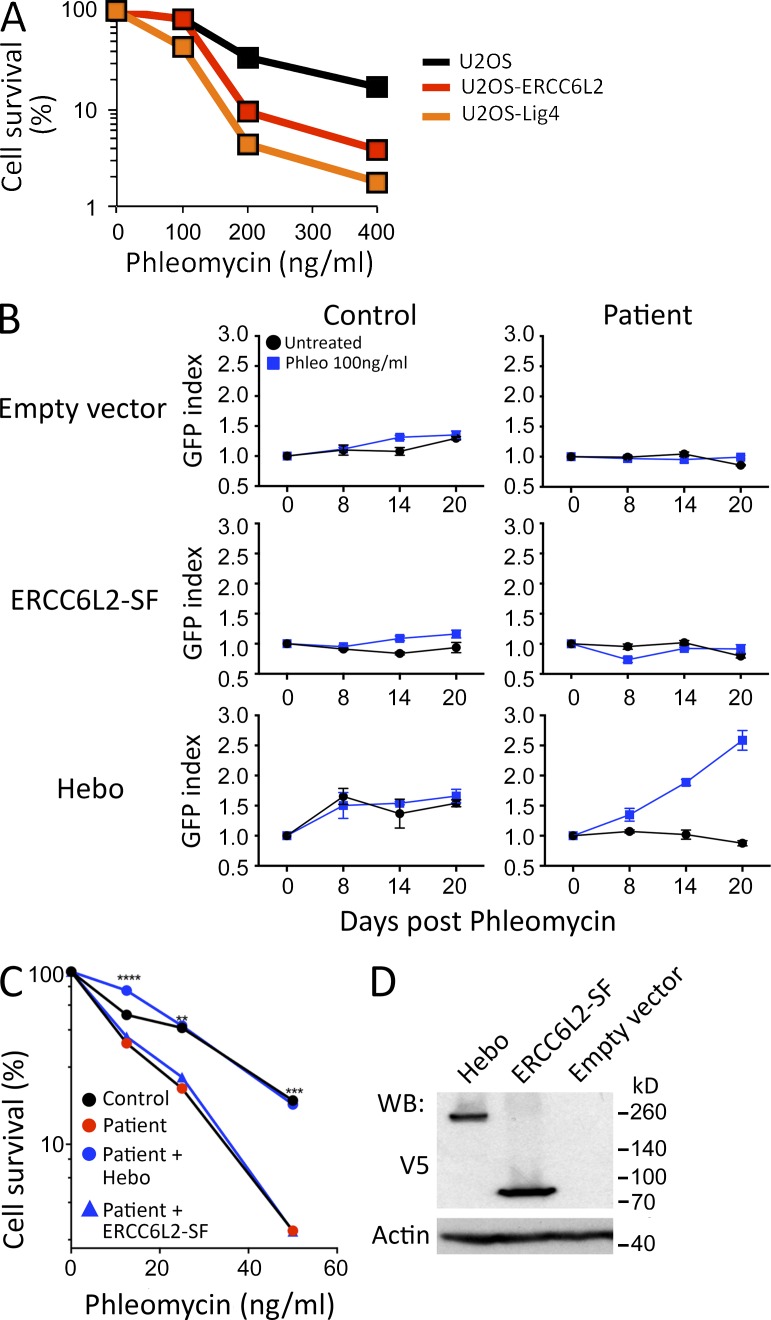
As a first mean to corroborate the possible implication of ERCC6L2 in the DDR after DNAdsb’s, we disrupted the ERCC6L2 gene in U2OS cells through CRISPR/Cas9 mutagenesis. Single molecule sequencing of PCR products spanning the guide RNA (gRNA) site from U2OS transfected with the dual spCas9 and gRNA pX330 expression plasmid revealed a level of 82% indels of various sizes (unpublished data). The bulk mutagenized U2OS cells presented increased phleo sensitivity comparable with that of U2OS cells in which DNA ligase IV had been disrupted by CRISPR/Cas9, well above the sensitivity observed in isogenic WT U2OS cells, attesting for the critical role of ERCC6L2 in DNA repair after phleo-induced DNA damage (Fig. 5 A).
Figure 5.
Functional complementation. (A) Phleo sensitivity of U20S cells, in which the ERCC6L2 gene has been disrupted through CRISPR/Cas9 technology, compared with the sensitivity of the WT parental cells and a U2OS line with mutated DNA ligase IV. This experiment was performed three times. (B) Functional complementation of phleo sensitivity provided by WT Hebo transduced into the patient’s cells as compared with the ERCC6L2-SF or empty vector. A mix population of transduced (GFP+) and untransduced (GFP−) was analyzed through multicolor competition assay (Smogorzewska et al., 2007). The selective growth advantage is scored as the increase in the index of GFP-positive cells/GFP-negative cells at various times compared with the initiation of the culture (index = 1). This experiment was performed two times. Data are mean ± SEM. (C) Sensitivity of the patient’s cells to phleo after Hebo, ERCC6L2-SF, or mock transduction. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 for a Mann-Whitney test. This experiment was performed three times. (D) Expression level of Hebo and ERCC6L2-SF after transient transfection into 293T cells, revealed with anti-V5 antibody. WB, Western blot.
To further validate the R655X mutation in the ERCC6L2 gene as being responsible for the DNA repair deficiency observed in the patient’s cells, we PCR amplified both the ERCC6L2-SF (Ensembl transcript ENST00000288985) and Hebo orf, cloned them into an Ires-GFP lentiviral vector, and transduced the patient’s fibroblasts. The resulting mix populations of transduced (GFP+) and untransduced (GFP−) cells were analyzed through multicolor competition assay after phleo treatment (Smogorzewska et al., 2007). Neither transduction with the empty vector nor with the ERCC6L2-SF–expressing vector conferred any selective advantage in culture, as evaluated by the GFP index of transduced over nontransduced cells upon treatment with 100 ng/ml phleo (Fig. 5 B). In contrast, introduction of WT Hebo into the patient’s fibroblasts conferred a strong selective advantage over cells transduced with the empty vector as judged by the 2.5-fold increase in GFP-expressing cells 3 wk after phleo treatment. In addition, GFP-sorted Hebo-transduced cells recovered an overall phleo sensitivity comparable with WT control cells, whereas the patient’s fibroblasts transduced with ERCC6L2-SF remained as sensitive as their untransduced counterparts (Fig. 5 C). The absence of complementation by ERCC6L2-SF was not the consequence of a reduced expression of the protein compared with Hebo (Fig. 5 D).
Altogether, we conclude that the DNA repair deficiency identified in the patient’s cells is caused by the R655X homozygous nonsense mutation in the ERCC6L2 gene that results in the truncation of about half of the new DNA repair factor Hebo.
Structure of Hebo
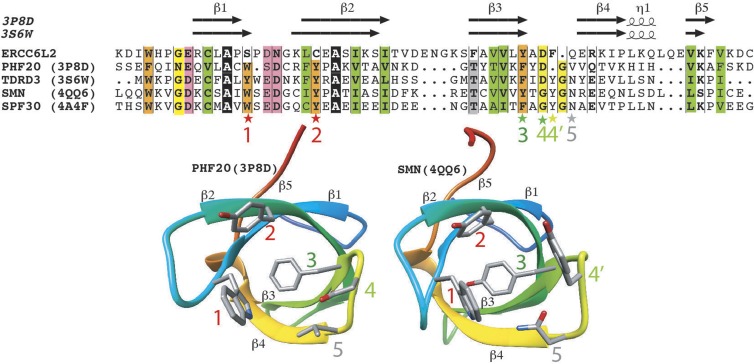
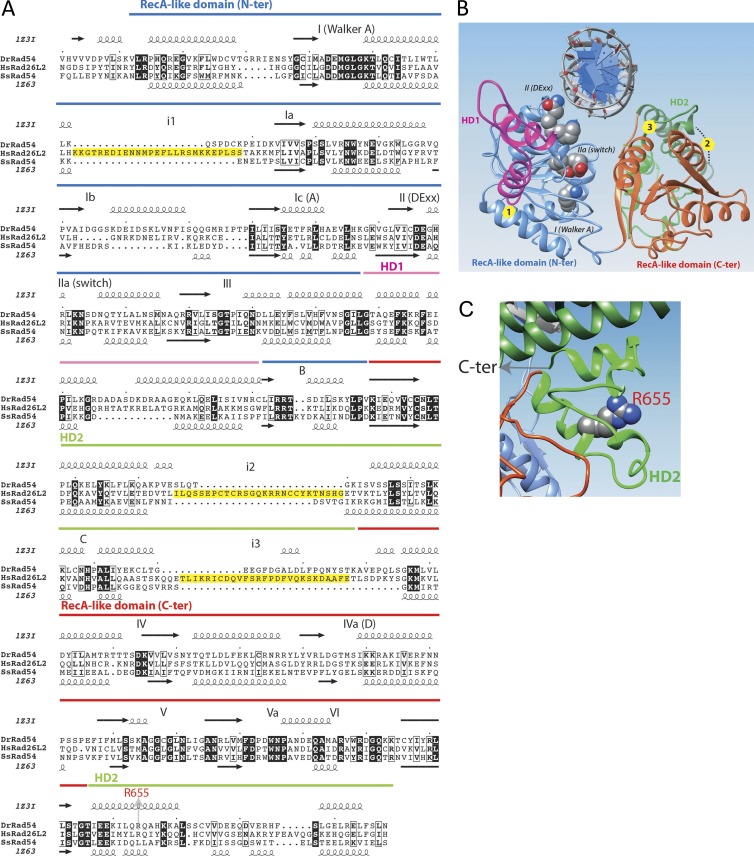
Analysis of the Hebo protein sequence revealed three structured domains, the first two of which are common with ERCC6L2-SF (Fig. 6 A). The C-terminal half of Hebo (R712–T1561), which is uniquely present in the ERCC6L2 long isoform, consists of 850 aa that contain large sequence segments that are predicted to be disordered. Using hydrophobic cluster analysis (Fig. 6 C), we identified here a novel domain conserved from human to Neurospora crassa, which we named HEBO after the name of the protein Hebo. This region, predicted to behave as a globular domain (or at least a domain that may fold upon interaction with a partner; Faure and Callebaut, 2013), encompasses ∼200 aa and is embedded within regions having a lower content in hydrophobic amino acids (disordered segments). Using the limits of this domain for searching sequence databases, we highlighted significant similarities with Hebo homologues in various species (Fig. 6 B) without, however, identifying this domain in other proteins. No significant similarities were detected with known three-dimensional (3D) structures using threading programs. This suggests that this domain is involved in a specific function of Hebo, likely related to its interaction with putative partners. The domain appears to be made of two distinct subdomains, separated by a variable hinge region. Moreover, highly conserved amino acids are observed in a few positions (Y, H, N, V, G, E, S, P, Q, M, A, A, L, and Y), suggesting that they play key roles in the function and/or structure of this domain. The HEBO domain matches the shorter profile described in the Pfam database (accession no. PF14773). Apart from the new HEBO domain, the N-terminal half of Hebo contains a Tudor domain (K26–C89) that shares the most striking sequence similarities with PHF20, TDRD3, SMN, and SPF30 (Fig. 7), which contain similar methyl-binding cages (Tripsianes et al., 2011; Cui et al., 2012; Liu et al., 2012). Three aromatic residues (Fig. 7, labeled 1–3) constitute Tudor domains, with a fourth additional one (Fig. 7, labeled 4′) existing in TDR3, SMN, and SPF30, whereas this last position is occupied by an aspartic acid in PHF20 (Fig. 7, labeled 4). The first two aromatic residues in positions 1 and 2, however, are not present in the Hebo Tudor domain, these being replaced by a serine (strand β1) and a cysteine (strand β2) residue, whereas an aromatic side chain is still present in positions 3 and 4′. This suggests that the Hebo Tudor domain, which is not conserved in yeast species, has evolved toward a different function than methyl binding. This contrasts with the Tudor domain present in other DNA repair factors such as 53BP1, which acts as a histone methylation reader at sites of DNAdsb’s (Fradet-Turcotte et al., 2013). The Tudor domain is followed by a helicase/ATPase domain (T129-H693), the 3D putative structure of which is represented in Fig. 8. The core of the ATPase domain contains, like Rad54, two RecA-like core domains, with an interface between them forming a deep groove accommodating the nucleotide-binding pocket and part of a DNA-binding site. Amino acids typical of the SF2 family, as well as of the SWI2/SNF2 family, are well conserved in Hebo and ERCC6L2-SF. Three large insertions (Fig. 8, A and B, labeled 1–3), however, are found in Hebo/ERCC6L2-SF relative to the Rad54 structures with which they were aligned, one of which (labeled 3) is oriented toward the groove. The R655X mutation shortens the ATPase domain of its last 38 residues. This fragment encompasses almost all of the second part of the discontinuous HD2 domain (Fig. 8 C), which participates in the structural integrity of the whole HD2 domain.
Figure 6.
The HEBO domain. (A) Schematic representation of the Hebo and ERCC6L2-SF domain architecture. The asterisk indicates the stop codon at R655 in the patient. (B) Multiple alignment of the HEBO domain sequence of Hebo proteins from representative species. Identical amino acids are represented as white on a black background, and amino acid similarities are boxed. Lines indicate the position of hydrophobic clusters, which are conserved within the whole family (Table S1). GenBank/NCBI RefSeq accession nos. are NP_064592.2 (Homo sapiens), AAH91795.1 (Danio rerio), XP_011663528 (Strongylocentrotus purpuratus), XP_006968442.1 (Trichoderma reesei), XP_002151909.1 (Talaromyces marneffei), CDP26308.1 (Podospora anserina), XP_001391908.1 (Aspergillus niger), and CAD37001.1 (Neurospora crassa). (C) Hydrophobic cluster analysis of the C-terminal region of ERCC6L6 reveals a globular domain (boxed), which is conserved in the ERCC6L2 orthologues. The sequences are shown on duplicated α-helical nets, on which hydrophobic amino acids are contoured and form clusters, which mainly correspond to the regular secondary structures forming globular domains. Conservation of hydrophobic clusters is indicated in green, whereas sequence identities are reported in orange. A more variable region (arrows) is highlighted in the middle of the domain, likely corresponding either to a large loop or to a linker separating two distinct subdomains. S, T, P, and G stand for serine, threonine, proline, and glycine, respectively. Orange and yellow colorings are used to designate identical and similar amino acids, respectively.
Figure 7.
ERCC6L2/Hebo TUDOR domain. (Top) Alignment of the TUDOR domain of human ERCC6L2 with sequences of TUDOR domains of known 3D structures. Protein Data Bank identifiers are given after the protein names. Secondary structures, as observed in the experimental 3D structures of PHF20 and TDRD3, are reported above the sequences. Colors are used to represent highly conserved positions: orange, aromatic; yellow, loop forming; green, hydrophobic; pink, acidic. Stars designate amino acids participating in the aromatic binding cage. Red stars, aromatic amino acids that are conserved in the experimental 3D structures but not in ERCC6L2. Dark green star, aromatic amino acid that is strictly conserved in all the sequences shown. Light green stars, two positions that can be substituted for each other in the cage depending on the considered domain. Gray star, nonaromatic position participating in the cage. (Bottom) Ribbon representations of the 3D structures of PHF20 and survival of motor neurons, highlighting the positions of the amino acids participating in the methyl-binding cage (labeled 1–5).
Figure 8.
ERCC6L2/Hebo ATPase domain. (A) Sequence alignment of the human ERCC6L2 SWI/SNF2 ATPase domain with those of the Rad54 proteins of known 3D structures (D. rerio, Protein Data Bank accession no. 1z3i [Thomä et al., 2005]; and Sulfolobus solfataricus, Protein Data Bank accession no. 1z63 [Dürr et al., 2005]). SWI2/SNF2-specific elements are depicted in magenta (HD1) and green (HD2), whereas the RecA-like helicase domains are blue (lobe1) and red (lobe2). Helicase-specific domains are shown with numbers, and SWI-SNF2–specific elements are depicted with letters (Thomä et al., 2005). Yellow boxes (alignment) indicate the positions of insertions in the ERCC6L2 structure relative to the Rad54 templates. (B) Global view of the 3D structure model (ribbon representation) of the human ERCC6L2 SWI/SNF2 ATPase domain, constructed on the basis of its alignment with sequences of known 3D structures (see A). The same colors are used as in A. Circles (3D structure) indicate the positions of insertions in the ERCC6L2 structure relative to the Rad54 templates (yellow boxes in A). (C) Focus on the HD2 domain showing the location of arginine 655. N-ter, N-terminal. C-ter, C-terminal.
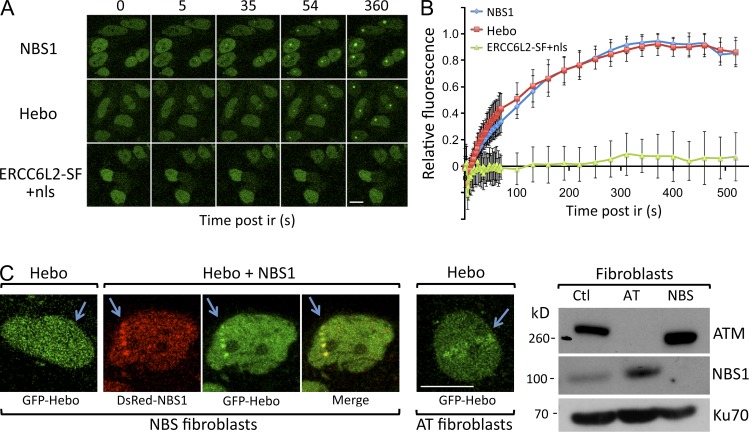
Hebo is recruited to DNA damages
As a first step in the description of the precise function of Hebo during DDR, we analyzed its recruitment at sites of DNA damage after laser microirradiation. GFP-Hebo and GFP-NBS1 fusion proteins were transfected into HeLa cells. 48 h after transfection, DNA damage was inflicted through laser irradiation, and the recruitment of GFP-Hebo and GFP-NBS1 was followed by live imaging. As shown in Fig. 9 A and quantified in Fig. 9 B, Hebo is recruited at the site of DNA damage with the same kinetics observed with NBS1. Interestingly, the forced expression of GFP–ERCC6L2-SF into the nucleus through the addition of the SV40–nuclear localization signal (nls) did not lead to the recruitment of this isoform to the DNA lesion. Altogether, these results further strengthen the notion that Hebo acts as a bona fide DNA repair factor and strongly suggests that the C-terminal–specific region of Hebo is critical for its localization in the nucleus and its recruitment to DNA lesions. The recruitment of NBS1 to DNAdsb’s represents one of the earliest events in the process of DDR (Lukas et al., 2003). The comparable early kinetics of Hebo and NBS1 recruitment after laser microirradiation prompted us to search for a possible functional relationship between these two events. For this, we transfected NBS1-deficient fibroblasts obtained from a Nijmegen breakage syndrome patient with GFP-Hebo and found that the absence of NBS1 precluded the recruitment of Hebo at the site of DNA lesions (Fig. 9 C). Conversely, the cotransfection of DsRed-NBS1 and GFP-Hebo in the NBS1-defective cells restored the Hebo recruitment to DNA damage. In contrast, the localization of Hebo to DNA damage does not depend on ataxia telangiectasia (AT) mutated (ATM) as shown by its detectable recruitment in fibroblasts from an AT patient. We conclude that the recruitment of Hebo at DNA breaks is a very initial event during DDR and is dependent on the presence of NBS1.
Figure 9.
Recruitment of Hebo to DNA damage. (A) Snapshots of recruitment of NBS1, Hebo, and nls–ERCC6L2-SF to DNA damage upon laser microirradiation of HeLa-transfected cells. This experiment was performed five times. (B) Kinetics of recruitment of NBS1 and Hebo to DNA damage upon laser microirradiation. This experiment was performed twice. Data are mean ± SEM. (C) Fibroblasts from an NBS1-deficient patient were transfected with GFP-Hebo with or without DsRed-NBS1 and subjected to laser microirradiation. No Hebo recruitment was detected in the absence of NBS1. GFP-Hebo transfected in ATM-deficient fibroblasts localized normally to DNA damage upon laser microirradiation. Both experiments were performed twice. The Western blot confirms the genetic status of NBS1- and ATM-deficient cell lines. Bars, 10 µm. Ctl, control. Arrows indicate the location of the laser microirradiation.
Hebo is not essential for DNAdsb repair by NHEJ or HR
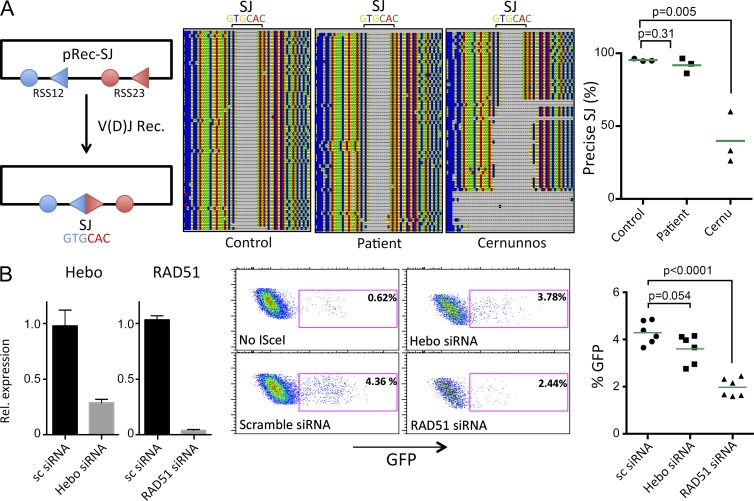
We analyzed the implication of Hebo during NHEJ using a V(D)J recombination assay, as this mechanism relies exclusively on NHEJ. The formation of signal joints (SJs) during V(D)J recombination consists of the error-free fusion of recombination sequences in WT cells but is highly imprecise in cells harboring NHEJ defects such as Cernunnos or DNA ligase IV mutants (Buck et al., 2006a,b). Fibroblasts from the Hebo patient were cotransfected with the RAG1/2 expression constructs together with the V(D)J reporter substrate pRec-SJ. SJs were PCR amplified and analyzed by single molecule sequencing on a personal genome machine (PGM; IonTorrent). As shown in Fig. 10 A, whereas 40% of SJs are imprecise in the case of Cernunnos cells with various lengths of deletions as previously described (Buck et al., 2006a), 92% of SJs recovered from Hebo cells were precise compared with the 95% in control cells, arguing against a major role of Hebo during NHEJ. The function of Hebo during HR was analyzed using the well established U20S-DRGFP experimental system (Pierce and Jasin, 2005). Expression of Hebo and RAD51 as controls was down-regulated by means of siRNA, and levels of HR were determined after introduction of DNAdsb’s through I-SceI. As shown in Fig. 10 B, whereas extinction of RAD51 severely impaired HR (1.97% GFP+ vs. 4.29% GFP+ for control; P < 0.0001), the efficacy of HR was only slightly (not statistically significant) diminished (3.60% GFP+; P = 0.054) in the condition with siRNA against Hebo. This suggests that Hebo is not critically required for HR either, although it may participate to some extent. These two results have to be considered in light of the mild clinical presentation of the patient, which does not support, despite the existence of a homozygous nonsense mutation, a major role of Hebo during NHEJ or HR. Further studies are now required to better understand the implication of Hebo during DNA repair in general.
Figure 10.
Role of Hebo in NHEJ and HR. (A) Analysis of SJ fidelity after V(D)J recombination (V[D]J rec.) of the pRec-SJ substrate in healthy control and patient’s fibroblasts and fibroblasts from a Cernunnos (Cernu)-deficient patient. SJs were PCR amplified and sequenced using single molecule sequencing by Next Generation Sequencing. The frequency of precise SJs was scored in three independent experiments. (B) Analysis of HR using the U2OS-DRGFP system. Hebo and RAD51 were down-regulated by siRNA transfection, and HR was evaluated in six independent experiments after transfection of I-SceI–expressing plasmid. HR is scored by determining the frequency of GFP-positive cells 48 h after transfection. Statistics were analyzed by Mann-Whitney t tests. Data are mean ± SEM. Rel., relative.
DISCUSSION
We analyzed the cells of a young patient presenting with mild bone marrow failure and microcephaly to demonstrate the existence of a general DNA repair defect in these cells that affects resolution of DNAdsb’s as judged by specific phleo and IR sensitivity. Genetic analyses uncovered a nonsense mutation (R655X) in the ERCC6L2 gene as the molecular cause of this deficiency. Tummala et al. (2014) previously reported the same mutation and concluded that the 712-aa protein encoded by ERCC6L2 participates in DDR. Our results only partly support this conclusion, primarily based on the fact that the 712-aa ERCC6L2-encoded protein does not complement the DNA repair defect in our patient’s cells, a point that was not addressed in Tummala’s study. Instead, we identified a new protein, Hebo, encoded by an alternative ERCC6L2 transcript, in which an additional 850-aa peptide is appended to the previously annotated ERCC6L2-SF. Hebo presents features of a bona fide DNA repair factor. It is ubiquitously expressed, localized primarily in the nucleus, and rapidly recruited to the site of DNA lesions in an NBS1-dependent manner. Most importantly, the introduction of WT Hebo in the patient’s cells rescues their DNA repair deficiency as judged by the recovery of a normal sensitivity to the DNAdsb inducer phleo. Whether the ERCC6L2-SF exerts any biological function or represents a nonfunctional alternative transcript remains to be investigated. Tummala et al. (2014) suggested a possible link to mitochondrial function, a facet not addressed in our study. Alternative splicing events have been previously described in the case of the RecQ5 helicase, for which the long form is localized in the nucleus, whereas the SF remains in the cytoplasm (Shimamoto et al., 2000). Both isoforms include the ATPase domain like in ERCC6L2-SF/Hebo. Although in the case of RecQ5 the different splicing forms appear to be tissue specific, we did not find strong evidence for such a regulation in the case of ERCC6L2/Hebo.
Hebo carries three noticeable structured regions, a Tudor domain, an ATPase domain, and HEBO, a new globular domain conserved through evolution but only present in direct homologues of Hebo. The presence of a Tudor domain adjacent to the ATPase fold suggested that this region might be responsible for the recruitment of Hebo to chromatin after DNA damage. One of the recognized functions of the Tudor domain is its interaction with methylated proteins, which makes it a histone mark reader, particularly within the 53BP1 DNA repair factor (Charier et al., 2004; Botuyan et al., 2006; Faure and Callebaut, 2013; Fradet-Turcotte et al., 2013). Indeed, the binding of the 53BP1 Tudor domain to histone H4 dimethylated on lysine 20 (H4K20me2) is essential during DDR (Mallette et al., 2012; Tang et al., 2013). Unexpectedly, the SF of ERCC6L2 containing only the Tudor domain and the ATPase domain is not recruited to DNA damage, even when its nuclear localization is enforced by the addition of the SV40-nls, which argues against a critical role of the Hebo Tudor domain in direct chromatin binding during DNA repair, as opposed to 53BP1. Structural differences in the Hebo Tudor domain, in particular the replacement of two and four aromatic residues critical for methyl binding by serine and cysteine, suggest that it has probably evolved to serve a different function. Although this would be largely unprecedented, a few examples exist, such as the Tudor domain in the Tejas (Tej) protein in Drosophila melanogaster, which drives interaction with its partner Aubergine (Aub) independently of arginine methylation (Patil and Kai, 2010). The search for specific protein partners of the Hebo Tudor domain should help clarify this interesting issue. If the Hebo Tudor domain is not directly involved in chromatin binding, what other regions of the protein are critical for Hebo’s recruitment to DNA lesions? Because only the full-length Hebo, as opposed to the ERCC6L2-SF, is recruited to DNA damage, this suggests that the C-terminal half of the protein, perhaps the newly described HEBO domain, is critical for this function. Whether this implies a direct binding of Hebo to chromatin or its conveyance through interaction with a cargo protein remains to be determined. The second noticeable region of Hebo is the ATPase domain adjacent to the Tudor domain, which suggests a possible helicase function for Hebo. DNA/RNA helicases are classified into six superfamilies that perform various functions (Singleton et al., 2007). The SF2 family is the largest and includes the DEAD-box helicases, the RecQ family, and the Snf2-related enzymes. Defects in the members of the RecQ family Bloom syndrome helicase, Werner syndrome helicase, and RECQL4, known to be critical for the repair of DNAdsb’s through HR, result in Bloom, Werner, and Rothmund-Thomson syndromes, respectively, which associate developmental anomalies and a clear cancer predisposition (Hickson, 2003; Suhasini and Brosh, 2013). Another family of DNA helicase, the iron-sulfur (Fe-S) cluster helicases, includes the XPD and XPB factors mutated in xeroderma pigmentosum, FANCJ, the mutations of which are responsible for one group of FA patients, the DDX11 factor involved in Warsaw breakage syndrome, and the more recently described RTEL1 helicase with a more dedicated function to telomere maintenance (Le Guen et al., 2013; Suhasini and Brosh, 2013). In contrast to RecQ and Fe-S helicases that exert their function directly on DNA through their ATP- and Mg2+-dependent unwinding activity, the helicases of the Snf2/Swi2 family lack classical helicase activity in the sense of DNA strand displacement but are motor proteins that act as chromatin remodelers (Ceballos and Heyer, 2011). One prototypical member of this family is the RAD54 protein, which together with RAD51 plays a major role during various phases of HR. Hebo most likely belongs to the Snf2/Swi2 family of chromatin remodelers given its sequence/structure similarities with RAD54. The third domain identified in Hebo is new. The 200-aa HEBO domain is well conserved during evolution but is only present in direct homologues of Hebo. To date, no clear function can be ascribed to this domain outside the fact that the 850-aa peptide to which it belongs is required for both the nuclear localization and the recruitment of Hebo to DNA damage. Identification of putative interacting partners of the HEBO domain should help clarify its function in the future.
One important question to understand is what the specific function of Hebo is among the panel of other chromatin remodelers in general and during the repair of DNAdsb’s in particular, and how and why its defect translates into clinical manifestations observed in the described patient here. The patient suffers from mild bone marrow failure that did not necessitate HSC transplantation up to now, in contrast to the much more severe aplasia conditions observed in FA or Hoyeraal-Hreidarsson syndrome. The premium physiological causes underlying IBMFS in FA and other related disorders have remained poorly understood, in particular given the fact that several KO mouse models do not recapitulate this phenotypic trait spontaneously (Parmar et al., 2009). Nevertheless, it was proposed that a chronic activation of the TP53 DDR was involved in the decline of HSCs in FA patients (Ceccaldi et al., 2012), although the source of DNA damages in HSCs remained unclear. Walter et al. (2015) recently demonstrated that DNA damages in HSCs is the direct consequence of their exit from the quiescent state as experimentally provoked by injection of polyinosinic:polycytidylic acid to mimic a viral infection and induce a type I IFN response. The same observations were made using more physiological stimuli such as IFN-α, G-CSF, or serial bleeding. Perhaps most importantly, Fanca−/− mice, but not WT or untreated Fanca−/− mice, subjected to several rounds of polyinosinic:polycytidylic acid–induced HSC activation succumbed to severe aplastic anemia, thus mimicking the bone marrow failure observed in human FA patients. In this context, it is of interest to note that the clinical symptoms of the patient we studied, in particular the thrombocytopenia, arose subsequent to a febrile episode of unknown etiology. It is therefore tempting to speculate that an acute triggering of HSCs during that episode resulted in DNA damages that require Hebo for their proper repair. IBMFS is not restricted to patients (or mouse models) harboring gene defects in the Fanconi pathway. Indeed, a recent study noticed a cytopenia requiring platelet transfusion in 80% of a dozen of DNA ligase IV–mutated patients (Murray et al., 2014). Likewise, one of the patients we initially described with a Cernunnos/Xlf deficiency (Buck et al., 2006a) subsequently developed bone marrow failure (unpublished data), a characteristic that can be explained in light of a study demonstrating a premature aging of HSCs in a Cernunnos/Xlf KO mouse model (Avagyan et al., 2014). Lastly, congenital bone marrow failure caused the premature death of mice harboring three alanine substitutions in the DNA-dependent protein kinase catalytic subunit in a cluster of threonine residues known to be phosphorylated as part of DDR (Zhang et al., 2011). One could argue that Hebo deficiency may be even more connected to these three examples of NHEJ defect–related bone marrow failure, as these conditions are associated with a DNAdsb repair defect (phleo) like in Hebo-deficient cells rather than the inability to cope with interstrand cross-links (MMC) as observed in FA cells.
FA patients, as other conditions of DNA repair deficiency, present with a high risk of developing hemopoietic malignancies and other types of solid cancers. It is therefore legitimate to consider that Hebo may represent yet another critical genome caretaker, although this question cannot obviously be addressed at the present time with so few affected patients. The development of a Hebo-deficient mouse model should be very valuable in the future to address these important questions by providing the possibility to analyze a large series of affected individuals, whereas our case is unique.
MATERIALS AND METHODS
Cells
Informed consent for our study was obtained from the family in accordance with the Helsinki Declaration. The Institut National de la Santé et de la Recherche Médicale Institutional Review Board also approved this study. Primary fibroblasts were obtained through culture of the patient’s skin biopsy and further transformed by transfection with SV40 large T expression plasmids. Control primary fibroblasts (GM05757 and GM08399) were from apparently healthy donors of 7 and 19 yr, respectively (Coriell Cell Repositories). A-T cells (GM05823) were from an 18-yr-old patient suffering from AT (Coriell Cell Repositories). Cernunnos-deficient cells were described previously (Buck et al., 2006a). DNA ligase IV–deficient EBV B cells were from our undescribed patient. FA fibroblasts were from our undescribed FANCG-mutated patient. AT and rad3-related protein–deficient fibroblasts were from patient F02-98 described previously (O’Driscoll et al., 2003).
Sensitivity to genotoxics and complementation survival assay
Cellular sensitivity was performed as previously described (Buck et al., 2006a). In brief, cells were seeded at low density in complete culture medium and incubated with increasing doses of genotoxic drugs or subjected to ionizing or UV radiations. Surviving cells were counted using flow cytometry with a constant amount of fluorescent beads. After 7 d, survival fraction was calculated relative to untreated cells. IRIF were analyzed using anti-53BP1 staining as described previously (Buck et al., 2006a). In brief, early passaged primary fibroblasts from the healthy control, patient, and Cernunnos/Xlf patient were X irradiated (2 Gy) on coverslips. Cells were harvested 1 and 24 h after irradiation, fixed in 4% paraformaldehyde, and permeabilized in 1× PBS, 1% BSA, 0.1% Triton X-100, and 1 mM EDTA before staining for 1 h with anti-53BP1 (Novus Biologicals). Cells were further incubated with Alexa Fluor 488 secondary antibodies (Molecular Probes and Invitrogen), stained with DAPI (Invitrogen), and mounted in FluorSave (EMD Millipore). Images were taken under the same quantity of laser power and exposure time with epifluorescence microscopy (Axioplan; ZEISS). Foci numbers were quantified for each cell with ImageJ software (National Institutes of Health). For complementation of phleo sensitivity, patients’ SV40-transformed fibroblasts were transduced with a bicistronic ERCC6L2-Ires-GFP–expressing lentivirus or empty vector and treated or not with 100 ng/ml phleo for up to 3 wk. The number of GFP-positive cells was scored at various time points to assess for relative selective advantage of transduced (GFP+) over untransduced (GFP−) cells. The absolute sensitivity of transduced cells was also determined by incubating cell-sorted GFP-positive transduced cells with increasing doses of phleo and determining the surviving fraction after 7 d. For G2M block analysis, EBV-transformed B cells were treated with increasing doses of MMC and collected 48 h later. The cells were washed in PBS, resuspended in 100 µl PBS, fixed in 1 ml of 70% EtOH, and kept overnight at −20°C. Fixed cells were washed in PBS, resuspended in 300 µl PBS with 5% Triton X-100 and 25 µg/ml propidium iodide, and analyzed by FACS.
Cytogenetic analysis
Primary fibroblasts of similar cell culture passages were seeded at a density of 4 × 105 cells per 75-cm2 flask. After adhesion of the cells, 50 ng/ml MMC was added for 24 h to the cell medium. Then, the medium was changed, and the cells were grown for another 28 h. During the last 8 h, 0.1 µg/ml colchicine (Sigma-Aldrich) was added to the cells. Trypsinized cell pellet was swollen in hypotonic KCl solution and incubated at 37°C for 20 min. Cells were fixed in ethanol/acetic acid (3:1) and spread on glass slides. After air drying, the slides were stained with 4% Giemsa for 10 min. After mounting, metaphase spreads were captured in a blind fashion using bright field microscopy (AX70; Olympus) with a 63× objective lens. Statistical analyses were performed with Prism software (GraphPad Software) using an unpaired Student’s t test.
SA–β-gal staining
SA–β-gal–positive cells were detected as previously described (Dimri et al., 1995). In brief, monolayers of cells were fixed with 4% paraformaldehyde, incubated at 37°C, and stained overnight at 37°C with a staining solution mix at pH 6.0 using a β-gal detection kit (Invitrogen). After overnight incubation, cells were counterstained with propidium iodide before observation under a fluorescence microscope (B51; Olympus) with a 20× objective lens. The number of positive cells with blue color was counted and normalized to the number of total cells in the same field. For each condition, cells from more than five randomly chosen fields were counted (to total at least 100 cells counted per condition). Similar results were obtained in two independent experiments.
Telomere length determination
Telomere length was determined using terminal restriction fragment analysis by Southern blotting of DNA extracted from whole blood from the patient, his siblings, and parents as described previously (Touzot et al., 2010).
WGHM genotyping and WES
In accordance with the Helsinki Declaration, informed consent for the genetic and functional investigations was obtained from the parents and investigated family members. WGHM was determined using a BeadChip (Human Linkage-24; Illumina) according to standard methodology as previously described (Le Guen et al., 2013). WES was performed through 100-bp paired-end reads on Illumina HiSeq after capture using an exome enrichment kit (TruSeq; Illumina) as previously described (Le Guen et al., 2013).
Real-time quantitative RT-PCR
2 µg RNA from a panel of tissues (Human Total RNA Panel II; Takara Bio Inc.) was treated with DNaseI (amplification grade; Invitrogen) before reverse transcription. 80 ng cDNA was used for each Taqman reaction (ViiA7) using sets of primers and probes specifically designed to distinguish ERCC6L2-SF and Hebo alternative transcripts. The expression level was normalized with the housekeeping gene PSMB2 (Hs01002946; Thermo Fisher Scientific) sequence of the following primers: ERCC6L2-SF forward, 5′-AGGCAGTTCAAGGAGCAATTG-3′; ERCC6L2-SF reverse, 5′-TTCCACCTCCGGTCAAGATG-3′; ERCC6L2-SF Taqman MGB Probe, 5′-TGTCCTTGAGGAACTTA-3′; Hebo forward, 5′-TGGATCGAGCAAAGCTGAAA-3′; Hebo reverse, 5′-CTGCTTCAACTCAAATACGTCATGT-3′; and Hebo Taqman MGB Probe, 5′-CACATGAGCCGATGGG-3′.
ERCC6L2 long form (Hebo) identification and cloning
Based on human (accession no. ENSG00000182150) and rat (accession no. ENSRNOG00000019175) genome annotation in the Ensembl database for the ERCC6L2 gene, primers were designed to amplify a putative transcript linking human transcripts 001, 006, and 007 (Fig. 4) as follows: forward exon 12, 5′-TGGACTAGGCCTCAATTTTGTCGGTGC-3′; reverse exon 15, 5′-GGCTCTTCATCACTGAAGTCACTGCAGAGA-3′; forward exon 16, 5′-CAACAGAAGCCAAAGATGCTGGTTGTGAG-3′; and reverse exon 16, 5′-ACTAGTTGATTCTTGGTCTGTGGCTCTGT-3′. The expression of a full-length ERCC6L2 long form was further validated by cDNA PCR amplification using the following primers: forward, 5′-TTTGCTGGGATCCCCCTCCTCCATCCT-3′; and reverse, 5′-GGCACTGGATTCCACACTGATTCTATTATCTTG-3′.
ERCC6L2-SF and ERCC6L2 long form (or HEBO) orf’s were PCR amplified and cloned into a bicistronic (IresGFP) lentiviral vector for complementation experiments and in fusion with GFP into EGFP-N2 vector for cellular localization and microirradiation experiments. To drive the ERCC6L2-SF in the nucleus, the SV40 large T antigen nls PPKKKRKV sequence was added at the N terminus of ERCC6L2-SF.
Laser microirradiation
NBS1-EGFP and DsRed plasmids were provided by C. Lukas (University of Copenhagen, Copenhagen, Denmark). HeLa cells were transfected with ERCC6L2-GFP and NBS1-GFP constructs using Jet Prime reagent (Polyplus transfection). 24 h after transfection, cells were presensitized with BrdU (BD) for another 28 h. Microirradiation was performed using a 405-nm UV laser focused through a 63× objective lens in a confocal microscope system (TCS sp8; Leica Biosystems). Laser settings (100% laser power for 10% of laser duration) were used to generate DNA damage specifically along the laser path (defined by a series of dots) in a BrdU-dependent manner. The cells were analyzed in time course mode with a 0.329-s interval between pictures for up to 215 pictures. The interval between pictures was then shifted to 30 s for another 15 pictures. Images were exported as LIF files and analyzed using ImageJ.
CRISPR/Cas9 gene inactivation
The 5′-TTGAGAGACTACCAAAGAGA-3′ located in ERCC6L2 exon 2 was used as gRNA and cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 plasmid (a gift from F. Zhang, the Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA; plasmid 42230; Addgene; Ran et al., 2013) for transfection into U2OS cells. The extent of mutagenesis was evaluated by single molecule sequencing using PGM (IonTorrent) after amplification of genomic DNA spanning the CRISPR target site.
Sequence analysis and molecular modeling
Phyre2 was used to select the best templates for the modeling of the 3D structure of the ATPase domain, which was then performed using Modeller (v9.10; Martí-Renom et al., 2000). The analysis of the C-terminal domain was made using hydrophobic cluster analysis (Callebaut et al., 1997; Faure and Callebaut, 2013). Profile–profile search methods, such as HH-PRED (Söding et al., 2005), were also used together with standard sequence similarity searches (Altschul et al., 1997) to highlight putative relationships of the HEBO domain with other protein sequences.
V(D)J and HR assays
V(D)J recombination assays were performed as previously described (Buck et al., 2006a). SJs were sequenced using single molecule sequencing on a PGM (IonTorrent). For HR assay, 5 µg I-Scel–expressing plasmid was electroporated in 106 U2OS-DRGFP cells (a gift from M. Jasin, Memorial Sloan Kettering Cancer Center, New York, NY). Cells were then reverse transfected with scramble, Hebo, and Rad51 siRNAs (SMARTpool ON-TARGET plus siRNA; GE Healthcare) according to manufacturer recommendations. The percentage of GFP-positive cells was determined by FACS analysis 72 h after transfection. The efficacy of siRNA was verified by Taqman real-time PCR.
Online supplemental material
Table S1 shows biological characteristics of the patient. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151183/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patient and his parents for participating in this study, the imaging facility of Institut Imagine for help with microlaser irradiation, the Bioinformatic Department of Institut Imagine for help with WES analysis, and the genomic facility of Institut Imagine for single molecule sequencing on the PGM. We thank Dr. Claudia Lukas for the pEGFP-NBS1 plasmid and Dr. Feng Zhang for the pX330 plasmid.
This work was supported by institutional grants from the Institut National de la Santé et de la Recherche Médicale, the Institut National du Cancer (PLBIO11-151), Imagine Institut (ANR-10-IAHU-01), the European Research Council (ERC-2009-AdG_20090506 n°FP7-249816), and Ligue Contre le Cancer (Equipe Labelisée LA LIGUE).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 3D
- three-dimensional
- AT
- ataxia telangiectasia
- ATM
- AT mutated
- CBC
- cell blood count
- DDR
- DNA damage response
- dsb
- double strand break
- FA
- Fanconi anemia
- gRNA
- guide RNA
- HR
- homologous recombination
- HSC
- hemopoietic stem cell
- IBMFS
- inherited bone marrow failure syndrome
- IR
- ionizing radiation
- IRIF
- irradiation-induced foci
- MMC
- mitomycin C
- NHEJ
- nonhomologous end-joining
- nls
- nuclear localization signal
- NSV
- nonsynonymous variant
- orf
- open reading frame
- PGM
- personal genome machine
- SA
- senescence-associated
- SF
- short isoform
- SJ
- signal joint
- WES
- whole exome sequencing
- WGHM
- whole genome homozygosity mapping
- Xlf
- XRCC4-like factor
References
- Alt F.W., Zhang Y., Meng F.L., Guo C., and Schwer B.. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 152:417–429. 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., and Lipman D.J.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach A.D. 2009. Fanconi anemia and its diagnosis. Mutat. Res. 668:4–10. 10.1016/j.mrfmmm.2009.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avagyan S., Churchill M., Yamamoto K., Crowe J.L., Li C., Lee B.J., Zheng T., Mukherjee S., and Zha S.. 2014. Hematopoietic stem cell dysfunction underlies the progressive lymphocytopenia in XLF/Cernunnos deficiency. Blood. 124:1622–1625. 10.1182/blood-2014-05-574863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan M.V., Lee J., Ward I.M., Kim J.E., Thompson J.R., Chen J., and Mer G.. 2006. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 127:1361–1373. 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck D., Malivert L., de Chasseval R., Barraud A., Fondanèche M.C., Sanal O., Plebani A., Stéphan J.L., Hufnagel M., le Deist F., et al. 2006a Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 124:287–299. 10.1016/j.cell.2005.12.030 [DOI] [PubMed] [Google Scholar]
- Buck D., Moshous D., de Chasseval R., Ma Y., le Deist F., Cavazzana-Calvo M., Fischer A., Casanova J.L., Lieber M.R., and de Villartay J.P.. 2006b Severe combined immunodeficiency and microcephaly in siblings with hypomorphic mutations in DNA ligase IV. Eur. J. Immunol. 36:224–235. 10.1002/eji.200535401 [DOI] [PubMed] [Google Scholar]
- Callebaut I., Labesse G., Durand P., Poupon A., Canard L., Chomilier J., Henrissat B., and Mornon J.P.. 1997. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell. Mol. Life Sci. 53:621–645. 10.1007/s000180050082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos S.J., and Heyer W.D.. 2011. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta. 1809:509–523. 10.1016/j.bbagrm.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Parmar K., Mouly E., Delord M., Kim J.M., Regairaz M., Pla M., Vasquez N., Zhang Q.S., Pondarre C., et al. 2012. Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell. 11:36–49. 10.1016/j.stem.2012.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charier G., Couprie J., Alpha-Bazin B., Meyer V., Quéméneur E., Guérois R., Callebaut I., Gilquin B., and Zinn-Justin S.. 2004. The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure. 12:1551–1562. 10.1016/j.str.2004.06.014 [DOI] [PubMed] [Google Scholar]
- Cui G., Park S., Badeaux A.I., Kim D., Lee J., Thompson J.R., Yan F., Kaneko S., Yuan Z., Botuyan M.V., et al. 2012. PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53. Nat. Struct. Mol. Biol. 19:916–924. 10.1038/nsmb.2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villartay J.P., Fischer A., and Durandy A.. 2003. The mechanisms of immune diversification and their disorders. Nat. Rev. Immunol. 3:962–972. 10.1038/nri1247 [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 92:9363–9367. 10.1073/pnas.92.20.9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal I., and Vulliamy T.. 2008. Inherited aplastic anaemias/bone marrow failure syndromes. Blood Rev. 22:141–153. 10.1016/j.blre.2007.11.003 [DOI] [PubMed] [Google Scholar]
- Dürr H., Körner C., Müller M., Hickmann V., and Hopfner K.P.. 2005. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 121:363–373. 10.1016/j.cell.2005.03.026 [DOI] [PubMed] [Google Scholar]
- Faure G., and Callebaut I.. 2013. Identification of hidden relationships from the coupling of hydrophobic cluster analysis and domain architecture information. Bioinformatics. 29:1726–1733. 10.1093/bioinformatics/btt271 [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A., Canny M.D., Escribano-Díaz C., Orthwein A., Leung C.C., Huang H., Landry M.C., Kitevski-LeBlanc J., Noordermeer S.M., Sicheri F., and Durocher D.. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 499:50–54. 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M.S., Timmers C., Hejna J., Grompe M., and D’Andrea A.D.. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 7:249–262. 10.1016/S1097-2765(01)00173-3 [DOI] [PubMed] [Google Scholar]
- Glousker G., Touzot F., Revy P., Tzfati Y., and Savage S.A.. 2015. Unraveling the pathogenesis of Hoyeraal-Hreidarsson syndrome, a complex telomere biology disorder. Br. J. Haematol. 170:457–471. 10.1111/bjh.13442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I.D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 3:169–178. 10.1038/nrc1012 [DOI] [PubMed] [Google Scholar]
- Hira A., Yoshida K., Sato K., Okuno Y., Shiraishi Y., Chiba K., Tanaka H., Miyano S., Shimamoto A., Tahara H., et al. 2015. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am. J. Hum. Genet. 96:1001–1007. 10.1016/j.ajhg.2015.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guen T., Jullien L., Touzot F., Schertzer M., Gaillard L., Perderiset M., Carpentier W., Nitschke P., Picard C., Couillault G., et al. 2013. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum. Mol. Genet. 22:3239–3249. 10.1093/hmg/ddt178 [DOI] [PubMed] [Google Scholar]
- Liu K., Guo Y., Liu H., Bian C., Lam R., Liu Y., Mackenzie F., Rojas L.A., Reinberg D., Bedford M.T., et al. 2012. Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS One. 7:e30375 10.1371/journal.pone.0030375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C., Falck J., Bartkova J., Bartek J., and Lukas J.. 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5:255–260. 10.1038/ncb945 [DOI] [PubMed] [Google Scholar]
- Mallette F.A., Mattiroli F., Cui G., Young L.C., Hendzel M.J., Mer G., Sixma T.K., and Richard S.. 2012. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31:1865–1878. 10.1038/emboj.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., and Sali A.. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291–325. 10.1146/annurev.biophys.29.1.291 [DOI] [PubMed] [Google Scholar]
- Moldovan G.L., and D’Andrea A.D.. 2009. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 43:223–249. 10.1146/annurev-genet-102108-134222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.E., Bicknell L.S., Yigit G., Duker A.L., van Kogelenberg M., Haghayegh S., Wieczorek D., Kayserili H., Albert M.H., Wise C.A., et al. 2014. Extreme growth failure is a common presentation of ligase IV deficiency. Hum. Mutat. 35:76–85. 10.1002/humu.22461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M., Ruiz-Perez V.L., Woods C.G., Jeggo P.A., and Goodship J.A.. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33:497–501. 10.1038/ng1129 [DOI] [PubMed] [Google Scholar]
- Parmar K., D’Andrea A., and Niedernhofer L.J.. 2009. Mouse models of Fanconi anemia. Mutat. Res. 668:133–140. 10.1016/j.mrfmmm.2009.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V.S., and Kai T.. 2010. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol. 20:724–730. 10.1016/j.cub.2010.02.046 [DOI] [PubMed] [Google Scholar]
- Pierce A.J., and Jasin M.. 2005. Measuring recombination proficiency in mouse embryonic stem cells. Methods Mol. Biol. 291:373–384. [DOI] [PubMed] [Google Scholar]
- Pinto F.O., Leblanc T., Chamousset D., Le Roux G., Brethon B., Cassinat B., Larghero J., de Villartay J.P., Stoppa-Lyonnet D., Baruchel A., et al. 2009. Diagnosis of Fanconi anemia in patients with bone marrow failure. Haematologica. 94:487–495. 10.3324/haematol.13592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman K.A., Lach F.P., Abhyankar A., Donovan F.X., Sanborn E.M., Kennedy J.A., Sougnez C., Gabriel S.B., Elemento O., Chandrasekharappa S.C., et al. 2015. Deficiency of UBE2T, the E2 ubquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T subtype of fanconi anemia. Cell Reports. 12:35–41. 10.1016/j.celrep.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., and Bonner W.M.. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868. 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Ruggero D., and Shimamura A.. 2014. Marrow failure: a window into ribosome biology. Blood. 124:2784–2792. 10.1182/blood-2014-04-526301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L.A., Unsal-Kaçmaz K., and Linn S.. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39–85. 10.1146/annurev.biochem.73.011303.073723 [DOI] [PubMed] [Google Scholar]
- Shimamoto A., Nishikawa K., Kitao S., and Furuichi Y.. 2000. Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3α and 3β. Nucleic Acids Res. 28:1647–1655. 10.1093/nar/28.7.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton M.R., Dillingham M.S., and Wigley D.B.. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76:23–50. 10.1146/annurev.biochem.76.052305.115300 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R. III, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D’Andrea A.D., and Elledge S.J.. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 129:289–301. 10.1016/j.cell.2007.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J., Biegert A., and Lupas A.N.. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248. 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhasini A.N., and Brosh R.M. Jr. 2013. Disease-causing missense mutations in human DNA helicase disorders. Mutat. Res. 752:138–152. 10.1016/j.mrrev.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Cho N.W., Cui G., Manion E.M., Shanbhag N.M., Botuyan M.V., Mer G., and Greenberg R.A.. 2013. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 20:317–325. 10.1038/nsmb.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomä N.H., Czyzewski B.K., Alexeev A.A., Mazin A.V., Kowalczykowski S.C., and Pavletich N.P.. 2005. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat. Struct. Mol. Biol. 12:350–356. 10.1038/nsmb919 [DOI] [PubMed] [Google Scholar]
- Touzot F., Callebaut I., Soulier J., Gaillard L., Azerrad C., Durandy A., Fischer A., de Villartay J.P., and Revy P.. 2010. Function of Apollo (SNM1B) at telomere highlighted by a splice variant identified in a patient with Hoyeraal-Hreidarsson syndrome. Proc. Natl. Acad. Sci. USA. 107:10097–10102. 10.1073/pnas.0914918107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzot F., Gaillard L., Vasquez N., Le Guen T., Bertrand Y., Bourhis J., Leblanc T., Fischer A., Soulier J., de Villartay J.P., and Revy P.. 2012. Heterogeneous telomere defects in patients with severe forms of dyskeratosis congenita. J. Allergy Clin. Immunol. 129:473–482.e3. 10.1016/j.jaci.2011.09.043 [DOI] [PubMed] [Google Scholar]
- Tripsianes K., Madl T., Machyna M., Fessas D., Englbrecht C., Fischer U., Neugebauer K.M., and Sattler M.. 2011. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat. Struct. Mol. Biol. 18:1414–1420. 10.1038/nsmb.2185 [DOI] [PubMed] [Google Scholar]
- Tummala H., Kirwan M., Walne A.J., Hossain U., Jackson N., Pondarre C., Plagnol V., Vulliamy T., and Dokal I.. 2014. ERCC6L2 mutations link a distinct bone-marrow-failure syndrome to DNA repair and mitochondrial function. Am. J. Hum. Genet. 94:246–256. 10.1016/j.ajhg.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virts E.L., Jankowska A., Mackay C., Glaas M.F., Wiek C., Kelich S.L., Lottmann N., Kennedy F.M., Marchal C., Lehnert E., et al. 2015. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum. Mol. Genet. 24:5093–5108. 10.1093/hmg/ddv227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P., et al. 2015. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 520:549–552. 10.1038/nature14131 [DOI] [PubMed] [Google Scholar]
- Wang A.T., and Smogorzewska A.. 2015. SnapShot: Fanconi anemia and associated proteins. Cell. 160:354–354.e1. 10.1016/j.cell.2014.12.031 [DOI] [PubMed] [Google Scholar]
- Zhang S., Yajima H., Huynh H., Zheng J., Callen E., Chen H.T., Wong N., Bunting S., Lin Y.F., Li M., et al. 2011. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J. Cell Biol. 193:295–305. 10.1083/jcb.201009074 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.