Lear et al. report a novel molecular pathway in which Fibrosis Inducing E3 Ligase 1 (FIEL1) regulates TGFβ and fibrosis pathway through SUMO-E3 ligase PIAS4. They also develop a small molecule inhibitor toward FIEL1 that is highly effective in ameliorating fibrosis in mice.
Abstract
The E3 small ubiquitin-like modifier (SUMO) protein ligase protein inhibitor of activated STAT 4 (PIAS4) is a pivotal protein in regulating the TGFβ pathway. In this study, we discovered a new protein isoform encoded by KIAA0317, termed fibrosis-inducing E3 ligase 1 (FIEL1), which potently stimulates the TGFβ signaling pathway through the site-specific ubiquitination of PIAS4. FIEL1 targets PIAS4 using a double locking mechanism that is facilitated by the kinases PKCζ and GSK3β. Specifically, PKCζ phosphorylation of PIAS4 and GSK3β phosphorylation of FIEL1 are both essential for the degradation of PIAS4. FIEL1 protein is highly expressed in lung tissues from patients with idiopathic pulmonary fibrosis (IPF), whereas PIAS4 protein levels are significantly reduced. FIEL1 overexpression significantly increases fibrosis in a bleomycin murine model, whereas FIEL1 knockdown attenuates fibrotic conditions. Further, we developed a first-in-class small molecule inhibitor toward FIEL1 that is highly effective in ameliorating fibrosis in mice. This study provides a basis for IPF therapeutic intervention by modulating PIAS4 protein abundance.
Idiopathic pulmonary fibrosis (IPF) is a fibrotic disease of unknown etiology, but it is characterized by deposition of extracellular matrix into the interstitium and destruction of alveolar architecture, resulting in progressive impairment in gas exchange and, ultimately, death (Gross and Hunninghake, 2001; King et al., 2011; Noble et al., 2012; Sheppard, 2013). IPF is the most common form of interstitial lung disease with a prevalence of 50 per 100,000 cases, and it almost exclusively affects patients older than 50 (Raghu et al., 2011). Despite its unknown etiology, the downstream effectors of IPF are well-characterized. Samples from IPF patients show increased levels of TGFβ across all three isoforms (Annes et al., 2003). TGFβ promotes several IPF-relevant phenotypes: the differentiation of fibroblasts to myofibroblasts, the activation of epithelial to mesenchymal cellular transition, and the promotion of lung epithelial cell apoptosis (Chapman, 2011; Kage and Borok, 2012; Massagué, 2012; Wick et al., 2013). TGFβ transduces downstream signaling in part through the mothers against decapentaplegic homologue (SMAD) protein family. SMAD proteins regulate a variety of cellular processes, such as differentiation, proliferation, tumorigenesis, and immune responses (Attisano and Wrana, 2000; Yingling et al., 2004; Bonniaud et al., 2005). The SMAD family is comprised of receptor-SMADs (R-SMAD), inhibitor SMADs (I-SMAD), and the common mediator SMAD (coSMAD; Derynck and Zhang, 2003). TGFβ signal transduction commences with the phosphorylation of R-SMADs, often SMAD2 or SMAD3, which form a trimeric structure with the coSMAD, SMAD4, and translocate to the nucleus. In the nucleus, the trimer binds to the SMAD-binding element in the JunB promoter to activate the transcription of pro-fibrotic genes (Jonk et al., 1998). Therefore, TGFβ is a major profibrotic growth factor through the downstream SMAD signaling pathway (Wang et al., 2006; Hecker et al., 2009).
Protein inhibitor of activated STAT (PIAS) proteins are a family of proteins that are known to negatively control and regulate gene transcription and inflammatory pathways in cells (Rytinki et al., 2009). There are four characterized PIAS family members—PIAS1, PIASx (PIAS2), PIAS3, and PIASy (PIAS4)—each with specificity toward different pathways (Gross et al., 2001). Specifically, PIAS4 has been shown to suppress TGFβ signaling by several mechanisms (Imoto et al., 2003; Long et al., 2003). First, TGFβ promotes the interaction of PIAS4 with SMAD3 and SMAD4 to form a ternary complex (Long et al., 2003). PIAS4 is known to possess small ubiquitin-like modifier (SUMO) E3 ligase activity within its RING-type domain (Imoto et al., 2004). It promotes the sumoylation of SMAD3, in turn stimulating its nuclear export and inhibiting SMAD3/4-driven pro-fibrotic transcription (Lee et al., 2003; Imoto et al., 2008). Furthermore, PIAS4 directly recruits and interacts with histone deacetylase 1 (HDAC1) to suppress SMAD3-driven transcriptional activation (Long et al., 2003). PIAS4 is an important negative regulator of TGFβ signaling.
Protein ubiquitination is the major mechanism of protein processing in cells. Ubiquitin flags a targeted protein for degradation through the 26s proteasome or the lysosome (Tanaka et al., 2008). Ubiquitin is conjugated to a target protein in a three-step process. First, an E1 ubiquitin-activating enzyme binds to ubiquitin via a thioester covalent bond. Next, the E1 transfers the ubiquitin to an E2 ubiquitin-conjugating enzyme. Finally, the C terminus of ubiquitin is attached to the ε-amino moiety of a substrate’s ubiquitin-accepting lysine (K) residue, mediated by an ubiquitin E3 ligase. There are several families of ubiquitin E3 ligases that include over 1,000 proteins (Hatakeyama et al., 2001; Jin et al., 2007). Of these, the E6-AP carboxyl terminus (HECT) domain E3 ligase family remains poorly characterized (Rotin and Kumar, 2009). There are ∼30 HECT E3 ligases in mammalian cells, and functional data are only available for a select few, including E6AP, Smurf, HECTD2, and NEDD4 (Rotin and Kumar, 2009; Coon et al., 2015). HECT E3 ligases possess a unique feature in which they accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester bond and directly transfer the ubiquitin to the substrate. An active site within the C terminal of the HECT domain containing a cysteine residue is required for ubiquitin-thioester formation (Huibregtse et al., 1995). A recently identified member of the HECT E3 ligase family, apoptosis-resistant E3 ligase 1 (AREL1), which is encoded by the KIAA0317 gene, regulates the ubiquitination of the apoptosis proteins SMAC, HtrA2, and ARTS (Kim et al., 2013).
In this study, we discovered a new protein isoform encoded by KIAA0317, termed fibrosis-inducing E3 ligase 1 (FIEL1). We also revealed a new molecular pathway, in which FIEL1 regulates TGFβ signaling through the ubiquitin-mediated degradation of PIAS4. Under TGFβ stimulation, two kinases, PKCζ and GSK3β, phosphorylate PIAS4 and FIEL1, respectively. Phosphorylated FIEL1 targets phosphorylated PIAS4 and mediates its ubiquitination and degradation, thereby exaggerating TGFβ signaling. These studies provide a new molecular model of fibrotic lung injury and lead to the development of a small molecule antagonist that exerts potent antifibrotic activity by regulating the abundance of PIAS4.
RESULTS
FIEL1–PIAS4 pathway in pulmonary fibrosis
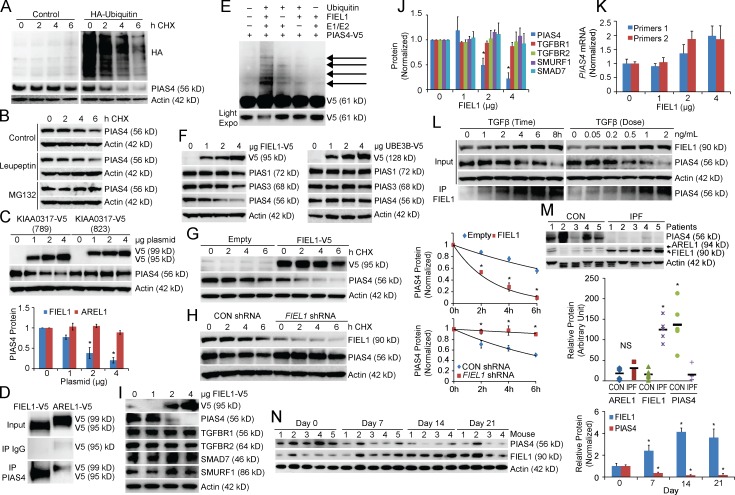
PIAS4 degradation occurs in an ubiquitin-dependent manner through the proteasome (Fig. 1, A and B). We screened our E3 ligase library and determined that the HECT domain E3 ligase KIAA0317 regulates PIAS4 protein stability (unpublished data). KIAA0317 encodes two major isoforms, 823 and 789 residues in length, respectively. The longer isoform has been previously characterized as AREL1 and contains an additional 34 residues in the C-terminal HECT-domain (Kim et al., 2013). However, we found that the shorter isoform (789 residues, termed FIEL1) behaves distinctly in cells. First, only FIEL1 overexpression significantly decreased PIAS4 protein levels (Fig. 1 C). Compared with AREL1, FIEL1 alone interacted with PIAS4 protein via coimmunoprecipitation (Fig. 1 D). Moreover, overexpressed FIEL1 in murine lung epithelial (MLE), HeLa, and 293T cells comigrated with the endogenous protein upon FIEL1 immunoblotting, which suggested that FIEL1 is the predominant KIAA0317 isoform in all of these cell lines (unpublished data). FIEL1 is sufficient for PIAS4 ubiquitination in vitro (Fig. 1 E). FIEL1 expression selectively decreased PIAS4, compared with other PIAS family members in MLE cells (Fig. 1 F). A randomly selected HECT E3 ligase, UBE3B, was also tested as a negative control (Fig. 1 F). FIEL1 expression in HeLa and 293T cells also decreased PIAS4 protein levels (unpublished data). Conditional expression of FIEL1 in MLE cells using a doxycycline-inducible plasmid resulted in PIAS4 protein degradation (not depicted). Further, FIEL1 expression significantly decreased PIAS4 protein levels, whereas FIEL1 knockdown using shRNA stabilized PIAS4 by extending its half-life (Fig. 1, G and H). FIEL1 also regulates PIAS4 protein levels in human fetal lung primary fibroblast MRC5 cells. As shown in Fig. 1 (I and J), expression of FIEL1 reduced PIAS4 protein levels in a dose-dependent manner, whereas other proteins such as TGFβR1, TGFβR2, SMURF1, and SMAD7 levels were unchanged. Moreover, FIEL1 expression did not reduce PIAS4 mRNA levels (Fig. 1 K). TGFβ treatment increased FIEL1 protein and decreased PIAS4 protein in MRC5 cells, while also increasing the association of PIAS4 and FIEL1 (Fig. 1 L). Last, TGFβ treatment drastically increased FIEL1 mRNA levels (unpublished data). We also determined that K31 is the ubiquitin acceptor site within PIAS4 (Fig. S1, A–C). We measured FIEL1 and PIAS4 protein levels in lung tissues from five control patients and five patients with IPF. Patients with IPF had significantly less PIAS4 protein and more immunoreactive FIEL1 protein in their lungs versus control patients (Fig. 1 M). We also tested this pathway in bleomycin-induced murine lung fibrosis (Tager et al., 2008; Jiang et al., 2010). Bleomycin challenge significantly increased FIEL1 protein levels and decreased PIAS4 protein levels in murine lung tissue with a maximum effect at day 14 (Fig. 1 N). These results suggest that the FIEL1–PIAS4 pathway is functional and important in individuals with IPF.
Figure 1.
FIEL1–PIAS4 pathway in pulmonary fibrosis. (A) PIAS4 protein half-life determination in MLE cells transfected with empty plasmid or ubiquitin plasmid (n = 2). (B) PIAS4 protein half-life determination with MG132 or leupeptin treatment (n = 3). (C) Immunoblots (top) showing levels of PIAS4 protein and V5 after KIAA0317 (823 aa, AREL1) and (789 aa, FIEL1) plasmid expression. PIAS4 protein quantification was normalized and graphed (bottom). Data represent mean values ± SEM (n = 3 independent experiments; *, P < 0.05 compared with 0 µg plasmid, Student’s t test). (D) PIAS4 protein was immunoprecipitated from cell lysate using a PIAS4 antibody and coupled to protein A/G beads. PIAS4 beads were then incubated with in vitro–synthesized products expressing HIS-V5-FIEL1 (789 aa) or HIS-V5-AREL1 (823 aa). After washing, proteins were eluted and processed for V5 immunoblotting (n = 2). (E) In vitro ubiquitination assay. Purified E1 and E2 components were incubated with V5-PIAS4 and FIEL1. The full complement of ubiquitination reaction components (second lane) showed polyubiquitinated PIAS4 proteins (n = 3). (F) Immunoblots showing levels of PIAS proteins and V5 after ectopic FIEL1 or UBE3B expression. (G and H) PIAS4protein half-life determination in MLE cells with empty plasmid or FIEL1 expression (G); PIAS4 protein half-life determination with CON shRNA or FIEL1 shRNA expression (H). Data represent mean values ± SEM (n = 3 independent experiments; *, P < 0.05 compared with Empty or to Control, Student’s t test). (I–J) Immunoblots (I) showing levels of PIAS4, TGFBR1, TGFBR2, SMAD7, Smurf1, and V5 after FIEL1 expression. Protein quantification was graphed (J). Data represent mean values ± SEM (n = 3 independent experiments; *, P < 0.05 compared with 0 µg FIEL1, Student’s t test). (K) mRNA levels of PIAS4 upon FIEL1 expression was measured using two sets of PIAS4 RT-PCR primers. Data represent mean values ± SEM (n = 3 independent experiments). (L) MRC5 cells were treated with TGFβ in a time- or dose-dependent manner; cells were collected and immunoblotted for FIEL1 and PIAS4. Endogenous FIEL1 was also immunoprecipitated and immunoblotted for PIAS4 (n = 3). (M) PIAS4 and FIEL1 immunoblotting from lung tissues samples from five control and five IPF patients. PIAS4 and both the shorter and longer forms of KIAA0317 were quantified using ImageJ and graphed. Data represent mean values (n = 5 patients; NS, not significant *, P < 0.05 compared with CON, Student’s t test). (N) C57BL/6J mice were treated i.t. with bleomycin (0.02 U) for up to 21 d. Mice were then euthanized, and lungs were isolated and assayed for PIAS4 and FIEL1 immunoblotting. Bands corresponding to each protein on immunoblots were quantified using ImageJ software, and the results are displayed graphically. Data represent mean values ± SEM (n = 4–5 mice per group; *, P < 0.05 compared with day 0, Student’s t test).
FIEL1 promotes TGFβ signaling
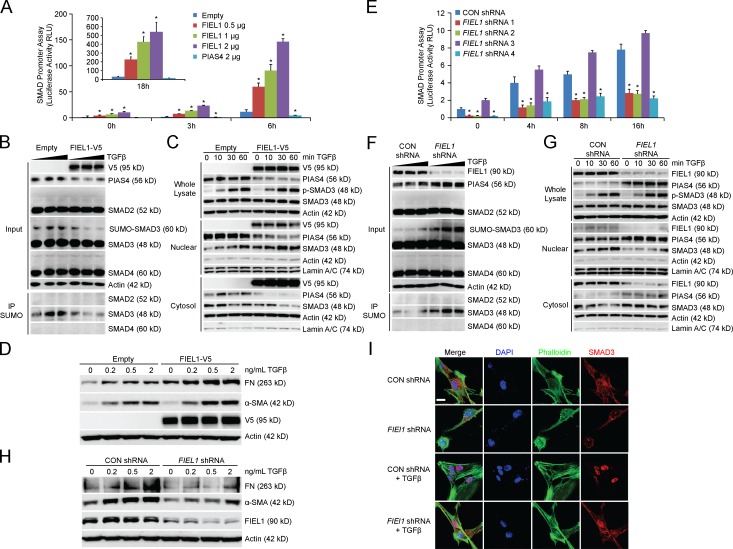
Using a SMAD reporter assay (QIAGEN), we determined that FIEL1 overexpression increased SMAD promoter-driven luciferase activity upon TGFβ stimulation (Fig. 2 A). By decreasing PIAS4 protein, expression of FIEL1 also decreased SMAD3 sumoylation (Fig. 2 B) as previously described (Lee et al., 2003; Imoto et al., 2008) and further promoted SMAD3 nuclear concentration (Fig. 2 C). FIEL1 ectopic expression did not change SMAD3 phosphorylation. FIEL1 expression increased the expression of fibrotic proteins fibronectin (FN) and α smooth muscle actin (α-SMA) in MRC5 cells (Fig. 2 D). When we knock down FIEL1 using several shRNAs, we observed a significantly reduced SMAD-driven luciferase activity, with shRNA 3 serving as a negative control (Fig. 2 E). FIEL1 knockdown increased PIAS4 protein, increased SMAD3 sumoylation (Fig. 2 F), and further decreased SMAD3 nuclear concentration (Fig. 2 G). FIEL1 knockdown also decreased the expression of fibrotic proteins FN and α-SMA in MRC5 cells (Fig. 2 H). To complement the biochemical approach, we immunostained MRC5 cells and observed that FIEL1 knockdown also reduced nuclear localization of SMAD3 upon TGFβ treatment (Fig. 2 I).
Figure 2.
FIEL1 promotes TGFβ signaling. (A and E) SMAD reporter assays. 293T cells were cotransfected with Cignal SMAD dual luciferase reporter plasmids along with empty, FIEL1, Con shRNA, or FIEL1 shRNA. 24 h later, cells were treated with TGFβ for 2–16 h. Cells were collected and assayed for luciferase activity to evaluate SMAD promoter activity. Data represent mean values ± SEM (n = 3; *, P < 0.05 compared with empty, Student’s t test). (B and F) 293T cells were transfected with empty, FIEL1, CON shRNA, or FIEL1 shRNA for 48 h before TGFβ treatment (0–2 ng/ml) for 1 h. Cells were then collected and immunoblotted. Cell lysates were immunoprecipitated using SUMO antibody before SMAD2, 3, and 4 immunoblotting (n = 2). (C and G) 293T cells were transfected with empty, FIEL1, CON shRNA, or FIEL1 shRNA for 48 h before TGFβ treatment (2 ng/ml) for up to 1 h. Cells were then collected and nuclear/cytosol fractions were isolated prior to immunoblotting. (D and H) MRC5 cells were transfected with empty, FIEL1, CON shRNA, or FIEL1 shRNA for 48 h before TGFβ dose course treatment for an additional 18 h. Cells were then collected and immunoblotted (n = 2). (I) MRC5 cells were seeded in 35-mm glass bottom dishes before being transfected with CON shRNA or FIEL1 shRNA for 48 h before TGFβ treatment for an additional 30 min. Cells were then fixed and immunostained with α-SMAD3.The nucleus was counterstained with DAPI and F-actin was counterstained with phalloidin (n = 3). Bar, 10 µm.
PIAS4 phosphorylation by PKCζ is required for FIEL1 binding
We next investigated the FIEL1-binding site within PIAS4. We determined that PIAS4 S18 and Q21 are both important for FIEL1 interaction (Fig. S1, D–F). We performed a kinase screen and determined that PKCζ interacts with PIAS4 via Co-IP (Fig. 3 A). PKCζ expression also decreased PIAS4 protein level, whereas JNK1 expression was unable to achieve such an effect (Fig. 3 B). Moreover, PKCζ knockdown using shRNA drastically stabilized PIAS4 protein in a half-life study (Fig. 3 C), whereas PKCζ expression decreased PIAS4 half-life to ∼2 h (Fig. 3 D). TGFβ stimulation also drastically increased PIAS4 serine phosphorylation and PKCζ association, but not PKCα association (Fig. 3, E and F). PKCζ directly phosphorylated PIAS4 in a kinase assay (Fig. 3 G). PKCζ knockdown also protected PIAS4 from phosphorylation and degradation during TGFβ treatment (Fig. 3 H). Compared with WT PIAS4, S18A and S14/S18A double mutants exhibited a dramatic decrease in phosphorylation and offered resistance to degradation during TGFβ treatment (Fig. 3 I). PIAS4 S18A, Q21A, and S18/Q21A double mutants also exhibited much longer half-lives (Fig. S1 G) and resisted degradation with FIEL1 coexpression (Fig. S1 H). Finally, we performed a peptide-binding experiment (Fig. 3 J). The peptide with S18 phosphorylation (P2) showed the strongest binding to FIEL1; the peptide with both S18 phosphorylation and Q21 mutation (P3) offered drastically decreased FIEL1 interaction. These experiments suggested that PKCζ is an authentic regulator of PIAS4 protein stability; Q21 and phosphorylated S18 of PIAS4 are both required for FIEL1 interaction.
Figure 3.
PIAS4 phosphorylation by PKCζ is required for FIEL1 binding. (A) Endogenous PIAS4 was immunoprecipitated and immunoblotted for Erk1, PKCα, and PKCζ (n = 2). (B) MLE cells were transfected with increasing amounts of PKCζ or JNK1 plasmids for 18 h before PIAS4 immunoblotting (n = 2). (C) PIAS4 protein half-life determination with CON shRNA or PKCζ shRNA expression (n = 3). (D) PIAS4 protein half-life determination with Empty or PKCζ plasmid overexpression (n = 3). (E and F) MRC5 cells were treated with TGFβ in a time or dose-dependent manner; cells were collected and immunoblotted for FIEL1, PIAS4, PKCζ, and p-PKCζ (Thr410). Endogenous PIAS4 was also immunoprecipitated and followed by PKCζ, PKCα, phosphoserine, and phosphothreonine immunoblotting (n = 2). (G) In vitro PKCζ kinase assay. Recombinant PKCζ (Enzo) was used as the kinase, and V5-tagged PIAS4 was synthesized via TnT in vitro kits (Promega), purified by HIS pulldown, and used as the substrate. The kinase reactions were incubated at 37°C for 2 h, and products were resolved by SDS-PAGE and processed for autoradiography either by using Personal Molecular Imager (Bio-Rad Laboratories) or immunoblotting for V5 to visualize the substrate input. *, heat inactivated PKCζ (n = 2). (H) Immunoblots showing levels of FIEL1, PKCζ, p-PKCζ (Thr410), and PIAS4 protein in 293T cells transfected with either CON shRNA or PKCζ shRNA, followed by a TGFβ dose treatment. Endogenous PIAS4 was also immunoprecipitated and followed by PKCζ, phosphoserine, and phosphothreonine immunoblotting. (I) 293T cells were transfected with WT, S14A, S18A, or S14/18A PIAS4 before being treated with a dose course of TGFβ. Cells were then collected and assayed for V5-PIAS4 immunoblotting. Overexpressed V5-PIAS4 was also immunoprecipitated using a V5 antibody and followed by phosphoserine immunoblotting (n = 2). (J) Four biotin-labeled PIAS4 peptides were bound to streptavidin and served as the bait for FIEL1 binding. After washing, proteins were eluted and immunoblotted for FIEL1-V5 (n = 2).
GSK3β phosphorylation of FIEL1 is required for PIAS4 targeting
We next investigated the PIAS4-binding site within FIEL1. A mapping study was conducted similarly to Fig. S1 D and we determined that both P779 and T783 are important for PIAS4 interaction (Fig. S1, I–N). Compared with WT FIEL1, neither P779A nor T783A mutant expression decreased PIAS4 protein levels (Fig. S1 O). We also determined that C770 is a potential active site of FIEL1 as the C770S mutant also failed to decrease PIAS4 protein level (Fig. S1 O). SNP database analysis indicated a naturally occurring polymorphism (rs371610162) within FIEL1 (P779L). We further tested this mutation in a binding assay and showed that T783A, P779L, and P779L/T783A double mutants all lost interaction with PIAS4 (Fig. S1 P). We performed a kinase screen and determined that GSK3β interacts with FIEL1 via Co-IP (Fig. 4 A). WT GSK3β overexpression decreased PIAS4 protein levels in a dose-dependent manner, and PIAS4 protein levels decreased more dramatically when we transfected cells with a constitutively activated GSK3β hyper mutant plasmid (Fig. 4 B). Moreover, a half-life study suggested that WT GSK3β ectopic expression decreased PIAS4 half-life to ∼4 h, whereas the more potent GSK3β hyper mutant further decreased PIAS4 half-life to ∼2 h (Fig. 4 C). TGFβ stimulation drastically increased FIEL1 threonine phosphorylation (Fig. 4 D), and GSK3β knockdown drastically stabilized PIAS4 in a half-life study (Fig. 4 E). Moreover, GSK3β knockdown also prevented FIEL1 threonine phosphorylation and protected PIAS4 from degradation with TGFβ treatment (Fig. 4 F).
Figure 4.
GSK3β regulates PIAS4 protein stability through FIEL1. (A) Endogenous FIEL1 was immunoprecipitated and followed by JNK2, PKCα, and GSK3β immunoblotting (n = 2). (B) MLE cells were transfected with increasing amounts of WT or constitutively activated GSK3β hyper mutant plasmids for 18 h before PIAS4 immunoblotting. The arrow indicates the overexpressed GSK3β (n = 2). (C) PIAS4 protein half-life determination with WT GSK3β or hyperactive GSK3β plasmid overexpression. The arrow indicates the overexpressed GSK3β (n = 2). (D) MRC5 cells were treated with TGFβ in a time or dose-dependent manner; cells were then collected and immunoblotted for FIEL1 and PIAS4. Endogenous FIEL1 was also immunoprecipitated and followed by phosphoserine and phosphothreonine immunoblotting (n = 2). (E) PIAS4 protein half-life determination with CON shRNA or GSK3β shRNA expression (n = 2). (F) Immunoblots showing levels of GSK3β, phospho-GSK3β (Ser9), PIAS4, and FIEL1 protein in 293T cells transfected with either CON shRNA or GSK3β shRNA followed by a TGFβ dose treatment. Endogenous FIEL1 was also immunoprecipitated, followed by phosphoserine and phosphothreonine immunoblotting. (G) Lung samples from Fig. 1 J were subjected to FIEL1 immunoprecipitation, followed by PIAS4, phosphothreonine, and phosphoserine immunoblotting. PIAS4 protein abundance was plotted as a function of p-Thr protein (n = 5 patients per group; *, P < 0.01, Pearson correlation).
Using the lung lysates from Fig. 1 M, we performed a Co-IP experiment and observed a positive and significant association between PIAS4 and the phosphothreonine signal, which suggested that FIEL1 threonine phosphorylation is essential for PIAS4 binding (Fig. 4 G). We further studied the role of FIEL1 T783 in regulating PIAS4 protein stability. FIEL1 T783A mutant overexpression completely failed to decrease PIAS4 protein level. However, in phosphorylation mimic T783D mutant, FIEL1 expression decreased PIAS4 protein level more dramatically compared with WT FIEL1 expression (Fig. 5 A). Moreover, a half-life study suggested that WT FIEL1 expression decreased PIAS4 half-life to ∼4 h, whereas the more potent phosphorylation mimic, T783D, further decreased PIAS4 half-life to ∼2 h (Fig. 5 B). The FIEL1 T783A mutant was resistant to GSK3β phosphorylation in vitro (Fig. 5 C) and in cells (Fig. 5 D). We also performed a peptide binding experiment (Fig. 5 E). The peptide phosphorylated at T783 (P2) showed the strongest binding to PIAS4; the peptide with no phosphorylation (P1) or T783A mutant peptide (P4) offered drastically decreased PIAS4 interaction. FIEL1 T783A and P779L expression both failed to decrease PIAS4 protein level (Fig. 5 F) or half-lives (Fig. 5 G). FIEL1 T783A/P779L double mutant expression showed a dominant negative phenotype by increasing PIAS4 protein levels (Fig. 5 F). We further used PIAS4 peptide 2 with S18 phosphorylation (Fig. 3 J) as bait and determined that both P779L and T783A FIEL1 drastically lost binding with PIAS4 (Fig. 5 H), which is similar to Fig. S1 P. Similarly, FIEL1 peptide 2 with phosphorylation at T783 (P2; Fig. 5 E) was used to reconfirm the importance of S18 phosphorylation and Q21 within PIAS4 for FIEL1 interaction (Fig. 5 I), which is similar to Fig. S1 F. Last, FIEL1 T783A/P779L dominant-negative mutant overexpression protected PIAS4 from TGFβ treatment (Fig. 5 J). These experiments suggested that GSK3β phosphorylation of FIEL1 is required for PIAS4 targeting, and FIEL1 residues P779 and phosphorylated T783 are both required for PIAS4 interaction (Fig. S1 Q).
Figure 5.
GSK3β phosphorylation of FIEL1 is required for PIAS4 targeting. (A) MLE cells were transfected with increasing amounts of WT, T783A, or T783D mutant FIEL1 plasmids for 18 h before PIAS4 immunoblotting (n = 2). (B) PIAS4 protein half-life determination with WT, T783A, or T783D mutant FIEL1 (n = 2). (C) In vitro GSK3β kinase assay. Recombinant GSK3β (Enzo) was used as the kinase, and V5-tagged FIEL1 were synthesized via TnT in vitro kits (Promega), purified by HIS pulldown, and used as the substrate. The kinase reactions were incubated at 37°C for 2 h, and products were resolved by SDS-PAGE and processed for autoradiography either by using Personal Molecular Imager (Bio-Rad Laboratories) or immunoblotting for V5 to visualize the substrate input. *heat inactivated GSK3β (n = 2). (D) 293T cells were transfected with empty, WT, or T783A FIEL1 for 24h. Cells were then collected and immunoblotted for V5-FIEL1 and PIAS4. Overexpressed V5-FIEL1 was also immunoprecipitated using V5 antibody and followed by phosphothreonine immunoblotting (n = 2). (E) Four biotin-labeled FIEL1 peptides were prebound to streptavidin and served as the bait for PIAS4 binding. After washing, proteins were eluted and processed for PIAS4 immunoblotting (n = 2). (F) MLE cells were transfected with increasing amounts of WT, T783A, P779L, or T783A/P779L double mutant FIEL1 plasmids for 18 h before PIAS4 immunoblotting. (G) PIAS4 protein half-life determination with WT, T783A, P779L, or T783A/P779L double mutant FIEL1 (n = 2). (H) PIAS4 peptide 2 (Biotin-MSFRVS(p)DLQM) was prebound to streptavidin and served as the bait for FIEL1 binding. After washing, proteins were eluted and processed for V5-FIEL1 immunoblotting (n = 2). (I) FIEL1 peptide 2 (Biotin-QIIAAPTHST(p)LPTA) was bound to streptavidin and served as the bait for PIAS4 binding. After washing, proteins were eluted and processed for V5-PIAS4 immunoblotting (n = 2). (J) 293T cells were transfected with WT, T783A, P779L, or T783A/P779L double mutant FIEL1 before being treated with TGFβ. Cells were then collected and assayed for PIAS4 immunoblotting. (K) MLE cells were transfected with increasing amounts of WT, I514V, or D207V mutant FIEL1 plasmids for 18 h before PIAS4 immunoblotting. (L) PIAS4 protein half-life determination with WT, I514V, or D207V mutant FIEL1 expression.
Gene transfer of FIEL1 exacerbates bleomycin-induced lung injury in vivo
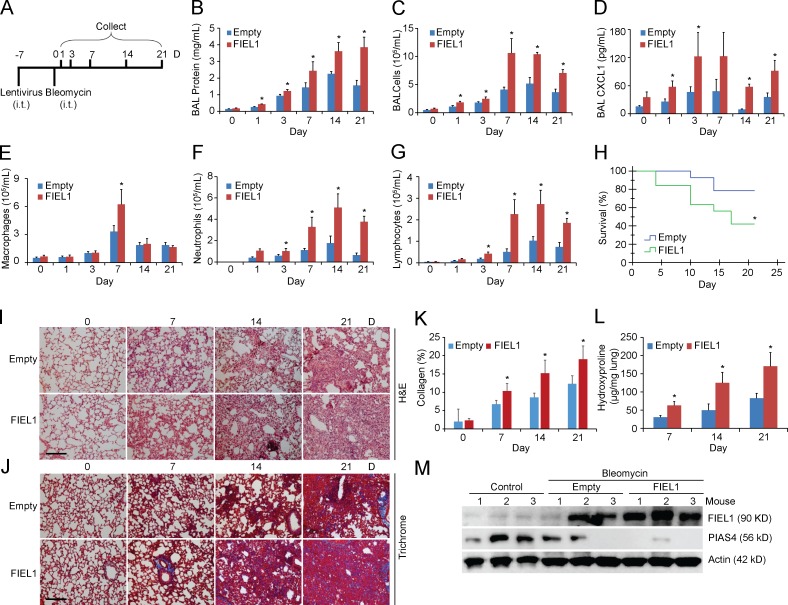
So far, our in vitro studies suggest that FIEL1 promotes TGFβ signaling in vitro. As exacerbated TGFβ signaling partakes in fibrotic formation, the results raise the possibility that expression of FIEL1 in vivo might alter host inflammatory responses and induce fibrotic lung injury. To extend the aforementioned observations in vivo, mice were infected with an empty lentivirus or lentivirus encoding FIEL1 for 144 h (107 CFU/mouse, i.t.). Mice were then challenged with bleomycin (0.02 U i.t.) for an additional 1–21 d (Fig. 6 A). Mice were euthanized to analyze parameters of fibrotic lung injury. Bleomycin injury is one of the most widely studied models of pulmonary fibrosis (Tager et al., 2008; Hecker et al., 2009; Jiang et al., 2010; Reed et al., 2015). As shown in Fig. 6 B, the increased bronchoalveolar lavage (BAL) total protein concentration that occurs after bleomycin injury in control mice was significantly higher in mice overexpressing FIEL1. Total inflammatory cells and chemokine CXCL1 levels in BALs were also significantly increased in mice overexpressing FIEL1 (Fig. 6, C and D). Specifically, the differential cell counts of the BALs revealed that the total increase in inflammatory cells was mostly a result of neutrophils and lymphocytes, with the exception of macrophages on day 7 (Fig. 6, E–G). FIEL1 expression in mice also significantly reduced survival (Fig. 6 H). Bleomycin challenge also showed changes consistent with peribronchiolar and parenchymal fibrosis in a time-dependent manner (Fig. 6, I and J). Elevated lung collagen deposition visualized by Masson Trichrome staining also suggested that FIEL1 expression exacerbates bleomycin-induced lung fibrosis (Fig. 6, J and K).We observed a marked increase in lung fibrosis in mice overexpressing FIEL1 as demonstrated by significantly increased hydroxyproline content (Fig. 6 L). The extent of these changes present in FIEL1 expression mice was substantially increased compared with the empty control (Fig. 6, I–L). Last, we analyzed FIEL1–PIAS4 protein levels in murine lung samples and observed that lentiviral FIEL1 expression completely depleted PIAS4 protein in the lung (Fig. 6 M).
Figure 6.
Gene transfer of FIEL1 exacerbates bleomycin-induced lung injury in vivo. (A)C57BL/6J mice weretreated i.t. with Lenti-Empty or Lenti-FIEL1 (107 PFU/mouse) for 144 h; mice were then treated i.t. with bleomycin (0.02 U). Mice were euthanized over the next 1–21 d, and lungs were lavaged with saline, harvested, and then homogenized. (B–D) Lavage protein, total cells, and CXCL1 concentrations were measured. Data represent mean values ± SEM (n = 4–6 mice per group, data are from one of the two experiments performed; *, P < 0.05 compared with empty, Student’s t test). (E–G) Lavage cells were then processed for Wright-Giemsa stain; lavage macrophages, neutrophils, and lymphocytes were counted and graphed. Data represent mean values ± SEM (n = 4–6 mice per group, *, P < 0.05 compared with empty, Student’s t test). (H) Survival studies of mice that were given bleomycin. Mice were carefully monitored over time; moribund, preterminal animals were immediately euthanized and recorded as deceased. Kaplan-Meier survival curves were generated using SPSS software (n = 9–11 mice per group; *, P < 0.05 compared with Empty, Log-rank test). Empty: n = 9, FIEL1: n = 11. (I and J) Hematoxylin and eosin (H&E) and Trichrome staining were performed on lung samples. Bars, 100 µm. (K) Collagen percentage quantification from Trichrome staining. Data represent mean values ± SEM (n = 4–6 mice per group, *, P < 0.05 compared with empty, Student’s t test). (L) Hydroxyproline content was measured in lungs from 7, 14, and 21 d after bleomycin challenge. Data represent mean values ± SEM (n = 4–6 mice per group, *, P < 0.05 compared with empty, Student’s t test). (M) Mice lungs were isolated and assayed for PIAS4 and FIEL1 immunoblotting.
FIEL1 knockdown ameliorates bleomycin-induced lung injury in vivo
To further confirm the role of FIEL1 in lung fibrosis and inflammation, we pursued in vivo knockdown studies. Mice were first infected with lentivirus encoding CON shRNA or FIEL1 shRNA for 144 h (107 CFU/mouse, i.t), and then challenged with bleomycin (0.05 U i.t.) for an additional 1–21 d (Fig. 7 A). FIEL1 knockdown significantly decreased BAL protein concentrations, total cell counts, and chemokine CXCL1 levels (Fig. 7, B–D). Specifically, the differential cell counts of BALs revealed that the total decrease in inflammatory cells was mostly a result of neutrophils and lymphocytes, with the exception of macrophages on day 3 and 21 (Fig. 7, E–G). FIEL1 knockdown in mice also significantly improved survival (Fig. 7 H). Peribronchiolar and parenchymal fibrosis were also substantially decreased in FIEL1 knockdown mice (Fig. 7, I and J), suggesting that FIEL1 knockdown ameliorates bleomycin-induced lung injury. Elevated lung collagen deposition visualized by Masson Trichrome staining also suggested that FIEL1 expression exacerbates bleomycin-induced lung fibrosis (Fig. 7, J and K). We observed a marked decrease in lung fibrosis in FIEL1 knockdown mice as demonstrated by a significant decrease in hydroxyproline content (Fig. 7 L). Last, we analyzed FIEL1 and PIAS4 protein levels in murine lung samples and showed that lentiviral FIEL1 knockdown reversed bleomycin-induced FIEL1 increase and rescued PIAS4 protein levels in the lung (Fig. 7 M).
Figure 7.
FIEL1 knockdown ameliorates bleomycin-induced lung injury in vivo. (A) C57BL/6J mice were treated i.t. with Lenti-CON shRNA or Lenti-FIEL1 shRNA (107 PFU/mouse) for 144 h; mice were then treated i.t. with bleomycin (0.05 U). Mice were euthanized over the next 1–21 d, and lungs were lavaged with saline, harvested, and then homogenized. (B–D) Lavage protein, total cells, and CXCL1 concentrations were measured. Data represent mean values ± SEM (n = 4–6 mice per group, data are from one of the two experiments performed; *, P < 0.05 compared with Control, Student’s t test). (E–G) Lavage cells were then processed for Wright-Giemsa stain; lavage macrophages, neutrophils, and lymphocytes were counted and graphed. Data represent mean values ± SEM (n = 4–6 mice per group, *, P < 0.05 compared with Control, Student’s t test). (H) Survival studies of mice that were given bleomycin. Mice were carefully monitored over time; moribund, preterminal animals were immediately euthanized and recorded as deceased. Kaplan-Meier survival curves were generated using SPSS software (n = 8 mice per group; *, P < 0.05 compared with Control, Log-rank test P < 0.05). Empty: n = 8, FIEL1: n = 8. (I and J) H&E and Trichrome staining was performed on lung samples. Bar indicates 100 µm. (K) Collagen percentage quantification from Trichrome staining. Data represent mean values ± SEM (n = 4–6 mice per group, *, P < 0.05 compared with Control, Student’s t test). (L) Hydroxyproline content was measured in lungs from 7, 14, and 21 d after bleomycin challenge. Data represent mean values ± SEM (n = 4–6 mice per group; *, P < 0.05 compared with Control, Student’s t test). (M) Mice lungs were isolated and assayed for PIAS4 and FIEL1 immunoblotting.
Antifibrotic activity of a FIEL1 small molecule inhibitor in vitro
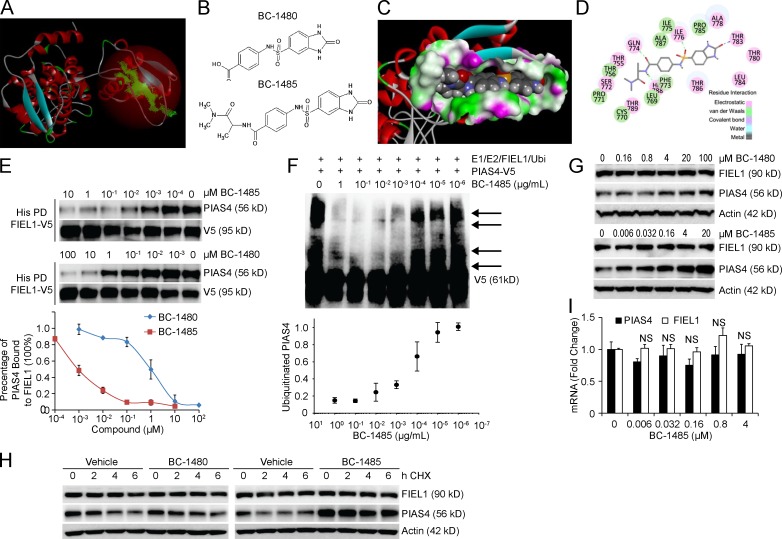
We first constructed a FIEL1 HECT domain homology model using the NEDD4 HECT domain structure (Umadevi et al., 2005; Kamadurai et al., 2009; Maspero et al., 2011 PDB: 2XBF; Fig. 8 A). We observed a major cavity within the C terminus of the FIEL1 HECT domain that is also required for PIAS4 binding (Fig. 8 A). Through the LibDock program from Discovery Studio 3.5, we were able to screen potential ligands for the FIEL1 cavity. We selected BC-1480 as a backbone to further develop new small molecule inhibitors (Fig. 8 B). BC-1485 was synthesized by reacting alaninamide with BC-1480 (unpublished data). BC-1485 fits in the FIEL1 cavity fairly well by having several electrostatic interactions with Gln774, His788, Ile776, and Thr783 (Fig. 8, C and D). Compared with BC-1480, BC-1485 exhibited >100-fold activity in disrupting the FIEL1–PIAS4 interaction (Fig. 8 E). BC-1485 also exhibited potent activity toward disrupting FIEL1-directed PIAS4 ubiquitination (Fig. 8 F). Finally, we tested BC-1485 in cells and observed a drastic dose-dependent increase in PIAS4 protein levels (Fig. 8 G). BC-1485 also decreased the expression of α-SMA in MRC5 cells (unpublished data). BC-1485 also stabilized PIAS4 by extending its half-life (Fig. 8 H) without altering its mRNA level (Fig. 8 I).
Figure 8.
Antifibrotic activity of a FIEL1 small molecule inhibitor in vitro. (A) Structural analysis of the FIEL1 HECT domain revealed a major cavity within the C terminus of the HECT domain. (B) Structures of the BC-1480 backbone (4-(2-Oxo-2,3-dihydro-1H-benzoimidazole-5-sulfonylamino)-benzoic acid) and lead compound BC-1485. (C and D) Docking studies of the lead compound, BC-1485, interacting with the FIEL1-HECT domain. (E) FIEL1 protein was HIS-purified from FIEL1 expression in 293T cells using cobalt beads. Beads were then extensively washed before exposure to BC-1480 or BC-1485 at different concentrations (10−4 to 100 µM). Purified PIAS4 protein was then incubated with drug-bound FIEL1 beads overnight. Beads were washed, and proteins were eluted and resolved on SDS-PAGE. The relative amounts of PIAS4 detected in the pull-downs was normalized to loading and quantified (n = 2). (F) In vitro ubiquitination assay. Purified FIEL1, E1, and E2 protein were incubated with purified V5-PIAS4, and the full complement of ubiquitination reaction components with increased concentrations of BC-1485 showed decreased levels of polyubiquitinated PIAS4 (arrows). (bottom) Levels of ubiquitinated PIAS4 as a function of BC-1485 concentration (n = 2). (G) MLE cells were exposed to BC-1480 or BC-1485 at various concentrations for 18 h. Cells were then collected and immunoblotted (n = 3). (H) PIAS4 protein half-life determination after BC-1480 or BC-1485 treatment at 5 µM for 18 h. (I) PIAS4 and FIEL1 mRNA analysis after BC-1485 treatment for 18 h. Data represent mean values ± SEM (n = 3 independent experiments; NS, not significant compared with 0 µM condition, Student’s t test).
Antifibrotic activity of a FIEL1 small molecule inhibitor in vivo
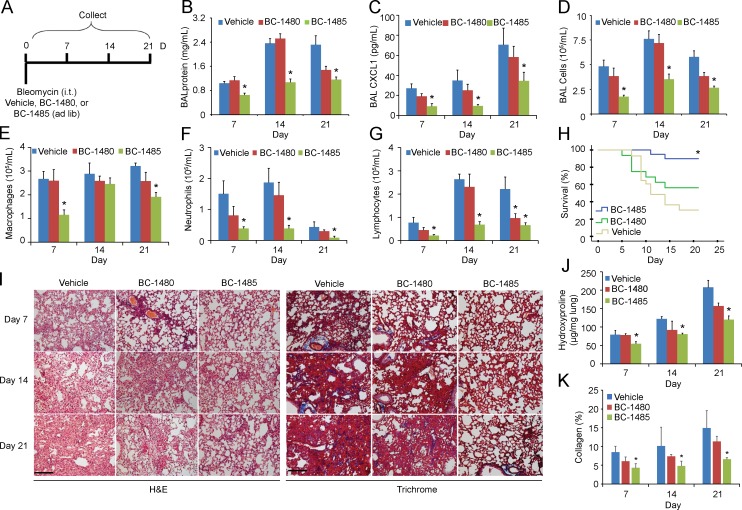
To further assess the kinetics of the antifibrotic activity of BC-1485, we first tested it in vivo after bleomycin injury. In brief, mice were challenged with bleomycin (0.05 U i.t.), and BC-1485 and control compound BC-1480 were given in the drinking water (∼5 mg/kg/d) for an additional 7–21 d (Fig. 9 A). BC-1485 significantly decreased BAL protein concentrations and CXCL1 levels (Fig. 9, B and C). BC-1485 also significantly decreased BAL total cell counts, specifically neutrophils and lymphocytes (Fig. 9, D–G), and significantly improved survival (Fig. 9 H). Peribronchiolar and parenchymal fibrosis were also substantially decreased by BC-1485 (Fig. 9 I). Decreased lung collagen visualized by Trichrome staining also suggested that BC-1485 ameliorates bleomycin-induced lung injury (Fig. 9, I and K). We observed a marked decrease in lung fibrosis in mice treated with the BC-1485 as demonstrated by a significant decrease in hydroxyproline content (Fig. 9, I and J). We further tested BC-1485 in a dose-dependent manner using a bleomycin model. This time, mice were challenged with bleomycin (0.05 U i.t.) for 10 d before being treated with BC-1485 (2 and 10 mg/kg/d in drinking water) for an additional 10 d before being sacrificed (Fig. 10 A). BC-1485 significantly decreased BAL protein concentrations and cell counts compared with vehicles (Fig. 10, B and C). Specifically, BC-1485 dose-dependently decreased macrophages, neutrophils, and lymphocytes (Fig. 10 D) and significantly improved survival (Fig. 10 E). We observed a marked decrease in lung fibrosis in mice treated with BC-1485, as demonstrated by a significant decrease in hydroxyproline content (Fig. 10 F). Peribronchiolar and parenchymal fibrosis were also substantially decreased by BC-1485 (Fig. 10 G). Decreased lung collagen visualized by Trichrome staining also suggested that BC-1485 ameliorates bleomycin-induced lung injury (Fig. 10, G and H). Lastly, BC-1485 rescued PIAS4 protein levels in bleomycin-treated lungs (Fig. 10 I). Hence, small molecule targeting of the FIEL1–PIAS4 pathway reduced the severity of fibrosis in a preclinical model (Fig. S1 R).
Figure 9.
Antifibrotic activity of a FIEL1 small molecule inhibitor in vivo. (A) C57BL/6J mice were treated i.t. with bleomycin (0.05 U). Compounds BC-1480 and BC-1485 were given to mice at the same time through drinking water with an estimated dose of 5 mg/kg/d. Mice were euthanized over the next 1–21 d, and lungs were lavaged with saline, harvested, and then homogenized. (B–D) Lavage proteins, CXCL1, and total cell count were measured. Data represent mean values ± SEM (n = 4–8 mice per group; data are from one of two experiments performed; *, P < 0.05 compared with Vehicle, Student’s t test). (E–G) Lavage cells were also processed for Wright-Giemsa stain; Lavage macrophages, neutrophils, and lymphocytes were counted and graphed. Data represent mean values ± SEM (n = 4–8 mice per group; *, P < 0.05 compared with Vehicle, Student’s t test). (H) Survival studies of mice that were given bleomycin and BC-compound treatments. Mice were carefully monitored over time; moribund, preterminal animals were immediately euthanized and recorded as deceased. Kaplan-Meier survival curves were generated using SPSS software (n = 12–24 mice per group; *, P < 0.05 compared with Vehicle, Log-rank test P < 0.05). Vehicle, n = 24, BC-1480; n = 13, BC-1485; n = 12. (I) H&E and Trichrome staining was performed on lung samples. Bar, 100 µm. (J) Hydroxyproline content were measured in lungs from 7, 14, and 21 d after bleomycin challenge. Data represent mean values ± SEM (n = 4–8 mice per group; *, P < 0.05 compared with Vehicle, Student’s t test). (K) Collagen percentage quantification from Trichrome staining. Data represent mean values ± SEM (n = 4–8 mice per group; *, P < 0.05 compared with Vehicle, Student’s t test).
Figure 10.
Antifibrotic activity of a FIEL1 small molecule inhibitor in vivo. (A) C57BL/6J mice were treated i.t. with bleomycin (0.05 U). 10 d later, compound BC-1485 was given to mice through the drinking water with an estimated dose of 2 or 10 mg/kg/d. Mice were euthanized 10 d later, and lungs were lavaged with saline, harvested, and then homogenized. (B and C) Lavage proteins and total cell count were measured. Data represent mean values ± SEM (n = 6–9 mice per group; *, P < 0.05 compared with Vehicle, Student’s t test). (D) Lavage cells were also processed for Wright-Giemsa stain; lavage macrophages, neutrophils, and lymphocytes were counted and graphed. Data represent mean values ± SEM (n = 6–9 mice per group; *, P < 0.05 compared with Vehicle, Student’s t test). (E) Survival studies of mice that were given bleomycin and BC-compound treatments. Mice were carefully monitored over time; moribund, preterminal animals were immediately euthanized and recorded as deceased. Kaplan-Meier survival curves were generated using SPSS software (n = 12–16 mice per group; *, P < 0.05 compared with Vehicle, Log-rank test P < 0.05). Vehicle, n = 16, BC-1485 (2 mg/kg/d); n = 12, BC-1485 (10 mg/kg/d); n = 12. (F) Hydroxyproline content was measured in lungs from 10 d after bleomycin challenge. Data represent mean values ± SEM (n = 6–9 mice per group; *, P < 0.05 compared with Vehicle, Student’s t test). (G) H&E and Trichrome staining were performed on lung samples from A. Bar, 100 µm. (H) Collagen percent quantification from Trichrome staining. Data represent mean values ± SEM (n = 6–9 mice per group; *, P < 0.05 compared with Vehicle, Student’s t test). (I) Mice lungs were isolated and assayed for PIAS4 and FIEL1 immunoblotting.
DISCUSSION
TGFβ signaling plays a key role in the pathogenesis of tissue fibrosis (Wilson and Wynn, 2009). PIAS4 suppresses the TGFβ pathway, in part, by SUMOylating SMAD3 and causing its nuclear export (Imoto et al., 2003, 2008; Long et al., 2003). Here we show that FIEL1 appears to promote TGFβ signaling and fibrosis by destabilizing PIAS4. Our data not only authenticate FIEL1 as a bona-fide E3 ligase, but also show that it triggers the site-directed ubiquitination and degradation of PIAS4 leading to exacerbated fibrotic signaling. MRC5 cells ectopically expressing FIEL1 increased PIAS4 mRNA level, supporting the belief that FIEL1 regulates PIAS4 protein stability (Fig. 1 K). This is a common compensatory mechanism used by cells to rescue protein loss. As an E3 ligase, it is quite possible that FIEL1 targets many other proteins for ubiquitination and degradation. We measured protein abundance of several pro-TGFβ and anti-TGFβ proteins such as TGFβR1, TGFβR2, SMAD7, and SMURF1 in cell lysates ectopically expressing FIEL1 (Fig. 1 I). However, the absence of any difference in their protein levels suggests that it is plausible that FIEL1 regulates SMAD signaling through PIAS4. However, we do not rule out the possibility that other proteins could also play a role linking FIEL1 to the SMAD pathway, given that all E3 ligases are known to target many substrates at the same time. We believe that the FIEL1–PIAS4 pathway is relevant in both epithelial cells and fibroblasts, as we have tested this pathway in both cell types (Fig. 1 C, I, L, Fig. 2 B-I); this matches the current understanding that both epithelial cells and fibroblasts may be involved in the formation of fibrosis (Jiang et al., 2005; Hecker et al., 2009; Li et al., 2011; Noble et al., 2012). However, future studies are needed to identify which cell type is more involved in the FIEL1–PIAS4 pathway.
We found that PKCζ is a regulator of PIAS4 protein stability and described a mechanism of PIAS4 basal protein turnover controlled constitutively by FIEL1. This provides an internal checkpoint for the cell to shut off the antifibrotic pathway, a mechanism that TGFβ hijacks to shut down PIAS4 and promote fibrosis. Moreover, we found that GSK3β is another regulator of PIAS4 protein stability through the phosphorylation of FIEL1. There appears to be a new double locking molecular interplay between FIEL1 and PIAS4. Specifically, both the P779 and GSK3β phosphorylated T783 residues within FIEL1 are required for PIAS4 binding; both the Q21 and PKCζ phosphorylated S18 residues within PIAS4 are required for FIEL1 binding. We believe that the FIEL1–PIAS4 molecular interplay is based on two pairs of interactions between pT783-Q21 and P779-pS18 (Fig. S1, Q and R). In this model, both PKCζ and GSK3β up-regulate TGFβ signaling, which correlates well with several previous studies (Takeda et al., 2001; Mulsow et al., 2005; Baarsma et al., 2013).
We observed a naturally occurring amino acid variant (rs371610162) in FIEL1 (P779L). This hypomorphic polymorphism exhibited a significant decrease in PIAS4 interaction. Thus, its expression did not decrease PIAS4 protein levels or half-life. We further characterized a naturally occurring amino acid variant (rs113140862) in FIEL1 (I514V). This polymorphism, conversely, exhibited hypermorphism as shown by accelerated PIAS4 degradation and shortened protein half-life (Fig. 5, K and L). FIEL1P779L may be an important and protective polymorphism in IPF, whereas FIEL1I514V may be a susceptible polymorphism in IPF via exacerbated TGFβ signaling. Further prospective studies in large IPF cohorts are required to validate this observation.
The FIEL1 HECT domain served as the mechanistic centerpiece and structural basis for designing first-in-class FIEL1 inhibitors. We had previously used a similar approach to design the inhibitors for the proinflammatory protein FBXO3 (Chen et al., 2013; Mallampalli et al., 2013). Because the FIEL1 inhibitor also docks at the same region within the HECT domain as the substrate PIAS4, the inhibitor works by disrupting the FIEL1–PIAS4 interaction. By adding an alaninamide group to the BC-1480 backbone we generated BC-1485, which improved its activity >100 fold. BC-1485 optimally interacts with FIEL1, exerts robust antifibrotic activity by stabilizing PIAS4, and inhibits TGFβ signaling in both human MRC5 cells and a murine model of pulmonary fibrosis. The effectiveness of this inhibitor underscores the physiological importance of the FIEL1–PIAS4 pathway in regulating TGFβ signaling. Currently, there is no cure for IPF, despite extensive research. Many failed clinical trials suggest the complexity of TGFβ signaling and lack of an authentic drug target (Denton et al., 2007; Akhmetshina et al., 2009; Daniels et al., 2010; Pope et al., 2011). Our study may be highly clinically relevant because subjects with IPF exhibited a significant increase in FIEL1 and decrease in PIAS4 protein. Further, FIEL1 threonine phosphorylation is also significantly increased in IPF lung tissue, which is positively associated with PIAS4 binding (Fig. 4 G). Interestingly, only the shorter isoform of KIAA0317 (789 aa, FIEL1) is significantly increased in IPF lung tissue; our FIEL1 inhibitor design was also based on the cavity within FIEL1. The longer form of KIAA0317 lacks the drug binding cavity that renders the FIEL1 inhibitor ineffective (unpublished data). Moreover, in all of the cell lines (MLE, 293T, and HeLa) and tissues tested here, we found very little, if any, longer isoform of KIAA0317 protein present (Fig. 1 M and not depicted). Thus, FIEL1 may be physiologically relevant in TGFβ signaling and IPF. Indeed, both FIEL1 knockdown and inhibition significantly ameliorates tissue fibrosis coupled with a very dramatic decrease in collagen level in a bleomycin model (Fig. 7 K, 9 K, 10 H). These results suggest that FIEL1 may be a promising new therapeutic target for fibrotic diseases, such as IPF.
MATERIALS AND METHODS
Materials
Sources of the murine lung epithelial (MLE) and 293T cell lines were described previously (Chen and Mallampalli, 2007; Ray et al., 2010). MRC5 cells were obtained from ATCC. Purified ubiquitin, E1, E2, MG132, leupeptin, and cyclohexamide were purchased from EMD Millipore. Mouse monoclonal V5 antibody, the pcDNA3.1D cloning kit, E. coli Top10 One Shot competent cells, the pENTR Directional TOPO cloning kits, and Gateway mammalian expression system were obtained from Invitrogen. The HECT domain E3 ligase cDNA, scramble shRNA, FIEL1, PKCζ, and GSK3β shRNA sets were purchased from OpenBiosystems. Nucleofector transfection kits were obtained from Amaxa. The lentiviral packaging system and cobalt beads were purchased from Takara Bio Inc. Immobilized protein A/G beads were purchased from Thermo Fisher Scientific. In vitro transcription and translation (TnT) kits were obtained from Promega. Cignal SMAD Reporter luciferase kit (CCS-017L) and mRNA isolation kit were obtained from QIAGEN. Complete protease inhibitors were purchased from Roche. KIAA0317 antibodies were obtained from Sigma-Aldrich, Antibody Verify, and Santa Cruz Biotechnology, Inc. PIAS and GSK3β antibodies were obtained from Cell Signaling Technology and Santa Cruz Biotechnology, Inc. CXCL1 and IL6 mouse ELISA kits and TGFβ protein were purchased from R&D Systems. Peptides were custom synthesized from CHI Scientific. DNA sequencing was performed by Genewiz. All small molecule compound analysis was performed by the University of Pittsburgh Mass Spectrometry and NMR facility (Pittsburgh, PA).
Human samples
This study was approved by the University of Pittsburgh Institutional Review Board. Lung tissues were obtained from the University of Pittsburgh lung transplant tissue bank. Lung tissues from IPF patients were obtained from excess pathological tissues after lung transplantation under a protocol approved by the University of Pittsburgh Institutional Review Board. Normal lung tissues were obtained from donor lungs that were not suitable for patient use and provided by the Center for Organ Recovery and Education (CORE; Pittsburgh, PA). The CORE lungs were only used if there were no considerable lung abnormalities based on high resolution computerized tomography scan. Multiple small sections of the parenchyma were collected and immediately frozen at −80°C for future use. Tissue lysates were used for the Western blot analysis.
Cell culture
MLE cells were cultured in Dulbecco’s Modified Eagle Medium-F12 (Gibco) supplemented with 10% fetal bovine serum (DMEM-F12-10). 293T cells were cultured in Dulbecco’s Modified Eagle Medium (Gibco) supplemented with 10% fetal bovine serum (DMEM-10). MRC5 cells were cultured in Eagle's Minimum Essential Medium (Gibco) supplemented with 10% fetal bovine serum (EMEM-10). For protein expression in MLE cells, nucleofection was used following Amaxa’s protocol. For protein overexpression in 293T cells, Fugene6HD transfection reagents were used following the manufacturer’s protocol. For protein expression in MRC5 cells, MRC-5 Cell Avalanche Transfection Reagent was used following the manufacturer’s protocol. Cells were treated with TGFβ at 0–2 ng/ml for 0–18 h. For FIEL1, PKCζ, or GSK3β knockdown studies in cells, scramble shRNA, FIEL1, PKCζ, or GSK3β shRNA were used to transfect cells for 48 h. For drug treatment, compounds were solubilized in DMSO before being added to the cells for up to 18 h. Cell lysates were prepared by brief sonication in 150 mM NaCl, 50 mM Tris, 1.0 mM EDTA, 2 mM dithiothreitol, 0.025% sodium azide, and 1 mM phenylmethylsulfonyl fluoride (Buffer A) at 4°C. For half-life study, MLE cells were exposed to cyclohexamide (40 µg/ml) in a time-dependent manner for up to 8 h. Cells were then collected and immunoblotted.
In vitro protein-binding assays
PIAS4 protein was immunoprecipitated from 1 mg cell lysate using PIAS4 antibody (goat) and coupled to protein A/G agarose resin. PIAS4 beads were then incubated with in vitro–synthesized products (50 µl) expressing V5-FIEL1 mutants. After washing, the proteins were eluted and processed for V5-FIEL1 immunoblotting. Similarly, FIEL1 was immunoprecipitated from 1 mg cell lysate using FIEL1 antibody (rabbit) and coupled to protein A/G agarose resin. FIEL1 beads were then incubated with in vitro–synthesized products (50 μl) expressing V5-PIAS4 mutants. After washing, the proteins were eluted and processed for V5-PIAS4 immunoblotting.
In vitro peptide-binding assays
Biotin-labeled peptides were first coupled to streptavidin agarose beads for 1 h. Beads were then incubated with in vitro–synthesized FIEL1 or PIAS4 for 18 h. After washing, proteins were eluted and processed for FIEL1 or PIAS4 immunoblotting.
In vitro drug-binding assays
FIEL1 protein was HIS-purified from FIEL1 expressed in 293T cells using Talon metal affinity resin. Resins were then extensively washed before exposure to BC-1480 or BC-1485 at different concentrations (10−4 to 100 µM). Purified recombinant PIAS4 protein was then incubated with drug-bound FIEL1 resins overnight. Resins were washed, and proteins were eluted and resolved on SDS-PAGE. The relative amounts of PIAS4 detected in the pull-downs were normalized to loading and quantified.
In vitro ubiquitin conjugation assays
The assay was performed in a volume of 20 µl containing 50 mM Tris, pH 7.6, 5 mM MgCl2, 0.6 mM DTT, 2 mM adenosine triphosphate (ATP), 400 µM MG132, 50 nM Ubiquitin activating enzyme, 0.5 µM UbcH5, 0.5 µM UbcH7, 2 µM ubiquitin, and 1 µM ubiquitin aldehyde. TnT coupled reticulocyte in vitro–synthesized FIEL1-V5 and PIAS4-V5 proteins were purified via Talon metal affinity resin, and reaction products were processed for V5 immunoblotting.
In vitro kinase assays
The assays were performed by combining 50 mM Tris, pH 7.6, 100 mM MgCl2, 25 mM β-Glycerolphosphate, and 5 mg/ml BSA, bringing them to a total volume of 25 µl using combinations of 0.5 mM ATP, 0.2 µCi γ-32P ATP (Perkin Elmer), 500 nM recombinant Aurora B (EMD Millipore), either 500 nM of recombinant PKCζ (Enzo) or 500 nM of recombinant GSK3β (Enzo), and V5-tagged PIAS4 or V5-tagged FIEL1 synthesized via TnT in vitro kits (Promega) and purifying them by HIS pull-down. Reaction products were incubated at 37°C for 2 h, resolved by SDS-PAGE, and processed for autoradiography either by using Personal Molecular Imager (Bio-Rad Laboratories) or immunoblotting for V5 to visualize substrate input.
Hydroxyproline assay
Murine lungs were dried and weighed before digestion with HCl. Hydroxyproline concentrations were measured using previously described methods (Neuman and Logan, 1950; Reddy and Enwemeka, 1996). Hydroxyproline content was normalized to dry lung weight.
SMAD reporter assay
Cignal SMAD Reporter luciferase plasmids were co-transfected with Empty, FIEL1, PIAS4, CON shRNA, or FIEL1 shRNA for 24-48 h before TGFβ treatment for an additional 2–18 h. Cells were then collected and assayed for firefly and Renilla luciferase activity. SMAD transcription activity was normalized by a firefly and Renilla luciferase activity ratio.
Immunostaining
MRC5 cells were seeded in 35-mm MatTek glass bottom dishes before the plasmid transfection, inhibitor, and TGFβ treatment. Cells were washed with PBS and fixed with 4% paraformaldehyde for 20 min, and then exposed to 2% BSA, 1:500 mouse α-SMA, goat FN, or SMAD antibodies, and 1:1,000 Alexa Fluor 488– or 567–labeled goat anti–rabbit, chicken anti–mouse, or donkey anti–goat secondary antibodies sequentially for immunostaining. The nucleus was counterstained with DAPI and F-actin was counterstained with Alexa Fluor 488-Phalloidin. Immunofluorescent cell imaging was performed on a Nikon A1 confocal microscope using 405 nm, 488 nm, or 567 nm wavelengths. All experiments were done with a 60× oil differential interference contrast objective lens.
Molecular docking studies and compound design
The docking experiments were performed using software from Discovery Studio 3.5. A library containing 500,000 approved or experimental drugs was first used to screen potential ligands for FIEL1. FIEL1-HECT domain structural analysis revealed a major drug-binding cavity within the C terminus of the HECT domain. The binding cavity was adopted into the LibDock algorithm to screen for the optimum inhibitor. Based on the docking and best-fit analysis of suitable ligands, BC-1480 was used as the backbone to synthesize other compounds.
BC-1485 synthesis
A mixture of 4-(2-oxo-2,3-dihydro-1H-benzo[d]imidazole-5-sulfonamido)benzoic acid (66 mg, 0.2 mmol), N-(3-Dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (38.3 mg, 0.2 mmol), and 1-Hydroxybenzotriazole hydrate (30.6 mg, 0.2 mmol) in DMF (3 ml) was stirred at room temperature for 10 min, followed by the addition of 2-amino-N,N-dimethylpropanamide hydrochloride (37 mg and 0.24 mmol), and triethylamine (24.3 mg and 0.24 mmol). The reaction was stirred at room temperature under nitrogen overnight and concentrated under vacuum. The residue was dissolved in dichloromethane (1 ml) and purified by flash chromatography (silica gel, toluene/2-propanol/ammonia hydroxide = 80/20/1, vol/vol/v) to obtain a sticky white solid. It was suspended in 2N HCl (2 ml), sonicated for 10 min, and filtered. The wet cake was washed with water several times, and then dried by vacuum suction to obtain the desired product as a white powder (30 mg, 35% yield): 1H NMR (400 MHz, DMSO-d6) δ 11.10 (s, 1H), 10.97 (s, 1H), 10.47 (s, 1H), 8.38 (d, J = 8.0 Hz, 1H), 7.72 (d, J = 8.4 Hz, 2H), 7.41 (d, J = 9.2 Hz, 1H),7.28 (s, 1H), 7.12 (d, J = 8.4 Hz, 2H), 7.01 (d, J = 8.4 Hz, 1H), 4.84 (m, 1H), 3.01 (s, 3H), 2.81 (s, 3H), 1.22 (d, J = 6.8 Hz, 3H); 13C NMR (100 MHz, DMSO-d6) δ 172.30 (C = O), 165.45 (C = O), 155.66 (C =O), 141.26, 133.95, 131.45, 130.10, 129.26, 129.13, 120.83, 118.62, 108.74, 107.02, 45.55, 36.89, 35.71, 17.42; HRMS (ESI) calculated for C19H21N5O5S: 431.12634, found: 432.13353 [M+H]+.
Quantitative RT-PCR, cloning, and mutagenesis
Total RNA was isolated and reverse transcription was performed, followed by real-time quantitative PCR with SYBR Green qPCR mixture as previously described (Butler and Mallampalli, 2010). All mutant PIAS4 and FIEL1 plasmid constructs were generated using PCR-based approaches and appropriate primers and subcloned into a pcDNA3.1D/V5-HIS vector.
Lentivirus construction
To generate lentivirus encoding FIEL1, Lenti-Plvx-FIEL1 plasmid was co-transfected with Lenti-X HTX packaging plasmids (Takara Bio Inc.) into 293T cells following the manufacturer’s instructions. 72 h later, virus was collected and concentrated using Lenti-X concentrator (Takara Bio Inc.).
Animal studies
All procedures were approved by the University of Pittsburgh Institutional Animal Care and Use Committee. For fibrosis studies, male C57BL/6J mice were deeply anesthetized using a ketamine/xylazine mixture, and the larynx was well visualized under a fiberoptic light source before endotracheal intubation with a 3/400 24-gauge plastic catheter. 107 CFU of lentivirus encoding genes for Empty (E), FIEL1, CON shRNA, or FIEL1 shRNA was given i.t. for 144 h before administration of bleomycin (0.02–0.05 U i.t.) for up to 21 d. Animals were euthanized and assayed for BAL protein, cell count, cytokines, and lung infiltrates. Survival studies were performed on mice that were given bleomycin (0.02–0.05 U i.t.). Mice were carefully monitored over time; moribund, preterminal animals were immediately euthanized and recorded as deceased. For time course drug studies, mice were deeply anesthetized as with fibrous studies. Bleomycin (0.05 U i.t.) was given i.t. before BC-1480 or BC-1485 (∼5 mg/kg/d) was administered to the mice through their drinking water. 7–21 d later, animals were euthanized and analyzed as above. For dose course drug studies, mice were deeply anesthetized as above. Bleomycin (0.05 U i.t.) was given i.t. 10 d before BC-1485 (∼2–10 mg/kg/d) was administered to the mice through their drinking water. Another 10 d later, animals were euthanized and analyzed as above.
Tissue staining
Murine lung samples were fixed in 10% neutral buffered formalin and embedded in paraffin and sectioned as previously described (Reed et al., 2015). Sections were stained with eosin and hematoxylin or Masson’s Trichrome. Images were acquired from 20× lens from random fields from each section. Fibrotic areas (blue channel) were isolated and determined by pixel area using ImageJ (National Institutes of Health).
Statistical analysis
Statistical comparisons were performed through SPSS (IBM) with P < 0.05 indicative of significance. Survival curves were generated through SPSS (IBM).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health R01 grants HL116472 (to B.B. Chen) and HL126990 (to D.J. Kass), P01 grant HL114453 (to B.B. Chen and Y. Zhang), and a University of Pittsburgh Vascular Medicine Institute seed fund.
A provisional patent application covering FIEL1 inhibitors and additional modifications was filed through the University of Pittsburgh. The authors declare no additional competing financial interests.
Author contributions: B.B. Chen designed the study, analyzed the data, and wrote the manuscript; T. Lear, A.C. McKelvey, S. Rajbhandari, S.R. Dunn., T.A. Coon, W. Connelly, J.Y. Zhao, and Y. Liu performed all in vitro experiments and animal experiments; Y. Zhang assisted with human studies; D.J. Kass helped revise the manuscript. B.B. Chen directed the study.
Footnotes
Abbreviations used:
- α-SMA
- α smooth muscle actin
- AREL1
- apoptosis-resistant E3 ligase 1
- ATP
- adenosine triphosphate
- BAL
- bronchoalveolar lavage
- FN
- fibronectin
- IPF
- idiopathic pulmonary fibrosis
- PIAS4
- protein inhibitor of activated stat
- SMAD
- mothers against decapentaplegic homolog
- SUMO
- small ubiquitin-like modifier
- TnT
- transcription and translation
References
- Akhmetshina A., Venalis P., Dees C., Busch N., Zwerina J., Schett G., Distler O., and Distler J.H.. 2009. Treatment with imatinib prevents fibrosis in different preclinical models of systemic sclerosis and induces regression of established fibrosis. Arthritis Rheum. 60:219–224. 10.1002/art.24186 [DOI] [PubMed] [Google Scholar]
- Annes J.P., Munger J.S., and Rifkin D.B.. 2003. Making sense of latent TGFbeta activation. J. Cell Sci. 116:217–224. 10.1242/jcs.00229 [DOI] [PubMed] [Google Scholar]
- Attisano L., and Wrana J.L.. 2000. Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 12:235–243. 10.1016/S0955-0674(99)00081-2 [DOI] [PubMed] [Google Scholar]
- Baarsma H.A., Engelbertink L.H., van Hees L.J., Menzen M.H., Meurs H., Timens W., Postma D.S., Kerstjens H.A., and Gosens R.. 2013. Glycogen synthase kinase-3 (GSK-3) regulates TGF-β₁-induced differentiation of pulmonary fibroblasts. Br. J. Pharmacol. 169:590–603. 10.1111/bph.12098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonniaud P., Margetts P.J., Ask K., Flanders K., Gauldie J., and Kolb M.. 2005. TGF-beta and Smad3 signaling link inflammation to chronic fibrogenesis. J. Immunol. 175:5390–5395. 10.4049/jimmunol.175.8.5390 [DOI] [PubMed] [Google Scholar]
- Butler P.L., and Mallampalli R.K.. 2010. Cross-talk between remodeling and de novo pathways maintains phospholipid balance through ubiquitination. J. Biol. Chem. 285:6246–6258. 10.1074/jbc.M109.017350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman H.A. 2011. Epithelial-mesenchymal interactions in pulmonary fibrosis. Annu. Rev. Physiol. 73:413–435. 10.1146/annurev-physiol-012110-142225 [DOI] [PubMed] [Google Scholar]
- Chen B.B., and Mallampalli R.K.. 2007. Calmodulin binds and stabilizes the regulatory enzyme, CTP: phosphocholine cytidylyltransferase. J. Biol. Chem. 282:33494–33506. 10.1074/jbc.M706472200 [DOI] [PubMed] [Google Scholar]
- Chen B.B., Coon T.A., Glasser J.R., McVerry B.J., Zhao J., Zhao Y., Zou C., Ellis B., Sciurba F.C., Zhang Y., and Mallampalli R.K.. 2013. A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat. Immunol. 14:470–479. 10.1038/ni.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon T.A., McKelvey A.C., Lear T., Rajbhandari S., Dunn S.R., Connelly W., Zhao J.Y., Han S., Liu Y., Weathington N.M., et al. 2015. The proinflammatory role of HECTD2 in innate immunity and experimental lung injury. Sci. Transl. Med. 7:295ra109 10.1126/scitranslmed.aab3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels C.E., Lasky J.A., Limper A.H., Mieras K., Gabor E., and Schroeder D.R.. Imatinib-IPF Study Investigators . 2010. Imatinib treatment for idiopathic pulmonary fibrosis: Randomized placebo-controlled trial results. Am. J. Respir. Crit. Care Med. 181:604–610. 10.1164/rccm.200906-0964OC [DOI] [PubMed] [Google Scholar]
- Denton C.P., Merkel P.A., Furst D.E., Khanna D., Emery P., Hsu V.M., Silliman N., Streisand J., Powell J., Akesson A., et al. Scleroderma Clinical Trials Consortium . 2007. Recombinant human anti-transforming growth factor beta1 antibody therapy in systemic sclerosis: a multicenter, randomized, placebo-controlled phase I/II trial of CAT-192. Arthritis Rheum. 56:323–333. 10.1002/art.22289 [DOI] [PubMed] [Google Scholar]
- Derynck R., and Zhang Y.E.. 2003. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 425:577–584. 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- Gross T.J., and Hunninghake G.W.. 2001. Idiopathic pulmonary fibrosis. N. Engl. J. Med. 345:517–525. 10.1056/NEJMra003200 [DOI] [PubMed] [Google Scholar]
- Gross M., Liu B., Tan J., French F.S., Carey M., and Shuai K.. 2001. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene. 20:3880–3887. 10.1038/sj.onc.1204489 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Yada M., Matsumoto M., Ishida N., and Nakayama K.I.. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276:33111–33120. 10.1074/jbc.M102755200 [DOI] [PubMed] [Google Scholar]
- Hecker L., Vittal R., Jones T., Jagirdar R., Luckhardt T.R., Horowitz J.C., Pennathur S., Martinez F.J., and Thannickal V.J.. 2009. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat. Med. 15:1077–1081. 10.1038/nm.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J.M., Scheffner M., Beaudenon S., and Howley P.M.. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA. 92:2563–2567. 10.1073/pnas.92.7.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto S., Sugiyama K., Muromoto R., Sato N., Yamamoto T., and Matsuda T.. 2003. Regulation of transforming growth factor-beta signaling by protein inhibitor of activated STAT, PIASy through Smad3. J. Biol. Chem. 278:34253–34258. 10.1074/jbc.M304961200 [DOI] [PubMed] [Google Scholar]
- Imoto S., Sugiyama K., Yamamoto T., and Matsuda T.. 2004. The RING domain of PIASy is involved in the suppression of bone morphogenetic protein-signaling pathway. Biochem. Biophys. Res. Commun. 319:275–282. 10.1016/j.bbrc.2004.04.161 [DOI] [PubMed] [Google Scholar]
- Imoto S., Ohbayashi N., Ikeda O., Kamitani S., Muromoto R., Sekine Y., and Matsuda T.. 2008. Sumoylation of Smad3 stimulates its nuclear export during PIASy-mediated suppression of TGF-beta signaling. Biochem. Biophys. Res. Commun. 370:359–365. 10.1016/j.bbrc.2008.03.116 [DOI] [PubMed] [Google Scholar]
- Jiang D., Liang J., Fan J., Yu S., Chen S., Luo Y., Prestwich G.D., Mascarenhas M.M., Garg H.G., Quinn D.A., et al. 2005. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat. Med. 11:1173–1179. 10.1038/nm1315 [DOI] [PubMed] [Google Scholar]
- Jiang D., Liang J., Campanella G.S., Guo R., Yu S., Xie T., Liu N., Jung Y., Homer R., Meltzer E.B., et al. 2010. Inhibition of pulmonary fibrosis in mice by CXCL10 requires glycosaminoglycan binding and syndecan-4. J. Clin. Invest. 120:2049–2057. 10.1172/JCI38644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Li X., Gygi S.P., and Harper J.W.. 2007. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 447:1135–1138. 10.1038/nature05902 [DOI] [PubMed] [Google Scholar]
- Jonk L.J., Itoh S., Heldin C.H., ten Dijke P., and Kruijer W.. 1998. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 273:21145–21152. 10.1074/jbc.273.33.21145 [DOI] [PubMed] [Google Scholar]
- Kage H., and Borok Z.. 2012. EMT and interstitial lung disease: a mysterious relationship. Curr. Opin. Pulm. Med. 18:517–523. 10.1097/MCP.0b013e3283566721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamadurai H.B., Souphron J., Scott D.C., Duda D.M., Miller D.J., Stringer D., Piper R.C., and Schulman B.A.. 2009. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol. Cell. 36:1095–1102. 10.1016/j.molcel.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.B., Kim S.Y., Kim B.M., Lee H., Kim I., Yun J., Jo Y., Oh T., Jo Y., Chae H.D., and Shin D.Y.. 2013. Identification of a novel anti-apoptotic E3 ubiquitin ligase that ubiquitinates antagonists of inhibitor of apoptosis proteins SMAC, HtrA2, and ARTS. J. Biol. Chem. 288:12014–12021. 10.1074/jbc.M112.436113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T.E. Jr., Pardo A., and Selman M.. 2011. Idiopathic pulmonary fibrosis. Lancet. 378:1949–1961. 10.1016/S0140-6736(11)60052-4 [DOI] [PubMed] [Google Scholar]
- Lee P.S., Chang C., Liu D., and Derynck R.. 2003. Sumoylation of Smad4, the common Smad mediator of transforming growth factor-beta family signaling. J. Biol. Chem. 278:27853–27863. 10.1074/jbc.M301755200 [DOI] [PubMed] [Google Scholar]
- Li M., Krishnaveni M.S., Li C., Zhou B., Xing Y., Banfalvi A., Li A., Lombardi V., Akbari O., Borok Z., and Minoo P.. 2011. Epithelium-specific deletion of TGF-β receptor type II protects mice from bleomycin-induced pulmonary fibrosis. J. Clin. Invest. 121:277–287. 10.1172/JCI42090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Matsuura I., He D., Wang G., Shuai K., and Liu F.. 2003. Repression of Smad transcriptional activity by PIASy, an inhibitor of activated STAT. Proc. Natl. Acad. Sci. USA. 100:9791–9796. 10.1073/pnas.1733973100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallampalli R.K., Coon T.A., Glasser J.R., Wang C., Dunn S.R., Weathington N.M., Zhao J., Zou C., Zhao Y., and Chen B.B.. 2013. Targeting F box protein Fbxo3 to control cytokine-driven inflammation. J. Immunol. 191:5247–5255. 10.4049/jimmunol.1300456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maspero E., Mari S., Valentini E., Musacchio A., Fish A., Pasqualato S., and Polo S.. 2011. Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 12:342–349. 10.1038/embor.2011.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. 2012. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13:616–630. 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulsow J.J., Watson R.W., Fitzpatrick J.M., and O’Connell P.R.. 2005. Transforming growth factor-β promotes pro-fibrotic behavior by serosal fibroblasts via PKC and ERK1/2 mitogen activated protein kinase cell signaling. Ann. Surg. 242:880–887. 10.1097/01.sla.0000189606.58343.cd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman R.E., and Logan M.A.. 1950. The determination of hydroxyproline. J. Biol. Chem. 184:299–306. [PubMed] [Google Scholar]
- Noble P.W., Barkauskas C.E., and Jiang D.. 2012. Pulmonary fibrosis: patterns and perpetrators. J. Clin. Invest. 122:2756–2762. 10.1172/JCI60323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope J., McBain D., Petrlich L., Watson S., Vanderhoek L., de Leon F., Seney S., and Summers K.. 2011. Imatinib in active diffuse cutaneous systemic sclerosis: Results of a six-month, randomized, double-blind, placebo-controlled, proof-of-concept pilot study at a single center. Arthritis Rheum. 63:3547–3551. 10.1002/art.30549 [DOI] [PubMed] [Google Scholar]
- Raghu G., Collard H.R., Egan J.J., Martinez F.J., Behr J., Brown K.K., Colby T.V., Cordier J.F., Flaherty K.R., Lasky J.A., et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis . 2011. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am. J. Respir. Crit. Care Med. 183:788–824. 10.1164/rccm.2009-040GL [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray N.B., Durairaj L., Chen B.B., McVerry B.J., Ryan A.J., Donahoe M., Waltenbaugh A.K., O’Donnell C.P., Henderson F.C., Etscheidt C.A., et al. 2010. Dynamic regulation of cardiolipin by the lipid pump Atp8b1 determines the severity of lung injury in experimental pneumonia. Nat. Med. 16:1120–1127. 10.1038/nm.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G.K., and Enwemeka C.S.. 1996. A simplified method for the analysis of hydroxyproline in biological tissues. Clin. Biochem. 29:225–229. 10.1016/0009-9120(96)00003-6 [DOI] [PubMed] [Google Scholar]
- Reed N.I., Jo H., Chen C., Tsujino K., Arnold T.D., DeGrado W.F., and Sheppard D.. 2015. The αvβ1 integrin plays a critical in vivo role in tissue fibrosis. Sci. Transl. Med. 7:288ra79 10.1126/scitranslmed.aaa5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D., and Kumar S.. 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 10:398–409. 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- Rytinki M.M., Kaikkonen S., Pehkonen P., Jääskeläinen T., and Palvimo J.J.. 2009. PIAS proteins: pleiotropic interactors associated with SUMO. Cell. Mol. Life Sci. 66:3029–3041. 10.1007/s00018-009-0061-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard D. 2013. ROCKing pulmonary fibrosis. J. Clin. Invest. 123:1005–1006. 10.1172/JCI68417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tager A.M., LaCamera P., Shea B.S., Campanella G.S., Selman M., Zhao Z., Polosukhin V., Wain J., Karimi-Shah B.A., Kim N.D., et al. 2008. The lysophosphatidic acid receptor LPA1 links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat. Med. 14:45–54. 10.1038/nm1685 [DOI] [PubMed] [Google Scholar]
- Takeda M., Babazono T., Nitta K., and Iwamoto Y.. 2001. High glucose stimulates hyaluronan production by renal interstitial fibroblasts through the protein kinase C and transforming growth factor-beta cascade. Metabolism. 50:789–794. 10.1053/meta.2001.24207 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Tanaka N., Saeki Y., Tanaka K., Murakami M., Hirano T., Ishii N., and Sugamura K.. 2008. c-Cbl-dependent monoubiquitination and lysosomal degradation of gp130. Mol. Cell. Biol. 28:4805–4818. 10.1128/MCB.01784-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umadevi N., Kumar S., and Narayana N.. 2005. Crystallization and preliminary X-ray diffraction studies of the WW4 domain of the Nedd4-2 ubiquitin-protein ligase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61:1084–1086. 10.1107/S174430910503767X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.M., Zhang Y., Kim H.P., Zhou Z., Feghali-Bostwick C.A., Liu F., Ifedigbo E., Xu X., Oury T.D., Kaminski N., and Choi A.M.. 2006. Caveolin-1: a critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J. Exp. Med. 203:2895–2906. 10.1084/jem.20061536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick G., Grundtman C., Mayerl C., Wimpissinger T.-F., Feichtinger J., Zelger B., Sgonc R., and Wolfram D.. 2013. The immunology of fibrosis. Annu. Rev. Immunol. 31:107–135. 10.1146/annurev-immunol-032712-095937 [DOI] [PubMed] [Google Scholar]
- Wilson M.S., and Wynn T.A.. 2009. Pulmonary fibrosis: pathogenesis, etiology and regulation. Mucosal Immunol. 2:103–121. 10.1038/mi.2008.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J.M., Blanchard K.L., and Sawyer J.S.. 2004. Development of TGF-beta signalling inhibitors for cancer therapy. Nat. Rev. Drug Discov. 3:1011–1022. 10.1038/nrd1580 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.