The ubiquitin E3 ligase March1 controls the turnover and surface levels of peptide–MHCII in GC B cells, contributing to optimal GC responses.
Abstract
Antibody affinity maturation occurs in germinal centers (GCs) through iterative rounds of somatic hypermutation and selection. Selection involves B cells competing for T cell help based on the amount of antigen they capture and present on their MHC class II (MHCII) proteins. How GC B cells are able to rapidly and repeatedly transition between mutating their B cell receptor genes and then being selected shortly after is not known. We report that MHCII surface levels and degradation are dynamically regulated in GC B cells. Through ectopic expression of a photoconvertible MHCII-mKikGR chimeric gene, we found that individual GC B cells differed in the rates of MHCII protein turnover. Fluctuations in surface MHCII levels were dependent on ubiquitination and the E3 ligase March1. Increases in March1 expression in centroblasts correlated with decreases in surface MHCII levels, whereas CD83 expression in centrocytes helped to stabilize MHCII at that stage. Defects in MHCII ubiquitination caused GC B cells to accumulate greater amounts of a specific peptide–MHCII (pMHCII), suggesting that MHCII turnover facilitates the replacement of old complexes. We propose that pMHCII complexes are periodically targeted for degradation in centroblasts to favor the presentation of recently acquired antigens, thereby promoting the fidelity and efficiency of selection.
Germinal centers (GCs) form in secondary lymphoid tissues after infections and immunizations and are the principle sites in which high-affinity antibodies to protein antigens develop. Antibodies generated via this pathway are essential for the sterilizing immunity provided by many vaccines and are needed for normal homeostasis at barrier sites. GC B cells refine and improve their B cell receptor (BCR) specificities through the random introduction of point mutations into their immunoglobulin variable region genes in a reaction catalyzed by the enzyme activation-induced cytidine deaminase (AID). GC B cells carrying beneficial mutations are then selected at the expense of their neighbors for their continued participation in the response as a result of their having an increased capacity to capture antigens from follicular DCs and to subsequently present peptides in complex with MHC class II (peptide–MHCII [pMHCII] complexes). Selection involves GC B cells competing for help in the form of coreceptor ligation and cytokine secretion from limiting numbers of GC follicular helper T cells (Tfh cells; Batista and Neuberger, 2000; Allen et al., 2007; Victora et al., 2010). In addition, GC B cells with greater amounts of surface pMHCII receive a better quality of help from Tfh cells; this in turn enhances their rates of proliferation and the accrual of further somatic mutations (Gitlin et al., 2014, 2015). Therefore, the nature and amount of peptides presented by GC B cells determines their fate.
GCs are polarized into two regions known as light and dark zones, between which GC B cells regularly transit. The movement of cells between these two compartments is associated with changes in phenotype and behavior that lead to the GC B cells of the light zone and dark zone being known as centrocytes and centroblasts, respectively. The transitioning of cells between centroblast and centrocyte states was recently shown to occur independently of positioning but correlate with it, leading to the proposal that GC B cell behavior is determined in large part by an intrinsic cellular program (Bannard et al., 2013). However, the spatial separation of certain cues and functions probably enhances the efficiency of the response. GC B cell selection is thought to occur at the centrocyte state in the light zone where the majority of antigen is located, whereas somatic hypermutation and mitosis occur in centroblasts (Allen et al., 2007; Victora et al., 2010; Calado et al., 2012; Dominguez-Sola et al., 2012). Up to 50% of GC B cells transition between centroblast and centrocyte stages every 4 h, with cells remaining as centroblasts for between one and six cellular divisions (Victora et al., 2010; Gitlin et al., 2014).
The repetitive and iterative nature of GC B responses poses unique demands on GC B cells. It is not known how GC B cells ensure that they are selected only on the basis of antigens acquired through their current BCR and are not influenced by older pMHCII complexes. Where they have been measured in other lineages, pMHCII complexes have often had long half-lives that might not be compatible with the requirements of GC B cells (Cella et al., 1997; Pierre et al., 1997; Lazarski et al., 2005; De Riva et al., 2013). We therefore hypothesized MHCII presentation may be subject to dynamic forms of regulation in GC B cells that correlate with the specific requirements of each state. The control of antigen presentation in other cell lineages involves regulation in the localization and turnover of MHCII proteins; pMHCII complexes are continuously internalized into early endosomes, but the fate of the protein is determined by ubiquitination (Shin et al., 2006; Oh and Shin, 2015). The addition of ubiquitin chains by the E3 ligase March1 prevents MHCII recycling back to the surface for further presentation and instead targets it for lysosomal degradation (Matsuki et al., 2007; Walseng et al., 2010; Cho et al., 2015). An earlier study evaluating the role of MHCII ubiquitination in promoting B cell responses was unable to identify a need for this pathway (McGehee et al., 2011). However, these assessments mostly involved looking at the magnitude rather than quality of the antibody response and so did not fully address whether this pathway might contribute to events occurring within GCs.
In this study, we report that MHCII levels and turnover are dynamically regulated in GC B cells as a result of changes in ubiquitination and March1 function at different stages of their program. MHCII complexes were found to be preferentially degraded in subsets of centroblasts displaying very low levels of MHCII protein but were stabilized in centrocytes. By staining for a specific pMHCII complex at different times after antigen delivery, we found that MHCII ubiquitination prevented the accumulation of older pMHCII complexes in GC B cells. We speculate that changes in MHCII turnover at different times in GC B cells may help refine the peptide repertoire to reflect recently captured antigens and to promote productive interactions with T cells at the time of selection.
RESULTS
Surface MHCII levels are high on centrocytes but vary greatly on centroblasts
To begin investigating whether MHCII-mediated antigen presentation might be differentially regulated in GC B cells as they transition between states, we compared surface MHCII levels on centroblasts and centrocytes. Splenic GC B cells (IgDlow CD95high GL7+) from mice that had been immunized with sheep RBCs (SRBCs) were costained with antibodies to CXCR4 and MHCII (I-Ab). These experiments revealed differences in the intensity of MHCII staining on different GC subsets (Fig. 1 a). Although centrocytes almost all expressed very high levels of surface MHCII, the centroblast population contained cells with MHCII levels that varied over a >1.5 log range in terms of florescent intensities. After this finding, we subgated GC B cells into four different populations based on their CXCR4 versus MHCII staining profiles; gate A, which represents centrocytes, and gates B–D, which contain centroblasts with high, medium, or low MHCII levels. The equivalent staining intensities of IgD, CD95, and GL7 on populations B–D confirmed that all the subsets are bona fide GC B cells (Fig. 1 b). All centroblast subpopulations expressed comparable levels of the centroblast-associated gene Cxcr4 and centrocyte gene Cd86 (Fig. 1 c). In contrast, CD86 (but not CD83) protein levels correlated with those of MHCII and varied in the different centroblast subsets, suggesting possible posttranscriptional regulation of CD86 (Fig. 1 d). These findings indicate that the abundance of surface MHCII and CD86 molecules is dynamically regulated in GC B cells.
Figure 1.
Individual centroblasts vary widely in their surface MHCII levels. (a) C57BL/6 mice were immunized with SRBCs by i.p. injection and GC B cells were gated using an IgDlow CD95high GL7+ gating scheme. GC B cells were subgated into four populations based on their surface MHCII (I-Ab) and CXCR4 levels. (b) Back gating on populations A–D confirmed that the CXCR4high MHCIIlow population displays staining patterns of GC B cells. Horizontal lines indicate means. (c) Populations A–D were FACS sorted and subjected to RT-PCR. Data were normalized to Hprt. (d) CD86 and CD83 surface staining was compared for the four GC subsets, color coded as in a. Points in c represent individual samples from separate mice, with data pooled from two experiments. Data in a, b, and d are representative of four mice analyzed in two experiments.
Fluctuations in MHCII abundance during the GC program involves changes in protein degradation and stabilization
The finding that individual GC B cells vary widely in their MHCII levels led us to ask whether rates of this protein’s degradation might be differentially controlled as cells pass through stages of the GC program. We speculated that such a mechanism might exist for the purpose of purging the cell of old pMHCII complexes after somatic hypermutation, but before reentry of the light zone for selection. In vitro experimentation with GC B cells is mostly not possible because of their propensity to die rapidly when cultured. Therefore, we established a new tool to track MHCII protein turnover in vivo with single-cell resolution. A chimeric gene composed of the monomeric photoconvertible protein mKikGR fused to the intracellular tail of the I-Ab β chain was expressed in B cells via retroviral transduction of BM progenitors. mKikGR undergoes a rapid and permanent change from green (mKikGRgreen) to red (mKiKGRred) states upon exposure to violet light (Habuchi et al., 2008), thereby allowing us to indelibly mark the preexisting fusion protein and to subsequently track its degradation (Fig. 2 a). After their reconstitution, BM chimeric mice were s.c. immunized with hapten 4-hydroxy-3-nitrophenylacetyl coupled to chicken gamma globulin (NP-CGG), precipitated in alum. Inguinal LNs (iLNs) were subsequently surgically exposed on days 8 or 9 of the response, and the entire node was photoconverted using an LED light source. As expected, violet light exposure caused the appearance of mKikGRred signal in almost all transduced GC B cells, as determined by FACS (Fig. 2 b). The intensity of the ectopic I-Ab–mKikGRred signal immediately after photoconversion was similar in all GC B cells regardless of their endogenous MHCII levels. Cohorts of mice were then recovered from surgery and analyzed at different time points. The extent of uniformity of the red signal changed over time; by 8 h after photoconversion, almost all GC B cells expressing low levels of endogenous MHCII also had low or undetectable I-Ab–mKikGRred, indicating that the fusion protein had been rapidly degraded in these cells. In contrast, the I-Ab–mKikGRred signal remained easily detectible in most MHCIIhigh GC B cells at this time point. Furthermore, we were able to identify I-Ab–mKikGRred cells within the MHCIIhigh population a full 24 h after their photoconversion, demonstrating that MHCII complexes are relatively stable and long lived in some GC B cell subsets but not others. The absolute frequency of detectable I-Ab–mKikGRred cells present at the late time varied between experiments because of differences in transduction efficiency. Findings from multiple mice and experiments are summarized in Fig. 2 c. Together, these findings indicate that MHCII complexes are degraded more rapidly in some GC B cells than in others and that this is reflected in the surface abundance of the protein. MHCII degradation is fastest in MHCIIlow centroblasts. Consequently, rates for completion of MHCII protein replacement vary by more than threefold in different GC subsets.
Figure 2.
MHCIIlow GC B cells degrade their MHCII molecules more rapidly. (a) A fusion construct encoding the photoconvertible protein mKiKGR fused to the intracellular tail of the I-Ab β chain was generated. Exposure of the encoded protein to violet light causes a permanent switch between green and red states. BM cells were transduced with the construct and used to reconstitute lethally irradiated recipients. (b) Reconstituted mice were immunized with NP-CGG/alum and iLNs photoconverted 8–9 d later. iLNs were harvested without conversion or at various time points after photoconversion, and transduced (Thy1.1+) GC B cells were interrogated for their expression of endogenous surface MHCII and I-AbmKiKGRred. (c) Cells were gated according to high and low levels of endogenous MHCII (dashed lines in b), and the relative difference in I-AbmKiKGRred signal in the MHCIIlow versus MHCIIhigh cells is plotted for multiple mice from three experiments (two to six mice per experiment), with each symbol representing individual animals. Horizontal lines indicate means. Analysis was performed using an unpaired two-tailed Student’s t test. ***, P < 0.001.
March1-mediated, CD83-regulated ubiquitination determines MHCII surface levels on GC B cells
The maturation of DCs involves increases in MHCII abundance as a result of decreases in ubiquitin-mediated MHCII protein degradation, and this is associated with enhanced antigen presentation efficiency (Shin et al., 2006). We therefore asked whether similar but reversible processes might occur in GC B cells as they transition between centroblast and centrocyte states. Splenic GC B cells were sorted based on the CXCR4 and MHCII gating strategy described in Fig. 1 and subjected to quantitative RT-PCR. These experiments revealed increased expression of the class II transactivator (Ciita) and its target MHCII genes H2-Aa, H2-Ab, H2-DMa, and H2-DMb in CXCR4low MHCIIhigh centrocytes relative to all centroblast subsets, consistent with an earlier study (Victora et al., 2012). However, we did not observe appreciable differences in expression of these genes within the different CXCR4high centroblast subsets that differ in their MHCII protein levels (Fig. 3). In contrast, the abundance of March1 transcripts varied even within the centroblast subsets. March1 is the principle E3 ligase that controls MHCII ubiquitination in DCs and in naive B cells (De Gassart et al., 2008). March1 mRNA abundance inversely correlated with surface MHCII expression in GC B cells, with mRNA for the E3 ligase being most abundant in MHCIIlow centroblasts and least abundant in centrocytes.
Figure 3.
Expression of MHCII-related genes and the E3 ligase March1 vary in different GC subsets. GC B cells were FACS sorted based on their CXCR4 and MHCII staining profile and subjected to RT-PCR analysis. Data were normalized to Hprt. Points represent individual samples from separate mice, with data pooled from two experiments each with two mice. Horizontal lines indicate means.
Given the aforementioned findings of changes in protein turnover and differences in March1 expression, we asked whether ubiquitination of MHCII plays any role in controlling surface protein abundance in GC B cells. Mixed BM chimeric mice were generated in which approximately half the cells were from a CD45.2+ donor encoding a form of MHCII that cannot be a ubiquitin substrate because of a cytoplasmic K>R mutation (I-Ab(K>R); Oh et al., 2013). The remaining ∼50% of donor cells were WT and CD45.1+. Reconstituted chimeric animals were immunized with SRBCs and the surface levels of MHCII on both genotypes of splenic GC B was determined (Fig. 4 a). I-Ab(K>R) GC B cells displayed uniformly high levels of surface MHCII regardless of their CXCR4 state, with staining intensities on all mutant cells being equivalent to (or marginally higher than) that on the very brightest of WT GC B cells, indicating an important role for ubiquitination in controlling abundance of this protein complex. This control was intrinsic to GC B cells because WT CD45.1+ cells from the same mice displayed normal CXCR4 versus MHCII staining patterns. Importantly, the uniformity of MHCII surface expression levels on I-Ab(K>R) GC B cells did not appear to be caused by a defect in switching between centrocyte and centroblast stages because WT and I-Ab(K>R) populations contained similar frequencies of centroblasts, as determined by CXCR4 and CD23 staining (Fig. 4 b; Victora et al., 2010). Small shifts toward lower CXCR4 staining intensities were observed in I-Ab(K>R) cells, perhaps suggesting subtle defects in selection or in CXCR4 membrane dynamics (not depicted).
Figure 4.
Surface abundance of MHCII in GC B cells is determined by March1-mediated ubiquitination and CD83 regulation. (a) Mixed BM chimeric mice containing WT CD45.1+ cells and I-Ab(K>R) CD45.2+ cells were generated and immunized with SRBCs. Surface levels of MHCII and CXCR4 were determined in IgDlow CD95+ GL7+ GC B cells of both origins. (b) Frequencies of CXCR4high CD23low centroblasts and CXCR4low CD23high centrocytes were determined in the BM chimeras. (c) The same analyses as in panel a were performed using BM chimeric mice containing March1−/− cells. (d) CD86 levels on GC B cells were determined. (e) The same analysis as in panel a was performed using BM chimeric mice containing Cd83−/− cells. (f) Comparisons of surface MHCII level (mean fluorescence intensity [MFI]) on CXCR4high and CXCR4low GC cells of the various genotypes. (g and h) Summary of MHCII staining data (g) and I-Ab–clip staining data (h) from multiple mice. a and c–e are representative plots. Points in b and f–h represent individual mice pooled from two to three experiments. Horizontal lines in b and f–h indicate means. Analysis was performed using an unpaired two-tailed Student’s t test. **, P < 0.01; ***, P < 0.001.
We next investigated the specific role of March1 in mediating changes in surface MHCII protein abundance in GC B cells. Mixed March1−/−/WT BM chimeric mice were generated and subjected to the same immunizations and assessments as those described in the previous paragraph (Fig. 4 c). These experiments revealed March1−/− mice phenocopied the I-Ab(K>R) mice in terms of surface MHCII abundance, with all March1−/− GC B cells displaying uniformly high levels. Again, the frequency of centroblasts and centrocytes was approximately normal in March1−/− GCs (Fig. 4 b). Consistent with the observation that CD86 protein abundance correlates with that of MHCII (Fig. 1 c), these experiments also revealed CD86 to be a target of March1 in GC B cells, as it is in other cell types (Fig. 4 d; Baravalle et al., 2011). Therefore, March1 is the principle E3 ligase controlling ubiquitin-mediated down-regulation of MHCII and CD86 in GC B cells.
The transmembrane protein CD83 is a key marker of centrocytes and has important functions in negatively regulating March1 in other cell types (Victora et al., 2010; Tze et al., 2011). We therefore examined its contributions to controlling fluctuations in MHCII levels in GC B cells using mixed BM chimeras. Cd83−/− GC B cell populations contained fewer MHCIIhigh cells (subsets A and B) than did the WT populations from the same mice, consistent with CD83 playing an important role in inhibiting the E3 ligase (Fig. 4 e). Despite this, the Cd83−/− population still contained within it centroblasts displaying a broad range of MHCII levels including a CXCR4high MHCIIhigh state, indicating that although CD83 promotes MHCII stabilization, other CD83-independent changes in March1 regulation also play important roles in controlling MHCII levels. These additional control mechanisms probably include the fluctuations in March1 transcript expression reported earlier (Fig. 2 b), noting that March1 has a very short half-life (Jabbour et al., 2009; Bourgeois-Daigneault and Thibodeau, 2012). The effect of CD83 deficiency was greatest in CXCR4low centrocytes, as indicated by CXCR4low and CXCR4high GC B cells becoming more similar in their MHCII staining intensities (Fig. 4, e and f), in accord with CD83 being most highly expressed in centrocytes (Figs. 1 d and 3). Interestingly, surface CD83 was itself increased in March1−/− (but not I-Ab(K>R)) GC B cells, indicating that it is probably a March1 target and suggesting the existence of a feedback loop (not depicted). A summary of the findings from the four mixed chimeras types and from multiple experiments is displayed in Fig. 4 (f and g). Finally, similar findings of conflicting March1- and CD83-mediated mechanisms of regulation were made using an antibody against MHCII-clip that preferentially detects newly synthesized MHCII complexes that have not yet been loaded with exogenous peptides (Fig. 4 h). Together, these experiments indicate that surface MHCII levels are subjected to cyclical fluctuations in degradation and stabilization in GC B cells caused by CD83 and transcript-level regulation of March1-mediated ubiquitination.
MHCIIlow GC B cells are enriched within populations that have divided extensively in the past two days
We next wished to determine whether any temporal relationship existed between when GC B cells enter their MHCIIlow state and a key centroblast process, cellular division. Costaining of GC B cells for DNA content and BrdU incorporation revealed that the MHCIIlow centroblast subset was enriched for cells in the G1 and early S phases of cell cycle (Fig. 5, a–c). Appreciably fewer G2/M phase cells were present in this population than within the CXCR4high MHCIIhigh centroblast and total GC B cell gates. Despite this, we were surprised to observe that equivalent frequencies of MHCIIlow and MHCIIhigh CXCR4high cells had recently completed mitosis (BrdU+ G1 cells) at 2 and 4 h after BrdU pulse and that by 6–8 h after BrdU pulse the CXCR4high MHCIIlow gate contained within it a disproportionately high fraction of postmitotic BrdU+ G1 cells (Fig. 5 d). Distinguishing G1 from early S phase cells by DNA content staining alone can be imprecise; therefore, we repeated these experiments but this time used a double EdU/BrdU pulse labeling strategy to identify cells that had completed mitosis and were no longer synthesizing DNA. EdU and BrdU injections were administered at 8-h and 30-min time points before tissue harvest, respectively. Consistent with the earlier experiments, the CXCR4high MHCIIlow GC B cell population was enriched for cells that had recently been in S phase but no longer were (EdU+, BrdU− cells; Fig. 5, e and f). Furthermore, these experiments also revealed that almost all CXCR4high MHCIIlow GC B cells that incorporated BrdU in the final 30 min had also been in S phase during the earlier EdU labeling period. This was contrasted by results from the total and CXCR4high MHCIIhigh GC B cell populations in as far as these contained at least as many BrdU+ cells that were EdU− as were EdU+. Together, these findings suggest that the CXCR4high MHCIIlow GC B cell population is enriched for cells in G1 and early S phase but that it contains a disproportionally large fraction of cells that have completed at least one cell division during the last 6–8-h period.
Figure 5.
Centroblasts with low MHCII are preferentially in G1 and early S phases of cell cycle but are underrepresented in populations that have completed fewer recent mitotic events. (a) Mice were immunized with NP-CGG/alum and then given a single i.p. BrdU treatment on day 9. Spleens were harvested at various time points, and CXCR4high MHCIIlow and CXCR4high MHCIIhigh CD95+ GL7+ GC B cells were gated. (b) BrdU incorporation and DNA content were assessed at 30 min after BrdU treatment to determine cell cycle status. (c) The proportion of cells in G2/M (4n, BrdU−) was determined for multiple mice. (d) The proportion of BrdU+ Dapilow (G1) cells was determined at the different time points. (e) NP-CGG/alum–immunized mice received treatments of EdU and BrdU by single i.p. injections 8 h (EdU) and 30 min (BrdU) before euthanasia. The respective IgDlow CD95+ GL7+ GC B cell populations were gated. Control mice, shown on top, received either EdU or BrdU alone. (f) Summarized data from multiple mice analyzed as in e. (g–l) Tet-off H2b-GFP mice were immunized with NP-CGG/alum and were subsequently treated with doxycycline (or saline) for the final 40 h before analysis on day 9. (g) GFP fluorescence was assessed in naive and GC B cells. WT nonfluorescent cells are shown as shaded areas. GFPdim and GFPbright, GC B cells (IgDlow CD95+ GL7+) from dox-treated mice were gated as shown in h, and the frequency of the different subsets was examined (i). Data from multiple experiments and mice are summarized in j. (k) The frequency of CXCR4low cells within the GFPdim and GFPbright gates was calculated. (l) A similar analysis to j was performed but pregating only on CXCR4high cells. Data in a–d are from one representative experiment of three, with each experiment containing one to three mice per time point. Means are shown in d (±SD). Data in e and f are from one representative experiment of two, with each experiment containing three double-labeled mice. Data in g–l are pooled from three experiments each containing two mice per condition. Each circle in c, f, and k represents individual mice. All FACS plots are from representative mice for the respective experiments. Analysis was performed using an unpaired two-tailed Student’s t test. Horizontal lines in c, f, and j–l indicate means. **, P < 0.01; ***, P < 0.001.
To further investigate this apparent correlation between cellular division and surface MHCII levels in GC B cells, we took advantage of a tet-off H2b-GFP mouse model that permits proliferation to be tracked over multiple divisions in nontransferable populations in a CFSE-like manner (Foudi et al., 2009; Gitlin et al., 2014). Treatment of transgenic mice with doxycycline leads to the switching off of H2b-GFP gene expression; subsequent decreases in H2b-GFP signal occur mostly as a consequence of histone segregation during cell division. The suitability of the model was confirmed by comparing H2b-GFP dilution in GC B cells from mice that had or had not received doxycycline treatment and by comparing H2b-GFP dilution by dividing GC B cells and quiescent naive B cells (Fig. 5 g). MHCII levels on GC B cells that had divided little (H2b-GFPbright) or more (H2b-GFPdim) during the previous 40 h of a response to NP-CGG/alum immunization were examined (Fig. 5 h). Consistent with the BrdU chase experiment, CXCR4high MHCIIlow GC B cells were underrepresented within the H2b-GFPbright gate despite their being preferentially in G1 and early S stages of cell cycle (Fig. 5, b, c, e, i, and j). This observation reflected there being fewer MHCIIlow cells within the brightest GFP+ population that had divided least rather than them being overrepresented in subsets with the dimmest versus intermediate GFP intensities (not depicted). The H2b-GFPbright gate is enriched for CXCR4low centrocytes as a result of mitosis occurring preferentially at the centroblast stage (Fig. 5 k; Gitlin et al., 2014). However, similar findings of CXCR4high MHCIIlow GC B cells being underrepresented in this gate were made even when only CXCR4high GC B cells were analyzed, indicating that these cells have recently completed more cell divisions even when compared with other centroblasts (Fig. 5 l).
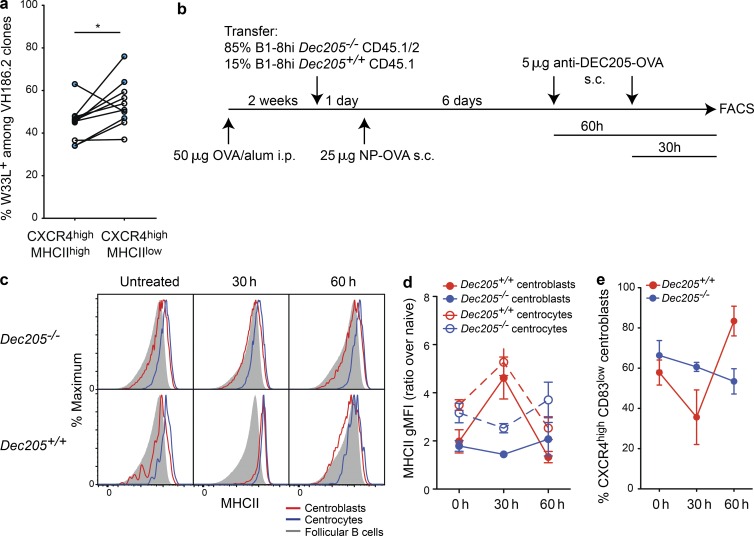
We considered possible explanations for why MHCIIlow centroblasts may have completed more recent cell divisions than their MHCIIhigh counterparts. It seemed plausible that the CXCR4high MHCIIlow population might be enriched for higher-affinity cells that are dividing faster because of their having received a better quality of T cell help (Gitlin et al., 2014, 2015). However, we failed to find evidence for such a mechanism because similar frequencies of NP-specific MHCIIhigh and MHCIIlow centroblasts carried the affinity-conferring W33L mutation (Fig. 6 a). Although the CXCR4high MHCIIlow GC B cell populations did contain marginally higher frequencies of cells with the W33L mutation relative to CXCR4high MHCIIlow cells from the same mice, the differences were very small (54% vs. 45%), and the MHCIIlow populations were far from being entirely made up of the high-affinity cells. As an alternative explanation, we examined the possibility that MHCII proteins may be preferentially down-regulated at later stages of the centroblast program. This could result in the observed correlation between MHCII levels and recent replicative history because late stage centroblasts would be temporally further from the stalling in cell division that occurs during the centrocyte-associated selection checkpoint. To begin testing this possibility, we took advantage of the anti–DEC205-OVA antigen delivery system as this allowed us to synchronize selection in a population of GC B cells (Victora et al., 2010). Two populations of allotypically distinguishable NP-specific (B1-8) B cells, on Dec205+/+ and Dec205−/− backgrounds, were transferred into Dec205−/− hosts that had previously been preimmunized with OVA in alum 2–4 wk earlier (Fig. 6 b). Dec205−/− hosts expressed a human DEC205 transgene under the control of the Cd11c promoter because this had been found to prevent the rejection of the Dec205+/+ donor cells but is not recognized by the anti–DEC205-OVA antibody (Pasqual et al., 2015). Recipient mice were then reimmunized with NP-OVA to drive the B1-8 B cells into GCs and later received the chimeric anti–DEC205-OVA antibody containing the T cell antigen OVA at different time points before analysis. This results in augmented antigen presentation by the Dec205+/+ but not the Dec205−/− GC B cells (Victora et al., 2010).
Figure 6.
MHCII down-regulation in centroblasts is not determined by affinity and occurs at late times after T cell mediated selection. (a) CXCR4high MHCIIhigh and CXCR4high MHCIIlow NP-binding GC B cells, gated as in Fig. 5 a, were sorted on days 11 or 12 of the response to NP-CGG (blue circles) or NP-KLH (white circles) from WT C57BL/6 mice, and the frequency of W33L mutations in the VH186.2 heavy chain was determined. Linked pairs indicate populations from individual mice. (b) Dec205−/− mice were preimmunized with 50 µg OVA in alum (i.p.) 2 wk before the transfer of Dec205−/− and Dec205+/+ NP-specific B1-8 B cells, mixed at a 85:15 ratio. Mice were subsequently reimmunized with 25 µg NP-OVA via s.c. footpad injection. Mice then received a single injection of 5 µg anti–Dec205-OVA at 30- or 60-h time points before analysis to promote interactions with T cells by the Dec205+/+ GC B cells. (c) CD38low CD95+ GC B cells were gated. The surface levels of MHCII on CXCR4high CD83low centroblasts and CXCR4low CD83high centrocytes were determined for each population. Representative histograms are shown. (d) Summary of data from multiple mice and experiments, showing the MHCII geometric mean fluorescent intensity (gMFI) for each population, relative to follicular B cells. (e) The frequency of CXCR4high CD83low centroblasts for each population and time point. Data in d and e are compiled from two experiments with a total of three to four mice per condition (±SD). Analysis was performed using unpaired two-tailed Student’s t test. *, P < 0.05.
Analysis of GC B cells at an early time point after treatment (30 h) revealed that the Dec205+/+ CXCR4high CD83low centroblast population displayed uniformly high levels of surface MHCII, with staining intensities on centroblasts and centrocytes being almost equivalent (Fig. 6, c and d). The effect was specific for the cells that bound the anti–DEC205-OVA chimeric antibody. The phenotype of the centroblasts changed over time after anti–DEC205-OVA treatment, however, with the Dec205+/+ CXCR4high CD83low GC B population returning to become more like the Dec205−/− cells by the late 60-h time point in terms of their MHCII levels. Importantly, 60 h is a time point at which the Dec205+/+ centroblast population is still expanded, presumably indicating that the majority of these cells have not yet transitioned back to become centrocytes (Fig. 6 e; Gitlin et al., 2014). When considered alongside the BrdU labeling and H2b-GFP dilution studies, these experiments suggest that MHCII may be high on centroblasts at periods immediately after selection but that GC B cells may be hardwired to transition between MHCIIhigh and MHCIIlow states at later stages of their centroblast program. In addition, this finding further supports the conclusion that MHCII down-regulation in a subset of centroblasts is unlikely the direct result of these cells having a higher affinity for the cognate antigen. We speculate that late-stage centroblasts may preferentially degrade their preexisting pMHCII complexes before moving back to the light zone, where they will test their newly mutated BCRs.
MHCII ubiquitination is required to prevent accumulation of pMHCII complexes in GC B cells and for optimal responses after influenza A infection
To directly ask whether the ubiquitination and subsequent degradation of MHCII complexes in GC B cells impacts antigen presentation, we intercrossed the I-Ab(K>R) mice with the Hy10 mouse line that carries knock-in and transgenic immunoglobulin heavy and light chains, respectively, that are specific for hen egg lysozyme (HEL; Allen et al., 2007). Small numbers of naive I-Ab(K>R) Hy10 B cells were transferred together with B cells from WT Hy10 mice and OVA-specific CD4+ T cells (OT-II) into B6-CD45.1 mice (Fig. 7 a). Recipient animals were immunized with HEL-OVA in adjuvant to induce GCs containing the donor cells; mutant and WT Hy10 B cells were distinguishable from each other and from the host by congenic CD45 alleles. 5–7 d after immunization, mice received a s.c. injection of HEL-EaGFP, a conjugate composed of cognate antigen (HEL) and a well characterized peptide (Ea52–68) that can be detected by antibody staining when presented in complex with I-Ab (Itano et al., 2003; Wang et al., 2014). This antigen conjugate also contains GFP, which provides a readout of the amount of antigen captured but does not depend on processing. Control mice received HEL coupled to BSA. Antigen presentation by WT and K>R Hy10 GC B cells was assessed on day 7 after immunization at times corresponding to 4.5, 24, and 48 h after the delivery of the HEL-EaGFP conjugate.
Figure 7.
Increased accumulation of pMHCII complexes in the absence of MHCII ubiquitination. (a) WT and I-Ab(K>R) HEL-specific Hy10 B cells were transferred, together with OT-II CD4+ T cells, into WT hosts. Recipient mice were subsequently immunized with HEL-OVA/Sigma (Ribi) adjuvant by s.c. injection. Mice later received HEL-EaGFP or HEL-BSA conjugates at the same sites at various time points before analysis. (b) Representative plots showing GFP florescence and I-Ab–Ea52–68 staining on the two populations of IgDlow CD95+ GC B cells. (c, left) GFP florescent intensity on each genotype of GC B cell at each time point. (right) I-Ab(Ea52–68) mean fluorescence intensity (MFI) with mean background staining from HEL-BSA treated mice (4.5-h time point) deducted to give the delta MFI for each condition and time point. The fold differences in delta MFI between I-Ab(K>R) and WT Hy10 cells are indicated above the plots. Each data point represents GC B cells from a single iLN, with two LNs analyzed per mouse. Data in c are from one experiment that is representative of two (48 h) and three (4.5 or 24 h) independent experiments. (d) The fold differences in delta MFI between WT and I-Ab(K>R) Hy10 GC B cells from all mice and experiments are shown. Horizontal lines in c and d indicate means. Analysis was performed using an unpaired two-tailed Student’s t test. *, P < 0.05; ***, P < 0.001.
I-Ab–Ea52–68 staining was observed on both WT and K>R GC B cell populations by 4.5 h after HEL-EaGFP injection (Fig. 7, b and c). I-Ab(K>R) GC B cells had slightly higher background staining, presumably reflecting their raised MHCII levels. However, Ea52–68 peptide presentation also appeared to be marginally increased in the I-Ab(K>R) cells; the relative florescent intensity was 1.3× greater even after the mean background staining intensity was deducted (Fig. 7 c). As expected, the gathering of antigen as determined by GFP intensity was not affected by the I-Ab(K>R) mutation at any of the time points (Fig. 7, b and c). By 24 h, both the I-Ab–Ea52–68 staining intensity and the difference in staining between mutant and WT cells had increased (1.5× greater in I-Ab(K>R) cells), indicating that I-Ab(K>R) GC B cells accumulated more pMHCII during this time (Fig. 7 c). The difference between WT and I-Ab(K>R) cells became even greater (2.2×) at 48 h because of I-Ab(K>R) GC B cells retaining appreciable amounts of the pMHCII complex at a time point when it had mostly been lost in WT cells. Findings from multiple experiments are summarized in Fig. 7 d. These findings suggest that the presentation of the Ea52–68 peptide by GC B cells occurs at least as efficiently in the absence of MHCII ubiquitination, but that pMHCII complexes accumulate for longer periods in this setting. This is likely to in turn impact the repertoire of peptides presented by GC B cells and may lead to cells presenting antigens for extended times after they have been gathered.
Given the apparent impact that MHCII ubiquitination has on antigen presentation by GC B cells, we next investigated whether ubiquitination of this protein is needed for B cells to compete effectively in a virally induced GC response. Mixed BM chimeras containing I-Ab(K>R) and CD45.1+ WT cells were infected with the HKx31 strain of influenza and the GC responses by the two cell types were tracked. We examined the frequency in the draining mediastinal LN of each genotype of cells within the GC B cell compartment and within the naive population from which they arise (Fig. 8). These experiments revealed I-Ab(K>R) B cells to be similarly efficient to their WT counterparts at receiving the necessary activation signals to allow them to seed the GC response. However, these experiments indicated a need for MHCII ubiquitination in GC B cells for optimal long-term responses because I-Ab(K>R) CD45.2+ cells were out-competed by their neighbors by 5 wk after infection. Together, these experiments indicate that ubiquitination of MHCII by GC B cells helps prevent the accumulation of old pMHCII complexes at their surface and facilitates effective responses during a viral infection.
Figure 8.
Reduced competitiveness of GC B cells when ubiquitin-mediated MHCII regulation is defective. (a and b) Mixed BM chimeric mice containing CD45.1+ and either I-Ab(K>R) or WT CD45.2+ cells were infected with influenza HKx31. The frequency of naive or GC B cells of CD45.2+ donor origin in mediastinal LNs was assessed on days 7 or 8 (a) and after 5 wk (b). Lines connect data from individual mice, and plots summarize data pooled from three experiments. Analysis was performed using an unpaired two-tailed Student’s t test. ***, P < 0.001.
DISCUSSION
The iterative and repetitive nature of antibody affinity maturation poses unique challenges for GC B cells. GC B cells are constantly switching back and forth between periods of mutating their immunoglobulin variable region genes and then undergoing selection based on the specificity of the new BCR generated through this process. Cells with higher affinity receptors capture and present more antigen in complex with MHCII to Tfh cells (Batista and Neuberger, 2000; Allen et al., 2007; Victora et al., 2010), yet how GC B cells ensure that this selection process is not influenced by older pMHCII complexes that were formed before the most recent change in receptor specificity is not known. Previous studies looking at MHCII turnover in other cell types have suggested these complexes have long half-lives, with estimates ranging from 6 to 150 h depending on the cell lineage, conditions, and mouse strain (Cella et al., 1997; Pierre et al., 1997; Lazarski et al., 2005; De Riva et al., 2013). However, a uniformly long half-life in GC B cells seems unlikely to be compatible with their need to frequently test new BCRs when up to 50% of GC B cells transition from the dark zone to the light zone for selection every 4–6 h (Victora et al., 2010). Equally, a very short protein half-life might not fit with the need of GC B cells to efficiently present peptides during times of limiting antigen availability, such as after an infection has been cleared. This led us to investigate whether GC B cells may differentially regulate the degradation and stabilization of MHCII complexes at different times within their cellular program to promote replacement and selection, respectively.
Consistent with the proposed hypothesis, individual centroblasts differ widely in their surface MHCII levels, with some cells displaying far lower levels than others. A similar MHCIIlow subset is present in human tonsil samples, indicating evolutionary conservation (Glazier et al., 2002; Victora et al., 2012). Through the generation and ectopic expression of a photoconvertible chimeric mKikGR–I-Ab protein, we have shown that MHCII degradation rates differ significantly within different GC subsets. MHCII protein turnover was fastest in the MHCIIlow centroblast population, providing a likely explanation for why they have low surface MHCII levels despite having similar levels of MHCII mRNA transcripts to the MHCIIhigh centroblasts. In unpublished experiments, we confirmed that the cellular abundance of the chimeric protein was subject to the same ubiquitin- and CD83-dependent regulatory processes as WT MHCII, indicating that the fluorophore attachment is not changing MHCII β chain behavior. This is in line with findings from published GFP–I-Ab chimeric protein studies (Boes et al., 2002; Chow et al., 2002; McGehee et al., 2011). Unlike in DCs, where endocytosed antigens may be loaded onto new or recycling MHCII proteins, BCR-captured antigens are thought to be preferentially loaded onto newly synthesized molecules (Forquet et al., 1999; Clark et al., 2004). Therefore, the lysosomal degradation of existing MHCII molecules provides the opportunity for replacement by newly formed complexes. As GC B cells transition back to the centrocyte stage, they enhance Ciita and MHCII gene expression to provide a burst of freshly synthesized protein that coincides with the timing of antigen acquisition through the newly mutated BCRs (Victora et al., 2012). Whether or not additional changes occurring in GC B cells, such as decreases in the H-20/H-2M ratio, contribute to determining the stability of specific pMHCII complexes remain important questions for future studies (Pathak et al., 2001; Glazier et al., 2002).
An important finding from our study is that similar mechanisms to those controlling MHCII abundance in DCs determine its regulation in GC B cells. In DCs, changes in MHCII posttranslational regulation contribute to ensuring that CD4+ T cell responses are primed only after the sensing of infection-associated danger signals. Then, upon toll-like receptor stimulation, DCs undergo a process of maturation that involves decreases in rates of MHCII ubiquitination and lysosomal degradation that lead to higher surface protein levels and improved antigen presentation capacity (Shin et al., 2006). Our findings suggest that similar, but reversible, changes occur in GC B cells as they transition between centroblast and centrocyte states. In GC B cells, as in immature DCs and follicular B cells, a single E3 ligase (March1) promotes MHCII degradation through the addition of ubiquitin moieties to a conserved lysine in the MHCII β chain (Matsuki et al., 2007; De Gassart et al., 2008; Walseng et al., 2010). In the absence of either March1 or the substrate lysine, MHCII levels were uniformly high in all GC B cells regardless of their centroblast/centrocyte states. Stepwise increases in March1 transcript abundance across the three CB subsets contrasted with the uniform up-regulation of Cxcr4 transcripts. Future studies will need to discern whether this reflects different modes of transcriptional induction or differential control of transcript stability.
A further similarity between DCs and GC B cells was the involvement of CD83 in controlling MHC abundance (Tze et al., 2011). CD83 is a transmembrane protein that has become a key identifying marker of centrocytes (Victora et al., 2010); we found that its expression in this stage contributes to stabilizing MHCII complexes. However, although increases in CD83 expression were important for enhancing MHCII levels in centrocytes, MHCII down-regulation in centroblasts does not occur only because of decreases in CD83. Instead, we detected increases in March1 expression in centroblasts that correlated with MHCII down-regulation, suggesting opposing pro-stabilization and pro-turnover mechanisms exist in centrocytes and centroblasts, respectively. Surprisingly, Cd83−/− centrocytes had slightly lower surface MHCII levels than did Cd83−/− MHCIIhigh centroblasts despite both populations expressing similar levels of March1 transcripts and centrocytes having slightly raised MHCII mRNA levels. This observation might indicate the involvement of additional forms of March1-mediated regulation, such as those that determine ubiquitin chain length and protein fate in DCs versus naive B cells (Ma et al., 2012), or instead, the differences could be related to the temporal order in which opposing CD83/March1 changes occur. The findings that CD83 itself appears to be a target of March1, that March1 autoubiquitinates and that March1 protein has a very short half-life together suggest that feedback regulatory loops may contribute to the dynamic nature of MHCII fluctuations in GC B cells (Jabbour et al., 2009; Tze et al., 2011; Bourgeois-Daigneault and Thibodeau, 2012).
It has become increasingly evident in recent years that centroblast populations contain significant heterogeneity in terms of clones that vary in their behavior because of differences in affinity and because of the existence of individual GC B cells at different stages of their centroblast program (Bannard et al., 2013; Gitlin et al., 2014; McHeyzer-Williams et al., 2015). Differences in MHCII abundance add to this heterogeneity. We investigated a possible correlation between MHCII levels and BCR affinity but observed only modest differences in the frequency of affinity-conferring mutations in MHCIIhigh and MHCIIlow subsets, suggesting that the differences in amounts of T cell help received are not the main determinant of variations in MHCII levels. Our cell cycle analysis indicated that MHCIIlow centroblasts are less likely to be in G2/M phases of cell cycle than their MHCIIhigh counterparts, but equally or more likely to have recently divided, suggesting that movement between states probably occurs, i.e., centroblasts may move from MHCIIhigh to MHCIIlow states soon after cellular division. This finding also suggests that cells may not be likely to reach the MHCIIlow state immediately after the centrocyte to centroblast selection checkpoint because mitosis is mostly restricted to centroblasts (Allen et al., 2007; Victora et al., 2010). Furthermore, MHCIIlow cells were underrepresented within GC B cell subsets that had divided the least number of times during a 40-h window. We therefore investigated whether any correlation existed between the timing since a cell has interacted with T cells and MHCII down-regulation. Strikingly, MHCII levels were high on centroblasts at early time points after the augmentation of T cell help but decreased later. Together, these findings raise the possibility that the MHCIIlow state may be reached late in the centroblast program as a result of increases in March1 expression after significant cellular division has occurred. However, new experimental approaches and further experimentation are needed to definitively determine whether this hypothesis is correct.
We have proposed that the destabilization of MHCII proteins in centroblasts helps purge cells of old complexes and thereby contributes to ensuring that centrocytes are selected only based on recently acquired antigens. According to such a model, increased MHCII ubiquitination in centroblasts at certain times promotes the degradation of pMHCII complexes that were processed before the most recent somatic hypermutation events, preventing them influencing subsequent B–T interactions. This would enhance the pMHCII signal/noise ratio, but also possibly prevent GC B cells being inappropriately selected if they have recently lost antigen-binding ability (or have recently acquired self-reactivity) as the result of them still displaying older pMHCII complexes. In the absence of such regulation, the temporary retention in the response of certain clones could cause delays in affinity maturation that would ultimately manifest in competition assays. Consistent with this model, I-Ab(K>R) cells showed a competitive defect within influenza-induced GCs. This occurs despite I-Ab(K>R) cells having higher MHCII levels which could otherwise enhance participation by promoting T–B interactions. In considering this model, it is important to note that ubiquitin-mediated mechanisms are not the only ways by which old pMHCII complexes are degraded or lost in GC B cells; MHCII proteins do not accumulate exponentially in its absence. Background degradation and dilution via cellular division also play roles; GC B cells may undergo between one and six cell divisions between selection events (Gitlin et al., 2014; Cho et al., 2015). Temporal March1-mediated MHCII purging may therefore help refine rather than dictate the pMHCII repertoire when favoring the presentation of recently acquired antigens. It is likely that such a mechanism will be more important in some settings than others; for example, its roles might be more evident during complex infections involving multiple antigens or in periods of limiting antigen availability. In line with this reasoning, we did not see any significant effect of the K>R mutation on the accumulation of affinity conferring W33L mutations during NP-protein conjugate responses (unpublished data), likely reflecting the simple nature of this antigen.
Although our findings are consistent with the proposed model, the accumulation of pMHCII complexes and defects in competitive fitness by I-Ab(K>R) GC B cells during influenza infection are correlative. It is therefore important to note that other explanations for why centroblasts increase March1 activity and decrease their MHCII levels remain possible. For example, in continuing the analogy with DCs, it may be that MHCII down-regulation contributes to sheltering subsets of GC B cells from T cell engagement at particular times. This would contribute to preventing the aberrant or premature selection of GC B cells during periods immediately after somatic hypermutation and before them being ready for optimal T cell interactions. The spatial separation of centroblasts and centrocytes in dark zones and light zones probably contributes to achieving this, but further efficiency may be provided through this mechanism (Bannard et al., 2013). In this regard, it is of interest that CD86 levels are synchronously regulated with MHCII in GC B cells. An intrinsic requirement for CD86 in GC B cells has been difficult to prove or disprove; however, its expression in B cells is required at earlier activation stages, and the blocking of this pathway can impact GC responses even after they are established (Salek-Ardakani et al., 2011). Therefore, MHCII and CD86 increases together may promote optimal selection at particular periods. Alternatively, fluctuations in MHCII ubiquitination may enhance the processing and presentation of certain types of antigens at particular times through yet unknown mechanisms (Walseng et al., 2010). We also cannot rule out at this time that MHCII ubiquitination in GC B cells enhances GC B cell responses by orchestrating the localization of the protein within the cell (Shin et al., 2006; Ma et al., 2012).
In summary, we have identified that the turnover and surface levels of MHCII are dynamically regulated in GC B cells due to fluctuations in March1 activity. We propose that temporal increases in MHCII degradation occur as a consequence of the centroblast cellular program and contribute to effective GC responses.
MATERIALS AND METHODS
Mice, immunizations, and infections
I-Ab(K>R), March1−/−, Cd83−/−, and Hy10 mice have been described previously (Fujimoto et al., 2002; Allen et al., 2007; Matsuki et al., 2007; Oh et al., 2013). I-Ab(K>R) mice were provided by I. Mellman (Genentech, South San Francisco, CA). Tet-off H2bGFP mice were generated by crossing intercrossing TetOP-H2B-GFP, ROSA:LNL:tTA, and Mb1-Cre mice (Hobeika et al., 2006; Wang et al., 2008; Foudi et al., 2009). To generate mixed BM chimeric mice, C57BL/6 or B6-CD45.1+ mice were lethally irradiated with a split dose of 1,100 rads γ-irradiation and then i.v. injected with the relevant mixture of BM cells. Mice were sometimes treated with Baytril for 4 wk after irradiation. Reconstituted mice were rested for >8 wk before use in experiments. In mixed BM chimera experiments, the degree of reconstitution by the two types of donor BM cells varied with each mixture, possibly as a result of cell counting variability or differences in the proportions of mature and hematopoietic stem cells in the donor BM. Animals were housed in specific pathogen–free enclosures at the University of Oxford Biomedical Sciences facility and at the University of California, San Francisco (UCSF) Laboratory Animal Research Center. All UK-based experiments were approved by a project license granted by the UK Home Office and were also approved by the Institutional Animal Ethics Committee Review Board at the University of Oxford, and all US based experiments complied with animal protocols that were approved by the UCSF Institutional Animal Care and Use Committee.
Mice were immunized with 2 × 108 SRBCs (Colorado serum) by i.p. injection on day 0 or with 2 × 107 SRBCs on day 0 and 2 × 108 SRBCs on day 4 to induce large GCs on days 10–12 for FACS sorting experiments (Dominguez-Sola et al., 2012); NP sequencing experiments involved immunizations with 100 µg NP20–29-CGG or NP32-KLH precipitated in equal volumes of alum (200 µl total; Alhydrogel; BRENNTAG Nordic). iLN GCs were induced by s.c. immunization with 40 µg NP20–29-CGG in alum and split between two sites, one each side of the LN. For Hy10 experiments, mixtures of 105 HEL-binding B cells and 105 OT-II T cells were transferred by i.v. injection 24 h before immunization. GCs were induced by immunizing with 50 µg HEL-OVA in an equal volume of Sigma-Aldrich adjuvant system (200 µl total; Allen et al., 2007). The EaGFP construct was provided by M. Jenkins (University of Minnesota, Minneapolis, MN). Hel-EaGFP was generated as described previously (Itano et al., 2003), whereas Hel-BSA was conjugated similarly but not purified. A total of 10 µg Hel-EaGFP was delivered per LN by s.c. injection, split between two sites. Two experiments were discarded because of I-Ab(Ea52–68) antibody-staining signals being below the level of detection as late time points (relative to background). Background staining by the Y-ae antibody has been observed in earlier studies (Viret and Janeway, 2000; Oh et al., 2013). For BrdU experiments, mice were given a single i.p. injection of 2.5 mg BrdU (Sigma-Aldrich) in saline. For double-labeling experiments, mice received 1 mg EdU i.p. followed by 2.5 mg BrdU i.p. at the indicated time point. For tet-off experiments, mice received a single i.p. injection of 1.6 mg doxycycline (Sigma-Aldrich) in saline and then maintained by including doxycycline (2 mg/ml) and sucrose (2%) in their drinking water.
For Dec205-OVA experiments, Dec205−/− CD45.2+ mice engineered to express human Dec205 (Cd11c-hDEC205) were primed by immunization with 50 µg OVA (Sigma-Aldrich) precipitated in alum (Thermo Fisher Scientific) 2 wk before the adoptive transfer of the indicated B1-8 populations (Pasqual et al., 2015). Dec205−/− and Dec205+/+ B1-8 cells were distinguishable by their CD45.1/2 and CD45.1 allotypic markers, respectively. A total of 5 × 105 B1-8 B cells were given. 1 d later, mice were reimmunized by s.c. footpad injection of 50 µg NP-OVA. Mice later received 5 µg anti–DEC205-OVA by s.c. footpad injection at the indicated times (Victora et al., 2010). Popliteal LNs were harvested at the indicated times.
Hkx/31 influenza was propagated and titred in MDCK cells. Anaesthetized mice were infected with 2 × 104 PFU virus in 30-µl volume by intranasal inhalation.
Retroviral constructs and transduction
The I-Ab–mKiKGR construct was generated by fusing a human codon–optimized mKiKGR gene in frame to the 3′ of I-Ab via a short flexible linker, GGGAAS, using overlap PCR. The chimeric gene was subcloned into the MSCV2.2 retrovirus followed by an internal ribosomal entry site and Thy1 expression marker. For transduction of BM, donor mice were injected with 3 mg 5-fluorouracil (Sigma-Aldrich) 4 d before BM collection. BM cells were plated for 20–24 h in DMEM containing 15% FBS (vol/vol), 10 mM Hepes (Cellgro), 50 IU/ml penicillin, 50 µg/ml streptomycin (Cellgro), and glutamine and supplemented with IL-3, IL-6, and stem cell factor (concentrations of 20, 50 and 100 ng/ml; PeproTech). Cells were spin-fected twice on consecutive days in the presence of 1,5-dimethyl-1,5-diazaundecamethylene polymethobromide (Sigma-Aldrich) and then transferred into irradiated C57BL/6 or B6-Ly5.2 mice. For analysis of reconstituted mice, Thy1.1bright cells were gated.
mKikGR photoconversion
Survival surgeries were performed with proper anesthetic, sterile technique, and analgesia. They were approved by and in accordance with the UCSF Institutional Animal Care and Use Committee guidelines. A mouse was anesthetized with isoflurane, shaved, and antiseptically prepared with 0.02% chlorhexidine gluconate. Mouse body temperatures were maintained by holding on an electric heated mat (Walgreens), and mice received appropriate pre- and post-surgical analgesia (0.1 mg/kg Buprenorphrine i.p. or s.c. and 8 mg/kg Bupivacaine local treatment). iLNs were surgically exposed through the making of an incision ∼2 cm in size on the underside of the abdomen. Skin around the LN was pinned back, and the integrity of the site was maintained by continuous bathing with warm saline. A Silver LED 415 (Prizmatix), set to maximum intensity, with a high numerical aperture polymer optical fiber (1.5-mm core diameter) light guide and fiber collimator, was used as a 415-nm violet light source. The light source was placed over the LN for a total of 10.5 min. The surgical site was closed using Autoclips (Thermo Fisher Scientific).
Flow cytometry
To analyze mice, single-cell suspensions were generated using a 70-µM strainer and cells were stained using empirically determined concentrations of antibodies for 25 min on ice. The antibodies used are listed in Table S1. To obtain samples for RNA analysis, cells were FACS sorted using a FACSAria II (BD). Sorted GC B cells populations were identified by B220+, IgDlow, CD95+, and GL7+ gating. Flow cytometry data were analyzed using FlowJo software (Tree Star). For BrdU experiments, antibody-stained cells were fixed and permeabilized with the BrdU Flow kit (BD) according to manufacturer’s instructions. DAPI (BioLegend) was added at a final concentration of 5 µM shortly before analysis, and samples were run on LOW setting. For EdU/BrdU double labeling experiments, EdU was stained using the Click-it plus EdU flow kit (Thermo Fisher Scientific), and BrdU was detected using the Mobu-1 antibody clone that does not cross-react with EdU.
Measurements of gene expression
Total RNA was isolated using an RNeasy kit and in-column DNA digestion according to the manufacturer’s instructions (QIAGEN). Gene expression levels were measured by real-time PCR using SYBR green PCR mix (Roche) on an ABI Prism 7300 sequence detection system (Applied Biosystems). Values were normalized to those of a housekeeping gene, Hprt. The primers used are listed in Table S2.
Immunoglobulin variable region sequencing
DNA was extracted from a few hundred to a few thousand sorted splenic GC B cells using the QIAamp DNA micro kit (QIAGEN) and eluted in 55 µl of water (majority of samples contained ∼2,000 cells). 50 µl DNA was then used as template for nested PCR using the following primers: PCR1 For, 5′-CATGGGATGGAGCTGTATCATGC-3′; PCR1 Rev, 5′-CTCACAAGAGTCCGATAGACCCTG-3′; PCR2 For, 5′-GGTGACAATGACATCCACTTTGC-3′; and PCR2 Rev, 5′-GACTGTGAGAGTGGTGCCTTG-3′. PCR1 was performed in 100 µl total volume with 25 cycles (98°C 30 s, 56°C 30 s, 72°C 90 s), followed by 5 min at 72°C. 5 µl of the product was used as template for the nested reaction with 30 cycles with a reduced elongation time of 30 s, in a total volume of 50 µl. Phusion high fidelity polymerase (New England Biolabs, Inc.) and the HF buffer were used throughout. Water controls were carried through all stages to ensure no contamination. Bands of approximately 400 bp were cut from agarose gels and purified using QIAquick gel extraction kit (QIAGEN). DNA was either ligated into blunt end TOPO (Thermo Fisher Scientific) and sent for colony sanger sequencing (NP-CGG samples) or MiSeq library preps were made (NP-KLH samples).
MiSeq library preps were generated using the NEBNext Ultra kit and paired-end barcoded adaptors (New England Biolabs, Inc.) as per the manufacturer’s protocol. MiSeq sequencing was performed using 8 pM of the library and the v2 2 × 250 kit, with 5% PhiX spike-in (Illumina) to provide nucleotide diversity. 20 barcoded samples were run on a single flow cell.
Sanger sequencing was analyzed by manually inspecting sequencing traces for quality and then trimming the ends according the region of interest and length. IGHV1-72*01 members were selected using IMGT V-QUEST and taken forward in analysis. Mutation frequencies and amino acid changes were identified using the SAIVGEM package (Messmer, 2005). MiSeq analysis was performed by first merging paired-end reads using PANDAseq (Masella et al., 2012). Sequences were then ordered according to quality score, and the top 149,000 sequences for each sample were taken forward for further analysis. IGHV1-72*01 members were identified using IMGT/HighV-QUEST (Alamyar et al., 2012). Output files containing unique sequences were dumped because of them likely occurring from sequencing errors. Sequences were aligned to the template using EMBOSS tool “water.” Amino acid changes in non–frame-shifted sequences were counted for each position in the IgH V region, and the frequency of W33L mutations was determined.
Online supplemental material
Table S1 lists the antibodies used for FACS staining. Table S2 lists the primers used for RT-PCR reactions. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151682/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. An and Y. Xu for expert technical assistance, A. Reboldi for help harvesting BM cells, C. Waugh in the flow cytometry facility at the Weatherall Institute of Molecular Medicine (WIMM) for cell sorting services, and I. Mellman (Genentech) for I-Ab(K>R) mice.
O. Bannard is a Sir Henry Dale Fellow (Wellcome Trust/Royal Society). J.G. Cyster is an Investigator of the Howard Hughes Medical Institute. This work was supported in part by National Institutes of Health grants AI45073 (to J.G. Cyster) and 5DP5OD012146 (to G.D. Victora), the Medical Research Council (MRC) through core funding to the MRC Human Immunology Unit and WIMM (to O. Bannard and S.J. McGowan), and by Wellcome Trust grant 105654/Z/14/Z (to O. Bannard).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BCR
- B cell receptor
- GC
- germinal center
- HEL
- hen egg lysozyme
- iLN
- inguinal LN
- pMHCII
- peptide–MHCII
- SRBC
- sheep RBC
References
- Alamyar E., Duroux P., Lefranc M.-P., and Giudicelli V.. 2012. IMGT® tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 882:569–604. 10.1007/978-1-61779-842-9_32 [DOI] [PubMed] [Google Scholar]
- Allen C.D.C., Okada T., Tang H.L., and Cyster J.G.. 2007. Imaging of germinal center selection events during affinity maturation. Science. 315:528–531. 10.1126/science.1136736 [DOI] [PubMed] [Google Scholar]
- Bannard O., Horton R.M., Allen C.D.C., An J., Nagasawa T., and Cyster J.G.. 2013. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 39:912–924. 10.1016/j.immuni.2013.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baravalle G., Park H., McSweeney M., Ohmura-Hoshino M., Matsuki Y., Ishido S., and Shin J.-S.. 2011. Ubiquitination of CD86 is a key mechanism in regulating antigen presentation by dendritic cells. J. Immunol. 187:2966–2973. 10.4049/jimmunol.1101643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista F.D., and Neuberger M.S.. 2000. B cells extract and present immobilized antigen: implications for affinity discrimination. EMBO J. 19:513–520. 10.1093/emboj/19.4.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boes M., Cerny J., Massol R., Op den Brouw M., Kirchhausen T., Chen J., and Ploegh H.L.. 2002. T-cell engagement of dendritic cells rapidly rearranges MHC class II transport. Nature. 418:983–988. 10.1038/nature01004 [DOI] [PubMed] [Google Scholar]
- Bourgeois-Daigneault M.-C., and Thibodeau J.. 2012. Autoregulation of MARCH1 expression by dimerization and autoubiquitination. J. Immunol. 188:4959–4970. 10.4049/jimmunol.1102708 [DOI] [PubMed] [Google Scholar]
- Calado D.P., Sasaki Y., Godinho S.A., Pellerin A., Köchert K., Sleckman B.P., de Alborán I.M., Janz M., Rodig S., and Rajewsky K.. 2012. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat. Immunol. 13:1092–1100. 10.1038/ni.2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M., Engering A., Pinet V., Pieters J., and Lanzavecchia A.. 1997. Inflammatory stimuli induce accumulation of MHC class II complexes on dendritic cells. Nature. 388:782–787. 10.1038/42030 [DOI] [PubMed] [Google Scholar]
- Cho K.-J., Walseng E., Ishido S., and Roche P.A.. 2015. Ubiquitination by March-I prevents MHC class II recycling and promotes MHC class II turnover in antigen-presenting cells. Proc. Natl. Acad. Sci. USA. 112:10449–10454. 10.1073/pnas.1507981112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A., Toomre D., Garrett W., and Mellman I.. 2002. Dendritic cell maturation triggers retrograde MHC class II transport from lysosomes to the plasma membrane. Nature. 418:988–994. 10.1038/nature01006 [DOI] [PubMed] [Google Scholar]
- Clark M.R., Massenburg D., Siemasko K., Hou P., and Zhang M.. 2004. B-cell antigen receptor signaling requirements for targeting antigen to the MHC class II presentation pathway. Curr. Opin. Immunol. 16:382–387. 10.1016/j.coi.2004.03.007 [DOI] [PubMed] [Google Scholar]
- De Gassart A., Camosseto V., Thibodeau J., Ceppi M., Catalan N., Pierre P., and Gatti E.. 2008. MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc. Natl. Acad. Sci. USA. 105:3491–3496. 10.1073/pnas.0708874105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Riva A., Varley M.C., Bluck L.J., Cooke A., Deery M.J., and Busch R.. 2013. Accelerated turnover of MHC class II molecules in nonobese diabetic mice is developmentally and environmentally regulated in vivo and dispensable for autoimmunity. J. Immunol. 190:5961–5971. 10.4049/jimmunol.1300551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D., Victora G.D., Ying C.Y., Phan R.T., Saito M., Nussenzweig M.C., and Dalla-Favera R.. 2012. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat. Immunol. 13:1083–1091. 10.1038/ni.2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forquet F., Barois N., Machy P., Trucy J., Zimmermann V.S., Leserman L., and Davoust J.. 1999. Presentation of antigens internalized through the B cell receptor requires newly synthesized MHC class II molecules. J. Immunol. 162:3408–3416. [PubMed] [Google Scholar]
- Foudi A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., and Hock H.. 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27:84–90. 10.1038/nbt.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y., Tu L., Miller A.S., Bock C., Fujimoto M., Doyle C., Steeber D.A., and Tedder T.F.. 2002. CD83 expression influences CD4+ T cell development in the thymus. Cell. 108:755–767. 10.1016/S0092-8674(02)00673-6 [DOI] [PubMed] [Google Scholar]
- Gitlin A.D., Shulman Z., and Nussenzweig M.C.. 2014. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 509:637–640. 10.1038/nature13300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin A.D., Mayer C.T., Oliveira T.Y., Shulman Z., Jones M.J.K., Koren A., and Nussenzweig M.C.. 2015. T cell help controls the speed of the cell cycle in germinal center B cells. Science. 349:643–646. 10.1126/science.aac4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier K.S., Hake S.B., Tobin H.M., Chadburn A., Schattner E.J., and Denzin L.K.. 2002. Germinal center B cells regulate their capability to present antigen by modulation of HLA-DO. J. Exp. Med. 195:1063–1069. 10.1084/jem.20012059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi S., Tsutsui H., Kochaniak A.B., Miyawaki A., and van Oijen A.M.. 2008. mKikGR, a monomeric photoswitchable fluorescent protein. PLoS One. 3:e3944 10.1371/journal.pone.0003944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., and Reth M.. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794. 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itano A.A., McSorley S.J., Reinhardt R.L., Ehst B.D., Ingulli E., Rudensky A.Y., and Jenkins M.K.. 2003. Distinct dendritic cell populations sequentially present antigen to CD4 T cells and stimulate different aspects of cell-mediated immunity. Immunity. 19:47–57. 10.1016/S1074-7613(03)00175-4 [DOI] [PubMed] [Google Scholar]
- Jabbour M., Campbell E.M., Fares H., and Lybarger L.. 2009. Discrete domains of MARCH1 mediate its localization, functional interactions, and posttranscriptional control of expression. J. Immunol. 183:6500–6512. 10.4049/jimmunol.0901521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarski C.A., Chaves F.A., Jenks S.A., Wu S., Richards K.A., Weaver J.M., and Sant A.J.. 2005. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 23:29–40. 10.1016/j.immuni.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Ma J.K., Platt M.Y., Eastham-Anderson J., Shin J.-S., and Mellman I.. 2012. MHC class II distribution in dendritic cells and B cells is determined by ubiquitin chain length. Proc. Natl. Acad. Sci. USA. 109:8820–8827. 10.1073/pnas.1202977109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella A.P., Bartram A.K., Truszkowski J.M., Brown D.G., and Neufeld J.D.. 2012. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 13:31 10.1186/1471-2105-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki Y., Ohmura-Hoshino M., Goto E., Aoki M., Mito-Yoshida M., Uematsu M., Hasegawa T., Koseki H., Ohara O., Nakayama M., et al. 2007. Novel regulation of MHC class II function in B cells. EMBO J. 26:846–854. 10.1038/sj.emboj.7601556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGehee A.M., Strijbis K., Guillen E., Eng T., Kirak O., and Ploegh H.L.. 2011. Ubiquitin-dependent control of class II MHC localization is dispensable for antigen presentation and antibody production. PLoS One. 6:e18817 10.1371/journal.pone.0018817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHeyzer-Williams L.J., Milpied P.J., Okitsu S.L., and McHeyzer-Williams M.G.. 2015. Class-switched memory B cells remodel BCRs within secondary germinal centers. Nat. Immunol. 16:296–305. 10.1038/ni.3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messmer B.T. 2005. SAIVGeM: spreadsheet analysis of immunoglobulin VH gene mutations. Biotechniques. 39:353–358. 10.2144/05393ST03 [DOI] [PubMed] [Google Scholar]
- Oh J., and Shin J.-S.. 2015. Molecular mechanism and cellular function of MHCII ubiquitination. Immunol. Rev. 266:134–144. 10.1111/imr.12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Wu N., Baravalle G., Cohn B., Ma J., Lo B., Mellman I., Ishido S., Anderson M., and Shin J.-S.. 2013. MARCH1-mediated MHCII ubiquitination promotes dendritic cell selection of natural regulatory T cells. J. Exp. Med. 210:1069–1077. 10.1084/jem.20122695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqual G., Angelini A., and Victora G.D.. 2015. Triggering positive selection of germinal center B cells by antigen targeting to DEC-205. Methods Mol. Biol. 1291:125–134. 10.1007/978-1-4939-2498-1_10 [DOI] [PubMed] [Google Scholar]
- Pathak S.S., Lich J.D., and Blum J.S.. 2001. Cutting edge: editing of recycling class II:peptide complexes by HLA-DM. J. Immunol. 167:632–635. 10.4049/jimmunol.167.2.632 [DOI] [PubMed] [Google Scholar]
- Pierre P., Turley S.J., Gatti E., Hull M., Meltzer J., Mirza A., Inaba K., Steinman R.M., and Mellman I.. 1997. Developmental regulation of MHC class II transport in mouse dendritic cells. Nature. 388:787–792. 10.1038/42039 [DOI] [PubMed] [Google Scholar]
- Salek-Ardakani S., Choi Y.S., Rafii-El-Idrissi Benhnia M., Flynn R., Arens R., Shoenberger S., Crotty S., Croft M., and Salek-Ardakani S.. 2011. B cell-specific expression of B7-2 is required for follicular Th cell function in response to vaccinia virus. J. Immunol. 186:5294–5303. 10.4049/jimmunol.1100406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.-S., Ebersold M., Pypaert M., Delamarre L., Hartley A., and Mellman I.. 2006. Surface expression of MHC class II in dendritic cells is controlled by regulated ubiquitination. Nature. 444:115–118. 10.1038/nature05261 [DOI] [PubMed] [Google Scholar]
- Tze L.E., Horikawa K., Domaschenz H., Howard D.R., Roots C.M., Rigby R.J., Way D.A., Ohmura-Hoshino M., Ishido S., Andoniou C.E., et al. 2011. CD83 increases MHC II and CD86 on dendritic cells by opposing IL-10–driven MARCH1-mediated ubiquitination and degradation. J. Exp. Med. 208:149–165. 10.1084/jem.20092203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., and Nussenzweig M.C.. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605. 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Dominguez-Sola D., Holmes A.B., Deroubaix S., Dalla-Favera R., and Nussenzweig M.C.. 2012. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 120:2240–2248. 10.1182/blood-2012-03-415380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viret C., and Janeway C.A. Jr. 2000. Functional and phenotypic evidence for presentation of Eα52–68 structurally related self-peptide(s) in I-Eα-deficient mice. J. Immunol. 164:4627–4634. 10.4049/jimmunol.164.9.4627 [DOI] [PubMed] [Google Scholar]
- Walseng E., Furuta K., Bosch B., Weih K.A., Matsuki Y., Bakke O., Ishido S., and Roche P.A.. 2010. Ubiquitination regulates MHC class II-peptide complex retention and degradation in dendritic cells. Proc. Natl. Acad. Sci. USA. 107:20465–20470. 10.1073/pnas.1010990107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sharma K., Deng H.-X., Siddique T., Grisotti G., Liu E., and Roos R.P.. 2008. Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiol. Dis. 29:400–408. 10.1016/j.nbd.2007.10.004 [DOI] [PubMed] [Google Scholar]
- Wang X., Rodda L.B., Bannard O., and Cyster J.G.. 2014. Integrin-mediated interactions between B cells and follicular dendritic cells influence germinal center B cell fitness. J. Immunol. 192:4601–4609. 10.4049/jimmunol.1400090 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.