Lambrecht et al. show that the transcription factor Zeb2 regulates commitment toward both the pDC and cDC2 lineages by repressing Id2.
Abstract
Plasmacytoid dendritic cells (DCs [pDCs]) develop from pre-pDCs, whereas two lineages of conventional DCs (cDCs; cDC1s and cDC2s) develop from lineage-committed pre-cDCs. Several transcription factors (TFs) have been implicated in regulating the development of pDCs (E2-2 and Id2) and cDC1s (Irf8, Id2, and Batf3); however, those required for the early commitment of pre-cDCs toward the cDC2 lineage are unknown. Here, we identify the TF zinc finger E box–binding homeobox 2 (Zeb2) to play a crucial role in regulating DC development. Zeb2 was expressed from the pre-pDC and pre-cDC stage onward and highly expressed in mature pDCs and cDC2s. Mice conditionally lacking Zeb2 in CD11c+ cells had a cell-intrinsic reduction in pDCs and cDC2s, coupled with an increase in cDC1s. Conversely, mice in which CD11c+ cells overexpressed Zeb2 displayed a reduction in cDC1s. This was accompanied by altered expression of Id2, which was up-regulated in cDC2s and pDCs from conditional knockout mice. Zeb2 chromatin immunoprecipitation analysis revealed Id2 to be a direct target of Zeb2. Thus, we conclude that Zeb2 regulates commitment to both the cDC2 and pDC lineages through repression of Id2.
DCs reside in almost all tissues in the body, where they function as immune sentinels. DCs can be subdivided into two main groups: plasmacytoid DCs (pDCs), which are specialized in the production of type I interferons to elicit an antiviral immune response (Swiecki and Colonna, 2015), and conventional DCs (cDCs), which are professional antigen-presenting cells. cDCs form the crucial link between the innate and adaptive arms of the immune system by sampling local antigens and subsequently migrating to their draining LNs, where they initiate appropriate responses from T cells (Merad et al., 2013). cDCs exist in two main subtypes, which were recently termed cDC1s and cDC2s (Guilliams et al., 2014). cDC1s, expressing XCR1 and usually CD103 and/or CD8α, are functionally specialized in cross-presentation of antigens to CD8+ T cells (Hildner et al., 2008; Bachem et al., 2010; Haniffa et al., 2012). cDC2s express CD172a (signal regulatory protein α [SIRPα]) and usually CD11b or CD4 and are functionally specialized in the presentation of antigens to CD4+ T cells (Merad et al., 2013).
DCs develop in the BM through a series of differentiation intermediates, each with a further degree of commitment to a specific lineage, that see the common myeloid progenitor (CMP) become a macrophage–DC progenitor (MDP) and then a common DC progenitor (CDP), with the penultimate step of differentiation resulting in generation of the pre-pDC and pre-cDCs (Naik et al., 2007; Liu et al., 2009; Merad et al., 2013; Onai et al., 2013). Furthermore, it has recently been shown that pre-cDCs can be further subdivided into pre-cDC1s and pre-cDC2s, which are committed to the cDC1 and cDC2 lineages, respectively (Grajales-Reyes et al., 2015; Schlitzer et al., 2015). Pre-pDCs develop into pDCs in the BM, which then migrate out to the periphery, whereas the pre-cDC subsets first migrate to the periphery and then undergo their final differentiation into cDCs (Liu et al., 2009; Onai et al., 2013; Schlitzer et al., 2015). The development of pDCs, cDC1s, and cDC2s requires the concerted action of several lineage-determining transcription factors (TFs). The main TF involved in pDC development and maintenance is the basic helix–loop–helix E protein E2-2 (Cisse et al., 2008; Ghosh et al., 2010), whereas cDC1s require Irf8, Batf3, and Id2 (Hacker et al., 2003; Tamura et al., 2005; Tailor et al., 2008; Edelson et al., 2011; Jackson et al., 2011; Grajales-Reyes et al., 2015). Notch2 (Lewis et al., 2011; Satpathy et al., 2013), Klf4 (Tussiwand et al., 2015), and more recently Irf4 (Bajaña et al., 2016) have been shown to be required for terminal differentiation into tissue-specific cDC2 subsets but do not seem to be involved in the early commitment toward the cDC2 lineage in the BM. Additionally, Irf4 has also been implicated in the survival of differentiated cDC2s, as well as in their migration to the LNs (Tamura et al., 2005; Persson et al., 2013b; Schlitzer et al., 2013; Williams et al., 2013). However, the TFs involved in the initial specification and commitment toward the cDC2 lineage have not yet been identified, and much remains unknown regarding how the decision to commit to a specific DC lineage is made (Murphy, 2013).
Zinc finger E box–binding homeobox 2 (Zeb2) is a TF primarily associated with the epithelial–mesenchymal transition, a process important in embryonic development, wound healing, and cancer progression (De Craene and Berx, 2013). However, Zeb2 has also been implicated in the development of the nervous system and is known to be required for normal mouse melanocyte differentiation and embryonic hematopoiesis (Goossens et al., 2011; Denecker et al., 2014; Hegarty et al., 2015). Although its role in the immune system is largely unknown, Zeb2 has recently been shown to be required for the maturation of NK cells (van Helden et al., 2015) and terminal differentiation of T cells (Dominguez et al., 2015; Omilusik et al., 2015). Zeb2 has also been identified in transcriptional analyses as a potential TF involved in DC development (Miller et al., 2012; Grajales-Reyes et al., 2015; Schlitzer et al., 2015). Here, we use mice either lacking or ectopically expressing Zeb2 in CD11c+ cells to examine the effects of manipulating Zeb2 expression in DCs. We demonstrate that Zeb2 is required for the development of pDCs and cDC2s in vivo, with Cd11cCrexZeb2fl/fl cells being more efficient at generating cDC1s, which is associated with a rise in the E protein inhibitor Id2. Reciprocally increasing the expression of Zeb2 resulted in a decrease in the cDC1 population with an associated increase in cDC2 development. Thus, Zeb2 is a previously uncharacterized key player in the regulation of the decision of DC precursors to commit to a specific DC lineage by mediating Id2 expression.
RESULTS AND DISCUSSION
High expression of Zeb2 beyond the CDP stage and in subsets of mature DCs
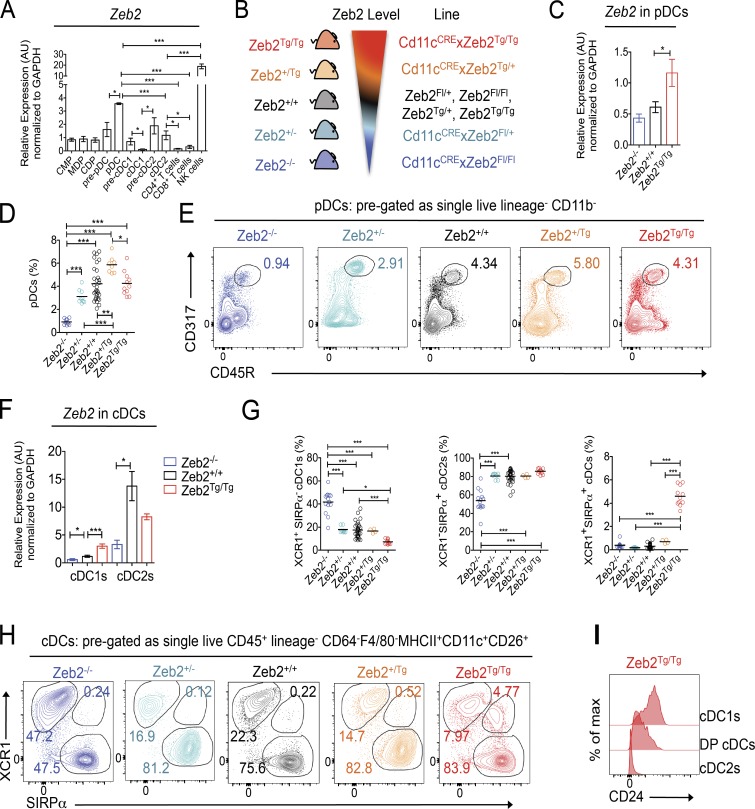
Zeb2 has recently been identified in several genome-wide microarrays and RNA-sequencing transcriptional analyses as a TF associated with cDC2 development and found to be expressed in pre-cDC2s and cDC2s (Miller et al., 2012; Schlitzer et al., 2015) but down-regulated in pre-cDC1s and cDC1s (Grajales-Reyes et al., 2015). To validate these results and determine at which stages in DC development Zeb2 could play a role, we FACS purified CMPs, MDPs, CDPs, and pre-pDCs from the BM and pDCs, pre-cDC1s, pre-cDC2s, cDC1s, and cDC2s from the spleen and performed RT–quantitative PCR (qPCR) analysis for Zeb2 expression. In agreement with published data, we found Zeb2 mRNA expression to be increased in pre-cDC2s and decreased in pre-cDC1s compared with CDPs (Fig. 1 A; Grajales-Reyes et al., 2015; Schlitzer et al., 2015). Strikingly, we also found Zeb2 to be up-regulated in pre-pDCs and pDCs, suggesting a role for this TF in pDCs (Fig. 1 A). As Zeb2 has recently been implicated in NK cell (van Helden et al., 2015) and T cell development (Dominguez et al., 2015; Omilusik et al., 2015), we also FACS purified NK cells and CD4+ and CD8+ T cells as a reference for Zeb2 expression levels. cDC2s and pDCs expressed lower levels of Zeb2 than NK cells but higher levels of Zeb2 than both T cell subsets (Fig. 1 A).
Figure 1.
Zeb2 expression levels regulate pDCs and cDCs. (A) RT-qPCR for Zeb2 expression in FACS-purified BM-derived CMPs, MDPs, CDPs, and pre-pDCs and splenic pDCs, pre-cDC1s, pre-cDC2s, cDC1s, cDC2s, CD4+ T cells, CD8+ T cells, and NK cells from WT mice. Results shown are expressed relative to GAPDH expression using the 2−ΔΔC(t) method with CMP set to 1. Data are pooled from two to three experiments, with at least n = 4 per cell type. Two-way Student’s t test was used between indicated groups. (B) Schematic showing range of Zeb2 levels and abbreviations for the transgenic mice lines used in the study. Zeb2+/+ = Zeb2fl/fl, Zeb2fl/+, Zeb2Tg/Tg, or Zeb2+/Tg; Zeb2+/− = Cd11cCrexZeb2fl/+; Zeb2−/− = Cd11cCrexZeb2fl/fl; Zeb2+/Tg = Cd11cCrexZeb2Tg/+; and Zeb2Tg/Tg = Cd11cCrexZeb2Tg/Tg. (C) Splenic pDCs were FACS purified from Zeb2−/−, Zeb2+/+, and Zeb2Tg/Tg mice, and Zeb2 levels were assessed by RT-qPCR. The results shown are expressed relative to GAPDH expression using the 2−ΔΔC(t) method with Zeb2+/+ pDCs set to 1. Data are pooled from two experiments, with at least n = 7 per group. Two-way Student’s t test was used. (D) Proportion of splenic pDCs as a percentage of live lineage−CD11b− cells in Zeb2+/+, Zeb2+/−, Zeb2−/−, Zeb2+/Tg, and Zeb2Tg/Tg mice. Data are pooled from two experiments with at least n = 8 per group. One-way ANOVA with Bonferroni posttest was used. (E) Representative FACS plots showing identification of CD317+CD45R+ pDCs in the spleen of Zeb2+/+, Zeb2+/−, Zeb2−/−, Zeb2+/Tg, and Zeb2Tg/Tg mice. Cells were pregated as single live lineage−CD11b− and were Ly6C+MHCIIintCD11cint. Numbers represent proportion of pDCs as a percentage of live Lin−CD11b− cells. (F) Splenic cDC1s and cDC2s were FACS purified from Zeb2+/+, Zeb2−/−, or Zeb2Tg/Tg mice, and Zeb2 levels were assessed by RT-qPCR. The results shown are expressed relative to GAPDH expression using the 2−ΔΔC(t) method with Zeb2+/+ cDC1s set to 1. Data are pooled from two to three experiments, with at least n = 7 per group. Two-way Student’s t test was used. (G) Proportion of splenic cDC1s, cDC2s, and DP cDCs as a percentage of total cDCs in Zeb2+/+, Zeb2+/−, Zeb2−/−, Zeb2+/Tg, and Zeb2Tg/Tg mice. Data are pooled from two to three experiments, with at least n = 4 per group. One-way ANOVA with Bonferroni posttest was used. (H) Representative FACS plots showing identification of XCR1+SIRPα− cDC1s, XCR1−SIRPα+ cDC2s, and XCR1+SIRPα+ DP cDCs in the spleen of Zeb2+/+, Zeb2+/−, Zeb2−/−, Zeb2+/Tg, and Zeb2Tg/Tg mice. Cells were pregated as single live lineage−CD26+CD11c+MHCII+F4/80−CD64−. Numbers represent proportion of cDC1s, cDC2s, and DP cDCs as a percentage of total cDCs. (I) Representative histogram showing CD24 expression by splenic cDC1s, cDC2s, and DP cDCs in Zeb2Tg/Tg mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent SEM. AU, arbitrary units.
Splenic pDCs and cDC2s are reduced in Cd11cCrexZeb2fl/fl mice, whereas splenic cDC1s are reduced in Cd11cCrexR26-Zeb2Tg/Tg mice
To examine whether Zeb2 expression has a functional role in cDCs and/or pDCs, we generated mice lacking or ectopically expressing either one or both alleles of Zeb2 in CD11c+ cells, including pre-cDCs, cDCs, and pDCs, by crossing Cd11cCre mice (Caton et al., 2007) with Zeb2fl/fl mice (Higashi et al., 2002) or with R26-Zeb2Tg/Tg mice (Tatari et al., 2014) to drive transgenic Zeb2 expression from the Rosa-26 promoter (Fig. 1 B). Analysis of Zeb2 mRNA levels in the splenic pDC populations of these mice revealed a slight down-regulation of Zeb2 in the Cd11cCrexZeb2fl/fl mice (Zeb2−/− mice) and a striking up-regulation of Zeb2 in the Cd11cCrexR26-Zeb2Tg/Tg mice (Zeb2Tg/Tg mice; Fig. 1 C and Fig. S1, A and B). Despite the only slight down-regulation of Zeb2 observed in the remaining pDCs in Zeb2−/− mice, there was a significant reduction in the pDC population in Zeb2−/− mice in the spleen and the blood (Fig. 1, D and E; and Fig. S1, C–E), suggesting Zeb2 expression is indeed down-regulated in pDCs. Interestingly, loss of only one allele of Zeb2 (Zeb2+/−) had only minimal effects on the pDC population (Fig. 1, D and E; and Fig. S1 D), demonstrating that although Zeb2 is a crucial TF for pDC homeostasis, haplo-sufficient expression is able to generate a normal pDC population. Increasing expression of Zeb2 in pDCs did not result in any significant changes in the proportion of pDCs, but we did observe an increase in terms of absolute cell numbers (Fig. 1, D and E; and Fig. S1 D). We hypothesize that this is because pDCs already express high levels of Zeb2 in steady state, and so the moderate increase in Zeb2 expression driven by the Rosa-26 promoter does not provide significant advantages to the cells.
Analysis of the cDC populations in Zeb2−/− and Zeb2Tg/Tg mice revealed a striking down-regulation of Zeb2 expression among cDC2s in Zeb2−/− mice and a significant up-regulation of Zeb2 expression among cDC1s in Zeb2Tg/Tg mice (Fig. 1 F and Fig. S2). Ablation of Zeb2 in cDC1s did not cause any significant decrease in Zeb2 expression, likely because WT cDC1 cells do not express Zeb2. There was also no significant increase in Zeb2 levels in cDC2s in Zeb2Tg/Tg mice (Fig. 1 F). Fitting with the high mRNA expression data for Zeb2 in cDC2s, we found that in addition to the severe reduction in pDCs, ablation of Zeb2 expression in CD11c+ cells resulted in a reduction in the proportion and absolute number of XCR1−SIRPα+ cDC2s (Fig. 1, G and H; and Fig. S3). Somewhat surprisingly, this reduction was coupled with an increase in both the proportion and absolute number of XCR1+SIRPα− cDC1s (Fig. 1, G and H; and Fig. S3). Similarly to the pDCs, this reduction in cDC2s was only apparent when both alleles of Zeb2 were targeted (Fig. 1, G and H; and Fig. S3). On the contrary, two alleles of the transgenic Zeb2 in CD11c+ cells led to a decrease in the proportion and number of splenic cDC1s, whereas the cDC2s were unaffected. As hypothesized with the pDCs, we believe that this is because the Rosa-26 promoter does not significantly increase the level of Zeb2 expression in this subset. Once again, we observed that only targeting one allele had limited effects on either subset (Fig. 1, G and H; and Fig. S3). Interestingly, the increased expression of Zeb2 in CD11c+ cells led to the presence of a new XCR1+SIRPα+ population of cDCs in the spleen (Fig. 1, G and H; and Fig. S3). To our knowledge, the presence of such a population has not previously been reported. These XCR1+SIRPα+ (double positive [DP]) cDCs also expressed intermediate levels of CD24, a marker typically associated with cDC1s (Fig. 1 I), suggesting these cells represent an intermediate subset between a cDC1 and a cDC2.
Zeb2 expression differentially affects subsets of tissue-resident cDC2s
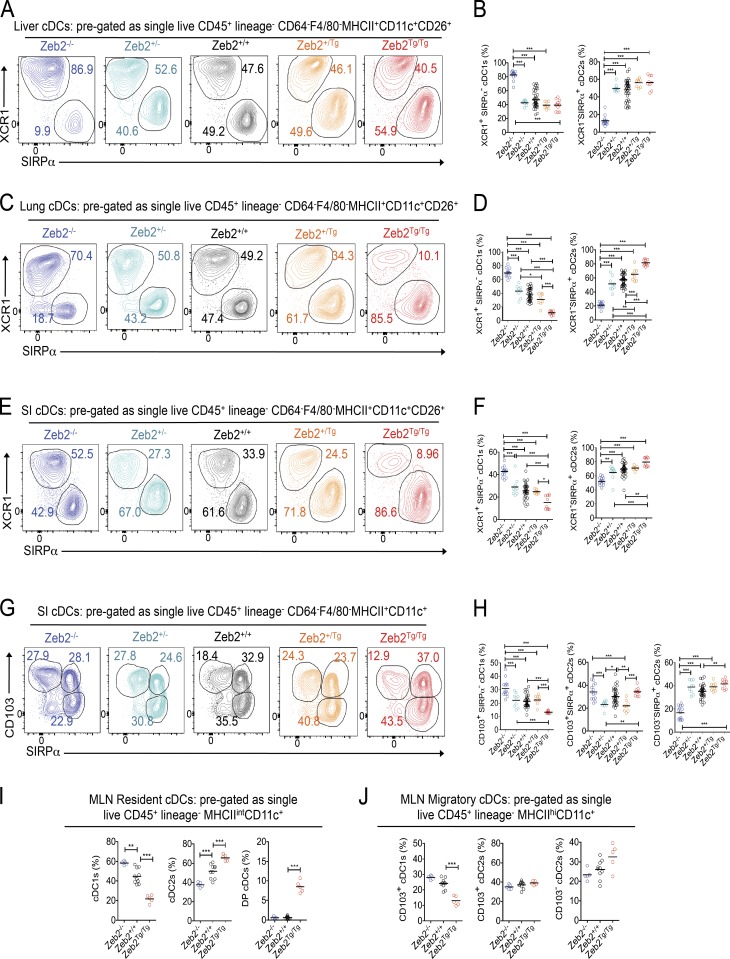
We next extended our analysis to the liver, lung, and small intestine (SI) lamina propria (LP) to determine whether Zeb2 expression regulates cDCs globally or whether its role is restricted to lymphoid tissues. Analysis of cDC1s and cDC2s on the basis of XCR1 and SIRPα expression in these tissues revealed, similarly to the spleen, an overall increase in cDC1s and decrease in cDC2s in Zeb2−/− mice and an overall decrease in cDC1s in Zeb2Tg/Tg mice (Fig. 2), demonstrating that Zeb2 expression in CD11c+ cells regulates cDC subset development across all these mouse tissues. However, the scale of these differences was tissue dependent, suggesting the involvement of some local tissue-imprinting factors. For example, we did not observe any effect of Zeb2 overexpression in the liver (Fig. 2, A and B; and Fig. S4 B), whereas the lung cDCs appeared to be extra sensitive to Zeb2 expression levels with effects of Zeb2 loss and overexpression being observed even when only one allele of Zeb2 was targeted (Fig. 2, C and D; and Fig. S4 C). Additionally, a population of XCR1−SIRPα− cDCs was identified in the lungs of the Zeb2−/− and Zeb2+/− mice (Fig. 2 C). In the SI LP, the same trends were observed in terms of total cDC1s and cDC2s as in the other tissues (Fig. 2, E and F). However, when the cDC2s were further subdivided on the basis of CD103 expression, a marker commonly used to define cDC subsets in the gut (Persson et al., 2013a; Scott et al., 2015), we found that the two subsets of cDC2s were not equally sensitive to Zeb2 expression. Intriguingly, we found the CD103− cDC2s to be highly susceptible to the loss of Zeb2 expression, whereas the CD103+ cDC2s appeared to be unaffected in Zeb2−/− mice. No effects were seen in either population in Zeb2Tg/Tg mice, with only the cDC1s (CD103+SIRPα−) being affected (Fig. 2, G and H; and Fig. S4, A and D). Analysis of the resident cDC populations in the mesenteric LN (MLN) found similar trends within the cDC1 and cDC2 populations in Zeb2+/+, Zeb2−/−, and Zeb2Tg/Tg mice (Fig. 2 I). As observed in the spleen, a DP cDC population was present among the resident cDCs in the Zeb2Tg/Tg mice (Fig. 2 I). Examination of the migratory cDCs in the MLN found similar trends to that observed in the SI LP (Fig. 2 J), confirming that Zeb2 expression does not affect mature cDC migration to the draining LNs.
Figure 2.
Zeb2 expression differentially affects subsets of tissue-resident cDC2s. (A, C, and E) Representative FACS plots showing identification of XCR1+SIRPα− cDC1s and XCR1−SIRPα+ cDC2s in the liver (A), lung (C), and SI LP (E) of Zeb2+/+, Zeb2+/−, Zeb2−/−, Zeb2+/Tg, and Zeb2Tg/Tg mice. Cells were pregated as single live CD45+ lineage−CD64−F4/80−MHCII+CD11c+CD26+. The numbers represent the proportion of cDC1s and cDC2s as a percentage of total cDCs. (B, D, and F) Proportion of liver (B), lung (D), and SI LP (F) cDC1s and cDC2s as a percentage of total cDCs in Zeb2+/+, Zeb2+/−, Zeb2−/−, Zeb2+/Tg, and Zeb2Tg/Tg mice. Data are pooled from two to three experiments, with at least n = 7 per group. (G) Representative FACS plots showing identification of CD103+SIRPα− cDC1s, CD103+SIRPα+ cDC2s, and CD103−SIRPα+ cDC2s in the SI LP of Zeb2+/+, Zeb2+/−, Zeb2−/−, Zeb2+/Tg, and Zeb2Tg/Tg mice. Cells were pregated as single live CD45+ lineage−CD64−F4/80−MHCII+CD11c+CD26+. The numbers represent the proportion of each cDC subset as a percentage of total cDCs. (H) Proportion of SI LP CD103+ cDC1s, CD103+ cDC2s, and CD103− cDC2s as a percentage of total cDCs in Zeb2+/+, Zeb2+/−, Zeb2−/−, Zeb2+/Tg, and Zeb2Tg/Tg mice. Data are pooled from two to three experiments, with at least n = 7 per group. (I) Proportion of MLN-resident XCR1+SIRPα− cDC1s, XCR1−SIRPα+ cDC2s, and XCR1+SIRPα+ DP cDCs as a percentage of total resident cDCs in Zeb2+/+, Zeb2−/−, and Zeb2Tg/Tg mice. Data are representative of two experiments, with n = 5 per group. (J) Proportion of MLN migratory CD103+ cDC1s, CD103+ cDC2s, and CD103− cDC2s as a percentage of total migratory cDCs in Zeb2+/+, Zeb2−/−, and Zeb2Tg/Tg mice. Data are representative of two experiments, with n = 5 per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001. One-way ANOVA with Bonferroni posttest was used.
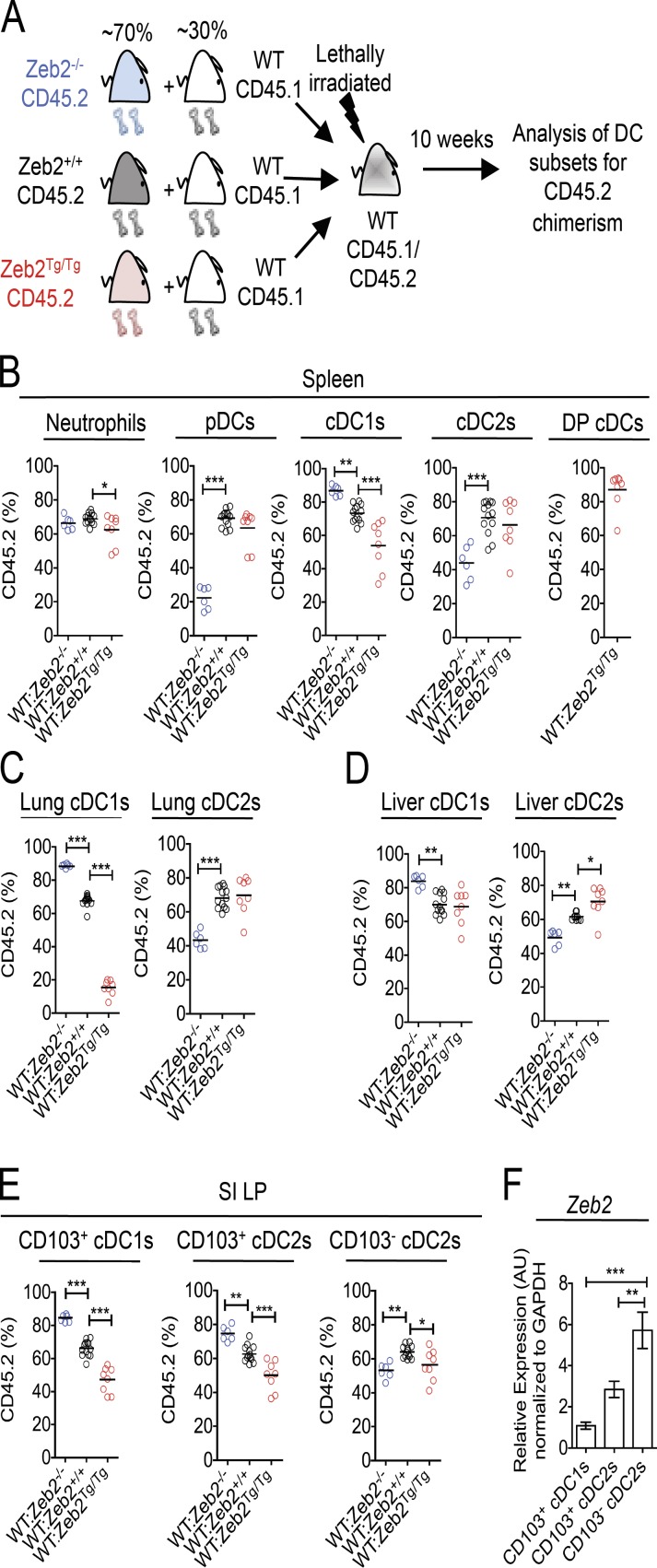
DC-intrinsic effects of Zeb2 expression on cDC commitment revealed by competitive BM chimerism
Having shown that Zeb2 expression in CD11c+ cells regulates both pDCs and cDCs, we next sought to determine whether these effects were cell intrinsic or caused by immune dysregulation. To this end, we generated competitive BM chimeric mice (Fig. 3 A), in which CD45.1/CD45.2 WT mice were lethally irradiated and reconstituted with an ∼70:30 mix (determined by analysis of neutrophils in the spleen; Fig. 3 B and Fig. S5 A) of Zeb2fl/fl/Zeb2Tg/Tg (Zeb2+/+), Cd11cCrexZeb2fl/fl (Zeb2−/−), or Cd11cCrexZeb2Tg/Tg (Zeb2Tg/Tg) CD45.2+ BM and WT CD45.1+ BM. 10–12 wk after reconstitution, the proportions of CD45.2+ cells among pDCs in the spleen and cDC1s and cDC2s in the spleen, lung, liver, and SI LP were analyzed. This analysis revealed that the defect in pDCs in the Zeb2−/− mice was indeed cell intrinsic, with the Zeb2−/− CD45.2+ cells dramatically losing the competition with the CD45.1+ WT BM to generate pDCs when compared with their Zeb2+/+ counterparts (Fig. 3 B and Fig. S5 B). Consistent with our earlier findings that increasing Zeb2 expression did not alter the pDC population, the WT and Zeb2Tg/Tg BM were equally capable of generating pDCs (Fig. 3 B and Fig. S5 B). Thus, we can conclude that pDCs, in addition to the TF E2-2 (Cisse et al., 2008; Ghosh et al., 2010; Murphy, 2013), require Zeb2 expression for their homeostasis. Concurrently, we also found the defect in cDC2 generation in the spleen, lung, and liver and CD103− cDC2 generation in the SI LP of Zeb2−/− mice to be cell intrinsic, demonstrating that Zeb2 expression is also required by cDC2s (Fig. 3, B–E; and Fig. S5, C–F). Analysis of the cDC1 populations in these tissues also found that the enhanced generation of cDC1s in Cd11cCrexZeb2fl/fl mice was cell intrinsic, as Zeb2−/− CD45.2+ BM had a competitive advantage over WT CD45.1+ BM in the generation of cDC1s compared with their Zeb2+/+ counterparts (Fig. 3, B–E; and Fig. S5, C–F). Similarly, we found that the decrease in spleen, lung, and SI LP cDC1s in Cd11cCrexR26-Zeb2Tg/Tg mice was cell intrinsic (Fig. 3, B, C, and E; and Fig. S5, C, D, and F), as was the generation of DP cDCs, with these being almost uniformly derived from Zeb2Tg/Tg BM (Fig. 3 B). However Zeb2Tg/Tg BM cells did not show a competitive advantage over WT BM in generating cDC2s in the chimeras (Fig. 3, B–E; and Fig. S5, C–F). Thus, collectively, we can conclude that Zeb2 is necessary for cDC subtype lineage commitment, with its absence skewing the cDC population away from the cDC2 subtype and its overexpression skewing the cDC population away from the cDC1 population.
Figure 3.
Zeb2 regulation of cDC subsets is cell intrinsic. (A) Competitive BM chimeric mice were generated by lethally irradiating CD45.1/CD45.2 WT mice and reconstituting with an ∼70:30 mix of Zeb2+/+, Zeb2−/−, or Zeb2Tg/Tg CD45.2 BM and WT CD45.1 BM. (B) Proportions of splenic neutrophils, pDCs, cDC1s, cDC2s, and DP cDCs deriving from CD45.2 donor BM. (C) Proportions of lung cDC1s and cDC2s deriving from CD45.2 donor BM. (D) Proportions of liver cDC1s and cDC2s deriving from CD45.2 donor BM. (E) Proportions of SI LP CD103+ cDC1s, CD103+ cDC2s, and CD103− cDC2s deriving from CD45.2 donor BM. Two-way Student’s t test was used. (F) SI LP cDCs were FACS purified from WT mice, and Zeb2 expression was assessed by RT-qPCR. Results shown are expressed relative to GAPDH expression using the 2−ΔΔC(t) method with CD103+ cDC1s set to 1. AU, arbitrary units. One-way ANOVA with Bonferroni posttest was used. Data are pooled from two experiments, with at least n = 6 per group. Error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Interestingly, analysis of the CD103+ cDC2s in the SI LP demonstrated that in addition to having a competitive advantage over WT BM to become cDC1s, Zeb2−/− BM also outcompeted WT BM to generate CD103+ cDC2s, whereas WT BM outcompeted Zeb2Tg/Tg BM to become both CD103+ cDC1s and CD103+ cDC2s (Fig. 3 E and Fig. S5 F). This finding in combination with the results in Fig. 2 (E–J) prompted us to examine Zeb2 expression in the three SI LP cDC populations. Fitting with our results, we found that the CD103+ cDC2s express significantly less Zeb2 than their CD103− counterparts, instead expressing similar levels of Zeb2 to the CD103+ cDC1s (Fig. 3 F). Together, this demonstrates that Zeb2 is not required for the generation of CD103+ cDC2s and that, in terms of Zeb2 dependence, these cells are more similar to the CD103+ cDC1s than their CD103− cDC2 counterparts. This is not the first example where the CD103+ cDC2s behave similarly to their cDC1 counterparts, as, for example, CSF2R2b−/− mice that have reduced cDC1 populations in the periphery also show a reduction in CD103+ cDC2s in the SI (Greter et al., 2012; Li et al., 2012; unpublished data). However, there is also ample evidence that the CD103+ cDC2s represent a unique cDC subset in the gut. For example, the CD103+ cDC2s are the only intestinal cDCs that have been reported to be Notch2 dependent (Lewis et al., 2011), targeted in hu-Langerin diptheria toxin A mice (Welty et al., 2013), and affected in mice that express a truncated form of SIRPα (Scott et al., 2014). Thus, it is clear that further research is required to fully understand the regulation of this cDC2 subset.
Zeb2 expression regulates cDC development
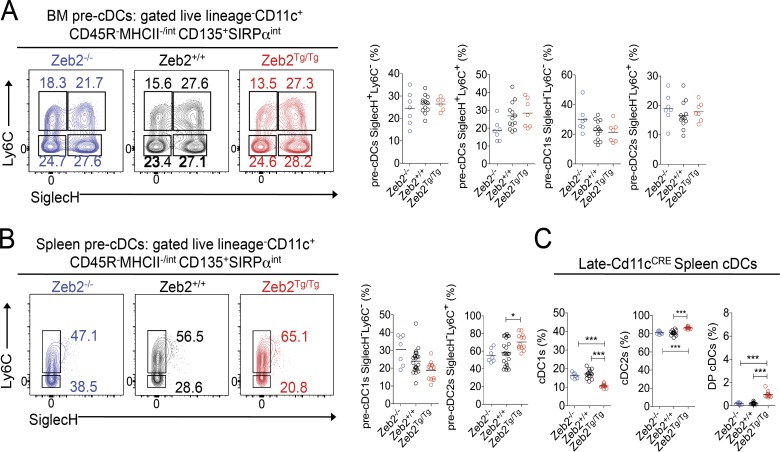
Having shown that Zeb2 expression levels skew the prevalence of the cDC subtypes present in multiple tissues, we next sought to determine whether Zeb2 functions during cDC2 development or whether, similar to the previously described cDC2 TFs, it functions in terminally differentiated cDC2s. It has recently been proposed that commitment to the cDC1 and cDC2 lineage is already apparent at the pre-cDC level such that pre-cDCs can be further subdivided into cDC1- and cDC2-committed pre-cDCs on the basis of Ly6C, SiglecH, and CD24 expression (Grajales-Reyes et al., 2015; Schlitzer et al., 2015). Thus, we first examined the proportions of these pre-cDCs in the BM of Zeb2−/−, Zeb2+/+, and Zeb2Tg/Tg mice (Fig. 4 A and Fig. S6). Although no significant differences were observed in any of the populations across the three genotypes, we did notice a trend toward less pre-cDC1s with increasing levels of Zeb2 expression (Fig. 4 A), consistent with Zeb2 functioning during cDC development. Examination of the pre-cDC subsets in the spleen revealed a similar trend in pre-cDC1s as observed in the BM, in addition to a significant increase in pre-cDC2s with increasing levels of Zeb2 expression (Fig. 4 B), further pointing toward a role for Zeb2 in controlling cDC2 development. To definitively demonstrate that Zeb2 functions during cDC2 development, we next crossed the recently described late-Cd11cCre mouse (Williams et al., 2013), in which the CRE is not active during pre-cDC development but only once the pre-cDCs mature into cDCs (Williams et al., 2013), with Zeb2fl/fl and Zeb2-R26Tg/Tg mice. Analysis of these mice revealed that the loss of Zeb2 late in cDC development does not affect the prevalence of the cDC subsets (Fig. 4 C). Additionally, the effects of overexpressing Zeb2 were minimized in these mice (Fig. 4 C). Thus, we can conclude that Zeb2 functions to maintain the balance between cDC1s and cDC2s during development.
Figure 4.
Zeb2 regulates cDC subset development. (A and B) Representative FACS plots showing identification of pre-cDC subsets and proportions of pre-cDC subsets in the BM (A) and spleen (B) of Zeb2+/+, Zeb2−/−, and Zeb2Tg/Tg mice. Cells were pregated as single live lineage−CD11c+CD45R−MHCII−/intCD135+SIRPαint. The numbers represent the proportion of each pre-cDC subset as a percentage of total pre-cDCs. (C) Proportions of cDC subsets as a percentage of total cDCs in the spleen of late-Cd11cCrexZeb2fl/fl (Zeb2−/−) and late-Cd11cCrexZeb2Tg/Tg (Zeb2Tg/Tg) compared with Cre− littermate controls (Zeb2+/+). Data are pooled from two experiments, with at least n = 7 per group. *, P < 0.05; ***, P < 0.005. One-way ANOVA with Bonferroni posttest was used.
Minimal changes in transcriptomes of mature cDC subsets
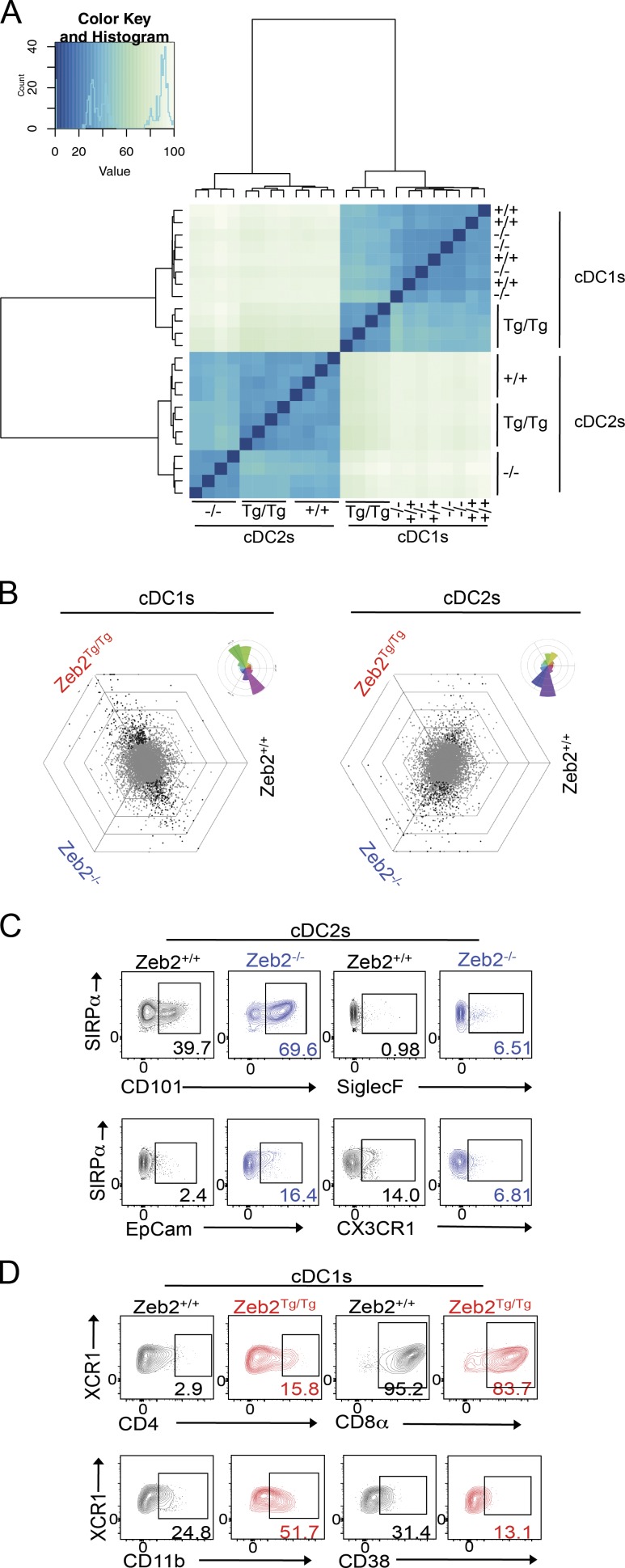
Having identified Zeb2 as a regulator of cDC development, we next sought to examine the consequences of differential Zeb2 expression levels in the mature cDCs. To this end, we FACS purified cDC1s and cDC2s from Zeb2−/−, Zeb2+/+, and Zeb2Tg/Tg mice and performed RNA-sequencing analysis (Fig. S2, gating strategies). Cluster analysis of this data demonstrated that, fitting with our earlier analysis, the ablation of Zeb2 expression in cDC1s and overexpression of Zeb2 in cDC2s did not have significant effects on the transcriptomes (Fig. 5 A). To confirm this, we used a visualization method in which each gene is plotted in a graph containing three axes (one axis per genotype) that are placed at a 120° angle, creating a hexagonal triwise plot (Fig. 5 B). In these hexagons, the direction of a point represents an up-regulation in one or two populations, whereas the distance from the origin represents the magnitude of the up-regulation. Each grid line represents a log2 fold change (FC). Plotting all uniquely annotated genes for cDC1s and cDC2s yielded a horizontal profile with most differentially expressed genes in cDC1s either being up-regulated specifically in Zeb2Tg/Tg mice or in both Zeb2−/− and Zeb2+/+ mice (Fig. 5 B). Conversely, in the cDC2s, differentially expressed genes were either up-regulated solely in Zeb2−/− mice or in both Zeb2+/+ and Zeb2Tg/Tg mice (Fig. 5 B), confirming the cluster analysis. Thus, we subsequently focused our analysis on cDC2s from the Zeb2−/− mice and the cDC1s from the Zeb2Tg/Tg mice compared with their Zeb2+/+ counterparts. Applying a stringency level where the adjusted p-value was equal to 0.01 and the log2 FC was less than −1 or greater than 1, we found that 263 genes were differentially expressed in Zeb2Tg/Tg cDC1s (Table S1), whereas 118 genes were differentially expressed in Zeb2−/− cDC2s (Table S2). To our surprise, however, most genes were only just over the thresholds, with only a few genes having a log2 FC less than −2 or greater than 2. This suggests that once these cDCs develop, they behave normally. Fitting with this, we did not observe any differences in the ability of these cells to induce naive CD4+ or CD8+ T cell proliferation in the steady state (unpublished data); however, further examination of the functional consequences of altering Zeb2 expression levels during infection and inflammation settings are still required. Examining the list of differentially expressed genes for surface receptors enabled us to validate some of the changes at the protein level by flow cytometry. In Zeb2−/− cDC2s, we were able to confirm changes in CD101, CX3CR1, SiglecF, and epithelial cell adhesion molecule protein (EpCam) expression (Fig. 5 C). However, some of the differentially expressed genes did not translate into altered protein expression, including CD69 and CCR2 (unpublished data). Protein analysis in cDC1s from Zeb2Tg/Tg mice validated changes in CD4, CD8α, CD11b, CD38 (Fig. 5 D), and CD115 (unpublished data), whereas the slight up-regulation of CD101 observed at the mRNA level did not result in increased protein expression (unpublished data).
Figure 5.
Altering Zeb2 expression levels affects cDC1 and cDC2 transcriptomes. (A) Cluster analysis of RNA-sequencing data from splenic cDC1s and cDC2s from Zeb2+/+, Zeb2−/−, and Zeb2Tg/Tg mice. (B) To visualize differential gene expression between Zeb2+/+, Zeb2−/−, and Zeb2Tg/Tg cDC1s or cDC2s, each gene was plotted in a hexagonal triwise diagram in which the direction of a point represents an up-regulation in one or two populations, whereas the distance from the origin represents the magnitude of this up-regulation. Genes that are ≥32-fold differentially expressed are plotted on the outer grid line. Rose diagrams (top right corner of each triwise plot) show the percentages of genes in each orientation. Gray dots represent genes that are not differentially expressed. Black dots represent statistically significant differentially expressed genes. (C) Representative FACS plots showing expression of CD101, SiglecF, epithelial cell adhesion molecule (EpCam), and CX3CR1 in Zeb2−/− cDC2s compared with Zeb2+/+ cDC2s. Numbers represent the percentage of cDC2s expressing each marker. (D) Representative FACS plots showing expression of CD4, CD8α, CD11b, and CD38 in Zeb2Tg/Tg cDC1s compared with Zeb2+/+ cDC1s. Numbers represent the percentage of cDC1s expressing each marker.
Zeb2 acts as a cDC subset fate switch by regulating Id2 expression
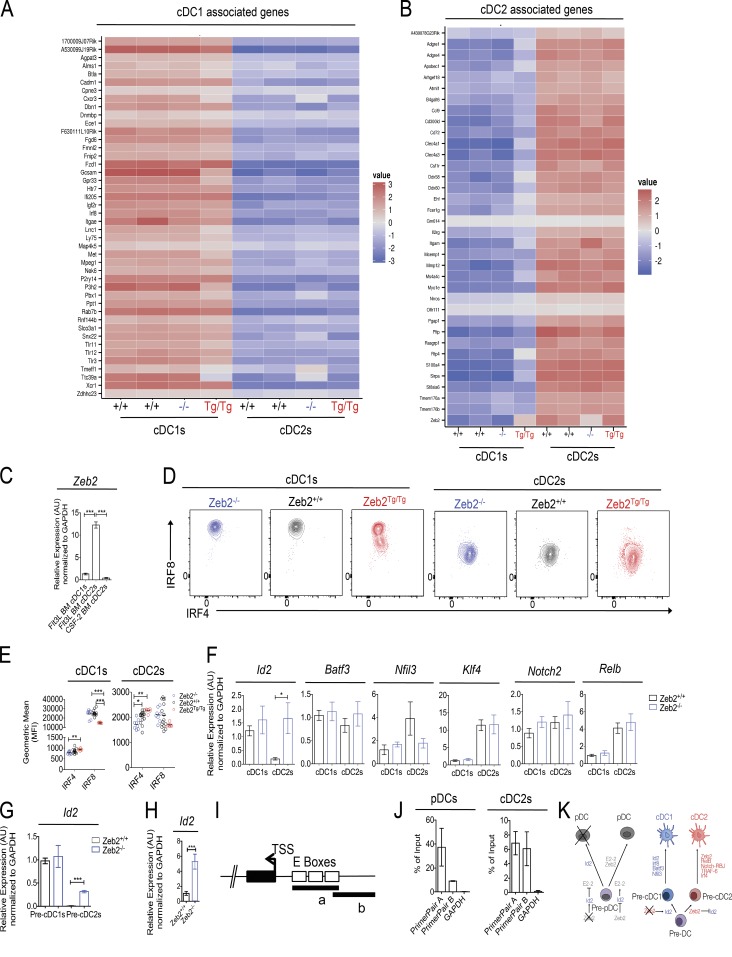
Given our findings that Zeb2 functions during development to determine the ratio between the two cDC subsets across a range of mouse tissues, we next sought to investigate whether this is caused by Zeb2 acting as a subset fate switch. To this end, we generated a list of cDC1- and cDC2-associated genes across a range of tissues by examining the transcriptomes of distinct cDC1 and cDC2 subsets available on the Immunological Genome Project consortium (DC.8+.Sp, DC.8+.MLN, DC.8+.SLN, DC.103+11b-.Lu, and DC.103+11b-.SI for cDC1s; and DC.4+.Sp, DC.4+.MLN, DC.4+.SLN, and DC.103-11b+24+.Lu for cDC2s). The expression of these genes in cDC1s and cDC2s across the range of Zeb2 expression levels was then examined. This analysis revealed that the Zeb2Tg/Tg cDC1s reduce their expression of some of the cDC1-associated genes including Alms1, Btla, Cxcr3, Gcsam, Gpr33, Lrrc1, Ly75, Met, Pbx1, and Tct39a while increasing their expression of some of the cDC2-associated genes including Apobec1, Clec4a1, Ddx58, Ehf, Itgam, Rtp4, and Sirpa (Fig. 6, A and B). Conversely, Zeb2−/− cDC2s increased their expression of some of the cDC1-associated genes including Cxcr3, Map4k5, Pbx1, Rnf144b, Snx22, Tmeff1, and Tct39a, whereas their expression of the cDC2-associated genes, with the exception of Zeb2 itself, was largely unaffected (Fig. 6, A and B). Together with our earlier findings that Zeb2 functions during development, this suggests that the absence or overexpression of Zeb2 switches the fate of the majority of pre-cDCs, resulting in the development of cDC1s or cDC2s, respectively. However, this is not 100% efficient, as some pre-cDCs do become either cDC2s or cDC1s in Zeb2−/− and Zeb2Tg/Tg mice, respectively, and consequently, these express some genes typically associated with the alternate subset, but these differences are not sufficient to confer functional differences to the subsets, at least in the steady state.
Figure 6.
cDC subset fate is dictated by Zeb2 expression levels. (A and B) Heat maps showing relative expression of cDC1 (A)- and cDC2 (B)-associated genes normalized per mean expression of each gene in cDC1s and cDC2s from Zeb2+/+, Zeb2−/−, and Zeb2Tg/Tg mice. (C) cDC1s from Flt3L WT BMDC cultures and cDC2s from WT Flt3L and WT GM-CSF BMDC cultures were FACS purified, and Zeb2 expression was assessed by RT-qPCR. The results shown are expressed relative to GAPDH expression using the 2−ΔΔC(t) method with cDC1s set to 1. Data are pooled from two experiments, with n = 6 per group. (D) Representative FACS plots showing Irf8 and Irf4 expression by splenic cDC subsets in Zeb2−/−, Zeb2+/+, and Zeb2Tg/Tg mice. (E) Geometric mean (mean fluorescence intensity [MFI]) of Irf8 and Irf4 expression by splenic Zeb2−/−, Zeb2+/+, and Zeb2Tg/Tg cDCs. Data are pooled from two experiments, with at least n = 6 per group. (C and E) One way ANOVA with Bonferroni posttest was used. (F) Splenic cDC1s and cDC2s were FACS purified from Zeb2+/+ or Zeb2−/− mice, and the indicated TF expression was assessed by RT-qPCR. The results shown are expressed relative to GAPDH expression using the 2−ΔΔC(t) method with Zeb2+/+ cDC1s set to 1. Data are pooled from three experiments, with at least n = 3 per group. Two-way Student’s t test was used. (G) Splenic pre-cDC1s and pre-cDC2s were FACS purified from Zeb2+/+ or Zeb2−/− mice, and Id2 expression was assessed by RT-qPCR. The results shown are expressed relative to GAPDH expression using the 2−ΔΔC(t) method with Zeb2+/+ pre-cDC1s set to 1. Data are pooled from two experiments, with at least n = 2 per group. (H) Splenic pDCs were FACS purified from Zeb2+/+ or Zeb2−/− mice, and Id2 expression was assessed by RT-qPCR. The results shown are expressed relative to GAPDH expression using the 2−ΔΔC(t) method with Zeb2+/+ pDCs set to 1. Data are pooled from two experiments, with at least n = 7 per group. (F–H) Two-way Student’s t test was used. (I) Schematic representation of the Id2 promoter with predicted E boxes. a and b represent regions amplified by qPCR with primer pair A and primer pair B, respectively. TSS, transcription start site. (J) qPCR analysis of ChIP performed against the FLAG tag present on recombinant Zeb2 in splenic pDCs and cDC2s of Zeb2Tg/Tg mice. Data are expressed as the percentage of input. Data are pooled from two independent experiments, with n = 2 per group. (K) Proposed model for the action of Zeb2 in cDC and pDC development. Error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. AU, arbitrary units.
With this finding in mind, we sought to focus further on the role of Zeb2 in cDC development. First, we examined how Zeb2 was induced during cDC development by analyzing Zeb2 expression levels in WT BM-derived cDC subsets after culture of total BM with the canonical cDC growth factors Flt3L or GM-CSF (CSF-2). As it has recently been shown that GM-CSF BM cultures consist of both monocyte-derived DCs and cDCs (Helft et al., 2015), we used MHCII, CD115 (CSF-1R), and CD26 expression to delineate cDCs and monocyte-derived DCs in these culture systems, concentrating our analysis on the CD115−CD11c+MHCIIhi cDCs (Fig. S7). Intriguingly, Zeb2 expression was only induced in cDC2s from the Flt3L cultures (Fig. 6 C), demonstrating that Zeb2 expression is induced by Flt3L but not by GM-CSF signaling. This is consistent with recent findings that cDC2s develop normally in mice lacking GM-CSF signaling (Greter et al., 2012).
Having shown that Zeb2 is induced during cDC development, we next investigated the effects of manipulating Zeb2 expression on the other TFs known to function in cDC development and terminal differentiation. Analysis of Irf8 and Irf4 protein expression in the cDCs of Zeb2−/−, Zeb2+/+, and Zeb2Tg/Tg mice revealed a striking decrease in Irf8 expression in Zeb2Tg/Tg cDC1s that was coupled with a significant increase in Irf4. We also observed a significant decrease in Irf4 expression in Zeb2−/− cDC2s, but this was not correlated with an increase in Irf8 expression (Fig. 6, D and E). Consistent with our early findings of an intermediate cDC1/cDC2 phenotype, the DP cDCs in the Zeb2Tg/Tg mice expressed intermediate levels of Irf8 and Irf4 (unpublished data). In addition to Irf8 and Irf4, the TFs Id2, Batf3, Nfil3, RelB, Klf4, and Notch2 have all been implicated in cDC development and/or terminal differentiation (Wu et al., 1998; Tamura et al., 2005; Caton et al., 2007; Tailor et al., 2008; Edelson et al., 2011; Jackson et al., 2011; Kashiwada et al., 2011; Lewis et al., 2011; Persson et al., 2013b; Schlitzer et al., 2013; Grajales-Reyes et al., 2015; Tussiwand et al., 2015). As the expression levels of these TFs cannot be analyzed with flow cytometry, we instead examined their mRNA expression by RT-qPCR in Zeb2−/− cDC1s and cDC2s compared with their Zeb2-sufficient counterparts. Strikingly, we found a significant up-regulation of the cDC1-associated TF Id2 among the Zeb2−/− cDC2 population (Fig. 6 F). Consistent with this, we found Id2 to be differentially expressed in the RNA-sequencing data from Zeb2Tg/Tg cDC1s and Zeb2−/− cDC2s with a log2 FC of −0.6 or 0.6, respectively. We did not detect any other significant changes in the other TFs examined, although there was a trend toward less Nfil3 expression in Zeb2−/− cDC2s (Fig. 6 F). To determine whether Id2 expression was also affected by the loss of Zeb2 early in cDC development, we next examined Id2 mRNA levels in the cDC1- and cDC2-committed splenic pre-cDC populations. Similar to the results seen in the mature cDC populations, we found an increase in Id2 expression among the Zeb2−/− pre-cDC2s (Fig. 6 G), suggesting that Zeb2 may function in cDC2s during development to repress Id2 expression. As Id2 is known to suppress pDC development by antagonizing the E protein TF E2-2 (Spits et al., 2000; Hacker et al., 2003; Cisse et al., 2008; Ghosh et al., 2010), we also checked Id2 expression in the remaining pDCs in the Zeb2−/− mice. As observed for the cDC2s, we found a significant increase in Id2 expression among the Zeb2−/− pDCs (Fig. 6 H). Thus, mechanistically, Zeb2 appears to function during DC development by repressing Id2 expression, facilitating both pDC and cDC2 development. As Zeb2 is itself a TF, we next examined whether this effect was through direct binding of Zeb2 to the Id2 promoter region or through an indirect mechanism. In silico analysis using PhysBinder (Broos et al., 2013) or ConTra (Broos et al., 2011) identified several conserved Zeb binding sites (Remacle et al., 1999) in Id2 and the surrounding regions in mice and humans (Fig. S8). Thus, we next designed two sets of primers to amplify two overlapping regions (A and B) in the Id2 promoter containing the predicted binding site (Fig. 6 I). qPCR was performed on chromatin extracted from splenic pDCs and cDC2s of Zeb2Tg/Tg mice after chromatin immunoprecipitation (ChIP) with an antibody recognizing the FLAG tag present on Zeb2 in these mice. For both pDCs and cDC2s, we observed a clear enrichment for region A, with ∼40% for pDCs and 6% for cDC2s, whereas region B was amplified with a percentage of enrichment of ∼7% and 5% in pDCs and cDC2s, respectively (Fig. 6 J). Thus, Zeb2 binds the E boxes present in the Id2 promoter, repressing its expression.
Collectively, these results highlight a previously uncharacterized role for the TF Zeb2 in regulating DC development through modulating Id2 expression (Fig. 6 K). In pDCs, Zeb2 expression represses Id2, allowing the pDC TF E2-2 to bind DNA and induce pDC development, whereas in cDC development, Zeb2 functions to regulate commitment toward the cDC2 lineage by repressing Id2. As such, Zeb2 represents the first TF to be described that is involved in the early lineage commitment of pre-cDCs toward the cDC2 lineage.
MATERIALS AND METHODS
Mice
The generation of Zeb2fl/fl and R26-Zeb2Tg/Tg mice was described previously (Higashi et al., 2002; Tatari et al., 2014). Mice were backcrossed to a C57BL/6 background for at least seven generations before crossing with the Cd11cCre mice (Caton et al., 2007) or late-Cd11cCre mice (Williams et al., 2013). All mice were bred and maintained at the Vlaams Instituut voor Biotechnologie (Ghent University) under specific pathogen-free conditions and were used between 6 and 12 wk of age. All experiments were performed in accordance with the ethical committee of the Faculty of Science of the Vlaams Instituut voor Biotechnologie.
Isolation of tissue leukocytes
For the isolation of liver leukocytes, livers were isolated from PBS-perfused mice, chopped finely, and incubated for 15–20 min with 1 mg/ml collagenase A (Sigma-Aldrich) and 10 U/ml DNase (Roche) in a shaking water bath at 37°C. For the isolation of lung and spleen leukocytes, lungs and spleens were isolated from PBS-perfused mice, chopped finely, and incubated for 30 min with 0.2 mg/ml Liberase TM (Roche) and 10 U/ml DNase (Roche) in a shaking water bath at 37°C. SI LP leukocytes were isolated as described previously (Cerovic et al., 2013). Pre-cDCs were isolated from spleens by gently pressing the spleens through a 70-µm filter (no enzymes) to allow for the detection of SiglecH. BM cells were obtained by flushing femurs and tibias with RPMI. In all instances, except for the SI, red blood cells were lysed before staining for flow cytometric analysis.
Flow cytometry and FACS
For flow cytometry, 3–4 × 106 cells were stained at 4°C in the dark with antibodies (Table S3). Intracellular staining of Irf4 and Irf8 was performed after fixing and permeabilizing of the cells with a Foxp3 TF-staining buffer set (eBioscience). Data were acquired on a cell analyzer (LSRFortessa; BD) and analyzed using FlowJo software (Tree Star). Cells were FACS purified using a flow cytometer (FACSAria II or FACSAria III; BD). After sorting, a purity check was performed for all samples.
RNA sequencing
25,000 cDC1s or cDC2s from Zeb2−/−, Zeb2+/+, and Zeb2Tg/Tg mice were FACS purified into 500 µl of buffer (RLT Plus; QIAGEN) and β-mercaptoethanol. RNA was isolated using a micro-RNA isolation kit (QIAGEN) and sent to the Vlaams Instituut voor Biotechnologie Nucleomics facility, where the RNA sequencing was performed using a NextSeq sequencer (Illumina). The preprocessing of the RNA sequencing data was done by Trimmomatic. The adapters were cut off, and reads were trimmed when the quality dropped below 20. Reads with a length <35 were discarded. All samples passed quality control based on the results of FastQC. Reads were mapped to the mouse reference genome via Tophat2 and counted via HTSeqCount. Samples were subsequently analyzed using R/Bioconductor, and the DESeq2 procedure was used to normalize the data.
Gene expression analysis by real-time RT-PCR
RNA was purified from sorted cells using an RNeasy Plus micro kit (QIAGEN). RNA was reverse transcribed to cDNA with an iScript Advanced cDNA Synthesis kit (Bio-Rad Laboratories). Gene expression was assayed by real-time RT-PCR using a SensiFast SYBR No-Rox kit (GC Biotech) on a PCR amplification and detection instrument (LightCycler 480; Roche) with the primers listed in Table S4. Gene expression was normalized to GAPDH, and the mean relative gene expression was calculated using the 2−ΔΔC(t) method.
BMDC cultures
Total BM was harvested from WT mice, and 2 × 106 cells were cultured in RPMI supplemented with Glutamax and Gentamicin in a 6-well plate for 7 d with either 250 ng/ml Flt3L and 10% FCS for Flt3L cultures or 20 ng/ml CSF-2 and 5% FCS for CSF-2 cultures. CSF-2 BMDCs were further supplemented with additional media, FCS, and CSF-2 at day 3 of culture.
Zeb2-FLAG ChIP and Id2 qPCR
FACS-purified pDC and cDC2 cells from Zeb2Tg/Tg mice were cross-linked with 1% paraformaldehyde in fixation buffer (Active Motive). After nuclei isolation, DNA was fragmented with 25 U micrococcal nuclease for 20 min at 37°C in micrococcal nuclease–digesting buffer (50 mM Tris-HCl, pH 7.6, 1 mM CaCl2, and 0.2% Triton X-100). DNA fragment size (150–500 bp) was confirmed after chromatin preparation in a 1.2% agarose gel. The fragmented chromatin was incubated with 10 µg FLAG-M2 (F3165; Sigma-Aldrich) antibody overnight followed by a pull-down assay using A/G-conjugated agarose beads (EMD Millipore). DNA was purified with an iPure kit (Diagenode), and the quality was measured using a 2100 Bioanalyzer system with a DNA kit (High Sensitivity; Agilent Technologies). Real-time qPCR was performed on a Zeb-predicted binding site identified in the Id2 promoter with the following primers: 5′-TACCTGACAAAGAGCTTCCC-3′ and 5′-TTACATACACTGCCCTTGGT-3′ (primer pair A) and 5′-ATGTGGCTGCATCTAGGAA-3′ and 5′-GGGAAGCTCTTTGTCAGGTA-3′ (primer pair B). Primers in the coding sequence of the GAPDH gene were used as a control of unspecific binding with the following primers: 5′-TTGAGCTAGGACTGGATAAGCAGG-3′ and 5′-AGTCCGTATTTATAGGAACCCGG-3′. The percentage of enrichment to the input was calculated and shown in a bar graph.
Statistical analysis
Groups were compared with a two-way Student’s t test, and multiple-group comparisons were performed using one-way ANOVA followed by a Bonferroni posttest with Prism Software (GraphPad Software). Samples were assumed to be normally distributed with similar variance between groups. No randomization was used to determine experimental groups, and no blinding of the investigator was performed. Group sizes were determined on the basis of previous experience.
Accession numbers
All RNA-sequencing data have been deposited in the Gene Expression Omnibus public database under accession no. GSE79903.
Online supplemental material
Fig. S1 shows splenic pDCs. Fig. S2 shows the splenic cDC sorting strategy and purities. Fig. S3 shows splenic cDC gating strategy and absolute numbers. Fig. S4 shows cDCs in other tissues. Fig. S5 shows BM chimeras. Fig. S6 shows the pre-cDC gating strategy. Fig. S7 shows BMDC gating strategies and sorting purities. Fig. S8 shows the predicted Zeb binding sites in the Id2 locus. Table S1 shows differentially expressed genes of Zeb2+/+ versus Zeb2Tg/Tg cDC1s. Table S2 shows differentially expressed genes of Zeb2+/+ versus Zeb2−/− cDC2s. Table S3 shows antibodies used for flow cytometry. Table S4 shows primers used for RT-qPCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151715/DC1.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Marie Curie Intra-European Fellowship as part of Horizon 2020 (H2020-MSCA-IF-2014 660448) to C.L. Scott, a Fonds Wetenschappelijk Onderzoek PhD fellowship (1102716N) to B. Soen, and an Interuniversity Attraction Poles grant (IUAP VII/03) from the federal government of Belgium to B.N. Lambrecht. G. Berx’s laboratory is supported by the Fonds Wetenschappelijk Onderzoek (G.0529.12N and G.0817.13N), the Geconcerteerde Onderzoeksacties Ghent University (GOA-01GB1013W), and the Belgian Federation for the Study Against Cancer (B/13590).
The authors declare no competing financial interests.
Author contributions: C.L. Scott conceived and performed experiments and co-wrote the manuscript. B. Soen conceived and performed experiments and co-wrote the manuscript. J. Taminau, L. Martens, N. Skrypek, G. Blancke, and G.V. Isterdael performed experiments. D. Huylebroeck, J. Haigh, M. Guilliams, Y. Saeys, B.N. Lambrecht, and G. Berx supervised the work.
Footnotes
Abbreviations used:
- cDC
- conventional DC
- CDP
- common DC progenitor
- ChIP
- chromatin immunoprecipitation
- CMP
- common myeloid progenitor
- DP
- double positive
- FC
- fold change
- LP
- lamina propria
- MDP
- macrophage–DC progenitor
- MLN
- mesenteric LN
- pDC
- plasmacytoid DC
- qPCR
- quantitative PCR
- SI
- small intestine
- SIRPα
- signal regulatory protein α
- TF
- transcription factor
References
- Bachem A., Güttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H.W., et al. 2010. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207:1273–1281. 10.1084/jem.20100348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaña S., Turner S., Paul J., Ainsua-Enrich E., and Kovats S.. 2016. IRF4 and IRF8 act in CD11c+ cells to regulate terminal differentiation of lung tissue dendritic cells. J. Immunol. 196:1666–1677. 10.4049/jimmunol.1501870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broos S., Hulpiau P., Galle J., Hooghe B., Van Roy F., and De Bleser P.. 2011. ConTra v2: a tool to identify transcription factor binding sites across species, update 2011. Nucleic Acids Res. 39:W74–W78. 10.1093/nar/gkr355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broos S., Soete A., Hooghe B., Moran R., van Roy F., and De Bleser P.. 2013. PhysBinder: Improving the prediction of transcription factor binding sites by flexible inclusion of biophysical properties. Nucleic Acids Res. 41:W531–W534. 10.1093/nar/gkt288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., and Reizis B.. 2007. Notch–RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 204:1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerovic V., Houston S.A., Scott C.L., Aumeunier A., Yrlid U., Mowat A.M., and Milling S.W.F.. 2013. Intestinal CD103− dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol. 6:104–113. 10.1038/mi.2012.53 [DOI] [PubMed] [Google Scholar]
- Cisse B., Caton M.L., Lehner M., Maeda T., Scheu S., Locksley R., Holmberg D., Zweier C., den Hollander N.S., Kant S.G., et al. 2008. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 135:37–48. 10.1016/j.cell.2008.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Craene B., and Berx G.. 2013. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 13:97–110. 10.1038/nrc3447 [DOI] [PubMed] [Google Scholar]
- Denecker G., Vandamme N., Akay O., Koludrovic D., Taminau J., Lemeire K., Gheldof A., De Craene B., Van Gele M., Brochez L., et al. 2014. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 21:1250–1261. 10.1038/cdd.2014.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C.X., Amezquita R.A., Guan T., Marshall H.D., Joshi N.S., Kleinstein S.H., and Kaech S.M.. 2015. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 212:2041–2056. 10.1084/jem.20150186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelson B.T., Bradstreet T.R., KC W., Hildner K., Herzog J.W., Sim J., Russell J.H., Murphy T.L., Unanue E.R., and Murphy K.M.. 2011. Batf3-dependent CD11blow/- peripheral dendritic cells are GM-CSF-independent and are not required for Th cell priming after subcutaneous immunization. PLoS One. 6:e25660 10.1371/journal.pone.0025660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh H.S., Cisse B., Bunin A., Lewis K.L., and Reizis B.. 2010. Continuous expression of the transcription factor E2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 33:905–916. 10.1016/j.immuni.2010.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens S., Janzen V., Bartunkova S., Yokomizo T., Drogat B., Crisan M., Haigh K., Seuntjens E., Umans L., Riedt T., et al. 2011. The EMT regulator Zeb2/Sip1 is essential for murine embryonic hematopoietic stem/progenitor cell differentiation and mobilization. Blood. 117:5620–5630. 10.1182/blood-2010-08-300236 [DOI] [PubMed] [Google Scholar]
- Grajales-Reyes G.E., Iwata A., Albring J., Wu X., Tussiwand R., Kc W., Kretzer N.M., Briseño C.G., Durai V., Bagadia P., et al. 2015. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α+ conventional DC clonogenic progenitor. Nat. Immunol. 16:708–717. 10.1038/ni.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M., Helft J., Chow A., Hashimoto D., Mortha A., Agudo-Cantero J., Bogunovic M., Gautier E.L., Miller J., Leboeuf M., et al. 2012. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 36:1031–1046. 10.1016/j.immuni.2012.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., Ginhoux F., Jakubzick C., Naik S.H., Onai N., Schraml B.U., Segura E., Tussiwand R., and Yona S.. 2014. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 14:571–578. 10.1038/nri3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker C., Kirsch R.D., Ju X.-S., Hieronymus T., Gust T.C., Kuhl C., Jorgas T., Kurz S.M., Rose-John S., Yokota Y., and Zenke M.. 2003. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 4:380–386. 10.1038/ni903 [DOI] [PubMed] [Google Scholar]
- Haniffa M., Shin A., Bigley V., McGovern N., Teo P., See P., Wasan P.S., Wang X.-N., Malinarich F., Malleret B., et al. 2012. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 37:60–73. 10.1016/j.immuni.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty S.V., Sullivan A.M., and O’Keeffe G.W.. 2015. Zeb2: A multifunctional regulator of nervous system development. Prog. Neurobiol. 132:81–95. 10.1016/j.pneurobio.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Helft J., Böttcher J., Chakravarty P., Zelenay S., Huotari J., Schraml B.U., Goubau D., and Reis e Sousa C.. 2015. GM-CSF mouse bone marrow cultures comprise a heterogeneous population of CD11c+MHCII+ macrophages and dendritic cells. Immunity. 42:1197–1211. 10.1016/j.immuni.2015.05.018 [DOI] [PubMed] [Google Scholar]
- Higashi Y., Maruhashi M., Nelles L., Van de Putte T., Verschueren K., Miyoshi T., Yoshimoto A., Kondoh H., and Huylebroeck D.. 2002. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis. 32:82–84. 10.1002/gene.10048 [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.T., Hu Y., Liu R., Masson F., D’Amico A., Carotta S., Xin A., Camilleri M.J., Mount A.M., Kallies A., et al. 2011. Id2 expression delineates differential checkpoints in the genetic program of CD8α+ and CD103+ dendritic cell lineages. EMBO J. 30:2690–2704. 10.1038/emboj.2011.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M., Pham N.-L.L., Pewe L.L., Harty J.T., and Rothman P.B.. 2011. NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood. 117:6193–6197. 10.1182/blood-2010-07-295873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K.L., Caton M.L., Bogunovic M., Greter M., Grajkowska L.T., Ng D., Klinakis A., Charo I.F., Jung S., Gommerman J.L., et al. 2011. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 35:780–791. 10.1016/j.immuni.2011.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.S., Yang C.Y., Nallaparaju K.C., Zhang H., Liu Y.J., Goldrath A.W., and Watowich S.S.. 2012. The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development. Blood. 120:4363–4373. 10.1182/blood-2012-07-441311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.-F., Randolph G.J., Rudensky A.Y., and Nussenzweig M.. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., and Mortha A.. 2013. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31:563–604. 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., et al. Immunological Genome Consortium . 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13:888–899. 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K.M. 2013. Transcriptional control of dendritic cell development. Adv. Immunol. 120:239–267. 10.1016/B978-0-12-417028-5.00009-0 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.-Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226. 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Omilusik K.D., Best J.A., Yu B., Goossens S., Weidemann A., Nguyen J.V., Seuntjens E., Stryjewska A., Zweier C., Roychoudhuri R., et al. 2015. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212:2027–2039. 10.1084/jem.20150194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N., Kurabayashi K., Hosoi-Amaike M., Toyama-Sorimachi N., Matsushima K., Inaba K., and Ohteki T.. 2013. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity. 38:943–957. 10.1016/j.immuni.2013.04.006 [DOI] [PubMed] [Google Scholar]
- Persson E.K., Scott C.L., Mowat A.M., and Agace W.W.. 2013a Dendritic cell subsets in the intestinal lamina propria: ontogeny and function. Eur. J. Immunol. 43:3098–3107. 10.1002/eji.201343740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson E.K., Uronen-Hansson H., Semmrich M., Rivollier A., Hägerbrand K., Marsal J., Gudjonsson S., Håkansson U., Reizis B., Kotarsky K., and Agace W.W.. 2013b IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 38:958–969. 10.1016/j.immuni.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Remacle J.E., Kraft H., Lerchner W., Wuytens G., Collart C., Verschueren K., Smith J.C., and Huylebroeck D.. 1999. New mode of DNA binding of multi-zinc finger transcription factors: deltaEF1 family members bind with two hands to two target sites. EMBO J. 18:5073–5084. 10.1093/emboj/18.18.5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Briseño C.G., Lee J.S., Ng D., Manieri N.A., KC W., Wu X., Thomas S.R., Lee W.-L., Turkoz M., et al. 2013. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14:937–948. 10.1038/ni.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W.S., See P., Shin A., Wasan P.S., et al. 2013. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 38:970–983. 10.1016/j.immuni.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., Sivakamasundari V., Chen J., Sumatoh H.R.B., Schreuder J., Lum J., Malleret B., Zhang S., Larbi A., Zolezzi F., et al. 2015. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16:718–728. 10.1038/ni.3200 [DOI] [PubMed] [Google Scholar]
- Scott C.L., Tfp Z.M., Beckham K.S.H., Douce G., and Mowat A.M.. 2014. Signal regulatory protein alpha (SIRPα) regulates the homeostasis of CD103+ CD11b+ DCs in the intestinal lamina propria. Eur. J. Immunol. 44:3658–3668. 10.1002/eji.201444859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.L., Bain C.C., Wright P.B., Sichien D., Kotarsky K., Persson E.K., Luda K., Guilliams M., Lambrecht B.N., Agace W.W., et al. 2015. CCR2+CD103− intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal Immunol. 8:327–339. 10.1038/mi.2014.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H., Couwenberg F., Bakker A.Q., Weijer K., and Uittenbogaart C.H.. 2000. Id2 and Id3 inhibit development of CD34+ stem cells into predendritic cell (pre-DC)2 but not into pre-DC1: Evidence for a lymphoid origin of pre-DC2. J. Exp. Med. 192:1775–1784. 10.1084/jem.192.12.1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., and Colonna M.. 2015. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 15:471–485. 10.1038/nri3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor P., Tamura T., Morse H.C. III, and Ozato K.. 2008. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 111:1942–1945. 10.1182/blood-2007-07-100750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Tailor P., Yamaoka K., Kong H.J., Tsujimura H., O’Shea J.J., Singh H., and Ozato K.. 2005. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 174:2573–2581. 10.4049/jimmunol.174.5.2573 [DOI] [PubMed] [Google Scholar]
- Tatari M.N., De Craene B., Soen B., Taminau J., Vermassen P., Goossens S., Haigh K., Cazzola S., Lambert J., Huylebroeck D., et al. 2014. ZEB2-transgene expression in the epidermis compromises the integrity of the epidermal barrier through the repression of different tight junction proteins. Cell. Mol. Life Sci. 71:3599–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R., Everts B., Grajales-Reyes G.E., Kretzer N.M., Iwata A., Bagaitkar J., Wu X., Wong R., Anderson D.A., Murphy T.L., et al. 2015. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 42:916–928. 10.1016/j.immuni.2015.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden M.J., Goossens S., Daussy C., Mathieu A.-L., Faure F., Marçais A., Vandamme N., Farla N., Mayol K., Viel S., et al. 2015. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J. Exp. Med. 212:2015–2025. 10.1084/jem.20150809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welty N.E., Staley C., Ghilardi N., Sadowsky M.J., Igyártó B.Z., and Kaplan D.H.. 2013. Intestinal lamina propria dendritic cells maintain T cell homeostasis but do not affect commensalism. J. Exp. Med. 210:2011–2024. 10.1084/jem.20130728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.W., Tjota M.Y., Clay B.S., Vander Lugt B., Bandukwala H.S., Hrusch C.L., Decker D.C., Blaine K.M., Fixsen B.R., Singh H., et al. 2013. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat. Commun. 4:2990 10.1038/ncomms3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., D’Amico A., Winkel K.D., Suter M., Lo D., and Shortman K.. 1998. RelB is essential for the development of myeloid-related CD8α− dendritic cells but not of lymphoid-related CD8α+ dendritic cells. Immunity. 9:839–847. 10.1016/S1074-7613(00)80649-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.