Abstract
Background
Standardized MedDRA Queries (SMQs) are sets of terms determined by experts that are used to identify adverse events (AEs) related to different disease processes. Their use can be challenging because most SMQs have 50 to 100 preferred terms and AE databases can have many thousands of events.
Aim
The aim of this study is to develop a technique where AEs corresponding to preferred terms in SMQs may be easily detected.
Methodology
The method I developed uses the Table Join function of the JMP® software program to quickly and easily probe clinical trial AE databases. The SMQ Severe cutaneous adverse reactions was used as a probe in a mock AE dataset. Potentially confounding demographic or study-specific factors were evaluated by combining these datasets with the dataset containing the AEs identified with the SMQs.
Results
AEs were successfully detected in an AE database using the method described. Cases with potential confounding factors, such as concomitant medications, were identified.
Conclusions
The method developed allows for AEs to be found in clinical trial databases and evaluated using software programs that are readily available to clinical researchers.
1 Introduction
The Medical Dictionary for Regulatory Activities (Med-DRA) is a standardized international terminology that is used by regulatory authorities and the biopharmaceutical industry to share regulatory information about medical products. Standardised MedDRA Queries (SMQs) are tools developed to facilitate retrieval of MedDRA-coded data as a first step in investigating drug safety issues in pharmacovigilance and clinical development. SMQs are validated, pre-determined sets of MedDRA terms (typically preferred terms; PTs) related to specific medical conditions and there are currently almost 100 SMQs in production in MedDRA [1]. Screening with SMQs is particularly useful to evaluate adverse event (AE) databases for early signs of complex diseases, since a subject is often discontinued from a clinical trial before a disorder has fully developed. However, the analysis of AE databases with SMQs may be quite cumbersome, since the latter may contain dozens of PTs and hundreds of their subordinate lowest level terms (LLTs). Furthermore, AE databases often have hundreds, or even thousands, of individual events. This short communication describes a method to perform analysis of AE databases using SMQs, some features of AE databases that need to be addressed to optimize results, and some advantages of this methodology.
2 Materials and Methods
2.1 Accessing SMQs and Assembling the Required Dataset in JMP
SMQs can be downloaded by two methods, both of which require subscription to MedDRA through the MedDRA Maintenance and Support Services Organization (MSSO). SMQs are bundled with each semi-annual MedDRA version release in an EXCEL® file named SMQ spreadsheet. The PTs making up an SMQ can be copied into the column of a JMP data table and labeled appropriately [e.g. Angioedema (SMQ) PTs]. The name of the data table optimally includes the version of MedDRA from which the SMQ information is derived. A second method of accessing SMQs is to export them from the MedDRA web-based or desktop browsers [2].
2.2 Creation of the Adverse Event, Demography, and Extra Tabulation Datasets of Interest for this Study
Mock datasets were created for the purposes of demonstration in this study in EXCEL and then imported into JMP. While much more abbreviated than a typical clinical trial dataset, each was formatted in a standard format similar to what would be produced in an actual clinical trial. These datasets were created for multiple projects and so may have features not needed for this demonstration.
2.2.1 Demography Tabulation Dataset (DM)
One hundred unique subjects (column variable USUBJID) were included in this model demography data set (DM) with identifiers ranging from 1001–001 to 1001–100. The DM dataset was created with column variables for Study, (STUDY), subject (SUBJ), Unique subject identifier (USUBJID), Treatment Arm (TREATMENT), race (RACE), age (AGE 20–40 years old), (BMI 20–40, ordinally arranged in ascending/descending order), weight (120–250 pounds; ordinally arranged in ascending/descending order in five-pound increments), and height (HEIGHT). The height was calculated from the BMI and weight using the formula Height = [(Weight/BMI) × 703]1/2.
2.2.2 Adverse Event Dataset (AE)
The AE dataset was created in EXCEL so that the 100 mock subjects had 9 or 10 AEs each. Terms from Severe cutaneous adverse reactions (SMQ) were included in the first 14 subjects in this list to ensure and to measure assay sensitivity. Randomly chosen terms from other SMQs were included as the remaining 985 AE terms of the AE database.
2.2.3 Assembly of Datasets to Evaluate Other Patient Attributes
A dataset of concomitant medication (CM) was created by serially assigning Greek alphabet names as mock medications. Anatomical Therapeutic Class (ATC) classifications were assigned for demonstration purposes [3]. An asterisk (*) was added to two of the mock medication names to designate that they also were thought to cause severe cutaneous adverse reactions (SCARs), to illustrate the utility of this methodology.
2.3 Screening the Adverse Event Database for Specific SMQ Terms
The database to be screened (e.g. JMP table AE) and the Demography table (JMP table DM) are joined using the Table Join function in JMP and the resulting table is named DM_AE (Fig. 1). The JMP dataset table containing the SMQ terms is opened. Under the Tables menu of the JMP tool bar, the Join function is elected (arrow 1). This will prompt the user to select a table to join to the first one opened. For example, if DM_AE were opened first, one would select the table containing the SMQ terms (arrow 2). One then selects the columns that should be matched between the two tables (arrow 3 and then 4). If the SMQ table contains multiple columns, e.g. PT terms, PT codes, LLT terms, and LLT codes, one would select the desired column, PT terms, and then the corresponding column containing PT terms from DM_AE. This Join function dialogue box also asks whether mismatches should be included; however, these boxes are not selected in this procedure. A name (DMAE_SMQ) is selected for the table that will contain the hybridized terms (arrow 5) and then OK is selected in the Join dialogue box (arrow 6). The resulting table (Fig. 2) will be a subset of the AE database corresponding to those cases where a potential AE corresponding to the term in the SMQ has occurred.
Fig. 1.
Demonstration of the Table Joining function to perform SMQ hybridization. AE adverse event, AE_1001 adverse event data set of Study 100, BMI body mass index, CM_1001 concomitant medications data set of Study 1001, DM_1001 demography data set of Study 1001, DM_AE demography data set of Study 1001 joined to the adverse event dataset, M male, PT MedDRA preferred term, SCARS Serious cutaneous adverse reactions (SMQ), SMQ Standardized MedDRA Query, SMQ SCARS 18.0 the dataset containing the Serious cutaneous adverse reactions (SMQ) preferred terms, USUBJID unique subject identification
Fig. 2.
The resulting AE table after the SMQ hybridization procedure has been performed. AE adverse event, DM_1001 demography data set of Study 1001, PT MedDRA preferred term, SMQ Standardized MedDRA Query, SMQ SCARS the dataset containing the Serious cutaneous adverse reactions (SMQ) preferred terms, USUBJID unique subject identification
2.4 Investigating Other Subject Attributes
JMP datasets with other attributes (e.g. concomitant medications, pharmacodynamics variables) can be added after the hybridization procedure by using the Table Join function, linking the datasets by the USUBJID variable. One can also join the other dataset to the AE_DM table before the SMQ hybridization procedure. In this study the CM dataset was joined to the DMAE_SMQ to form the dataset DMAESMQ_CM (Fig. 3).
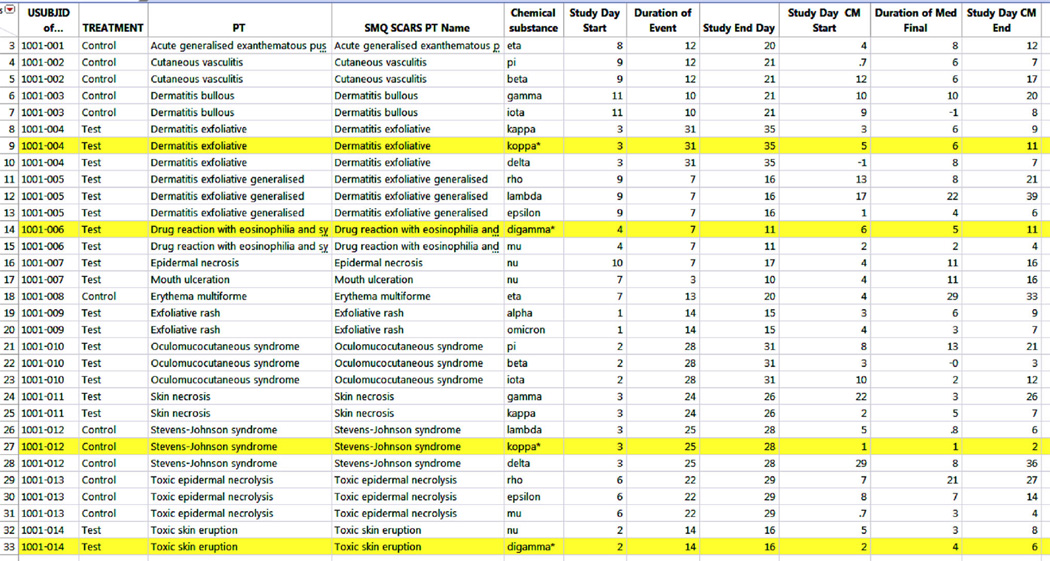
Fig. 3.
The SMQ hybridized AE Table joined to a Concomitant Medication Database. The highlighted rows are for cases where subjects were taking hypothetical drugs (marked with an asterisk) with a prior association with causing serious cutaneous adverse reactions. AE adverse event, CM concomitant medications, PT MedDRA preferred term, SMQ Standardized MedDRA Query, SMQ SCARS the dataset containing the Serious cutaneous adverse reactions (SMQ) preferred terms, USUBJID unique subject identification
3 Results
3.1 SMQ Hybridization
Joining the AE to the SMQ dataset reduced the 999-row AE set to a 15-row dataset DMAE_SMQ, where each row had an event that was also included in the Severe cutaneous adverse reactions (SMQ) set of PTs. While 14 SCAR terms were included in the AE list to ensure there was a target for the SMQ to hybridize, PT Mouth ulceration was included in the AE dataset as a target for Systemic lupus erythematosus (SMQ). This term is also in Severe cutaneous adverse reactions (SMQ) and so it was also picked up with the hybridization technique.
3.2 Investigating Other Subject Attributes
Joining a CM dataset allowed for simulation of a realistic scenario where one would look for concomitant medications that could confound the association of the test drug and the AE (Fig. 3). In this case, four subjects with SCARs (1001–004, –006, –012, and –016) were taking medications known to cause SCARs. For subjects 1001–004 and –006, the concomitant medication was taken after the AE, so it does not confound the relation of the test drug and the event. Subject 1001–012 took the concomitant medication prior to the event and subject 1001–014 took the concomitant medication on the same day as the event, so the events of these two subjects are confounded by the concomitant medications.
4 Discussion
In this communication, a method is described for searching AE databases using SMQs to identify potential cases of interest. The method is reminiscent of RNA hybridization in that the SMQ terms function like a probe and the Table Join function acts like RNase, which eliminates the unhybridized terms leaving only those complementary to the probe. An illustrative example is provided to demonstrate the potential use and advantages of this technique. A limitation of this study is that it was based on a hypothetical, versus an actual clinical trial dataset; however, since the datasets created for this study were formatted in the same manner as actual datasets the utility of this methodology was not compromised.
4.1 Availability of Tools for SMQ Searches
While there are numerous statistical packages that allow linkage and partitioning of AE tables, there are few publications describing tools that may be used for SMQ searches. A program known as MAED (MedDRA-based Adverse Event Diagnostics) was developed and is used at the FDA to search for SMQ terms in adverse event databases [4]. This tool allows the user to specify test and reference treatments from an appropriate database and MedDRA levels from the AE database. Different statistical comparisons may be made between the test and control treatments. Numerator analyses may be performed on single-arm studies, such as open-label extensions. One may also drill down on results to identify patients identified with AEs corresponding to terms in SMQs. Directions for downloading and setting up the MAED program locally have been published [5].
Using SMQs to find terms is advantageous because the SMQ standardized term lists have been assembled though the collective efforts of the Council for International Organizations of Medical Sciences (CIOMS) and the MedDRA Maintenance and Support Services Organization. However, one may also augment the list of terms by adding or deleting PTs depending on their clinical relevance, although the resulting term list would then not be considered an SMQ [6]. Similarly, using this technique, one can easily merge terms of SMQs that describe a common clinical concept, as was done in the New Drug Application #22271 for Nesina (alogliptin), which combined the SMQs of Serious cutaneous adverse reactions, Angioedema, and Anaphylactic reactions to form a probe for systemic allergic reactions [7].
4.2 Useful Features of the SMQ Hybridization Technique for Detecting Potential AEs using SMQs in Databases
The detection of potential AEs with SMQs using the hybridization technique is convenient because it uses a readily available software program and is easy to perform.
As demonstrated in the illustrative example, the ability to link other database tables to the resulting SMQ-matched terms allows the investigator to perform useful analyses. In the example in this study, the potential for concomitant medication to be considered a confounding factor was eliminated because the event onset preceded the administration of a concomitant medication. One may investigate other demographic factors, such as medical history and genetic data, or study events, such as laboratory and vital signs that may be associated with the AE.
A critical step in the identification of AEs with SMQs is the review and adjudication of cases to verify whether they actually had the medical condition represented by the SMQ used to retrieve the terms. Verification often includes evaluating laboratory values, prior and concomitant medications, other AEs from the subject’s medical history, and whether the event preceded or followed potentially confounding factors. The hybridization technique allows the investigation of all of these basic data structures to support the safety analyses.
5 Conclusions
A method for detecting AEs that correspond to SMQ PTs is described. This method simplifies the difficult task of sorting through very large clinical trial databases during safety analyses. It can be done with software that is readily available and the methodology adapted so that the cases that are identified can be further evaluated using demographic and other clinical trial databases.
Key Points.
A method is described for detecting adverse events in clinical trial databases that correspond to Standardised MedDRA Query preferred terms.
This method can be adapted for personalized lists of adverse events of special interest and for investigating potentially associated demographic or confounding factors.
Acknowledgments
The author would like to thank Drs. Judy Harrison, Chief Medical Officer, MedDRA MSSO and Anna Zhao-Wang, Deputy Director, MedDRA MSSO for their critical reading of the manuscript and suggestions.
Funding No funding was received for the conduct or publication of this study.
Footnotes
Compliance with Ethical Standards
Ethical approval This study did not require approval by an ethics committee because it did not involve human subjects or actual data from human subjects.
Conflict of interest The author is a medical officer with the United States Food and Drug administration. The views expressed in this paper represent the opinion of the author and do not necessarily represent the views of the FDA.
The author is a Deputy Topic Leader for the FDA in the ICH M1 Points to Consider Group and also directs the FDA’s MedDRA Coordinating Working Group.
The author has no affiliation or financial interest in the software vendor (SAS® Institute).
References
- 1.MedDRA Maintenance and Service Organization. Standardised MedDRA Queries. [Accessed 10 Oct 2015]; Available from: http://www.meddra.org/how-to-use/tools/smqs.
- 2.MedDRA Maintenance and Service Organization. MedDRA Web-Based Browser and MedDRA Desktop Browser. [Accessed 10 Oct 2015]; Available from: http://www.meddra.org/browsers.
- 3.World Health Organization. ATC/DDD Index 2015. [Accessed 10 Oct 2015]; Available from: http://www.whocc.no/atcddd/
- 4.Li XJ, Cooper C, Luo Z. MAED service: FDA-developed tool for clinical AE Data Signal Detection; Computaional Science Symposium; March 18th–19th, 2013; Silver Spring, MD. 2013. [Google Scholar]
- 5.Holland PC. MAED Service: A SAS Tool for Implementing SMQs and Performing MedDRA-Based Analyses of AE Data. Pharmaceutical SAS Users Group 2010; Paper AD13; May 23–26, 2010; Orlando, Florida. [Google Scholar]
- 6.MedDRA Maintenance and Service Organization. MedDRA Web-based Browser User Guide. 2015
- 7.Medical Officer review for Nesina (alogliptin), NDA 22271S000, Drugs@FDA. [Accessed 20 Dec 2015];2013 http://www.accessdata.fda.gov/drugsatfda_docs/nda/2013/022271Orig1s000MedRedt2.pdf.