Abstract
Introduction
Escherichia coli (E. coli) is the most common cause of urinary infections in hospitals. The aim of this study was to evaluate the ERIC-PCR method for molecular typing of uropathogenic E. coli strains isolated from hospitalized patients.
Methods
In a cross sectional study, 98 E. coli samples were collected from urine samples taken from patients admitted to Baqiyatallah Hospital from June 2014 to January 2015. The disk agar diffusion method was used to determine antibiotic sensitivity. DNA proliferation based on repetitive intergenic consensus was used to classify the E. coli strains. The products of proliferation were electrophoresed on 1.5% agarose gel, and their dendrograms were drawn. The data were analyzed by online Insillico software.
Results
The method used in this research proliferated numerous bands (4–17 bands), ranging from 100 to 3000 base pairs. The detected strains were classified into six clusters (E1–E6) with 70% similarity between them.
Conclusion
In this study, uropathogenic E. coli strains belonged to different genotypic clusters. It was found that ERIC-PCR had good differentiation power for molecular typing of uropathogenic E. coli strains isolated from the patients in the study.
Keywords: uropathogenic E. coli, urinary tract infection, ERIC-PCR, molecular typing
1. Introduction
E. coli, an abbreviation for Escherichia coli, is a gram negative bacillus of the Enterobacteriaceae family, and it is commonly found in the intestines of warm-blooded animals. Most strains of E. coli are harmless, but some strains cause food poisoning, diarrhea, and urinary tract infections (1). Many bacteria can cause infection in the urinary tract. In fact, 80 to 90% of urinary tract infections in the world are caused by E. coli. Extraintestinal pathogenic strains of E. coli that cause urinary tract infections in humans are known as uropathogenic E.coli (2–7). Knowledge of the epidemiology of microbial populations is important in the field of medical microbiology. It is possible to identify infection reservoirs, examine the prevalence of hospitalization infections, and identify the type of microbial pathogenic agents via molecular typing (8). Enterobacterial Repetitive Intergenic Consensus (ERIC) has conserved sequences and repeated sequences that can be seen in bacteria or in some fungi. These sequences are generally found in the whole chromosome in the inter-genome (9–14). In the ERIC-PCR technique, which is based on amplification of random distribution of inter-genome parts, various organisms can be distinguished from one another. This means that bacteria differ in the frequency of repeated sequences, and, therefore, this method produces different patterns of specific primers in electrophoresis. In this technique, primers are designed to be able to attach to repeated sequences and to amplify the distances among those attached primers to repeated sequences (9–14). The population type of patients referred to Baqiyatallah Hospital is different from other hospitals in Tehran, so we aimed to do this study to examine the ERIC-PCR molecular technique in order to classify strains of E. coli isolated from clinical cases in Baqiyatallah Hospital.
2. Material and Methods
2.1. Study population and bacterial isolation
In a cross-sectional study over a period of eight months, from June 2014 to January 2015, all 98 E. coli strains isolated from patients who suffered from urinary tract infection in Baqiyatallah Hospital were subjected. These patients were of different age groups and were examined according to standard methods of bacteriology and biochemistry.
2.2. Antibiogram Test
A disc agar-diffusion method, as recommended by Clinical and Laboratory Standards Institute (CLSI) (2013), was used to determine antibiotic sensitivity (15). The antibiogram test was performed using the antibiotics Ampicillin (AM) - Amikacin (AN) - Ciprofloxacin (CP) - Ceftazidime (CAZ) - Ceftriaxone (CRO) - Cotrimoxazole (SXT) - Norfloxacin (NOR) - Nalidixic acid (NA) - Gentamicin (GM) - Nitrofurantoin (FM), and the ATCC 25922 strain of E. coli was used as the quality control.
2.3. DNA preparation
After bacterial isolation, E. coli isolates were cultured in Luria Bertani (LB) broth. After 16–18 hours of incubation at 37 °C, DNA extraction was performed using the alkaline lysis method. The DNA concentration was determined by nanodrop.
2.4. Primers
The oligonucleotide primers used in this study were: 5′-ATGTAAGCTCCTGGGGATTCAC-3′ ERIC 1 and5′-AAGTAAGTGACTGGGGTGAGCG-3′ ERIC 2 (13).
2.5. ERIC-PCR
ERIC-PCR reactions were performed in 20 μl volumes containing 1 μl of each primer, 10 μl of the master mix, 1.5 μl of MgCl2, 2 μl of template DNA, and 3.5 μl of deionized water. The ERIC-PCR reaction was as follows: initial denaturation at 94 °C for 1 minute, with the next 30 cycles consisting of a denaturation step at 94 °C for 30 seconds, annealing at 52 °C for 35 seconds, extension at 72 °C for 4 minutes, and a final extension for 5 minutes at 72 °C. PCR products were electrophoresed using a 1.5% agarose (Merck) gel electrophoresis at 80 V for 180 minutes then visualized under UV in a gel documentation system. The zero-one manual method was used to count the bands, and, then, the data were entered on the following site: http://insilico.ehu.es/dice_upgma/, and the dendrograms were drawn.
3. Results
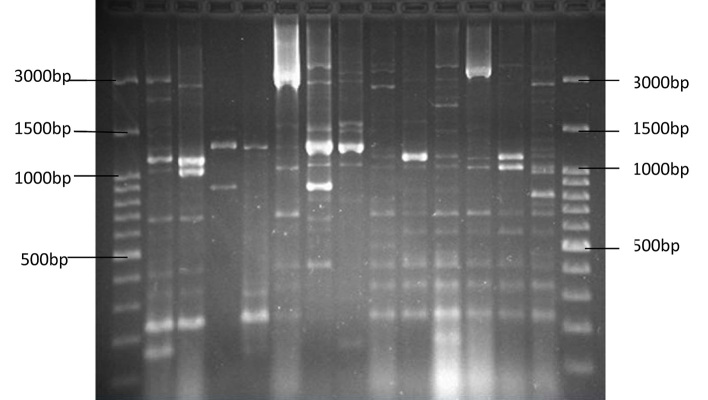
Of all 98 samples examined, 18 were from men and 80 were from women. The age and gender distribution of the patients with urinary tract infection were as follows: six girls under one year of age, five girls between the ages of 1–10, three girls between the ages of 10–20, 14 women between the ages of 20–30, 12 women between the ages of 30–40, 12 women and two men between the ages 40–50, 10 women and four men between the ages 50–60, six women and four men between the ages 60–70, 10 women and five men between the ages 70–80, and two women and three men between the ages of 80–90. The results of the study on antibiotic resistance indicated the maximum resistance was shown to Ampicillin and the minimum to Nitrofurantoin. ERIC-PCR patterns were used to calculate the visibility and placement of the gels according to their molecular weights and molecular markers. Bands for each sample were counted (Figure 1). After dendrogram analysis, it was shown that ERIC-PCR differentiated the isolates into six clusters, E1 – E6, with 70% similarity (Figure 2). In the strains that were studied, the maximum number, 62, belonged to the E2 cluster, and the minimum, 1, belonged to the E6 cluster. Twenty-one of the studied strains were in E1, 3 strains in E3, 2 strains in E4, 5 strains in E5, and 1 strain in the E6 cluster. Three of the 98 strains did not have bands and, therefore, could not be genotyped. The most frequent band was 1350 bp, which was found in 70 strains, and the least frequent was 170 bp, which was observed in two strains. Study and comparison of the results obtained from the antibiogram test with dendrogram yielded noteworthy results, i.e., samples in the E1 cluster were sensitive to Gentamicin and Nitrofurantoin, those in the E5 cluster were female and resistant to Ampicillin and sensitive to Nitrofurantoin, and those in E3 cluster had completely similar antibiotic patterns and were in the 70–90 age range.
Figure 1.
Results of ERIC PCR on some of the E. coli strains, with the first column in both sides showing Ladder 100 bp
Figure 2.
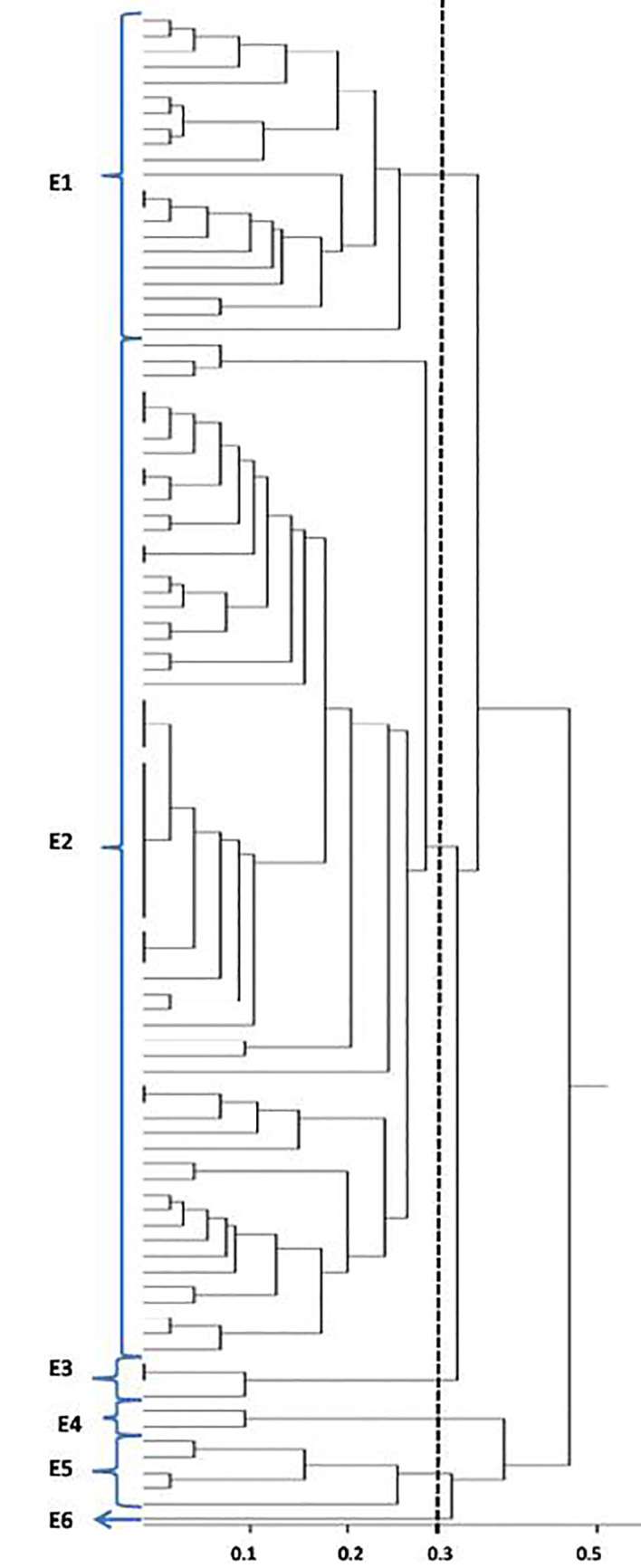
Dendrogram drawn for the strains studied in this research
4. Discussion
This study was conducted on 98 isolates recovered from urine samples of patients with urinary infections admitted to Baqiyatallah Hospital. All of the isolates were studied genetically using the ERIC PCR. The most frequent band was 1350 bp, which was found in 70 strains, and the least frequent band was 170 bp, which was observed in two strains. The maximum number, 62, belonged to the E2 cluster, and the minimum, 1, belonged to the E6 cluster. The comparison of the results obtained from the antibiogram test with dendrogram showed that samples in the E1 cluster were sensitive to Gentamicin and Nitrofurantoin, those in the E5 cluster were female and resistant to Ampicillin and sensitive to Nitrofurantoin, and those in the E3 cluster had completely similar antibiotic patterns and were in the 70–90 age range. Versalvic et al. concluded that fingerprinting techniques were very powerful tools for identification and classification of bacteria and in molecular epidemiology of human and plant pathogens (13).
In 2002, Wanderley Dias da Silveira et al. isolated 49 E. coli strains in chickens with septicemia, swollen head syndrome, omphalitis, and in some without any clinical symptoms (16). The ERIC PCR technique determined their types. These isolates were classified into four main groups, A to D, and the survey results also showed that the ERIC PCR method can be a good alternative to molecular techniques, such as RAPD PCR and RFLP (16). Meacham et al., at the University of Michigan evaluated genotyping of a large number of E. coli samples and concluded that more than one ERIC-PCR method should be used in determining genetic similarities through genotyping (11). In 2006, Lindsay et al. studied ERIC sequence distribution in the whole genomes of intestinal bacteria, including E. coli and Shigella. They concluded that ERIC sequence in regions close to genes with high expression levels was more inclined to be copied, and that the ERIC-PCR was widely used for genotyping because the ERIC sequence was small (12). VivekPrabhu et al. used the ERIC-PCR method for genotyping 40 samples of E. coli isolated from cows infected with mastitis, and 37 of their samples could be genotyped. Seven isolates were in the phylogenetic group A, 17 isolates were in the phylogenetic group B, and 13 isolates were not in any groups because no similarity with other groups. They reached the conclusion that ERIC-PCR was a quick, reliable, and suitable method for differentiating E. coli strains (17). In 2011, Ramazanzadeh et al. used ERIC-PCR to evaluate the genetic diversity of E. coli isolated from hospital samples. They studied 230 clinical samples and found that 205 of them could be genotyped in 20 clusters, with most of them being in the phylogenetic group D. The high genetic diversity in the isolated E. coli indicated the low rate of infection at the hospital (18). Xiu-Yan Lang et al. used ERIC-PCR for genotyping enterotoxigenic E. coli isolated from cows and found most isolates belonged to the phylogenetic group D. They concluded that the ERIC-PCR is a fast, efficient, and repeatable method for genotyping (19). The findings indicated that the ERIC-PCR method is a fast and excellent way to describe the diversity of phylogenetic classification and evolution racial structure. Using this method, most of the isolates were classified and separated into different categories. Another result is that E. coli were obtained from a variety of distinct clones that have been studied from a variety of bacteria in clinical samples.
The choice of a sub typing technique is depend on the skill level and resources of the laboratory and the purpose of the study (20). In our previous studies, we evaluated ERIC-PCR for subtyping of sporadic and epidemic strains of different foodborne bacterial pathogens in Iran (20–26) and found this technique had a good subtyping power. As one of the most important and prevalent uropathogen organism in Iran (27–30), E. coli was subjected here to ERIC-PCR. As indicated in previous studies, our finding showed here an acceptable discriminatory power of this methods for subtyping of uropathogenic E. coli.
5. Conclusions
Our results showed that the ERIC PCR technique was a rapid method for describing then genetic diversity of E. coli strains. The strains of E. coli were composed of various distinct colonies that show the variety of these bacteria in clinical samples. For future studies, we suggest the use of more samples from different hospitals and the comparison of the discrimination of the ERIC PCR using other methods, such as PFGE. We hope the results of our study will be useful for molecular epidemiology of uropathogenic E. coli and upgrading the public health in our country.
Acknowledgments
This research was supported by the Pharmaceutical Sciences Branch of Islamic Azad University, Tehran, Iran and the Molecular Biology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran. We thank Dr. Najafi, Mr. Daryoosh Ghasemi, and Ms. Fatemeh Pourali for their technical assistance.
Footnotes
iThenticate screening: January 28, 2016, English editing: March 20, 2016, Quality control: April 04, 2016
Conflict of Interest: There is no conflict of interest to be declared.
Authors' contributions: All authors contributed to this project and article equally. All authors read and approved the final manuscript.
References
- 1.Kaper JB, Nataro JP, Mobley HL. Pathogenic escherichia coli. Nature Reviews Microbiology. 2004;2(2):123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Sheerin NS. Urinary tract infection. Medicine. 2011;39(7):384–9. doi: 10.1016/j.mpmed.2011.04.003. [DOI] [Google Scholar]
- 3.Lee JB, Neild GH. Urinary tract infection. Medicine. 2007;35(8):423–8. doi: 10.1016/j.mpmed.2007.05.009. [DOI] [Google Scholar]
- 4.Farshad S, Anvarinejad M, Tavana AM, Ranjbar R, Japoni A, Zadegan RM, et al. Molecular epidemiology of Escherichia coli strains isolated from children with community acquired urinary tract infections. African J Microbiol Res. 2011;5(26):4476–83. doi: 10.5897/AJMR11.285. [DOI] [Google Scholar]
- 5.Momtaz H, Karimian A, Madani M, Safarpoor Dehkordi F, Ranjbar R, Sarshar M, et al. Uropathogenic Escherichia coli in Iran: serogroup distributions, virulence factors and antimicrobial resistance properties. Ann Clin Microbiol Antimicrob. 2013;12(8) doi: 10.1186/1476-0711-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farshad S, Ranjbar R, Japoni A, Hosseini M, Anvarinejad M, Mohammadzadegan R. Microbial susceptibility, virulence factors, and plasmid profiles of uropathogenic Escherichia coli strains isolated from children in Jahrom, Iran. Archives of Iranian medicine. 2012;15(5):312–6. doi: 012155/AIM.0013. [PubMed] [Google Scholar]
- 7.Anvarinejad M, Farshad S, Ranjbar R, Giammanco GM, Alborzi A, Japoni A. Genotypic analysis of E. coli strains isolated from patients with cystitis and pyelonephritis. Iran Red Crescent Med J. 2012;14(7):408–16. [PMC free article] [PubMed] [Google Scholar]
- 8.Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, et al. Microbial DNA typing by automated repetitive-sequence-based PCR. Journal of clinical microbiology. 2005;43(1):199–207. doi: 10.1128/JCM.43.1.199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulton CS, Higgins CF, Sharp PM. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol Microbiol. 1991;5(4):825–34. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 10.Sharples GJ, Lloyd RG. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990;18(22):6503–8. doi: 10.1093/nar/18.22.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meacham KJ, Zhang L, Foxman B, Bauer RJ, Marrs CF. Evaluation of genotyping large numbers of Escherichia coli isolates by enterobacterial repetitive intergenic consensus-PCR. J Clin Microbiol. 2003;41(11):5224–6. doi: 10.1128/JCM.41.11.5224-5226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson LA, Sharp PM. Enterobacterial repetitive intergenic consensus (ERIC) sequences in Escherichia coli: Evolution and implications for ERIC-PCR. Mol Biol Evol. 2006;23(6):1156–68. doi: 10.1093/molbev/msj125. [DOI] [PubMed] [Google Scholar]
- 13.Versalovic J, Koeuth T, Lupski R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Research. 1991;19(24):6823–31. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Versalovic J, Schneider M, De Bruijn FJ, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods in molecular and cellular biology. 1994;5(1):25–40. [Google Scholar]
- 15.Performance standards for antimicrobial susceptibility testing twenty second informational supplement USA. Clinical and Laboratory Standards Institute; 2013. [Google Scholar]
- 16.da Silveira WD, Ferreira A, Lancellotti M, Barbosa IA, Leite DS, de Castro AF, et al. Clonal relationships among avian Escherichia coli isolates determined by enterobacterial repetitive intergenic consensus (ERIC)–PCR. Veterinary Microbiology. 2002;89(4):323–8. doi: 10.1016/S0378-1135(02)00256-0. [DOI] [PubMed] [Google Scholar]
- 17.Prabhu V, Isloor S, Balu M, Suryanarayana VVS, Rathnamma D. Genotyping by ERIC-PCR of Escherichia coli isolated from bovine mastitis cases. Indian Journal of Biotechnology. 2010;9(3):298–301. [Google Scholar]
- 18.Ramazanzadeh R, Zamani S, Zamani S. Genetic diversity in clinical isolates of Escherichia coli by enterobacterial repetitive intergenic consensus (ERIC)–PCR technique in Sanandaj hospitals. Iran J Microbiol. 2013;5(2):126. [PMC free article] [PubMed] [Google Scholar]
- 19.Lang XY, Zhang YL, Jiang HT, Liu J NIHB. Development of an enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) to detect and genotype enterotoxigenic Escherichia coli of calf origin. Afr J Microbiol Res. 2013;7(31):4001–5. doi: 10.5897/AJMR2013.5748. [DOI] [Google Scholar]
- 20.Ranjbar R, Karami A, Farshad S, Giammanco GM, Mammina C. Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiol. 2014;37(1):1–15. [PubMed] [Google Scholar]
- 21.Ranjbar R, Naghoni A, Yousefi S, Ahmadi A, Jonaidi N, Panahi Y. The Study of Genetic Relationship Among Third Generation Cephalosporin-resistant Salmonella enterica Strains by ERIC-PCR. Open Microbiol J. 2013;7:142–5. doi: 10.2174/1874285801307010142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ranjbar R, Mirsaeed Ghazi F. Antibiotic Sensitivity Patterns and Molecular Typing of Shigella sonnei Strains Using ERIC-PCR. Iran J Public Health. 2013;42(10):1151–7. [PMC free article] [PubMed] [Google Scholar]
- 23.Ranjbar R, Rahbar M, Naghoni A, Farshad S, Davari A, Shahcheraghi F. A cholera outbreak associated with drinking contaminated well water. Arch Iran Med. 2011;14(5):339–40. [PubMed] [Google Scholar]
- 24.Ranjbar R, Hosseini MJ, Kaffashian AR, Farshad S. An outbreak of shigellosis due to Shigella flexneri serotype 3a in a prison in Iran. Arch Iran Med. 2010;13(5):413–6. [PubMed] [Google Scholar]
- 25.Pourshafie MR, Bakhshi B, Ranjbar R, Sedaghat M, Sadeghifard N, Zaemi Yazdi J, Parzadeh M, Raesi J. Dissemination of a single Vibrio cholerae clone in cholera outbreaks during 2005 in Iran. J Med Microbiol. 2007;56(Pt 12):1615–9. doi: 10.1099/jmm.0.47218-0. [DOI] [PubMed] [Google Scholar]
- 26.Khakabimamaghani S, Najafi A, Ranjbar R, Raam M. GelClust: a software tool for gel electrophoresis images analysis and dendrogram generation. Comput Methods Programs Biomed. 2013;111(2):512–8. doi: 10.1016/j.cmpb.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Tajbakhsh E, Khamesipour F, Ranjbar R, Ugwu IC. Prevalence of class 1 and 2 integrons in multi-drug resistant Escherichia coli isolated from aquaculture water in Chaharmahal Va Bakhtiari province, Iran. Ann Clin Microbiol Antimicrob. 2015 Jul 31;14:37. doi: 10.1186/s12941-015-0096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jahandeh N, Ranjbar R, Behzadi P, Behzadi E. Uropathogenic Escherichia coli virulence genes: invaluable approaches for designing DNA microarray probes. Cent European J Urol. 2015;68(4):452–8. doi: 10.5173/ceju.2015.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behzadi P, Najafi A, Behzadi E, Ranjbar R. Microarray long oligo probe designing for Escherichia coli: an in-silico DNA marker extraction. Cent European J Urol. 2016;69(1):105–11. doi: 10.5173/ceju.2016.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranjbar R, Haghi-Ashtiani MT, Jafari NJ, Abedini M. The prevalence and antimicrobial susceptibility of bacterial uropathogens isolated from pediatric patients. Iran J Publ Health. 2009;38(2):134–138. [Google Scholar]