Abstract
Background:
The catechol-O-methyltransferase (COMT) enzyme plays a crucial role in dopamine degradation, and the COMT Val158Met polymorphism (rs4680) is associated with significant differences in enzymatic activity and consequently dopamine concentrations in the prefrontal cortex. Multiple studies have analyzed the COMT Val158Met variant in relation to antipsychotic response. Here, we conducted a meta-analysis examining the relationship between COMT Val158Met and antipsychotic response.
Methods:
Searches using PubMed, Web of Science, and PsycInfo databases (03/01/2015) yielded 23 studies investigating COMT Val158Met variation and antipsychotic response in schizophrenia and schizo-affective disorder. Responders/nonresponders were defined using each study’s original criteria. If no binary response definition was used, authors were asked to define response according to at least 30% Positive and Negative Syndrome Scale score reduction (or equivalent in other scales). Analysis was conducted under a fixed-effects model.
Results:
Ten studies met inclusion criteria for the meta-analysis. Five additional antipsychotic-treated samples were analyzed for Val158Met and response and included in the meta-analysis (ntotal=1416). Met/Met individuals were significantly more likely to respond than Val-carriers (P=.039, ORMet/Met=1.37, 95% CI: 1.02–1.85). Met/Met patients also experienced significantly greater improvement in positive symptoms relative to Val-carriers (P=.030, SMD=0.24, 95% CI: 0.024–0.46). Posthoc analyses on patients treated with atypical antipsychotics (n=1207) showed that Met/Met patients were significantly more likely to respond relative to Val-carriers (P=.0098, ORMet/Met=1.54, 95% CI: 1.11–2.14), while no difference was observed for typical-antipsychotic-treated patients (n=155) (P=.65).
Conclusions:
Our findings suggest that the COMT Val158Met polymorphism is associated with response to antipsychotics in schizophrenia and schizo-affective disorder patients. This effect may be more pronounced for atypical antipsychotics.
Keywords: COMT, Val158Met, antipsychotics, schizophrenia, clinical response
Introduction
Schizophrenia (SCZ) is a debilitating neurodevelopmental disorder with high heritability (~0.8) (McGuffin et al., 1995). While antipsychotic drugs are currently the most effective treatment, clinical response varies significantly between patients, with an overall response rate of only 50% (Lieberman et al., 2005). The field of pharmacogenetics aims to optimize treatment at the individual level by leveraging our knowledge of how genetic variants influence clinical response. Since all antipsychotics target the dopamine (DA) system to various extents by binding to dopamine D2 receptors, pharmacogenetics studies have largely focused on DA system-related genes, including those encoding the dopamine D2 receptor (DRD2) and catechol-O-methyltransferase (COMT). Previous studies have identified associations for genes such as DRD2 and DRD3 (dopamine D3 receptor) in relation to response to antipsychotics (Hwang et al., 2010; Zhang et al., 2010). Other genes implicated in response include 5-HT2A and 5-HT2C (Pouget and Müller, 2014).
The COMT gene is located within the 22q11 chromosomal locus, a region highly implicated in SCZ risk (Badner and Gershon, 2002; Lewis et al., 2003). The COMT enzyme plays a significant role in cortical DA degradation (Lachman et al., 1996). The majority of pharmacogenetics studies examining COMT in relation to antipsychotic response have focused on the functional variant Val158Met (rs4680). Other putatively functional variants have also been examined, though relatively infrequently (Chen et al., 2004; Molero et al., 2007). The Val158Met variant is located within exon 4 of the COMT gene and involves a G-to-A nucleotide switch, leading to a valine (Val) to methionine (Met) substitution at codon 158. The Met-allele form of the enzyme has lower thermostability and has been observed to exhibit reduced enzymatic activity, contributing to increased levels of cortical dopamine (Chen et al., 2004); the Met/Met homozygote has 3- to 4-fold lower enzymatic activity than the Val/Val homozygote, while the heterozygote has intermediate activity (Chen et al., 2004).
Studies of COMT Val158Met and antipsychotic response in SCZ and schizo-affective disorder have yielded mixed results. A number of studies have observed that the Met-allele is associated with better overall clinical response as well as greater improvement in negative symptoms and cognitive abilities such as working memory (Bertolino et al., 2004, 2007; Molero et al., 2007; Woodward et al., 2007; Pelayo-Terán et al., 2011). However, other studies have either been unable to replicate these findings (Yamanouchi et al., 2003; Gao et al., 2012; Tybura et al., 2012; Bishop et al., 2015) or observed the opposite effect, with the Met-allele predicting nonresponse to antipsychotic therapy (Illi et al., 2003; Porcelli et al., 2009). In an effort to clarify the COMT Val158Met variant’s role in antipsychotic response, we conducted a meta-analysis of the existing literature plus additional data. In our primary analysis, we examined the association between this variant and both binary and continuous measures of response to antipsychotics. The meta-analysis focused solely on clinical response to antipsychotics, since the overwhelming majority of Val158Met pharmacogenetics studies examined this phenotype.
Methods and Materials
Literature Search
A literature search was conducted for articles written in English published up to March 1, 2015, examining the COMT Val158Met variant and antipsychotic response in SCZ and schizo-affective disorder patients. We performed searches on PubMed, PsychInfo, and Web of Science using all combinations of the key terms ‘COMT,’ ‘Catechol O-methyltransferase,’ and ‘Val158Met,’ with the phrases ‘antipsychotic response’ and ‘SCZ treatment response.’ Criteria for inclusion in the current study consisted of: (1) the association between the COMT Val158Met polymorphism and antipsychotic response was assessed; (2) the patients were diagnosed with SCZ or schizoaffective disorder based on DSM-III, DSM-IV, DSM-V, or ICD-10 criteria and their diagnoses were verified through a standardized structured clinical interview; (3) antipsychotic response was evaluated according to a standardized clinical rating scale, such as the Brief Psychiatric Rating Scale (BPRS) or the Positive and Negative Syndrome Scale (PANSS); (4) drug response (symptom severity) was evaluated at a minimum of 2 time points, one of which was directly prior to starting antipsychotic therapy; and (5) patients were evaluated for a minimum of 2 weeks for response to treatment. Studies investigating nonclinical aspects of response, such as cognitive response, were excluded. We also searched through the reference sections of articles meeting these criteria to find other articles fit for inclusion. Authors were contacted for data when the data necessary for analysis was not reported in articles that met inclusion criteria.
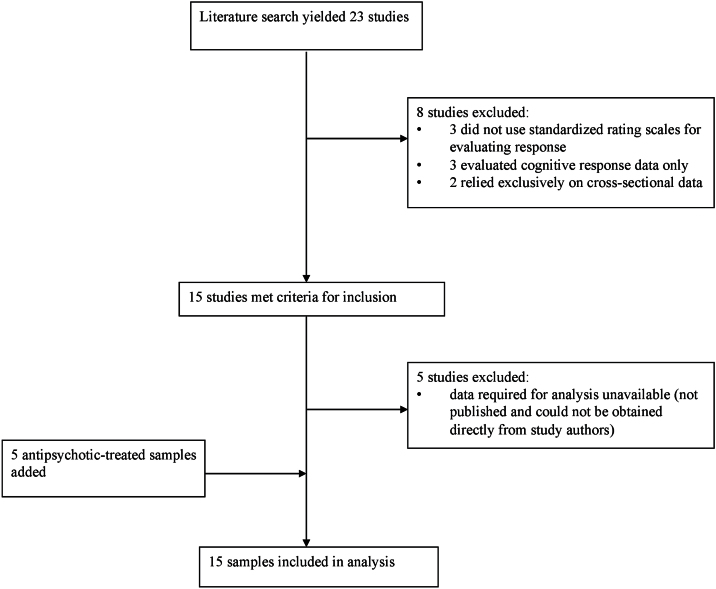
In total, the literature search yielded 23 peer-reviewed articles published before March 1, 2015. Two of these articles were identified through searching the reference sections of the other 21 articles (Porcelli et al., 2009; Kang et al., 2010). The literature search process is outlined in Figure 1. In total, 15 articles met inclusion criteria for the current study. However, 11 of the 15 studies did not have group-level data (ie, number of responders/nonresponders) available online (Molero et al., 2007; Ikeda et al., 2008; Fijal et al., 2009; Need et al., 2009; Porcelli et al., 2009; Kang et al., 2010; Gao et al., 2012; Tybura et al., 2012; Zhao et al., 2012; Bishop et al., 2015; Bosia et al., 2015). Another 2 studies focused on first-episode psychosis and did not provide the response/nonresponse data for its subset of SCZ or schizo-affective disorder patients (Pelayo-Terán et al., 2011; Prata et al., 2012). We attempted to contact each of these groups and obtained data from 7 of these studies (Molero et al., 2007; Ikeda et al., 2008; Porcelli et al., 2009; Pelayo-Terán et al., 2011; Gao et al., 2012; Tybura et al., 2012; Bishop et al., 2015). The studies for which we were unable to retrieve the required data were excluded (Fijal et al., 2009; Need et al., 2009; Kang et al., 2010; Prata et al., 2012; Zhao et al., 2012; Bosia et al., 2015). We added data from 5 additional samples that our group has published on previously (Hwang et al., 2012; Zai et al., 2012; Tiwari et al., 2013; Gonçalves et al., 2014) to these 10 studies’ samples to give a total of 15 samples (ntotal=1416). Two of these samples had previously been analyzed for Val158Met and antipsychotic response (Nolan et al., 2006), while the other 3 had not been. Figure 1 illustrates the literature search process. The demographic and clinical characteristics of the 15 samples are described in Table 1. All 15 samples included data for the number of responders and nonresponders. Ten of these samples also had data for change in total score (%). Of these 10 samples, 6 further included positive and negative symptom subscale score change data. We later contacted the authors for studies with a mix of atypical and typical-antipsychotic-treated patients to obtain response data for each antipsychotic type separately.
Figure 1.
Literature search process results.
Table 1.
Characteristics of Studies Investigating the Association between COMT Polymorphism Val158Met and Antipsychotic Drug Response in Schizophrenia/Schizo-Affective Disorder Patients
| Study/Sample | N | Ethnicity | MAF | Design | Medication | Response Criteria |
Responders/
Nonresponders |
|---|---|---|---|---|---|---|---|
| Bertolino et al., 2004 | 30 | EU | 0.45 | Prospective | Olanzapine | PANSS 30% ↓ | 14 / 16 |
| Bertolino et al., 2007 | 29 | EU | 0.59 | Prospective | Olanzapine | PANSS 30% ↓ | 14 / 15 |
| Bishop et al., 2014* | 61 | EU, AA, AS, HS, O | 0.41 | Prospective | Various | BPRS 35% ↓ | 10 / 51 |
| Toronto Sample 1 | 58 | EU, AA, O | 0.38 | Prospective, Randomized | Various | PANSS 30% ↓ | 10 / 48 |
| Gao et al., 2012 | 83 | AS | 0.39 | Prospective | Risperidone | PANSS 30% ↓ | 69 / 14 |
| Toronto Sample 2 | 89 | EU, AA | 0.51 | Naturalistic | Various | PANSS 30% ↓ | 59 / 30 |
| Gupta et al., 2009 | 117 | SI | 0.37 | Naturalistic | Risperidone | CGI 2 or less | 69 / 48 |
| Toronto Sample 3* | 90 | EU, AA | 0.44 | Prospective | Various | BPRS 35% ↓ | 31 / 59 |
| Toronto Sample 4 | 90 | EU, AA | 0.43 | Prospective | Clozapine | BPRS 20% ↓ | 55 / 35 |
| Molero et al., 2007 | 205** | EU | 0.46 | Naturalistic, Prospective | Various | PANSS 30% ↓ | 148 / 57 |
| Pelayo-Teran et al., 2011 | 99 | EU | 0.40 | Prospective, Randomized | Various | BPRS 35% ↓ | 134 / 27 |
| Porcelli et al., 2009* | 144 | EU | 0.45 | Prospective | Various | PANSS 30% ↓ | 37 / 107 |
| Toronto Sample 5* | 35 | EU, AA | 0.24 | Prospective | Clozapine | BPRS 35% ↓ | 13 / 22 |
| Tybura et al., 2012* | 179** | EU | 0.44 | Naturalistic, Randomized | Various | PANSS 30% ↓ | 90 / 89 |
| Ikeda et al., 2008* | 107** | AS | 0.30 | Prospective | Risperidone | PANSS 30% ↓ | 58 / 49 |
Abbreviations: AA, African American; AS, Asian; BPRS, Brief Psychiatric Rating Scale; CGI, Clinical Global Impressions Scale; EU, European; HS, Hispanic; MAF, minor allele frequency; O, other; PANSS, Positive and Negative Symptom Scale; SI, South Indian.
*Positive and negative subscale scores available.
**N reflects additional subjects not included in original publication.
Subjects from Toronto Samples
The 5 samples from Toronto in this analysis included Toronto samples (TS)1–5. TS1 (Lieberman, N=58) was collected at 4 psychiatric state hospitals in New York and North Carolina. These patients met DSM-IV criteria for chronic SCZ and or schizo-affective disorder. They exhibited suboptimal response to prior treatment, defined primarily by persistent positive symptoms and poor functioning for the past 2 years. The patients were recruited as part of a 14-week, randomized, double-blinded study and response was evaluated using the PANSS scale. This sample is also described in greater detail elsewhere (Volavka et al., 2004). TS2 (Müller, N=89) was collected at Charité University Medicine, Berlin, Germany. Patients were diagnosed with SCZ or schizo-affective disorder according to DSM-IV or ICD-10 criteria and treated with several different antipsychotics. This sample is described in more detail elsewhere (Müller et al., 2012). TS3 (Meltzer, N=90), 4 (Lieberman, N=90), and 5 (Potkin, N=32) were collected from Case Western Reserve University in Cleveland, Ohio, Zucker Hillside Hospital in Glen Oaks, New York, and University of California at Irvine, respectively. Subjects from TS3-5 had DSM-III-R or DSM-IV diagnoses of SCZ and met criteria for treatment refractoriness or intolerance to traditional antipsychotic therapy. After a 2- to 4-week washout period, patients were treated with clozapine and evaluated prospectively for 6 months, during which blood levels of clozapine were monitored to confirm treatment adherence. Treatment response was evaluated using the 18-item BPRS (Overall and Gorham, 1962) at the beginning and end of treatment. These samples are described in greater detail elsewhere (Hwang et al., 2005). Analysis of response in each of these samples is discussed in the section “Definition of Response.” The ethnicities of patients from all 5 samples are described in Table 1.
Ethical Considerations
The scientific work described in this article is in compliance with the ethical standards stated in the 1964 Declaration of Helsinki. Informed consent was obtained prior to patient participation, and this work was approved by the Ethics Committee of the Centre for Addiction and Mental Health.
Definition of Clinical Response
Clinical responders and nonresponders were defined based on the criteria applied within each original study separately. If a study did not report the antipsychotic response data in binary responder/nonresponder form, study investigators were contacted and requested to apply a response threshold of 30% change in PANSS score or the equivalent in the clinical rating scale used in their study. The 30% cutoff was adopted for the current study, as it was the most commonly applied criteria in the studies included in the meta-analysis. We used the conversion scale of Leucht et al. (2013) to calculate corresponding response thresholds for other rating scales. For instance, according to this scale, the equivalent score change in the BPRS scale is 35%. For TS4, only responder/nonresponder data were available, for which response was defined as >20% reduction in overall BPRS score after 6 months of treatment based on criteria proposed by Kane et al. (1988). Thus, we had no option but to utilize this response definition for this sample. Overall, 14 of the 15 samples used the 30% PANSS score change (or equivalent) response criteria. Nonclinical dimensions of response, such as improvement in cognition, were not analyzed.
Genotyping
For the Toronto samples, genomic data were extracted using the high-salt method from venous blood (Lahiri and Nurnberger, 1991). The COMT Val158Met (rs4680) variant was genotyped using Taqman 5’ nuclease assay (Applied Biosystems; Foster City, CA). All ambiguous genotypes were retyped. Genotypes that remained ambiguous were excluded from further analyses. Genotyping accuracy was assessed by repeating genotyping for 10% of the sample, and results showed 100% concordance.
Statistical Analysis
All data were analyzed using R statistical software (v3.1.1). Meta-analysis was performed using the ‘meta’ package, version 4.1-0 (Guido Schwarzer, University of Freiburg). Fixed effects models were used for each analysis, consistent with approaches commonly used in pharmacogenetic meta-analyses (Serretti et al., 2007; Zhang et al., 2010). In our primary analysis, separate meta-analyses were conducted assuming three genetic models: recessive (Met/Met vs Val-allele carriers), dominant (Val/Val vs Met-allele carriers), and allelic (Val vs Met allele). All 3 models were used to test the association of COMT Val158Met with numbers of treatment responders vs nonresponders. Odds ratios and 95% CIs were computed and pooled using the Mantel-Haenszel method to determine overall effect sizes. We also examined each genetic model in relation to total symptom score change (%), positive subscale score change (%), and negative subscale score change (%) in a subset of study samples for which data were available (total: studies=10, n=1085; subscales: studies=6, n=616). Effect sizes were analyzed by computing SMD. Heterogeneity was assessed using Cochran’s Q test. Meta-regression was used to check for confounding effects due to the following variables: self-reported ethnicity, SCZ type (first-episode vs non-first episode), antipsychotic type (atypical-only vs mixed; specific drugs), age of onset, sex, study design (eg, prospective, naturalistic), duration of study, drug naïve vs prior exposure, and criteria for response. Publication bias was evaluated using the Harbord test (Harbord et al., 2006) and the funnel plot method (“metabias” and “funnel” commands, respectively). Sensitivity analyses were conducted on any significant findings (P<.05) by removing the studies with the largest and smallest effect sizes, as in other antipsychotic pharmacogenetic meta-analyses (Zhang et al., 2010).
To dissect the factors (eg, ancestry, antipsychotic type) contributing to the observed genetic effect, we examined the association between genotype and number of responders/nonresponders in samples of European ancestry (studies=10, ntotal=929). We focused on this ancestry group, as it constituted the majority of the samples. Ancestry may be important due to significant differences in Val158Met allele frequency between ethnicities. Moreover, since antipsychotic type showed a trend-level association with response, we examined the association between Val158Met and number of responders vs nonresponders in atypical and typical antipsychotic-treated patients separately.
With regards to correction for multiple testing, our approach is in line with that of prior pharmacogenetic meta-analyses in that we did not correct for the multiple genetic models tested when analyzing single genetic variants due to high correlations between the models (Bakker et al., 2006, 2008; Zhang et al., 2010).
Results
Primary Analyses
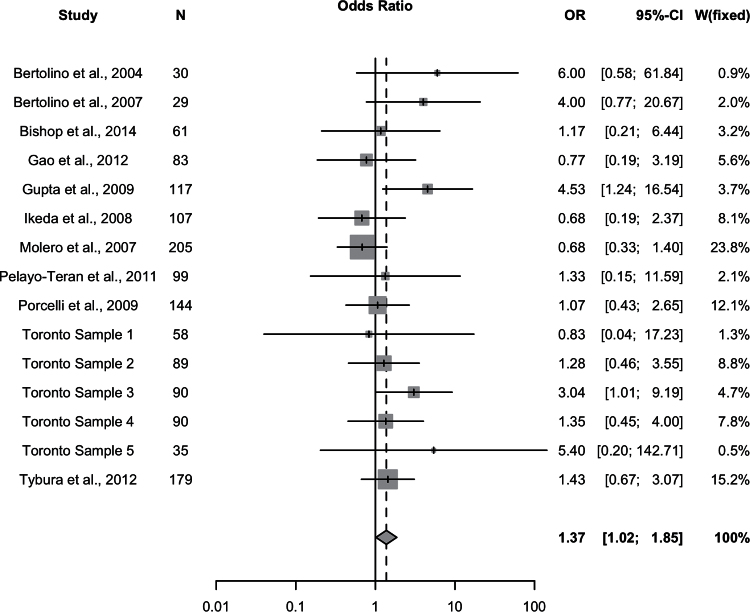
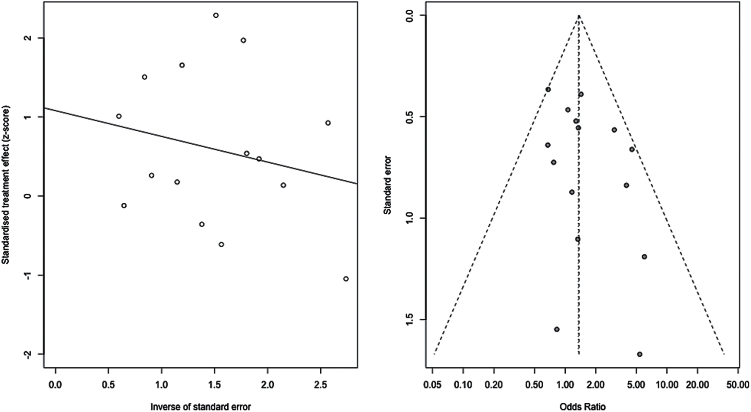
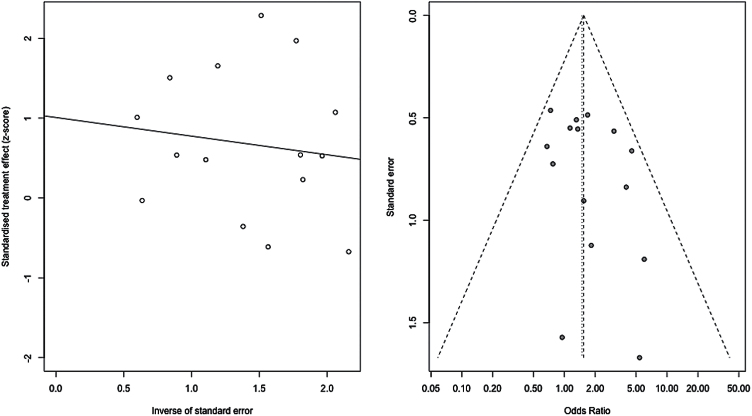
We observed a significant association with number of responders/non-responders under the recessive model (Met/Met vs Val-allele carriers) (P=.039, ORMet/Met=1.37, 95% CI: 1.02–1.85) (Figure 2; Table 2). We also observed a trend-level association for total score change (P=.087, SMD=0.14, 95% CI: -0.020–0.30) under a recessive model. Upon examination of the subscales, we observed a significant effect of Val158Met on positive symptom response also under a recessive model (P=.030, SMD=0.24, 95% CI: 0.024–0.46). No significant associations of Val158Met genotype were observed for any other response measures under any genetic model (all P>.05). The Cochran chi-squared test did not reveal any significant heterogeneity for the recessive model in relation to number of responders/non-responders (P=.37, Q=15.07, df=14, I2 = 7.1%) or for the other genetic models and response measures (all Cochran test P>.05). Funnel plot analysis and the Harbord test revealed no evidence of publication bias for the recessive model in relation to number of responders/nonresponders (P=.35) (Figure 3) or for the other genetic models.
Figure 2.
All studies. Meta-analysis of the association between the Val158Met polymorphism (Met/Met vs Val carrier) and antipsychotic drug response: forest plot.
Table 2.
Primary Analysis Results: All Studies (n=15; total sample size=1416).
| Genetic Model | Association Effect Size (Odds Ratio, 95% CI) | Significance (P-Value) |
|---|---|---|
| Recessive (M/M vs V/M and V/V) |
1.37 (1.02–1.85) | 0.039* |
| Dominant (M/M and V/M vs V/V) |
1.07 (0.83–1.37) | 0.58 |
| Allelic (M vs V) | 1.13 (0.96–1.34) | 0.14 |
Abbreviations: M/M, Met/Met genotype; V/M, Val/Met genotype; V/V, Val/Val genotype.
*Denotes significance (P<.05).
Figure 3.
All studies. Meta-analysis of the association between the Val158Met polymorphism (Met/Met vs Val carrier) and antipsychotic drug response: funnel plot and regression plot.
To screen for potentially undetected heterogeneity, we employed the sensitivity analysis conducted by Zhang et al. (2010) in their pharmacogenetic meta-analysis (Zhang et al., 2010), excluding the 2 studies with the largest (Bertolino et al., 2004) and smallest effect sizes (Ikeda et al., 2008). Sensitivity analysis was conducted for the association between the Val158Met variant and number of responders/nonresponders under the recessive model. This revealed no undetected heterogeneity and no significant influence on overall effect. After exclusion of the 2 studies, the significance of the association increased slightly (P=.012), while the effect size remained the same (OR=1.54, 95% CI: 1.10–2.15) and the heterogeneity decreased (I2=0%, P=.65).
Meta-regressions including potential confounding variables revealed a trend for antipsychotic type (atypical-only vs mixed) as a predictor of response rate across studies (P=.055). We could not include the variable of atypical vs typical antipsychotic-treated samples, as there were no studies with patients exclusively treated with typical antipsychotics. None of the remaining potential confounders tested were associated with response rate: gender, ethnicity, first episode vs non-first episode, treatment duration, study design, age at onset, drug-naïve vs prior exposure, and criteria for response.
Posthoc Analyses
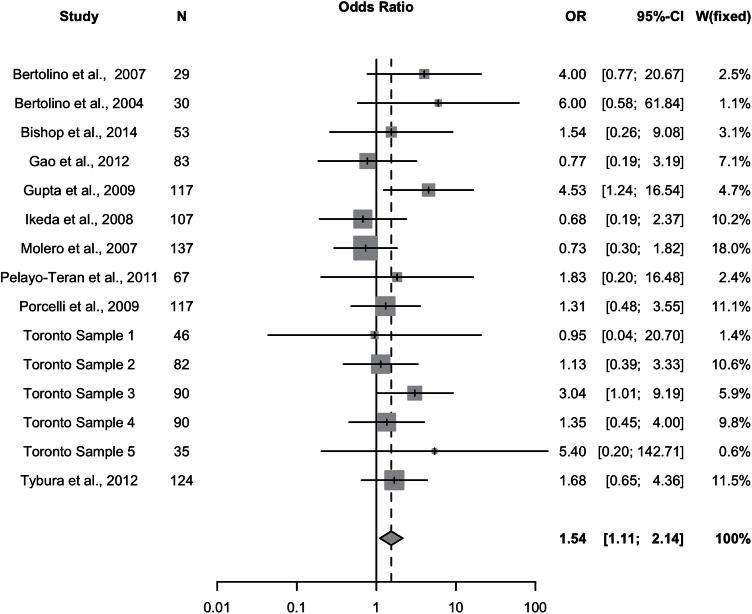
Since the meta-regression revealed antipsychotic type (atypical vs mixed) to be a potential predictor of response (P=.055), analyses were conducted separately on patients treated with atypical antipsychotics (studies=15; ntotal=1207) and typical antipsychotics (studies=7; ntotal=155). The 7 studies with typical antipsychotic-treated patients also included patients treated with atypical antipsychotics, but these were excluded for this analysis. In the subsample of patients treated with atypical antipsychotics, significant differences in the number of responders/nonresponders were observed under a recessive genetic model (Met/Met vs Val-allele carriers), whereby patients with Met/Met genotype experienced improved clinical response relative to Val carriers (P=.0098, OR=1.54, 95% CI: 1.11–2.14) (Figure 4; Table 2). Additionally, allelic analyses revealed that the Met-allele was associated with greater likelihood of response compared to the Val-allele (P=.043, OR=1.20, 95% CI: 1.01–1.44) (supplementary Figure 1; Table 2). No association was observed between Val158Met and response/nonresponse in the subsample of patients treated with typical antipsychotics (Table 3).
Figure 4.
Atypical antipsychotic-treated samples. Meta-analysis of the association between the Val158Met polymorphism (Met/Met vs Val carrier) and antipsychotic drug response: forest plot.
Table 3.
Post Hoc Analysis Results – Atypical and Typical-Antipsychotic Patient Subsamples
| Genetic Model |
Atypical
Association Effect Size (Odds Ratio, 95% CI) (n=1207) |
Significance
(P-Value) |
Typical
Association Effect Size (Odds Ratio, 95% CI) (n=155) |
Significance
(P-Value) |
|---|---|---|---|---|
| Recessive (M/M vs V/M and V/V) |
1.54 (1.11–2.14) | 0.0098* | 0.78 (0.36–1.70) | 0.54 |
| Allelic (M vs V) |
1.20 (1.01–1.44) | 0.043* | 0.78 (0.49–1.22) | 0.27 |
Abbreviations: M/M, Met/Met genotype; V/M, Val/Met genotype; V/V, Val/Val genotype.
*Denotes significance (P<.05).
In the subsample of atypical antipsychotic-treated patients, Cochran’s chi-square test revealed that heterogeneity was not significant under the recessive (I2= 0%, P=.52) or allelic models (I2 = 0%, P=.50). After excluding the 2 studies with the largest (Bertolino et al., 2004) and smallest effect sizes (Molero et al., 2007), sensitivity analyses revealed no undetected heterogeneity and no significant influence on overall effect under the recessive or allelic models. Further analyses revealed no significant publication bias (supplementary Material; Figure 5). The effect remained significant when analyzing only samples for which response was defined as 30% PANSS score reduction (studies=14, n=1317) (I2=13.8%, P=.046).
Figure 5.
Atypical antipsychotic-treated samples. Meta-analysis of the association between the Val158Met polymorphism (Met/Met vs Val carrier) and antipsychotic drug response: funnel plot and regression plot.
Due to significant differences in Val158Met allele frequency between ethnicities (ASN: 0.29, EUR: 0.52, AFR: 0.31), we conducted an analysis on studies with samples of European-ancestry (studies=10, ntotal=929). No significant effects were observed for binary response/nonresponse under the recessive model (P=.29, OR=1.21, 95% CI=0.85–1.72); however, consistent with whole group analyses, a trend was observed for atypical antipsychotic-treated patients of European ancestry (studies=10, ntotal=745) (P=.061, OR=1.45, 95% CI=0.98–2.15), with Met/Met patients experiencing better response than Val-allele carriers. The Cochran test showed no significant heterogeneity under any of the genetic models (all tests P>.05). Furthermore, funnel plot analysis and the Harbord test did not reveal any significant publication bias (supplementary Figure 2).
Discussion
To our knowledge, this study is the most comprehensive meta-analysis to investigate the relationship between COMT Val158Met and antipsychotic clinical response. Our meta-analysis includes data from 8 studies not reported in the original manuscripts and kindly provided to us by the authors (Molero et al., 2007; Ikeda et al., 2008; Gupta et al., 2009; Porcelli et al., 2009; Pelayo-Terán et al., 2011; Gao et al., 2012; Tybura et al., 2012; Bishop et al., 2015), as well as 3 samples previously not investigated for Val158Met and antipsychotic response (TS2, TS3, TS5). Thus, our study provides a much-expanded sample compared with a previous meta-analysis (Chen et al., 2015). More importantly, all samples included in our meta-analysis were analyzed for response/nonresponse according to uniform criteria (PANSS 30% reduction or equivalent), with the exception of one (TS4), minimizing inter-study heterogeneity.
Overall, significant effects of Val158Met on response vs nonresponse, as well as positive symptom improvement, were observed. The association was stronger when restricting the analysis to studies with atypical antipsychotic-treated samples (eg, clozapine, olanzapine, risperidone). Overall, Met-allele homozygotes experienced improved response compared with Val-allele carriers.
These results are consistent with previous studies finding that the Met-allele is associated with improved cognitive response to atypical antipsychotics (Bertolino et al., 2004; Weickert et al., 2004; Woodward et al., 2007). These studies evaluated cognitive response using tests such as the Wisconsin Card Sorting Test that evaluate cognitive abilities including working memory, executive function, and attention (Weickert et al., 2004). Conceivably, improvements in clinical symptoms, particularly positive symptoms, may contribute to cognitive improvement. Alternatively, improvements in clinical symptoms may be mediated by improvement in cognitive function (Censits et al., 1997; Velligan et al., 1997; Gold et al., 1999; Hoff et al., 1999); in particular, improved working memory and executive function could lead to improved treatment compliance and adherence to psychiatrist recommendations, resulting in improved clinical outcome. Cognitive response data were available in 3 of the studies included in the meta-analysis. Consistent with previous findings, Bertolino et al. (2004) observed that the Met-allele predicted better working memory response to olanzapine as measured by the N-back task (Bertolino et al., 2004). The 2 other studies did not observe an association between Val158Met and improvement in memory function or cognitive flexibility (Gao et al., 2012) and spatial working memory (Bishop et al., 2015) in response to antipsychotic treatment, respectively. The reasons for this are not clear but may relate to antipsychotic drug-type (both these samples were treated either largely or entirely with risperidone) and cognitive assessment methods (ie, Bishop et al. assessed spatial working memory). However, it should be noted that Gao et al. (2012) observed the same direction effect as previous studies, with the Met-allele appearing to experience greater improvement in orientation, comprehension, and experiential and number memory (Gao et al., 2012).
The findings are also consistent with the tonic-phasic DA hypothesis as applied to antipsychotic response. According to this hypothesis, DA regulation in limbic striatal regions occurs through transient, high-amplitude, phasic DA release and persistent low-level tonic DA release modulated by corticostriatal glutamatergic afferents. In these regions, tonic DA release also decreases phasic release through the stimulation of D2 auto-receptors at the synapse (Bilder et al., 2004). Importantly, COMT is involved in modulating the tonic, extracellular pool of DA subcortically, as it breaks down DA that is not reabsorbed by dopamine transporters. Thus, the Met-allele leads to lower COMT activity, higher tonic DA, and lower phasic DA release (more negative symptoms), while the Val-allele gives rise to lower tonic and higher phasic DA release (more positive symptoms) (Bilder et al., 2004). Bilder et al. (2004) have also suggested that D1 receptors are more important for modulating tonic release, while D2 receptors are more critical for regulating phasic DA release (Bilder et al., 2004). Therefore, the COMT Val158Met variant may have different effects on response to different classes of antipsychotics. Specifically, they suggest that the Met-allele may predict better response to agents enhancing D1-transmission (5-HT2A-antagonists and/or 5-HT1A-agonists or partial agonists), since they are thought to increase tonic DA transmission (Bilder et al., 2004). These agents include the majority of atypical antipsychotics. The Val-allele, in contrast, is hypothesized to predict better response to conventional D2-blocking agents (ie, typical antipsychotics), as they decrease phasic DA transmission.
Our findings for positive symptoms may also be consistent with the tonic-phasic DA hypothesis. Based on the predictions of Bilder et al., the Met-allele may also predict improved response in positive symptoms due to lower phasic DA transmission subcortically, which could potentiate the effects of antipsychotic-induced D2 receptor blockade. This prediction is supported by previous findings that the Met-allele is associated with greater dopamine synthesis in the midbrain and thus lower striatal dopaminergic stimulation (Meyer-Lindenberg et al., 2005). Moreover, consistent with this theory, reduced COMT activity has been observed to potentiate clozapine-induced DA release in the prefrontal cortex (PFC) (Tunbridge et al., 2004). By the tonic-phasic DA hypothesis, increased DA release in the PFC leads to greater D1 receptor activation, stimulating greater glutamate release from pyramidal neurons onto the nucleus accumbens. This, in turn, increases tonic DA release, subsequently decreasing phasic DA release and thus reducing positive symptom severity (Bilder et al., 2004). Specific atypical antipsychotics such as risperidone and olanzapine have also been observed to upregulate COMT mRNA expression in the rat PFC (Chen and Chen, 2007), suggesting a direct effect of these drugs on COMT activity.
The observed null genetic effect in typical-antipsychotic-treated patients may be a product of lack of statistical power, as the majority of patients were treated with atypical antipsychotics (typical: n=155, atypical: n=1207). Ultimately, it is possible that analyses conducted in larger samples could reveal associations of Val158Met with response in subjects treated with typical antipsychotics. Further, we acknowledge that differences among specific drugs within each class of antipsychotics, rather than broader class differences between atypical and typical antipsychotics, may be driving the results. However, due to limited data, this could not be tested.
The direction of genetic effect was consistent across 11 of the 15 samples in both the analysis of total sample as well as of atypical-antipsychotic-treated patients alone. The four samples observing a genetic effect in the opposite direction included TS1, Molero et al. (2007), Gao et al. (2007), and Ikeda et al. (2008). Notably, these 4 samples’ effects were all nonsignificant (95% CI of OR encompasses 1), leaving open the possibility that the opposite directions of effect may be due to random chance. It is possible that ancestry-related differences in the frequencies of COMT genetic variants other than Val158Met may explain these results. For instance, the functional variant Ala72Ser (rs6267), which is not in LD with Val158Met, also alters COMT activity and exists only in East-Asian populations (MAF: 0.07–0.11) (Gupta et al., 2011). Functional investigations suggest that this variant may exert its effect additively together with Val158Met, whereby presence of both low-activity alleles results in lower COMT activity than possession of either allele alone (Lee et al., 2005). Thus, while ancestry-related differences in MAF and COMT genetic variation may provide insight into the opposite direction of effect observed, we did not have sufficient samples of Asian ancestry to test for ancestry-specific associations. Another sample showing an opposite direction of effect, TS1, was highly treatment-resistant and included only 4 responders compared with 54 nonresponders; this lack of statistical power likely contributed to the null genetic effect observed in this sample. Finally, none of the 4 samples deviated from Hardy-Weinberg equilibrium (HWE) (P<.05), ruling this out as a possible explanation.
In addition, Molero et al. (2007) may have observed a nonsignificant effect for Val158Met in their sample due to several reasons. First, their sample differed from the majority of other samples in that the study specifically included patients who had recently experienced an acute worsening of psychotic symptoms, leading to uniquely high initial scores (average PANSS score: ~90). Second, the Val-allele carriers had significantly higher PANSS scores compared with Met-allele homozygotes at the study’s beginning. Since baseline score has been observed to predict overall response, this score differential may have contributed to greater score change for Val-allele carriers and thus a higher number of responders. Importantly, Met-allele homozygotes had significantly lower PANSS scores than Val-allele carriers at the end of the 6 months of treatment, which is consistent with findings from other studies in the meta-analysis.
The present meta-analysis had several limitations. First, while we attempted to test and control for possible confounding factors, several factors could not be accounted for due to lack of data availability. These include pharmacological aspects of treatment response, such as drug dose or concentrations, as well as environmental factors, such as significant stress or lifetime exposure to psychosis-inducing drugs. Nevertheless, it should be noted that no confounding effects were observed for the variables tested, except for antipsychotic type (trend observed). Moreover, heterogeneity across studies was not significant for any of the genetic models or response measures evaluated, suggesting minimal confounding by unidentified factors. Second, although we did not observe a significant influence of ethnicity in our samples, it should be noted non-European ethnicities were underrepresented. This imbalance may be important, since, as mentioned previously, Val158Met allele frequencies differ considerably among ethnic groups. Nevertheless, the fact that the sample consisted of mostly patients of European ancestry helps reduce heterogeneity in the sample. Third, it should also be noted that the genotypes in the sample from Pelayo-Teran et al. (2011) were not in HWE (P<.05). However, the skewing effect due to non-HWE should be minimal, as removing this sample did not impact the overall genetic effect observed or the level of heterogeneity. Lastly, if we were to apply a Bonferroni correction threshold, the association observed between Val158Met and number of responders/nonresponders in the total sample would be nonsignificant. However, given the high correlation between the genetic models applied and phenotype measures of response, this method for correction would be overly conservative.
Overall, our meta-analysis provides evidence that COMT Val158Met is associated with response to antipsychotics, with a stronger effect observed for atypical antipsychotics. Prospective studies with complete categorical and continuous response measure data that differentiate between atypical and typical antipsychotics would help in strengthening the evidence of this functional variant’s relationship with antipsychotic response. Future studies of COMT should also include other putatively functional variants such as rs6267, rs737865, and rs4818 (SZGene Data Base, Nature Genetics, 2008) (Bray et al., 2003; Chen et al., 2004) in order to obtain a more comprehensive understanding of how genetically predicted COMT function may influence antipsychotic response. Moreover, given the complexity of the antipsychotic response phenotype, it is likely that other genetic, environmental, and potentially epigenetic factors are involved, which were beyond the scope of this study. Notably, several findings provide evidence for multiple gene-gene interaction and gene-environment interactions involving COMT (Nicodemus et al., 2007). Moving forward, investigations of epistatic and gene-environment interactions involving COMT should further improve our understanding of this gene’s effect on antipsychotic response. Nevertheless, our findings suggest that COMT Val158Met may warrant closer consideration in randomized clinical trials. Since this variant appears to associate with response, balancing each arm of a RCT so that the same percentage of Met/Met are represented in each arm may prevent COMT genetic imbalance from confounding the efficacy results.
Statement of Interest
H.Y.M. is a shareholder in SureGene, ACADIA, and Glaxo Smith Kline. J.A.L. serves on the Advisory Board of Intracellular Therapies and does not receive direct financial compensation or salary support for his participation. He receives grant support from Alkermes, Biomarin, EnVivo/Forum, Genentech, Novartis/Novation, and Sunovion; is a member of the Advisory Board of Clintara and Pear Therapeutics and holds a financial interest; and holds a patent from Repligen. J.L.K. has acted as a consultant or received honoraria from GlaxoSmith-Kline, Sanofi-Aventis, Dainippon-Sumitomo, Novartis, Shire, Eli Lilly, and Roche; he is on a Scientific Advisory Board member (unpaid) of AssureRx Health Inc. J.M.P. has received lecture honoraria and travel support form Janssen Johnson & Johnson, Lundbeck, Otsuka Pharmaceuticals, GlaxoSmithkline and Eli Lilly. J.R.B. serves on the advisory board or is a consultant for Physician’s Choice Laboratory Services and Janssen Scientific Affairs. P.M. has been a clinical consultant for MedAvante and has received lecture support from Scienta (once) and AB-Biotics (once). Without any relevance to this work, S.G.P. declares that he has received grant/research support from Alkermes, Amgen, Eisai, Eli Lilly, FORUM, Otsuka, Thuris, and Toyama and has served as a consultant or on advisory boards for Alkermes, Concert, FORUM, Otsuka/Lundbeck, Roche/Genentech, Takeda, Teva, and Vanda, and has served on speakers’ bureaus for Forest, Merck, Novartis, Otsuka, and Sunovion. The remaining authors have no potential conflicts of interests to disclose.
Supplementary Material
Acknowledgments
C.C.Z. is supported by the Brain and Behavior Research Foundation, American Foundation for Suicide Prevention and Eli Lilly. D.F. is supported by the Vanier Canada Graduate Scholarship. D.J.M. has been or is supported by the Canadian Institute of Health Research (CIHR) Operating Grant: “Genetics of antipsychotic-induced metabolic syndrome,” Michael Smith New Investigator Salary Prize for Research in Schizophrenia, NARSAD Independent Investigator Award by the Brain & Behavior Research Foundation, and Early Researcher Award from Ministry of Research and Innovation of Ontario. E.H. is supported by the Canada Graduate Scholarship. H.Y.M. has grant support from Sumitomo Dainippon, Sunovion, Boehringer Ingelheim, Eli Lilly, Janssen, Reviva, Alkermes, Auspex, and FORUM. J.A.L. has received research funding from Alkermes, Biomarin, EnVivo/Forum, Genentech, and Novartis. J.L.K. is supported the CIHR grant “Strategies for gene discovery in schizophrenia: subphenotypes, deep sequencing and interaction.” J.R.B. is supported by NIH grant MH083888. A.K.T. is supported by a NARSAD Young Investigator Award. J.S. is supported by a Pfizer independent grant. P.M. receives salary from Clinica Universidad de Navarra and has received research grants from the Ministry of Education (Spain), the Government of Navarra (Spain), the Spanish Foundation of Psychiatry and Mental Health, and Astrazeneca. S.G. is supported by the Ningbo Medical Technology Project Fund (No. 2004050), the Natural Science Foundation of Ningbo (No. 2009A610186, No. 2013A610249), and the Zhejiang Provincial Medical and Health Project Fund (No. 2015127713). S.G.P. has received research support from Otsuka, Lundbeck, FORUM, and Alkermes.
References
- Anttila S, Illi A, Kampman O, Mattila KM, Lehtimäki T, Leinonen E. (2004) Interaction between NOTCH4 and catechol-O-methyltransferase genotypes in schizophrenia patients with poor response to typical neuroleptics. Pharmacogenet Genomics 14:303–307. [DOI] [PubMed] [Google Scholar]
- Badner J, Gershon E. (2002) Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 7:405–411. [DOI] [PubMed] [Google Scholar]
- Bakker P, Van Harten P, Van Os J. (2008) Antipsychotic-induced tardive dyskinesia and polymorphic variations in COMT, DRD2, CYP1A2 and MnSOD genes: a meta-analysis of pharmacogenetic interactions. Mol Psychiatry 13:544–556. [DOI] [PubMed] [Google Scholar]
- Bakker PR, van Harten PN, van Os J. (2006) Antipsychotic-induced tardive dyskinesia and the Ser9Gly polymorphism in the DRD3 gene: a meta analysis. Schizophr Res 83:185–192. [DOI] [PubMed] [Google Scholar]
- Bertolino A, Caforio G, Blasi G, De Candia M, Latorre V, Petruzzella V, Altamura M, Nappi G, Papa S, Callicott JH. (2004) Interaction of COMT Val108/158 Met genotype and olanzapine treatment on prefrontal cortical function in patients with schizophrenia. Am J Psychiatry 161:1798–1805. [DOI] [PubMed] [Google Scholar]
- Bertolino A, Caforio G, Blasi G, Rampino A, Sinibaldi L, Douzgou S, Nardini M, Weinberger DR, Dallapiccola B. (2007) COMT Val 158 Met polymorphism predicts negative symptoms response to treatment with olanzapine in schizophrenia. Schizophr Res 95:253–255. [DOI] [PubMed] [Google Scholar]
- Bilder RM, Volavka J, Lachman HM, Grace AA. (2004) The catechol-O-methyltransferase polymorphism: relations to the tonic-phasic dopamine hypothesis and neuropsychiatric phenotypes. Neuropsychopharmacology. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Reilly JL, Harris MS, Patel SR, Kittles R, Badner JA, Prasad KM, Nimgaonkar VL, Keshavan MS, Sweeney JA. (2015) Pharmacogenetic associations of the type-3 metabotropic glutamate receptor (GRM3) gene with working memory and clinical symptom response to antipsychotics in first-episode schizophrenia. Psychopharmacology (Berl) 232:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosia M, Lorenzi C, Pirovano A, Guglielmino C, Cocchi F, Spangaro M, Bramanti P, Smeraldi E, Cavallaro R. (2015) COMT Val158Met and 5-HT1A-R-1019 C/G polymorphisms: effects on the negative symptom response to clozapine. Pharmacogenomics 16:35–44. [DOI] [PubMed] [Google Scholar]
- Bray NJ, Buckland PR, Williams NM, Williams HJ, Norton N, Owen MJ, O’Donovan MC. (2003) A haplotype implicated in schizophrenia susceptibility is associated with reduced COMT expression in human brain. Am J Hum Genet 73:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Censits DM, Ragland JD, Gur RC, Gur RE. (1997) Neuropsychological evidence supporting a neurodevelopmental model of schizophrenia: a longitudinal study. Schizophr Res 24:289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tu J, Ni P, Zhang W, Xu L. (2015) [COMT genetic variation and clinical response to antipsychotic drug treatment: A Meta-analysis]. Zhong Nan Da Xue Xue Bao Yi Xue Ban 40:623–631. [DOI] [PubMed] [Google Scholar]
- Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S, Kolachana BS, Hyde TM, Herman MM, Apud J. (2004) Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet 75:807–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-L, Chen C-H. (2007) Chronic antipsychotics treatment regulates MAOA, MAOB and COMT gene expression in rat frontal cortex. J Psychiatr Res 41:57–62. [DOI] [PubMed] [Google Scholar]
- Fijal B, Kinon B, Kapur S, Stauffer V, Conley R, Jamal H, Kane J, Witte M, Houston J. (2009) Candidate-gene association analysis of response to risperidone in African-American and white patients with schizophrenia. Pharmacogenomics J 9:311–318. [DOI] [PubMed] [Google Scholar]
- Gao S, Hu Z, Cheng J, Zhou W, Xu Y, Xie S, Liu S, Li Z, Guo J, Dong J. (2012) Impact of catechol-o-methyltransferase polymorphisms on risperidone treatment for schizophrenia and its potential clinical significance. Clin Biochem 45:787–792. [DOI] [PubMed] [Google Scholar]
- Gold S, Arndt S, Nopoulos P, O’Leary DS, Andreasen NC. (1999) Longitudinal study of cognitive function in first-episode and recent-onset schizophrenia. Am J Psychiatry 156:1342–1348. [DOI] [PubMed] [Google Scholar]
- Gonçalves VF, Zai CC, Tiwari AK, Brandl EJ, Derkach A, Meltzer HY, Lieberman JA, Müller DJ, Sun L, Kennedy JL. (2014) A hypothesis-driven association study of 28 nuclear-encoded mitochondrial genes with antipsychotic-induced weight gain in schizophrenia. Neuropsychopharmacology 39:1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood K, Hung C-F, Tropeano M, McGuffin P, Wykes T. (2011) No association between the catechol-O-methyltransferase (COMT) val158met polymorphism and cognitive improvement following cognitive remediation therapy (CRT) in schizophrenia. Neurosci Lett 496:65–69. [Google Scholar]
- Gupta M, Bhatnagar P, Grover S, Kaur H, Baghel R, Bhasin Y, Chauhan C, Verma B, Manduva V, Mukherjee O. (2009) Association studies of catechol-O-methyltransferase (COMT) gene with schizophrenia and response to antipsychotic treatment. Pharmacogenomics 10:385–397. [DOI] [PubMed] [Google Scholar]
- Gupta M, Kaur H, Jajodia A, Jain S, Satyamoorthy K, Mukerji M, Thirthalli J, Kukreti R. (2011) Diverse facets of COMT: from a plausible predictive marker to a potential drug target for schizophrenia. Curr Mol Med 11:732–743. [DOI] [PubMed] [Google Scholar]
- Harbord RM, Egger M, Sterne JA. (2006) A modified test for small‐study effects in meta‐analyses of controlled trials with binary endpoints. Stat Med 25:3443–3457. [DOI] [PubMed] [Google Scholar]
- Hoff AL, Sakuma M, Wieneke M, Horon R, Kushner M, DeLisi LE. (1999) Longitudinal neuropsychological follow-up study of patients with first-episode schizophrenia. Am J Psychiatry 156:1336–1341. [DOI] [PubMed] [Google Scholar]
- Hwang R, Shinkai T, De Luca V, Muller DJ, Ni X, Macciardi F, Potkin S, Lieberman JA, Meltzer HY, Kennedy JL. (2005) Association study of 12 polymorphisms spanning the dopamine D(2) receptor gene and clozapine treatment response in two treatment refractory/intolerant populations. Psychopharmacology (Berl) 181:179–187. [DOI] [PubMed] [Google Scholar]
- Hwang R, Zai C, Tiwari A, Müller D, Arranz M, Morris A, McKenna P, Munro J, Potkin S, Lieberman J. (2010) Effect of dopamine D3 receptor gene polymorphisms and clozapine treatment response: exploratory analysis of nine polymorphisms and meta-analysis of the Ser9Gly variant. Pharmacogenomics J 10:200–218. [DOI] [PubMed] [Google Scholar]
- Hwang R, Tiwari AK, Zai CC, Felsky D, Remington E, Wallace T, Tong RP, Souza RP, Oh G, Potkin SG. (2012) Dopamine D4 and D5 receptor gene variant effects on clozapine response in schizophrenia: replication and exploration. Prog Neuropsychopharmacol Biol Psychiatry 37:62–75. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Yamanouchi Y, Kinoshita Y, Kitajima T, Yoshimura R, Hashimoto S, O’Donovan MC, Nakamura J, Ozaki N, Iwata N. (2008) Variants of dopamine and serotonin candidate genes as predictors of response to risperidone treatment in first-episode schizophrenia. [DOI] [PubMed]
- Illi A, Mattila KM, Kampman O, Anttila S, Roivas M, Lehtimäki T, Leinonen E. (2003) Catechol-O-methyltransferase and monoamine oxidase A genotypes and drug response to conventional neuroleptics in schizophrenia. J Clin Psychopharmacol 23:429–434. [DOI] [PubMed] [Google Scholar]
- Illi A, Kampman O, Hänninen K, Anttila S, Mattila KM, Katila H, Rontu R, Hurme M, Lehtimäki T, Leinonen E. (2007) Catechol‐O‐methyltransferase val108/158met genotype and response to antipsychotic medication in schizophrenia. Hum Psychopharmacol 22:211–215. [DOI] [PubMed] [Google Scholar]
- Kane J, Honigfeld G, Singer J, Meltzer H. (1988) Clozapine for the treatment-resistant schizophrenic. A double-blind comparison with chlorpromazine. Arch Gen Psychiatry 45:789–796. [DOI] [PubMed] [Google Scholar]
- Kang CY, Xu XF, Liu H, Shi ZY, Xu HH, Yang JZ. (2010) Interaction of catechol-O-methyltransferase (COMT) Val108/158 Met genotype and risperidone treatment in Chinese Han patients with schizophrenia. Psychiatry Res 176:94–95. [DOI] [PubMed] [Google Scholar]
- Lachman HM, Papolos DF, Saito T, Yu Y-M, Szumlanski CL, Weinshilboum RM. (1996) Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenet Genomics 6:243–250. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Nurnberger JI., Jr (1991) A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res 19:5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-G, Joo Y, Kim B, Chung S, Kim H-L, Lee I, Choi B, Kim C, Song K. (2005) Association of Ala72Ser polymorphism with COMT enzyme activity and the risk of schizophrenia in Koreans. Hum Genet 116:319–328. [DOI] [PubMed] [Google Scholar]
- Leucht S, Rothe P, Davis J, Engel R. (2013) Equipercentile linking of the BPRS and the PANSS. Eur Neuropsychopharmacol 23:956–959. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV. (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet 73:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, Keefe RS, Davis SM, Davis CE, Lebowitz BD. (2005) Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med 353:1209–1223. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Owen M, Farmer A. (1995) Genetic basis of schizophrenia. The Lancet 346:678–682. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Kohn PD, Kolachana B, Kippenhan S, McInerney-Leo A, Nussbaum R, Weinberger DR, Berman KF. (2005) Midbrain dopamine and prefrontal function in humans: interaction and modulation by COMT genotype. Nat Neurosci 8:594–596. [DOI] [PubMed] [Google Scholar]
- Molero P, Ortuno F, Zalacain M, Patino-Garcia A. (2007) Clinical involvement of catechol-O-methyltransferase polymorphisms in schizophrenia spectrum disorders: influence on the severity of psychotic symptoms and on the response to neuroleptic treatment. Pharmacogenomics J 7:418–426. [DOI] [PubMed] [Google Scholar]
- Müller D, Zai C, Sicard M, Remington E, Souza R, Tiwari A, Hwang R, Likhodi O, Shaikh S, Freeman N. (2012) Systematic analysis of dopamine receptor genes (DRD1–DRD5) in antipsychotic-induced weight gain. Pharmacogenomics J 12:156–164. [DOI] [PubMed] [Google Scholar]
- Need AC, Keefe RS, Ge D, Grossman I, Dickson S, McEvoy JP, Goldstein DB. (2009) Pharmacogenetics of antipsychotic response in the CATIE trial: a candidate gene analysis. Eur J Hum Genet 17:946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemus KK, Kolachana BS, Vakkalanka R, Straub RE, Giegling I, Egan MF, Rujescu D, Weinberger DR. (2007) Evidence for statistical epistasis between catechol-O-methyltransferase (COMT) and polymorphisms in RGS4, G72 (DAOA), GRM3, and DISC1: influence on risk of schizophrenia. Hum Genet 120:889–906. [DOI] [PubMed] [Google Scholar]
- Nolan KA, Czobor P, Citrome LL, Krakowski M, Lachman HM, Kennedy JL, Ni X, Lieberman J, Chakos M, Volavka J. (2006) Catechol-O-methyltransferase and monoamine oxidase-A polymorphisms and treatment response to typical and atypical neuroleptics. J Clin Psychopharmacol 26:338–340. [DOI] [PubMed] [Google Scholar]
- Overall JE, Gorham DR. (1962) The brief psychiatric rating scale. Psychol Rep 10:799–812. [Google Scholar]
- Pelayo-Terán JM, Pérez-Iglesias R, Vázquez-Bourgon J, Mata I, Carrasco-Marín E, Vázquez-Barquero JL, Crespo-Facorro B. (2011) Catechol-O-methyltransferase Val158Met polymorphism and negative symptoms after acute antipsychotic treatment in first-episode non-affective psychosis. Psychiatry Res 185:286–289. [DOI] [PubMed] [Google Scholar]
- Porcelli S, Bosia M, Marino E, Angelone S, Smeraldi E, Cavallaro R. (2009) p. 3.14 Association study of COMT Val108/158Met polymorphism and treatment response to haloperidol, risperidone and clozapine. Eur Neuropsychopharmacol 19:S73–S74. [Google Scholar]
- Pouget JG, Müller DJ. (2014) Pharmacogenetics of antipsychotic treatment in schizophrenia. In: Pharmacogenomics in Drug Discovery and Development, pp 557–587: Springer. [DOI] [PubMed] [Google Scholar]
- Prata DP, Gafoor R, Kay V, Arranz M, Munro J, McGuire P. (2012) Dopaminergic Genes Influence Early Response to Atypical Antipsychotics in Patients With First Presentation of Psychosis. J Clin Psychopharmacol 32:566–569. [DOI] [PubMed] [Google Scholar]
- Serretti A, Kato M, De Ronchi D, Kinoshita T. (2007) Meta-analysis of serotonin transporter gene promoter polymorphism (5-HTTLPR) association with selective serotonin reuptake inhibitor efficacy in depressed patients. Mol Psychiatry 12:247–257. [DOI] [PubMed] [Google Scholar]
- Tiwari AK, Brandl EJ, Weber C, Likhodi O, Zai CC, Hahn MK, Lieberman JA, Meltzer HY, Kennedy JL, Müller DJ. (2013) Association of a functional polymorphism in neuropeptide Y with antipsychotic-induced weight gain in schizophrenia patients. J Clin Psychopharmacol 33:11–17. [DOI] [PubMed] [Google Scholar]
- Tunbridge E, Bannerman D, Sharp T, Harrison P. (2004) Catechol-o-methyltransferase inhibition improves set-shifting performance and elevates stimulated dopamine release in the rat prefrontal cortex. J Neurosci 24:5331–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tybura P, Samochowiec A, Beszlej A, Grzywacz A, Mak M, Frydecka D, Bieńkowski P, Mierzejewski P, Potemkowski A, Samochowiec J. (2012) Some dopaminergic genes polymorphisms are not associated with response to antipsychotic drugs in schizophrenic patients. Pharmacol Rep 64:528–535. [DOI] [PubMed] [Google Scholar]
- Vehof J, Burger H, Wilffert B, Al Hadithy A, Alizadeh BZ, Snieder H. (2012) Clinical response to antipsychotic drug treatment: Association study of polymorphisms in six candidate genes. Eur Neuropsychopharmacol 22:625–631. [DOI] [PubMed] [Google Scholar]
- Velligan DI, Mahurin RK, Diamond PL, Hazleton BC, Eckert SL, Miller AL. (1997) The functional significance of symptomatology and cognitive function in schizophrenia. Schizophr Res 25:21–31. [DOI] [PubMed] [Google Scholar]
- Volavka J, Czobor P, Sheitman B, Lindenmayer J-P, Citrome L, McEvoy JP, Cooper TB, Chakos M, Lieberman JA. (2004) Clozapine, olanzapine, risperidone, and haloperidol in the treatment of patients with chronic schizophrenia and schizoaffective disorder. Focus. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fang Y, Shen Y, Xu Q. (2010) Analysis of association between the catechol-O-methyltransferase (COMT) gene and negative symptoms in chronic schizophrenia. Psychiatry Res 179:147–150. [DOI] [PubMed] [Google Scholar]
- Weickert TW, Goldberg TE, Mishara A, Apud JA, Kolachana BS, Egan MF, Weinberger DR. (2004) Catechol-O-methyltransferase val108/158met genotype predicts working memory response to antipsychotic medications. Biol Psychiatry 56:677–682. [DOI] [PubMed] [Google Scholar]
- Woodward ND, Jayathilake K, Meltzer HY. (2007) COMT val108/158met genotype, cognitive function, and cognitive improvement with clozapine in schizophrenia. Schizophr Res 90:86–96. [DOI] [PubMed] [Google Scholar]
- Yamanouchi Y, Iwata N, Suzuki T, Kitajima T, Ikeda M, Ozaki N. (2003) Effect of DRD2, 5-HT2A, and COMT genes on antipsychotic response to risperidone. Pharmacogenomics J 3:356–361. [DOI] [PubMed] [Google Scholar]
- Zai GC, Zai CC, Chowdhury NI, Tiwari AK, Souza RP, Lieberman JA, Meltzer HY, Potkin SG, Müller DJ, Kennedy JL. (2012) The role of brain-derived neurotrophic factor (BDNF) gene variants in antipsychotic response and antipsychotic-induced weight gain. Prog Neuropsychopharmacol Biol Psychiatry 39:96–101. [DOI] [PubMed] [Google Scholar]
- Zhang JP, Lencz T, Malhotra AK. (2010) D2 receptor genetic variation and clinical response to antipsychotic drug treatment: a meta-analysis. Am J Psychiatry 167:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q-Z, Liu B-C, Zhang J, Wang L, Li X-W, Wang Y, Ji J, Yang F-P, Wan C-L, Xu Y-F. (2012) Association between a COMT polymorphism and clinical response to risperidone treatment: a pharmacogenetic study. Psychiatr Genet 22:298–299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.