Abstract
Allosteric modulation of metabotropic glutamate (mGlu) receptors offers a promising pharmacological approach to normalize neural circuit dysfunction associated with various psychiatric and neurological disorders. As mGlu receptor allosteric modulators progress through discovery and clinical development, both technical advances and novel tool compounds are providing opportunities to better understand mGlu receptor pharmacology and neurobiology. Recent advances in structural biology are elucidating the structural determinants of mGlu receptor–negative allosteric modulation and supplying the means to resolve active, allosteric modulator-bound mGlu receptors. The discovery and characterization of allosteric modulators with novel pharmacological profiles is uncovering the biological significance of their intrinsic agonist activity, biased mGlu receptor modulation, and novel mGlu receptor heterodimers. The development and exploitation of optogenetic and optopharmacological tools is permitting a refined spatial and temporal understanding of both mGlu receptor functions and their allosteric modulation in intact brain circuits. Together, these lines of research promise to provide a more refined understanding of mGlu receptors and their allosteric modulation that will inform the development of mGlu receptor allosteric modulators as neurotherapeutics in the years to come.
Keywords: allosteric modulators, brain disorders, GPCR biased signaling, GPCR heterodimers, mGlu receptor
Introduction
Metabotropic glutamate (mGlu) receptors are Class C 7-transmembrane domain (7-TMD) G-protein coupled receptors (GPCRs) involved in the modulation of neurotransmission throughout the nervous system. mGlu receptor dimers bind the major excitatory neurotransmitter, glutamate, which leads to the receptor’s subsequent conformational change and initiation of intracellular signaling. The mGlu receptors differentially regulate synaptic transmission and other aspects of neuronal function based on their G-protein coupling and synaptic localization. Group I mGlu receptors, mGlu1 and mGlu5, are expressed primarily at postsynaptic sites and modulate neuronal excitability and synaptic transmission through Gq/11-mediated signaling. In contrast, Group II mGlu receptors, mGlu2 and mGlu3, are expressed at both presynaptic and postsynaptic sites, and often reduce synaptic transmission through Gi/o-mediated signaling. Group III mGlu receptors, mGlu4 and mGlu6-8, are expressed predominantly in presynaptic terminals and inhibit synaptic transmission through Gi/o-mediated inhibition of neurotransmitter release (Niswender and Conn, 2010). Due to their localization and functionality, much research has focused on targeting mGlu receptors to reverse pathological activity of identified brain circuits in brain disorders.
Based on both pharmacological and genetic studies, mGlu receptors are promising therapeutic targets for a variety of brain disorders. Modulation of mGlu receptor activity has demonstrated therapeutic efficacy in preclinical models of schizophrenia, depression, anxiety disorders, Alzheimer’s disease, Parkinson’s disease, addiction, chronic pain, and other brain disorders (Nickols and Conn, 2014). As a result of this therapeutic potential, drug discovery efforts initially focused on developing novel agonists and antagonists in order to modulate mGlu receptor–mediated function. However, many of these efforts failed to develop subtype-selective agonists and antagonists, likely due to the high degree of conservation at the mGlu receptor orthosteric binding site (Kunishima et al., 2000). More recently, an alternative strategy has emerged that is focused on selectively targeting mGlu receptors using allosteric modulators (Annoura et al., 1996; Litschig et al., 1999; Varney et al., 1999).
Novel allosteric modulators of mGlu receptors offer theoretical advantages over conventional agonists and antagonists (Conn et al., 2009, 2014). Akin to agonists and antagonists, mGlu receptor activity can be increased or decreased by positive allosteric modulators (PAM) and negative allosteric modulators (NAM), respectively; however, there are important distinctions between allosteric modulators and agonists/antagonists. PAMs and NAMs target allosteric sites on the 7-TMD that are less highly conserved across the eight mGlu receptor subtypes. Recent efforts have developed selective PAMs and NAMs for each of the eight mGlu receptor subtypes (Sheffler et al., 2011; Urwyler, 2011; Engers and Lindsley, 2013; Conn et al., 2014; Walker et al., 2015). Allosteric modulators also have the potential to maintain both the spatial and temporal aspects of mGlu receptor signaling, and in many cases are inactive in the absence of the endogenous agonist, glutamate. Furthermore, such modulators could theoretically avoid problems such as receptor desensitization and/or down-regulation, and also exhibit ceiling effects that could reduce the likelihood of accidental overdose. Thus, due to superior selectivity and potential advantages, drug discovery efforts have developed a range of novel mGlu allosteric modulators, some of which are currently being investigated as clinical candidates to treat various brain disorders.
In parallel with their development as potential neurotherapeutics, novel allosteric modulators have provided excellent tool compounds for basic research on mGlu receptor pharmacology and neurobiology. Recent studies using high–affinity binding NAMs and X-ray crystallography have now resolved the 3D, inactive conformation of the 7-TMD for both mGlu1 and mGlu5. These studies and further advances in structural biology provide the tools necessary to better model allosteric modulator binding and to potentially resolve the active, PAM-bound mGlu receptor 7-TMD (Figure 1A). Allosteric modulators with unique modes of efficacy have provided important new insights into the biological significance of intrinsic agonist activity, biased mGlu receptor modulation, and mGlu receptor heterodimerization. Yet, these initial findings only provide a limited insight as to the biological importance of the diverse pharmacological profiles observed with allosteric modulators for mGlu receptors. Finally, at a neural circuit level, recent studies have revealed the exciting potential for elucidating how mGlu receptors and their allosteric modulation affect neural circuit function and in vivo behavioral outcomes. When combined with new technologies, such as optogenetic and optopharmacological approaches, highly selective allosteric modulators promise to refine our understanding of mGlu receptor function and their modulation in identified brain circuits that affect specific in vivo behavioral outcomes.
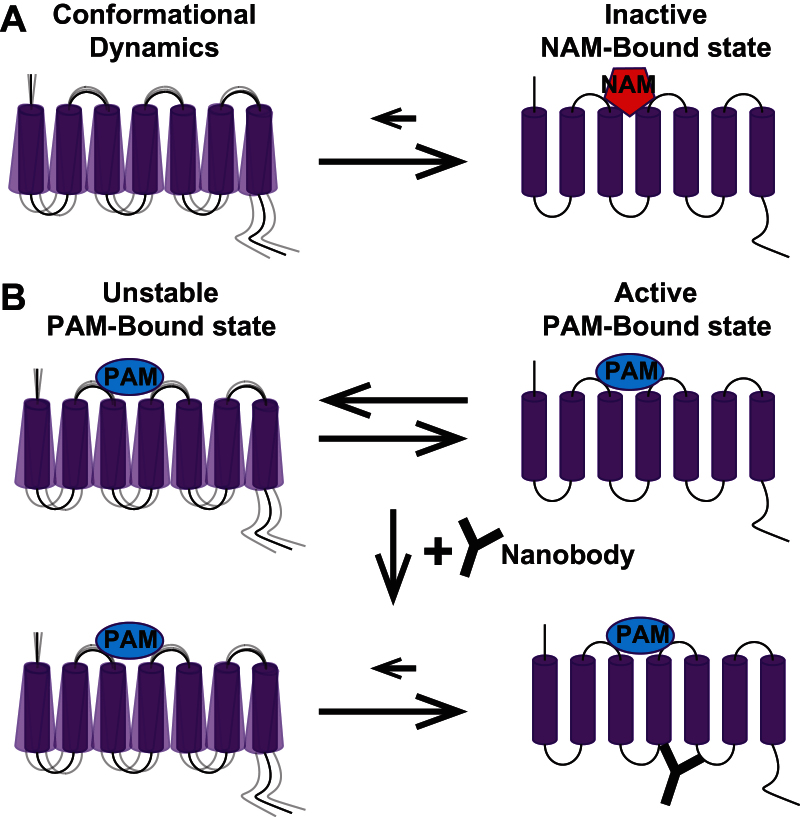
Figure 1.
Conformational dynamics of metabotropic glutamate (mGlu) receptor 7-transmembrane domain (7-TMD) are important for crystallization and resolution of 7-TMD structures. (A) Upon binding of high affinity negative allosteric modulators to the mGlu receptor 7-TMD, the inactive state is stabilized over other conformations, allowing for crystallization and resolution of this 3D, inactive conformation. (B) However, the positive allosteric modulators (PAM)-bound active state for mGlu receptors is likely unstable, thereby complicating the crystallization and resolution of this active structure. The recent emergence of nanobodies has permitted the stabilization and subsequent crystallization and resolution of active G-protein coupled receptor structures, raising the possibility of elucidating the active PAM-bound mGlu receptor 7-TMD by nanobody-mediated stabilization of this conformation.
Structural Insights into mGlu Receptor Allosteric Modulation
Previous studies have defined both the general makeup of mGlu receptors and the crystal structure of the glutamate orthosteric binding site. Like most other class C GPCRs, mGlu receptors are comprised of three discrete domains: a large N-terminal Venus fly-trap domain (VFD), a linker cysteine-rich domain (CRD), and a 7-TMD. In the hypothesized model of activation, glutamate binds in the VFD, stabilizes its closed conformation, and propagates this conformational change through the 7-TMD by way of the linker CRD. Indeed, the crystal structure of the mGlu1 VFD dimer has demonstrated significant rearrangements within the VFD protomer and dimer that could be involved in this proposed activation mechanism (Kunishima et al., 2000). However, without a crystal structure of the 7-TMD, it is unclear how this 7-TMD changes its conformation upon activation or inactivation in order to regulate intracellular signaling. Furthermore, without the mGlu receptor 7-TMD bound to an allosteric modulator, it is unclear how allosteric modulators may act to preferentially stabilize specific conformations.
Recent resolution of both mGlu1 and mGlu5 7-TMD structures elucidated their inactive, NAM-bound conformations and surprisingly demonstrated many similarities between these Class C GPCRs and other GPCR classes (Doré et al., 2014; Wu et al., 2014). Despite <15% sequence homology with other GPCR superfamilies, both mGlu1 and mGlu5 7-TMDs shared similar topology with class A and B GPCRs. The mGlu1 and mGlu5 7-TMDs also exhibited conserved features seen in other classes of GPCRs. Both mGlu1 and mGlu5 possessed a salt bridge between a Lys residue in TMD3 and a Glu residue in TMD6 forming an “ionic lock,” presumably to stabilize the inactive state. Notably, both of the residues involved in this salt bridge are highly conserved among Class C GPCRs, suggesting that this “ionic lock” may be a common mechanism of stabilizing Class C GPCRs in the inactive state. Moreover, in both inactive structures, the NAM binds in an allosteric site that is analogous to the ligand-binding site in Class A GPCRs. In contrast to other GPCRs, the allosteric binding cavity in both mGlu1 and mGlu5 is narrowed due to an inward shift of helices V and VII, and the formation of a lid over the cavity by the extracellular loop (ECL) 2 (Doré et al., 2014; Wu et al., 2014). Despite this distinction, Class C GPCR 7-TMDs are strikingly similar to other GPCR classes, thereby providing some face validity to the previous use of homology modeling in order to generate hypotheses about mGlu receptor allosteric modulation.
Homology models of mGlu receptors previously were constructed using class A GPCR templates in order to form and test hypotheses about allosteric modulator binding to mGlu receptor 7-TMDs. Using such homology models for mGlu receptors constructed from the inactive conformation of either rhodopsin or the β-adrenergic receptor (β-AR) 7-TMDs, modeling experiments have generated hypotheses about allosteric modulator binding that have been tested using site-directed mutagenesis. Using this experimental design, modeling experiments identified the 2-methyl-6-(phenylethynyl)-pyridine (MPEP) binding site in mGlu5 and investigated the binding modes of novel allosteric modulators (Pagano et al., 2000; Malherbe et al., 2003, 2006; Kaae et al., 2012; Molck et al., 2012). However, these mGlu receptor homology models still possessed major shortfalls due to their inability to explain all experimental data and differing explanations of experimental findings. Most notably, two unique homology models predicted different binding sites for MPEP in mGlu5, highlighting the importance of subtle differences between homology models generated with distinct templates (Pagano et al., 2000; Kaae et al., 2012; Molck et al., 2012). More recently, pairing homology modeling with an extensive studies of structure-activity relationships (SAR) obtained from diverse chemical scaffolds of mGlu5 allosteric modulators was successful in iteratively improving homology model predictions (Gregory et al., 2014). Indeed, further experimentation validated this homology model’s hypothesis that structurally unrelated mGlu5 allosteric modulators can bind identical sites in a fully competitive manner, thereby fully displacing the mGlu5 NAM positron emission tomography (PET) ligand [18F]FPEB. More importantly, these experiments validated and highlighted the importance of the homology model’s hypothesis that closely related mGlu5 allosteric modulators can bind nonidentical sites. As a result of their noncompetitive binding, structural analogs of [18F]FPEB failed to displace the mGlu5 NAM PET ligand, demonstrating no measureable receptor occupancy at doses that achieve full efficacy in rodent models (Rook et al., 2014). As such, prior to studies of in vivo receptor occupancy, in vitro binding assays must verify that allosteric ligands interact in a fully competitive manner in order to accurately establish a relationship between receptor occupancy and efficacy. Overall, this example highlights the power of homology modeling at generating critical and testable hypotheses, especially when the models are iteratively improved based on data from both site-directed mutagenesis and SAR from diverse chemical scaffolds. Moving forward, new homology models of other mGlu receptors should also utilize the inactive mGlu1 and mGlu5 7-TMD structures as templates, since higher sequence homology between mGlu receptors likely will result in improved homology models.
The resolved mGlu1 and mGlu5 7-TMD structures can also be used to both predict how novel allosteric modulators may interact with either mGlu1 or mGlu5 and perform in silico drug screening. For example, based on the mavoglurant-bound mGlu5 structure, it was postulated that mode switches observed in this scaffold may arise from disruption of a hydrogen bond network around a water molecule (Doré et al., 2014). Future experiments can directly test whether novel mavoglurant analogs predicted to disrupt this network actually induce a mode switch from NAM to neutral binder or PAM. Similarly, the mGlu1 7-TMD structure elucidated precise ligand-binding interactions between mGlu1 and its NAM, 4-fluoro-N-(4-(6-(isopropylamino)pyrimidin-4-yl)-N-methylbenzamide (FITM) that may be important for understanding allosteric modulator SAR. In tandem with primary sequence analysis and site-directed mutagenesis, the mGlu1 7-TMD bound to the NAM, FITM, suggested which amino acid residues are involved in the mGlu receptor subtype selectivity of FITM for mGlu1 over other mGlu receptors. Using this knowledge, subsequent in silico docking experiments predicted decreases in bond strength between mGlu1 and FITM analogs that could account for the observed decreases in the analogs’ affinity and/or negative cooperativity in functional assays (Wu et al., 2014). Thus, the inactive conformations of these 7-TMDs may aid in drug discovery efforts by informing allosteric modulator SAR and allowing for in silico structure-based drug screening.
Unfortunately, for other GPCRs, structure-based drug screens using previously-resolved GPCR inactive structures have yielded only antagonists and inverse agonists, suggesting that resolution of the active GPCR conformation is necessary for understanding ligand interactions stabilizing this active state (Shoichet and Kobilka, 2012). Recent advances in structural biology have facilitated the stabilization and subsequent crystallization of unstable, active receptor conformations (Figure 1B). In particular, crystallographers have utilized nanobodies, or single domain antibody fragments generated by immunizing camelids with agonist-bound GPCR structures, in order to stabilize and crystallize active receptor conformations (Steyaert and Kobilka, 2011). Using this strategy, the β-AR structure bound to a synthetic agonist was resolved both alone and in complex with its G-protein, Gs (Rasmussen et al., 2011a, 2011b). Importantly, the β-AR structures did not differ significantly when resolved using these different strategies, exhibiting only minor differences at the cytoplasmic end of β-AR where the G-protein interacts. This structural similarity provides more validity to using nanobodies in order to resolve the active receptor complex with or without the G-protein. Thus, a nanobody stabilizing the PAM-bound mGlu receptor could allow for crystallization and resolution of an active, PAM-bound state. Such studies will provide a better understanding of mGlu receptor-positive allosteric modulation and an active receptor template for structure-based drug screens for novel mGlu receptor PAMs.
Regulation of Signaling in the CNS by mGlu Receptor Allosteric Modulators
Beyond understanding allosteric modulator binding, drug discovery efforts aimed at optimizing mGlu receptor PAMs and NAMs must recognize the diversity of pharmacology profiles exhibited by different mGlu receptor allosteric modulators. In particular, such drug discovery efforts must understand the biological consequences of these diverse and often subtle differences in allosteric modulator effects on mGlu receptor signaling. Importantly, preclinical research utilizing tool compounds have identified modes of pharmacology that may be critical for both preclinical efficacy and side effect profiles of mGlu receptor allosteric modulators. Further research is necessary to elucidate whether these preclinical findings translate to humans since much research has highlighted the species differences in mGlu receptor pharmacology. If these modes of pharmacology demonstrate translatability from mice to men, drug discovery efforts should optimize allosteric modulators to either possess or avoid certain modes of pharmacology. However, even if there is not direct translatability to humans, these diverse modes of pharmacology must be identified and studied in preclinical drug discovery efforts in order to determine if such species-specific pharmacology contributes to either preclinical efficacy or adverse side effects.
Functional Impact of Allosteric Agonist Activity
In addition to potentiating physiological responses to orthosteric agonists, mGlu receptor PAMs can exhibit intrinsic allosteric agonist activity (Figure 2A). Theoretically, ago-PAMs likely would not maintain the physiological pattern of mGlu receptor activity, thereby nullifying one potential advantage of allosteric modulation and possibly leading to unintended side effects resulting from aberrant mGlu receptor activity (Conn et al., 2014). Recent studies using mGlu5 ago-PAMs explored whether ago-PAM activity contributes to either mGlu5 PAMs’ therapeutic efficacy or their adverse side effects. Although many structurally distinct mGlu5 PAMs previously exhibited antipsychotic efficacy in rodent models, many of the early generation PAMs possessed ago-PAM activity in vitro, raising the possibility that their ago-PAM activity was required for efficacy (Kinney et al., 2005; Liu et al., 2008; Rodriguez et al., 2010). However, Noetzel et al (2012) found that the presence or absence of allosteric agonist activity is highly influenced by levels of mGlu5 expression in cell lines. Thus, many of the early compounds identified at mGlu5 ago-PAMs did not display allosteric agonist activity in cell lines expressing lower levels of mGlu5 or in native systems. Furthermore, this study demonstrated that optimized pure mGlu5 PAMs that had no agonist activity, even in systems with high receptor expression, displayed comparable efficacy in a rodent model of antipsychotic activity, suggesting that ago-PAM activity is not necessary for mGlu5 PAM antipsychotic activity. In a subsequent study, medicinal chemistry optimization yielded mGlu5 ago-PAMs with intrinsic activity in cell lines expressing low levels of either rat or human mGlu5, and in native systems (Rook et al., 2013). Interestingly, an optimized mGlu5 ago-PAM, but not a pure mGlu5 PAM, induced convulsions and seizure activity in vivo reminiscent of established adverse effects of Group I mGlu receptor orthosteric agonists. These studies established the importance of avoiding true glutamate-independent ago-PAM activity of mGlu5 PAMs in order to reduce severe adverse effects associated with excessive activation of this mGlu receptor subtype (Tizzano et al., 1995; Conn and Pin, 1997; Rook et al., 2013).
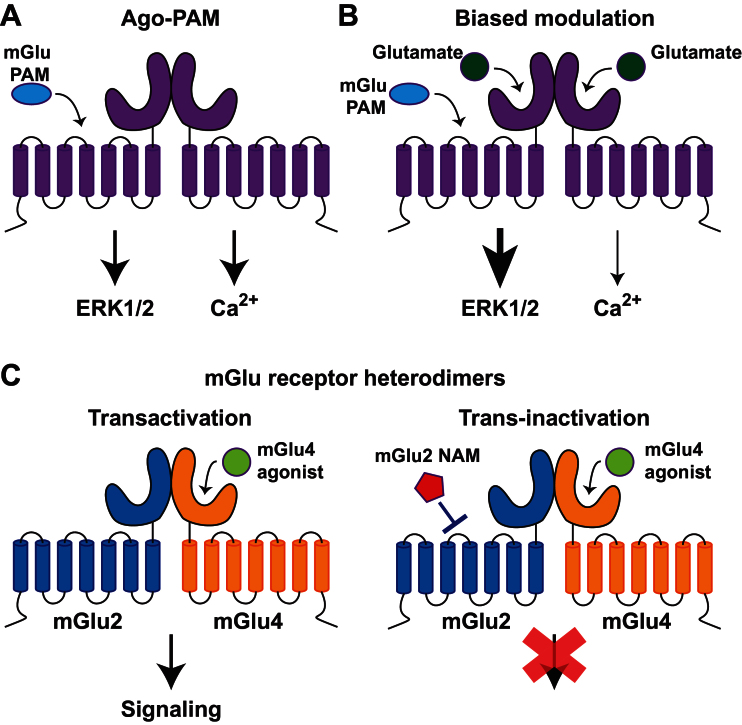
Figure 2.
Diverse pharmacological profiles exhibited by metabotropic glutamate (mGlu) receptor allosteric modulators. (A) In the absence of the orthosteric agonist, glutamate, some positive allosteric modulators (PAMs) demonstrate intrinsic agonist activity by inducing downstream signaling and are thus referred to as ago-PAMs. (B) In the presence of glutamate and other orthosteric agonists, other allosteric modulators can induce biased modulation of downstream signaling from the mGlu receptor, where the PAM selectively potentiates extracellular signal-regulated kinase 1/2 (ERK1/2) signaling. (C) Allosteric modulators can also act at novel mGlu receptor heterodimers, leading to novel pharmacology wherein transactivation of mGlu2/4 heterodimers by an mGlu4 agonist can be blocked by an mGlu2 negative allosteric modulators through trans-inactivation.
As discussed above, it should be emphasized that many mGlu5 PAMs with agonist activity in cell lines that overexpress mGlu5 do not demonstrate agonist activity in native tissues (Noetzel et al., 2012). Thus, it is critical to establish appropriate cell lines for assessing ago-PAM versus pure PAM activity and to directly relate findings in cell lines to studies in native tissues. While this can be readily achieved in preclinical species, this can present a challenge in drug discovery efforts since it is often impossible to evaluate physiological effects of mGlu5 PAMs in human central nervous system (CNS) preparations in situ. Thus, for preclinical studies of effects on the human receptor, one must rely on assessing effects of novel compounds in cell lines expressing different levels of the human receptor. While we do not know whether the functional impact of ago-PAM activity will translate to humans, a conservative approach is to avoid compounds with allosteric agonist activity at the human receptor, even in cell lines with high receptor expression (Rook et al., 2013).
Another critical factor to consider in avoiding ago-PAM activity is the possibility that metabolism of mGlu5 PAMs will generate active metabolites with a different mode of efficacy than the parent compound. For instance, P450 metabolism of the mGlu5 PAM, VU0403602, produced an active metabolite with robust allosteric agonist activity in native brain tissue that induced intense seizure activity in vivo. Importantly, administration of a P450 inhibitor prevented both formation of the ago-PAM metabolite and induction of seizure activity by VU0403602, suggesting that the activity of this metabolite may contribute to the adverse effect liability (Bridges et al., 2013). Thus, ago-PAM activity can be physiologically relevant at least in preclinical models and studies of agonist activity should employ cell lines expressing low levels of mGlu receptors in order to more accurately predict PAMs that will demonstrate ago-PAM activity in vivo.
Importantly, ago-PAM activity may not be the only factor that contributes to adverse effects of mGlu5 PAMs, since a novel, pure mGlu5 PAM also induced neurotoxicity and seizure-like activity upon high dosing for 4 days (Parmentier-Batteur et al., 2014). Regardless, mGlu5 ago-PAM activity is not necessary for antipsychotic efficacy and should be avoided in drug discovery efforts due to its potential for adverse side effects.
Allosteric Modulator-Induced Bias of mGlu Receptor Signaling
Biased modulation is another pharmacological property observed in mGlu receptor allosteric modulators that is beginning to be appreciated for its biological importance. Biased modulation is characterized by allosteric modulators that selectively modulate coupling of the receptor to specific signaling pathways (Figure 2B). In vitro, several mGlu1 and mGlu5 PAMs previously exhibited biased modulation, wherein PAMs differentially modulated extracellular signal-regulated kinase 1/2 (ERK1/2), cyclic adenosine monophosphate (cAMP), and Ca2+ signaling (Zhang et al., 2005; Sheffler and Conn, 2008). More recently, a novel mGlu5 PAM, NCFP (N-(4-chloro-2-((4-fluoro-1,3 dioxoisoindolin-2yl)methyl)phenyl)picolinamide), was identified that potentiates mGlu5-mediated signaling in both cells and brain slices without affecting mGlu5-dependent long-term depression (LTD) and long-term potentiation (LTP) in the hippocampus (Noetzel et al., 2013). Further characterization of biased mGlu receptor modulation also revealed biased mGlu7 modulation wherein the NAM, MMPIP (6-(4-methoxyphenyl)-5-methyl-3-pyridin-4-ylisoxazolo[4,5-c]pyridin-4(5H)-one), inhibited some, but not all, responses to mGlu7 activation and did not inhibit mGlu7–dependent modulation of synaptic transmission (Niswender et al., 2010). Although both Group I and Group III mGlu receptor allosteric modulators displayed biases in native tissues (Niswender et al., 2010; Noetzel et al., 2013), none of the early studies provided evidence for any in vivo relevance of biased mGlu receptor modulation. However, more recently a biased mGlu5 PAM, VU0409551, was identified that specifically potentiates Gαq-signaling without modulating mGlu5 coupling to N-methyl-D-aspartate receptor (NMDAR) currents and NMDAR-dependent synaptic plasticity (Rook et al., 2015). Despite the known importance of NMDAR hypofunction in schizophrenia pathophysiology (Field et al., 2011; Coyle et al., 2012; Timms et al., 2013), VU0409551 exhibited both antipsychotic and pro-cognitive effects in animal models, suggesting that mGlu5 potentiation of NMDAR currents is not necessary for its therapeutic efficacy, contrary to the field’s previous view. The paradigm shift resulting from an improved understanding of biased mGlu5 modulation emphasizes the importance of further research into biased mGlu receptor modulation. To establish an in vivo relevance for biased modulation of other mGlu receptor subtypes, medicinal chemistry efforts should be focused on intentionally optimizing other allosteric modulators that induce specific biased modulation of mGlu receptor signaling and have properties required for systemic dosing and use in in vivo studies. One major challenge with translating this to clinical studies is similar to the challenge outlined for ago-PAMs above. While it is possible to fully evaluate potential stimulus bias in brain slices in preclinical studies, this may not be possible in human tissue. Thus, if biased effects on specific physiological responses are important for efficacy or adverse effect liability, it will be difficult to have complete confidence that the stimulus bias established in rodent CNS preparations will also be present in the human brain.
Effects of Allosteric Modulators on mGlu Receptor Heterodimers
In recent years, novel allosteric modulators have helped to verify the existence and establish the functional relevance of mGlu receptor heterodimers (Figure 2C; Doumazane et al., 2011; Yin et al., 2014). Although mGlu receptors form obligate dimers in order to function, previous studies suggested that these mGlu receptors formed strict homodimers (Romano et al., 1996). An in vitro fluorescence resonance energy transfer (FRET) study challenged this long-standing hypothesis, demonstrating that mGlu receptors exhibit both intra- and inter-group heterodimerization, wherein heterodimers form between Group I mGlu receptors and between Group II and Group III mGlu receptors (Doumazane et al., 2011). Recently, in vitro studies verified the presence of the mGlu2/4 heterodimer and characterized its distinct functional properties (Kammermeier, 2012; Yin et al., 2014). Importantly, our lab also found compelling evidence for the presence and functionality of mGlu2/4 heterodimers in the striatum at the cortico-striatal synapse (Yin et al., 2014). At this synapse, only mGlu4 PAMs active at the mGlu2/4 heterodimer blunted neurotransmission, suggesting that mGlu2/4 heterodimers, but not mGlu4 homodimers, exist at this synapse. Despite this intriguing finding, to date there is no known in vivo function attributed to a specific heterodimer combination, and it remains unknown what the in vivo consequences of heterodimer modulation are.
The initial characterizations of mGlu2/4 heterodimer pharmacology also raised basic questions as to the mechanism of heterodimer activation and inactivation. In cultured neurons co-expressing mGlu2 and mGlu4, data suggested that neither mGlu2- nor mGlu4-selective PAMs potentiated mGlu2/4 heterodimer-mediated inhibition of voltage-gated Ca2+ channels when applied either alone or together (Kammermeier, 2012). However, in line with previous literature suggesting that PAM binding is only required and observed on one mGlu receptor protomer to produce its potentiation (Goudet et al., 2005; Lundstrom et al., 2011), mGlu2 PAMs alone and some mGlu4 PAMs alone potentiated mGlu2/4-mediated G protein-coupled inwardly-rectifying potassium (GIRK) channel activity and depression of synaptic transmission at the cortico-striatal synapse (Yin et al., 2014). Similarly, data from these two experimental lines generated conflicting views on whether or not mGlu2/4 heterodimers require agonist binding to one or both orthosteric sites in order to initiate downstream signaling of mGlu2/4 (Kammermeier, 2012; Yin et al., 2014). Our findings demonstrated that binding of either a selective Group II or Group III agonist is sufficient for receptor activity, consistent with previous findings supporting transactivation between mGlu receptor dimers (Figure 2C; Kniazeff et al., 2004; Yin et al., 2014). However, data on mGlu2/4 heterodimer-mediated inhibition of voltage-gated Ca2+ channels indicated that transactivation does not occur upon binding of an agonist to a single protomer, which is consistent with previous findings in this neuron culture system (Kammermeier and Yun, 2005; Kammermeier, 2012). These discrepancies likely reflect either a context-dependent difference in mGlu2/4 signaling or unique requirements for receptor activation to initiate specific downstream signaling events. A more surprising finding was that a Group II mGlu receptor NAM attenuated L-AP4-mediated mGlu2/4 signaling both in vitro and in brain slice preparations (Figure 2C; Yin et al., 2014). While this finding directly contradicts the present view in the field that a NAM must bind both protomers in order to attenuate mGlu receptor dimer-mediated signaling (Hlavackova et al., 2005; Kammermeier, 2012), the Group II NAM may prevent transactivation of mGlu2 by a Group III agonist rather than attenuating signaling through trans-inactivation. In order to resolve these discrepancies and better understand mGlu receptor heterodimer pharmacology, in vitro molecular pharmacology experiments should directly measure heterodimer activation using heteromer identification technology (HIT) or complemented donor-acceptor resonance energy transfer (CODA-RET) that assess immediate downstream recruitment of β-arrestin or G protein to the heterodimer, respectively (See et al., 2011; Urizar et al., 2011; Vischer et al., 2015).
Effects of mGlu Receptor Allosteric Modulators on Activity of Identified Brain Circuits
Although the aforementioned research informs efforts to further optimize binding and signaling properties of PAMs and NAMs, it remains critical for neuroscience drug discovery efforts to understand how allosteric modulators achieve therapeutic efficacy through modulation of neural circuit function and dysfunction. Using selective mGlu receptor allosteric modulators, previous slice electrophysiology studies have uncovered roles for mGlu receptors at specific synapses. As mentioned above, electrophysiology studies have also dissected how ago-PAMs, biased mGlu receptor modulators, and heterodimer-active allosteric modulators affect neural circuit function (Noetzel et al., 2013; Rook et al., 2013, 2015; Yin et al., 2014). Additionally, the recent discovery of selective NAMs for mGlu2, mGlu3, and mGlu7 facilitated experimentation on the necessity of each mGlu receptor at specific synapses for neurotransmission (Wenthur et al., 2013; Kalinichev et al., 2014; Walker et al., 2015). Following the optimization of mGlu2-selective and mGlu3-selective NAMs, Walker et al. (2015) found that LTD in the medial prefrontal cortex (mPFC) depends on mGlu3, but not mGlu2, activity. Likewise, use of the novel mGlu7 NAM, ADX71743, allowed for confirmation that mGlu7 activation reduces neural transmission at the SC-CA1 synapse; however, subsequent electrophysiology experiments using this mGlu7 NAM indicated that, contrary to the current view of mGlu7 function at this synapse, mGlu7 did not function as an autoreceptor on glutamatergic neurons (Klar et al., 2015). With the help of optogenetics in slice electrophysiology experiments, the mechanism of mGlu7-mediated synaptic depression was definitively resolved, demonstrating how optogenetic tools used in concert with a pharmacological tool compound could elucidate the compound’s precise site of action (Figure 3A).
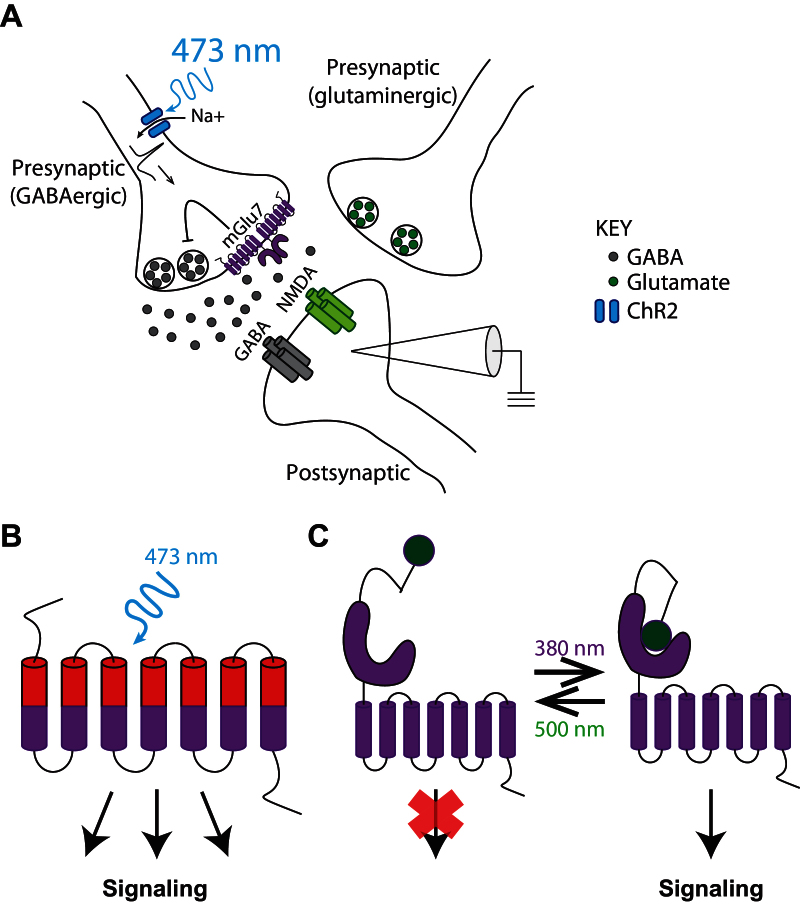
Figure 3.
Optogenetic and optopharmacological dissection of metabotropic glutamate (mGlu) receptor function in defined neural circuits. (A) In the depiction of the experimental paradigm from Klar et al (2015), parvalbumin-expressing GABAergic neurons were selectively stimulated using blue light to activate channelrhodopsin, expressed only in this neuronal population. Electrophysiological recordings from the postsynaptic neuron demonstrated that mGlu7 activation on presynaptic GABAergic interneurons decreases inhibitory postsynaptic currents (IPSCs) through modulation of presynaptic GABA release. (B) Cartoon of opto-mGlu6 illustrates the chimeric receptor composed of the extracellular domain of melanopsin (red) and intracellular domain of mGlu6 (purple). Through this design, mGlu6-mediated intracellular signaling can be induced by a blue light activating the melanopsin extracellular domain. (C) Light-activated (pictured) and -inactivated (not shown) mGlu receptors, or LimGluRs, utilize a tethered ligand that, through light-induced isomerization, can bind to the orthosteric site in order to activate or inhibit mGlu receptor signaling, respectively.
The advent of optogenetics has provided complementary tools for refining our understanding of mGlu receptor modulation and its effects in defined neural circuits. Optogenetics utilizes the light-activated, non-selective cation channel, channelrhodopsin (ChR2) in order to induce a depolarizing current and subsequent action potential initiation in response pulses of blue light (Tye and Deisseroth, 2012). Using either viral vectors or transgenic mice, ChR2 can be expressed in defined neuronal populations, thus allowing for activation of specific neurons with pulses of blue light. As alluded to above and illustrated in Figure 3A, this complimentary technique was used to directly test whether presynaptic mGlu7 activation on GABAergic neurons led to attenuation of GABA-mediated inhibitory postsynaptic currents (IPSCs) in CA1 pyramidal neurons (Klar et al., 2015). Although initial experiments supported this hypothesis by assessing the effect of mGlu7 activation on pharmacologically-isolating monosynaptic IPSCs in CA1 pyramidal neurons, the electrical stimulation used to evoke IPSCs in this experimental paradigm could cause release of other neuromodulators involved in the regulation of GABA release. Using ChR2 expressed in parvalbumin-expressing GABAergic interneurons to specifically activate this neuronal subpopulation at the SC-CA1 synapse, slice electrophysiology experiments demonstrated that presynaptic mGlu7 activation attenuated optically-induced IPSCs (Figure 3A). Thus, this optogenetic experiment verified the proposed mechanism of action behind mGlu7 modulation of neurotransmission at this synapse, thereby demonstrating the value of using optogenetics in order to determine the site of action at which mGlu receptor modulation exerts its neuromodulatory effect.
Novel optogenetic tools are also facilitating characterization of the spatial and temporal dynamics of modulating mGlu receptor signaling in defined neural circuits in vivo. Using photoswitch molecules tethered to mGlu receptors, Levitz et al (2013) designed light-agonized and light-antagonized mGlu receptors (LimGluRs; Figure 3C). In both in vitro and in vivo systems, these LimGluRs were bistable, thus permitting precise spatial and temporal control of their activation or inhibition. Importantly, in vivo expression and light-agonism of LimGluR2 in zebrafish revealed a behavioral role for mGlu2 signaling in the acoustic startle response, thus displaying the potential for further research with LimGluRs to tease apart the behavioral function of specific mGlu receptors in defined neural circuits. To add to the optogenetic toolbox for mGlu receptors, a light-activated, opto-mGlu6 receptor was developed, consisting of the extracellular light-sensitive melanopsin and the intracellular domain of mGlu6 in order to induce light-activated mGlu6 signaling (Figure 3B; van Wyk et al., 2015). Expression and activation of opto-mGlu6 in retinal ON-bipolar cells generated light-evoked current in these cells and partially restored visual performance in a mouse model of blindness. Thus, both optogenetic tools offer the possibility to precisely control mGlu receptor signaling and identify the in vivo behavioral effect of activating specific mGlu receptors in defined neural circuits. However, further development of transgenic mice or viral vectors expressing LimGluRs and opto-mGluRs is necessary to facilitate research into the role of specific mGlu receptors in distinct neuronal populations known to natively express the mGlu receptor of interest, thereby informing our understanding on potential in vivo effects of mGlu receptor allosteric modulation.
Novel optopharmacological tools may also permit parallel lines of experimentation in order to better understand the spatial and temporal effects of allosteric modulation. Through integration of a photochromatic group within a ligand, Pittolo et al (2014) developed a novel light-activated and light-inactivated mGlu5 NAM called Alloswitch-1. Under green light, Alloswitch-1 exhibits nanomolar potency at mGlu5; however, the introduction of violet light induces an isomerization that diminishes its activity on mGlu5. This optopharamcological tool compound displays reversible effects on both cell signaling in vitro and tadpole locomotor behavior in vivo. This proof-of-concept study demonstrated the potential for tissue-specific, reversible activation and inactivation of an allosteric modulator as a therapeutic. Again, following the necessary further optimization and development of optopharmacological tool compounds for use in rodents, future experiments could elucidate the precise spatial and temporal effect of mGlu receptor allosteric modulation in preclinical models of brain disorders.
Conclusion
While much research on mGlu receptor allosteric modulators focuses on identifying potent clinical candidates, innovative basic research using mGlu receptor allosteric modulator tool compounds can improve our understanding of mGlu receptor neurobiology and subsequently aid further drug discovery efforts. Using high-affinity binding mGlu receptor allosteric modulators, structural biologists resolved the inactive, NAM-bound 7-TMD structure of mGlu receptors and now possess the tools to resolve the active, PAM-bound 7-TMD structure. Allosteric modulators with intrinsic agonist activity, biased mGlu receptor modulation, or mGlu receptor heterodimer activity have helped to elucidate the biological importance of these diverse pharmacological profiles. And, finally, using optogenetic and optopharmacological tools aimed at understanding mGlu receptor modulation at the neural circuit level, basic research has gained and will continue to gain insights on how mGlu receptor modulation affects both neurotransmission in brain slices and in vivo behavioral outcomes. Thus, these studies with allosteric modulator tool compounds have elucidated mGlu receptor neurobiology and subsequently informed drug discovery efforts, thereby demonstrating the necessity for continuing these basic research studies in tandem with allosteric modulator drug discovery efforts.
Disclosures
Dr Conn has received funding from the National Institutes of Health, Johnson & Johnson, AstraZeneca, Bristol-Myers Squibb, Michael J Fox Foundation, and Seaside Therapeutics. He has consulted for Pfizer, Cambridge, and Millipore Corporation and received compensation over the past 3 years. Additionally, he has served on the Scientific Advisory Boards of Seaside Therapeutics, Michael J Fox Foundation, Stanley Center for Psychiatric Research Broad Institute (MIT/Harvard), Karuna Pharmaceuticals, Lieber Institute for Brain Development Johns Hopkins University, Clinical Mechanism (POCM) and Proof of Concept (POC) Consortium, and Neurobiology Foundation for Schizophrenia and Bipolar Disorder over the past 3 years. Dr O’Brien has no financial interests to disclose.
Acknowledgments
This work was supported by the National Institutes of Health (R01 MH062646 and R37 NS031373 to Dr Conn and T32 MH093366 to Dr O’Brien). The authors thank Drs Molly Altman, Rocco Gogliotti, and Vijay Samineni for their critical comments on this review.
References
- Annoura H, Fukunaga A, Uesugi M, Tatsuoka T, Horikawa Y. (1996) A novel class of antagonists for metabotropic glutamate receptors, 7-(Hydroxyimino)cyclopropa[b]chromen-1a-carboxylates. Bioorg Med Chem Lett 6:763–766. [Google Scholar]
- Bridges TM, Rook JM, Noetzel MJ, Morrison RD, Zhou Y, Gogliotti RD, Vinson PN, Xiang Z, Jones CK, Niswender CM, Lindsley CW, Stauffer SR, Conn PJ, Daniels JS. (2013) Biotransformation of a novel positive allosteric modulator of metabotropic glutamate receptor subtype 5 contributes to seizure-like adverse events in rats involving a receptor agonism-dependent mechanism. Drug Metab Dispos 41:1703–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn PJ, Pin JP. (1997) Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol 37:205–237. [DOI] [PubMed] [Google Scholar]
- Conn PJ, Christopoulos A, Lindsley CW. (2009) Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov 8:41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn PJ, Lindsley CW, Meiler J, Niswender CM. (2014) Opportunities and challenges in the discovery of allosteric modulators of GPCRs for treating CNS disorders. Nat Rev Drug Discov 13:692–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle JT, Basu A, Bennyworth M, Balu D, Konopaske G. (2012) Glutamatergic synaptic dysregulation in schizophrenia: therapeutic implications. In: Novel antischizophrenia treatments (Geyer MA, Gross G, eds), pp 267–295. Berlin: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doré AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, Errey JC, Jazayeri A, Khan S, Tehan B, Weir M, Wiggin GR, Marshall FH. (2014) Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511:557–562. [DOI] [PubMed] [Google Scholar]
- Doumazane E, Scholler P, Zwier JM, Trinquet E, Rondard P, Pin JP. (2011) A new approach to analyze cell surface protein complexes reveals specific heterodimeric metabotropic glutamate receptors. FASEB J 25:66–77. [DOI] [PubMed] [Google Scholar]
- Engers DW, Lindsley CW. (2013) Allosteric modulation of Class C GPCRs: a novel approach for the treatment of CNS disorders. Drug Discov Today Technol 10:e269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field JR, Walker AG, Conn PJ. (2011) Targeting glutamate synapses in schizophrenia. Trends Mol Med 17:689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet C, Kniazeff J, Hlavackova V, Malhaire F, Maurel D, Acher F, Blahos J, Prezeau L, Pin JP. (2005) Asymmetric functioning of dimeric metabotropic glutamate receptors disclosed by positive allosteric modulators. J Biol Chem 280:24380–24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory KJ, Nguyen ED, Malosh C, Mendenhall JL, Zic JZ, Bates BS, Noetzel MJ, Squire EF, Turner EM, Rook JM, Emmitte KA, Stauffer SR, Lindsley CW, Meiler J, Conn PJ. (2014) Identification of specific ligand–receptor interactions that govern binding and cooperativity of diverse modulators to a common metabotropic glutamate receptor 5 allosteric site. ACS Chem Neurosci 5:282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavackova V, Goudet C, Kniazeff J, Zikova A, Maurel D, Vol C, Trojanova J, Prezeau L, Pin JP, Blahos J. (2005) Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J 24:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaae BH, Harpsøe K, Kvist T, Mathiesen JM, Mølck C, Gloriam D, Jimenez HN, Uberti MA, Nielsen SM, Nielsen B, Bräuner-Osborne H, Sauerberg P, Clausen RP, Madsen U. (2012) Structure-activity relationships for negative allosteric mGluR5 modulators. ChemMedChem 7:440–451. [DOI] [PubMed] [Google Scholar]
- Kalinichev M, Le Poul E, Bolea C, Girard F, Campo B, Fonsi M, Royer-Urios I, Browne SE, Uslaner JM, Davis MJ, Raber J, Duvoisin R, Bate ST, Reynolds IJ, Poli S, Celanire S. (2014) Characterization of the novel positive allosteric modulator of the metabotropic glutamate receptor 4 ADX88178 in rodent models of neuropsychiatric disorders. J Pharm Exp Ther 350:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier PJ. (2012) Functional and pharmacological characteristics of metabotropic glutamate receptors 2/4 heterodimers. Mol Pharmacol 82:438–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier PJ, Yun J. (2005) Activation of metabotropic glutamate receptor 1 dimers requires glutamate binding in both subunits. J Pharm Exp Ther 312:502–508. [DOI] [PubMed] [Google Scholar]
- Kinney GG, O’Brien JA, Lemaire W, Burno M, Bickel DJ, Clements MK, Chen TB, Wisnoski DD, Lindsley CW, Tiller PR, Smith S, Jacobson MA, Sur C, Duggan ME, Pettibone DJ, Conn PJ, Williams DL., Jr (2005) A novel selective positive allosteric modulator of metabotropic glutamate receptor subtype 5 has in vivo activity and antipsychotic-like effects in rat behavioral models. J Pharm Exp Ther 313:199–206. [DOI] [PubMed] [Google Scholar]
- Klar R, Walker AG, Ghose D, Grueter BA, Engers DW, Hopkins CR, Lindsley CW, Xiang Z, Conn PJ, Niswender CM. (2015) Activation of metabotropic glutamate receptor 7 is required for induction of long-term potentiation at SC-CA1 synapses in the hippocampus. J Neurosci 35:7600–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeff J, Bessis AS, Maurel D, Ansanay H, Prezeau L, Pin JP. (2004) Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat Struct Mol Biol 11:706–713. [DOI] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. (2000) Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407:971–977. [DOI] [PubMed] [Google Scholar]
- Levitz J, Pantoja C, Gaub B, Janovjak H, Reiner A, Hoagland A, Schoppik D, Kane B, Stawski P, Schier AF, Trauner D, Isacoff EY. (2013) Optical control of metabotropic glutamate receptors. Nat Neurosci 16:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litschig S, Gasparini F, Ruegg D, Stoehr N, Flor PJ, Vranesic I, Prezeau L, Pin JP, Thomsen C, Kuhn R. (1999) CPCCOEt, a noncompetitive metabotropic glutamate receptor 1 antagonist, inhibits receptor signaling without affecting glutamate binding. Mol Pharmacol 55:453–461. [PubMed] [Google Scholar]
- Liu F, et al. (2008) ADX47273 [S-(4-fluoro-phenyl)-{3-[3-(4-fluoro-phenyl)-[1,2,4]-oxadiazol-5-yl]-piperidin-1- yl}-methanone]: a novel metabotropic glutamate receptor 5-selective positive allosteric modulator with preclinical antipsychotic-like and procognitive activities. J Pharm Exp Ther 327:827–839. [DOI] [PubMed] [Google Scholar]
- Lundstrom L, Bissantz C, Beck J, Wettstein JG, Woltering TJ, Wichmann J, Gatti S. (2011) Structural determinants of allosteric antagonism at metabotropic glutamate receptor 2: mechanistic studies with new potent negative allosteric modulators. Br J Pharmacol 164:521–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malherbe P, Kratochwil N, Knoflach M, Kew J, Kratzeisen C, Maerki H, Adam G, Mutel V. (2003) Mutational analysis and molecular modeling of the allosteric binding site of a novel, selective, noncompetitive antagonist of the metabotropic glutamate 1 receptor. J Biol Chem 278:8340–8347. [DOI] [PubMed] [Google Scholar]
- Malherbe P, Kratochwil N, Muhlemann A, Zenner M-T, Fischer C, Stahl M, Gerber PR, Jaeschke G, Porter RHP. (2006) Comparison of the binding pockets of two chemically unrelated allosteric antagonists of the mGlu5 receptor and identification of crucial residues involved in the inverse agonism of MPEP. J Neurochem 98:601–615. [DOI] [PubMed] [Google Scholar]
- Molck C, Harpsoe K, Gloriam DE, Clausen RP, Madsen U, Pedersen LO, Jimenez HN, Nielsen SM, Mathiesen JM, Brauner-Osborne H. (2012) Pharmacological characterization and modeling of the binding sites of novel 1,3-Bis(pyridinylethynyl)benzenes as metabotropic glutamate receptor 5-selective negative allosteric modulators. Mol Pharmacol 82:929–937. [DOI] [PubMed] [Google Scholar]
- Nickols HH, Conn PJ. (2014) Development of allosteric modulators of GPCRs for treatment of CNS disorders. Neurobiol Dis 61:55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, Conn PJ. (2010) Metabotropic Glutamate Receptors: Physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol 50:295–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niswender CM, Johnson KA, Miller NR, Ayala JE, Luo Q, Williams R, Saleh S, Orton D, Weaver CD, Conn PJ. (2010) Context-dependent pharmacology exhibited by negative allosteric modulators of metabotropic glutamate receptor 7. Mol Pharmacol 77:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noetzel MJ, Rook JM, Vinson PN, Cho HP, Days E, Zhou Y, Rodriguez AL, Lavreysen H, Stauffer SR, Niswender CM, Xiang Z, Daniels JS, Jones CK, Lindsley CW, Weaver CD, Conn PJ. (2012) Functional impact of allosteric agonist activity of selective positive allosteric modulators of metabotropic glutamate receptor subtype 5 in regulating central nervous system function. Mol Pharmacol 81:120–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noetzel MJ, Gregory KJ, Vinson PN, Manka JT, Stauffer SR, Lindsley CW, Niswender CM, Xiang Z, Conn PJ. (2013) A novel metabotropic glutamate receptor 5 positive allosteric modulator acts at a unique site and confers stimulus bias to mGlu5 signaling. Mol Pharmacol 83:835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano A, Ruegg D, Litschig S, Stoehr N, Stierlin C, Heinrich M, Floersheim P, Prezeau L, Carroll F, Pin JP, Cambria A, Vranesic I, Flor PJ, Gasparini F, Kuhn R. (2000) The non-competitive antagonists 2-methyl-6-(phenylethynyl)pyridine and 7-hydroxyiminocyclopropan[b]chromen-1a-carboxylic acid ethyl ester interact with overlapping binding pockets in the transmembrane region of group I metabotropic glutamate receptors. J Biol Chem 275:33750–33758. [DOI] [PubMed] [Google Scholar]
- Parmentier-Batteur S, et al. (2014) Mechanism based neurotoxicity of mGlu5 positive allosteric modulators – development challenges for a promising novel antipsychotic target. Neuropharmacology 82:161–173. [DOI] [PubMed] [Google Scholar]
- Pittolo S, Gomez-Santacana X, Eckelt K, Rovira X, Dalton J, Goudet C, Pin JP, Llobet A, Giraldo J, Llebaria A, Gorostiza P. (2014) An allosteric modulator to control endogenous G protein-coupled receptors with light. Nat Chem Biol 10:813–815. [DOI] [PubMed] [Google Scholar]
- Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. (2011a) Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunahara RK, Kobilka BK. (2011b) Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477:549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AL, Grier MD, Jones CK, Herman EJ, Kane AS, Smith RL, Williams R, Zhou Y, Marlo JE, Days EL, Blatt TN, Jadhav S, Menon UN, Vinson PN, Rook JM, Stauffer SR, Niswender CM, Lindsley CW, Weaver CD, Conn PJ. (2010) Discovery of novel allosteric modulators of metabotropic glutamate receptor subtype 5 reveals chemical and functional diversity and in vivo activity in rat behavioral models of anxiolytic and antipsychotic activity. Mol Pharmacol 78:1105–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano C, Yang WL, O’Malley KL. (1996) Metabotropic glutamate receptor 5 is a disulfide-linked dimer. J Biol Chem 271:28612–28616. [DOI] [PubMed] [Google Scholar]
- Rook JM, Noetzel MJ, Pouliot WA, Bridges TM, Vinson PN, Cho HP, Zhou Y, Gogliotti RD, Manka JT, Gregory KJ, Stauffer SR, Dudek FE, Xiang Z, Niswender CM, Daniels JS, Jones CK, Lindsley CW, Conn PJ. (2013) Unique signaling profiles of positive allosteric modulators of metabotropic glutamate receptor subtype 5 determine differences in in vivo activity. Biol Psychiatry 73:501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook JM, Tantawy MN, Ansari MS, Felts AS, Stauffer SR, Emmitte KA, Kessler RM, Niswender CM, Daniels JS, Jones CK, Lindsley CW, Conn PJ. (2014) Relationship between in vivo receptor occupancy and efficacy of metabotropic glutamate receptor subtype 5 allosteric modulators with different in vitro binding profiles. Neuropsychopharmacology 40:755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook JM, et al. (2015) Biased mGlu5-positive allosteric modulators provide in vivo efficacy without potentiating mGlu5 modulation of NMDAR currents. Neuron 86:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See HB, Seeber RM, Kocan M, Eidne KA, Pfleger KD. (2011) Application of G protein-coupled receptor-heteromer identification technology to monitor beta-arrestin recruitment to G protein-coupled receptor heteromers. Assay Drug Dev Technol 9:21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffler DJ, Conn PJ. (2008) Allosteric potentiators of metabotropic glutamate receptor subtype 1a differentially modulate independent signaling pathways in baby hamster kidney cells. Neuropharmacology 55:419–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffler DJ, Gregory KJ, Rook JM, Conn PJ. (2011) Allosteric modulation of metabotropic glutamate receptors. Adv Pharmacol 62:37–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet BK, Kobilka BK. (2012) Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol Sci 33:268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyaert J, Kobilka BK. (2011) Nanobody stabilization of G protein-coupled receptor conformational states. Curr Opin Struct Biol 21:567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms AE, Dorschner MO, Wechsler J, Choi KY, Kirkwood R, Girirajan S, Baker C, Eichler EE, Korvatska O, Roche KW, Horwitz MS, Tsuang DW. (2013) Support for the N-methyl-D-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA Psychiatry 70:582–590. [DOI] [PubMed] [Google Scholar]
- Tizzano JP, Griffey KI, Schoepp DD. (1995) Receptor subtypes linked to metabotropic glutamate receptor agonist-mediated limbic seizures in mice. Ann NY Acad Sci 756:230–235. [DOI] [PubMed] [Google Scholar]
- Tye KM, Deisseroth K. (2012) Optogenetic investigation of neural circuits underlying brain disease in animal models. Nat Rev Neurosci 13:251–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urizar E, Yano H, Kolster R, Gales C, Lambert N, Javitch JA. (2011) CODA-RET reveals functional selectivity as a result of GPCR heterodimerization. Nat Chem Biol 7:624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwyler S. (2011) Allosteric modulation of family C G-protein-coupled receptors: from molecular insights to therapeutic perspectives. Pharmacol Rev 63:59–126. [DOI] [PubMed] [Google Scholar]
- van Wyk M, Pielecka-Fortuna J, Lowel S, Kleinlogel S. (2015) Restoring the ON switch in blind retinas: opto-mGluR6, a next-generation, cell-tailored optogenetic tool. PLOS Biol 13:e1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varney MA, Cosford NDP, Jachec C, Rao SP, Sacaan A, Lin F-F, Bleicher L, Santori EM, Flor PJ, Allgeier H, Gasparini F, Kuhn R, Hess SD, Velicelebi G, Johnson EC. (1999) SIB-1757 and SIB-1893: selective, noncompetitive antagonists of metabotropic glutamate receptor type 5. J Pharm Exp Ther 290:170–181. [PubMed] [Google Scholar]
- Vischer HF, Castro M, Pin JP. (2015) G protein-coupled receptor multimers: a question still open despite the use of novel approaches. Mol Pharmacol 88:561–571. [DOI] [PubMed] [Google Scholar]
- Walker AG, Wenthur CJ, Xiang Z, Rook JM, Emmitte KA, Niswender CM, Lindsley CW, Conn PJ. (2015) Metabotropic glutamate receptor 3 activation is required for long-term depression in medial prefrontal cortex and fear extinction. Proc Natl Acad Sci USA 112:1196–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenthur CJ, Morrison R, Felts AS, Smith KA, Engers JL, Byers FW, Daniels JS, Emmitte KA, Conn PJ, Lindsley CW. (2013) Discovery of (R)-(2-fluoro-4-((-4-methoxyphenyl)ethynyl)phenyl) (3-hydroxypiperidin-1-yl)methanone (ML337), an mGlu3 selective and CNS penetrant negative allosteric modulator (NAM). J Med Chem 56:5208–5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, Niswender CM, Katritch V, Meiler J, Cherezov V, Conn PJ, Stevens RC. (2014) Structure of a class C GRPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344:58–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Noetzel MJ, Johnson KA, Zamorano R, Jalan-Sakrikar N, Gregory KJ, Conn PJ, Niswender CM. (2014) Selective actions of novel allosteric modulators reveal functional heteromers of metabotropic glutamate receptors in the CNS. J Neurosci 34:79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rodriguez AL, Conn PJ. (2005) Allosteric potentiators of metabotropic glutamate receptor subtype 5 have differential effects on different signaling pathways in cortical astrocytes. J Pharm Exp Ther 315:1212–1219. [DOI] [PubMed] [Google Scholar]