Abstract
Dystonia, a common and genetically heterogeneous neurological disorder, was recently defined as “a movement disorder characterized by sustained or intermittent muscle contractions causing abnormal, often repetitive, movements, postures, or both.” Via the application of whole-exome sequencing, the genetic landscape of dystonia and closely related movement disorders is becoming exposed. In particular, several “novel” genetic causes have been causally associated with dystonia or dystonia-related disorders over the past 2 years. These genes include PRRT2 (DYT10), CIZ1 (DYT23), ANO3 (DYT24), GNAL (DYT25), and TUBB4A (DYT4). Despite these advances, major gaps remain in identifying the genetic origins for most cases of adult-onset isolated dystonia. Furthermore, model systems are needed to study the biology of PRRT2, CIZ1, ANO3, Gαolf, and TUBB4A in the context of dystonia. This review focuses on these recent additions to the family of dystonia genes, genotype-phenotype correlations, and possible cellular contributions of the encoded proteins to the development of dystonia.
Keywords: Dystonia, PRRT2, CIZ1, GNAL, ANO3, TUBB4A, Whole-exome sequencing
Introduction
Along with essential tremor, restless legs syndrome, and Parkinson disease, dystonia is a movement disorder commonly seen by neurologists in outpatient clinics. Genetic factors play an important role in the development of primary or isolated dystonia since more than 10 % of affected individuals report a positive family history [1•]. In 2013, dystonia was newly defined by an international consensus committee consisting of investigators with years of experience in this field as a “movement disorder characterized by sustained or intermittent muscle contractions causing abnormal, often repetitive, movements, postures, or both.” Dystonic movements are typically patterned with twisting and may be tremulous. Dystonia is often initiated or worsened by voluntary actions and associated with overflow muscle activation [2, 3]. The group classified dystonia along two axes: clinical characteristics, including age at onset, body distribution, temporal pattern, and associated features (additional movement disorders or neurological features); and causes, which include inherited (dystonia forms of proven genetic origin), acquired, and idiopathic (unknown causes) [2]. Future improvements in phenotypic (e.g., functional neuroimaging and postmortem pathology) and genetic resolution will serve to challenge the new definition and classification of dystonia. In fact, the new classification scheme was largely driven by recent developments in the genetics of dystonia and related neurological disorders [1•, 2, 4].
To date, more than 20 monogenic inherited dystonias and dystonia-related disorders (DYT1-DYT25) have been reported in Online Mendelian Inheritance in Man (OMIM), and many of the casual genes have been identified (Table 1). These include autosomal dominant forms due to mutations in TOR1A (DYT1), TUBB4A (DYT4), GCH1 (DYT5a/ DYT14), THAP1 (DYT6), PNKD (DYT8), SLC2A1 (DYT9/DYT18), PRRT2 (DYT10), SGCE (DYT11), ATP1A3 (DYT12), CIZ1 (DYT23), ANO3 (DYT24), and GNAL (DYT25); autosomal recessive forms due to mutations in TH and SPR (DYT5) and PRKRA (DYT16); and an X-linked recessive form due to mutations affecting expression of TAF1 (DYT3) (Table 1) [5, 6••, 7–12, 13••, 14••, 15••, 16–19, 20••]. Until the advent of whole-exome sequencing, the genetics of dystonia had progressed slowly after identification of dopa-responsive dystonia being caused by mutations in GCH1 in 1994 [7], and isolation of the classic TOR1A ΔGAG mutation as the first sequence variant (SV) associated with isolated dystonia in 1997 [5]. In less than 2 years, five additional genes associated with isolated dystonia and dystonia-related disorders have been reported. These include PRRT2, CIZ1, ANO3, GNAL, and TUBB4A [6••, 11, 13••, 14••, 15••, 20••, 21••]. In this article, we focus on these latest members of the dystonia family of genes. More extensive reviews of TOR1A, THAP1 (DYT6), and other dystonia-associated genes are available elsewhere [1•, 22•, 23].
Table 1.
Types of dystonia, modes of inheritance, and associated loci
| HUGO/OMIM | Common name | Locus/gene | Inheritance | Protein affected |
|---|---|---|---|---|
| DYT1 (OMIM 128100) | Oppenheim dystonia | 9q34.11/TOR1A | AD | Torsin A [5] |
| DYT2 (OMIM 224500) | Autosomal recessive dystonia | Unknown | AR | Unknown |
| DYT3 (OMIM 314250) | Lubag (X-linked dystonia parkinsonism) | Xql3.1/TAF1 | XLR | TATA box binding protein associated factor 1 [19, 68] |
| DYT4 (OMIM 128101) | Australian whispering dysphonia family | 19p13.3/TUBB4A | AD | Tubulin, beta 4A class IVa [6••, 21••] |
| DYT5 (OMIM 128230) | Dopa-responsive dystonia | 14q22.2/GCH1 | AD | GTP cyclohydrolase I [7] |
| (OMIM 605407) | Dopa-responsive dystonia | 11p.15.5/TH | AR | Tyrosine hydroxylase [16] |
| (OMIM 612716) | Dopa-responsive dystonia | 2q13.2/SPR | AR | Sepiapterin reductase [17] |
| DYT6 (OMIM 602629) | Mixed-type dystonia | 8p11.21/THAP1 | AD | THAP domain containing, apoptosis associated protein 1 [8] |
| DYT7 (OMIM 602124) | Familial torticollis | 18p | AD | Unknown |
| DYT8 (OMIM 118800) | Paroxysmal nonkinesigenic dyskinesia | 2q35/PNKD | AD | Paroxysmal nonkinesigenic protein [9] |
| DYT9 (OMIM 601042) | Paroxysmal choreoathetosis/spasticity | 1p34.2/SLC2A1 | AD | Glucose transporter 1 [69] |
| DYT10 (OMIM 128200) | Paroxysmal kinesigenic dyskinesia | 16p11.2/PRRT2 | AD | Proline-rich transmembrane protein 2 [11] |
| DYT11 (OMIM 159900) | Myoclonus-dystonia syndrome | 7q21.3/SGCE | AD | ε-Sarcoglycan [12] |
| DYT12 (OMIM 128235) | Rapid-onset dystonia parkinsonism | 19ql3.2/ATP1A3 | AD | Na+/K+-ATPase α3 subunit [70] |
| DYT13 (OMIM 607671) | Italian family primary torsion dystonia | 1p36.32-p36.13 | AD | Unknown |
| DYT15 (OMIM 607488) | Myoclonus dystonia, Canadian family | 18p11 | AD | Unknown |
| DYT16 (OMIM 612067) | Young-onset dystonia parkinsonism | 2q31.2/PRKRA | AR | Protein kinase, interferon-inducible double-stranded RNA dependent activator [18] |
| DYT17 (OMIM 612406) | Lebanese family with generalized dystonia, dysarthria, and dysphonia |
20p11.2-ql3.12 | AR | Unknown |
| DYT18 (OMIM 61216) | Paroxysmal exertional dyskinesia associated with hemolytic anemia |
1p34.2/SLC2A1 | AR | Glucose transporter 1 [10] |
| DYT19 (OMIM 611031) | Paroxysmal kinesigenic dyskinesia | 16ql3-q22.1 | AD | Unknown |
| DYT20 (OMIM 611147) | Paroxysmal nonkinesigenic dyskinesia 2 | 2q31 | AD | Unknown |
| DYT21 | Swedish family with adult-onset mixed dystonia | 2ql4.3-q21.3 | AD | Unknown |
| DYT23 (OMIM 614860) | Adult-onset cervical dystonia | 9q34.1l/CTZ1 | AD | Cyclin-dependent kinase inhibitor 1A interacting zinc finger protein 1 [13••] |
| DYT24 (OMIM 615034) | Adult-onset cervical and segmental dystonia | 11p14.2/ANO3 | AD | Anoctamin 3 [14••] |
| DYT25 (OMIM 615073) | Adult-onset cervical and segmental dystonia | 18p11.21/GNAL | AD | Guanine nucleotide binding protein, alpha activating activity polypeptide, olfactory type [15••, 20••] |
AD autosomal dominant, AR autosomal recessive
TUBB4A/DYT4
DYT4 (OMIM 128101) was assigned to the unique Australian-English “whispering dysphonia” kindred, which was first described by Parker [24] in 1985. This pedigree exhibits an autosomal dominant inheritance pattern of a neurological syndrome that includes dysphonia, a gait disorder, increased prevalence of psychiatric disorders, dysmorphic facies, and variable cognitive impairment. The dystonia may become generalized [6••]. Some patients exhibit a striking “hobbyhorse” ataxic gait which is unlike anything seen in primary or isolated dystonia. Despite the presence of Wilson disease in several family members, the two ATP7B mutations do not segregate with dystonia [25]. Despite the highly penetrant dysphonic phenotype in the large multigenerational pedigree, successful linkage analysis was not reported until 2012, when two independent groups simultaneously identified a pathogenic variant in TUBB4A using different samples from the same kindred [6••, 21••]. Hersheson et al. [6••] performed genome-wide linkage analysis in 19 family members with SNP chips and mapped DYT4 to chromosome locus 19p13.2-3. Then, with whole-exome sequencing in two definitely affected patients, a single missense mutation (c.4C>G, p.R2G) was localized to exon 1 of TUBB4A, which encodes tubulin, beta 4A class IVa (TUBB4A) [6••]. These findings were confirmed by an independent team using a similar strategy in 14 family members from the same kindred [21••]. Lohmann et al. [21••] also reported a missense variant (c.2723 G>A, p.R908H) in DOT1L (DOT1-like histone H3K79 methyltransferase) which co-segregated with dystonia but was also present in four of nine unaffected family members. Sequencing of TUBB4A in 394 unrelated subjects with dystonia revealed a missense variant (c.811G>A, p. A271T) in a subject with late-onset segmental dystonia that included spasmodic dysphonia [21••]. However, cosegregation in one other putatively affected family member was not established for the A271T variant.
TUBB4A encodes TUBB4A, a member of the β-tubulin family. TUBB4A is spliced into multiple transcripts. The major transcript encodes a 495-amino acid protein that is expressed in brain. Tubulins are globular proteins and include six family members: α-tubulin, β-tubulin, γ-tubulin, δ-tubulin, ε-tubulin, and ζ-tubulin. The most prevalent members are α-tubulin and β-tubulin, which associate with each other as heterodimers to form microtubules, an essential component of the cytoskeleton. Microtubules are critical for a variety of cellular processes, such as mitosis, intracellular transport, neuron morphology and ciliary and flagellar motility [26]. In humans, six β-tubulins with high sequence similarity (93–98 % to TUBB4A) have been characterized and associated with a variety of neurodevelopmental disorders, including microcephaly with structural brain abnormalities (TUBB), lissencephaly (TUBA1A), polymicrogyria (TUBA1A, TUBB2B, TUBB3), and ocular motility disorders (TUBB3) [27].
Consistent with a vital role for TUBB4A in the structure of the central nervous system, a recurrent de novo missense mutation (p.Asp249Asn) was identified in patients with hypomyelination with atrophy of the basal ganglia and cerebellum (H-ABC) [28]. This mutation affects a highly conserved asparagine that interfaces with α-tubulin and could disrupt dimerization, microtubule polymerization, or microtubule stability [28]. H-ABC is a rare leukodystrophy characterized by onset in infancy or childhood, developmental delay, movement disorders, and spasticity. H-ABC movement disorders may include dystonia, ataxia, and choreoathetosis. Magnetic resonance imaging shows evidence of hypomyelination, cerebellar atrophy, and volume loss in the putamen. A similar phenotype with slowly progressive spastic paraparesis, segmental dystonia, intellectual disability, behavioral problems, incomplete myelination, and progressive cerebellar atrophy has been linked to a p.E410K variant in TUBB4A [29].
The similar and overlapping phenotypes seen with R2G (DYT4), D249N (H-ABC), and E410K missense mutations in TUBB4A suggest that DYT4 is a forme fruste of H-ABC [30•]. In comparison with the DYT4 pedigree, the more severe and complex clinical phenotypes seen with the D249N and E410K mutations limit reproductive capacity [31]. Given the fundamental role of TUBB4A in neuronal biology and its highly conserved amino acid sequences, it is not surprising that only eight missense variants appear in the Exome Variant Server database and that only three of these eight are predicted to be deleterious (Table 2).
Table 2.
Newly reported dystonia genes and associated variants along with in silico predictions of pathogenicity
| Gene | Variants | SIFT | PolyPhen-2 | MutationTaster | Publication | Missensea variants (pathogenic)b |
|
|---|---|---|---|---|---|---|---|
| Coding | Protein | ||||||
| GNAL | c.409G>A | V137M | Tolerated | Probably damaging | Polymorphism | Fuchs et al. [15••] | 26 (16) |
| c.878C>A | S293* | NP | NP | Disease causing | |||
| c.463G>A | E155K | Damaging | Probably damaging | Disease causing | |||
| c.283-284insT | S95Ffs*16 | NP | NP | Disease causing | |||
| c.591-592insAc | R198Tfs*13 | NP | NP | Disease causing | |||
| c.61C>T | R21* | NP | NP | Disease causing | |||
| c.304 312delCCTCCAGTT | P1 02 104del3 | NP | NP | Polymorphism | |||
| c.682G>T | V228F | Damaging | Probably damaging | Disease causing | Vemula et al. [20••] | ||
| c.591dupAc | R198Tfs*13 | NP | NP | Disease causing | |||
| c.733C>T | R245* | NP | NP | Disease causing | |||
| c.3G>A | M1? | NP | NP | Disease causing | |||
| c.284C>T | S95* | NP | NP | Disease causing | Miao et al. [55] | ||
| c.436G>A | V146M | Damaging | Probably damaging | Disease causing | Zech et al. [47] | ||
| c.733C>T | R245* | NP | NP | Disease causing | Dufke et al. [57] | ||
| c.514G>A | V172I | Tolerated | Benign | Disease causing | Saunders-Pullman et al. [59] |
||
| c.637G>A | G213S | Damaging | Probably damaging | Disease causing | Kumar et al. [58] | ||
| c.1057G>A | A353T | Damaging | Probably damaging | Disease causing | |||
| c.214C>T | P72S | Tolerated | Benign | Polymorphism | Charlesworth et al. [14••] | ||
| CIZ1 | c.790A>G | S264G | Tolerated | Probably damaging | Disease causing | Xiao et al. [13••] | 60 (45) |
| c.2015G>T | R672M | Damaging | Probably damaging | Disease causing | |||
| c.139C>T | P47S | Damaging | Probably damaging | Polymorphism | |||
| TUBB4A | c.4C>G | R2G | Damaging | Polymorphism | Disease causing | Lohmann et al. [21••] | 8 (3) |
| c.811G>A | A271T | Damaging | Benign | Disease causing | |||
| c.745G>A | D249N | Damaging | Damaging | Disease causing | Simons et al. [28] | ||
| c.1228G>Ad | E410K | Damaging | Damaging | Disease causing | Blumkin et al. [29] | ||
| ANO3 | c.161C>T | T54I | Tolerated | Benign | Polymorphism | Charlesworth et al. [56] | 68 (31) |
| c.1470G>C | W490C | Damaging | Damaging | Disease causing | |||
| c.1480A>T | R494W | Damaging | Damaging | Disease causing | |||
| c.2053A>G | S685G | Tolerated | Benign | Disease causing | |||
| c.2586G>T | K862N | Tolerated | Benign | Disease causing | |||
| c.2497A>G | I833V | Tolerated | Benign | Disease causing | Zech et al. [47] | ||
| c.2917G>C | G973R | Tolerated | Probably damaging | Disease causing | |||
NP not predictable
Exome Variant Server, NHLBI Exome Sequencing Project (http://evs.gs.washington.edu/EVS/)
Exome Variant Server uses PolyPhen-2 to predict pathogenicity
Same variant
Corrected variant location (the variant in the original article was misspelled as c.1218G>A)
To determine the contribution of TUBB4A to the pathogenesis of primary or isolated dystonia, in general, high-resolution melting and Sanger sequencing were used to interrogate TUBB4A in a cohort of 575 subjects with laryngeal, segmental, or generalized dystonia [30•]. No pathogenic variants in TUBB4A were identified in any of the subjects. Therefore, given the allelic association of DYT4 with H-ABC, we concluded that clinical testing for TUBB4A mutations is not justified in spasmodic dysphonia or other forms of primary dystonia [30•].
PRRT2/DYT10
DYT10 (OMIM 128200) was assigned to paroxysmal dystonia with kinesigenic choreoathetosis in 1998 [32], and later to paroxysmal kinesigenic dyskinesia (PKD) or episodic kinesigenic dyskinesia. The paroxysmal dyskinesias are an important category of movement disorders characterized by the sudden onset of involuntary movements that may include one or more of the following: dystonia, chorea, athetosis, and ballism. The paroxysmal dyskinesias are divided into four major types: PKD, paroxysmal nonkinesigenic dyskinesias (PNKD), paroxysmal exertion-induced dyskinesias (PED), and paroxysmal hypnogenic dyskinesias. Precipitating factors are included within the name of each type: kinesigenic (sudden movements), nonkinesigenic (triggered by factors such as alcohol, caffeine, fatigue, and anxiety), exertion- or exercise-induced (running or prolonged walking), and hypnogenic (non-rapid-eye-movement sleep). The paroxysmal dyskinesias may be familial (hereditary), sporadic, or secondary to other neurological or metabolic disorders (e.g., head trauma, multiple sclerosis, hypoparathyroidism). Typically, dystonia is the foremost involuntary movement manifested by patients with PKD, PNKD, PED, and paroxysmal hypnogenic dyskinesia. Compared with primary dystonia, PKD is relatively rare. In general, familial cases of PKD follow an autosomal dominant inheritance mode with incomplete penetrance. In patients with PKD, recurrent and brief attacks of involuntary movement are usually triggered by sudden voluntary movement. These attacks usually begin in childhood or early adulthood and may include various combinations of dystonia, chorea, and athetosis affecting the face, trunk, arms, and legs. In many patients, paroxysmal dystonia is the sole or predominant clinical manifestation [1•]. PKD often improves with age, and the vast majority of patients show a favorable response to relatively low doses of anticonvulsant medications, particularly carbamazepine and phenytoin [1•, 33].
Most patients with PKD harbor loss-of-function mutations in PRRT2, which encodes proline-rich transmembrane protein 2 (PRRT2). A PRRT2 hotspot mutation (c.649dupC, p.R217Pfs*8) likely causes haploinsufficiency via nonsense-mediated decay of mutant transcripts [33]. PKD, infantile convulsions and choreoathetosis (ICCA), and benign familial infantile epilepsy (BFIE) may appear in the same family. Moreover, PKD, ICCA, and BFIE had been linked to chromosome locus 16p11.2 for over 10 years [34]. With the advent of whole-exome sequencing, this was confirmed in 2011 after the causal gene, PRRT2, was first identified in both familial and sporadic PKD, ICCA, and BFIE in Han Chinese, Japanese, Caucasians, and African-Americans [11, 33, 35]. PRRT2 mutations also cause rare cases of PED, PNKD-like syndromes, hemiplegic migraine, episodic ataxia, febrile seizures, and childhood-absence epilepsy. Over the past 3 years, more than 60 PRRT2 mutations have been found in PKD, ICCA, and BFIE. Some of the mutations, including the hot spot mutation (c.649dupC), were shared among PKD, ICCA, and BFIE phenotypes [35]. PRRT2 is a four-exon gene that encodes PRRT2, a 340 amino acid protein with two putative transmembrane domains near its C-terminal end. PRRT2 is highly expressed in the developing nervous system, particularly the cerebellum [11]. PRRT2 may interact with SNAP-25, a component of the molecular machinery involved in the release of neurotransmitters at the presynaptic membrane [36]. Most PRRT2 mutations are frameshift or nonsense mutations which result in truncated proteins and lead to haploinsufficiency and, possibly, dysregulation of neurotransmitter release. An intensive review by Heron and Dibbens [35] indicated that mutations in PRRT2 account for 62–100 % of familial cases of PKD, 40–100 % of familial cases of BFIE, and 33–100 % of familial cases of ICCA; although the percentages are lower in the sporadic cases, with mutations found in 27–50 % of sporadic PKD patients and 29–100 % of patients with sporadic benign infantile seizures. Genetic testing for mutations in PRRT2 is straightforward and should be pursued in patients with typical PKD, ICCA, and BFIE phenotypes, particularly when supported by a positive family history.
CIZ1/DYT23
Cervical dystonia, also known as spasmodic torticollis, is the commonest type of focal dystonia, affecting over one million people worldwide, with most cases being of the late-onset type (more than 20 years of age). Genetic factors play a major role in late-onset cervical dystonia given that 10 % of probands have one or more affected family members [13••]. However, the rarity of large kindreds suggests that causal genetic variants are incompletely penetrant. In 2012, using linkage and haplotype analyses in combination with whole-exome sequencing, Xiao et al. [13••] mapped the causal SV in an American autosomal dominant cervical dystonia pedigree [37] to microsatellite marker D9S159 on chromosome locus 9q34.11. Whole-exome sequencing exposed an SV (c.790A>G, p.S264G) in exon 7 of CIZ1 which encodes cyclin-dependent kinase inhibitor 1A interacting zinc finger protein 1 (CIZ1). This SV is predicted, “in silico,” to be pathogenic. Moreover, in vitro studies showed that this variant altered splicing of CIZ1 and nuclear localization of CIZ1.
The potential causality of CIZ1 SVs as a cause of cervical dystonia has come under scrutiny. First of all, an SV in SETX (c.2385_2388delAAAG) also co-segregated with dystonia in the index family. It is possible that combined mutations in CIZ1 and SETX were required for the development of dystonia in the index pedigree. Second, no other pedigrees with CIZ1 mutations have been reported in the available literature. In our original report, only two of 308 additional cervical dystonia probands harbored a variant in CIZ1 (Table 2). Moreover, these patients had sporadic dystonia, precluding cosegregation analysis. In addition, no CIZ1 mutations were found in 12 Chinese pedigrees with autosomal dominant cervical dystonia [38]. Third, there are dozens of CIZ1 missense variants in normal populations, and some of these are predicted to be pathogenic by one or more in silico programs.
Given concerns regarding the possibility of other pathogenic SVs in the American pedigree with linkage to chromosome locus 9q34.11 [13••, 39], whole-exome sequencing was repeated in one definitely affected subject. In brief, 98.44 % of exons were covered at a coverage of 20-fold or greater. On chromosome 9, 98.39 % of exons (3,371/3,426) were covered at a coverage of tenfold or greater, and 99 % or more were covered at a coverage of twofold or greater. Most importantly, no newly identified potentially pathogenic SVs were identified in repeated analyses, strongly supporting the causality of CIZ1 c.790A>G (p.S264G). In particular, no pathogenic variants were identified in TOR1A [39]. Using high-resolution melting of CIZ1 exon 2 (exon 1 is noncoding) in over 1,200 Caucasian subjects with dystonia followed by confirmatory Sanger sequencing, we have identified several novel and potentially pathogenic SVs within the QD1 domain of CIZ1 in subjects with dystonia (Fig. 1). None of these SVs appear in existing variant databases (Exome Variant Server, dbSNP, 1000 Genomes).
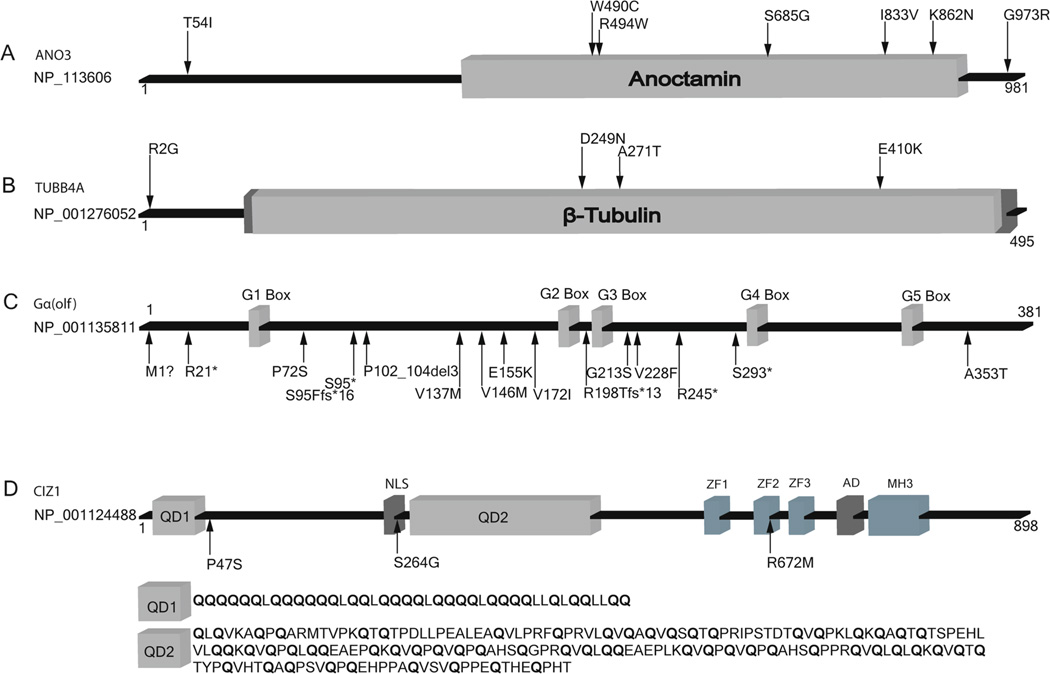
Fig. 1.
Dystonia-associated proteins and localization of disease-associated pathogenic sequence variants. G1-G5 are GTP binding domains, and ZF1-ZF3 are zinc-finger domains. AD acidic domain, ANO3 anoctamin 3, CIZ1 cyclin-dependent kinase inhibitor 1A interacting zinc finger protein 1, Gα(olf) guanine nucleotide binding protein, alpha activating activity polypeptide, olfactory type, MH3 matrin 3 homologous domain 3, NLS nuclear localization signal, QD1 glutamine-rich domain 1, QD2 glutamine rich domain 2, TUBB4A tubulin, beta 4A class IVa
CIZ1 is an 898 amino acid DNA replication factor with neural expression patterns similar to those of THAP1 (DYT6) and TOR1A (DYT1). Robust expression of CIZ1, THAP1, and TOR1A is noted in fetal brain and adult cerebellum, particularly in Purkinje cells [13••, 40, 41]. CIZ1 was first recognized through its interaction with p21Cip1/Waf1, a cyclin-dependent kinase inhibitor involved in G1/S cell-cycle regulation and cellular differentiation. The cellular role and neural localization of CIZ1 are compatible with current themes in dystonia research as four other dystonia-associated proteins [THAP1 (DYT6), TAF1 (DYT3), ataxia telangiectasia mutated, and guanine nucleotide binding protein, alpha activating activity polypeptide, olfactory type (Gαolf)] are also involved in G1/S cell-cycle regulation [42]. The role of wild-type and mutant CIZ1 in terminally differentiated neurons requires further elucidation.
ANO3/DYT24
A British family with autosomal dominant adult-onset craniocervical dystonia was first reported in 2000 [43]. All five definitely affected family members exhibit tremulous cervical dystonia with a variable degree of associated upper-limb dystonic tremor, and most show spread to segmental dystonia over time. In addition, some subjects manifest spasmodic dysphonia and/or blepharospasm, but none exhibit generalized dystonia [14••, 44]. SNP-based linkage analysis on 15 samples identified five loci with identical logarithm of odds scores of 2.01 on chromosomes 4, 5, 6, 7, and 11. Whole-exome sequencing was performed in two affected individuals and identified several candidate causal variants. The two exomes received 83 % and 90 % coverage with read depths of 2 or more. A heterozygous missense mutation (c.1480A>T, p.Arg494Trp) in exon 15 of ANO3 on chromosome 11 co-segregated with dystonia in the index pedigree (family A) [14••]. Focused screening of exon 15 in 384 subjects with mainly cervical dystonia with or without upper-limb dystonic tremor exposed a second co-segregating SV (c.1470G>C, p.Trp490Cys) in family B. Targeted sequencing was used to identify additional variants in exons 2, 21, and 25 and the 5′-untranslated region of ANO3. The SVs in exons 2,21, and 25 are predicted as tolerated and benign by SIFT and PolyPhen-2, respectively.
ANO3, a 27-exon gene, encodes anoctamin 3 (ANO3), a 981 amino acid transmembrane protein that belongs to a family of Ca2+ -activated chloride channels with highest expression in the striatum. Some members of the ANO family (ANO1-ANO10) have been linked to disease (ANO5, ANO6, and ANO10), mainly via autosomal recessive mechanisms. For example, homozygous mutations in ANO10 are found in a form of autosomal recessive spinocerebellar ataxia [45]. Functional studies using mutation-bearing fibroblasts from family A (p.Arg494Trp) showed defective endoplasmic-reticulum Ca2+ handling [14••]. Ano3 knockout rats show diminished Slack expression, broadened action potentials, increased excitability and enhanced KNa channel activity in dorsal root ganglia neurons [46]. These findings suggest that ANO3 plays a role in regulating the processing of pain in the peripheral nervous system. Although the Ano3 knockout rats showed increased thermal and mechanical sensitivity, they did not manifest dystonia [46].
Results from knockout rats question the role of ANO3 in dystonia. In the EVS database, more than 30 deleterious rare variants in ANO3 are reported (0.48 % of available samples). Moreover, only two novel missense SVs (0.6 %) were found in a screening study of 342 German dystonia patients, and neither variant was predicted to be deleterious via in silico analyses. In addition, two novel missense variants were also found in 376 normal controls (0.5 %) [47]. Finally, rare variants in ANO3 are not associated with essential tremor [48]. Taken together, these data suggest that the role of ANO3 in dystonia and tremor is questionable and demands additional study.
GNAL/DYT25
Although without strong linkage data from whole-genome genotyping owing to relatively small pedigrees, two independent teams used whole-exome sequencing to identify 11 heterozygous GNAL mutations in 12 dystonia families with mainly adult-onset cervical or craniocervical dystonia [15••, 20••]. GNAL is located on chromosome locus 18p11.22-p11.21, within the 18p deletion syndrome (OMIM 146390) region. The 18p deletion syndrome is characterized by mental retardation, growth retardation, craniofacial dysmorphism, and in many patients, dystonia [49, 50]. This region also harbors loci for DYT7 (OMIM 602124, autosomal dominant adult-onset cervical dystonia) and DYT15 (OMIM 607488, autosomal dominant alcohol-responsive myoclonic dystonia) [51, 52]. At present, the potential relationships among DYT7, DYT15, and DYT25 are unclear. Moreover, the DYT7 gene locus for cervical dystonia on chromosome locus 18p is now questionable [53].
To date, more than 15 different GNAL mutations have been found to be associated with dystonia in a wide range of racial/ ethnic groups, including American Caucasians, Amish-Mennonites, European Caucasians, African-Americans, Chinese, and Japanese. These mutations include seven missense mutations, four nonsense mutations, two frameshift mutations, one small deletion, and one mutation (c.3G>A) which alters the start codon (Table 2). Data from a total of 43 patients from 17 families (16 males and 27 females) show that the average age of onset is 35±13 (7–63) years. Over 83 % of the subjects (36 of 43) had onset in the neck as cervical dystonia, and in 42 % (18 of 43) the dystonia has remained focally distributed as cervical dystonia, with the remaining subjects showing spread to the cranial musculature or arms as segmental dystonia. Only 12 % of the subjects (five of 43) developed generalized dystonia. Ultimately, the neck is affected in over 95 % of subjects (41 of 43) [15••, 20••, 47, 54–59]. Although GNAL genotype–phenotype correlations are limited by sample size, currently available data appear to distinguish GNAL dystonia from dystonia associated with mutations in TOR1A and THAP1. DYT1 caused by mutations of TOR1A is usually characterized by early onset of dystonia in one limb with progression to a segmental or generalized distribution. DYT1 dystonia is rarely isolated to the cervical region [1•]. The mean age of onset for DYT6 dystonia caused by mutations of THAP1 is 16.8 years. The arms and neck are the commonest sites of onset for THAP1-associated dystonia, with the neck and arms being ultimately affected in 77 % and 71 % of patients, respectively [22•]. In addition, the larynx is affected in 44 % of patients with pathogenic SVs in THAP1. Therefore, in comparison with TOR1A and THAP1, mutations in GNAL are more commonly associated with adult-onset cervical dystonia and are less likely to generalize. Similarly to other genes associated with isolated or primary dystonia, penetrance with GNAL mutations is incomplete.
The initial report by Fuchs et al. [15••] suggested that almost 15 % of families (six of 39) with adult-onset cervical dystonia carried a GNAL mutation, but follow-up studies showed that the frequency is less than 0.5 % in mainly adult-onset dystonia patients [20••, 47, 55, 56]. The frequency of GNAL mutations in primary or isolated dystonia is in line with that in other genetic forms of dystonia such as DYT1 (0.2 %) and DYT6 (0.5 %) [22•, 23, 60].
GNAL encodes Gαolf. The major variant (NP_001135811) contains 381 amino acids and is highly expressed in the olfactory bulb, striatum, and cerebellar Purkinje cells, and colocalizes with corticotropin-releasing hormone receptors in the latter [20••]. Gαolf plays a role in olfaction, coupling dopamine D1 and adenosine A2A receptors to adenylyl cyclase, and histone H3 phosphorylation. Gnal knockout mice and some patients with mutations in GNAL exhibit hyposmia [20••, 61].
Microarray data of lymphoblastoid cell lines from subjects with a p.V228F mutation in GNAL showed upregulation of genes involved in cell-cycle control and development, consistent with cellular pathways that have been implicated in dystonia [20••, 41, 62]. In vivo administration of the dopamine D2 receptor blocker haloperidol to rodents induces a prolonged increase in the levels of Ser10 phosphorylated histone H3 in dopamine D2 receptor expressing neurons of the dorsomedial and the dorsolateral striatum, and this effect is mediated through adenosine A2A receptor mediated activation of Gαolf [63]. Histone H3 phosphorylation at Ser10 inhibits checkpoint kinase 1, a key component of the ataxia telangiectasia mutated/ataxia telangiectasia and Rad3 related arm of the G1/S checkpoint pathway [64]. Therefore, it is conceivable that loss of Gαolf function could manifest itself as dystonia via prolonged disruption of G1/S cell-cycle control in striatal medium spiny neurons or cerebellar Purkinje cells.
Conclusions
GNAL is a bona fide primary or isolated dystonia gene, DYT4 is not a primary or isolated dystonia gene but, instead, may be a forme fruste of H-ABC, and the suggested pathogenic roles of CIZ1 and ANO3 in dystonia will require confirmation by independent groups. At this time, clinical genetic testing for GNAL mutations is justified in subjects with familial adult-onset dystonia mainly manifested as cervical dystonia. However, clinicians must recognize that in silico analyses are imperfect and the pathogenicity of missense variants must be interpreted with caution [39]. For example, Fuchs et al. [15••] reported a normal control with a p.V16F variant in GNAL, whereas Charlesworth et al. [14••] reported a p. P149S variant in ANO3 which did not segregate with dystonia. Dozens of putatively pathogenic missense variants in CIZ1, ANO3, and GNAL are present in public domain databases (Table 2). On the other hand, the mutation spectra of DYT4, DYT23, DYT24, and DYT25 may be much broader than currently appreciated given that most screening studies largely ignored the promoter, untranslated, and intronic regions of the disease-associated genes.
Genetics opens doors for deeper understanding of dystonia and related movement and neurodegenerative disorders. Importantly, all five dystonia-associated genes discussed in this review were identified by whole-exome sequencing, which largely bypasses the limitations of traditional positional cloning. The next wave of genetics is upon us with relatively inexpensive whole-genome sequencing along with longer and more accurate reads that should expose increasingly larger portions of the dystonia genetic landscape.
With rapid advances come challenges. A recent report of an Amish pedigree described a proband with typical DYT6 manifestations and an associated THAP1 mutation, whereas her distant cousin harbored a de novo mutation in TOR1A and her aunt with blepharospasm was negative for TOR1A, THAP1, and GNAL mutations [59]. The reduced penetrance and variable expressivity of dystonia along with important phenotypic overlap among the different forms of dystonia should guide clinical judgment and genetic counseling.
The recently identified dystonia genes broaden our understanding of pathological cellular and systems networks. CIZ1 and Gαolf are tied to the G1/S cell-cycle checkpoint [62]. PRRT2, ANO3, and TUBB4A can be linked to other cellular mechanisms believed to play a role in dystonia and related disorders, such as control of synaptic function, the endoplasmic reticulum, and nuclear envelope function. Gαolf, CIZ1, PRRT2, and TUBB4A show high expression levels in brain structures, especially striatum and cerebellum, which have been implicated in the pathogenesis of dystonia [65–67].
Acknowledgments
Our dystonia research has been supported by grants from the National Institutes of Health (K08NS001593, R01EY012232, R01NS048458, R01NS050185, R01NS069936, and U54NS065701), the Dystonia Medical Research Foundation, the Bachmann-Strauss Dystonia & Parkinson Foundation, Tyler’s Hope for a Dystonia Cure, and the University of Tennessee Health Science Center Neuroscience Institute.
Mark S. LeDoux has received consultancy fees from Teva and Lundbeck. He has also received grants from the National Institutes of Health, the Cure Huntington Disease Initiative, and Prana. He has also received honorarium payments from Teva, Lundbeck, and UCB Pharma, as well as royalty payments from Elsevier.
Footnotes
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
Conflict of Interest Jianfeng Xiao and Satya R. Vemula declare that they have no conflict of interest.
Contributor Information
Jianfeng Xiao, Email: jxiao@uthsc.edu, Department of Neurology, University of Tennessee Health Science Center, 855 Monroe Avenue, Link Building Suite 415, Memphis, TN 38163, USA; Department of Anatomy and Neurobiology, University of Tennessee Health Science Center, Memphis, TN 38163, USA.
Satya R. Vemula, Email: svemula@uthsc.edu, Department of Neurology, University of Tennessee Health Science Center, 855 Monroe Avenue, Link Building Suite 415, Memphis, TN 38163, USA; Department of Anatomy and Neurobiology, University of Tennessee Health Science Center, Memphis, TN 38163, USA.
Mark S. LeDoux, Email: mledoux@uthsc.edu, Department of Neurology, University of Tennessee Health Science Center, 855 Monroe Avenue, Link Building Suite 415, Memphis, TN 38163, USA; Department of Anatomy and Neurobiology, University of Tennessee Health Science Center, Memphis, TN 38163, USA.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1. LeDoux MS. The genetics of dystonias. Adv Genet. 2012;79:35–85. doi: 10.1016/B978-0-12-394395-8.00002-5. This is an encyclopedic review of dystonia genetics.
- 2.Albanese A, Bhatia K, Bressman SB, et al. Phenomenology and classification of dystonia: a consensus update. Mov Disord. 2013;28:863–873. doi: 10.1002/mds.25475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeDoux MS. Dystonia: phenomenology. Parkinsonism Relat Disord. 2012;18(Suppl 1):S162–S164. doi: 10.1016/S1353-8020(11)70050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fahn S. Classification of movement disorders. Mov Disord. 2011;26:94–57. doi: 10.1002/mds.23759. [DOI] [PubMed] [Google Scholar]
- 5.Ozelius LJ, Hewett JW, Page CE, et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- 6. Hersheson J, Mencacci NE, Davis M, et al. Mutations in the autoregulatory domain of beta-tubulin 4a cause hereditary dystonia. Ann Neurol. 2013;73:546–553. doi: 10.1002/ana.23832. The authors used linkage analysis and whole-exome sequencing to identify a TUBB4A mutation in the “whispering dysphonia” pedigree.
- 7.Ichinose H, Ohye T, Takahashi E, et al. Hereditary progressive dystonia with marked diurnal fluctuation caused by mutations in the GTP cyclohydrolase I gene. Nat Genet. 1994;8:236–242. doi: 10.1038/ng1194-236. [DOI] [PubMed] [Google Scholar]
- 8.Fuchs T, Gavarini S, Saunders-Pullman R, et al. Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nat Genet. 2009;41:286–288. doi: 10.1038/ng.304. [DOI] [PubMed] [Google Scholar]
- 9.Rainier S, Thomas D, Tokarz D, et al. Myofibrillogenesis regulator 1 gene mutations cause paroxysmal dystonic choreoathetosis. Arch Neurol. 2004;61:1025–1029. doi: 10.1001/archneur.61.7.1025. [DOI] [PubMed] [Google Scholar]
- 10.Weber YG, Storch A, Wuttke TV, et al. GLUT1 mutations are a cause of paroxysmal exertion-induced dyskinesias and induce hemolytic anemia by a cation leak. J Clin Invest. 2008;118:2157–2168. doi: 10.1172/JCI34438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen WJ, Lin Y, Xiong ZQ, et al. Exome sequencing identifies truncating mutations in PRRT2 that cause paroxysmal kinesigenic dyskinesia. Nat Genet. 2011;43:1252–1255. doi: 10.1038/ng.1008. [DOI] [PubMed] [Google Scholar]
- 12.Zimprich A, Grabowski M, Asmus F, et al. Mutations in the gene encoding epsilon-sarcoglycan cause myoclonus-dystonia syndrome. Nat Genet. 2001;29:66–69. doi: 10.1038/ng709. [DOI] [PubMed] [Google Scholar]
- 13. Xiao J, Uitti RJ, Zhao Y, et al. Mutations in CIZ1 cause adult onset primary cervical dystonia. Ann Neurol. 2012;71:458–469. doi: 10.1002/ana.23547. With use of linkage analysis and whole-exome sequencing, a pathogenic variant in CIZ1 was identified as the first cause of familial cervical dystonia. This work pointed out the potential role of the G1/S checkpoint pathway in the pathogenesis of primary dystonia.
- 14. Charlesworth G, Plagnol V, Holmstrom KM, et al. Mutations in ANO3 cause dominant craniocervical dystonia: ion channel implicated in pathogenesis. Am J Hum Genet. 2012;91:1041–1050. doi: 10.1016/j.ajhg.2012.10.024. Mutations in ANO3 were identified as a potential cause of adult-onset dystonia, mainly cervical and segmental dystonia with dystonic tremors. Next-generation targeted sequencing was used to screen patients with familial and sporadic dystonia for SVs in ANO3.
- 15. Fuchs T, Saunders-Pullman R, Masuho I, et al. Mutations in GNAL cause primary torsion dystonia. Nat Genet. 2013;45:88–92. doi: 10.1038/ng.2496. Fuchs et al. used whole-exome sequencing to identify GNAL mutations in relatively small pedigrees with mainly adult-onset cervical and segmental dystonia. Screening of primary dystonia pedigrees exposed additional putatively pathogenic variants. Impaired function of several of the mutations was shown by bioluminescence resonance energy transfer assays.
- 16.Ludecke B, Dworniczak B, Bartholome K. A point mutation in the tyrosine hydroxylase gene associated with Segawa’s syndrome. Hum Genet. 1995;95:123–125. doi: 10.1007/BF00225091. [DOI] [PubMed] [Google Scholar]
- 17.Bonafe L, Thony B, Penzien JM, et al. Mutations in the sepiapterin reductase gene cause a novel tetrahydrobiopterin-dependent monoamine-neurotransmitter deficiency without hyperphenylalaninemia. Am J Hum Genet. 2001;69:269–277. doi: 10.1086/321970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camargos S, Scholz S, Simon-Sanchez J, et al. DYT16, a novel young-onset dystonia-parkinsonism disorder: identification of a segregating mutation in the stress-response protein PRKRA. Lancet Neurol. 2008;7:207–215. doi: 10.1016/S1474-4422(08)70022-X. [DOI] [PubMed] [Google Scholar]
- 19.Nolte D, Niemann S, Muller U. Specific sequence changes in multiple transcript system DYT3 are associated with X-linked dystonia parkinsonism. Proc Natl Acad Sci U S A. 2003;100:10347–10352. doi: 10.1073/pnas.1831949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vemula SR, Puschmann A, Xiao J, et al. Role of Gα(olf) in familial and sporadic adult-onset primary dystonia. Hum Mol Genet. 2013;22:2510–2519. doi: 10.1093/hmg/ddt102. Whole-exome sequencing identified a GNAL missense mutation (V228F) in an African-American pedigree with clinical phenotypes that include cervical, laryngeal ,and hand- forearm dystonia. Screening of 760 subjects with familial or sporadic primary dystonia identified three Caucasian pedigrees with GNAL mutations. Immunohistochemical studies showed that the encoded protein, Gαolf, is highly expressed in striatum and cerebellar Purkinje cells, and colocalized with corticotropin-releasing hormone receptors in Purkinje cells.
- 21. Lohmann K, Wilcox RA, Winkler S, et al. Whispering dysphonia (DYT4 dystonia) is caused by a mutation in the TUBB4 gene. Ann Neurol. 2013;73:537–545. doi: 10.1002/ana.23829. The large pedigree was adequately powered for generation of a multipoint logarithm of odds score of 5.3. Then, whole-exome sequencing identified a missense mutation in TUBB4A as the cause of DYT4. Sequencing of TUBB4A in 394 unrelated dystonia patients identified one missense variant possibly associated with a case of segmental dystonia with spasmodic dysphonia.
- 22. LeDoux MS, Xiao J, Rudzinska M, et al. Genotype-phenotype correlations in THAP1 dystonia: molecular foundations and description of new cases. Parkinsonism Relat Disord. 2012;18:414–425. doi: 10.1016/j.parkreldis.2012.02.001. This is a detailed and comprehensive analysis of coding and noncoding variants in THAP1 and exacting statistical analysis of THAP1 genotype-phenotype correlations.
- 23.Xiao J, Bastian RW, Perlmutter JS, et al. High-throughput mutational analysis of TOR1A in primary dystonia. BMC Med Genet. 2009;10:24. doi: 10.1186/1471-2350-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parker N. Hereditary whispering dysphonia. J Neurol Neurosurg Psychiatry. 1985;48:218–224. doi: 10.1136/jnnp.48.3.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilcox RA, Winkler S, Lohmann K, et al. Whispering dysphonia in an Australian family (DYT4): a clinical and genetic reappraisal. Mov Disord. 2011;26:2404–2408. doi: 10.1002/mds.23866. [DOI] [PubMed] [Google Scholar]
- 26.Leandro-Garcia LJ, Leskela S, Landa I, et al. Tumoral and tissue-specific expression of the major human β-tubulin isotypes. Cytoskeleton (Hoboken) 2010;67:214–223. doi: 10.1002/cm.20436. [DOI] [PubMed] [Google Scholar]
- 27.Breuss M, Heng JI, Poirier K, et al. Mutations in the β-tubulin gene TUBB5 cause microcephaly with structural brain abnormalities. Cell Rep. 2012;2:1554–1562. doi: 10.1016/j.celrep.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simons C, Wolf NI, McNeil N, et al. A de novo mutation in the β-tubulin gene TUBB4A results in the leukoencephalopathy hypomyelination with atrophy of the basal ganglia and cerebellum. Am J Hum Genet. 2013;92:767–773. doi: 10.1016/j.ajhg.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blumkin L, Halevy A, Ben-Ami-Raichman D, et al. Expansion of the spectrum of TUBB4A-related disorders: a new phenotype associated with a novel mutation in the TUBB4A gene. Neurogenetics. 2014;15:107–113. doi: 10.1007/s10048-014-0392-2. [DOI] [PubMed] [Google Scholar]
- 30. Vemula SR, Xiao J, Bastian RW, et al. Pathogenic variants in TUBB4A are not found in primary dystonia. Neurology. 2014;82:1227–1230. doi: 10.1212/WNL.0000000000000294. High-resolution melting and Sanger sequencing excluded TUBB4A coding region mutations in 575 subjects with primary laryngeal, segmental, or generalized dystonia.
- 31.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 32.Muller U, Steinberger D, Nemeth AH. Clinical and molecular genetics of primary dystonias. Neurogenetics. 1998;1:165–177. doi: 10.1007/s100480050025. [DOI] [PubMed] [Google Scholar]
- 33.Hedera P, Xiao J, Puschmann A, et al. Novel PRRT2 mutation in an African-American family with paroxysmal kinesigenic dyskinesia. BMC Neurol. 2012;12:93. doi: 10.1186/1471-2377-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caraballo R, Pavek S, Lemainque A, et al. Linkage of benign familial infantile convulsions to chromosome 16p12-q12 suggests allelism to the infantile convulsions and choreoathetosis syndrome. Am J Hum Genet. 2001;68:788–794. doi: 10.1086/318805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heron SE, Dibbens LM. Role of PRRT2 in common paroxysmal neurological disorders: a gene with remarkable pleiotropy. J Med Genet. 2013;50:133–139. doi: 10.1136/jmedgenet-2012-101406. [DOI] [PubMed] [Google Scholar]
- 36.Stelzl U, Worm U, Lalowski M, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 37.Uitti RJ, Maraganore DM. Adult onset familial cervical dystonia: report of a family including monozygotic twins. Mov Disord. 1993;8:489–494. doi: 10.1002/mds.870080413. [DOI] [PubMed] [Google Scholar]
- 38.Ma L, Chen R, Wang L, et al. No mutations in CIZ1 in twelve adult-onset primary cervical dystonia families. Mov Disord. 2013;28:1899–1901. doi: 10.1002/mds.25542. [DOI] [PubMed] [Google Scholar]
- 39.LeDoux MS. Exome sequencing for gene discovery: time does not stand still. Ann Neurol. 2012;72:628–629. doi: 10.1002/ana.23660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y, Xiao J, Gong S, et al. Neural expression of the transcription factor THAP1 during development in rat. Neuroscience. 2013;231:282–295. doi: 10.1016/j.neuroscience.2012.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao J, Gong S, Zhao Y, et al. Developmental expression of rat torsinA transcript and protein. Brain Res Dev Brain Res. 2004;152:47–60. doi: 10.1016/j.devbrainres.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 42.Ledoux MS. Non-Parkinson movement disorders: five new things. Neurol Clin Pract. 2013;3:22–29. doi: 10.1212/CPJ.0b013e318283ff2d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munchau A, Valente EM, Davis MB, et al. AYorkshire family with adult-onset cranio-cervical primary torsion dystonia. Mov Disord. 2000;15:954–959. doi: 10.1002/1531-8257(200009)15:5<954::aid-mds1028>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 44.Stamelou M, Charlesworth G, Cordivari C, et al. The phenotypic spectrum of DYT24 due to ANO3 mutations. Mov Disord. 2014 doi: 10.1002/mds.25802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vermeer S, Hoischen A, Meijer RP, et al. Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am J Hum Genet. 2010;87:813–819. doi: 10.1016/j.ajhg.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang F, Wang X, Ostertag EM, et al. TMEM16C facilitates Na+-activated K+ currents in rat sensory neurons and regulates pain processing. Nat Neurosci. 2013;16:1284–1290. doi: 10.1038/nn.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zech M, Gross N, Jochim A, et al. Rare sequence variants in ANO3 and GNAL in a primary torsion dystonia series and controls. Mov Disord. 2014;29:143–147. doi: 10.1002/mds.25715. [DOI] [PubMed] [Google Scholar]
- 48.Hopfner F, Bungeroth M, Pendziwiat M, et al. Rare variants in ANO3 are not a susceptibility factor in essential tremor. Parkinsonism Relat Disord. 2014;20:134–135. doi: 10.1016/j.parkreldis.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 49.Postma AG, Verschuuren-Bemelmans CC, Kok K, et al. Characteristics of dystonia in the 18p deletion syndrome, including a new case. Clin Neurol Neurosurg. 2009;111:880–882. doi: 10.1016/j.clineuro.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Esposito F, Addor MC, Humm AM, et al. GNAL deletion as a probable cause of dystonia in a patient with the 18p- syndrome. Parkinsonism Relat Disord. 2014;20:351–352. doi: 10.1016/j.parkreldis.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 51.Leube B, Hendgen T, Kessler KR, et al. Evidence for DYT7 being a common cause of cervical dystonia (torticollis) in central Europe. Am J Med Genet. 1997;74:529–532. doi: 10.1002/(sici)1096-8628(19970919)74:5<529::aid-ajmg15>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 52.Han F, Racacho L, Lang AE, et al. Refinement of the DYT15 locus in myoclonus dystonia. Mov Disord. 2007;22:888–892. doi: 10.1002/mds.21400. [DOI] [PubMed] [Google Scholar]
- 53.Winter P, Kamm C, Biskup S, et al. DYT7 gene locus for cervical dystonia on chromosome 18p is questionable. Mov Disord. 2012;27:1819–1821. doi: 10.1002/mds.25219. [DOI] [PubMed] [Google Scholar]
- 54.Kaur A. Rare autosomal dominant mutations in GNAL are associated with primary torsion dystonia. Clin Genet. 2013;84:211–212. doi: 10.1111/cge.12178. [DOI] [PubMed] [Google Scholar]
- 55.Miao J, Wan XH, Sun Y, et al. Mutation screening of GNAL gene in patients with primary dystonia from northeast China. Parkinsonism Relat Disord. 2013;19:910–912. doi: 10.1016/j.parkreldis.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Charlesworth G, Bhatia KP, Wood NW. No pathogenic GNAL mutations in 192 sporadic and familial cases of cervical dystonia. Mov Disord. 2014;29:154–155. doi: 10.1002/mds.25713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dufke C, Sturm M, Schroeder C, et al. Screening of mutations in GNAL in sporadic dystonia patients. Mov Disord. 2014 doi: 10.1002/mds.25794. [DOI] [PubMed] [Google Scholar]
- 58.Kumar KR, Lohmann K, Masuho I, et al. Mutations in GNAL: a novel cause of craniocervical dystonia. JAMA Neurol. 2014;71:490–494. doi: 10.1001/jamaneurol.2013.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saunders-Pullman R, Fuchs T, San Luciano M, et al. Heterogeneity in primary dystonia: lessons from THAP1, GNAL, and TOR1A in Amish-Mennonites. Mov Disord. 2014;29:812–818. doi: 10.1002/mds.25818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao J, Zhao Y, Bastian RW, et al. Novel THAP1 sequence variants in primary dystonia. Neurology. 2010;74:229–238. doi: 10.1212/WNL.0b013e3181ca00ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo AH, Cannon EH, Wekesa KS, et al. Impaired olfactory behavior in mice deficient in the α subunit of Go . Brain Res. 2002;941:62–71. doi: 10.1016/s0006-8993(02)02566-0. [DOI] [PubMed] [Google Scholar]
- 62.Ledoux MS, Dauer WT, Warner TT. Emerging common molecular pathways for primary dystonia. Mov Disord. 2013;28:968–981. doi: 10.1002/mds.25547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bertran-Gonzalez J, Hakansson K, Borgkvist A, et al. Histone H3 phosphorylation is under the opposite tonic control of dopamine D2 and adenosine A2A receptors in striatopallidal neurons. Neuropsychopharmacology. 2009;34:1710–1720. doi: 10.1038/npp.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liokatis S, Stutzer A, Elsasser SJ, et al. Phosphorylation of histone H3 Ser10 establishes a hierarchy for subsequent intramolecular modification events. Nat Struct Mol Biol. 2012;19:819–823. doi: 10.1038/nsmb.2310. [DOI] [PubMed] [Google Scholar]
- 65.LeDoux MS, Brady KA. Secondary cervical dystonia associated with structural lesions of the central nervous system. Mov Disord. 2003;18:60–69. doi: 10.1002/mds.10301. [DOI] [PubMed] [Google Scholar]
- 66.LeDoux MS, Lorden JF, Ervin JM. Cerebellectomy eliminates the motor syndrome of the genetically dystonic rat. Exp Neurol. 1993;120:302–310. doi: 10.1006/exnr.1993.1064. [DOI] [PubMed] [Google Scholar]
- 67.Neychev VK, Gross RE, Lehericy S, et al. The functional neuro-anatomy of dystonia. Neurobiol Dis. 2011;42:185–201. doi: 10.1016/j.nbd.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Makino S, Kaji R, Ando S, et al. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am J Hum Genet. 2007;80:393–406. doi: 10.1086/512129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber YG, Kamm C, Suls A, et al. Paroxysmal choreoathetosis/ spasticity (DYT9) is caused by a GLUT1 defect. Neurology. 2011;77:959–964. doi: 10.1212/WNL.0b013e31822e0479. [DOI] [PubMed] [Google Scholar]
- 70.De Carvalho P, Sweadner KJ, Penniston JT, et al. Mutations in the Na+/K+-ATPase α3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron. 2004;43:169–175. doi: 10.1016/j.neuron.2004.06.028. [DOI] [PubMed] [Google Scholar]