Abstract
To date, fMRI research has been concerned primarily with evincing generic principles of brain function through averaging data from multiple subjects. Given rapid developments in both hardware and analysis tools, the field is now poised to study fMRI-derived measures in individual subjects, and to relate these to psychological traits or genetic variations. We discuss issues of validity and reliability that arise when the focus shifts to individual subjects and that are widely applicable across imaging modalities. We also emphasize that individual assessment of neural function with fMRI presents specific challenges and necessitates careful consideration of anatomical and vascular between-subject variability, sources of within-subject variability, and statistical power.
Keywords: BOLD fMRI, individual differences, neurometrics, validity, reliability, prediction
From the group to the individual
Brain imaging with blood oxygen level dependent (BOLD) fMRI has been used extensively since the early 1990s to understand generic aspects of brain function, typically by averaging data across individuals in order to improve the signal-to-noise ratio (SNR). The statistical benefits of averaging across subjects have also been leveraged in-group comparisons, e.g. in studies of clinical populations. Yet these studies have historically fallen short of a proper characterization of brain function at the level of the individual. While the importance of a fully personalized investigation of brain function has now been recognized for several years [1,2], it is only quite recent technological advances that make it possible. For example, there are advances already at the acquisition level, such as higher field strength and faster acquisition, which have led to substantial SNR improvements [3]. Attempts at interpreting individual subjects’ fMRI measurements have become a major focus in the past five years or so, partly driven by the rise of “resting-state” fMRI (Box 1). There is, for example, interest in examining individual differences in relation to healthy ageing [4,5], personality [6], intelligence [7,8], mood [9] and genetic polymorphism [10]. On the clinical side, there are considerable efforts to use fMRI to classify individual subjects as patient or control ([11,12]; reviewed in [13,14]), to select treatment [15], or predict future outcome ([16]; reviewed in [17]).
Box 1. Resting-state fMRI: the workhorse of individual differences research.
Resting-state fMRI, or RS-fMRI, entails imaging subjects while they lie in the scanner doing nothing and trying not to think of anything in particular [155]. Spontaneous fluctuations in activity show reproducible correlations across brain regions; regions with correlated spontaneous activity also tend to be co-activated in task fMRI, thus establishing the relevance of spontaneous correlations (functional connectivity) to brain function [156,157]. RS-fMRI has now established itself as a leading approach to study brain organization [155].
A major reason for the widespread adoption of the RS-fMRI approach is its minimal requirements. Most subjects can lie quietly in the scanner for 5 or more minutes [158] (for more advanced analyses, up to 100 min of data may be required for best results [159]). Of course, careful investigations have pointed out “details” that influence the data collected during the resting-state, such as whether the subject’s eyes are open or closed [160], whether they are completely awake [124], what they are actually thinking about during the run [161–163] or what task they performed immediately preceding the run [164]. Changes in the strength and directionality of functional connections have been described between runs in the same session, but also at much faster timescales (seconds to minutes) during a run (reviewed in [165]). Functional connectivity can also be estimated from task fMRI data, usually following the removal of stimulus-evoked activity [72], and shares about half its variance with the functional connectivity estimated from RS-fMRI data [166]. The remaining half of unshared variance could complicate the interpretation of individual differences in functional connectivity [94].
It remains the case that RS-fMRI is the easiest functional data to collect and aggregate, across subject populations and sites [145], as is now done in several large efforts, such as those of the Human Connectome Project [149,167]. Individual differences research requires large sample sizes (see also main text) and thus RS-fMRI, despite imperfection on several fronts, is likely to remain the workhorse of individual differences fMRI research for the years to come (for a glimpse of the future, see Box 4).
Several issues arise when the focus shifts from group averaging to the comparison of individual subjects’ statistics. The issues can be framed in terms of key concepts from behavioral research on individual differences, namely validity and reliability. Validity asks whether the individual differences we measure with BOLD-fMRI really reflect what we intend to measure. One specific concern for validity is whether we are indeed comparing functionally homologous regions across subjects. Another is whether we are indeed comparing neural function, as BOLD fMRI only provides an indirect measure of neural activity. Reliability, on the other hand, asks whether a finding is stable in the face of variations that should not matter. Reliability is notably hindered by relatively well-understood noise sources such as motion and subject physiology, as well as less well-understood ones such as neuro- or vaso-active substances that we might not properly take into account. Both validity and reliability can be enhanced through the use of readily available tools, but there is still considerable work to be done to ensure a valid and reliable fMRI science of individual subjects. Though correlation analysis (between individual fMRI-derived statistics and other measures of the same individuals) is overwhelmingly used in the literature, it is subject to overfitting and findings need not generalize to other samples. The out-of-sample predictive value of an fMRI-derived statistic with respect to another individual difference measure must be established. The typical sample size for fMRI studies (N=15–30) needs to be scaled up (to N>100) for individual differences research; larger sample sizes not only increase statistical power in general, but also allow for more complex models to be fit. We discuss the latest advances with respect to these issues and provide some specific recommendations for how the field can best advance.
VALIDITY: are individual differences attributable to brain function?
A common space for mapping function
How can we tell whether individual differences in metrics such as BOLD activations or functional connectivity are actually related to differences in the underlying neural activity or communication, respectively? One ubiquitous problem arises in matching different brains so that functionally meaningful comparisons across subjects are possible in the first place. In a typical fMRI analysis pipeline, both structural and functional data from individual brains are spatially warped to a common anatomical space. The most widely used common space [18] is the MNI152 atlas, to which subjects’ brains are anatomically warped via a volumetric transform (for a review, see [19]). Such registration is appropriate for subcortical structures which are inherently volumetric; yet the cortex is a folded two-dimensional structure, and volumetric alignment does not properly align folding patterns across subjects. Switching to cortical folding-based inter-subject alignment [20,21] has been shown to somewhat reduce functional mismatch [22,23] (but, see [24]), yet has not become common practice. There are several reasons for this, from the burden of generating accurate cortical surfaces for each brain, to the additional complexities of working with surface-based data (which until recently were not handled well by leading fMRI analysis software). Recent improvements to Freesurfer’s automated cortical surface reconstruction pipeline [25], together with the release of a new file format that combines surface and volumetric data (Connectivity InFormatics Technology Initiative, CIFTI) and software for visualization and analysis (the Human Connectome Project’s Connectome Workbench), could foster wider adoption of surface analysis. Yet there is still no guarantee that functionally similar voxels correspond spatially across subjects after such alignment: individual differences may thus invalidly arise from incorrect alignment of function across subjects.
A multimodal common cortical topography
Watching a movie with a rich variety of visual, auditory and social percepts elicits time-locked brain activity that is correlated across subjects in many brain regions (inter-subject correlation (ISC) [26]). After cortical-folding-based inter-subject alignment, the cortical mesh can be further warped to maximize inter-subject correlations during movie viewing [27]. In the absence of brain activity that is time-locked to a shared external stimulus, it is also possible to instead improve the vertex-wise match of functional connectivity patterns across subjects [28] (see also [29,30]). A multimodal surface matching (MSM) framework [31] was recently introduced that not only performs similarly to, and faster than, state-of-the-art algorithms based solely on cortical folding [21], but that can also flexibly accept other types of data (e.g., fMRI) to further inform registration (Figure 2(a)). The framework can, for example, accept any combination of geometric (shape-based), myelin [32], task activation, retinotopy [33], and functional and structural connectivity features. In theory, the combination of all this information in the MSM framework should yield the best possible inter-subject alignment, with the constraint that neighboring vertices must remain neighbors. However, the optimal set of modalities to include, the associated cost functions, and the relative weights assigned to each modality, remain under investigation, and further validation of an improved alignment of function will be required before widespread use can be prescribed [31].
Figure 2. Multimodal surface matching and hyperalignment.
(a) The Multimodal Surface Matching framework [31] accepts multiple sources of information to align subjects: not only sulcal patterns, but also cortical thickness, myelin maps, RS-fMRI derived functional connectivity, etc. (b) After selecting a region of interest in a common anatomical space, hyperalignment [34] aligns representational spaces across subjects; it is usually based on movie data.
From a common cortical topography to a common representational space
Another recently introduced technique, “hyperalignment”, goes one step further, ignoring topological constraints altogether and using only functional representational geometry across subjects - i.e. matching multivariate spatial patterns of activity (Figure 2(b)) [34]. As before, the fMRI data used in this study was obtained in response to a full-length movie. The selection of voxels/vertices that are fed into the hyperalignment algorithm is the sole spatial constraint; data from each subject is eventually projected into a common space that can violate topology. The original implementation of the technique relied on pre-selecting a Region-Of-Interest (ROI) [34]; a more recent whole-brain version of hyperalignment uses a searchlight centered on each vertex, then combines the resulting transformation matrices into a large, whole-brain transformation matrix that can be used to project the cortical data from each individual subject to a common representational space [35]. Ongoing work is extending the hyperalignment algorithm to make use of resting-state data instead of movie data, in an effort to maximize applicability to extant datasets. In theory, whole-brain hyperalignment provides the most thorough correction for anatomical variability. Because hyperalignment is based on an invertible transformation of the data, individual differences are not lost at any stage of the process. Yet, the validity of individual differences measured in the common space is not assured, because individual differences of interest may also be mixed into the transformation matrix for each subject. We would encourage individual difference studies to explore hyperalignment for their datasets, and to report results obtained with and without hyperalignment, so that we can accumulate further evidence for its most appropriate application. In general, we would recommend that investigators try more than one approach to alignment, and report all of them so we can see which might work best for which kinds of questions.
Region-of-interest analysis
Many of the problems with alignment can be largely circumvented if a functional region of interest (ROI) is isolated in each individual subject. Functional localizers have traditionally been used to improve validity [36]: in an independent functional MRI run, a task designed to activate a ROI is performed, and the ROI is defined in each individual from a statistical parametric map as one of the clusters that exceeds a given threshold. However, the choice of a statistical threshold in single subjects is often a thorny issue; recent approaches can avoid arbitrary thresholds by using mixture models that fit the noise in each subject’s data [37] or by using multiple spatial scales to define clusters of activation (threshold-free cluster enhancement (TFCE) [38]). Another issue is that of ensuring that the individually defined ROIs are indeed well matched across subjects, a problem for which approaches such as the group-constrained subject-specific (GCSS) algorithm [39] provide elegant solutions. Functional localizers have a long and successful track record and can provide functional ROIs in individual subjects with only a few minutes of scanning, for processes ranging from face perception [40] to theory of mind [41,42]. However, they quickly become inefficient if one wants to map many different processes in a study [43]. At present, we would encourage investigators to continue using well-vetted localizers when studying specific psychological processes; in other contexts, emerging methods for single-subject functional parcellation from resting-state data and other modalities (Box 2) should be considered. Pending further validation against known function, these methods may replace functional localizers altogether in the not-too-distant future.
Box 2. Functional parcellation of individual subjects’ brains.
It is now well established that resting-state functional connectivity (RSFC) can extract networks in the brain that subserve shared psychological functions [145]. Accordingly, there has been interest in using RSFC to define functional parcels or ROIs in single subjects’ brains. There are two schools of thought when it comes to functional parcellation: those who allow the parcels to overlap spatially, and those who prefer to tile the brain with non-overlapping parcels. The first school typically uses Independent Component Analysis (ICA) to decompose the RS-fMRI signal into statistically independent non-Gaussian sources (spatial maps) via a linear and instantaneous mixing process corrupted by additive Gaussian noise components [168]. The second school typically uses some variation of a clustering or region growing algorithm that results in parcels that have homogeneous patterns of whole-brain connectivity [169–173], or an algorithm that detects transitions in whole-brain RSFC [174,175].
There are two approaches to establishing a one-to-one correspondence between the parcels in different subjects that enhance validity beyond whole-brain alignment techniques. One is to perform RSFC-based parcellation in individual subjects independently with no prior information [171,172], then use some clustering or matching algorithm to establish one-to-one correspondence between parcels; this can prove a rather difficult problem, with e.g. splitting of parcels. The other, preferred approach is to start from a set of group-level parcels and discover their instantiation in individual subjects. The initial group parcellation is of course critical, and may come from analyzing averaged or concatenated individual data, or from a previously published result. Projecting it into individual brains is typically done using dual regression in the case of IC components [176], and different approaches have been proposed for other parcellation schemes, e.g. a Multi Layer Perceptron neural network which assigns each voxel from an individual subject to a parcel [177], or an iterative assignment procedure as in [178].
Including information from multiple modalities has proved beneficial to refine inter-subject whole-brain alignment, as demonstrated in the MSM framework [31]. Using multiple modalities can also be instrumental in parcellating an individual brain, as demonstrated recently: after defining a multimodal group parcellation from the HCP data (including architecture, such as thickness and myelin content; task fMRI; RS-fMRI connectivity; and RS-fMRI derived topographic maps), a machine-learning approach can be used to find the instantiation of each group-defined parcel in single subjects, correctly accounting for variations in anatomy [200].
The ever-lurking plumbing issue
Differences in vasculature cause differences in the hemodynamic response, as do any differences in baseline cerebral blood flow (CBF), hematocrit level, vessel volume or vessel elasticity. A review of all the potential effects of vasculature differences is outside the scope of this article [44–46]. Here we focus instead on existing solutions at the level of acquisition and data analysis that are rarely implemented yet may prove critical for valid individual differences research.
Capturing response shape variability
The hemodynamic response function (HRF) is a model of the BOLD activity triggered by a neural event of infinitesimal duration. Most fMRI analyses assume a fixed-shape HRF. However, the shape of the HRF is well-known to vary across subjects and brain regions [44,47]. Factors such as vasodilatory signaling, blood vessel stiffness, neurovascular coupling delay, venous transit time and the time constant of autoregulatory feedback contribute to this variability [48,49]; these factors are known to be particularly affected by ageing and disease [50]. Inferences about individual differences in BOLD response magnitude are not valid when the true shape of the HRF differs across subjects. To account for these variations, the use of multiple, orthogonal basis functions to more accurately model the HRF has been proposed (reviewed in [51]). Popular constrained basis sets include the canonical HRF plus its temporal and dispersion derivatives [52] or a basis function set based on singular value decomposition [53]. Releasing all constraints, the finite impulse response (FIR) basis set has one free parameter for every time point following stimulation for every event type modeled [54]. A caveat of increasing the number of free parameters is the concurrent increase in variance of the HRF produced, as well as the cost to compute it.
When the shape of the HRF is fixed, the amplitude of the BOLD response is easily compared across subjects on the basis of a single parameter estimate. Modeling HRF shape faithfully complicates the comparison of response amplitude across subjects: the BOLD response is now represented across several parameters, which must be jointly compared. Some solutions have been proposed for this joint comparison, yet none is widely accepted yet [51,55]. Recent work combining estimation of the HRF shape with detection of activation in the same optimization seems a step in the right direction [56,57], though these approaches currently assume that HRF shape is similar across subjects in a given brain region, which may not be a valid assumption when investigating individual differences.
HRF shape variability is problematic even for resting-state fMRI, since differences in response shape across subjects can result in differences in functional connectivity estimates. With current analytical practices of low-pass filtering the data (considering only fluctuations slower than 0.1Hz), the effects of small differences in response shape should be small [45]. However, as investigators start looking at higher frequency fluctuations [58], HRF shape variability may seriously limit validity in interpreting individual differences. Methods are emerging to deconvolve the spontaneous signal [59,60] and thus possibly account for these variations. In our view, further development of how best to quantify and incorporate HRF variability should be of the highest priority for individual differences research, since all subsequent measures depend on getting this right in the first place.
Accounting for vascular differences across subjects: calibration and normalization
The BOLD signal is a function of changes in cerebral blood flow (CBF), cerebral blood volume and the cerebral metabolic rate of oxygen consumption (CMRO2) [61]; it also depends on the baseline physiological state (hematocrit, oxygen extraction fraction, blood pressure). Both hemodynamic coupling and baseline physiology differ across subjects [45,62]. Two main approaches have been put forward to try and control for differences in these parameters across subjects: calibration and normalization.
The calibrated fMRI technique has been around for many years, and can, in theory, control for differences in both baseline physiology and hemodynamic coupling across subjects. The technique has not seen widespread adoption, chiefly because it is difficult to implement (e.g., requiring concurrent measurement of BOLD and CBF and inhalation of CO2 [63,64]) and still imperfect [65].
Several approaches have been proposed to normalize for differences in baseline physiology across subjects. Many rely on a companion scan. For example, the BOLD response to hypercapnia, induced through administration of carbon dioxide [66] or using a breath-hold challenge [67], can been used as a normalization factor (Figure 3(a)). Alternatively, whole-brain venous oxygenation levels can be measured with a special pulse sequence and used to normalize the BOLD response [68]. A more easily applicable option is to use the amplitude of low-frequency fluctuations in resting-state fMRI data (RS-ALFF) [69,70] as a normalization factor; indeed RS-ALFF reflects naturally occurring variations in cardiac rhythm and respiratory rate and depth [71] and approximates the BOLD response to a hypercapnic challenge (Figure 3(a)). In fact, one does not even need to acquire a separate resting-state scan here. In the same way that functional connectivity can be derived from the residuals of a General Linear Model (GLM) for task-based fMRI data [72], the amplitude of low frequency fluctuations in the residuals of task-based fMRI data (GLMres-ALFF) can also be used to rescale the BOLD signal change; this “vascular autorescaling” (VasA) technique was even shown to outperform RS-ALFF based normalization [73] (Figure 3(b)). Using data from the same fMRI run, as VasA does, is also desirable because it avoids contamination of the data with noise from a separate run.
Figure 3. Post-hoc vascular normalization techniques.
(a) Traditionally, hypercapnia has been used to elicit a BOLD signal change with no change in neural activity or consumption of oxygen; this response can be used to normalize the BOLD response measured in task runs. Breathholding (BH, top) can be used to induce hypercapnia. Recently, it was found that the Amplitude of Low Frequency Fluctuations from RS-fMRI (RS-ALFF, bottom) mimics the hypercapnic BOLD response [69], and thus may be used instead of a hypercapnic run. Figure reproduced with permission from [154]. (b) The ALFF of the residuals of task fMRI data (GLMres-ALFF) also looks very similar to RS-ALFF (left) and can be used to rescale the BOLD signal change (vascular autorescaling, or VasA). Figure modified with permission from [73]. (c) Decrease in BOLD response to a sensorimotor task as a function of increasing age in the occipital lobe (blue) is abolished after scaling the sensorimotor-task with RS-ALFF (red), where each point represents an individual. Figure adapted with permission from [74].
While all these methods were developed with the aim of improving group statistics by reducing inter-subject vascular variance, they should be considered for a valid assessment of neural individual differences (see Figure 3(c) [74]). As with any manipulation of the data, there are potential caveats: calibration and normalization methods may remove individual differences of neural origin, for instance if baseline blood oxygenation were linked to neural activity [62]. Before such techniques are routinely applied, more examples of their successful application, ideally vetted by an independent measure of neural function [74], are needed.
RELIABILITY: individual differences or unmodeled noise?
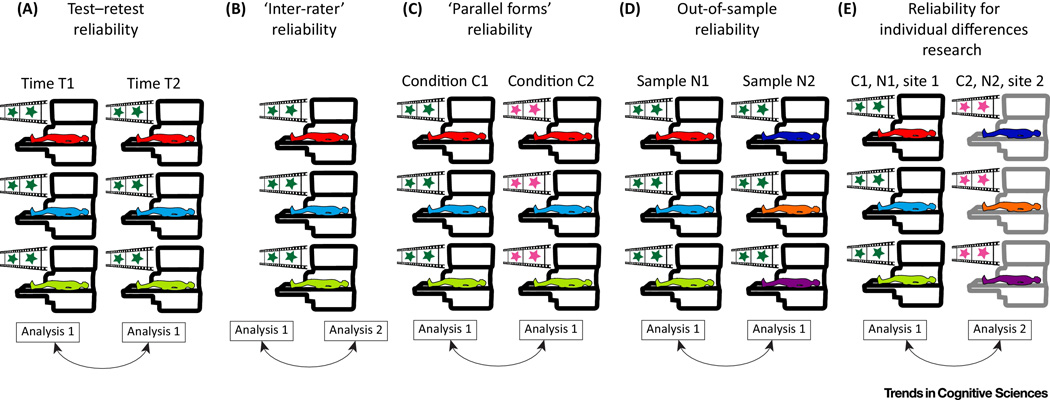
Once validity is maximized (inasmuch as current technology allows), it is also crucial to ensure that individual differences measured with fMRI are not merely attributable to unaccounted-for noise in the measurements. The reliability of fMRI has been inspected closely in recent years [75–77]. Of course there is no such thing as “the” reliability of fMRI because different derived statistics are differentially affected by noise in the raw data and thus have different reliabilities [78] (for an overview of studies addressing the reliability of specific fMRI-derived measures, see Table S1). Moreover, although a general sense of the reliability of the most widely used fMRI measures can be derived from extant literature, details of the experimental procedures [79] and of the preprocessing applied to the raw data [80,81] matter. For these reasons, we believe that it is critical to provide an assessment of reliability in each and every study that looks at individual differences, under the specific pipeline used for the analysis. At present, individual differences in fMRI are always studied with respect to an independent measure of the same individuals. The reliability of interest is that of the relationship between the fMRI-derived statistic and the independent variable (see also [78,82]). There are several flavors of reliability which are relevant to individual differences research, with direct analogs in behavioral testing. Test-retest reliability quantifies how variable the established relationship is in the same sample of subjects, under the same conditions (stimulus, scanner, time of day, analysis) at an appropriate time interval (Figure 4(a)). It is also important to ensure that the relationship is robust to the exact preprocessing performed on the raw data, which can be conceived of as “inter-rater” reliability (Figure 4(b)). In addition, since we are interested in brain function rather than the low-level properties of a given stimulus, the relationship must also be robust to the exact experimental conditions, known in psychology as “parallel forms” reliability (Figure 4(c)). The relationship should further hold for a different sample of subjects (from the same population) (Figure 4(d)), and across scanners.
Figure 4. Reliability for individual differences research.
(a) Test-retest reliability. The same subjects are tested with the same stimuli in the same scanner, at an appropriate time interval given the function of interest. (b) Inter-rater reliability. The same data is preprocessed in slightly different ways which are thought to be interchangeable (e.g., using physiological regressors or ICA to remove physiological noise). (c) Parallel forms reliability. The same subjects view slightly different stimuli which are thought to involve the same brain processing. (d) Out-of-sample reliability. A different set of subjects undergo the same experiment in the same scanner, and the data is analyzed in the same way. (e) Putting it all together for individual differences research. A result (e.g. relationship between fMRI-derived statistic and neuropsychological score) should be reliably obtained for different subjects at different sites, possibly using slightly different stimuli and preprocessing steps.
Though many measures of reliability have been proposed over the years [75], some directly addressing the ratio of inter- to intra-subject variance [83], a predictive framework inspired by machine-learning is best suited to establish the reliability needed for individual differences research as we discuss further below (see “choosing prediction over correlation” section). Reliability is ensured when a relationship discovered in a sample of subjects at site 1 using stimulus 1 and analysis 1 generalizes to a different sample of subjects at site 2 using stimulus 2 and analysis 2 (Figure 4(e)).
Sources of within-subject variance we acknowledge and strive to correct for
There are a number of sources of within-subject, inter-session variance that are well-known, and which fMRI researchers strive to eliminate at acquisition time and/or correct for during preprocessing of the data [84,85]. Scanner-related noise, artifacts and drift are unavoidable, yet fairly simple to address through artifact rejection and temporal filtering. Here we briefly review two main sources of within-subject variance which are more problematic and the state-of-the-art in addressing them: subject motion, and subject bodily physiology.
Motion
Subject motion in the scanner leads to poor data quality, for obvious reasons: in fast echo-planar imaging (EPI) sequences, motion not only disrupts spatial encoding, it also disrupts the physical phenomena that the MR signal relies on (e.g., resonance frequency, relaxation time between samples). Artifacts occur with frame-to-frame movements of a few tenths of a millimeter or less [86], meaning that they affect all datasets to some extent. The classical approach to correcting for motion artifacts has been to first estimate subject motion through rigid-body realignment of brain volumes, then to regress the calculated head motion parameters out of the fMRI time course to correct for any residual effects of motion [87]. Other researchers also include the global signal time course, the white matter time course and the CSF time course as confound regressors [88]. This regression approach was recently shown to incompletely correct for motion artifacts, in the context of resting-state fMRI analyses [86,89,90]. Additional corrections have been proposed, e.g. the removal of independent components related to motion [91], or the complete removal of motion-affected frames known as scrubbing [86] (see [92] for a comprehensive review).
Thoroughly correcting for motion artifacts is important to ensure test-retest reliability: a given subject may move more in one session than in another. Yet, a discussion of motion would have been appropriate in the previous section on validity as well: some subjects are generally more fidgety than others in the scanner (some have argued that this constitutes a trait in and of itself, with a neurobiological basis [93]). Motion arguably contributes more to inter-subject variance than to intra-subject variance (e.g., men tend to exhibit more head movements than women [90]; older people move more than younger people [94]; and people with autism move more than controls [95]). Motion artifacts have complex effects on fMRI statistics, and incompletely correcting for them can lead to erroneous conclusions in individual differences research [95–97].
Physiology
In a previous section we pointed out how differences in vasculature may affect the validity of inter-individual comparisons. To complicate matters further, breathing and heart rate affect fMRI measurements. For instance, motion of the chest wall with respiration results in magnetic field changes [98]; normal alterations of the depth or rate of breathing lead to variations in arterial CO2 [99] and subsequent blood flow changes [100]; and pulsatile blood flow due to heartbeat leads to fluctuations in signal intensity in arteries, arterioles, and other large vessels [101].
A common approach to correcting for these artifacts is to collect independent measurements of the heart rate (with a pulse oximeter) and respiration (with a respiratory belt), and regress them out of the fMRI signal, using one of several available algorithms (reviewed in [85]); yet these corrections are not routinely applied. Interestingly, it has been found that many of these model-based corrections actually reduce test-retest reliability for functional connectivity analyses [102,103]. This has been interpreted to mean that a large fraction of functional connectivity reflects basic physiological signals [102] (another validity issue, e.g. [104]). A more optimistic take is that these model-based approaches still need some tweaking - either the models are not physiologically accurate, or the measurements of heart rate and respiration are suboptimal (too noisy, or not measuring the variable of interest). Further development of the models (e.g. [105]) and of new, better MR compatible measurement devices such as a continuous blood pressure monitoring device is needed [85].
Other approaches only rely on the fMRI data without the need for separate measurements (reviewed in [85]; see also [106–108]). Most of them are based on decomposing the data into components (but see [109]). Some of these methods have been shown to improve reliability [110]. However a fair comparison with model-based physiological regression has not been conducted yet -- typically, decomposition-based denoising techniques target a wider range of artifacts, including motion.
In decomposition-based methods, identifying noise components is not always trivial [111]; some components may be a mixture of signal and noise and there is a risk of “throwing the baby out with the bathwater”. Automated algorithms, like ICA-FIX [108], rely on prior manual classification of components and are therefore not immune to this criticism. A promising new approach to teasing apart signals of neural origin from artifacts was recently introduced: using multi-echo EPI [112], it appears possible to distinguish a genuine BOLD effect from artifacts by looking at the time course of MR decay. The technique could lead to improvements in both the validity and reliability of measurements [113]. Finally, its combination with recent developments in simultaneous multi-slice acquisition [114] could make it a viable imaging protocol in terms of temporal and spatial resolution [115]; we are eagerly waiting for further validation and improvements of this interesting technique (see also [116] for a related approach).
Other sources of intra-subject variance
Another physiological rhythm: vasomotion
Vasomotion refers to the spontaneous changes in tone of blood vessels, independently of heart rate and respiration. Vasomotion leads to low frequency oscillations in the BOLD signal [117]. There is some evidence that vasomotion may be localized to specific regions of cortex [118], making it a potentially serious confound. As with previous artifacts, vasomotion may differ systematically across subjects (validity).
Baseline physiological state
Baseline physiological state was already discussed in the validity section; yet baseline physiology is also variable for a given subject. Going for a run an hour or less prior to scanning may lead to alterations of baseline physiology [119,120]. Anxiety or stress [121], extended exposure to high altitudes [122], recent sleep quality [123,124], or phase of the menstrual cycle (for women) [125] are all examples of poorly understood vascular and neural factors that may hinder the reliability of fMRI measurements if they are not properly measured and incorporated into a full model [126].
Neuromodulators and vasoactive substances
The effects of caffeine on fMRI measurements have been studied extensively. While it was once held that a dose of caffeine was a way to enhance the BOLD response [127], subsequent studies have muddied this picture [128,129]. Many complex effects of caffeine have been reported over the years, such as enhanced linearity of visually evoked BOLD responses [130]. The complexity likely stems from the double action of caffeine on neural activity and on haemodynamics (for a review see [131]).
Several other routinely consumed substances such as alcohol, nicotine, illicit drugs and prescription medication, and even foods and food supplements, also have complex neural and vascular effects on fMRI measurements, which are beyond the scope of this review. Using arterial spin labeling (ASL) and fMRI to get a better understanding of these effects, a field of research known as pharmacological MRI [132], may be yet another important ingredient towards a reliable fMRI science of individual subjects.
Further considerations for a fMRI science of individual differences
Choosing prediction over correlation
Currently the only way to interpret a fMRI-derived statistic is to relate it to another individual measure in the same set of subjects, such as their age, gender, test scores indexing aspects of intelligence and personality, or other measures of behavior (Table S2). By far the majority of fMRI studies of individual differences use correlation analysis to establish such a relationship. Correlation analysis relies on in-sample population inference and does not directly ensure the generalizability of the established relationship to out-of-sample individual subjects [133,134]. Shifting to a predictive framework is necessary to ensure generalizability and to interpret fMRI-derived statistics at the individual subject level [17,135] (for a more in-depth discussion of adopting a predictive machine-learning inspired framework, and the proper use of training, validation and test datasets, see [136]). To fully control for remaining confounds, fMRI-derived statistics should be included in a full model alongside other potential predictors, and the unique predictive power of fMRI features should then be assessed through their selective removal from the model (as demonstrated in [16]) (see Figure 1(e)).
Figure 1. Proposed generic analytical pipeline for individual differences research in fMRI.
This pipeline may not fit all situations, given the multidimensionality and multi-component nature of fMRI data. (a) Data Yraw is acquired while a subject does a particular task (movie with stars). Minimal preprocessing is applied (e.g., HCP pipelines [25] shown in the box to the right). (b) Subcortical areas are volumetrically warped to match the MNI template, while the cortex is registered to a surface template using cortical folding patterns or a multi-modal surface matching approach. A choice is then made between conducting a region-of-interest or whole-brain analysis. Several options for group-informed definitions of functional parcels in individual subjects are available. Finally, hyperalignment can be performed to project data into a common representational space. (c) The preprocessed data Ypreproc is modeled as a weighted linear combination of known signal (except for RS-fMRI) and confounds of no interest, plus unmodeled noise. All parameter estimates are normalized for vascular differences before further analysis. A statistic S (possibly multidimensional) is derived for this subject. (d) More than 100 subjects participate in the experiment, possibly with slight variations (e.g., at two different sites, black and gray scanners). (e) The fMRI-derived individual statistic is used together with confound variables to predict an individual measure of interest IV, for example the subject’s IQ (prediction) or patient/control (classification). The same analysis is conducted without the fMRI-derived statistic. Performance for the full model is compared to performance for the null model, using a permutation test, to establish the unique predictive power of the fMRI-derived statistic. Text in gray: awaiting further validation.
Increasing sample size
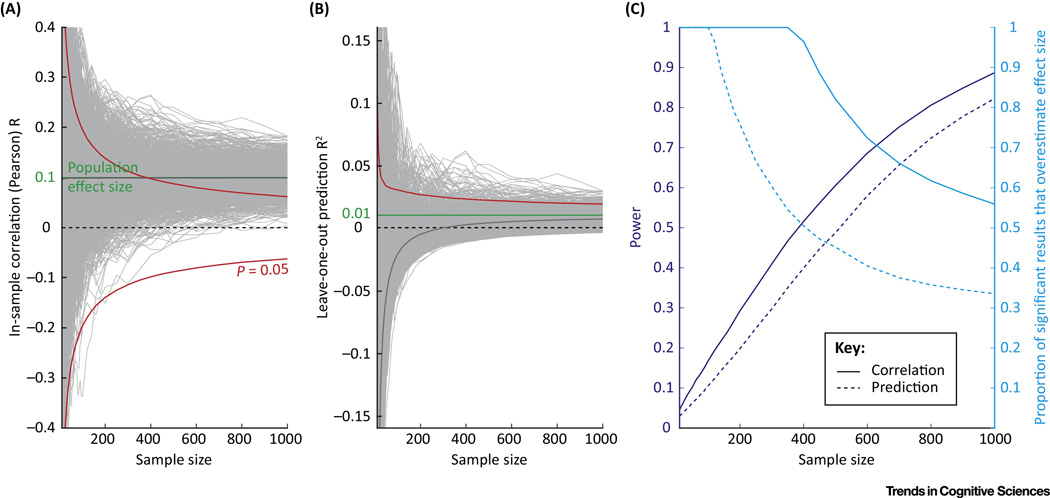
It is now widely recognized that the small sample sizes routinely used in fMRI studies (N=15–30) have low statistical power; given current reporting practices, this leads to inflated estimates of effect size [137,138] (Figure 5c) and thus poor replicability (see also BOX 3 for an overview of replicability-enhancing practices). The current heuristic for studies using correlation analysis is a sample size of at least 100 (see [139] for a more quantitative recommendation). In addition replication in an independent sample is a worthy precaution, which should be a requirement for studies that are not based on a strong prior hypothesis. Larger samples are also beneficial in a predictive framework: a larger number of examples guards somewhat against overfitting and mitigates the “curse of dimensionality”. Replication in an independent sample is built in for a predictive framework, provided that model selection is completely blind to the testing examples; in practice, this means entirely setting aside part of the data until the model is finalized. Only then can the fully trained model be tested, once and once only, on the test data to establish generalizability. This is the way machine-learning competitions are usually set up [140].
Figure 5. A predictive framework for a more reproducible science.
These plots are provided for illustrative purposes only. They are based on simple simulations described in the supplemental material. (a) The “cone of confidence” for correlation analysis, as in [139]. The true effect size in the population is r=0.1 (green), with SNR=0.1. The light gray traces (1,000 shown, out of 100,000 generated) are bootstrap samples from this population. As sample size increases, the range of correlation values observed in the samples decreases. In red, the 95% confidence interval for the null hypothesis r=0. (b) Same representation for the R2 of a leave-one-out prediction analysis, using the same simulated samples. The average of all bootstrap samples is shown in dark gray; it asymptotes to the true effect size (green line) for larger sample sizes. (c) Power of the correlation and prediction analysis (dark blue), for α=0.05. Proportion of significant results (p<0.05) which overestimates the effect size (light blue). Prediction analysis is less likely to overestimate effect size than correlation analysis; it even tends to underestimate it, making it a conservative analysis.
Box 3. Increasing the reproducibility of fMRI research.
The replicability of fMRI research has recently been called into question. Culprits for the lack of replicability are small sample sizes [138], analytical flexibility [179] which fosters p-hacking [180] without appropriate control for the overall risk of false positives [181], and the bias to publish only statistically significant results (a.k.a. the file drawer problem [182]). This means that we are often building new research on fragile grounds (previous false positive results), and thus wasting a large amount of resources [183]. A shift in research practices is necessary.
Reproducibility consists in obtaining the same results using the same code and data [184] (Figure I) (see also [183] for a slightly different terminology); it is a lower scientific standard compared to a fully independent replication using a different dataset and similar analysis, yet ensuring reproducibility in the first place is a surefire way to enhance replicability (though see [185] for a differing view). We note that failure to replicate a finding need not always imply that the finding was a false positive: a difference in outcome may also stem from any number of effects in execution or analysis that, unbeknownst to the investigators attempting the replication, have actually changed the psychological process under investigation. Replication attempts help establish the conditions under which a given finding is robust.
Following earlier recommendations for reporting fMRI methods [186], a recent white paper draft (from the OHBM Committee on Best practices in Data Analysis & Sharing, a.k.a. COBIDAS) is likely to be adopted by the imaging community. It identifies transparency, detailed reporting of methods, and importantly sharing of data and analysis pipelines (Figure I) as key ingredients for reproducible fMRI research. Data sharing platforms were discussed in the main text [146]; there are also platforms to share code, and guidelines on how to use scripting and pipelining to foster reproducibility [183]. An example of how to implement these recommendations is to release all the data along with a virtual machine environment to allow others to execute identical analyses [126]. At this stage such diligence still requires a large amount of technical overhead and cannot be made mandatory. The enforcement of these recommendations will initially largely depend on journals following suit and requiring these ingredients for publication, or on reviewers taking them into account when evaluating their peers’ research. However, there is a growing set of resources that may soon make this effort much more feasible and thus more widely adopted; in particular, we are closely following the efforts of the Stanford Center for Reproducible Neuroscience (http://reproducibility.stanford.edu) and of the Scientific Transparency project (http://post.stanford.edu), both aiming at providing web-based platforms to generate reproducible workflows and leverage high-performance computing for the analysis of neuroimaging data.
Figure I. From reproducibility to replicability: sharing of data and executable analysis pipelines. Adapted from [184].
Sample sizes in excess of 100 are still difficult to achieve for a small research group funded by a typical research grant; thus, despite this recommendation, underpowered correlation studies without internal replication will continue to be published. While it might seem that the cumulative output from many underpowered studies should eventually converge to a reliable conclusion through meta-analysis, this is not the case because of the strong bias to publish only significant findings [136,141] (Figure 5c). The reporting of all results regardless of their significance in the null hypothesis significance testing (NHST) framework, which could be implemented through initial pre-registration to ensure the quality of the methods [142], would be a way for studies with small sample sizes to contribute unbiased information for meta-analysis. FMRI research is not the first to face this challenge: in genetics, similar issues [143] were eventually resolved through the formation of large-scale consortia to generate the sample sizes required for adequate statistical power.
To achieve larger datasets in fMRI, data collected at different institutions can be aggregated in a shared online resource [144], as pioneered by the 1000 functional connectomes project [145]. Several data-sharing platforms are currently available (see [146]), for example openfMRI [147]. For the accumulation of data across sites, standardized procedures must however be implemented. Resting-state fMRI is an obvious candidate for data aggregation due to minimal instructions and requirements, but as we noted earlier, “small” details must still be carefully controlled (BOX 1). Although simple task fMRI paradigms (e.g., finger tapping [171]) would be good candidates as well, they may be too inefficient in that they are restricted to a narrow set of processes. We view movie fMRI as particularly well-suited for data aggregation, given its ability to capture richer representational information while better matching subjects’ mental states compared to the resting-state (BOX 4), and at the same time preserving similarly minimal instructions and requirements.
Box 4. Individual assessment with naturalistic, condition-rich designs.
One promising class of stimuli, which affords tighter control on internal states than the resting-state while still being scalable (e.g., for aggregation of data across centers), is movies [26] or narrated stories [187]. These are naturalistic, highly engaging stimuli, which are very effective at driving brain activations and encompass a great diversity of mental states. Public datasets relying on story/movie data are in the making: for example, the studyforrest project recently released a high-quality 7T fMRI dataset of subjects listening to the narrated movie “Forrest Gump” (1994) [188]. Efforts to extract and distribute annotations that can be used in analyses have also begun [189]. Here we present three main methods to study the representations triggered by such stimuli across subjects.
Inter-subject correlation
Since all subjects watch the same movie, the time course of stimulus-related activity in their brains should look similar [190]. As one would expect, activity in early sensory cortices is quite similar across subjects, whereas activity in association cortices is less similar [26]. Dynamic inter-subject correlation analysis in light of a rich featural description of the stimuli is a promising avenue of research to capture rich information about individual differences in brain representations. Representational geometry. Different stimuli are represented in the brain along a number of dimensions, and representational geometry denotes the arrangement of stimuli in that space and the distances between them [191]. While representational geometry and representational similarity analysis have so far been exploited mostly to assess different models of brain function [192–194], they can also be leveraged to compare representational geometry across individual subjects[195,196]. Representational geometry can be derived from movie or story data, as long as the events of interest are properly labeled [189].
Voxel-wise modeling
Voxel-wise modeling consists in using high-dimensional featural models of natural stimuli as encoding models to make sense of fMRI data [197]. When a model is found to satisfactorily predict the data out-of-sample, it can then be interpreted. A recent study using movies with annotated semantic features generated detailed semantic maps of each subject’s brain [198]. These maps represented those movie features that were most predictive of the activations observed at each voxel in the brain. These semantic maps capture a tremendous amount of information about the brain activity of each subject, and thus interrogating them with respect to individual differences is a very exciting endeavor. Such work is already well under way [199].
Standardized procedures are of course easiest to implement in the context of large-scale projects such as the IMAGEN project (2000 subjects) [148], the WashU-UMinn Human Connectome Project (1200 subjects) [149], or the Cambridge Center for Ageing and Neuroscience (approximately 700 subjects) [150]. The latter two projects are aimed at acquiring state-of-the-art data and distributing it as a data-mining resource for fMRI researchers around the world. Projects like these are akin to the accelerators used by particle physicists, or the large telescopes used by astronomers: a few sites in the world acquire the best possible data, and this data is subsequently probed (for many years) by the best analysts around the world.
Towards normative fMRI research?
A key concept in psychology is that of a “norm”, i.e. the distribution of a score in the population of interest. Scores on psychological tests are usually standardized with respect to their distribution in a large normative sample (the size of which depends on the score’s distribution [151]), which allows their direct interpretation without recourse to another measure. In the future, we may be able to interpret an individual subject’s fMRI-derived statistic in light of its distribution in a normative sample, provided absolute reliability is achieved; this approach could lead, for clinical imaging, to a more biologically informed science of human neuropsychiatric disease [152,153] and a better basis for personalized medicine.
Concluding remarks
Individual differences in brain function are critical to understanding healthy differences based on personality, gender, age, or culture. They are also critical for personalized medicine approaches to neuropsychiatric diseases. Recent technical advances have increased the sensitivity of functional MRI and set the stage for a characterization of brain activity at the level of momentary mental events in individual subjects. We now face key challenges of reliability and validity on the path to an fMRI-informed science of individual differences. We already have some tools to address these challenges; we propose a pipeline for the measurement and analysis of individual differences with fMRI as shown in Figure 1, on the basis of current knowledge (but see Outstanding Questions). While interpretation of fMRI-derived measures currently relies on independent measures of behavior, psychological scores and neuropsychiatric diagnosis, we are hopeful that in the near future they will stand on their own in light of normative data. No matter the strategy, increasing sample size dramatically is necessary, and data sharing efforts together with standardized procedures and reproducibility-enhancing practices will be key to the process. Beyond the current emphasis on resting-state fMRI data, which is easy to perform and aggregate, the use of naturalistic stimuli that capture rich inter-individual differences in cognition is a direction worth exploring.
Supplementary Material
Outstanding Questions.
Will the newer methods that we describe here and recommend (highlighted in gray in Figure 1) stand the test of time?
Which of the concerns we reviewed here (inter-subject alignment, hemodynamic variability across subjects, sources of noise) are the most important, and which can often be ignored in practice?
How do the many corrections we have reviewed here interact with one another? For example, does normalization become superfluous when the HRF is properly modeled?
How long will BOLD-fMRI still be the norm for non-invasive functional imaging of the whole brain in awake humans? Are other more valid non-invasive imaging techniques conceivable in the future?
What other modalities can be combined with fMRI to provide a richer set of measures and enhance validity, besides physiological recordings? Options include eye-tracking, EEG, and fNIRS.
Should pre-registration be required for fMRI studies of individual differences below a minimum sample size?
What role will behavioral and genotypic data play in the future? Will these always be required, or can we explore individual differences in a data-driven way based on fMRI data alone?
Will fMRI-derived individual differences lead us to revise clinical diagnostic categories?
Trends Box.
Interpretation of fMRI data at the level of individual brains is essential for characterizing brain function in health and disease.
Two core challenges are validity (Do we measure what we intend to measure?) and reliability (Is our measure stable in the face of variations that should not matter?) of fMRI-derived individual differences; these challenges can be partly addressed with recent tools.
Interpretation of single-subject fMRI measures relies on establishing a relationship with an independent measure in the same subjects. Out-of-sample prediction should be used over correlation analysis.
Accumulation of large samples through consortia and data sharing, as well as careful attention to statistical power issues, are critical for reproducible research.
Whole-brain characterization in naturalistic conditions, such as while watching a movie or listening to a story, may provide an alternative to resting-state data that permits a rich link to sensory and semantic stimulus variables.
Acknowledgments
This work was funded in part by a NARSAD grant from the Brain and Behavior Research Foundation (JD) and a Conte Center from NIMH (RA).
Glossary
- Preprocessing
operations performed on raw fMRI data before statistical analysis
- Realignment
one of the operations performed during preprocessing of fMRI data. It consists in spatially aligning all collected volumes to a reference volume chosen from the same experimental run
- Slice-timing correction
one of the operations optionally performed during preprocessing of fMRI data. It consists in interpolating and resampling the data so that the signal in the whole brain volume corresponds to the exact same time point. This step is often skipped with faster sequences (less than 2s repetition time)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller MB, Van Horn JD. Individual variability in brain activations associated with episodic retrieval: a role for large-scale databases. Int. J. Psychophysiol. 2007;63:205–213. doi: 10.1016/j.ijpsycho.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 2.Van Horn JD, et al. Individual Variability in Brain Activity: A Nuisance or an Opportunity? Brain Imaging Behav. 2008;2:327–334. doi: 10.1007/s11682-008-9049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirilina E, et al. The quest for the best: The impact of different EPI sequences on the sensitivity of random effect fMRI group analyses. Neuroimage. 2016;126:49–59. doi: 10.1016/j.neuroimage.2015.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dosenbach NUF, et al. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geerligs L, et al. A Brain-Wide Study of Age-Related Changes in Functional Connectivity. Cereb. Cortex. 2015;25:1987–1999. doi: 10.1093/cercor/bhu012. [DOI] [PubMed] [Google Scholar]
- 6.Yarkoni T. Neurobiological substrates of personality: A critical overview. In: Mikulincer, Mario Shaver, Phillip R, Cooper M, Lynne Larsen, Randy J., editors. Personality processes and individual differences. Vol. 4. American Psychological Association; 2015. [Google Scholar]
- 7.van den Heuvel MP, et al. Efficiency of functional brain networks and intellectual performance. J. Neurosci. 2009;29:7619–7624. doi: 10.1523/JNEUROSCI.1443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn ES, et al. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci. 2015;18:1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith SM, et al. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat. Neurosci. 2015;18:1565–1567. doi: 10.1038/nn.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy SE, et al. The effect of the serotonin transporter polymorphism (5-HTTLPR) on amygdala function: a meta-analysis. Mol. Psychiatry. 2013;18:512–520. doi: 10.1038/mp.2012.19. [DOI] [PubMed] [Google Scholar]
- 11.Arbabshirani MR, et al. Classification of schizophrenia patients based on resting-state functional network connectivity. Front. Neurosci. 2013;7:133. doi: 10.3389/fnins.2013.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yahata N, et al. A Small Number of Abnormal Brain Connections Predicts Adult Autism Spectrum Disorder. Nature Communications. 2016 doi: 10.1038/ncomms11254. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orrù G, et al. Using Support Vector Machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci. Biobehav. Rev. 2012;36:1140–1152. doi: 10.1016/j.neubiorev.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Wolfers T, et al. From estimating activation locality to predicting disorder: A review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neurosci. Biobehav. Rev. 2015;57:328–349. doi: 10.1016/j.neubiorev.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Williams LM, et al. Amygdala Reactivity to Emotional Faces in the Prediction of General and Medication-Specific Responses to Antidepressant Treatment in the Randomized iSPOT-D Trial. Neuropsychopharmacology. 2015;40:2398–2408. doi: 10.1038/npp.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whelan R, et al. Neuropsychosocial profiles of current and future adolescent alcohol misusers. Nature. 2014;512:185–189. doi: 10.1038/nature13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabrieli JDE, et al. Prediction as a humanitarian and pragmatic contribution from human cognitive neuroscience. Neuron. 2015;85:11–26. doi: 10.1016/j.neuron.2014.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laird AR, et al. Comparison of the disparity between Talairach and MNI coordinates in functional neuroimaging data: validation of the Lancaster transform. Neuroimage. 2010;51:677–683. doi: 10.1016/j.neuroimage.2010.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein A, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischl B, et al. II: Inflation, Flattening, and a Surface-Based Coordinate System. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 21.Yeo BTT, et al. Spherical Demons: Fast Diffeomorphic Landmark-Free Surface Registration. IEEE Trans. Med. Imaging. 2010;29:650–668. doi: 10.1109/TMI.2009.2030797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein A, et al. Evaluation of volume-based and surface-based brain image registration methods. Neuroimage. 2010;51:214–220. doi: 10.1016/j.neuroimage.2010.01.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frost MA, Goebel R. Measuring structural-functional correspondence: spatial variability of specialised brain regions after macro-anatomical alignment. Neuroimage. 2012;59:1369–1381. doi: 10.1016/j.neuroimage.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 24.Langers DRM. Assessment of tonotopically organised subdivisions in human auditory cortex using volumetric and surface-based cortical alignments. Hum. Brain Mapp. 2014;35:1544–1561. doi: 10.1002/hbm.22272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glasser MF, et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasson U, et al. Reliability of cortical activity during natural stimulation. Trends Cogn. Sci. 2010;14:40–48. doi: 10.1016/j.tics.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabuncu MR, et al. Function-based Intersubject Alignment of Human Cortical Anatomy. Cereb. Cortex. 2010;20:130–140. doi: 10.1093/cercor/bhp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conroy BR, et al. Inter-subject alignment of human cortical anatomy using functional connectivity. Neuroimage. 2013;81:400–411. doi: 10.1016/j.neuroimage.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khullar S, et al. ICA-fNORM: Spatial Normalization of fMRI Data Using Intrinsic Group-ICA Networks. Front. Syst. Neurosci. 2011;5:93. doi: 10.3389/fnsys.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Çetin MS, et al. Enhanced disease characterization through multi network functional normalization in fMRI. Front. Neurosci. 2015;9:95. doi: 10.3389/fnins.2015.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson EC, et al. MSM: a new flexible framework for Multimodal Surface Matching. Neuroimage. 2014;100:414–426. doi: 10.1016/j.neuroimage.2014.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J. Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdollahi RO, et al. Correspondences between retinotopic areas and myelin maps in human visual cortex. Neuroimage. 2014;99:509–524. doi: 10.1016/j.neuroimage.2014.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haxby JV, et al. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron. 2011;72:404–416. doi: 10.1016/j.neuron.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guntupalli JS, et al. A Model of Representational Spaces in Human Cortex. Cereb. Cortex. 2016 doi: 10.1093/cercor/bhw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saxe R, et al. Divide and conquer: a defense of functional localizers. Neuroimage. 2006;30:1088–1096. doi: 10.1016/j.neuroimage.2005.12.062. discussion 1097–9. [DOI] [PubMed] [Google Scholar]
- 37.Gorgolewski KJ, et al. Adaptive thresholding for reliable topological inference in single subject fMRI analysis. Front. Hum. Neurosci. 2012;6:245. doi: 10.3389/fnhum.2012.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith SM, Nichols TE. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44:83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- 39.Nieto-Castañón A, Fedorenko E. Subject-specific functional localizers increase sensitivity and functional resolution of multi-subject analyses. Neuroimage. 2012;63:1646–1669. doi: 10.1016/j.neuroimage.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanwisher N, Yovel G. The fusiform face area: a cortical region specialized for the perception of faces. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:2109–2128. doi: 10.1098/rstb.2006.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxe R, Powell LJ. It’s the thought that counts: specific brain regions for one component of theory of mind. Psychol. Sci. 2006;17:692–699. doi: 10.1111/j.1467-9280.2006.01768.x. [DOI] [PubMed] [Google Scholar]
- 42.Spunt RP, Adolphs R. Validating the Why/How contrast for functional MRI studies of Theory of Mind. Neuroimage. 2014;99:301–311. doi: 10.1016/j.neuroimage.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friston KJ, et al. A critique of functional localisers. Neuroimage. 2006;30:1077–1087. doi: 10.1016/j.neuroimage.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Handwerker DA, et al. The continuing challenge of understanding and modeling hemodynamic variation in fMRI. Neuroimage. 2012;62:1017–1023. doi: 10.1016/j.neuroimage.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu TT. Neurovascular factors in resting-state functional MRI. Neuroimage. 2013;80:339–348. doi: 10.1016/j.neuroimage.2013.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandettini PA. Neuronal or hemodynamic? Grappling with the functional MRI signal. Brain Connect. 2014;4:487–498. doi: 10.1089/brain.2014.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shan ZY, et al. Genes influence the amplitude and timing of brain hemodynamic responses. Neuroimage. 2016;124:663–671. doi: 10.1016/j.neuroimage.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buxton RB, et al. Modeling the hemodynamic response to brain activation. Neuroimage. 2004;(23 Suppl 1):S220–S233. doi: 10.1016/j.neuroimage.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Stephan KE, et al. Comparing hemodynamic models with DCM. Neuroimage. 2007;38:387–401. doi: 10.1016/j.neuroimage.2007.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D’Esposito M, et al. Alterations in the BOLD fMRI signal with ageing and disease: a challenge for neuroimaging. Nat. Rev. Neurosci. 2003;4:863–872. doi: 10.1038/nrn1246. [DOI] [PubMed] [Google Scholar]
- 51.Lindquist MA, et al. Modeling the hemodynamic response function in fMRI: efficiency, bias and mis-modeling. Neuroimage. 2009;45:S187–S198. doi: 10.1016/j.neuroimage.2008.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henson RNA, et al. Detecting latency differences in event-related BOLD responses: application to words versus nonwords and initial versus repeated face presentations. Neuroimage. 2002;15:83–97. doi: 10.1006/nimg.2001.0940. [DOI] [PubMed] [Google Scholar]
- 53.Woolrich MW, et al. Constrained linear basis sets for HRF modelling using Variational Bayes. Neuroimage. 2004;21:1748–1761. doi: 10.1016/j.neuroimage.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 54.Glover GH. Deconvolution of Impulse Response in Event-Related BOLD fMRI1. Neuroimage. 1999;9:416–429. doi: 10.1006/nimg.1998.0419. [DOI] [PubMed] [Google Scholar]
- 55.Calhoun VD, et al. fMRI analysis with the general linear model: removal of latency-induced amplitude bias by incorporation of hemodynamic derivative terms. Neuroimage. 2004;22:252–257. doi: 10.1016/j.neuroimage.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 56.Degras D, Lindquist MA. A hierarchical model for simultaneous detection and estimation in multi-subject fMRI studies. Neuroimage. 2014;98:61–72. doi: 10.1016/j.neuroimage.2014.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pedregosa F, et al. Data-driven HRF estimation for encoding and decoding models. Neuroimage. 2015;104:209–220. doi: 10.1016/j.neuroimage.2014.09.060. [DOI] [PubMed] [Google Scholar]
- 58.Chen JE, Glover GH. BOLD fractional contribution to resting-state functional connectivity above 0.1 Hz. Neuroimage. 2015;107:207–218. doi: 10.1016/j.neuroimage.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu G-R, et al. A blind deconvolution approach to recover effective connectivity brain networks from resting state fMRI data. Med. Image Anal. 2013;17:365–374. doi: 10.1016/j.media.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 60.Sreenivasan KR, et al. Nonparametric hemodynamic deconvolution of FMRI using homomorphic filtering. IEEE Trans. Med. Imaging. 2015;34:1155–1163. doi: 10.1109/TMI.2014.2379914. [DOI] [PubMed] [Google Scholar]
- 61.Griffeth VEM, Buxton RB. A theoretical framework for estimating cerebral oxygen metabolism changes using the calibrated-BOLD method: modeling the effects of blood volume distribution, hematocrit, oxygen extraction fraction, and tissue signal properties on the BOLD signal. Neuroimage. 2011;58:198–212. doi: 10.1016/j.neuroimage.2011.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu TT, et al. An introduction to normalization and calibration methods in functional MRI. Psychometrika. 2013;78:308–321. doi: 10.1007/s11336-012-9309-x. [DOI] [PubMed] [Google Scholar]
- 63.Pike GB. Quantitative functional MRI: concepts, issues and future challenges. Neuroimage. 2012;62:1234–1240. doi: 10.1016/j.neuroimage.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 64.Blockley NP, et al. Calibrating the BOLD response without administering gases: comparison of hypercapnia calibration with calibration using an asymmetric spin echo. Neuroimage. 2015;104:423–429. doi: 10.1016/j.neuroimage.2014.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blockley NP, et al. A review of calibrated blood oxygenation level-dependent (BOLD) methods for the measurement of task-induced changes in brain oxygen metabolism. NMR Biomed. 2013;26:987–1003. doi: 10.1002/nbm.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bandettini PA, Wong EC. A hypercapnia-based normalization method for improved spatial localization of human brain activation with fMRI. NMR Biomed. 1997;10:197–203. doi: 10.1002/(sici)1099-1492(199706/08)10:4/5<197::aid-nbm466>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 67.Handwerker DA, et al. Reducing vascular variability of fMRI data across aging populations using a breathholding task. Hum. Brain Mapp. 2007;28:846–859. doi: 10.1002/hbm.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu H, et al. Improving fMRI sensitivity by normalization of basal physiologic state. Hum. Brain Mapp. 2010;31:80–87. doi: 10.1002/hbm.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kannurpatti SS, Biswal BB. Detection and scaling of task-induced fMRI-BOLD response using resting state fluctuations. Neuroimage. 2008;40:1567–1574. doi: 10.1016/j.neuroimage.2007.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalcher K, et al. RESCALE: Voxel-specific task-fMRI scaling using resting state fluctuation amplitude. Neuroimage. 2013;70:80–88. doi: 10.1016/j.neuroimage.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Golestani AM, et al. Mapping the end-tidal CO2 response function in the resting-state BOLD fMRI signal: Spatial specificity, test-retest reliability and effect of fMRI sampling rate. Neuroimage. 2015;104:266–277. doi: 10.1016/j.neuroimage.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 72.Fair DA, et al. A method for using blocked and event-related fMRI data to study “resting state” functional connectivity. Neuroimage. 2007;35:396–405. doi: 10.1016/j.neuroimage.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kazan SM, et al. Vascular autorescaling of fMRI (VasA fMRI) improves sensitivity of population studies: A pilot study. Neuroimage. 2016;124:794–805. doi: 10.1016/j.neuroimage.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsvetanov KA, et al. The effect of ageing on fMRI: Correction for the confounding effects of vascular reactivity evaluated by joint fMRI and MEG in 335 adults. Hum. Brain Mapp. 2015;36:2248–2269. doi: 10.1002/hbm.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bennett CM, Miller MB. How reliable are the results from functional magnetic resonance imaging? Ann. N. Y. Acad. Sci. 2010;1191:133–155. doi: 10.1111/j.1749-6632.2010.05446.x. [DOI] [PubMed] [Google Scholar]
- 76.Zuo X-N, Xing X-X. Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: a systems neuroscience perspective. Neurosci. Biobehav. Rev. 2014;45:100–118. doi: 10.1016/j.neubiorev.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 77.Andellini M, et al. Test-retest reliability of graph metrics of resting state MRI functional brain networks: A review. J. Neurosci. Methods. 2015;253:183–192. doi: 10.1016/j.jneumeth.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 78.Barch DM, Mathalon DH. Using brain imaging measures in studies of procognitive pharmacologic agents in schizophrenia: psychometric and quality assurance considerations. Biol. Psychiatry. 2011;70:13–18. doi: 10.1016/j.biopsych.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bennett CM, Miller MB. fMRI reliability: influences of task and experimental design. Cogn. Affect. Behav. Neurosci. 2013;13:690–702. doi: 10.3758/s13415-013-0195-1. [DOI] [PubMed] [Google Scholar]
- 80.Churchill NW, et al. Optimizing preprocessing and analysis pipelines for single-subject fMRI: 2. Interactions with ICA, PCA, task contrast and inter-subject heterogeneity. PLoS One. 2012;7:e31147. doi: 10.1371/journal.pone.0031147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aurich NK, et al. Evaluating the reliability of different preprocessing steps to estimate graph theoretical measures in resting state fMRI data. Front. Neurosci. 2015;9:48. doi: 10.3389/fnins.2015.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raemaekers M, et al. Test-retest reliability of fMRI activation during prosaccades and antisaccades. Neuroimage. 2007;36:532–542. doi: 10.1016/j.neuroimage.2007.03.061. [DOI] [PubMed] [Google Scholar]
- 83.Caceres A, et al. Measuring fMRI reliability with the intra-class correlation coefficient. Neuroimage. 2009;45:758–768. doi: 10.1016/j.neuroimage.2008.12.035. [DOI] [PubMed] [Google Scholar]
- 84.Greve DN, et al. A survey of the sources of noise in fMRI. Psychometrika. 2013;78:396–416. doi: 10.1007/s11336-012-9294-0. [DOI] [PubMed] [Google Scholar]
- 85.Murphy K, et al. Resting-state fMRI confounds and cleanup. Neuroimage. 2013;80:349–359. doi: 10.1016/j.neuroimage.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Power JD, et al. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Power JD, et al. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fox MD, et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Satterthwaite TD, et al. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van Dijk KRA, et al. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pruim RHR, et al. Evaluation of ICA-AROMA and alternative strategies for motion artifact removal in resting state fMRI. Neuroimage. 2015;112:278–287. doi: 10.1016/j.neuroimage.2015.02.063. [DOI] [PubMed] [Google Scholar]
- 92.Power JD, et al. Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage. 2015;105:536–551. doi: 10.1016/j.neuroimage.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zeng L-L, et al. Neurobiological basis of head motion in brain imaging. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6058–6062. doi: 10.1073/pnas.1317424111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Geerligs L, et al. State and Trait Components of Functional Connectivity: Individual Differences Vary with Mental State. J. Neurosci. 2015;35:13949–13961. doi: 10.1523/JNEUROSCI.1324-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tyszka JM, et al. Largely typical patterns of resting-state functional connectivity in high-functioning adults with autism. Cereb. Cortex. 2014;24:1894–1905. doi: 10.1093/cercor/bht040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Satterthwaite TD, et al. Heterogeneous impact of motion on fundamental patterns of developmental changes in functional connectivity during youth. Neuroimage. 2013;83:45–57. doi: 10.1016/j.neuroimage.2013.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turner BO, et al. One dataset, many conclusions: BOLD variability’s complicated relationships with age and motion artifacts. Brain Imaging Behav. 2015;9:115–127. doi: 10.1007/s11682-014-9351-7. [DOI] [PubMed] [Google Scholar]
- 98.Raj D, et al. Respiratory effects in human functional magnetic resonance imaging due to bulk susceptibility changes. Phys. Med. Biol. 2001;46:3331–3340. doi: 10.1088/0031-9155/46/12/318. [DOI] [PubMed] [Google Scholar]
- 99.Wise RG, et al. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 100.Chang C, Glover GH. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. Neuroimage. 2009;47:1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dagli MS, et al. Localization of cardiac-induced signal change in fMRI. Neuroimage. 1999;9:407–415. doi: 10.1006/nimg.1998.0424. [DOI] [PubMed] [Google Scholar]
- 102.Birn RM, et al. The influence of physiological noise correction on test-retest reliability of resting-state functional connectivity. Brain Connect. 2014;4:511–522. doi: 10.1089/brain.2014.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lipp I, et al. Understanding the contribution of neural and physiological signal variation to the low repeatability of emotion-induced BOLD responses. Neuroimage. 2014;86:335–342. doi: 10.1016/j.neuroimage.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kassner A, et al. Blood-oxygen level dependent MRI measures of cerebrovascular reactivity using a controlled respiratory challenge: reproducibility and gender differences. J. Magn. Reson. Imaging. 2010;31:298–304. doi: 10.1002/jmri.22044. [DOI] [PubMed] [Google Scholar]
- 105.Särkkä S, et al. Dynamic retrospective filtering of physiological noise in BOLD fMRI: DRIFTER. Neuroimage. 2012;60:1517–1527. doi: 10.1016/j.neuroimage.2012.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kay K, et al. GLMdenoise: a fast, automated technique for denoising task-based fMRI data. Front. Neurosci. 2013;7 doi: 10.3389/fnins.2013.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Churchill NW, Strother SC. PHYCAA+: an optimized, adaptive procedure for measuring and controlling physiological noise in BOLD fMRI. Neuroimage. 2013;82:306–325. doi: 10.1016/j.neuroimage.2013.05.102. [DOI] [PubMed] [Google Scholar]
- 108.Salimi-Khorshidi G, et al. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anderson JS, et al. Network anticorrelations, global regression, and phase-shifted soft tissue correction. Hum. Brain Mapp. 2011;32:919–934. doi: 10.1002/hbm.21079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pruim RHR, et al. ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. Neuroimage. 2015;112:267–277. doi: 10.1016/j.neuroimage.2015.02.064. [DOI] [PubMed] [Google Scholar]
- 111.Kelly RE, Jr, et al. Visual inspection of independent components: defining a procedure for artifact removal from fMRI data. J. Neurosci. Methods. 2010;189:233–245. doi: 10.1016/j.jneumeth.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kundu P, et al. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage. 2012;60:1759–1770. doi: 10.1016/j.neuroimage.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kundu P, et al. Robust resting state fMRI processing for studies on typical brain development based on multi-echo EPI acquisition. Brain Imaging Behav. 2015;9:56–73. doi: 10.1007/s11682-014-9346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moeller S, et al. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn. Reson. Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Olafsson V, et al. Enhanced identification of BOLD-like components with multi-echo simultaneous multi-slice (MESMS) fMRI and multi-echo ICA. Neuroimage. 2015;112:43–51. doi: 10.1016/j.neuroimage.2015.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bright MG, Murphy K. Removing motion and physiological artifacts from intrinsic BOLD fluctuations using short echo data. Neuroimage. 2013;64:526–537. doi: 10.1016/j.neuroimage.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tong Y, Frederick BD. Studying the Spatial Distribution of Physiological Effects on BOLD Signals Using Ultrafast fMRI. Front. Hum. Neurosci. 2014;8:196. doi: 10.3389/fnhum.2014.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rayshubskiy A, et al. Direct, intraoperative observation of ~0.1 Hz hemodynamic oscillations in awake human cortex: implications for fMRI. Neuroimage. 2014;87:323–331. doi: 10.1016/j.neuroimage.2013.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.MacIntosh BJ, et al. Impact of a single bout of aerobic exercise on regional brain perfusion and activation responses in healthy young adults. PLoS One. 2014;9:e85163. doi: 10.1371/journal.pone.0085163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rajab AS, et al. A single session of exercise increases connectivity in sensorimotor-related brain networks: a resting-state fMRI study in young healthy adults. Front. Hum. Neurosci. 2014;8:625. doi: 10.3389/fnhum.2014.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Keulers EHH, et al. The association between cortisol and the BOLD response in male adolescents undergoing fMRI. Brain Res. 2015;1598:1–11. doi: 10.1016/j.brainres.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 122.Levin JM, et al. Influence of baseline hematocrit and hemodilution on BOLD fMRI activation. Magn. Reson. Imaging. 2001;19:1055–1062. doi: 10.1016/s0730-725x(01)00460-x. [DOI] [PubMed] [Google Scholar]
- 123.Yoo S-S, et al. The human emotional brain without sleep—a prefrontal amygdala disconnect. Curr. Biol. 2007;17:R877–R878. doi: 10.1016/j.cub.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 124.Tagliazucchi E, Laufs H. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron. 2014;82:695–708. doi: 10.1016/j.neuron.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 125.Dreher J-C, et al. Menstrual cycle phase modulates reward-related neural function in women. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2465–2470. doi: 10.1073/pnas.0605569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Poldrack RA, et al. Long-term neural and physiological phenotyping of a single human. Nat. Commun. 2015;6 doi: 10.1038/ncomms9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mulderink TA, et al. On the use of caffeine as a contrast booster for BOLD fMRI studies. Neuroimage. 2002;15:37–44. doi: 10.1006/nimg.2001.0973. [DOI] [PubMed] [Google Scholar]
- 128.Laurienti PJ, et al. Relationship between Caffeine-Induced Changes in Resting Cerebral Perfusion and Blood Oxygenation Level-Dependent Signal. Am J Neuroradiol. 2003;24:1607–1611. [PMC free article] [PubMed] [Google Scholar]
- 129.Addicott MA, et al. The Effects of Dietary Caffeine Use and Abstention on Blood Oxygen Level-Dependent Activation and Cerebral Blood Flow. Journal of caffeine research. 2012;2 doi: 10.1089/jcr.2011.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu TT, Liau J. Caffeine increases the linearity of the visual BOLD response. Neuroimage. 2010;49:2311–2317. doi: 10.1016/j.neuroimage.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koppelstaetter F, et al. Caffeine and cognition in functional magnetic resonance imaging. J. Alzheimers. Dis. 2010;(20 Suppl 1):S71–S84. doi: 10.3233/JAD-2010-1417. [DOI] [PubMed] [Google Scholar]
- 132.Wang DJJ, et al. Potentials and challenges for arterial spin labeling in pharmacological magnetic resonance imaging. J. Pharmacol. Exp. Ther. 2011;337:359–366. doi: 10.1124/jpet.110.172577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Whelan R, Garavan H. When Optimism Hurts: Inflated Predictions in Psychiatric Neuroimaging. Biol. Psychiatry. 2014;75:746–748. doi: 10.1016/j.biopsych.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 134.Lo A, et al. Why significant variables aren’t automatically good predictors. Proc. Natl. Acad. Sci. U. S. A. 2015;112:13892–13897. doi: 10.1073/pnas.1518285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Linden DEJ. The challenges and promise of neuroimaging in psychiatry. Neuron. 2012;73:8–22. doi: 10.1016/j.neuron.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 136.Yarkoni T, Westfall J. Choosing prediction over explanation in psychology: Lessons from machine learning. figshare. 2016 doi: 10.1177/1745691617693393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yarkoni T, Braver T. Cognitive neurosciences approaches to individual differences in working memory and executive control: conceptual and methodological issues. In: Gruszka A, editor. Handbook of individual differences in cognition: attention, memory and executive control. pp. 87–107. [Google Scholar]
- 138.Button KS, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013;14:365–376. doi: 10.1038/nrn3475. [DOI] [PubMed] [Google Scholar]
- 139.Schönbrodt FD, Perugini M. At what sample size do correlations stabilize? J. Res. Pers. 2013;47:609–612. [Google Scholar]
- 140.Silva RF, et al. The tenth annual MLSP competition: Schizophrenia classification challenge. Machine Learning for Signal Processing (MLSP), 2014 IEEE International Workshop on. 2014:1–6. [Google Scholar]
- 141.Ioannidis JPA. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nosek BA, Lakens D. Registered Reports. Soc. Psychol. 2014;45:137–141. [Google Scholar]
- 143.Siontis KCM, et al. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur. J. Hum. Genet. 2010;18:832–837. doi: 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Van Horn JD, Gazzaniga MS. Why share data? Lessons learned from the fMRIDC. Neuroimage. 2013;82:677–682. doi: 10.1016/j.neuroimage.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Biswal BB, et al. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Eickhoff S, et al. Sharing the wealth: Neuroimaging data repositories. Neuroimage. 2016;124:1065–1068. doi: 10.1016/j.neuroimage.2015.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Poldrack RA, Gorgolewski KJ. OpenfMRI: Open sharing of task fMRI data. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schumann G, et al. The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol. Psychiatry. 2010;15:1128–1139. doi: 10.1038/mp.2010.4. [DOI] [PubMed] [Google Scholar]
- 149.Van Essen DC, et al. The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Taylor JR, et al. The Cambridge Centre for Ageing and Neuroscience (Cam-CAN) data repository: Structural and functional MRI, MEG, and cognitive data from a cross-sectional adult lifespan sample. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Crawford JR, Garthwaite PH. On the “optimal” size for normative samples in neuropsychology: capturing the uncertainty when normative data are used to quantify the standing of a neuropsychological test score. Child Neuropsychol. 2008;14:99–117. doi: 10.1080/09297040801894709. [DOI] [PubMed] [Google Scholar]
- 152.Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126. doi: 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Insel TR, Cuthbert BN. Medicine. Brain disorders? Precisely. Science. 2015;348:499–500. doi: 10.1126/science.aab2358. [DOI] [PubMed] [Google Scholar]
- 154.Di X, et al. Calibrating BOLD fMRI activations with neurovascular and anatomical constraints. Cereb. Cortex. 2013;23:255–263. doi: 10.1093/cercor/bhs001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Power JD, et al. Studying brain organization via spontaneous fMRI signal. Neuron. 2014;84:681–696. doi: 10.1016/j.neuron.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Biswal B, et al. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 157.Smith SM, et al. Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Birn RM, et al. The effect of scan length on the reliability of resting-state fMRI connectivity estimates. Neuroimage. 2013;83:550–558. doi: 10.1016/j.neuroimage.2013.05.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Laumann TO, et al. Functional System and Areal Organization of a Highly Sampled Individual Human Brain. Neuron. 2015;87:657–670. doi: 10.1016/j.neuron.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Patriat R, et al. The effect of resting condition on resting-state fMRI reliability and consistency: a comparison between resting with eyes open, closed, and fixated. Neuroimage. 2013;78:463–473. doi: 10.1016/j.neuroimage.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Christoff K, et al. Experience sampling during fMRI reveals default network and executive system contributions to mind wandering. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8719–8724. doi: 10.1073/pnas.0900234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Delamillieure P, et al. The resting state questionnaire: An introspective questionnaire for evaluation of inner experience during the conscious resting state. Brain Res. Bull. 2010;81:565–573. doi: 10.1016/j.brainresbull.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 163.Hurlburt RT, et al. What goes on in the resting-state? A qualitative glimpse into resting-state experience in the scanner. Front. Psychol. 2015;6:1535. doi: 10.3389/fpsyg.2015.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tung K-C, et al. Alterations in resting functional connectivity due to recent motor task. Neuroimage. 2013;78:316–324. doi: 10.1016/j.neuroimage.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hutchison RM, et al. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cole MW, et al. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83:238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Smith SM, et al. Resting-state fMRI in the Human Connectome Project. Neuroimage. 2013;80:144–168. doi: 10.1016/j.neuroimage.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Beckmann CF, et al. Investigations into resting-state connectivity using independent component analysis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yeo BTT, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Craddock RC, et al. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 2012;33:1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Blumensath T, et al. Spatially constrained hierarchical parcellation of the brain with resting-state fMRI. Neuroimage. 2013;76:313–324. doi: 10.1016/j.neuroimage.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wig GS, et al. Parcellating an individual subject’s cortical and subcortical brain structures using snowball sampling of resting-state correlations. Cereb. Cortex. 2014;24:2036–2054. doi: 10.1093/cercor/bht056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Arslan S, et al. Joint Spectral Decomposition for the Parcellation of the Human Cerebral Cortex Using Resting-State fMRI. Inf. Process. Med. Imaging. 2015;24:85–97. doi: 10.1007/978-3-319-19992-4_7. [DOI] [PubMed] [Google Scholar]
- 174.Wig GS, et al. An approach for parcellating human cortical areas using resting-state correlations. Neuroimage. 2014;93(Pt 2):276–291. doi: 10.1016/j.neuroimage.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gordon EM, et al. Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb. Cortex. 2014 doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Beckmann CF, et al. Group comparison of resting-state FMRI data using multi-subject ICA and dual regression. Neuroimage. 2009;47:S148. [Google Scholar]
- 177.Hacker CD, et al. Resting state network estimation in individual subjects. Neuroimage. 2013;82:616–633. doi: 10.1016/j.neuroimage.2013.05.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang D, et al. Parcellating cortical functional networks in individuals. Nat. Neurosci. 2015;18:1853–1860. doi: 10.1038/nn.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Carp J. On the plurality of (methodological) worlds: estimating the analytic flexibility of FMRI experiments. Front. Neurosci. 2012;6:149. doi: 10.3389/fnins.2012.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Neuroskeptic. The Nine Circles of Scientific Hell. Perspect. Psychol. Sci. 2012;7:643–644. doi: 10.1177/1745691612459519. [DOI] [PubMed] [Google Scholar]
- 181.Simmons JP, et al. False-Positive Psychology: Undisclosed Flexibility in Data Collection and Analysis Allows Presenting Anything as Significant. Psychol. Sci. 2011;22:1359–1366. doi: 10.1177/0956797611417632. [DOI] [PubMed] [Google Scholar]
- 182.Simonsohn U, et al. P-curve: a key to the file-drawer. J. Exp. Psychol. Gen. 2014;143:534–547. doi: 10.1037/a0033242. [DOI] [PubMed] [Google Scholar]
- 183.Pernet C, Poline J-B. Improving functional magnetic resonance imaging reproducibility. Gigascience. 2015;4:15. doi: 10.1186/s13742-015-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Peng RD. Reproducible Research in Computational Science. Science. 2011;334:1226–1227. doi: 10.1126/science.1213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Drummond C. Proceedings of the Evaluation Methods for Machine Learning Workshop 26th International Conference for Machine Learning. Montreal: 2009. Replicability is not reproducibility: nor is it good science; p. 4. [Google Scholar]
- 186.Poldrack RA, et al. Guidelines for reporting an fMRI study. Neuroimage. 2008;40:409–414. doi: 10.1016/j.neuroimage.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Regev M, et al. Selective and invariant neural responses to spoken and written narratives. J. Neurosci. 2013;33:15978–15988. doi: 10.1523/JNEUROSCI.1580-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hanke M, et al. A high-resolution 7-Tesla fMRI dataset from complex natural stimulation with an audio movie. Sci. Data. 2014;1 doi: 10.1038/sdata.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Labs A, et al. Portrayed emotions in the movie “Forrest Gump.”. F1000Res. 2015;4:92. doi: 10.12688/f1000research.6230.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hasson U, et al. Intersubject synchronization of cortical activity during natural vision. Science. 2004;303:1634–1640. doi: 10.1126/science.1089506. [DOI] [PubMed] [Google Scholar]
- 191.Kriegeskorte N, Kievit RA. Representational geometry: integrating cognition, computation, and the brain. Trends Cogn. Sci. 2013;17:401–412. doi: 10.1016/j.tics.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nili H, et al. A toolbox for representational similarity analysis. PLoS Comput. Biol. 2014;10:e1003553. doi: 10.1371/journal.pcbi.1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Skerry AE, Saxe R. Neural representations of emotion are organized around abstract event features. Curr. Biol. 2015;25:1945–1954. doi: 10.1016/j.cub.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tamir DI, et al. Neural evidence that three dimensions organize mental state representation: Rationality, social impact, and valence. Proc. Natl. Acad. Sci. U. S. A. 2015 doi: 10.1073/pnas.1511905112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Charest I, et al. Unique semantic space in the brain of each beholder predicts perceived similarity. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14565–14570. doi: 10.1073/pnas.1402594111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Charest I, Kriegeskorte N. The brain of the beholder: honouring individual representational idiosyncrasies. Language, Cognition and Neuroscience. 2015;30:367–379. [Google Scholar]
- 197.Naselaris T, et al. Encoding and decoding in fMRI. Neuroimage. 2011;56:400–410. doi: 10.1016/j.neuroimage.2010.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Huth AG, et al. A continuous semantic space describes the representation of thousands of object and action categories across the human brain. Neuron. 2012;76:1210–1224. doi: 10.1016/j.neuron.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Huth AG, et al. Natural speech reveals the semantic maps that tile human cerebral cortex. Nature. 2016 doi: 10.1038/nature17637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Glasser MF, et al. A Multi-modal parcellation of human cerebral cortex. Nature. doi: 10.1038/nature18933. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.