ABSTRACT
During 2014, a subclade 2.3.4.4 highly pathogenic avian influenza (HPAI) A(H5N8) virus caused poultry outbreaks around the world. In late 2014/early 2015, the virus was detected in wild birds in Canada and the United States, and these viruses also gave rise to reassortant progeny, composed of viral RNA segments (vRNAs) from both Eurasian and North American lineages. In particular, viruses were found with N1, N2, and N8 neuraminidase vRNAs, and these are collectively referred to as H5Nx viruses. In the United States, more than 48 million domestic birds have been affected. Here we present a detailed structural and biochemical analysis of the surface antigens of H5N1, H5N2, and H5N8 viruses in addition to those of a recent human H5N6 virus. Our results with recombinant hemagglutinin reveal that these viruses have a strict avian receptor binding preference, while recombinantly expressed neuraminidases are sensitive to FDA-approved and investigational antivirals. Although H5Nx viruses currently pose a low risk to humans, it is important to maintain surveillance of these circulating viruses and to continually assess future changes that may increase their pandemic potential.
IMPORTANCE The H5Nx viruses emerging in North America, Europe, and Asia pose a great public health concern. Here we report a molecular and structural study of the major surface proteins of several H5Nx influenza viruses. Our results improve the understanding of these new viruses and provide important information on their receptor preferences and susceptibilities to antivirals, which are central to pandemic risk assessment.
INTRODUCTION
Influenza A viruses are encoded by eight segments of negative-sense RNA (vRNAs), which enable rapid evolution via an error-prone RNA-dependent RNA polymerase and gene transfer by reassortment of vRNAs during coinfections. Human infections with zoonotic influenza A virus subtypes continue to be a global concern. A highly pathogenic avian influenza (HPAI) A(H5N1) virus caused its first human infection in Hong Kong in 1997 (1). To date, more than 800 human cases in 16 countries, with an overall fatality rate of 53%, have been reported since 2003 (2). The H5 hemagglutinin (HA) vRNA continues to evolve into diverse clades and subclades (3, 4). In early 2014, a novel subtype of HPAI A(H5N8) virus of subclade 2.3.4.4 caused poultry outbreaks in South Korea and subsequently spread to China, Japan, the Russian Federation, and Europe. Another novel HPAI A(H5N6) virus subtype of the same H5 subclade caused multiple outbreaks in Southeast Asia and resulted in one fatal human infection in China in April 2014 (5).
At the end of 2014, commercial turkey farms in southern British Columbia, Canada, reported increased mortality in their flocks. Subsequent investigation revealed the presence of an HPAI A(H5N2) virus containing five Eurasian-lineage vRNA segments of A(H5N8) origin and three vRNA segments from North American-lineage viruses (6). During this time, U.S. authorities also detected an HPAI A(H5N2) virus (northern pintail/Washington/40964/2014) with the same vRNA constellation as the Canadian virus, an HPAI A(H5N8) virus with all vRNAs of Eurasian lineage (gyrfalcon/Washington/41088-6/2014), and an A(H5N1) virus (American green-winged teal/Washington/195750/2014) composed of four Eurasian- and four North American-lineage vRNAs (including a North American neuraminidase [NA] vRNA) (7). These virus reassortants are collectively referred to as H5Nx viruses and have dispersed throughout the Pacific, Central, and Mississippi flyways. As of September 2015, the U.S. Department of Agriculture (USDA) reported 219 detections across 15 states, affecting more than 48 million domestic birds (www.aphis.usda.gov). The fast movement of the HPAI H5N8 virus across Eurasia and North America, and its ability to reassort with circulating viruses, generates serious concerns among the poultry industry and the public health community.
Two surface proteins of the influenza A virus, hemagglutinin and neuraminidase, play essential roles during virus entry into host cells and release from those cells (8). Influenza A viruses attach to cells through HA binding to terminal sialic acids of glycoproteins on the surfaces of respiratory epithelial cells. The host range of influenza A viruses is dictated mainly by their affinity for different sialosides; avian viruses preferentially bind to sialic acid linked to galactose via an α2-3 linkage, and human viruses preferentially bind to sialic acid linked to galactose via an α2-6 linkage (9, 10). NA catalyzes the hydrolysis of terminal sialic acid residues from cell receptors and from newly formed virions and therefore helps release the virus from cells for the spread of infection, as well as prevention of the aggregation of virus particles (11). The indispensable role of NA during the virus life cycle also makes it an important target for antiviral therapeutics (12). In order to understand the molecular characteristics of the HPAI H5Nx viruses and their pandemic potential, the HA and NA proteins from A/Sichuan/26221/2014 (H5N6) (5), A/gyrfalcon/Washington/41088-6/2014 (H5N8), A/northern pintail/Washington/40964/2014 (H5N2) (6), and A/American green-winged teal/Washington/195750/2014 (H5N1) (7) were structurally and functionally analyzed. Specifically, we expressed HA and NA proteins from these H5Nx viruses and determined their 3-dimensional atomic structures by X-ray crystallography. In addition, the receptor binding of these recombinant HAs (RecHAs) was analyzed by both glycan microarray and biolayer interferometry (BLI). Finally, the enzymatic activities of the N8, N2, and N1 NAs, as well as their sensitivities to neuraminidase inhibitors (NAIs), were tested. Changes observed in the HAs close to the receptor binding site (RBS) in recent strains were also studied. While our results indicate that these H5Nx viruses currently pose a low risk to humans, it is important to carry out surveillance and pandemic risk assessment of continually evolving H5Nx viruses.
MATERIALS AND METHODS
Cloning and expression of recombinant HAs and NAs.
The cDNAs for the ectodomains of the H5Nx HA (residues 1 to 503 in mature protein numbering) and NA (residues 80 to 470) were synthesized (GenScript USA Inc.) as codon-optimized genes for insect cell expression and were subcloned into the baculovirus transfer vector pAcGP67B (BD Biosciences). To aid in purification, the HA had an additional C-terminal thrombin site, followed by a foldon trimerization sequence from bacteriophage T4 fibritin and a His tag, incorporated into the final construct (13). The recombinant NA (RecNA) protein contained an N-terminal His tag, a tetramerization domain from the human vasodilator-stimulated phosphoprotein (14), and a thrombin cleavage site (15). Both secreted proteins were recovered from the culture supernatant and were purified by metal affinity chromatography and size exclusion chromatography (SEC). For structural analyses, proteins were further subjected to trypsin cleavage and were repurified by SEC. Trypsin-treated RecHA and RecNA were buffer exchanged into 10 mM Tris-HCl–50 mM NaCl (pH 8.0) and 10 mM Tris-HCl–50 mM NaCl–1 mM CaCl2 (pH 8.0), respectively, and were concentrated for crystallization trials.
Crystallization and data collection.
Initial crystallization trials were set up using Oryx4, a crystallization robot for the sitting drop procedure (Douglas Instruments Ltd., Berkshire, United Kingdom). The conditions under which crystals were observed were optimized at 20°C using a modified method for microbatch screening under oil (16). All crystals were flash-cooled at 100 K, and data sets were collected and were processed with the DENZO-SCALEPACK suite (17). More-specific information for each target is included in Table 1.
TABLE 1.
Data collection and refinement statistics for the H5Nx crystal structures
| Condition or parameter | Setup or value for the following influenza virus (PDB code) protein: |
|||||
|---|---|---|---|---|---|---|
| A/Sichuan/26221/2014 (5HU8) H5 HA | A/gyrfalcon/Washington/41088-6/2014 (5HUF) H5 HA | A/American green-winged teal/Washington/195750/2014 (5HUG) N1 NA | A/Northern pintail/Washington/40964/2014 (5HUK) N2 NA | A/Sichuan/26221/2014 (5HUM) N6 NA | A/gyrfalcon/Washington/41088-6/2014 (5HUN) N8 NA | |
| Exptl conditions | ||||||
| Protein concn (mg/ml) | 18 | 14 | 20 | 16 | 12 | 15 |
| Crystallization conditionsa | 0.1 M imidazole (pH 7), 22.5% PEG (cutoff, 0.3 to 10 kDa; Polypure) | 0.1 M sodium citrate–citric acid (pH 5.5), 20% (wt/vol) PEG 3000 | 0.2 M magnesium chloride, 0.1 M HEPES–NaOH (pH 7.5), 30% (vol/vol) PPG P400 | 0.1 M sodium citrate–citric acid (pH 5.5), 40% PEG (vol/vol) 600 | 0.2 M sodium acetate, 0.1 M Tris-HCl (pH 8.5), 16% (wt/vol) PEG 4000 | 0.16 M calcium acetate, 0.08 M sodium cacodylate–HCl (pH 6.5), 14.4% (wt/vol) PEG 8000, 20% (vol/vol) glycerol |
| Cryoprotectant | None | None | None | 20% PEG 200 | 20% glycerol | None |
| Data collection statisticsb | ||||||
| Beamline collected | APS, 22-ID | APS, 22-BM | APS, 22-ID | APS, 22-ID | APS, 22-ID | APS, 22-ID |
| Space group | P212121 | P212121 | P4212 | P212121 | P21 | I4 |
| Cell dimensions (Å) | 89.51, 104.42, 215.69 | 125.33, 252.39, 70.62 | 91.64, 91.64, 108.97 | 116.23, 122.79, 176.85 | 117.34, 76.28, 117.21 | 90.65, 90.65, 110.81 |
| Cell angle (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90.2, 90 | 90, 90, 90 |
| Resolution range (Å) | 50–2.45 (2.54–2.45) | 50–2.81 (2.91–2.81) | 50–1.85 (1.92–1.85) | 50–2.45 (2.54–2.45) | 50–1.60 (1.66–1.60) | 50–2.30 (2.38–2.30) |
| Rsym (%) | 10.9 (87.0) | 11.7 (71.9) | 11.7 (45.7) | 12.6 (64.9) | 9.2 (36.5) | 7.5 (17.0) |
| I/σ | 25.8 (2.0) | 12.3 (1.6) | 27.9 (5.4) | 24.9 (3.6) | 22 (3.3) | 47.8 (24.4) |
| Completeness (%) | 100.0 (100.0) | 96.5 (96.2) | 99.8 (100.0) | 98.3 (95.8) | 99.9 (100.0) | 99.8 (99.9) |
| Redundancy | 7.3 (6.7) | 5.2 (4.9) | 6.9 (5.8) | 6.2 (6.4) | 3.7 (3.7) | 6.3 (7.0) |
| Refinement | ||||||
| No. of reflections | ||||||
| Total | 74,675 | 53,618 | 40,235 | 91,882 | 272,309 | 19,878 |
| Test | 7,154 | 5,033 | 3,949 | 8,819 | 26,255 | 1,900 |
| Rwork/Rfree | 19.1/22.7 | 22.8/25.6 | 16.4/19.5 | 15.7/18.4 | 19.8/22.0 | 20.1/23.7 |
| No. of atoms | 12,524 | 12,219 | 3,232 | 12,932 | 13,870 | 3,110 |
| RMSD | ||||||
| Bond length (Å) | 0.014 | 0.012 | 0.021 | 0.018 | 0.018 | 0.013 |
| Bond angle (°) | 1.69 | 1.71 | 1.90 | 1.94 | 1.69 | 1.57 |
| MolProbity scores (%)c | ||||||
| Favored regions | 94 | 90 | 97 | 96 | 97 | 95 |
| Outliers | 0.4 | 1.6 | 0 | 0 | 0 | 0 |
PEG, polyethylene glycol; PPG, polypropylene glycol.
Numbers in parentheses refer to the highest-resolution shell.
See reference 27.
Structure determination and refinement.
The HA and NA structures were determined by molecular replacement with Phaser (18). For the HA, the influenza virus A/Anhui/5/2005 (H5N1) structure (PDB code 4MWQ) was used as a search model (19). The sequence for the model was then mutated to the correct sequence, rebuilt by Coot (20), and refined with REFMAC (21) using both transition/libration/screw (TLS) refinement (22) and Phenix.refine (23).
The N1, N2, N6 and N8 structures were solved using NA structures from the influenza A/Brevig Mission/1/1918 (H1N1) (PDB code 3B7E), A/RI/5+/1957 (H2N2) (PDB code 3TIA), A/chicken/Nanchang/7-010/2000 (H3N6) (PDB code 4QN4), and A/harbor seal/Massachusetts/1/2011 (H3N8) (PDB code 4WA3) viruses, respectively, as search models (15, 24–26). The NA structures were refined using the same strategy as the HA structures. All final models were assessed using MolProbity (27), and statistics for data processing and refinement are presented in Table 1.
Glycan binding analyses.
Glycan microarray printing and recombinant HA analyses have been described previously (13, 28–32). Table 2 lists the glycans used in these experiments as well as a tabulated qualitative assessment of binding for each protein analyzed. For kinetic studies, biotinylated receptor analogs, Neu5Ac(α2-3)Gal(β1-4)GlcNAc(β1-3)Gal(β1-4)GlcNAcb-biotin (3SLNLN-b) and Neu5Ac(α2-6)Gal(β1-4)GlcNAc(β1-3)Gal(β1-4)GlcNAcb-biotin (6SLNLN-b), were obtained from the Consortium for Functional Glycomics (www.functionalglycomics.org) through the resource request program. Glycans were precoupled to streptavidin-coated biosensors (ForteBio, Inc.), and the binding of recombinant HA, diluted to 5.4 μM trimer in kinetics buffer (phosphate-buffered saline [PBS] containing 0.02% Tween 20, 0.005% sodium azide, and 100 μg/ml bovine serum albumin), was analyzed by biolayer interferometry (BLI) using an Octet Red instrument (ForteBio, Inc.) according to the manufacturer's instructions. Data were analyzed using the system software and were fitted to a 1:1 binding model.
TABLE 2.
Glycan microarray for H5Nx HAs
| Glycan no. by category | Structurea | Bindingb |
||
|---|---|---|---|---|
| A/northern pintail/ Washington/ 40964/14 |
A/Sichuan/ 26221/14 |
A/gyrfalcon/ Washington/ 41088-6/14 |
||
| Sialic acid | ||||
| 1 | Neu5Acα | NB | NB | NB |
| 2 | Neu5Acα | NB | NB | NB |
| 3 | Neu5Acβ | NB | NB | NB |
| α2-3 sialosides | ||||
| 4 | Neu5Acα2-3(6-O-Su)Galβ1-4GlcNAcβ | +++ | +++ | +++ |
| 5 | Neu5Acα2-3Galβ1-3[6OSO3]GalNAcα | +++ | +++ | +++ |
| 6 | Neu5Acα2-3Galβ1-4[6OSO3]GlcNAcβ | +++ | +++ | +++ |
| 7 | Neu5Acα2-3Galβ1-4(Fucα1-3)[6OSO3]GlcNAcβ-propyl-NH2 | +++ | +++ | +++ |
| 8 | Neu5Acα2-3Galβ1-3[6OSO3]GlcNAc-propyl-NH2 | +++ | +++ | +++ |
| 9 | Neu5Acα2-3Galβ1-3(Neu5Acα2-3Galβ1-4)GlcNAcβ | +++ | +++ | +++ |
| 10 | Neu5Acα2-3Galβ1-3(Neu5Acα2-3Galβ1-4GlcNAcβ1-6)GalNAcα | +++ | +++ | +++ |
| 11 | Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | +++ | +++ | +++ |
| 12 | Neu5Acα(2-3)-Galβ(1-4)-GlcNAcβ(1-3)-Galβ(1-4)-GlcNAcβ(1-2)-Manα(1-3)-[Neu5Acα(2-3)-Galβ(1-4)-GlcNAcβ(1-3)-Galβ(1-4)-GlcNAcβ(1-2)-Manα(1-6)]-Manβ(1-4)-GlcNAcβ(1-4)-GlcNAcβ | +++ | +++ | +++ |
| 13 | Neu5Acα2-3Galβ | +++ | +++ | ++ |
| 14 | Neu5Acα2-3Galβ1-3GalNAcα | +++ | +++ | +++ |
| 15 | Neu5Acα2-3Galβ1-3GlcNAcβ | +++ | +++ | ++ |
| 16 | Neu5Acα2-3Galβ1-3GlcNAcβ | +++ | +++ | +++ |
| 17 | Neu5Acα2-3Galβ1-4Glcβ | +++ | +++ | +++ |
| 18 | Neu5Acα2-3Galβ1-4Glcβ | +++ | +++ | +++ |
| 19 | Neu5Acα2-3Galβ1-4GlcNAcβ | +++ | +++ | +++ |
| 20 | Neu5Acα2-3Galβ1-4GlcNAcβ | +++ | +++ | +++ |
| 21 | Neu5Acα2-3GalNAcβ1-4GlcNAcβ | NB | NB | ++ |
| 22 | Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | +++ | +++ | +++ |
| 23 | Neu5Aca2-3Galβ1-3GlcNAcβ1-3Galβ1-4GlcNAcβ | +++ | +++ | +++ |
| 24 | Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | +++ | +++ | +++ |
| 25 | Neu5Acα2-3Galβ1-4GlcNAcβ1-3Galβ1-3GlcNAcβ | +++ | +++ | +++ |
| 26 | Neu5Acα2-3Galβ1-3GalNAcα | +++ | +++ | +++ |
| 27 | Galβ1-3(Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-6)GalNAcα | NB | + | NB |
| 28 | Neu5Acα2-3Galβ1-3(Fucα1-4)GlcNAcβ | +++ | +++ | +++ |
| 29 | Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ | +++ | +++ | +++ |
| 30 | Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ | +++ | +++ | +++ |
| 31 | Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ | +++ | +++ | +++ |
| 32 | Neu5Acα2-3Galβ1-3[Fucα1-4]GlcNAcβ1-3Galβ1-4[Fucα1-3]GlcNAcβ | +++ | +++ | +++ |
| 33 | Neu5Acα2-3Galβ1-3[Fucα1-3]GlcNAcβ1-3Galβ1-4[Fucα1-3]GlcNAcβ | +++ | +++ | +++ |
| 34 | Neu5Acα2-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ | +++ | +++ | +++ |
| 35 | Neu5Acα2-3(GalNAcβ1-4)Galβ1-4GlcNAcβ | NB | NB | NB |
| 36 | Neu5Acα2-3(GalNAcβ1-4)Galβ1-4GlcNAcβ | NB | NB | NB |
| 37 | Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glcβ | NB | NB | NB |
| 38 | Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ | NB | NB | NB |
| 39 | Fucα1-2Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ | NB | NB | NB |
| 40 | Fucα1-2Galβ1-3GalNAcβ1-4(Neu5Acα2-3)Galβ1-4Glcβ | NB | NB | NB |
| α2-6 sialosides | ||||
| 41 | Neu5Acα2-6Galβ1-4[6OSO3]GlcNAcβ | NB | NB | NB |
| 42 | Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3(Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB | NB |
| 43 | Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB | NB |
| 44 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-2Manα1-3[Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-2Manα1-6]Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB | NB |
| 45 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-2Manα1-3[Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-2Manα1-6]-Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | + | NB |
| 46 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3[Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-6]GalNAca | NB | NB | NB |
| 47 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3[Neu5Acα2-6Galβ1-4GlcNAcβ1-6]GalNAca | NB | NB | NB |
| 48 | Neu5Acα2-6GalNAcα | NB | NB | NB |
| 49 | Neu5Acα2-6Galβ | NB | NB | NB |
| 50 | Neu5Acα2-6Galβ1-4Glcβ | NB | NB | NB |
| 51 | Neu5Acα2-6Galβ1-4Glcβ | NB | NB | NB |
| 52 | Neu5Acα2-6Galβ1-4GlcNAcβ | NB | NB | NB |
| 53 | Neu5Acα2-6Galβ1-4GlcNAcβ | NB | NB | NB |
| 54 | Neu5Acα2-6GalNAcβ1-4GlcNAcβ | NB | NB | NB |
| 55 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3GalNAcα | NB | NB | NB |
| 56 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | NB | NB | NB |
| 57 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3GalNAcα | NB | NB | NB |
| 58 | Neu5Aca2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | NB | NB | NB |
| 59 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ1-3Galβ1-4(Fucα1-3)GlcNAcβ | NB | NB | NB |
| 60 | Galβ1-3(Neu5Acα2-6)GlcNAcβ1-4Galβ1-4Glcβ-Sp10 | NB | NB | NB |
| 61 | Neu5Acα2-6[Galβ1-3]GalNAca | NB | NB | NB |
| 62 | Neu5Acα2-6Galβ1-4GlcNAcβ1-6[Galβ1-3]GalNAca | NB | NB | NB |
| 63 | Neu5Acα2-6Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-6[Galβ1-3]GalNAca | NB | NB | NB |
| Mixed α2-3 and α2-6 biantennaries | ||||
| 64 | Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB | NB |
| 65 | Neu5Acα2-6Galβ1-4GlcNAcβ1-2Manα1-3(Neu5Acα2-3Galβ1-4GlcNAcβ1-2Manα1-6)Manβ1-4GlcNAcβ1-4GlcNAcβ | +++ | +++ | +++ |
| 66 | Neu5Acα2-3Galβ1-3(Neu5Acα2-6)GalNAcα | +++ | +++ | +++ |
| 67 | Neu5Acα2-3(Neu5Acα2-6)GalNAcα | NB | NB | NB |
| N-Glycolylneuraminic acid glycans | ||||
| 68 | Neu5Gcα | NB | NB | NB |
| 69 | Neu5Gcα2-3Galβ1-3(Fucα1-4)GlcNAcβ | NB | NB | NB |
| 70 | Neu5Gcα2-3Galβ1-3GlcNAcβ | NB | NB | NB |
| 71 | Neu5Gcα2-3Galβ1-4(Fucα1-3)GlcNAcβ | ++ | +++ | +++ |
| 72 | Neu5Gcα2-3Galβ1-4GlcNAcβ | NB | NB | NB |
| 73 | Neu5Gcα2-6GalNAcα | NB | NB | NB |
| 74 | Neu5Gcα2-6Galβ1-4GlcNAcβ | NB | NB | NB |
| α2-8-linked sialosides | ||||
| 75 | Neu5Acα2-8Neu5Acα | NB | NB | NB |
| 76 | Neu5Acα2-8Neu5Acα2-8Neu5Acα | NB | NB | NB |
| 77 | Neu5Acα2-8Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glcβ | NB | NB | NB |
| 78 | Neu5Acα2-8Neu5Acα2-3Galβ1-4Glcβ | NB | NB | NB |
| 79 | Neu5Acα2-8Neu5Acα2-8Neu5Acα2-3(GalNAcβ1-4)Galβ1-4Glcβ | NB | NB | NB |
| 80 | Neu5Acα2-8Neu5Acα2-8Neu5Acα2-3Galβ1-4Glcβ | NB | NB | NB |
| 81 | Neu5Acα2-8Neu5Acβ-Sp17 | NB | NB | NB |
| 82 | Neu5Acα2-8Neu5Acα2-8Neu5Acβ | NB | NB | NB |
| β2-6-linked and 9-O-acetylated sialic acids | ||||
| 83 | Neu5Acβ2-6GalNAcα | NB | NB | NB |
| 84 | Neu5Acβ2-6Galβ1-4GlcNAcβ | NB | NB | NB |
| 85 | Neu5Gcβ2-6Galβ1-4GlcNAc | NB | NB | NB |
| 86 | Galβ1-3(Neu5Acβ2-6)GalNAcα | NB | NB | NB |
| 87 | [9NAc]Neu5Acα | NB | NB | NB |
| 88 | [9NAc]Neu5Acα2-6Galβ1-4GlcNAcβ | NB | NB | NB |
| Asialoglycans | ||||
| 89 | Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ1-3Galβ1-4GlcNAcβ | NB | NB | NB |
| 90 | Galβ1-3GlcNAcβ1-3Galβ1-3GlcNAcβ | NB | NB | NB |
| 91 | Galβ1-4GlcNAcβ1-2Manα1-3[Galβ1-4GlcNAcβ1-2Manα1-6]Manβ1-4GlcNAcβ1-4GlcNAcβ | NB | NB | NB |
| 92 | GalNAcα1-3(Fucα1-2)Galβ1-3GlcNAcβ | NB | NB | NB |
| 93 | GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAcβ | NB | NB | NB |
| 94 | Galα1-3(Fucα1-2)Galβ1-3GlcNAcβ | NB | NB | NB |
| 95 | Galα1-3(Fucα1-2)Galβ1-4(Fucα1-3)GlcNAcβ | NB | NB | NB |
| 96 | Galβ1-3GalNAcα | NB | NB | NB |
Neu5Ac, sialic acid; Neu5Gc, N-glycolylneuraminic acid; OSO3, sulfate; Gal, galactose; Fuc, fucose; Glc, d-glucose; GlcNAc, N-acetyl-d-glucosamine; GalNAc, N-acetyl-d-galactosamine; Man, d-mannose; 9NAc, 9-O-acetyl.
Significant binding of samples to glycans was estimated qualitatively based on the relative strength of the signal for the data shown in Fig. 3. Fluorescence intensity is indicated as follows: +++, >3,000; ++, 2,000 to 2,999; +, 1,000 to 1,999; NB, <1,000 (no binding observed).
NA activity and drug susceptibility assays.
RecNA activities were assessed using the fluorescent compound 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA; Sigma-Aldrich Inc.) as the substrate (33). Briefly, RecNA protein was mixed with MUNANA in a reaction buffer containing 32.5 mM 2-(N-morpholino)ethanesulfonic acid (MES), 4 mM CaCl2, and 50 μg/ml of bovine serum albumin. The reaction mixture was incubated at 37°C for 30 min, and the reaction was terminated by the addition of a stop solution containing 25% ethanol and 0.1 M glycine. The fluorescence of the enzyme-cleaved product was measured using a Synergy H1 hybrid multimode microplate reader (BioTek) with excitation and emission wavelengths of 360 nm and 460 nm, respectively. To determine the enzyme kinetics of RecNA, measured as a function of the amount of the substrate, a series of 2-fold-diluted RecNAs were mixed with 200 μM MUNANA, and a sigmoidal curve of NA activity was generated. The NA concentration corresponding to the midpoint of the linear section of the curve was chosen for the kinetics assay. Different concentrations of MUNANA in a series of 2-fold dilutions were mixed with 31.25 ng/ml, 20 ng/ml, 156.3 ng/ml, and 19.5 ng/ml of the N1, N2, N6, and N8 RecNAs, respectively. Reaction parameters (Km, Vmax, and kcat) were calculated by fitting the data to Michaelis-Menten equations using GraphPad Prism software (GraphPad Software, Inc.).
The susceptibilities of RecNAs to neuraminidase inhibitors were tested in the fluorescent neuraminidase inhibition (NI) assay by the use of the NA-Fluor influenza neuraminidase assay kit (Life Technologies), as described previously (26). The NA inhibitors zanamivir (GlaxoSmithKline), oseltamivir carboxylate (Roche Diagnostics GmbH), peramivir (BioCryst Pharmaceuticals), and laninamivir (Biota) were kindly provided by the respective manufacturers. The IC50 value (the drug concentration required to inhibit enzyme activity by 50%), expressed as a nanomolar concentration, was determined using JASPR curve-fitting software, version 1.2 (26). Mean IC50s and standard deviations (SD) were calculated based on the results of three independent tests, each conducted in triplicate.
Protein structure accession numbers.
The atomic coordinates and structure factors of influenza virus A(H5N6) HA, A(H5N8) HA, A(H5N1) NA, A(H5N2) NA, A(H5N6) NA, and A(H5N8) NA are available from the RCSB PDB under accession codes 5HU8, 5HUF, 5HUG, 5HUK, 5HUM, and 5HUN, respectively.
RESULTS AND DISCUSSION
Overall structure of the H5Nx HAs.
In order to understand the structural features of HA from the novel HPAI virus H5Nx subtypes and to compare them to human influenza virus H5 HA, the 3-dimensional HA structures of trimeric ectodomains from A/gyrfalcon/Washington/41088-6/2014 (H5N8), isolated in the first case of the poultry outbreak in the United States in December 2014, and A/Sichuan/26221/2014 (H5N6), isolated from a human patient with a fatal infection in China in April 2014, were determined by X-ray crystallography at 2.4 Å and 2.8 Å resolution, respectively (Table 1). The HA protein is synthesized as a single-chain precursor (HA0) during viral replication and is subsequently cleaved by host proteases into the functional/infectious HA1/HA2 form. The recombinant HAs (RecHAs), overexpressed in a baculovirus expression system, were designed to be produced in the HA0 form. However, both RecHAs were found to be partially digested into the HA1/HA2 form after purification, due to the presence of polybasic sequences at the HA1/HA2 cleavage site in both HAs. This polybasic sequence is the central virulence determinant of HAs from HPAI viruses, because it increases the susceptibility of HA to a range of cellular proteases and therefore changes the tissue tropism from localized epithelial infection to systemic infection (34, 35). The RecHAs were trypsin digested into the functional HA1/HA2 form before the crystallization trial. In the final structures, the N terminus of HA2 of H5N6 HA has poor electron density, while in H5N8 HA, the N-terminal loop of HA2 is tucked into the fusion pocket.
The overall structure of the HA monomer for both viruses is composed of a globular head containing the receptor binding site (RBS), a membrane-proximal domain that includes a central helical stalk, and the HA1/HA2 cleavage site (Fig. 1A and B) (13, 29–32, 36–45). Both HAs have seven potential N-linked glycosylation sites (NXS/T), at amino acid residues 10, 11, 23, 38, 165, and 286 on HA1 and at residue 154 on HA2 (residue 483 in HA0 numbering). For H5N6 HA, residue 193 (NPT) was predicted not to be glycosylated due to the presence of proline at the X position in the sequon (46). In the final structures, interpretable carbohydrate electron density was observed at three sites on HA1 (Asn23, Asn165, and Asn286) for both HAs and at Asn154 on HA2 for H5N8 HA only (Fig. 1A).
FIG 1.
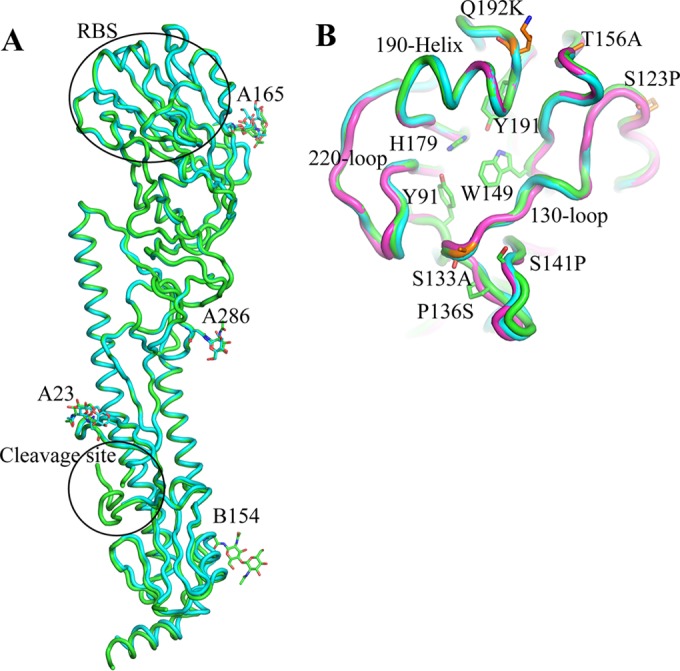
Structure of H5Nx HA. (A) HA monomer for H5N6 HA (cyan) and H5N8 HA (green). The occupied glycosylation sites, A23, A165, and A286 on HA1 and B154 on HA2, are labeled and are shown as sticks. (B) Comparison of the H5N6 HA RBS (green) with equivalent, overlapping structures from H5N8 HA (cyan) and Anhui HA (magenta). The three structural elements making up the binding site—the 130-loop, the 190-helix, and the 220-loop—are labeled. Conserved residues are shown as green sticks. The amino acids discussed in the text are shown as orange sticks. All structural figures were generated with MacPyMol (75).
Comparison of the HA monomers from the H5N6 and H5N8 viruses to that of a clade 2.3.4 HA of influenza virus A/Anhui/1/2005 (H5N1) (PDB code 4KWM) (37) reveals highly similar structures for the viruses, with the Cα atoms superimposing to give root mean square deviations (RMSD) of 1.47 Å and 1.28 Å, respectively. There are only 16 amino acid differences between H5N6 and H5N8 HAs, and their structures are almost identical, with an RMSD of 0.72 Å.
H5Nx antigenic sites.
While human seasonal influenza virus A(H1N1) HAs have four distinct antigenic sites (Sa, Sb, Ca, and Cb [47]), five antigenic sites (A, B, C, D, and E) have been described for human seasonal influenza virus A(H3N2) HAs (48). Antigenic and escape mutation analyses of older influenza virus A(H5N1) HAs identified antigenic residues that map onto regions similar to these A(H1N1) and A(H3N2) antigenic sites (19, 49). Based on structural alignment, positions equivalent to both A(H1N1) and A(H3N2) antigenic sites were identified on current U.S. H5Nx HAs and are highlighted in Fig. 2. Relative to the older Anhui/1/2005 clade 2.3.4 virus HA used in the alignment, a number of amino acid substitutions are found within these antigenic sites, suggesting that these viruses are antigenically distinct. The sequence identities of the positions equivalent to influenza virus A(H1N1) antigenic sites on the U.S. influenza virus A(H5Nx) HAs with corresponding positions on the Anhui virus HA ranged from 69 to 85%, while those for positions equivalent to influenza virus A(H3N2) antigenic sites ranged from 66 to 91%. The lowest sequence identities were found for A(H3N2) antigenic sites A (66%) and B (68%), both of which are close to the receptor binding site (Fig. 2). Analysis by a hemagglutination inhibition assay revealed a low reactivity of sera raised against these U.S. H5Nx viruses to the clade 2.3.4 Anhui/1/2005 virus, prompting the WHO to initiate the development of candidate vaccine viruses (50).
FIG 2.
Structure-based sequence alignment of H5Nx HA. The secondary structure (ss) is highlighted in cyan for the β-sheet and in magenta for the α-helix. The locations equivalent to H1 and H3 antigenic sites are labeled with the antigenic site designation (Sa [1], Sb [2], Ca [3], or Cb [4] for H1 antigenic sites and A, B, C, D, or E for H3 antigenic sites). Residues around the RBS are shaded.
Receptor binding analyses of the H5Nx HAs.
The HA RBS contributes to the host range (51) and is at the membrane distal end of each HA monomer (Fig. 1A). The consensus RBS in all influenza A HAs is composed of three structural elements: a 190-helix (residues 184 to 190), a 220-loop (residues 215 to 224), and a 130-loop (residues 126 to 135) (Fig. 1B). In addition, the highly conserved residues Tyr91, Trp149, His179, and Tyr191 form the base of the pocket (Fig. 1B). Double mutations in the HA receptor binding domains of A(H1N1) (Glu190Asp and Gly225Asp) and A(H2N2)/A(H3N2) (Gln226Leu and Gly228Ser) subtypes have been shown to be critical for the adaptation of these avian viruses into human pandemic viruses (10, 52, 53). Recent gain-of-function studies on Eurasian H5N1 viruses also identified a number of substitutions in the HA, including the RBS, that are important for airborne transmission of avian influenza virus A(H5N1) in ferrets (54), the best available animal model for mammalian transmission. Sequence alignment of the H5N8 and H5N6 HAs with those from the HPAI A(H5N1) viruses showed that these H5Nx HAs possess an avian influenza virus-like RBS (Fig. 2) and do not harbor any of the residues known to be critical for the adaptation of avian influenza virus H1, H2, and H3 to pandemic strains, or any residues shown to be important in the transmission of avian H5 influenza viruses among ferrets. However, four amino acid substitutions of A(H5N1) HAs—Ser123Pro, Ser133Ala, Thr156Ala, and Gln192Lys—have been reported to be associated with increased binding of the virus to mammalian receptors (55–58), and all of these were present in the H5Nx HAs (Fig. 1B). Ser133Ala is located on the 130-loop and may affect the binding properties of the RBS, while Gln192Lys is near the 190-helix and also resides in antigenic site Sb in A(H1N1) viruses.
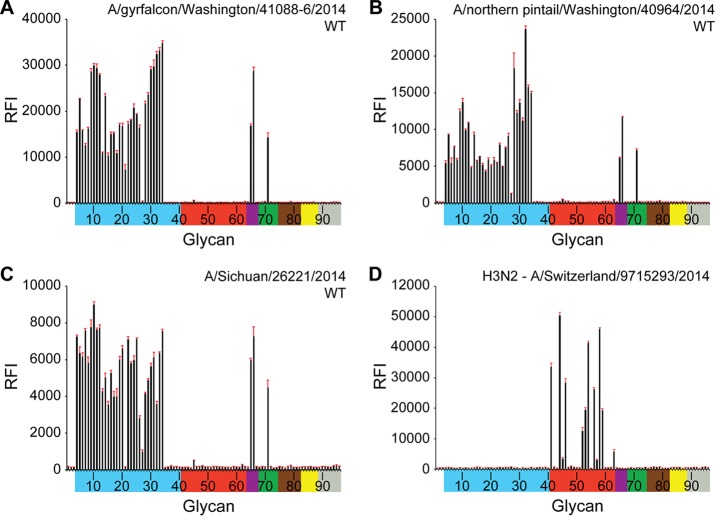
Glycan microarray analysis helps provide a detailed profile of influenza virus receptor specificity (28) and has been used as part of ongoing public health risk assessment activities (26, 59). To gain further insight into species specificity, the interactions of H5Nx viruses with various host receptors were examined using glycan-binding analyses with the various H5Nx RecHAs. Glycan microarray analyses of H5N6, H5N2, and H5N8 RecHAs were compared to that for RecHA from a human influenza virus A(H3N2) that has circulated in people for 40 years (Fig. 3A to D). The data show that all three H5 HAs had an avian influenza virus-like receptor binding preference. The HAs bound to a range of α2-3-linked sialosides (glycans 3 to 26 and 28 to 34), including linear and biantennary sialosides with or without fucosylation/sulfation (Fig. 3A to C; Table 2). These results suggest that the H5 viruses that were first detected in the Northwest U.S. states in late 2014 and early 2015 encode an HA that would not favor human-to-human transmission.
FIG 3.
Glycan array analysis of RecHAs. (A) H5N2 HA; (B) H5N6 HA; (C) H5N8 HA; (D) A(H3N2) HA. Glycans on the microarray are grouped according to sialoside linkage: α2-3 sialosides (blue), α2-6 sialosides (red), α2-6/α2-3 mixed sialosides (purple), N-glycolyl sialosides (green), α2-8 sialosides (brown), β2-6 and 9-O-acetyl sialosides (yellow), and asialoglycans (gray). Error bars reflect the standard error in each signal for six independent replicates on the array. The structure of each of the numbered glycans is found in Table 2. The specific glycan structures that were used in BLI assays are glycans 22 and 56 on the array.
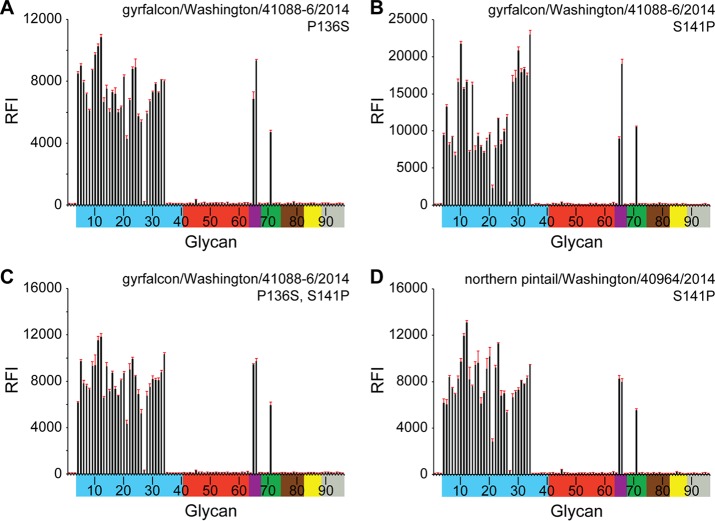
During the following weeks and months, H5Nx viruses continued to spread along the Pacific flyway and across into both the Central and Mississippi flyways. As they moved east, these viruses were detected in wild birds as well as in poultry flocks. Both A(H5N2) and A(H5N8) viruses have been found in recent poultry outbreaks, and all these Eurasian clade 2.3.4.4 HA viruses analyzed to date are highly pathogenic for poultry. Interestingly, while the majority of viruses isolated from wild birds are identical across the HA1 component of the HA protein, differences have been observed in poultry viruses. In particular, a number of turkey A(H5N2) viruses have been found to contain a Ser141Pro substitution relative to the gyrfalcon/Washington/41088-6/2014 A(H5N8) virus. A smaller number of viruses also contain an additional Pro136Ser substitution (60). While both changes were located in/near the known H1 (Ca) and H3 (site A) antigenic sites (Fig. 2), they reside at either end of the 130-loop (Fig. 1B), and the gain/loss of proline could affect the flexibility of this loop. Turkeys and other terrestrial poultry express α2-6-linked sialosides in their respiratory tracts, and the proximity of these proline substitutions to the RBS suggested that they could affect the receptor binding preference. To determine if these natural substitutions impacted receptor utilization, site-directed mutants were generated on both the gyrfalcon/Washington/41088-6/2014 A(H5N8) and northern pintail/Washington/40964/2014 HAs and were subjected to both glycan microarray and biolayer interferometry analyses (Fig. 4 and 5; Table 3). In both analyses, in contrast to wild-type (WT) RecHAs (Fig. 3 and 5; Table 3), HAs with Pro/Ser substitutions at residues 136 and/or 141 bound only avian influenza virus-type receptors. There was no indication of any detectable binding to human influenza virus-type receptors.
FIG 4.
Glycan array analysis of mutant RecHAs. (A) H5N8 Pro136Ser HA; (B) H5N8 Ser141Pro HA; (C) H5N8 Pro136Ser Ser141Pro HA; (D) H5N2 Ser141Pro HA. The glycans on the microarray are described in the legend to Fig. 3. The structure of each of the numbered glycans is found in Table 2. The specific glycan structures that were used in BLI assays are glycans 22 and 56 on the array.
FIG 5.
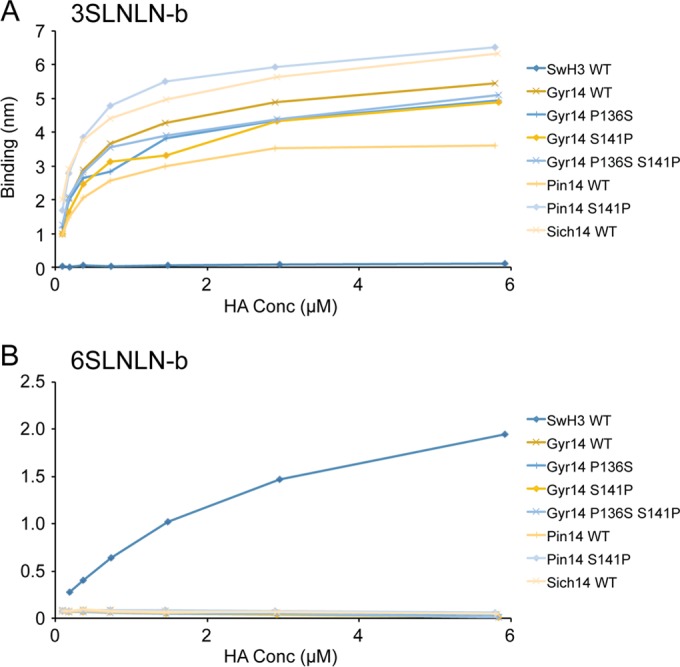
The binding of WT and mutant H5Nx RecHA proteins to specific biotinylated glycans, 3-SLNLN-b and 6-SLNLN-b, immobilized on streptavidin-coated biosensors was analyzed by BLI. Twofold dilutions of HA were analyzed, and their binding, as measured by the shift in the interference pattern (nm), was plotted. Binding kinetics are shown in Table 2.
TABLE 3.
Kinetic results for glycan binding to WT and mutant H5Nx recHAsa
| RecHA | Glycan | Apparent KD (μM) | kon (ms−1) | kobs ± SE (10−2 s−1) | koff ± SE (10−2 s−1) |
|---|---|---|---|---|---|
| Gyr14_WT | 3SLNLN-b | 1.26 | 75,286 | 50.71 ± 2.61 | 9.52 ± 0.65 |
| 6SLNLN-b | NB | NB | NB | NB | |
| Gyr14_136 | 3SLNLN-b | 1.32 | 84,740 | 57.88 ± 3.24 | 11.18 ± 0.71 |
| 6SLNLN-b | NB | NB | NB | NB | |
| Gyr14_141 | 3SLNLN-b | 1.10 | 81,281 | 53.75 ± 2.85 | 8.97 ± 0.60 |
| 6SLNLN-b | NB | NB | NB | NB | |
| Gyr14_136_141 | 3SLNLN-b | 1.43 | 86,241 | 59.88 ± 3.49 | 12.36 ± 0.79 |
| 6SLNLN-b | NB | NB | NB | NB | |
| pin14_WT | 3SLNLN-b | 1.88 | 94,675 | 69.39 ± 4.16 | 17.59 ± 0.87 |
| 6SLNLN-b | NB | NB | NB | NB | |
| pin14_141 | 3SLNLN-b | 0.30 | 85,474 | 49.34 ± 2.40 | 2.58 ± 0.21 |
| 6SLNLN-b | NB | NB | NB | NB | |
| Sich14 | 3SLNLN-b | 0.93 | 81,637 | 52.55 ± 2.84 | 7.63 ± 0.51 |
| 6SLNLN-b | NB | NB | NB | NB | |
| SwH3_WT | 3SLNLN-b | NB | NB | NB | NB |
| 6SLNLN-b | 141.2 | 2,040 | 30.23 ± 1.95 | 28.81 ± 1.78 |
KD, equilibrium dissociation constant; kon, kobs, and koff, association, observed, and dissociation rates, respectively; NB, no detectable binding.
Structural analysis of the H5Nx NAs.
Multiple recombinant H5Nx NAs (N1, N2, N6 and N8) were also expressed using a baculovirus expression system and were subjected to structural analyses. Despite variable sequence identities among these different NA subtypes (identities between the H5Nx NAs in this study range from 45% to 56%) (Fig. 6), the overall NA structure is very similar, with the typical “box-shaped” tetrameric association of identical monomers, containing six four-stranded, antiparallel β-sheets that form a propeller-like arrangement (Fig. 7A) (25, 61–68). The nine subtypes of influenza A virus NA found in birds have been classified into two groups according to their sequences (69). Group 1 comprises N1, N4, N5, and N8, and group 2 comprises N2, N3, N6, N7, and N9 (70). Comparison of the four H5Nx NAs shows that the loops are the most variable regions of the NA structures (Fig. 7B). The NA subtypes within the same groups have similar features in the 150-loop (residues 147 to 152), the 270-loop (residues 267 to 276), and the 430-loop (residues 429 to 433). The numbering follows the N2 numbering (for mature protein numbering for each subtype, see the structure-based sequence alignment in Fig. 6). H5N1 NA and H5N8 NA have the 150-cavity, which is a group 1 specific conformation/feature (63). H5N2 NA and H5N6 NA are typical group 2 NAs without the 150-cavity. The N6 NA possesses a unique 340-cavity similar to the previously published N6 and N7 structures (Fig. 8) (25). This small cavity has not been observed in any other NA structures, and given its close proximity to the active site and the conserved Ca2+ binding site, it is likely to play a role in NA function.
FIG 6.
Structure-based sequence alignment of H5Nx NA. The secondary structure (ss) is highlighted in cyan for the β-sheet and in magenta for the α-helix. Residues involved in substrate binding are represented by green letters, and surrounding conserved residues by red letters.
FIG 7.
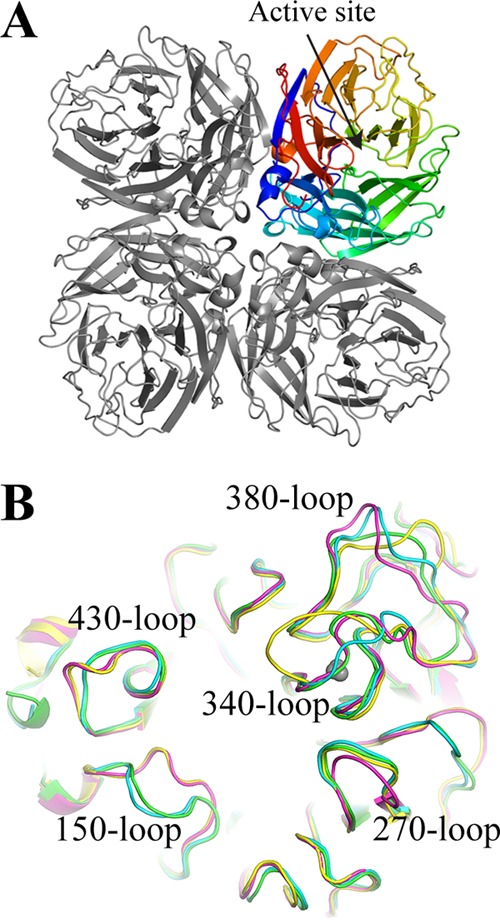
Structure of H5Nx NA. (A) The overall structure of the NA tetramer is shown. One monomer is highlighted in rainbow colors, with the position of the enzyme active site indicated by an arrow. (B) Overlap of the NA active site in N1 NA (green), N8 NA (cyan), N2 NA (magenta), and N6 NA (yellow),\with surrounding variable loops.
FIG 8.

Surface representations of NA monomers.
One calcium ion binding site, which is conserved in all known influenza A and influenza B virus NAs, has a Ca2+ ion in each of the four H5Nx NAs. Ca2+ is bound through interactions with four backbone carbonyl oxygen atoms from Asn293, Gly297, Gly345, and Tyr347, one of the carboxyl oxygens from Asp324, and a water molecule (Fig. 9A). Calcium ions have been shown to be critical for the thermostability and activity of influenza virus NAs (71, 72), and this conserved metal site has been proposed to be important in stabilizing a reactive conformation of the active site by otherwise flexible loops (73). Interestingly, influenza virus A(H5N1) NA has an extra Ca2+ binding site in the 380-loop. Ca2+ is bound through interactions with carboxyl oxygens from Asp379, Asn381, Asp387, Ser389, and two water molecules (Fig. 9A). This observation is similar to that for calcium binding site 3 of the 1918 H1N1 virus NA (15). However, the contribution of this binding site to the function of N1 NA is not yet clear.
FIG 9.
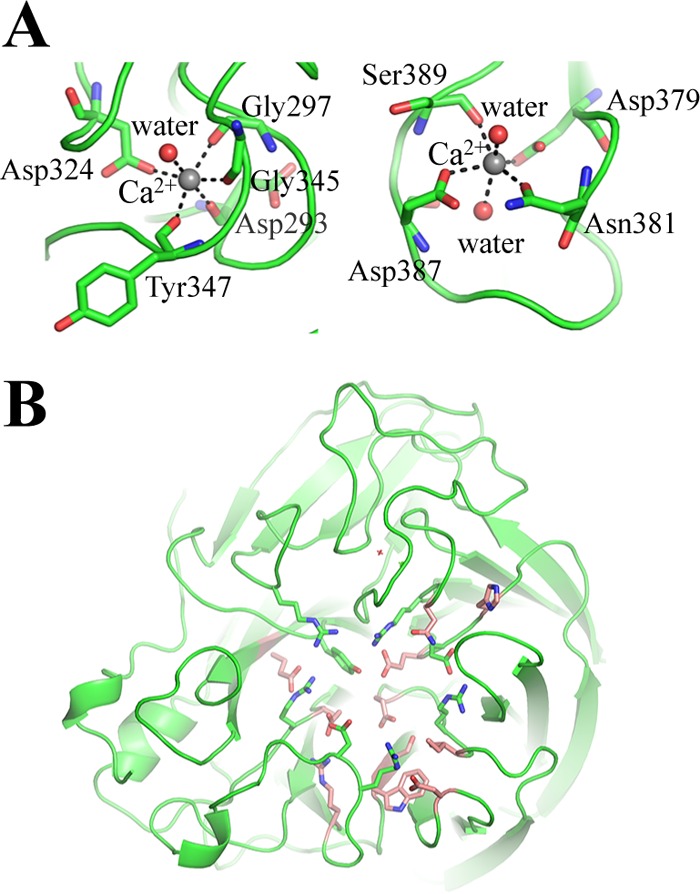
(A) Two Ca2+ binding sites of N1 NA, with interacting residues shown as sticks. (B) The active site of NA. Highly conserved residues involved in direct substrate binding are shown as green sticks, and surrounding conserved residues are shown as orange sticks.
Among all predicted potential N-linked glycosylation sites, Asn146, which is situated on the membrane-distal surface close to the active site, is the only glycosylation sequon conserved among all other influenza virus A and B NAs (61, 63, 74). At this position, electron density for a glycan was observed only in the N1, N2, and N6 NAs, but only one N-acetylglucosamine could be interpreted in each model. N1 NA has two additional potential glycosylation sites at Asn88 and Asn234, and both had interpretable glycan density in the final model. N2 NA has additional potential glycosylation sites at Asn86, Asn200, Asn234, and Asn402, but interpretable glycan density was observed only at Asn200 and Asn234. Interestingly, density for a biantennary glycan was observed at Asn200 in this model. N6 NA also has putative glycosylation sites at Asn86, Asn200, and Asn402, but only Asn86 had interpretable glycan density for one N-acetylglucosamine. Finally, while N8 NA has additional glycosylation sites at Asn86, Asn295, and Asn400, none of them had interpretable glycan density. There is a suggestion that N-glycosylation might be one of the factors to differentiate individual NAs within each subtype (25); however, more data are needed to confirm this correlation.
Active site of H5Nx NAs and antiviral drug susceptibility to NAIs.
In all the NA subtypes, the enzyme active site includes eight highly conserved residues: Arg118, Asp151, Arg152, Arg224, Glu276, Arg292, Arg371, and Tyr406. These are all charged/polar residues that interact directly with the substrate in the catalytic site. The geometry of the catalytic site is structurally stabilized through a network of hydrogen bonds and salt bridges by a constellation of largely conserved framework residues: Glu119, Arg156, Trp178, Ser179, Asp/Asn198, Ile222, Glu227, His274, Glu277, Asn294, and Glu425 (Fig. 9B) (65).
The NA activities of H5Nx NAs were assessed using the fluorescent reagent 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (MUNANA) as the substrate (33). Despite the structural difference near the active site between group 1 and group 2 NAs, all recombinant H5Nx NAs had similarly high activities compared to human influenza virus NAs (Table 4). In the NI assay, the activities of the recombinant N1, N2, N6, and N8 NAs of various H5Nx viruses were effectively inhibited by the antivirals currently approved by the FDA (oseltamivir carboxylate, zanamivir, peramivir) and an investigational NAI (laninamivir), indicating a neuraminidase inhibitor-susceptible phenotype for these H5Nx viruses (Table 4).
TABLE 4.
Drug susceptibility assessment in the fluorescent neuraminidase inhibition assay
| Virus name | NA subtype | Parameter |
Mean IC50 ± SD (nM)a |
|||||
|---|---|---|---|---|---|---|---|---|
| kcat ± SD (s−1) | Km ± SD (μM) | kcat/Km (M−1 s−1) | Oseltamivir carboxylate | Zanamivir | Peramivir | Laninamivir | ||
| A/American green-winged teal/Washington/195750/2014 | N1 | 134,254 ± 2,841 | 5.08 ± 0.41 | 3.29 × 1011 | 3.45 ± 0.35 | 0.33 ± 0.10 | 0.10 ± 0.03 | 0.19 ± 0.02 |
| A/northern pintail/Washington/40964/2014 | N2 | 115,191 ± 2,523 | 5.48 ± 0.45 | 2.56 × 1011 | 0.18 ± 0.06 | 0.75 ± 0.22 | 0.20 ± 0.03 | 1.05 ± 0.10 |
| A/Sichuan/26221/2014 | N6 | 147,235 ± 3,693 | 7.00 ± 0.63 | 2.36 × 1011 | 0.98 ± 0.18 | 0.98 ± 0.18 | 0.21 ± 0.03 | 1.95 ± 0.24 |
| A/gyrfalcon/Washington/41088-6/2014 | N8 | 173,671 ± 3,018 | 5.98 ± 0.38 | 4.55 × 1011 | 0.99 ± 0.20 | 0.35 ± 0.12 | 0.06 ± 0.02 | 0.22 ± 0.01 |
From the results of three independent experiments run in triplicate.
Conclusion.
Here we report the molecular characterization of 3 HAs and 4 NAs from the H5Nx subclade 2.3.4.4. Even though H5N6 HA is an isolate from a fatal human case, results for both avian influenza virus H5N8 HA and human influenza virus H5N6 HA show that they possess an avian receptor binding preference, thus reducing their potential to efficiently infect humans. The H5Nx NAs are also sensitive to FDA-approved (zanamivir, oseltamivir, and peramivir) and investigational (laninamivir) neuraminidase inhibitors. It is important to monitor H5Nx viruses in poultry and to continue to assess changes in circulating H5Nx viruses and their pandemic potential. Collectively, the data indicate that additional changes in the H5 HA would be needed for efficient transmission among humans and that currently available antivirals would be an effective tool to combat these viruses.
ACKNOWLEDGMENTS
We thank the staff of SER-CAT sector 22 at the Advanced Photon Source (APS) for help in data collection. The U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract DE-AC02-06CH11357, supports the use of the Advanced Photon Source at the Argonne National Laboratory. Glycan microarray slides were produced under contract for the Centers for Disease Control and Prevention using a glycan library generously provided by the Consortium for Functional Glycomics (CFG) (www.functionalglycomics.org), funded by National Institute of General Medical Sciences grant GM62116. We also thank the CFG for the supplying the biotinylated glycans used here through their resource request program. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
REFERENCES
- 1.Yuen KY, Chan PK, Peiris M, Tsang DN, Que TL, Shortridge KF, Cheung PT, To WK, Ho ET, Sung R, Cheng AF. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467–471. doi: 10.1016/S0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 2.WHO. 17 July 2015. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2015. World Health Organization, Geneva, Switzerland: http://www.who.int/influenza/human_animal_interface/EN_GIP_20150717cumulativeNumberH5N1cases.pdf?ua=1. [Google Scholar]
- 3.Vijaykrishna D, Bahl J, Riley S, Duan L, Zhang JX, Chen H, Peiris JS, Smith GJ, Guan Y. 2008. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog 4:e1000161. doi: 10.1371/journal.ppat.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith GJ, Donis RO, World Health Organization/World Organisation for Animal Health/Food and Agriculture Organization H5 Evolution Working Group. 2015. Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013–2014. Influenza Other Respir Viruses 9:271–276. doi: 10.1111/irv.12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan M, Gao R, Lv Q, Huang S, Zhou Z, Yang L, Li X, Zhao X, Zou X, Tong W, Mao S, Zou S, Bo H, Zhu X, Liu L, Yuan H, Zhang M, Wang D, Li Z, Zhao W, Ma M, Li Y, Li T, Yang H, Xu J, Zhou L, Zhou X, Tang W, Song Y, Chen T, Bai T, Zhou J, Wang D, Wu G, Li D, Feng Z, Gao GF, Wang Y, He S, Shu Y. 2015. Human infection with a novel highly pathogenic avian influenza A (H5N6) virus: virological and clinical findings. J Infect doi: 10.1016/j.jinf.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 6.World Organisation for Animal Health. 2014. Summary of immediate notifications and follow-ups—2014. Highly pathogenic avian influenza. OIE, Paris, France: http://www.oie.int/wahis_2/public/wahid.php/Diseaseinformation/Immsummary. [Google Scholar]
- 7.Ip HS, Torchetti MK, Crespo R, Kohrs P, DeBruyn P, Mansfield KG, Baszler T, Badcoe L, Bodenstein B, Shearn-Bochsler V, Killian ML, Pedersen JC, Hines N, Gidlewski T, DeLiberto T, Sleeman JM. 2015. Novel Eurasian highly pathogenic avian influenza A H5 viruses in wild birds, Washington, USA, 2014. Emerg Infect Dis 21:886–890. doi: 10.3201/eid2105.142020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, Karlsson KA. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233:224–234. doi: 10.1006/viro.1997.8580. [DOI] [PubMed] [Google Scholar]
- 10.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Eichelberger MC, Compans RW, Air GM. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol 69:1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moscona A. 2005. Neuraminidase inhibitors for influenza. N Engl J Med 353:1363–1373. doi: 10.1056/NEJMra050740. [DOI] [PubMed] [Google Scholar]
- 13.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 14.Kuhnel K, Jarchau T, Wolf E, Schlichting I, Walter U, Wittinghofer A, Strelkov SV. 2004. The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc Natl Acad Sci U S A 101:17027–17032. doi: 10.1073/pnas.0403069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Zhu X, Dwek R, Stevens J, Wilson I. 2008. Structural characterization of the 1918 influenza H1N1 neuraminidase. J Virol 82:10493–10501. doi: 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chayen NE. 2007. Optimization techniques for automation and high throughput. Methods Mol Biol 363:175–190. doi: 10.1007/978-1-59745-209-0_9. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 19.Shore DA, Yang H, Balish AL, Shepard SS, Carney PJ, Chang JC, Davis CT, Donis RO, Villanueva JM, Klimov AI, Stevens J. 2013. Structural and antigenic variation among diverse clade 2 H5N1 viruses. PLoS One 8:e75209. doi: 10.1371/journal.pone.0075209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.Winn MD, Isupov MN, Murshudov GN. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57:122–133. doi: 10.1107/S0907444900014736. [DOI] [PubMed] [Google Scholar]
- 23.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vavricka CJ, Li Q, Wu Y, Qi J, Wang M, Liu Y, Gao F, Liu J, Feng E, He J, Wang J, Liu H, Jiang H, Gao GF. 2011. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathog 7:e1002249. doi: 10.1371/journal.ppat.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X, Li Q, Wu Y, Wang M, Liu Y, Qi J, Vavricka CJ, Gao GF. 2014. Structure of influenza virus N7: the last piece of the neuraminidase “jigsaw” puzzle. J Virol 88:9197–9207. doi: 10.1128/JVI.00805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang H, Nguyen HT, Carney PJ, Guo Z, Chang JC, Jones J, Davis CT, Villanueva JM, Gubareva LV, Stevens J. 2015. Structural and functional analysis of surface proteins from an A(H3N8) influenza virus isolated from New England harbor seals. J Virol 89:2801–2812. doi: 10.1128/JVI.02723-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. 2006. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol 355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 30.Yang H, Carney P, Stevens J. 2010. Structure and receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr 2:RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Carney PJ, Donis RO, Stevens J. 2012. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. J Virol 86:8645–8652. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. 2010. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog 6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-d-N-acetylneuraminate) substrate. Anal Biochem 94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 34.Klenk HD, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol 2:39–43. doi: 10.1016/0966-842X(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 35.Hulse-Post DJ, Sturm-Ramirez KM, Humberd J, Seiler P, Govorkova EA, Krauss S, Scholtissek C, Puthavathana P, Buranathai C, Nguyen TD, Long HT, Naipospos TS, Chen H, Ellis TM, Guan Y, Peiris JS, Webster RG. 2005. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc Natl Acad Sci U S A 102:10682–10687. doi: 10.1073/pnas.0504662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2001. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci U S A 98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2003. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology 309:209–218. doi: 10.1016/S0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. 2009. Structures of receptor complexes formed by hemagglutinins from the Asian influenza pandemic of 1957. Proc Natl Acad Sci U S A 106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell RJ, Gamblin SJ, Haire LF, Stevens DJ, Xiao B, Ha Y, Skehel JJ. 2004. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325:287–296. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 40.Russell RJ, Kerry PS, Stevens DJ, Steinhauer DA, Martin SR, Gamblin SJ, Skehel JJ. 2008. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A 105:17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 42.Lu X, Qi J, Shi Y, Wang M, Smith DF, Heimburg-Molinaro J, Zhang Y, Paulson JC, Xiao H, Gao GF. 2013. Structure and receptor binding specificity of hemagglutinin H13 from avian influenza A virus H13N6. J Virol 87:9077–9085. doi: 10.1128/JVI.00235-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu X, Shi Y, Gao F, Xiao H, Wang M, Qi J, Gao GF. 2012. Insights into avian influenza virus pathogenicity: the hemagglutinin precursor HA0 of subtype H16 has an α-helix structure in its cleavage site with inefficient HA1/HA2 cleavage. J Virol 86:12861–12870. doi: 10.1128/JVI.01606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science 303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409–417. doi: 10.1016/S0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 46.Shakin-Eshleman SH, Spitalnik SL, Kasturi L. 1996. The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J Biol Chem 271:6363–6366. doi: 10.1074/jbc.271.11.6363. [DOI] [PubMed] [Google Scholar]
- 47.Brownlee GG, Fodor E. 2001. The predicted antigenicity of the haemagglutinin of the 1918 Spanish influenza pandemic suggests an avian origin. Philos Trans R Soc Lond B Biol Sci 356:1871–1876. doi: 10.1098/rstb.2001.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiley DC, Wilson IA, Skehel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 49.Velkov T, Ong C, Baker MA, Kim H, Li J, Nation RL, Huang JX, Cooper MA, Rockman S. 2013. The antigenic architecture of the hemagglutinin of influenza H5N1 viruses. Mol Immunol 56:705–719. doi: 10.1016/j.molimm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 50.Anonymous. 2015. Antigenic and genetic characteristics of zoonotic influenza viruses and development of candidate vaccine viruses for pandemic preparedness. Wkly Epidemiol Rec 90:109–120. [PubMed] [Google Scholar]
- 51.Neumann G, Kawaoka Y. 2006. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg Infect Dis 12:881–886. doi: 10.3201/eid1206.051336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 53.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74:8502–8512. doi: 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russell CA, Fonville JM, Brown AE, Burke DF, Smith DL, James SL, Herfst S, van Boheemen S, Linster M, Schrauwen EJ, Katzelnick L, Mosterin A, Kuiken T, Maher E, Neumann G, Osterhaus AD, Kawaoka Y, Fouchier RA, Smith DJ. 2012. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 336:1541–1547. doi: 10.1126/science.1222526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang ZY, Wei CJ, Kong WP, Wu L, Xu L, Smith DF, Nabel GJ. 2007. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317:825–828. doi: 10.1126/science.1135165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W, Lu B, Zhou H, Suguitan AL Jr, Cheng X, Subbarao K, Kemble G, Jin H. 2010. Glycosylation at 158N of the hemagglutinin protein and receptor binding specificity synergistically affect the antigenicity and immunogenicity of a live attenuated H5N1 A/Vietnam/1203/2004 vaccine virus in ferrets. J Virol 84:6570–6577. doi: 10.1128/JVI.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada S, Suzuki Y, Suzuki T, Le MQ, Nidom CA, Sakai-Tagawa Y, Muramoto Y, Ito M, Kiso M, Horimoto T, Shinya K, Sawada T, Kiso M, Usui T, Murata T, Lin Y, Hay A, Haire LF, Stevens DJ, Russell RJ, Gamblin SJ, Skehel JJ, Kawaoka Y. 2006. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444:378–382. doi: 10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 58.Chen LM, Blixt O, Stevens J, Lipatov AS, Davis CT, Collins BE, Cox NJ, Paulson JC, Donis RO. 2012. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology 422:105–113. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. 2013. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol 87:12433–12446. doi: 10.1128/JVI.01854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.USDA Animal and Plant Health Inspection Service. 15 June 2015. Epidemiologic and other analyses of HPAI-affected poultry flocks. APHIS, Riverdale Park, MD: https://www.aphis.usda.gov/animal_health/animal_dis_spec/poultry/downloads/Epidemiologic-Analysis-June-15-2015.pdf. [Google Scholar]
- 61.Varghese JN, Laver WG, Colman PM. 1983. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 62.Baker AT, Varghese JN, Laver WG, Air GM, Colman PM. 1987. Three-dimensional structure of neuraminidase of subtype N9 from an avian influenza virus. Proteins 2:111–117. doi: 10.1002/prot.340020205. [DOI] [PubMed] [Google Scholar]
- 63.Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 64.Wang M, Qi J, Liu Y, Vavricka CJ, Wu Y, Li Q, Gao GF. 2011. Influenza A virus N5 neuraminidase has an extended 150-cavity. J Virol 85:8431–8435. doi: 10.1128/JVI.00638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burmeister WP, Ruigrok RW, Cusack S. 1992. The 2.2 Å resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J 11:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Q, Qi J, Wu Y, Kiyota H, Tanaka K, Suhara Y, Ohrui H, Suzuki Y, Vavricka CJ, Gao GF. 2013. Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance. J Virol 87:10016–10024. doi: 10.1128/JVI.01129-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu X, Yang H, Guo Z, Yu W, Carney PJ, Li Y, Chen LM, Paulson JC, Donis RO, Tong S, Stevens J, Wilson IA. 2012. Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc Natl Acad Sci U S A 109:18903–18908. doi: 10.1073/pnas.1212579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Air GM. 2012. Influenza neuraminidase. Influenza Other Respir Viruses 6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Air GM, Laver WG. 1989. The neuraminidase of influenza virus. Proteins 6:341–356. doi: 10.1002/prot.340060402. [DOI] [PubMed] [Google Scholar]
- 71.Chong AK, Pegg MS, Taylor NR, von Itzstein M. 1992. Evidence for a sialosyl cation transition-state complex in the reaction of sialidase from influenza virus. Eur J Biochem 207:335–343. doi: 10.1111/j.1432-1033.1992.tb17055.x. [DOI] [PubMed] [Google Scholar]
- 72.Burmeister WP, Cusack S, Ruigrok RW. 1994. Calcium is needed for the thermostability of influenza B virus neuraminidase. J Gen Virol 75(Part 2):381–388. doi: 10.1099/0022-1317-75-2-381. [DOI] [PubMed] [Google Scholar]
- 73.Smith BJ, Huyton T, Joosten RP, McKimm-Breschkin JL, Zhang JG, Luo CS, Lou MZ, Labrou NE, Garrett TP. 2006. Structure of a calcium-deficient form of influenza virus neuraminidase: implications for substrate binding. Acta Crystallogr D Biol Crystallogr 62:947–952. doi: 10.1107/S0907444906020063. [DOI] [PubMed] [Google Scholar]
- 74.Janakiraman MN, White CL, Laver WG, Air GM, Luo M. 1994. Structure of influenza virus neuraminidase B/Lee/40 complexed with sialic acid and a dehydro analog at 1.8-Å resolution: implications for the catalytic mechanism. Biochemistry 33:8172–8179. doi: 10.1021/bi00193a002. [DOI] [PubMed] [Google Scholar]
- 75.DeLano WL. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, Palo Alto, CA: www.pymol.org. [Google Scholar]