ABSTRACT
The interferon (IFN)-mediated antiviral response is a central aspect of host defense; however, viruses have evolved multiple strategies to counteract IFN-mediated responses in order to successfully infect the host. Herpes simplex virus 1 (HSV-1), a typical human-restricted DNA virus, is capable of counteracting host immune responses via several distinct viral proteins, thus establishing a lifelong latent infection. In this study, we demonstrate that the VP24 protein, a serine protease of HSV-1 essential for the formation and maturation of capsids, is a novel antagonist of the beta interferon (IFN-β) pathway. Here, VP24 was shown for the first time to dampen interferon stimulatory DNA (ISD)-triggered IFN-β production and inhibit IFN-β promoter activation induced by cyclic GMP-AMP synthase (cGAS) and stimulator of interferon genes (STING) and by STING, respectively. Further study demonstrated that ectopic expression of VP24 selectively blocked IFN regulatory factor 3 (IRF3) but not NF-κB promoter activation. In addition, VP24 was demonstrated to downregulate ISD-induced phosphorylation and dimerization of IRF3 during HSV-1 infection with a VP24 stable knockdown human foreskin fibroblast cell line. The underlying molecular mechanism is that VP24 abrogates the interaction between TANK-binding kinase 1 (TBK1) and IRF3, hence impairing IRF3 activation. These results illustrate that VP24 is able to block the production of IFN-β by inhibiting IRF3 activation, which may represent a critical adaptation to enable viral effective replication within the host.
IMPORTANCE This study demonstrated that HSV-1 protein VP24 could inhibit IFN-β production and promoter activation triggered by ISD, cGAS and STING and by STING, respectively. VP24 selectively blocked IRF3 promoter activation and ISD-induced phosphorylation and dimerization of IRF3 without affecting the NF-κB promoter activation during viral infection. VP24 also inhibited IRF3 activation by impeding the interaction between TBK1 and IRF3 during viral infection. This study provides new insights into the immune evasion mediated by HSV-1 and identifies VP24 as a crucial effector for HSV-1 to evade the host DNA-sensing signal pathway.
INTRODUCTION
The innate immune response of the host is the first line of defense against viral infection and also plays a major part in the subsequent activation of the adaptive immune response. Viral components, such as viral nucleic acids, can be detected by many pathogen recognition receptors (PRRs) which subsequently lead to the induction of type I interferons (IFNs), including alpha/beta IFN (IFN-α/β), and triggering the expression of numerous antiviral proteins (1, 2). PRRs can be divided into membrane-associated receptors, such as the Toll-like receptors, and cytosolic receptors, including retinoic acid-inducible gene I (RIG-I)-like receptors and nucleotide-binding oligomerization domain (NOD)-like receptors (3). Recently, a novel class of cytosolic DNA-sensing receptors has been identified: cyclic GMP-AMP synthase (cGAS), IFN-γ-inducible protein 16 (IFI16), DEAD box polypeptide 41 (DDX41), DNA-dependent activator of IFN regulatory factors (IRFs) (DAI), and several proteins involved in the DNA damage response (4–8).
During the past few years, cGAS has emerged as a predominant cytosolic DNA sensor, which induces IFN-β production by activating stimulator of interferon genes (STING) (9, 10). Upon binding DNA, cGAS is activated and produces cyclic GMP-AMP also known as cGAMP (9–11). The latter interacts with STING and recruits TANK-binding kinase 1 (TBK1), which phosphorylates interferon regulatory factor 3 (IRF3), which then leads to nuclear localization of phosphorylated IRF3 and beta interferon (IFN-β) production (4, 9, 11–14). In turn, newly synthesized IFN-β, which is considered a hallmark of the antiviral response, induces the expression of numerous interferon-stimulated genes (ISGs), such as viperin and interferon-induced transmembrane proteins, which are responsible for the establishment of an antiviral state in infected cells and in neighboring noninfected cells (2, 15, 16).
Viruses have evolved various strategies to evade host innate immune responses and establish a persistent infection. During productive infection, cellular proteins as well as the viral gene expression could be modulated by virus to avoid host immune responses. Herpes simplex virus 1 (HSV-1), an archetypal member of the alphaherpesvirus subfamily, has evolved multiple strategies to target distinct steps in the signaling events that lead to IFN-β induction (17–19).
HSV-1 infection triggers a host type I IFN response, which is subsequently shut down by HSV-encoded gene products (20, 21). Previous studies showed that ICP0, an immediate early gene product of HSV-1, could inhibit the activation of IRF3 and NF-κB via its RING finger domain (22–26). ICP27, also an immediate early gene product, was shown to antagonize type I IFN signaling by inhibiting STAT-1 phosphorylation and reducing the accumulation of STAT-1 in the nucleus (27). US3, a serine/threonine protein kinase that is also conserved across the Herpesviridae family, was shown to block the expression of IFN-β-dependent genes by hyperphosphorylating IRF3 and p65, respectively (28–30). US11, an RNA-binding tegument protein of HSV-1, interacts with endogenous RIG-I and melanoma differentiation-associated protein 5 (MDA5), resulting in reduced production of IFN-β (31). UL36, the largest tegument protein of HSV-1, could block IFN-β production via its deubiquitinase activity (32). UL42, a DNA polymerase processivity factor, was found to interact with and retain p65 and p50 in the cytoplasm, thus inhibiting NF-κB activation (33). Our previous research also revealed that VP16, an abundant 65-kDa virion phosphoprotein, could inhibit IRF3 from recruiting its coactivator CREB-binding protein, thereby blocking its transactivation activity (34). Last, the virion host shutoff protein triggers degradation of host mRNAs, for example, viperin and zinc finger antiviral protein, and contributes to an overall decrease in host protein synthesis (35–39).
The scaffold proteins of HSV-1 encoded by overlapping in-frame UL26 and UL26.5 transcripts are essential for formation and maturation of capsids (40). During capsid maturation, the UL26-encoded (31) protease processes itself to release the N-terminal protease domain VP24 (40, 41). However, the role of VP24 in immune evasion is still unknown.
In this study, a serine protease VP24 of HSV-1 was shown for the first time to dampen interferon stimulatory DNA (ISD)- and cGAS-STING-mediated IFN-β production. The underlying molecular mechanism is that VP24 abrogates the interaction between TBK1 and IRF3, thus inhibiting the activation of IRF3 by blocking its phosphorylation and dimerization.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
HEK 293T cells and human foreskin fibroblast (HFF) cells were cultured in Dulbecco's modified Eagle medium (DMEM) (Gibco-BRL) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml of penicillin and streptomycin as described previously (31, 42). The wild-type (WT) HSV-1 F strain was propagated in Vero cells and titrated as described previously (42).
The protease inhibitor mixture cocktail was purchased from Cell Signaling Technology (CST) (Boston, MA). RIPA lysis buffer was purchased from Beyotime (Shanghai, China). Mouse antihemagglutinin (anti-HA) monoclonal antibody (MAb) was purchased from Abmart (Shanghai, China). Mouse anti-TBK1 polyclonal antibody (PAb) and mouse anti-β-actin MAb were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-IRF3 PAb and rabbit antibody against HSV-1 VP24 were made by GL Biochem Ltd. (Shanghai, China). Rabbit antiserum against IRF3-S396 (IRF3 phosphorylated at Ser396) was from Rongtuan Lin (McGill University, Canada).
Plasmid construction.
All enzymes for cloning procedures were purchased from Vazyme (Nanjing, China). To construct Flag-tagged VP24 (VP24-Flag), the VP24 gene was amplified from the HSV-1 genome as described in our previous study (43) and cloned into the HindIII and EcoRI sites of the pCMV-Flag (CMV stands for cytomegalovirus) plasmid (Beyotime, Shanghai, China). Small hairpin RNA specific for VP24 (shVP24) or scrambled small hairpin RNA (shNC) (NC stands for negative control) was cloned into pSIREN-RetroQ-ZsGreen (Clontech) to yield pSIREN-shVP24 and pSIREN-shNC plasmids, respectively. Commercial reporter plasmids included NF-κB-Luc (Luc stands for luciferase) (Stratagene, La Jolla, CA) and pRL-TK (RL stands for Renilla luciferase, and TK stands for thymidine kinase) (Promega). Other plasmids used that we were given include the following: IRF3-Luc (44), pcDNA3.1-Flag-TBK1 (25), IRF3/5D (45), and IFN-β promoter reporter plasmid (46).
Establishment of VP24 stably knockdowned HFF cells.
HFF cells were transfected with pSIREN-shVP24 (HFF-shVP24) or pSIREN-shNC plasmid (HFF-shNC), and flow cytometry was applied to enrich for cells containing the recombinant shRNA vector. The stably transfected HFF-shNC and HFF-shVP24 cells were then cultured with G418 (500 ng/ml).
RNA isolation and quantitative real-time reverse transcription-PCR (qRT-PCR).
Total RNA was extracted using TRIzol (Invitrogen, California) according to the manufacturer's manual. Samples were digested with DNase I and subjected to reverse transcription as previously described (38). The cDNA was used as a template for real-time PCR to investigate the expression patterns of human IFN-β. Detailed protocols have been described previously (38).
Transfection and dual-luciferase reporter (DLR) assays.
HEK 293T cells were cotransfected with reporter plasmids, such as IFN-β-Luc, NF-κB-Luc, and IRF3-Luc, and internal control plasmid pRL-TK, with or without expression plasmids, as indicated, by standard calcium phosphate precipitation (47, 48). At 24 h posttransfection, luciferase assays were performed with a dual-specific luciferase assay kit (Promega, Madison, WI) as described in our previous studies (31, 49).
Coimmunoprecipitation assay and Western blot analysis.
Coimmunoprecipitation (co-IP) assays and Western blot (WB) analysis were performed as described in our previous studies (18, 19). Briefly, HFF-shNC and HFF-shVP24 cells (∼1 × 106) were infected with HSV-1. The cells were then transfected with ISD at 2 h postinfection (hpi) for an additional 16 h and lysed on ice with 600 μl of lysis buffer. HEK 293T cells (∼5 × 106) were cotransfected with 10 μg of each of the indicated expression plasmids carrying HA tags. For each IP, a 500-μl aliquot of lysate was incubated with the anti-IRF3 PAb, anti-TBK1 or nonspecific mouse monoclonal antibody, and 30 μl of a 1:1 slurry of protein A/G Plus-agarose (Santa Cruz Biotechnology, Santa Cruz, CA) for at least 4 h or overnight at 4°C. The beads were washed four times with 1 ml of lysis buffer containing 500 mM NaCl and then subjected to WB analysis. All IP assays were repeated at least three times, and similar data were obtained.
Native PAGE.
Native polyacrylamide gel electrophoresis (PAGE) was performed using ReadyGels (7.5%; Bio-Rad) as described in our previous study (18). In brief, the gel was prerun with 25 mM Tris and 192 mM glycine, pH 8.4, with 1% deoxycholate (DOC) in the cathode chamber for 30 min at 40 mA. Samples in native sample buffer (10 μg protein, 62.5 mM Tris-Cl [pH 6.8], 15% glycerol, and 1% DOC) were size fractionated by electrophoresis for 60 min at 25 mA and transferred to nitrocellulose membranes for WB analysis.
RESULTS
VP24 inhibits ISD-mediated production of IFN-β and the activation of IFN-β promoter induced by cGAS-STING or STING alone.
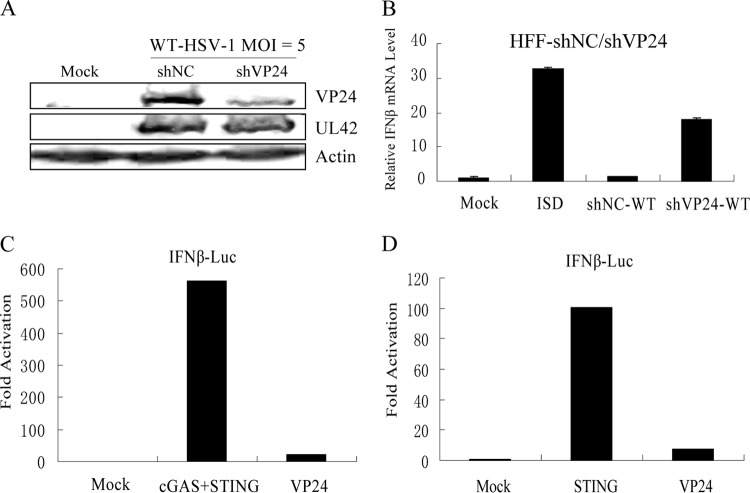
To determine whether VP24 affected the production of IFN-β induced by ISD, HFF were transfected with a shRNA specific for VP24 (HFF-shVP24) or a scrambled shRNA as a control (HFF-shNC) to screen for stably transfected cell lines. HFF-shVP24 and HFF-shNC were infected with HSV-1 for 16 h, and then the cells were harvested and subjected to WB to analyze the knockdown efficiency of VP24 small interfering RNA (siRNA). As shown in Fig. 1A, a relatively low level of VP24 was detected in shVP24-transfected cells (shVP24 cells) compared with shNC-transfected cells (shNC cells) during HSV-1 infection. Protein level of UL42 was measured in shNC and shVP24 cell lines as a control, which indicated that other viral proteins were not affected in VP24 knockdown cells (Fig. 1A). Meanwhile, HFF-shVP24 and HFF-shNC were infected with HSV-1 for 2 h before ISD transfection. The cells were then harvested at 8 hpi and subjected to qRT-PCR to analyze IFN-β mRNA. As a result, mRNA of endogenous IFN-β was strongly upregulated by ISD transfection, whereas VP24 reduced the accumulation of endogenous IFN-β mRNA and knockdown of VP24 recovered its mRNA to a certain extent (Fig. 1B) (18S rRNA was used as an internal control). In HEK 293T cells, ectopic expression of cGAS or a minimal amount of STING alone failed to activate the IFN-β promoter, while cotransfection of the same amounts of cGAS and STING plasmids activated the IFN-β promoter (11). However, ectopic expression of a large amount of STING alone successfully activates the IFN-β promoter (50). To further determine the role of the VP24 protein in the inhibition of cGAS-STING-induced IFN-β promoter activation, HEK 293T cells were cotransfected with a Flag-tagged VP24 expression plasmid and an IFN-β promoter construct and subjected to dual-luciferase reporter (DLR) assays to detect IFN-β promoter activity. As expected, transfection with cGAS and a minimal amount of STING plasmids or a large amount of STING plasmid resulted in strong induction of IFN-β reporter activity; in contrast, ectopic expression of VP24 inhibited the activation of the IFN-β promoter (Fig. 1C and D). Taken together, these results demonstrated that VP24 expression dramatically reduced ISD-mediated production of endogenous IFN-β and blocked IFN-β promoter activation induced by cGAS-STING or STING.
FIG 1.
VP24 inhibits ISD-mediated production of IFN-β and the activation of IFN-β promoter induced by cGAS-STING or STING alone. (A) HFF-shNC/shVP24 cells were infected with HSV-1 for 16 h at a multiplicity of infection (MOI) of 5. Cells were harvested and subjected to WB to analyze the VP24 and UL42 protein. (B) HFF-shNC/shVP24 cells were infected with HSV-1 for 2 h before ISD transfection at an MOI of 5. Cells were harvested at 8 hpi and subjected to qRT-PCR to analyze the IFN-β mRNA. (C and D) VP24-Flag plasmid was cotransfected into HEK 293T cells with an IFN-β reporter plasmid (200 ng), cGAS (15 ng), and a minimal amount of STING plasmid (2.5 ng) or a large amount of STING plasmid (200 ng), respectively. Renilla luciferase reporter plasmid (pRL-TK; 50 ng) was introduced into each transfection as an internal control to normalize transfection efficiency. Cells were harvested at 24 h posttransfection and subjected to the DLR assay to detect the IFN-β reporter activity. The data represent results from one of the triplicate experiments.
VP24 inhibits activation of the IFN-β promoter through IRF3 elements.
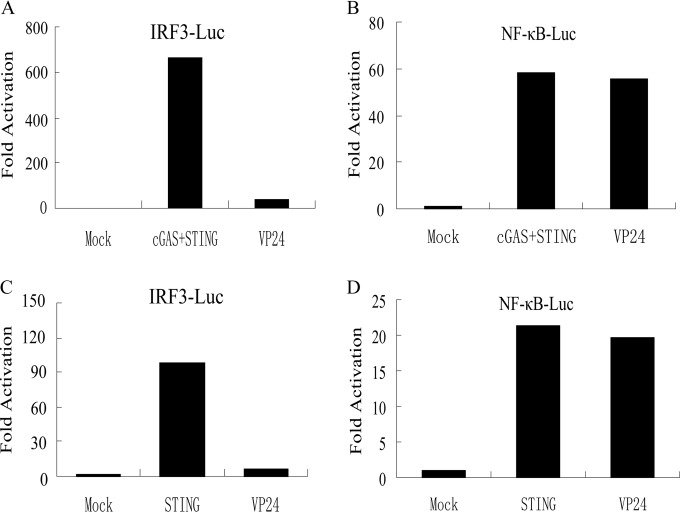
The transcription of IFN-β depends on synergistic interactions of NF-κB and IRFs that bind to distinct regulatory domains in the promoter. To examine the role of VP24 in inhibition of cGAS-STING- or STING-mediated activation of IRFs and NF-κB, the expression of luciferase reporter genes driven by IRF3 or NF-κB element (IRF3-Luc or NF-κB-Luc) in the IFN-β promoter were measured. As a result, ectopic expression of cGAS and a minimal amount of STING or a large amount of STING alone resulted in strong induction of both IRF3-Luc and NF-κB-Luc promoter activity; however, ectopic expression of VP24 inhibited IRF3-Luc, but not NF-κB-Luc, promoter activity (Fig. 2A to D). Taken together, these results demonstrated that VP24 selectively blocked IRF3 promoter activation.
FIG 2.
VP24 inhibits the activation of IFN-β promoter through IRF3 elements. (A to D) VP24-Flag plasmid was cotransfected into HEK 293T cells with a NF-κB reporter plasmid (200 ng) (A and C) or an IRF3 reporter plasmid (200 ng) (B and D) with cGAS (15 ng) and a minimal amount of STING (2.5 ng) plasmid or a large amount of STING plasmid (200 ng), respectively. Cells were harvested 24 h posttransfection and subjected to the DLR assay. The data represent results from one of the triplicate experiments.
VP24 inhibits IFN-β production by targeting IRF3.
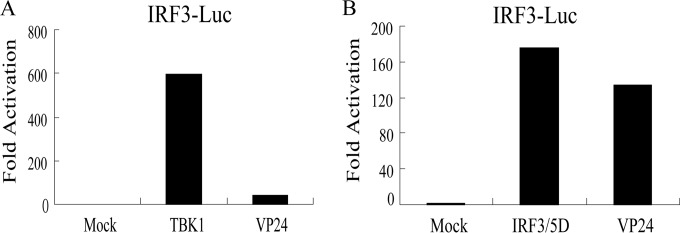
To determine which level in the pathway VP24 blocks IFN-β expression, HEK 293T cells were cotransfected with VP24 plasmids and plasmids expressing adaptor protein downstream of STING, including TBK1 kinase and the active form of IRF3 (IRF3/5D). All expression constructs resulted in a 180- to 600-fold induction of IRF3-Luc reporter activity (Fig. 3A and B). Activation driven by TBK1 was inhibited more than 90% (Fig. 3A), and activation driven by IRF3/5D was not affected (Fig. 3B). Collectively, these results suggested that VP24 inhibited the IFN antiviral response at the IRF-3 level.
FIG 3.
VP24 inhibits IFN-β production by targeting IRF3. HEK 293T cells were transfected with plasmids expressing TBK1 (200 ng) (A) or IRF3/5D (200 ng) (B), together with an IRF3 reporter plasmid. The DLR assay was performed 24 h posttransfection. All cells were transfected with pRL-TK (50 ng) as an internal control to normalize transfection efficiency. The data represent results from one of the triplicate experiments.
VP24 blocks the phosphorylation and dimerization of IRF3.
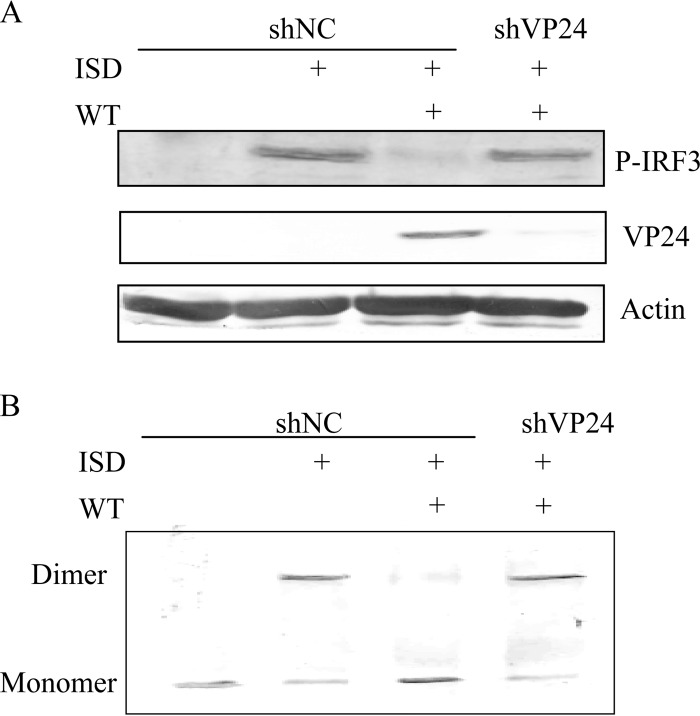
IRF3 is a crucial transcription factor in the IFN-β signaling pathway; in response to cellular stimulation, IRF3 is phosphorylated by TBK1, leading to its homodimerization and translocation into the nucleus. To investigate whether VP24 inhibited the phosphorylation of IRF-3, HFF transfected with pSIREN-shNC or pSIREN-shVP24 (HFF-shNC/shVP24 cells) were infected with WT HSV-1 before ISD transfection, and robust phosphorylation of IRF-3 was observed in HFF-shNC cells following ISD stimulation (Fig. 3A). However, HSV-1 infection abrogated the phosphorylation of IRF-3, and knockdown of VP24 restored its phosphorylation (Fig. 4A). Similarly, IRF-3 dimerization was markedly reduced during HSV-1 infection, and knockdown of VP24 recovered its dimerization (Fig. 4B). Taken together, these results demonstrated that VP24 dampened the activation of IRF-3 induced by ISD.
FIG 4.
VP24 blocks the phosphorylation and dimerization of IRF3. (A) HFF-shNC/shVP24 cells were infected with WT HSV at an MOI of 5 for 2 h before transfection with ISD. Western blot analysis was performed 16 h posttransfection to detect IRF3 phosphorylation. (B) HFF-shNC/shVP24 cells were mock infected or infected with WT HSV at an MOI of 5 for 2 h before transfection with ISD. Cells were harvested at 18 hpi, and native PAGE assays were performed to detect IRF3 dimerization. The data represent results from one of the triplicate experiments.
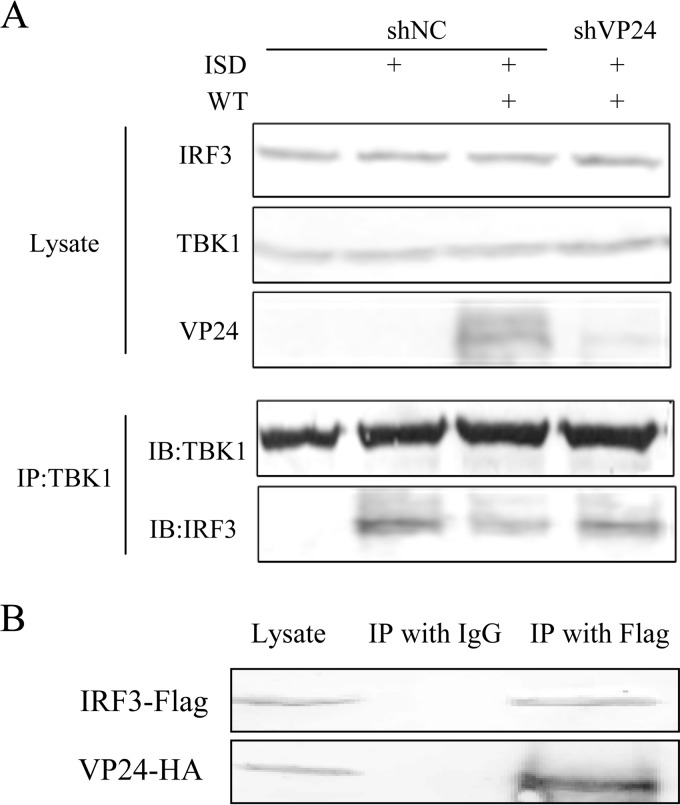
VP24 prevents the interaction between endogenous TBK1 and IRF3 during HSV-1 infection by interacting with IRF3.
On the basis of these data, we speculate that VP24 might affect the interaction between TBK1 and IRF3. To test our hypothesis, we next determined ISD-induced interaction between TBK1 and IRF3 in HFF-shNC and HFF-shVP24 cells infected with HSV-1. HFF-shNC/shVP24 cells were infected with HSV-1 for 2 h before ISD transfection and subjected to the co-IP assay to detect the interaction between endogenous TBK1 and IRF3. As shown in Fig. 5A, HSV-1 infection reduced the interaction between TBK1 and IRF3 induced by ISD, while knockdown of VP24 restored their interaction. These data suggested that VP24 blocked the interaction of TBK1 and IRF3. In order to further explore the underlying mechanisms, HEK 293T cells were transfected with VP24-HA plasmid and 36 h after transfection, cells were harvested and subjected to the co-IP assay to detect the interaction between VP24 and IRF3. As a result, VP24 was efficiently co-IPed with IRF-3 by anti-IRF3 PAb, but not by nonspecific mouse monoclonal antibody IgG (Fig. 5B). These results demonstrated that VP24 could prevent the interaction between endogenous TBK1 and IRF3 during HSV-1 infection by interacting with IRF3.
FIG 5.
VP24 prevents the interaction between endogenous TBK1 and IRF3 during HSV-1 infection by interacting with IRF3. (A) HFF-shNC/shVP24 cells were infected with WT HSV at an MOI of 5 for 2 h before transfection with ISD. Cells were harvested at 18 hpi, and the extracts were analyzed by immunoprecipitation using anti-TBK1 PAb (IP:TBK1), and Western blotting with anti-TBK1, anti-IRF3, and anti-VP24. The data represent results from one of the triplicate experiments. (B) HEK 293T cells were transfected with VP24-HA plasmid and harvested at 36 h posttransfection, and the extracts were analyzed by immunoprecipitation using IRF3 PAb (IP:IRF3) and WB with anti-HA and anti-IRF3. The data represent results from one of the triplicate experiments.
DISCUSSION
HSV-1 is a highly successful human pathogen that has evolved multiple evasion mechanisms to facilitate its proliferation. To date, a number of HSV-1 proteins have been reported to inhibit host innate antiviral responses, and all these proteins may function cooperatively during HSV-1 infection. During HSV-1 capsid maturation, the UL26-encoded protease processes itself to release the N-terminal protease domain VP24. However, the role of VP24 in the innate immune response of the host is still largely unknown.
The type I IFN signal pathway is the first line of host innate defense against viral infection, and it is responsible for the induction of numerous ISGs. Recent research reveals that the type I IFN signal pathway is activated upon recognition of viral nucleic acids by several cytosolic DNA sensors, such as cGAS, DAI, IFI16, and DDX41 (51), among which cGAS was considered the major DNA sensor. Upon binding the viral DNA segment, cGAS is activated and produces cGAMP to activate adaptor protein STING, followed by recruitment of TBK1, phosphorylation and nuclear localization of IRF3, and eventually, the series of events lead to IFN-β production. Growing evidence shows that the cGAS-STING DNA-sensing signal pathway plays a pivotal role in host defense against HSV-1 infection. One can speculate that viruses might have evolved certain mechanisms to impede the cGAS-STING signal pathway. Ma et al. reported that several Kaposi's sarcoma-associated herpesvirus (KSHV) proteins could block IFN-β promoter activity induced by the cGAS-STING signal pathway. One KSHV protein, viral interferon regulatory factor 1, was identified to inhibit the cytosol cGAS-STING signal by blocking the STING-TBK1 interaction (11). A recent study by Wu et al. revealed that KSHV ORF52, a gammaherpesvirus-specific tegument protein, could prevent cGAS-mediated DNA-sensing signal by directly inhibiting cGAS enzymatic activity (52). However, until now, little has been known about the countermeasures by DNA viruses against the cGAS-STING signal pathway, and no HSV-1 gene product has been found to target this pathway yet. IRF3 also plays a crucial role in the cGAS-STING signal pathway, as all signals ultimately converge at IRF3 or IRF7. Therefore, it is not surprising that viruses have evolved strategies to counteract innate immune responses by targeting IRF3.
Taken together, our results provide further information on the mechanism by which HSV-1 antagonizes the host antiviral innate immune response. In the present study, we have demonstrated that the HSV-1 VP24 protein inhibited cGAS-STING-mediated IFN-β signaling pathway by blocking the interaction between TBK1 and IRF3 during HSV-1 infection. The findings in this study expand our knowledge on the molecular mechanisms by which HSV-1 counteracts the antiviral innate immunity of the host to ensure its replication and spread.
ACKNOWLEDGMENTS
We thank Rongtuan Lin for STING-HA plasmid, Yi-Ling Lin for IRF3/5D plasmid, and Takashi Fujita for IFN-β-Luc.
Work in the Zheng laboratory relevant to this article was supported by grants from the National Natural Science Foundation of China (81371795 and 81571974) and the Innovative Research Team at Soochow University (PCSIRT IRT1075).
C.Z. is an adjunct professor of the Department of Microbiology, Immunology and Infectious Diseases, University of Calgary, Calgary, Alberta, Canada.
REFERENCES
- 1.Pichlmair A, Reis e Sousa C. 2007. Innate recognition of viruses. Immunity 27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. 2009. Innate immunity to virus infection. Immunol Rev 227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vandevenne P, Sadzot-Delvaux C, Piette J. 2010. Innate immune response and viral interference strategies developed by human herpesviruses. Biochem Pharmacol 80:1955–1972. doi: 10.1016/j.bcp.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 9.Abe T, Barber GN. 2014. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol 88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dempsey A, Bowie AG. 2015. Innate immune recognition of DNA: a recent history. Virology 479-480:146–152. doi: 10.1016/j.virol.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. 2015. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A 112:E4306–E4315. doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diner EJ, Vance RE. 2014. Taking the STING out of cytosolic DNA sensing. Trends Immunol 35:1–2. doi: 10.1016/j.it.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 13.Bhat N, Fitzgerald KA. 2014. Recognition of cytosolic DNA by cGAS and other STING-dependent sensors. Eur J Immunol 44:634–640. doi: 10.1002/eji.201344127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 89:1–47. [DOI] [PubMed] [Google Scholar]
- 16.Sarkar SN, Sen GC. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther 103:245–259. doi: 10.1016/j.pharmthera.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Leib DA. 2002. Counteraction of interferon-induced antiviral responses by herpes simplex viruses. Curr Top Microbiol Immunol 269:171–185. [DOI] [PubMed] [Google Scholar]
- 18.Mossman KL, Ashkar AA. 2005. Herpesviruses and the innate immune response. Viral Immunol 18:267–281. doi: 10.1089/vim.2005.18.267. [DOI] [PubMed] [Google Scholar]
- 19.Paladino P, Mossman KL. 2009. Mechanisms employed by herpes simplex virus 1 to inhibit the interferon response. J Interferon Cytokine Res 29:599–607. doi: 10.1089/jir.2009.0074. [DOI] [PubMed] [Google Scholar]
- 20.Mossman KL, Macgregor PF, Rozmus JJ, Goryachev AB, Edwards AM, Smiley JR. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J Virol 75:750–758. doi: 10.1128/JVI.75.2.750-758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholl MJ, Robinson LH, Preston CM. 2000. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J Gen Virol 81:2215–2218. doi: 10.1099/0022-1317-81-9-2215. [DOI] [PubMed] [Google Scholar]
- 22.Lin R, Noyce RS, Collins SE, Everett RD, Mossman KL. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J Virol 78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melroe GT, DeLuca NA, Knipe DM. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J Virol 78:8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mossman K. 2005. Analysis of anti-interferon properties of the herpes simplex virus type I ICP0 protein. Methods Mol Med 116:195–205. [DOI] [PubMed] [Google Scholar]
- 25.Paz S, Vilasco M, Arguello M, Sun Q, Lacoste J, Nguyen TL, Zhao T, Shestakova EA, Zaari S, Bibeau-Poirier A, Servant MJ, Lin R, Meurs EF, Hiscott J. 2009. Ubiquitin-regulated recruitment of IkappaB kinase epsilon to the MAVS interferon signaling adapter. Mol Cell Biol 29:3401–3412. doi: 10.1128/MCB.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Wang K, Wang S, Zheng C. 2013. Herpes simplex virus 1 E3 ubiquitin ligase ICP0 protein inhibits tumor necrosis factor alpha-induced NF-kappaB activation by interacting with p65/RelA and p50/NF-kappaB1. J Virol 87:12935–12948. doi: 10.1128/JVI.01952-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson KE, Song B, Knipe DM. 2008. Role for herpes simplex virus 1 ICP27 in the inhibition of type I interferon signaling. Virology 374:487–494. doi: 10.1016/j.virol.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang L, Roizman B. 2008. Expression of gamma interferon-dependent genes is blocked independently by virion host shutoff RNase and by US3 protein kinase. J Virol 82:4688–4696. doi: 10.1128/JVI.02763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Ni L, Wang S, Zheng C. 2014. Herpes simplex virus 1 protein kinase US3 hyperphosphorylates p65/RelA and dampens NF-kappaB activation. J Virol 88:7941–7951. doi: 10.1128/JVI.03394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol 87:12814–12827. doi: 10.1128/JVI.02355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xing J, Wang S, Lin R, Mossman KL, Zheng C. 2012. Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J Virol 86:3528–3540. doi: 10.1128/JVI.06713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Wang K, Li J, Zheng C. 2013. Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J Virol 87:11851–11860. doi: 10.1128/JVI.01211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Wang S, Wang K, Zheng C. 2013. Herpes simplex virus 1 DNA polymerase processivity factor UL42 inhibits TNF-alpha-induced NF-kappaB activation by interacting with p65/RelA and p50/NF-kappaB1. Med Microbiol Immunol 202:313–325. doi: 10.1007/s00430-013-0295-0. [DOI] [PubMed] [Google Scholar]
- 34.Xing J, Ni L, Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-kappaB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J Virol 87:9788–9801. doi: 10.1128/JVI.01440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elgadi MM, Hayes CE, Smiley JR. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J Virol 73:7153–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwong AD, Frenkel N. 1987. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci U S A 84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smiley JR. 2004. Herpes simplex virus virion host shutoff protein: immune evasion mediated by a viral RNase? J Virol 78:1063–1068. doi: 10.1128/JVI.78.3.1063-1068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen G, Wang K, Wang S, Cai M, Li ML, Zheng C. 2014. Herpes simplex virus 1 counteracts viperin via its virion host shutoff protein UL41. J Virol 88:12163–12166. doi: 10.1128/JVI.01380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su C, Zhang J, Zheng C. 2015. Herpes simplex virus 1 UL41 protein abrogates the antiviral activity of hZAP by degrading its mRNA. Virol J 12:203. doi: 10.1186/s12985-015-0433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, Roizman B. 1992. Differentiation of multiple domains in the herpes simplex virus 1 protease encoded by the UL26 gene. Proc Natl Acad Sci U S A 89:2076–2080. doi: 10.1073/pnas.89.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Person S, Laquerre S, Desai P, Hempel J. 1993. Herpes simplex virus type 1 capsid protein, VP21, originates within the UL26 open reading frame. J Gen Virol 74:2269–2273. doi: 10.1099/0022-1317-74-10-2269. [DOI] [PubMed] [Google Scholar]
- 42.Xing J, Wang S, Lin F, Pan W, Hu CD, Zheng C. 2011. Comprehensive characterization of interaction complexes of herpes simplex virus type 1 ICP22, UL3, UL4, and UL20.5. J Virol 85:1881–1886. doi: 10.1128/JVI.01730-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xing J, Wu F, Pan W, Zheng C. 2010. Molecular anatomy of subcellular localization of HSV-1 tegument protein US11 in living cells. Virus Res 153:71–81. doi: 10.1016/j.virusres.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ehrhardt C, Kardinal C, Wurzer WJ, Wolff T, von Eichel-Streiber C, Pleschka S, Planz O, Ludwig S. 2004. Rac1 and PAK1 are upstream of IKK-epsilon and TBK-1 in the viral activation of interferon regulatory factor-3. FEBS Lett 567:230–238. doi: 10.1016/j.febslet.2004.04.069. [DOI] [PubMed] [Google Scholar]
- 45.Chang TH, Liao CL, Lin YL. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-kappaB activation. Microbes Infect 8:157–171. doi: 10.1016/j.micinf.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 46.Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S, Wilkinson P, Julkunen I, Vitour D, Meurs E, Hiscott J. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol 80:6072–6083. doi: 10.1128/JVI.02495-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jordan M, Schallhorn A, Wurm FM. 1996. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res 24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Zhu H, Zheng C, Xing J, Wang S, Li S, Lin R, Mossman KL. 2011. Varicella-zoster virus immediate-early protein ORF61 abrogates the IRF3-mediated innate immune response through degradation of activated IRF3. J Virol 85:11079–11089. doi: 10.1128/JVI.05098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keating SE, Baran M, Bowie AG. 2011. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol 32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, Konrad A, Neipel F, Sturzl M, Whitby D, Li H, Zhu F. 2015. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]