ABSTRACT
While the role of high-risk human papillomavirus (HPV) oncoproteins E6 and E7 in targeting p53 and retinoblastoma (Rb) has been intensively studied, how E6 and E7 manipulate cellular signaling cascades to promote the viral life cycle and cancer development is less understood. Keratinocytes containing the episomal HPV-16 genome had decreased activation of AKT, which was phenocopied by HPV-16 E7 expression alone. Attenuation of phosphorylated AKT (pAKT) by E7 was independent of the Rb degradation function of E7 but could be ablated by a missense mutation in the E7 carboxy terminus, H73E, thereby defining a novel structure-function phenotype for E7. Downstream of AKT, reduced phosphorylation of p70 S6K and 4E-BP1 was also observed in E7-expressing keratinocytes, which coincided with an increase in internal ribosomal entry site (IRES)-dependent translation that enhanced the expression of several cellular proteins, including MYC, Bax, and the insulin receptor. The decrease in pAKT mediated by E7 is in contrast to the widely observed increase of pAKT in invasive cervical cancers, suggesting that the activation of AKT signaling could be acquired during the progression from initial productive infections to invasive carcinomas.
IMPORTANCE HPV causes invasive cervical cancers through the dysregulation of the cell cycle regulators p53 and Rb, which are degraded by the viral oncoproteins E6 and E7, respectively. Signaling cascades contribute to cancer progression and cellular differentiation, and how E6 and E7 manipulate those pathways remains unclear. The phosphoinositol 3-kinase (PI3K)/AKT pathway regulates cellular processes, including proliferation, cell survival, and cell differentiation. Surprisingly, we found that HPV-16 decreased the phosphorylation of AKT (pAKT) and that this is a function of E7 that is independent of the Rb degradation function. This is in contrast to the observed increase in AKT signaling in nearly 80% of cervical cancers, which typically show an acquired mutation within the PI3K/AKT cascade leading to constitutive activation of the pathway. Our observations suggest that multiple changes in the activation and effects of AKT signaling occur in the progression from productive HPV infections to invasive cervical cancers.
INTRODUCTION
The causative link between human papillomavirus 16 (HPV-16) infection and the development of cervical cancer is well established (reviewed in reference 1). High-risk alpha genera HPV E6 and E7 oncoproteins interact with and degrade p53 and retinoblastoma (Rb), respectively, to alter cell cycle regulation (reviewed in references 2, 3, and 4). However, less is known about the interaction of E6 and E7 with cellular proteins that manipulate cellular signaling cascades. We sought to examine the role of HPV-16, and specifically E7 (here 16E7), in manipulating cellular signaling pathways critical to the survival of the cell and initially focused upon the phosphoinositol 3-kinase (PI3K)/AKT pathway.
AKT was originally identified as the causative agent in the acute transforming retrovirus AKT8, which causes spontaneous lymphomas in mice (5). Human homologues of v-akt were identified as AKT1 and AKT2, and further studies found AKT1 to be upregulated in gastric adenocarcinomas, further validating the oncogenic potential of AKT (5). Taken together, these results showed that AKT alone could act as a transforming oncogene.
AKT can be activated by several upstream signaling receptors that result in the activation of PI3K (6). Once activated, PI3K phosphorylates the inositol PIP2 to PIP3. The presence of PIP3 in the cellular membrane recruits PH domain-containing proteins, including both AKT and PDK1. AKT activation is achieved through the sequential phosphorylation of two AKT sites: T308 (located within the catalytic domain) and S473 (located within the regulatory domain). Once recruited to the plasma membrane by PIP3, AKT undergoes a conformational change whereby the PH domain no longer covers the catalytic domain, leaving T308 accessible to be phosphorylated. Two kinases are responsible for the phosphorylation of the activation sites: PDK1 phosphorylates T308, and mammalian target of rapamycin complex 2 (mTORC2) phosphorylates S473, both of which are needed for full activation (6). Once fully activated, AKT plays a role in multiple downstream cellular processes, including cell survival, protein translation, metabolism, and proliferation (reviewed in reference 7). AKT both activates and inhibits multiple proteins directly to alter its downstream signaling cascade. The vast array of cellular processes that AKT manipulates informs its importance in the overall fate of the cell.
AKT regulates protein translation through phosphorylation of the downstream target mTORC1 and subsequent activation and inhibition of p70 S6K and 4E-BP1, respectively (8). While the role of S6 phosphorylation by S6K remains unclear, 4E-BP1 has a clear role in the regulation of translation, as it binds eukaryotic initiation factor 4E (eIF4E) to prevent its association with the translation initiation complex, thereby inhibiting cap-dependent translation (8, 9). In the unphosphorylated state, 4E-BP1 is active and binds to eIF4E. However, once phosphorylated, 4E-BP1 is inhibited, and cap-dependent translation can be initiated through the interaction of eIF4E with the translation initiation complex and the mRNA cap (8).
Cap-dependent translation is activated when growth factors bind and activate upstream signals feeding through the PI3K/AKT/mTOR signaling cascade. In times of nutrient deprivation or under stress conditions, cap-dependent translation is inhibited by 4E-BP1. However, a subset of cellular proteins is still translated during these times through cap-independent mechanisms via internal ribosomal entry sites (IRES) (10). IRES elements were first described in poliovirus, where viral proteins degrade translational regulators and inhibit cap-dependent translation, which then promotes the translation of viral proteins under the influence of the IRES sequence (11). Later studies utilized poliovirus's inhibition of cap-dependent translation to determine if there was a set of cellular proteins that are translated during poliovirus infections. These data demonstrated that cellular proteins associated with the polysomes even in the presence of poliovirus, suggesting that IRES-dependent translation was not exclusive to viral mRNAs (12). Using a cDNA microarray, mRNAs that associated with polysomes in the presence of poliovirus were identified, including c-MYC. Further studies showed diverse cellular mRNAs that contain IRES elements within their 5′ untranslated regions (UTRs), further demonstrating the potential of IRES-dependent translation to regulate cellular protein expression (13).
Given the importance of the AKT signaling pathway on cellular processes, we investigated if HPV-16, and specifically E7, manipulated the AKT signaling cascade.
MATERIALS AND METHODS
Cell culture.
Normal immortalized keratinocytes (NIKS) are spontaneously immortalized foreskin keratinocytes that are both feeder cell and growth factor dependent for proliferation and support of the complete HPV life cycle (14). NIKS were cocultured with mitomycin C-treated 3T3 cells in F medium as described previously (14). NIKS were retrovirally transduced with replication-defective murine retroviruses or transfected with the HPV-16 genome as previously described (14, 15). To investigate AKT phosphorylation, 4 × 105 trypsinized NIKS cells were plated on 6-well plates (day 1) together with mitomycin C-treated feeder cells. Approximately 48 h later, 3T3 cells were removed by light trypsinization, and the remaining keratinocytes were cultured in F medium for another 10 h (day 3, morning). The subconfluent, transduced NIKS were then placed into starvation medium (F medium without fetal bovine serum [FBS], epidermal growth factor [EGF], or insulin) for 12 h. Transduced NIKS were then stimulated with complete F medium for 5, 10, 15, 20, 30, or 360 min. Cells were then washed with ice-cold phosphate-buffered saline (PBS) and then lysed in 1% SDS. Primary keratinocytes were obtained from neonatal foreskins collected anonymously from the University of Virginia Medical Center following a previously described protocol (16). Primary keratinocytes were cultured in F medium with mitomycin C-treated 3T3 cells.
Western blotting.
SDS-lysed NIKS cell lysates were equalized for protein concentration with Bio-Rad protein assay reagents. Equal amounts of protein-normalized samples were loaded onto SDS-acrylamide gels, electrophoresed, and transferred onto polyvinylidene difluoride (PVDF) membranes. Blots were probed with the indicated antibodies: from Cell Signaling (antibody number in parentheses), pAKT T308 (2965), panAKT (2920), c-MYC (2276), insulin receptor (3020), Rb 4H1 (9309), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (3683), p70 S6K T389 (9205), p4EBP1 S65 9451, and 4EBP1 9644; from Sigma, tubulin (T9026); from BD Biosciences, p16 (550834) and Bax (610982); from Thermo Scientific, actin (ACTN05) (MS-1295-P1) and p53 (MA1-19055). HPV-16 E7 was obtained from 2 sources: Santa Cruz (sc-6981) and Invitrogen (catalog no. 28-0006). The Total S6K antibody was a gift from Janet Cross at the University of Virginia.
Luciferase assays.
Retrovirally transduced NIKS were plated as described above and transfected with the CrPV IRES Reporter (17) on day 2. Forty-eight hours later, subconfluent, transduced NIKS, with feeder cells removed, were starved for 12 h and then harvested in 1× passive lysis buffer; lysates were analyzed using the Promega dual-luciferase reporter assay system (catalog no. E1960).
RT-PCR.
Retrovirally transduced NIKS were plated as described above, but harvested following a TRIzol RNA harvest protocol (Invitrogen). cDNA was then generated via first-strand cDNA synthesis using a Moloney murine leukemia virus (M-MLV) reverse transcription (RT) protocol. Quantitative real-time PCR (qPCR) was prepared using the previously synthesized cDNA and the SSO Advanced SYBR green Supermix (Bio-Rad catalog no. 1725264). The primers utilized were Bax (5′-CATGTTTTCTGACGGCAACTTC-3′ and 5′-AGGGCCTTGAGCACCAGTTT-3′), insulin receptor (5′-CTGCACAACGTGGTTTTCGT-3′ and 5′-ACGGCCACCGTCACATTC-3′), c-MYC (5′-TCAAGAGGCGAACACACAAC-3′ and 5′-GGCCTTTTCATTGTTTTCCA-3′), hypoxanthine phosphoribosyltransferase (HPRT) (5′-TGACACTGGCAAAACAATGCA-3′ and 5′-GGTCCTTTTCACCAGCAAGCT-3′), AKT1 (5′-ATGAGCGACGTGGCTATTGTGAAT-3′ and 5′-GAGGCCGTCAGCCACAGTCTGGATG-3′), AKT2 (5′-ATGAATGAGGTGTCTGTCATCAAAGAAGGC-3′ and 5′-TGCTTGAGGCTGTTGGCGACC-3′). Relative values were analyzed using the ΔΔCT method (where CT is threshold cycle) and using HPRT as a control.
7-Methyl-GTP binding.
The ability of 4EBP-1 to bind the mRNA cap structure was assessed through a 7-methyl-GTP binding assay as described previously (18). Briefly, NIKS were plated as described for previous experiments and lysed in buffer D (50 mM HEPES [pH 7.4], 40 mM NaCl, 2 mM EDTA, and 0.1% Triton X-100). Cell lysates were incubated with 7-methyl-GTP-Sepharose Beads (a gift from Thurl Harris at the University of Virginia) for 1 h at 4°C. 7-Methyl beads were washed three times, and bound proteins were analyzed via SDS-PAGE and Western blotting.
RESULTS
HPV-16E7 attenuates the activation of AKT.
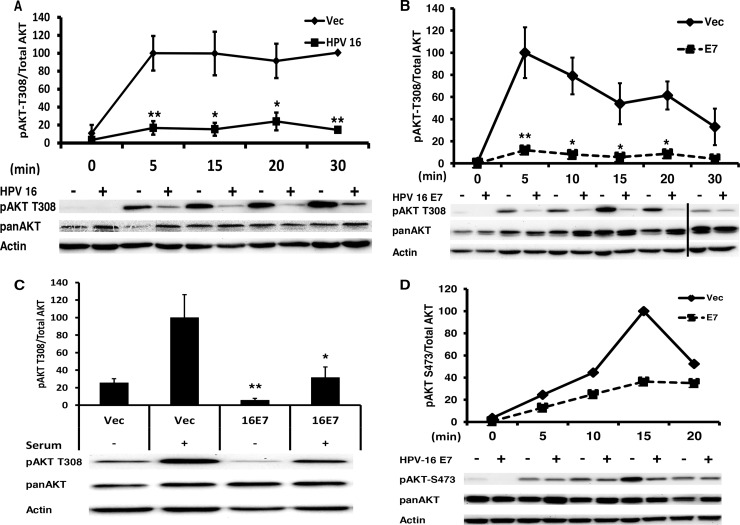
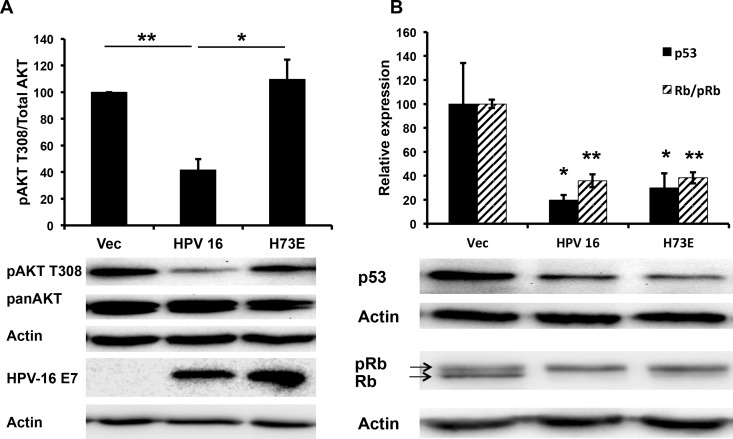
AKT signaling is typically activated in invasive squamous cell carcinomas, including cervical cancer (19). Surprisingly, we observed that in normal immortalized human foreskin keratinocytes (NIKS) transfected with the episomal HPV-16 genome, phosphorylation of AKT at T308 (pAKT) was attenuated upon stimulation with F medium (Fig. 1A).
FIG 1.
Papillomavirus E7 attenuates pAKT T308. (A) HPV-16 attenuates pAKT T308. Immortalized keratinocytes (NIKS) transfected with the HPV-16 genome were plated in coculture with mitomycin C-treated NIH 3T3 cells; 48 h later, NIH 3T3 cells were removed, and HPV-16 NIKS were placed back in F medium. Approximately 10 h later, HPV-16 NIKS were starved in F medium lacking FBS, insulin, and EGF for 12 h and then stimulated with complete F medium for the indicated time, harvested in 1% SDS, protein normalized, and then analyzed via Western blotting. Samples were normalized to actin and the highest value. Statistics were calculated from four independent experiments (*, P < 0.05; **, P < 0.01), with Western blots from a single representative experiment shown. (B) NIKS retrovirally transduced with 16E7 attenuate pAKT T308. NIKS transduced with vector or 16E7 were plated and harvested as described for panel A. Samples were normalized to actin and the highest value. Statistics were calculated from three independent experiments. The vertical bar in the Western blot represents the removal of the of a redundant time point (*, P < 0.05; **, P < 0.01). (C) E7 attenuates pAKT T308 in primary keratinocytes. Primary keratinocytes retrovirally transduced with Vec or 16E7 were plated and harvested as described for panel A in F medium. Statistics were calculated from four independent experiments. *, P < 0.05; **, P < 0.01. (D) pAKT S473 is attenuated by 16E7 in keratinocytes. Retrovirally transduced NIKS were plated and harvested as described for panel A. The Western blot shown is representative of three independent experiments.
To elucidate if HPV-16E7 was playing a role in the attenuation of AKT signaling, we retrovirally transduced NIKS with HPV-16 E7 (16E7) alone and found that pAKT T308 was again attenuated (Fig. 1B), demonstrating that 16E7 alone could phenocopy the attenuation of pAKT observed in NIKS transfected with the complete HPV-16 genome. This phenotype was replicated in primary keratinocytes maintained in F medium (Fig. 1C). The second AKT activation site, pAKT S473, showed a similar attenuation trend (Fig. 1D).
Culture conditions modulate the effect of E6 upon AKT activation.
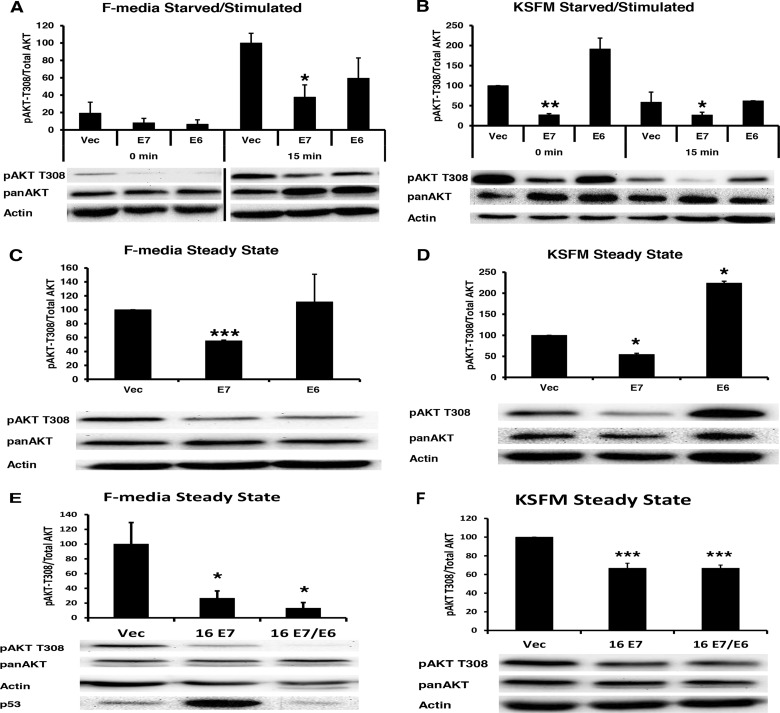
The attenuation of pAKT T308 was unexpected, since it was previously reported that HPV-16 E6 increased pAKT T308 (18). To examine the role of E6 and the contributions that culture conditions might play in AKT activation, NIKS cells expressing 16E6 or 16E7 alone or together were examined for AKT activation in either F medium or keratinocyte serum-free medium (KSFM). Our studies were performed in F medium containing serum, EGF, insulin, feeder cells, and higher calcium levels than KSFM (20). 16E7 attenuated pAKT T308 in both KSFM and F medium (Fig. 2A to F), while 16E6 activated pAKT-T308 only in KSFM and not in F medium or in the presence of E7 (Fig. 2A to F). In NIKS, pAKT T308 levels remained elevated even after starvation in KSFM medium (Fig. 2B), while in F medium, pAKT T308 levels were attenuated after starvation (Fig. 2A). An initial increase above the basal level of pAKT T308 was observed at 5 min in KSFM medium (not shown in Fig. 2B); however, at 15 min the levels had dropped below the basal level (Fig. 2B). Thus, basal activation and the kinetics of pAKT T308 activation and subsequent inactivation are different in F medium versus KSFM. Because KSFM does not allow for adherens junction formation, keratinocyte stratification (21), or papillomavirus replication, our remaining experiments were performed in F medium.
FIG 2.
16E6 does not alter E7-mediated pAKT attenuation. (A) Retrovirally transduced keratinocytes with 16E7 or 16E6 were plated and harvested as described for Fig. 1A. Statistics were calculated from three independent experiments. Bar represents the removal of a redundant time point. (B) Retrovirally transduced keratinocytes with 16E7 or 16E6 were plated and harvested as described for Fig. 1A, except that cells were maintained in KSFM. Statistics were calculated from three independent experiments. (C) Retrovirally transduced keratinocytes with 16E7 or 16E6 were plated and subsequently harvested 3 days later in 1% SDS. Statistics were calculated from three independent experiments. (D) Retrovirally transduced keratinocytes with 16E7 or 16E6 were plated and harvested as described for panel B. Statistics were calculated from three independent experiments. (E) Retrovirally transduced keratinocytes with 16E7 or 16E7/16E6 were plated and harvested as described for Fig. 1A. Statistics were calculated from three independent experiments. (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (F) Retrovirally transduced keratinocytes with 16E7 or 16E7/16E6 were plated and harvested as described for Fig. 2B. Statistics were calculated from four independent experiments. (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
E7 proteins from various papillomavirus genera attenuate AKT phosphorylation.
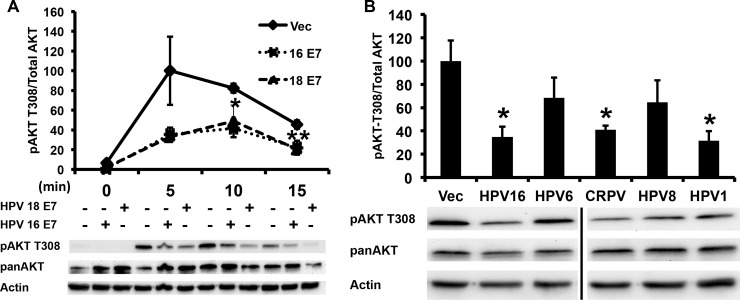
To determine if other high-risk HPV E7 proteins also attenuated pAKT-T308, we compared 16E7 with HPV-18 E7 (18E7) and found that both repressed pAKT T308 (Fig. 3A). Examining other papillomavirus genera, 16E7, HPV-1 E7, and cotton-tailed rabbit papillomavirus all significantly repressed pAKT-T308 while HPV-6 and HPV-8 E7 proteins, while trending down, did not reach statistical significance (Fig. 3B).
FIG 3.
The attenuation of pAKT by E7 varies across papillomavirus genera. (A) The AKT attenuation phenotype is conserved in high-risk 18E7. NIKS retrovirally transduced with either HPV-16 E7 or HPV-18 E7 were plated and harvested as described for Fig. 1A. Samples were normalized to actin and the highest value. Statistics were calculated from three independent experiments (*, P < 0.05; **, P < 0.01). (B) The AKT attenuation phenotype is not exclusive to high-risk E7 types. NIKS retrovirally transduced with the E7 genes from the indicated papillomavirus types were stimulated for 6 h following 12 h of starvation and harvested as described for Fig. 1A. Samples were normalized to actin and the highest value. Statistics were calculated from four independent experiments (*, P < 0.05).
Attenuation of pAKT is independent of the Rb degradation function of E7.
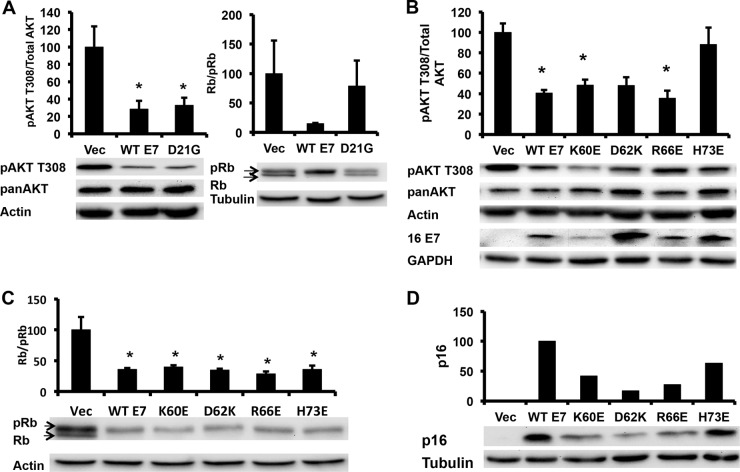
The CR2 region of E7 that binds to retinoblastoma (Rb) family proteins is well conserved between HPV-16 and HPV-1 E7, and the two demonstrate a similar binding affinity for Rb (22). However, unlike HPV-16 E7, HPV-1 E7 does not degrade unphosphorylated Rb (22). Cottontail rabbit papillomavirus E7, while cancer associated, has a lower binding affinity LxCxE motif than does HPV-16 E7, suggesting that the Rb-binding function of E7 might be independent of the pAKT attenuation phenotype (22). To explore how E7 decreased the phosphorylation of AKT, we utilized several 16E7 mutants and determined their effect on AKT phosphorylation. 16E7 D21G converts the high-affinity 16E7 Rb-binding site to the low-affinity Rb-binding site found in the low-risk HPV-6 E7 (23). 16E7 D21G no longer degraded Rb but retained the ability to decrease pAKT T308 (Fig. 4A). Next, we examined the effect of mutating surface-exposed side chains within the carboxyl zinc-structured domain of 16E7. Three 16E7 mutants (K60E, D62K, and R66E) decreased pAKT T308 to the same extent as wild-type (WT) 16E7; however, a single mutant, H73E, restored pAKT T308 to that of vector-transduced NIKS (Fig. 4B). The 16E7_H73E mutant retained the ability to degrade unphosphorylated Rb, similar to WT 16E7 (Fig. 4C).
FIG 4.
Attenuation of pAKT is independent of either p16 induction or Rb degradation by E7. (A) The pAKT attenuation phenotype is independent of a high-affinity LxCxE Rb-binding motif. NIKS retrovirally transduced with WT 16E7 or 16E7 D21G were plated, starved for 12 h, stimulated for 6 h, and harvested as described for Fig. 1A. Samples were normalized to actin and the highest value. Statistics were calculated from four independent experiments (*, P < 0.05). (B) The 16E7 pAKT attenuation phenotype can be ablated by mutation of the C terminus of E7. Results show the Western blot analysis of the C-terminal domain E7 mutants retrovirally transduced into NIKS that were processed as described for Fig. 1A. Samples were normalized to actin and the highest value. Statistics were calculated from three independent experiments (*, P < 0.05). (C) The AKT attenuation phenotype is independent of the E7-mediated degradation of Rb. Results show the Western blot analysis of NIKS retrovirally transduced with E7 zinc-structured domain mutants that were plated and harvested as described for Fig. 1A. Samples were normalized to actin and the highest value. Statistics were calculated from three independent experiments (*, P < 0.05). (D) The AKT attenuation phenotype is independent of the E7-mediated induction of p16. Results show the Western blot analysis of 16E7 mutants retrovirally transduced in NIKS that were plated and harvested as described for Fig. 3A. Samples were normalized to actin and the highest value. Shown is a Western blot that was representative of two independent experiments.
In cervical carcinomas, p16 is utilized as a marker of the presence of high-risk HPV, as studies have shown that high-risk HPV E7 induces overexpression of p16 in keratinocytes and dependence upon p16 expression for viability of E7-expressing keratinocytes (24, 25). A recent study demonstrated that this phenotype was independent of Rb degradation by E7 and could be mapped to the E7 induction of KDM6A and KDM6B to induce demethylation at the p16 promoter, resulting in increased p16 expression (26). In order to determine if decreased pAKT T308 was associated with the induction of p16, lysates from NIKS transduced with Vec, WT 16E7, or 16E7_H73E were examined for p16 expression; WT E7 and 16E7_H73E equally induced p16 compared to vector-transduced NIKS (Fig. 4D). This demonstrated that neither the degradation of Rb nor the induction of p16 was associated with the attenuation of pAKT T308.
Finally, to make sure that 16E7_H73E retained its phenotype when expressed from the complete HPV-16 genome, NIKS transfected with the HPV-16 genome harboring the 16E7 H73E mutation reversed the pAKT phenotype of WT HPV-16 and activated pAKT T308 to the levels of untransfected NIKS upon stimulation with F medium (Fig. 5A). HPV-16_H73E retained Rb and p53 degradation similar to HPV-16 (Fig. 5B). The expression levels of 16E7 and 16E7_H73E from the complete genome were similar (Fig. 5A). Taken together, these results demonstrated that the attenuation of pAKT was independent of the other viral early gene products, was independent of the Rb degradation function of E7, was independent of p16 induction, mapped to a previously uncharacterized site in the zinc-structured domain of E7, and could be specifically ablated by the 16E7_H73E mutation.
FIG 5.
HPV-16E7_H73E reverses pAKT attenuation mediated by HPV-16. (A) NIKS cells transfected with the HPV-16 genome or HPV-16 E7_H73E genome were plated and harvested as described for Fig. 1A. Samples were normalized to actin and the highest value. Statistics were calculated from three independent experiments (*, P < 0.05). (B) HPV-16 E7_H73E NIKS degrade p53 and unphosphorylated Rb to the same extent as HPV-16 NIKS. NIKS cells transfected with the HPV-16 genome or HPV-16 E7_H73E genome were plated and harvested as described for Fig. 1A. Samples were normalized to actin and the highest value. Statistics were calculated from five independent experiments (*, P < 0.05; **, P < 0.01).
HPV-16 E7 alters protein translation.
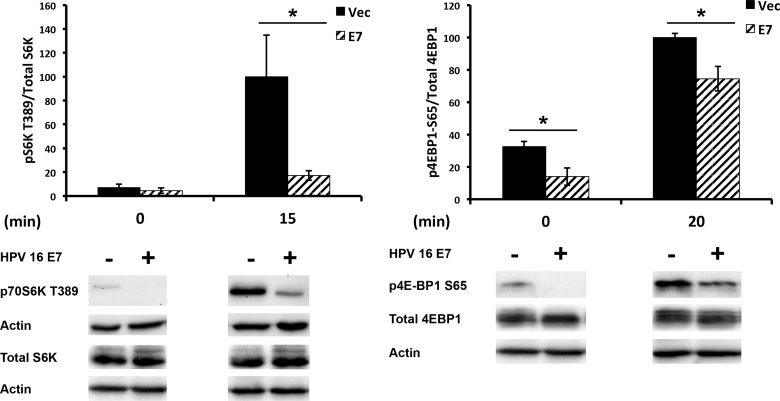
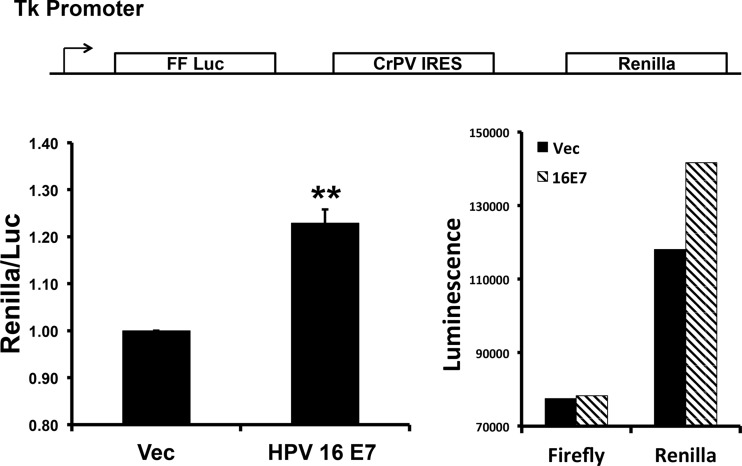
We next investigated the downstream effects of the E7-mediated attenuation of pAKT T308. AKT regulates protein translation through phosphorylation of the downstream effector mTORC1, which subsequently activates and inhibits p70 S6K and 4E-BP1, respectively. The activating phosphorylation site of S6K T389 and the inhibitory phosphorylation site of 4E-BP1 S64 were reduced in 16E7-expressing NIKS (Fig. 6). Decreased phosphorylation of 4E-BP1 results in its binding to and sequestrating eIF4E away from the translational initiation complex, resulting in the inhibition of cap-dependent translation (27). Thus, we wanted to determine if E7 altered the ratio of IRES to cap-dependent translation. To elucidate the ratio of IRES- to cap-dependent translation, we used a bicistronic reporter containing the cricket paralysis virus (CrPV) IRES (17). In NIKS expressing 16E7, compared to vector-transduced NIKS, the ratio of IRES to cap-dependent reporter was significantly shifted toward more-IRES-dependent translation (Fig. 7), suggesting that the decrease in AKT activation, mediated by E7, shifted cellular translation toward IRES-mediated translation.
FIG 6.
HPV-16 E7 decreases phosphorylation of downstream AKT effectors. Results show the Western blot analysis of immortalized keratinocytes retrovirally transduced with HPV-16 E7. Transduced NIKS were processed as described for Fig. 1A. Western blots are representative of 5 independent experiments. Western blots shown are a single blot exposure with the removal of redundant time points. Statistics were calculated from three independent experiments (*, P < 0.05).
FIG 7.
HPV-16 E7 alters cap- versus IRES-dependent translation. NIKS that were retrovirally transduced and selected to express either vector or 16E7 were transfected with a bicistronic-reporter plasmid (shown at the top) and 48 h later were starved for 12 h. Following starvation, transduced NIKS were assayed for firefly and Renilla luciferase activity. Values were normalized to vector-transduced cells. Statistics were calculated from three independent experiments each performed in duplicate (**, P < 0.005).
HPV-16E7 increases protein expression of IRES containing mRNAs.
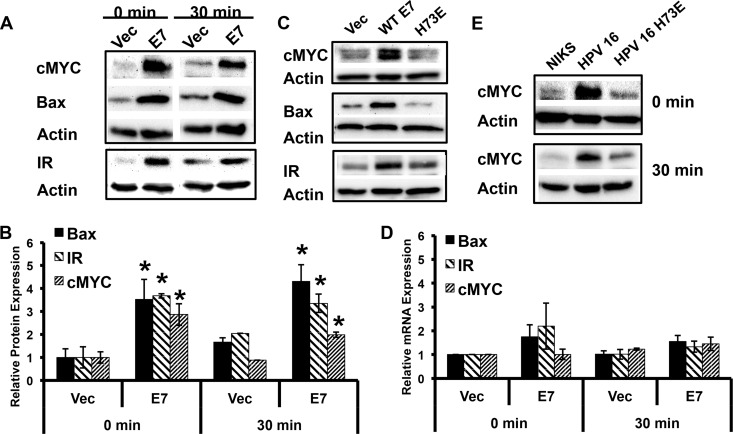
Many cellular proteins translated under the influence of IRES elements are proteins involved in processes such as apoptosis and mitosis, when signaling from growth factor receptors is low. When cap-dependent translation is decreased, translation machinery is readily available for IRES-dependent translation. IRES containing mRNAs include c-MYC and the insulin receptor (13), both of which were increased in 16E7 NIKS compared to vector-transduced NIKS (Fig. 8A and B). The levels of the c-MYC protein were also increased in NIKS containing the complete HPV-16 genome in a comparison of vector-transduced NIKS with NIKS harboring HPV-16 E7_H73E (Fig. 8E). While the protein levels were significantly increased, there was no significant change in the RNA levels of insulin receptor or c-MYC (Fig. 8D). 16E7_H73E, which reversed the pAKT attenuation phenotype, did not enhance the expression levels of c-MYC (Fig. 8C). Taken together, these data demonstrate that the pAKT attenuation mediated by 16E7 enhanced the expression of this set of cellular proteins whose mRNAs contain an IRES element. We found that Bax, a Bcl-2 family member, was also enhanced at the translational level in E7-expressing NIKS in a manner similar to that of insulin receptor and c-MYC (Fig. 8A to D). Although Bax has not yet been described as having an IRES element, our data suggest this possibility.
FIG 8.
HPV-16 E7 enhances protein expression from cellular IRES containing mRNAs. (A) 16E7 enhances the protein expression from three IRES-containing mRNAs. Results show the Western blot analysis of transduced NIKS plated and harvested as described for Fig. 1A. Results from a representative experiment in a cohort of three independent experiments are shown. (B) IRES containing mRNAs have significantly enhanced protein levels in 16E7 NIKS cells. Values were normalized to vector-transduced cells. Results shown represent the quantitation of three independent experiments (*, P < 0.05). (C) A single point mutation in E7 reverses the enhanced expression of IRES-containing proteins. Results show the Western blot analysis of retrovirally transduced NIKS that were plated and harvested as described for Fig. 1A. Results show a representative experiment at 30 min from a set of three independent experiments. (D) Relative RNA levels of IRES-containing proteins remain unchanged in 16E7 NIKS. Transduced NIKS were plated as described for Fig. 1A, and RNA was harvested according to a TRIzol protocol. Values were normalized to vector-transduced cells. Statistics were calculated from three independent experiments each performed in duplicate and showed no significant differences between samples. (E) HPV-16 E7_H73E reverses the enhancement of the c-MYC protein observed in HPV-16-transfected keratinocytes. NIKS containing the complete HPV-16 genome or the HPV-16 E7_H73E genome were processed as described for Fig. 1A. Shown is a representative figure from two independent experiments.
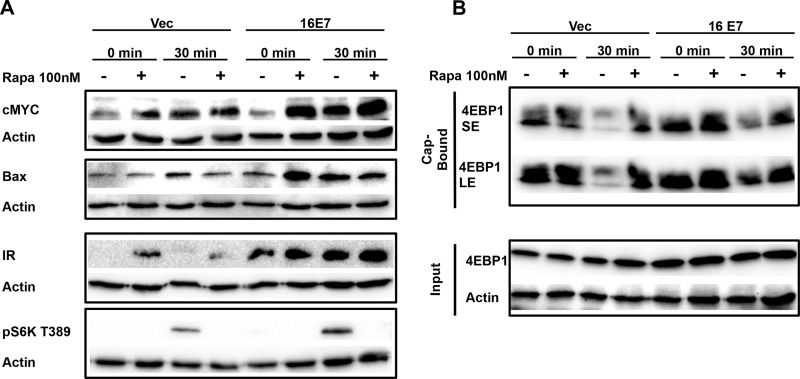
Rapamycin treatment does not alter the enhanced expression of IRES-containing mRNAs.
Rapamycin treatment inhibits cap-dependent translation through inhibition of mTORC1, leading to the dephosphorylation of both S6K and 4EBP-1 (28, 29). Treatment of 16E7-transduced NIKS in the presence or absence of growth factors with rapamycin did not blunt the enhanced expression of cMYC, insulin receptor, or Bax (Fig. 9A). Of note, rapamycin treatment of vector-transduced NIKS slightly induced the protein expression of cMYC and insulin receptor, over the time course of this experiment, further suggesting that inhibition of cap-dependent translation enhances IRES-dependent translation of these proteins.
FIG 9.
16E7 enhanced IRES-dependent translation of cellular mRNAs is not blunted by rapamycin (Rapa) treatment. (A) NIKS were plated and harvested as described for Fig. 1A, except that at 9 h of starvation cells were treated with rapamycin for 3 h and subsequently stimulated in the presence or absence of rapamycin. Blots are representative of three independent experiments. (B) 4EBP1 cap binding is increased in 16E7 samples. NIKS were plated and harvested as described for Fig. 1A. Blots are representative of two independent experiments. SE, short exposure; LE, long exposure.
When 4EBP-1 is bound to eIF4E, eIF4G is blocked from interacting with the complex to initiate cap-dependent translation. 16E7 increased 4EBP-1 binding to the 7-methyl RNA cap structure, through interaction with eIF4E (Fig. 9B), which corresponded to our data showing decreased p4EBP-1 S65 phosphorylation in cells expressing 16E7 (Fig. 6). Taken together, these data suggest that E7 attenuated p4EBP-1 S65, which led to increased cap association of 4EBP-1, inhibition of cap-dependent translation, and enhanced expression of IRES-containing mRNAs.
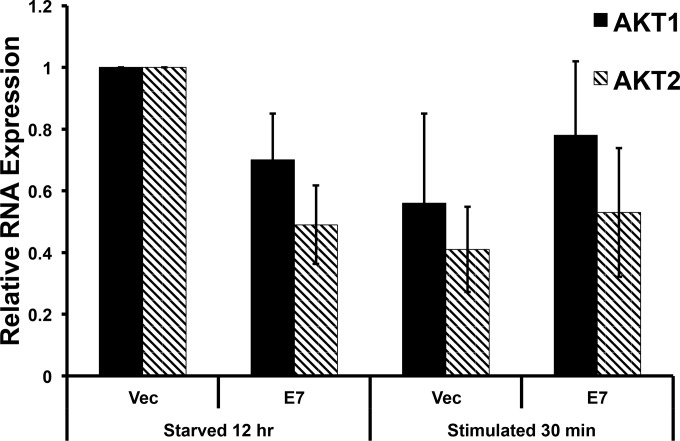
AKT transcription is unchanged in E7-expressing keratinocytes.
Enhanced AKT signaling promotes keratinocyte differentiation, and pharmacologic inhibition of AKT signaling reduces keratinocyte differentiation (30, 31). Paralleling our results, another group has shown a downregulation in AKT1 mRNA mediated by cutaneous HPV-8 E2 (32). They hypothesized that this decrease in AKT1 altered the normal keratinocyte differentiation pathway that weakened the top epidermal layer to promote viral release. However, we did not observe a significant decrease in the expression of AKT1 mRNAs or a significant increase in the expression of AKT2 mRNAs (Fig. 10).
FIG 10.
AKT RNA levels are unchanged in E7 keratinocytes. Retrovirally transduced keratinocytes with 16E7 were plated and harvested as described for Fig. 1A, except that total RNA was harvested following a TRIzol protocol. Statistics were calculated from three independent experiments and showed no significant difference between the samples.
DISCUSSION
Because activated PI3K has been described as characteristic of invasive cervical cancers (33), we expected to observe activation of AKT by the HPV-16 genome. To our surprise, we have shown that HPV-16 attenuates pAKT T308 and that this phenotype can be mapped to E7 alone (Fig. 1). Several other human and animal papillomavirus E7s also exhibit this phenotype, including HPV-18 E7, HPV-1 E7, and cottontail rabbit papillomavirus E7 (Fig. 3).
In contrast to our results, previous studies have shown that 16E7 augments the activation of AKT, promoting an increase in downstream signaling (34, 35). However, in our hands we observe that high-risk and some low-risk E7s attenuate AKT signaling. We realize that experimental differences such as culturing keratinocytes in medium with serum, mitomycin C-treated 3T3 cells, and calcium (F medium) compared to serum-free and low-calcium medium formulations (KSFM) may contribute to the variance in observations. In the first study (34), the effects of E7 were assessed in HEK 293 cells transiently transfected with E7; thus, the effects of E7 in that system must be interpreted in the context of E7 time course, E7 expression levels, the coexpression of adenoviral oncoproteins, and nonkeratinocyte cell lines (34). In the second study (35), the effects of E7 alone in organotypic cultures were examined and pAKT was assessed using phosphospecific antibodies to serine 473 (35). Although we show parallel effects on pAKT S473 versus T308 (Fig. 1), we chose to ascertain T308 phosphorylation because it is the initial activating phosphorylation that enables subsequent S473 phosphorylation. Since both T308 and S473 phosphorylations are required for full AKT activation (6), the decrease of T308 phosphorylation in E7-expressing cells represents the activation state. Reinforcing our interpretation is the observed decrease in the phosphorylation of both S6K and 4E-BP1 (both targets of AKT signaling) in cells expressing 16E7 (Fig. 6). Studies by another group showed that HPV-16 E6 augments AKT and mTORC1 signaling to promote cap-dependent translation (18, 36). As noted in Results, the ability of 16E6 to augment pAKT is manifested in low-calcium medium (KSFM), a medium formula that alters cell adhesion and ablates adherens junctions and stratification. In our system, the pAKT attenuation phenotype from E7 persists even in the presence of E6 (Fig. 2E and F) and in keratinocytes expressing the full HPV-16 genome (Fig. 1A). This suggests that any role that E6 may be playing in augmenting mTORC1 signaling and cap-dependent translation can be overcome by the presence of E7 in our system.
Our mutational analysis revealed a single point mutant, H73E, that reversed the pAKT attenuation phenotype independently of the Rb degradation function of E7 (Fig. 4). In other studies, 16E7 H73E has been shown to dimerize in vivo and transform baby rat kidney cells in cooperation with activated Ras to the same extent as WT E7; however, no biologic function has been associated with this residue until our findings (37, 38). H73E is conserved only with HPV-1 E7, while HPV-18 and cottontail rabbit papillomavirus E7, which also suppress pAKT, have alternate amino acids at this position. This suggests that H73E could be one among several amino acids that contribute to the phenotype. Further studies are required to fully delineate the surfaces of E7 involved in the AKT attenuation phenotype.
While there are many cellular functions regulated by AKT signaling, our initial focus was on translational regulation. We showed that two indirect downstream targets of AKT, S6K and 4E-BP1, had reduced phosphorylation at their activation and inhibitory sites, respectively (Fig. 6). 16E7 alters the translation of a specific subset of proteins, including c-MYC and insulin receptor (Fig. 8), which were identified as containing 5′ IRES elements (13). Other groups have shown that in cells with increased phosphatase and tensin homolog (PTEN) activity, AKT phosphorylation is reduced and c-MYC translation under the influence of its IRES elements is increased (39), which is consistent with our results. We hypothesize that induction of c-MYC could alter the expression of a c-MYC target gene set, which could influence metabolism, cell survival, protein translation, or proliferation (7). If metabolism genes are in fact upregulated by the enhanced protein expression of c-MYC, this could contribute to the induction of the Warburg effect mediated by E7 (40). The enhanced expression of Bax could also contribute to the known phenotype of E7 sensitization of keratinocytes to apoptosis (41). Like c-MYC, the vascular endothelial growth factor (VEGF) also contains a 5′ IRES element in its mRNA. 16E7-transduced keratinocytes secrete more VEGF than vector-transduced keratinocytes (42), and our work suggests a potential mechanism for increased protein translation of VEGF in 16E7 keratinocytes. Increased AKT activity has also been shown to decrease IRES-dependent translation of VEGF (43), which corresponds to our model of attenuated AKT signaling augmenting IRES-dependent translation. Taken together, these studies suggest that VEGF could also be an IRES-influenced protein augmented by the E7-mediated AKT attenuation phenotype that we have described.
Beyond papillomaviruses, there is precedence for viral oncogenes manipulating AKT activation. In polyomavirus, small t (PyST) has recently been shown to block differentiation by binding to and recruiting the activity of PP2A to AKT (44). In contrast, simian virus 40 (SV40) small and large T antigens together immortalize keratinocytes, promote anchorage-independent growth, and activate AKT; the SV40 large T antigen has been implicated as well in the activation of AKT (45). These disparate effects upon AKT signaling must be interpreted in light of the variable and combined effects of these viral oncoproteins. Although HPV-16 E7 has been reported to interact with PP2A leading to the activation of AKT (34), these results have been controversial, with a recent study finding no association of 16E7 with PP2A and no findings of PP2A components as E7-associated proteins (46–48). Finally, the manipulation of AKT by HPV-8 E2 has been reported (32); in that study, E2 repressed the mRNA abundance of AKT. Although this is an interesting parallel observation, we did not observe significant changes in AKT1 or AKT2 RNA levels in E7-expressing cells (Fig. 10).
As noted above, PI3K is activated in most cervical cancers (49). However, our data present a paradox, in that the enhanced AKT activation that is observed in invasive cancers would be predicted to drive keratinocyte differentiation. We can thus speculate that during the progression of HPV-16-infected keratinocytes from low-grade to high-grade lesions, changes in the effect of AKT upon cellular differentiation might precede any enhancement of AKT activation, changing AKT from a driver of keratinocyte differentiation to a driver of cancer cell phenotypes. More experiments are required to fully develop this hypothesis.
We have shown that HPV-16 attenuates pAKT T308 and that this can be mapped to 16E7. We have also shown that both high- and low-risk E7 types attenuate pAKT and that this phenotype is independent of the Rb degradation function of E7. The AKT attenuation phenotype can be ablated by a single point mutation within the carboxy terminus of E7 and is not dependent upon the E7-mediated induction of demethylases to induce epigenetic reprogramming. The phosphorylation of two downstream proteins of AKT (S6K and 4E-BP1) is attenuated, leading to a shift in protein translation toward IRES-dependent translation. The shift enhances the protein translation of several cellular proteins without altering their transcription, including c-MYC, insulin receptor, and Bax. The AKT attenuation is a novel phenotype of E7 and demonstrates a role for E7 in the manipulation of signal transduction cascades, thereby altering the regulation of cellular translation.
ACKNOWLEDGMENTS
We thank Thurl Harris at the University of Virginia for use of the 4E-BP1 antibodies and meaningful discussions and guidance. We also thank Nicole Brimer for help with technical challenges faced during the course of this project.
Funding Statement
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.zur Hausen H. 2009. Papillomaviruses in the causation of human cancers—a brief historical account. Virology 384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 2.McLaughlin-Drubin ME, Munger K. 2009. The human papillomavirus E7 oncoprotein. Virology 384:335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howie HL, Katzenellenbogen RA, Galloway DA. 2009. Papillomavirus E6 proteins. Virology 384:324–334. doi: 10.1016/j.virol.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vande Pol SB, Klingelhutz AJ. 2013. Papillomavirus E6 oncoproteins. Virology 445:115–137. doi: 10.1016/j.virol.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staal SP. 1987. Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: amplification of AKT1 in a primary human gastric adenocarcinoma. Proc Natl Acad Sci U S A 84:5034–5037. doi: 10.1073/pnas.84.14.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao Y, Hung MC. 2010. Physiological regulation of Akt activity and stability. Am J Transl Res 2:19–42. [PMC free article] [PubMed] [Google Scholar]
- 7.Manning BD, Cantley LC. 2007. AKT/PKB signaling: navigating downstream. Cell 129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 1998. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev 12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruvinsky I, Meyuhas O. 2006. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 10.Rubsamen D, Blees JS, Schulz K, Doring C, Hansmann ML, Heide H, Weigert A, Schmid T, Brune B. 2012. IRES-dependent translation of egr2 is induced under inflammatory conditions. RNA 18:1910–1920. doi: 10.1261/rna.033019.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 12.Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci U S A 96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokrejs M, Masek T, Vopalensky V, Hlubucek P, Delbos P, Pospisek M. 2010. IRESite—a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res 38:D131–D136. doi: 10.1093/nar/gkp981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert PF, Ozbun MA, Collins A, Holmgren S, Lee D, Nakahara T. 2005. Using an immortalized cell line to study the HPV life cycle in organotypic “raft” cultures. Methods Mol Med 119:141–155. [DOI] [PubMed] [Google Scholar]
- 15.Miller AD, Rosman GJ. 1989. Improved retroviral vectors for gene transfer and expression. Biotechniques 7:980–982, 984–986, 989–990. [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada S. 2004. Preparation of human epidermal keratinocyte cultures. Curr Protoc Cell Biol Chapter 2:Unit 2.6. doi: 10.1002/0471143030.cb0206s21. [DOI] [PubMed] [Google Scholar]
- 17.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. 2006. Short RNAs repress translation after initiation in mammalian cells. Mol Cell 21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 18.Spangle JM, Munger K. 2010. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J Virol 84:9398–9407. doi: 10.1128/JVI.00974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bertelsen BI, Steine SJ, Sandvei R, Molven A, Laerum OD. 2006. Molecular analysis of the PI3K-AKT pathway in uterine cervical neoplasia: frequent PIK3CA amplification and AKT phosphorylation. Int J Cancer 118:1877–1883. doi: 10.1002/ijc.21461. [DOI] [PubMed] [Google Scholar]
- 20.Boyce ST, Ham RG. 1983. Calcium-regulated differentiation of normal human epidermal keratinocytes in chemically defined clonal culture and serum-free serial culture. J Investig Dermatol 81:33s–40s. doi: 10.1111/1523-1747.ep12540422. [DOI] [PubMed] [Google Scholar]
- 21.Lewis JE, Jensen PJ, Wheelock MJ. 1994. Cadherin function is required for human keratinocytes to assemble desmosomes and stratify in response to calcium. J Investig Dermatol 102:870–877. doi: 10.1111/1523-1747.ep12382690. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt A, Harry JB, Rapp B, Wettstein FO, Iftner T. 1994. Comparison of the properties of the E6 and E7 genes of low- and high-risk cutaneous papillomaviruses reveals strongly transforming and high Rb-binding activity for the E7 protein of the low-risk human papillomavirus type 1. J Virol 68:7051–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heck DV, Yee CL, Howley PM, Munger K. 1992. Efficiency of binding the retinoblastoma protein correlates with the transforming capacity of the E7 oncoproteins of the human papillomaviruses. Proc Natl Acad Sci U S A 89:4442–4446. doi: 10.1073/pnas.89.10.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SS, Trunk M, Schiffman M, Herrero R, Sherman ME, Burk RD, Hildesheim A, Bratti MC, Wright T, Rodriguez AC, Chen S, Reichert A, von Knebel Doeberitz C, Ridder R, von Knebel Doeberitz M. 2004. Validation of p16INK4a as a marker of oncogenic human papillomavirus infection in cervical biopsies from a population-based cohort in Costa Rica. Cancer Epidemiol Biomarkers Prev 13:1355–1360. [PubMed] [Google Scholar]
- 25.Nakao Y, Yang X, Yokoyama M, Ferenczy A, Tang SC, Pater MM, Pater A. 1997. Induction of p16 during immortalization by HPV 16 and 18 and not during malignant transformation. Br J Cancer 75:1410–1416. doi: 10.1038/bjc.1997.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLaughlin-Drubin ME, Crum CP, Munger K. 2011. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci U S A 108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komar AA, Hatzoglou M. 2011. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10:229–240. doi: 10.4161/cc.10.2.14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J 15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 29.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163–175. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 30.Calautti E, Li J, Saoncella S, Brissette JL, Goetinck PF. 2005. Phosphoinositide 3-kinase signaling to Akt promotes keratinocyte differentiation versus death. J Biol Chem 280:32856–32865. doi: 10.1074/jbc.M506119200. [DOI] [PubMed] [Google Scholar]
- 31.Thrash BR, Menges CW, Pierce RH, McCance DJ. 2006. AKT1 provides an essential survival signal required for differentiation and stratification of primary human keratinocytes. J Biol Chem 281:12155–12162. doi: 10.1074/jbc.M512116200. [DOI] [PubMed] [Google Scholar]
- 32.O'Shaughnessy RF, Akgul B, Storey A, Pfister H, Harwood CA, Byrne C. 2007. Cutaneous human papillomaviruses down-regulate AKT1, whereas AKT2 up-regulation and activation associates with tumors. Cancer Res 67:8207–8215. doi: 10.1158/0008-5472.CAN-07-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC, Whang-Peng J, Liu JM, Yang DM, Yang WK, Shen CY. 2000. PIK3CA as an oncogene in cervical cancer. Oncogene 19:2739–2744. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- 34.Pim D, Massimi P, Dilworth SM, Banks L. 2005. Activation of the protein kinase B pathway by the HPV-16E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene 24:7830–7838. doi: 10.1038/sj.onc.1208935. [DOI] [PubMed] [Google Scholar]
- 35.Menges CW, Baglia LA, Lapoint R, McCance DJ. 2006. Human papillomavirus type 16E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res 66:5555–5559. doi: 10.1158/0008-5472.CAN-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spangle JM, Ghosh-Choudhury N, Munger K. 2012. Activation of cap-dependent translation by mucosal human papillomavirus E6 proteins is dependent on the integrity of the LXXLL binding motif. J Virol 86:7466–7472. doi: 10.1128/JVI.00487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Todorovic B, Hung K, Massimi P, Avvakumov N, Dick FA, Shaw GS, Banks L, Mymryk JS. 2012. Conserved region 3 of human papillomavirus 16E7 contributes to deregulation of the retinoblastoma tumor suppressor. J Virol 86:13313–13323. doi: 10.1128/JVI.01637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todorovic B, Massimi P, Hung K, Shaw GS, Banks L, Mymryk JS. 2011. Systematic analysis of the amino acid residues of human papillomavirus type 16E7 conserved region 3 involved in dimerization and transformation. J Virol 85:10048–10057. doi: 10.1128/JVI.00643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y, Sharma A, Wu H, Lichtenstein A, Gera J. 2005. Cyclin D1 and c-myc internal ribosome entry site (IRES)-dependent translation is regulated by AKT activity and enhanced by rapamycin through a p38 MAPK- and ERK-dependent pathway. J Biol Chem 280:10964–10973. doi: 10.1074/jbc.M407874200. [DOI] [PubMed] [Google Scholar]
- 40.Mazurek S, Zwerschke W, Jansen-Durr P, Eigenbrodt E. 2001. Effects of the human papilloma virus HPV-16E7 oncoprotein on glycolysis and glutaminolysis: role of pyruvate kinase type M2 and the glycolytic-enzyme complex. Biochem J 356(Part 1):247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, Munger K. 2009. Expression of the human papillomavirus type 16E7 oncoprotein induces an autophagy-related process and sensitizes normal human keratinocytes to cell death in response to growth factor deprivation. Virology 385:192–197. doi: 10.1016/j.virol.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker J, Smiley LC, Ingram D, Roman A. 2011. Expression of human papillomavirus type 16E7 is sufficient to significantly increase expression of angiogenic factors but is not sufficient to induce endothelial cell migration. Virology 410:283–290. doi: 10.1016/j.virol.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frost P, Shi Y, Hoang B, Lichtenstein A. 2007. AKT activity regulates the ability of mTOR inhibitors to prevent angiogenesis and VEGF expression in multiple myeloma cells. Oncogene 26:2255–2262. doi: 10.1038/sj.onc.1210019. [DOI] [PubMed] [Google Scholar]
- 44.Hwang JH, Jiang T, Kulkarni S, Faure N, Schaffhausen BS. 2013. Protein phosphatase 2A isoforms utilizing Abeta scaffolds regulate differentiation through control of Akt protein. J Biol Chem 288:32064–32073. doi: 10.1074/jbc.M113.497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y, Alwine JC. 2008. Interaction between simian virus 40 large T antigen and insulin receptor substrate 1 is disrupted by the K1 mutation, resulting in the loss of large T antigen-mediated phosphorylation of Akt. J Virol 82:4521–4526. doi: 10.1128/JVI.02365-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White EA, Sowa ME, Tan MJ, Jeudy S, Hayes SD, Santha S, Munger K, Harper JW, Howley PM. 2012. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc Natl Acad Sci U S A 109:E260–E267. doi: 10.1073/pnas.1116776109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White EA, Kramer RE, Hwang JH, Pores Fernando AT, Naetar N, Hahn WC, Roberts TM, Schaffhausen BS, Livingston DM, Howley PM. 2015. Papillomavirus E7 oncoproteins share functions with polyomavirus small T antigens. J Virol 89:2857–2865. doi: 10.1128/JVI.03282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]