ABSTRACT
The nucleolar subcompartment of the nucleus is increasingly recognized as an important target of RNA viruses. Here we document for the first time the ability of dengue virus (DENV) polymerase, nonstructural protein 5 (NS5), to accumulate within the nucleolus of infected cells and to target green fluorescent protein (GFP) to the nucleolus of live transfected cells. Intriguingly, NS5 exchange between the nucleus and nucleolus is dynamically modulated by extracellular pH, responding rapidly and reversibly to pH change, in contrast to GFP alone or other nucleolar and non-nucleolar targeted protein controls. The minimal pH-sensitive nucleolar targeting region (pHNTR), sufficient to target GFP to the nucleolus in a pH-sensitive fashion, was mapped to NS5 residues 1 to 244, with mutation of key hydrophobic residues, Leu-165, Leu-167, and Val-168, abolishing pHNTR function in NS5-transfected cells, and severely attenuating DENV growth in infected cells. This is the first report of a viral protein whose nucleolar targeting ability is rapidly modulated by extracellular stimuli, suggesting that DENV has the ability to detect and respond dynamically to the extracellular environment.
IMPORTANCE Infections by dengue virus (DENV) threaten 40% of the world's population yet there is no approved vaccine or antiviral therapeutic to treat infections. Understanding the molecular details that govern effective viral replication is key for the development of novel antiviral strategies. Here, we describe for the first time dynamic trafficking of DENV nonstructural protein 5 (NS5) to the subnuclear compartment, the nucleolus. We demonstrate that NS5's targeting to the nucleolus occurs in response to acidic pH, identify the key amino acid residues within NS5 that are responsible, and demonstrate that their mutation severely impairs production of infectious DENV. Overall, this study identifies a unique subcellular trafficking event and suggests that DENV is able to detect and respond dynamically to environmental changes.
INTRODUCTION
Although primarily known as the site within the nucleus of ribosome-subunit biogenesis, the nucleolus is involved in many diverse cellular functions, including regulation of mitosis, cell cycle control and proliferation, cellular response to stress and the assembly of various ribonucleoprotein particles (1, 2). Importantly, many viruses, including RNA viruses that replicate in the cell cytoplasm, target the host cell nucleolus and usurp its function in order to aid viral replication (3).
Protein targeting to organelles such as the nucleus typically relies on short stretches of amino acids, which are necessary and sufficient to direct a protein to its respective compartment. The nuclear import of proteins, for example, depends on a nuclear localization signal (NLS), generally consisting of a short stretch of basic residues that confer interaction with the nuclear import proteins, importins, mediating entry into the nucleus via the nuclear pore complex (4). Analogously, protein export from the nucleus requires a hydrophobic/leucine-rich sequence (a nuclear export signal [NES]) that allows interaction between the target protein and nuclear export proteins such as CRM1 (5, 6). In contrast, signals targeting proteins to the nucleolus are not well understood (7), with nucleolar localization largely attributed to direct or indirect association with nucleolar components such as other nucleolar proteins or rDNA (7, 8). Although many different sequences that are capable of modulating nucleolar accumulation of specific proteins have been described, there is no consensus (9). Importantly, the composition of the nucleolus changes dynamically according to different metabolic conditions (10) and cell cycle status (11).
Interestingly, while most positive-sense RNA viruses replicate in the cytoplasm of host cells, the gene products of many viral proteins have been shown to localize, at least at some stage during infection, within the nucleus (12), and in many cases, more specifically, in the nucleolus (13). Examples include the viral capsid/nucleoproteins from coronaviruses (14), arteriviruses (15), alphaviruses (16), and flaviviruses (17, 18). Viral proteins may subsequently commandeer nucleolar functions in various ways, for example, hepatitis C virus RNA-dependent RNA polymerase (RdRp) nonstructural protein 5B (NS5B) binds to the abundant nucleolar protein nucleolin, mislocalizing it to the cytoplasm, leading to enhanced viral replication (19). Although the precise cellular modifications induced by viral protein-nucleolar association are largely unclear, the interactions generally appear to facilitate, rather than hinder, viral replication (13).
Dengue virus (serotypes 1 to 4; DENV-1 to -4) is a single-stranded, positive-sense RNA flavivirus that causes almost 400 million infections per year (20). DENV replication and virion packaging occurs in the cytoplasm of infected cells in association with endoplasmic reticulum-derived membranes, but the virus encodes two proteins that translocate to the nucleus during infection, including capsid protein that localizes to the nucleolus and interacts with nucleolin (17, 18), and NS5 (DENV RdRp) (21, 22). NS5 is the largest and most conserved flaviviral protein, encompassing the RdRp and methyltransferase domains required for replication and capping of the viral RNA genome, respectively (23). We and others showed previously that NS5 from all four DENV serotypes localizes within the nucleus of infected cells but is able to shuttle between the nucleus and cytoplasm dependent on two NLSs and an NES (21, 22, 24–28). The ability of NS5 to localize in the nucleus has been shown to correlate with efficient DENV replication based on reverse genetics experiments (21, 27, 29), as well as studies with general and specific inhibitors of nuclear import (22, 30, 31).
Here, we report for the first time that NS5 from both DENV-1 and -2 is able to localize within the nucleolus of NS5-transfected and virus-infected cells. Importantly, we demonstrate that NS5 nucleolar accumulation is dynamically regulated by extracellular pH, and identify the key residues (the pH-dependent nucleolar targeting region [pHNTR]) responsible for this novel targeting ability. Mutation of key residues of the DENV-2 pHNTR results in a virus that cannot be recovered without compensating mutations, underlining the likely physiological importance of the sequence and the function it confers to the DENV infectious cycle.
MATERIALS AND METHODS
Cell lines and virus strains.
Vero and C6/36 cells were cultured as described previously (32). Human embryonic kidney cells (HEK-293T) were maintained in Dulbecco modified eagle medium (DMEM; Invitrogen) with 10% fetal calf serum and 2 mM l-glutamine. Virus strains DENV-1 (accession number EU081230) and DENV-2 (accession numbers EU081177 and AF038403) were prepared and titered by plaque assay as described previously (33).
Antibodies.
Rabbit polyclonal anti-nucleolin (Abcam), anti-fibrillarin (Abcam), anti-STAT2 (Santa Cruz), and mouse anti-GFP (Roche) were from commercial sources as indicated. DENV serotype cross-reactive human anti-NS5 antibody (34) was a gift from Subhash Vasudevan (Duke National University of Singapore). Alexa Fluor 568-goat anti-rabbit and Alexa Fluor 488-goat anti-human IgG secondary antibodies were used as required.
Cell culture transfection.
DNA transfection of Vero cells was performed using Lipofectamine 2000 or 3000 (Invitrogen) according to the manufacturer's instructions. Live confocal laser scanning microscopy (CLSM; see below) was performed 16 to 24 h posttransfection.
Generation of mammalian expression vectors.
pEGFP-NS5 (DENV-1 and DENV-2) (22) and pEPI-GFP-NS5-1-900 (DENV-2 Townsville strain, TSV01, accession AY037116) (27) mammalian expression constructs have been described. pmCherry-NS5 (DENV-2, TSV01) was produced by subcloning NS5 from pEGFP-NS5 using restriction sites XhoI and HpaI (New England BioLabs). pEPI-GFP-NS5-1-244 was generated for this study as previously using Gateway cloning technology (27). Briefly, NS5 PCR fragments from TSV01 were generated using primers, including attB1 or attB2 sites to allow integration into the Gateway system. NS5 truncations were inserted into the pDONR207 vector by using BP Recombination (Invitrogen) and then inserted into the mammalian green fluorescent protein (GFP) fusion construct, pEPI-GFP (35) by LR Recombination (Invitrogen), according to the manufacturer's instructions.
Site-directed Mutagenesis.
Mutagenesis of the pEPI-GFP-NS5-1-900 construct was performed using overlap extension PCR. Primers were designed to include appropriate mutations to encode L165A/L167A/V168A (termed pHNTRm mutations).
Mutagenesis of the pDVWSK601 vector encoding New Guinea C DENV-2 (accession number AF038403) (21) was performed as for pEPI-GFP-NS5-1-900, but cloned through unique restriction sites within NS5, MluI, and StuI (New England BioLabs).
Immunoprecipitation.
HEK-293T cells transfected with GFP, GFP-NS5 wild type (WT), or pHNTRm were lysed 36 h posttransfection, and immunocomplexes were precipitated and eluted using GFP-Trap resin (ChromoTek) according to the manufacturer's instructions. Input and immunoprecipitations were analyzed by Western blotting.
Generation of pHNTR mutant virus.
Viral RNA was in vitro transcribed from the WT and pHNTR mutant pDVWS601 constructs using an mMESSAGE mMACHINE T7 in vitro transcription kit (Ambion), according to the manufacturer's instructions. Vero cells were transfected with 6 μg of viral RNA/well of a six-well plate, using DMRIE-c reagent (Invitrogen) and maintained in DMEM–2% fetal calf serum (FCS) for 7 days posttransfection. Culture supernatant was collected and clarified through a 0.45-μm-pore size filter prior to passaging twice through C6/36 cells for 5 days and the titer determined by plaque assay. Viral RNA was extracted from a portion of harvested virus (Qiagen viral RNA minikit), with the remaining virus stored at −80°C. Viral RNA was converted to cDNA (Invitrogen Superscript III) using the reverse primer 5′-GGAACTACGAGTACACGACC, which anneals to the 3′ end of DENV-2 RNA (from amino acid residue 726). The cDNA product was amplified using Phusion high-fidelity polymerase (New England BioLabs) and then sequenced between MluI and StuI restrictions sites in NS5.
Live cell and indirect immunofluorescence microscopy and analysis.
Live imaging of Vero cells transfected with GFP-NS5 constructs was performed in phenol-free (PF) DMEM containing 10% FCS modified to the indicated pH using HCl and NaOH, with pH-adjusted medium added to the cells 10 min prior to imaging. Where indicated, the cells were stressed by adding 100 μM CoCl2 for 6 h (hypoxia) or incubated in hyperosmotic (0.5 M sorbitol in PF-DMEM) or hypo-osmotic (10% PF-DMEM) medium for 15 min prior to live imaging.
For indirect immunofluorescence, DENV-infected, or GFP-NS5 transfected Vero cells were incubated with pH-adjusted DMEM for 5 min prior to fixation with cold methanol-acetone (1:1 [vol/vol]) for 2 min and then immunostained with anti-NS5 (34) or anti-nucleolin (Abcam) primary antibodies, together with Alexa Fluor 488-goat anti-human IgG and Alexa Fluor 568-goat anti-rabbit secondary antibodies (Molecular Probes), respectively. Immunostained cells were mounted in ProLong Gold Antifade Mountant with DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes). All images were captured by CLSM (Nikon C1 invert microscope), using a ×60 oil immersion objective lens (Nikon; Monash Micro Imaging) with live-cell imaging performed on a 37°C heated stage. NIS-Elements version 4.10 software was used for image acquisition. Images were analyzed using ImageJ software (version 1.46; National Institutes of Health), whereby the fluorescence intensity in the nucleolus (Fnu), nucleus (Fn), and cytoplasm (Fc) was determined with the background fluorescence (untransfected cells) subtracted to enable the nucleolar/nuclear (Fnu/n) or the nuclear/cytoplasmic (Fn/c) ratio to be determined. To assess the extent of colocalization of nucleolin and NS5, the ImageJ plot profile tool was used to measure pixel intensity (fluorescence) along a line of interest that bisected the nucleus and nucleolar regions. The resulting data were exported to Microsoft Excel, and the graphs were then plotted using Prism 6.
FRAP.
Fluorescent recovery after photobleaching (FRAP) was performed as previously (36). Briefly, Vero cells transiently expressing either GFP-NS5, -nucleolin, or -fibrillarin were imaged using an Olympus Fluoview 1000 microscope (×100 oil immersion lens). Prebleach images were collected using 4% total laser power with excitation at 488 nm (×2 zoom, scanned 8 μs/pixel). Bleaching of the nucleolus was performed by subjecting a small area covering the nucleolus to an argon laser at 100% power for 10 scans (12.5 μs/pixel). Fluorescence recovery was followed over a period of 600 s at intervals of 20 s. The relative level of Fnu/n was determined at each time point, and % recovery determined as a proportion to prebleach Fnu/n. Data were fitted to an exponential function, and the half-maximal recovery (t½) and the maximal recovery determined for each protein as previously (36).
Virus Infections.
Vero cells were infected with the indicated DENV strain, at a multiplicity of infection (MOI) of 4, unless otherwise stated. For indirect immunofluorescence experiments, cells were incubated with pH-adjusted DMEM at 24 h postinfection (p.i.) for 5 min prior to fixation and staining. For replication analysis of the pHNTR mutant virus, mutant and WT virus titers were normalized to infect Vero cells at an MOI of 0.03. Virus was applied to cells for 2 h prior to removal of unbound virus and the addition of fresh DMEM–2% FCS. The culture supernatant was collected at 72 h p.i., viral RNA was extracted (Bioline Isolate II RNA minikit), and the amount of infectious virus produced was estimated using quantitative reverse transcriptase PCR (30, 32).
RESULTS
NS5 accumulates within the nucleolus of DENV-infected and transfected cells.
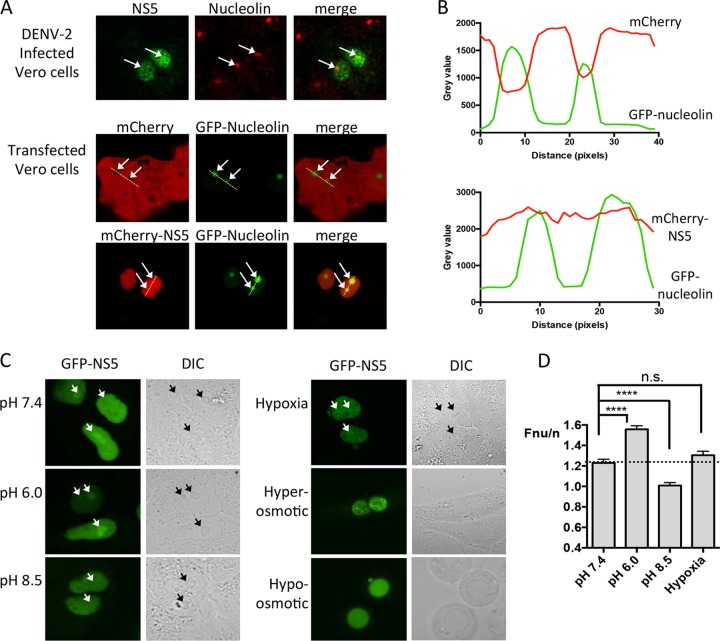
Based on observations that DENV NS5 localizes to the nucleus in infected cells, we set out to examine NS5 subnuclear localization in detail. Vero cells were infected with DENV-2, fixed, immunostained for NS5 and/or the nucleolar marker nucleolin at 24 h p.i., and imaged by CLSM. Consistent with previous observations, NS5 showed obvious nuclear localization but was also found in regions of the nucleus that also stained positive for nucleolin (Fig. 1A, top panel). Vero cells ectopically expressing mCherry-tagged NS5 (DENV-2) and GFP-nucleolin (37) and imaged live by CLSM also showed clear colocalization of NS5 with nucleolin, indicating that NS5 nucleolar accumulation is independent of other viral proteins (Fig. 1A, bottom panel). This was in contrast to mCherry alone cotransfected with GFP-nucleolin, where mCherry was clearly excluded from the nucleolar regions (Fig. 1A, middle panel). To confirm colocalization of mCherry-NS5 and GFP-nucleolin, the plot profile of fluorescence across the nucleus, including the nucleolus, for cells coexpressing GFP-nucleolin and either mCherry or mCherry-NS5 was analyzed (Fig. 1B). These data clearly show the fluorescence for mCherry alone troughs at the points where GFP-nucleolin peaks, a finding consistent with nucleolar exclusion of mCherry. In contrast, fluorescent levels of mCherry-NS5 (DENV-2) remained consistent across the nucleus even as it intersected with GFP-nucleolin peaks. Clearly, mCherry-NS5 is present in nucleolar regions colocalized with nucleolin.
FIG 1.
NS5 localizes within the nucleolus during DENV infection and in response to low pH. (A) CLSM images of DENV-2-infected Vero cells fixed and immunostained for endogenous nucleolar marker nucleolin and DENV-2 NS5 (top) or Vero cells cotransfected to express GFP-nucleolin and either mCherry or DENV-2 mCherry-NS5 (bottom) and imaged live. White arrows indicate nucleoli. The yellow dashed line indicates the region where the Plot Profile was performed to obtain the graphs in panel B. (B) The ImageJ plot profile tool was used to measure pixel intensity (fluorescence) along the line of interest (in panel A), with the resulting data plotted using Prism 6. (C) Vero cells transfected to express GFP-NS5 (DENV-2), treated with various stresses, including acidic pH (6.0), basic pH (8.5), 100 μM CoCl2 (hypoxia), or hyper- or hypo-osmotic stress prior to live cell imaging. DIC images are shown to the right to identify the nucleoli (white and black arrows in GFP and DIC images, respectively). (D) Quantitative analysis of images such as those in panel C to determine the ratio of fluorescence intensity in the nucleolus (Fnu) compared with the nucleus (Fn); the “Fnu/n ratio.” The results represent the means + the standards error of the mean (SEM), where n ≥ 22. Statistical analysis by Student t test was performed using GraphPad Prism software (****, P < 0.0001; n.s., not significant).
NS5 nucleolar accumulation is modulated by extracellular pH.
In establishing the ability of NS5 to localize within the nucleolus, we noted variation in the extent to which nucleolar accumulation occurred between infected samples and speculated that an environmental factor(s) may be responsible (38). To test this, we subjected Vero cells expressing GFP-NS5 (DENV-2) to various extracellular stress agents, including acidic pH (6.0), basic pH (8.5), hypoxia (CoCl2), and hyper- or hypo-osmotic stress (Fig. 1C). Cells were examined live by CLSM, and the extent of nucleolar accumulation of NS5 was determined, where the nucleoli were identified by differential interference contrast (DIC) microscopy (39). Under neutral pH (7.4), NS5 was found to accumulate in the nucleolus relative to the nucleus at a ratio of 1.2 (Fnu/n) (Fig. 1D). When the extracellular pH was lowered to 6.0, this ratio increased to 1.5 (P < 0.0001). Interestingly, increasing the extracellular pH to 8.5 resulted in a decrease in Fnu/n to 1.0 (P < 0.0001). Exposure of the cells to hypoxia or hyper- or hypo-osmotic agents did not appear to induce nucleolar accumulation of GFP-NS5, although it should be noted that, based on the DIC images, hyper- and hypo-osmotic media disrupted nucleolar structure so that quantitative analysis was not performed. Thus, it appears that the ability of NS5 to accumulate in intact nucleoli can be influenced by extracellular pH.
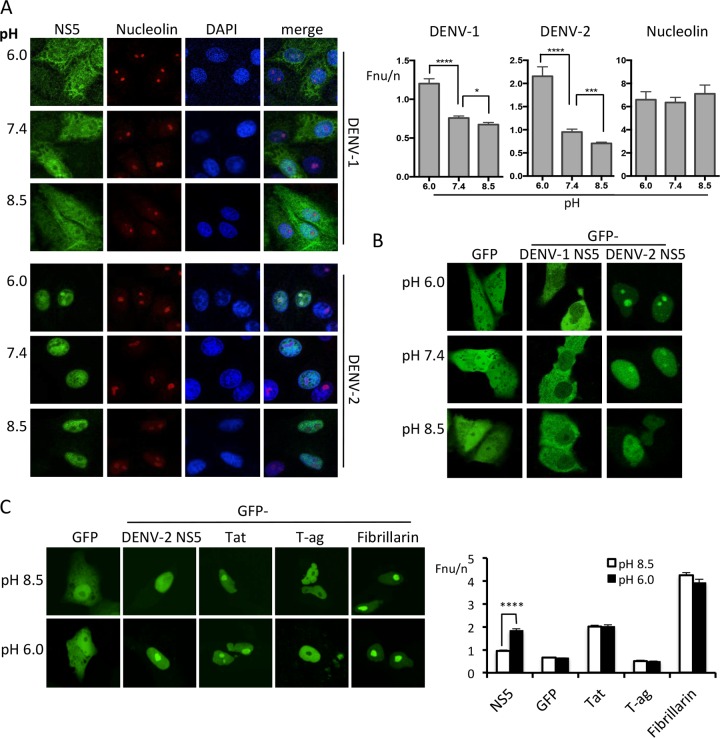
pH-induced nucleolar trafficking of NS5 is conserved between DENV-1 and DENV-2 serotypes and is a phenomenon specific to NS5.
We previously showed that the extent to which NS5 localizes in the nucleus of DENV-infected cells varies between serotypes, with NS5 from DENV-2 and -3 demonstrating almost exclusively nuclear localization, whereas NS5 from DENV-1 and -4 shows more diffuse localization throughout the nucleus and cytoplasm (22). As representatives of these distinct NS5 profiles, we examined Vero cells infected with DENV-1 or DENV-2. Cells were infected with an MOI of 4 and at 24 h p.i. were treated with cell culture medium with an adjusted pH of 6.0, 7.4, or 8.5 for 5 min prior to fixation and immunostaining for NS5 and nucleolin (Fig. 2A). Consistent with initial experiments using GFP-NS5 (DENV-2), quantitative analysis of the Fnu/n indicated that NS5 from DENV-2-infected cells was localized throughout the nucleus, including colocalizing with nucleolin in the nucleolus, when exposed to pH 7.4 (Fnu/n of 0.95, Fig. 2A). Decreasing the pH to 6.0 resulted in a significant increase in nucleolar accumulation (Fnu/n of 2.2, P < 0.0001), while under basic conditions NS5 became excluded from the nucleolus (Fnu/n of 0.7, P < 0.001 compared to at pH 7.4). Similar to previous observations, DENV-1 NS5 was localized throughout the nucleus and cytoplasm under neutral pH conditions (22). Analysis of the Fnu/n showed that NS5 was largely excluded from the nucleolus in DENV-1 infected cells at neutral pH (Fnu/n of 0.8), evident as punctate regions devoid of NS5 where nucleolin staining is clearly present. However, significant nucleolar accumulation of DENV-1 NS5 was evident under acidic conditions such that DENV-1 NS5 was diffusely localized throughout the nucleus, including regions that colocalized with nucleolin staining (Fnu/n of 1.2, P < 0.0001). At pH 8.5, the Fnu/n for DENV-1 was similar to that at pH 7.4, suggesting NS5 exclusion from the nucleolus is maximal at neutral pH. The Fnu/n ratio for nucleolin did not alter significantly in cells exposed to acidic or basic pH (Fig. 2A). Together, these results suggest that the ability of NS5 to accumulate in the nucleolus under acidic conditions is conserved between DENV serotypes 1 and 2. Similar effects were observed in Vero cells transfected with GFP-NS5 from DENV-1 and -2 and imaged live using CLSM (Fig. 2B). While the apparent distribution of NS5 in the nucleus of transfected, live-imaged cells appeared to vary slightly from that of NS5 in fixed DENV-infected cells, the trend of pH-dependent nucleolar accumulation of NS5 was clearly conserved. Importantly, GFP alone remained excluded from the nucleolus independent of pH.
FIG 2.
pH-dependent nucleolar accumulation of NS5 is conserved between DENV-1 and DENV-2 but is specific to NS5. (A) DENV-1- and DENV-2-infected Vero cells were incubated in cell culture medium at the indicated pH, 5 min prior to fixation and immunostaining for NS5 and nucleolin and imaging by CLSM. Quantitative analysis of images such as those shown was performed to determine the Fnu/n. The results represent the means + the SEM, where n ≥ 25. Statistical analysis (by Student t test) was performed using GraphPad Prism software (****, P < 0.0001; ***, P < 0.001; *, P < 0.05; n.s., not significant). (B) CLSM images of live GFP, GFP-NS5 (DENV-1 or -2 as indicated) expressed in Vero cells were imaged <10 min after the cell culture medium was changed to the indicated pH. (C) CLSM images of live GFP-tagged proteins expressed and pH-treated as in panel B, with corresponding quantitative analysis performed on images such as those shown. The results represent the means + the SEM, where n ≥ 25. Statistical analysis by Student t test was performed using GraphPad Prism software (****, P < 0.0001).
To establish whether pH-sensitive nucleolar trafficking was unique to NS5, the localization of GFP-tagged proteins with diverse subcellular trafficking abilities were examined following changes in extracellular pH; GFP-Tat (human immunodeficiency virus transactivator protein) which under neutral conditions is nucleolar and nuclear localized (40), GFP-T-ag (SV40 large T-antigen; strongly nucleoplasmic and excluded from the nucleolus) (31), GFP-fibrillarin (nucleolar) (37), and GFP itself which shows diffuse nuclear/cytoplasmic distribution and nucleolar exclusion. The localization of each of these proteins was insensitive to extracellular pH change, with nucleolar accumulation/exclusion not changing in response to exposure to pH extremes (Fig. 2C), implying that the ability of NS5 to accumulate in the nucleolus at low pH is likely due to specific sequences in NS5.
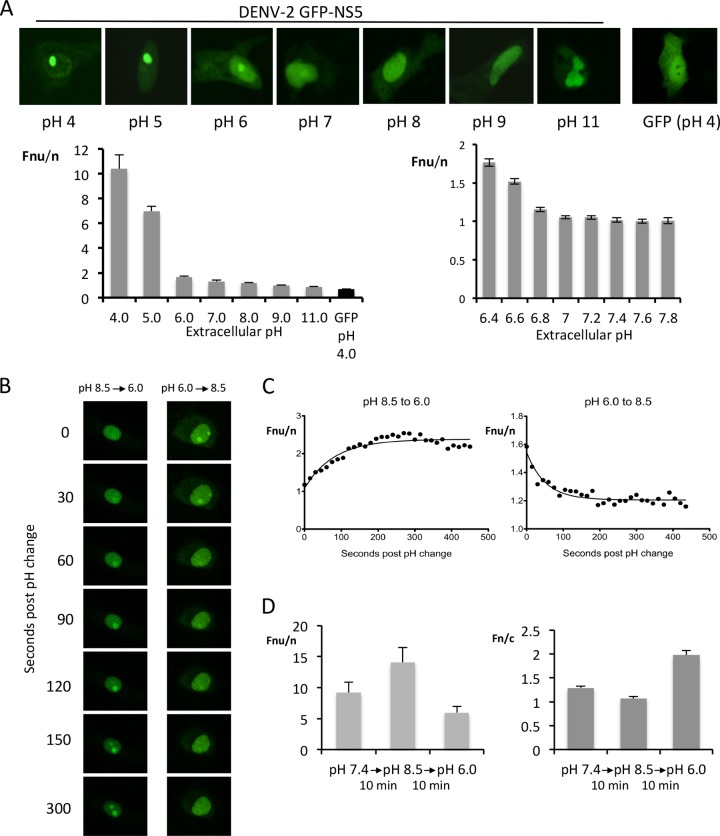
Nucleolar accumulation of NS5 occurs rapidly and reversibly in response to extracellular pH.
To characterize nucleolar accumulation of NS5 in more detail, a range of extracellular pHs were tested. Nucleolar accumulation of GFP-NS5 (DENV-2) increased progressively as the pH was lowered, reaching a maximal Fnu/n value of >10 at pH 4, while GFP alone remained excluded from the nucleolus (Fig. 3A, images and left graph). At pH 11, the concentration of GFP-NS5 was lower in the nucleolus than the nucleus (Fnu/n of 0.86). Nucleolar accumulation was also modulated consistently with changes in pH over the physiologically relevant pH range of 6.4 to 7.8 (Fig. 3A, right graph).
FIG 3.
Nucleolar accumulation of NS5 responds rapidly and reversibly in response to extracellular pH. (A) CLSM images of live Vero cells transfected to express GFP or DENV-2 GFP-NS5 and imaged <5 min after transfer to cell culture medium at the indicated pH. The corresponding quantitative analysis performed using images such as those shown to determine the Fnu/n across a wide (left) or narrow (right) pH range is shown below. (B) CLSM of an individual Vero cell imaged over time upon media change (t = 0) from pH 8.5 to 6.0 (left) or 6.0 to 8.5 (right); cells shown are from the same coverslip. (C) Fnu/n values were determined over time post-pH change for the individual cells shown in panel B. Data are fitted to an exponential one-phase decay curve (GraphPad Prism 5). (D) Fnu/n and Fn/c for n > 15 cells (mean + the SEM) from the same sample as for panel B.
The dynamics of nucleolar accumulation were investigated by initially exposing GFP-NS5 (DENV-2)-expressing cells to basic pH (8.5), before switching to pH 6, or vice versa, with live monitoring of single cells over time by CLSM. Nucleolar accumulation was induced very rapidly upon transition of cells from pH 8.5 to pH 6, with increased fluorescence in the nucleoli becoming evident <1 min after the pH change (Fig. 3B and C, left-hand panels). This occurred in both Vero and Cos-7 cells (data not shown). Significantly, nucleolar accumulation was reversible, whereby cells transferred from pH 8.5 to 6 and then back to 8.5 showed a rapid reduction in nucleolar accumulation (Fig. 3B and C; right-hand panels). Furthermore, the pH could be changed several times with responses repeated to a similar extent (data not shown). Analysis of fluorescent levels within each subcellular compartment (nucleolus, nucleus and cytoplasm) indicated that upon exposure to low pH, fluorescence intensity increased in the nucleolus and decreased in the nucleus (Fnu/n increased as pH decreases), whereas the ratio of nuclear to cytoplasmic fluorescence (Fn/c) showed the opposite, decreasing with acidic pH, implying that accumulation of NS5 in the nucleolus is likely the result of rapid exchange between the nucleus and nucleolus and not the result of increased transport from the cytoplasm to the nucleus (Fig. 3D). NLS-mutated (21), as well as nuclear export-inhibited (27), NS5 both displayed the same levels of nucleolar accumulation and pH-sensitivity as WT NS5 (data not shown), implying that rapid translocation between nucleus and nucleolus is largely independent of the absolute levels of NS5 in either the nucleus or the cytoplasm, a finding consistent with our observations for DENV-1 and DENV-2 NS5, which display distinct nuclear accumulation profiles and yet both demonstrate nucleolar accumulation in response to low pH.
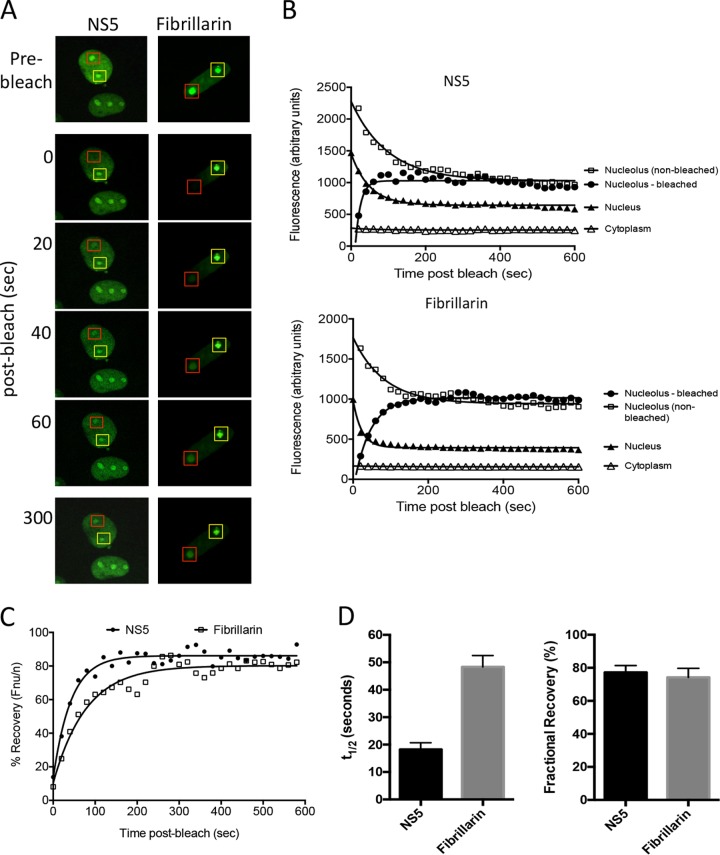
NS5 traffics dynamically between the nucleus and nucleolus.
Interestingly, the tumor suppressor, von Hippel-Lindau protein (VHL), has previously been reported to be retained in the nucleolus in an H+-dependent manner (38). Its mechanism of nucleolar accumulation involves sequestration of VHL, in which very little exchange occurs between the nucleolus and nucleus. In stark contrast, analysis of NS5 using FRAP determined NS5 nucleolar trafficking to be highly dynamic (Fig. 4). After bleaching of a region of the nucleolus containing GFP-tagged NS5 (red box), NS5 levels rapidly increased in the nucleolus and decreased in the nucleus, while remaining constant in the cytoplasm, consistent with intranuclear trafficking by NS5 (Fig. 4A and B). Furthermore, analysis of nonbleached nucleoli within the same cell (yellow box) showed a reduction in nucleolar NS5 over time until fluorescent levels were identical to those in the bleached nucleoli, a finding consistent with the dynamic movement of NS5 into and out of the nucleolus. Similar results were obtained for nucleolar markers nucleolin (not shown) and fibrillarin. Our data demonstrate that NS5 undergoes rapid exchange (t1/2 < 15 s) between the nucleolus and nucleoplasm with a high mobile fraction (approximately 80%), indicating very little binding or immobilization in the nucleolus (Fig. 4C and D). This rapid intranuclear trafficking of NS5, in addition to the progressive accumulation of NS5 in response to varied extracellular pH (in contrast to VHL, which only accumulates rapidly within the nucleolus below ∼ pH 6.4) (41), suggests that NS5 pH-dependent trafficking occurs in a unique manner.
FIG 4.
NS5 traffics dynamically between the nucleus and nucleolus. (A) FRAP analysis was performed on GFP-NS5 and -fibrillarin-expressing Vero cells. Images of representative cells prebleach and postbleach are shown with red squares indicating the bleach area, and yellow squares denoting the nonbleached nucleoli. (B) Image analysis of representative cells measuring the bleached and nonbleached nucleolar, nuclear, and cytoplasmic fluorescence levels over time after bleaching for the indicated GFP-tagged proteins shown in panel A. (C) The percent recovery postbleaching was determined by measuring Fnu/n values over time compared with initial (prebleach) Fnu/n values. (D) The time postbleaching to reach half-maximal recovery (t½) and fractional recovery were determined for each protein. The results represent the means + the SEM (n = 5).
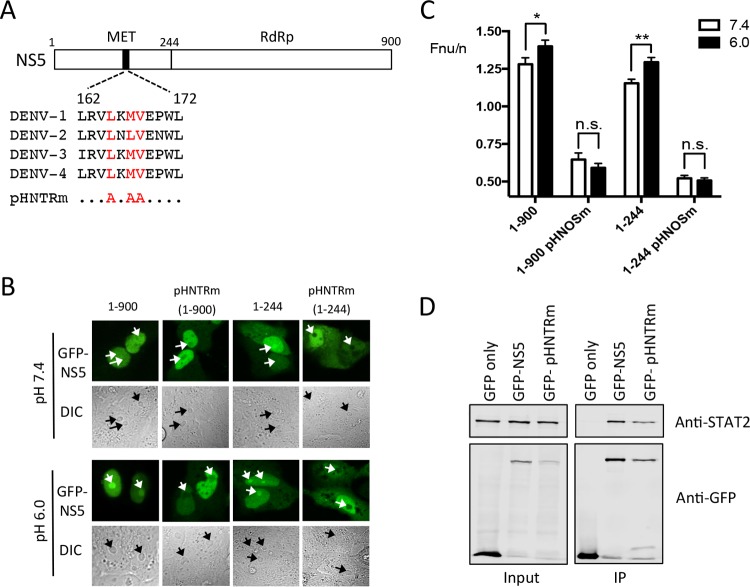
Identification of the pH-sensitive nucleolar-targeting region (pHNTR) in NS5.
Analysis of the subcellular localization of GFP-tagged truncation derivatives of DENV-2 NS5 expressed in Vero cells was used to map the pH-dependent nucleolar targeting region (pHNTR) of NS5 to residues 1 to 244 of NS5 (Fig. 5 and data not shown). Interestingly, GFP-NS5 1-244 was found to accumulate in the nucleus of transfected cells, albeit to a substantially lesser degree than full-length NS5, despite lacking the previously described NLSs (21), perhaps suggestive of further, unidentified NLS-containing elements in the N-terminal residues of NS5. Alternatively, the smaller size of GFP-NS5 1-244 may enable the protein to undergo passive diffusion into the nucleus.
FIG 5.
NS5 residues 1 to 244 contain the pH-sensitive nucleolar-targeting region (pHNTR). (A) Schematic diagram of NS5, highlighting a stretch of hydrophobic amino acids predicted to contribute to nucleolar targeting of NS5. (B) CLSM and DIC images (below) of full-length (residues 1 to 900) and truncated (residues 1 to 244) NS5, and corresponding pHNTRm versions (NS5 containing Ala substitutions at Leu-165, Leu-167, and Val-168), fused to GFP, expressed in Vero cells, and imaged live in cell culture medium at the indicated pH. Nucleoli are indicated by white and black arrows in GFP and DIC images, respectively. (C) Results for quantitative analysis for images such as those in panel B. The results represent the means + the SEM, where n > 25. Statistical analysis (by Student t test) was performed using GraphPad Prism software (**, P < 0.01; *, P < 0.05; n.s., not significant). (D) HEK-293T cells transfected to express GFP, GFP-NS5, or GFP-NS5 pHNTRm were lysed 36 h posttransfection and immunoprecipitated (IP) using GFP-trap (ChromoTek). Input or IP samples were probed by Western blotting using mouse anti-GFP (Roche) or rabbit anti-STAT2 (Santa Cruz) antibodies and then imaged using Odyssey infrared imager (LI-COR).
Mekhail et al. previously described a H+-dependent nucleolar detention signal (NoDSH+) within VHL protein, whereby hydrophobic [L(ϕ/N)(V/L)] and arginine-rich [RR(I/L)X3r] (phi symbolizes any hydrophobic residue; R, critical arginines; r, less critical arginine; and X, any amino acid) subnuclear targeting domains confer nucleolar targeting of VHL upon exposure to low pH (42). Importantly, we identified a sequence within NS5 1-244 possessing limited similarity to the hydrophobic [L(ϕ/N)(V/L)] region of the NoDSH+ (Fig. 5A) within residues 162 to 172, which is highly conserved between all 4 DENV serotypes. Accordingly, we targeted this region in DENV-2 GFP-NS5 for mutagenesis, substituting Leu-165, Leu-167, and Val-168 with Ala. Intriguingly, these mutations in the predicted pHNTR resulted in significantly reduced nucleolar accumulation of NS5, and in contrast to our observations for DENV-1 NS5 (above), where NS5 was absent from the nucleolus at neutral pH but accumulated under acidic conditions, nucleolar trafficking of GFP-NS5-1-900-pHNTRm and the truncated version GFP-NS5-1-244-pHNTRm, did not occur following acidification of the medium (Fig. 5B and C). The clear implication is that Leu-165/167 and Val-168 are critical elements of the DENV-2 NS5 pHNTR. To test the impact of the substitutions on other functions of NS5 such as in interferon antagonism, we decided to test whether the pHNTR mutations may impact the ability of NS5 to bind STAT2 (43). Immunoprecipitation experiments showed that both NS5 and NS5 pHNTRm were able to associate with comparable levels of STAT2 (Fig. 5D), suggesting that the key residues of the pHNTR and that NS5 nucleolar localization in general are not critical for this activity.
Mutation of critical pHNTR residues severely attenuates DENV-2.
We next examined whether the predicted pHNTR was required for efficient DENV replication. L165T, L167A, and V168A mutations were introduced into the DENV-2 cDNA infectious clone (21, 44). Sequencing of three independent cultures of passaged virus indicated a complete inability to recover a mutant virus carrying the pHNTR mutations, instead revealing mutation of residue Thr165 to Ile. Mutation of Leu167 to Ala was evident in all three cultures, whereas Val168 substitution by Ala was detected in cultures 1 and 2 but reverted to Val168 in culture 3 (Table 1). The clear implication is of strong selective pressure to retain the WT pHNTR sequence, with mutation thereof resulting in poor viral viability. Consistent with this, even after passaging of mutated virus three times, the titers remained 100- to 1,000-fold lower than that obtained for WT DENV-2, indicating the pHNTR-mutated virus was severely attenuated in growth (Table 1). Although the pHNTR is within the NS5 MTase domain, which is important for RNA capping activity, molecular modeling of full-length DENV NS5 (45) indicates that mutation of L165/L167/V168 will not either directly affect or impact through proximity any of the catalytic residues of the MTase domain (Lys61, Asp146, Lys180, and Glu216), nor will the orientation or number of polar contacts between the pHNTR mutations and nearby residues be influenced. Hence, it is very unlikely that mutation of L165/L167/V168 would reduce DENV replication as a result of impaired MTase activity. Infection of Vero cells with a normalized concentration of WT or pHNTR mutant virus (I165/A167/A168; MOI of 0.03) revealed a 71% reduction in viral RNA copies present in the culture supernatant at 72 h p.i. for the mutant virus, clearly demonstrating that pHNTR mutation is detrimental to viral growth and that the pHNTR provides an essential role for efficient virus replication. Due to the extremely low titers of pHNTR virus, it did not prove possible to assess the nucleolar localization of NS5 in pHNTR-mutant DENV-2-infected cells. Additional experiments were attempted using Vero cells directly transfected with high levels of viral RNA (29), but this failed to result in cells expressing pHNTRm NS5 (data not shown). This further underlines the critical importance of the pHNTR to DENV infection, but it remains not formally possible to discard the potential role of the pHNTR in additional functions of NS5 in the context of viral infection.
TABLE 1.
Effects of mutations within the DENV-2 pHNTR on virus production
| Culture | pHNTR motif | Passaged virus titer (PFU/ml) | RNA copies in culture medium (%)a |
|---|---|---|---|
| Wild type | L165, L167, V168 | 3.3 × 106 | (5.9 ± 1.4) × 105 (100) |
| pHNTRm culture 1 | I165, A167, A168 | 2.0 × 104 | (1.7 ± 0.2) × 105 (28.8) |
| pHNTRm culture 2 | I165, A167, A168 | 1.2 × 104 | ND |
| pHNTRm culture 3 | I165, A167, V168 | 4.8 × 103 | ND |
The data represent the means ± the standard deviations for two independent experiments performed in duplicate at an MOI of 0.03. ND, not done.
DISCUSSION
The nucleolus is increasingly recognized as a target for gene products from many diverse viruses, including DNA viruses (46), retroviruses (47, 48), and RNA viruses (15, 19, 39, 49). Here, we characterize the nucleolar localization of NS5 from DENV-1 and -2, demonstrating its dynamic and rapid trafficking between the nucleus and nucleolus in response to changes in extracellular pH, identifying DENV NS5 as the first viral protein whose nucleolar targeting is regulated by the extracellular environment. We define the pH responsive element responsible in NS5, the pHNTR, establishing its unique nature, and demonstrate its essential function in effecting viral replication.
The nucleolus is a key player in the cellular stress response, including to extracellular pH (50), with recent reports showing that cell stress can induce nucleolar remodelling, including redistribution of nucleolar factors (51), suggesting one way in which pH may influence NS5 localization. It is quite conceivable that NS5's ability to traffic reversibly and rapidly between the nucleus and nucleolus may be important in rapidly countering or modulating cellular responses to viral infection. Of note, it has been observed that metabolic acidosis is a common feature of severe DENV infections (52), perhaps suggesting a physiological relevance of the results obtained here. Furthermore, nucleolar localization of NS5 from the DENV-related flavivirus yellow fever virus appears to be reduced in pathogenic compared with vaccine strains (53), supporting the idea of an important link between NS5 nucleolar targeting and pathogenesis and/or immune responses. Exploring this possibility will be the focus of continued studies in this laboratory.
ACKNOWLEDGMENTS
We acknowledge Monash Micro Imaging for support with real-time imaging and Chunxiao Wang for assistance with cell culture.
This study was supported by the National Health and Medical Research Council Australia.
Funding Statement
This work, including the efforts of David Jans, was funded by Department of Health | National Health and Medical Research Council (NHMRC) (project grant 606409, project grant APP1059137, Senior Principal Research Fellowship award APP1002486/1103050).
REFERENCES
- 1.Mayer C, Grummt I. 2005. Cellular stress and nucleolar function. Cell Cycle 4:1036–1038. doi: 10.4161/cc.4.8.1925. [DOI] [PubMed] [Google Scholar]
- 2.Pederson T. 2011. The nucleolus. Cold Spring Harb Perspect Biol 3:a000638. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawlinson SM, Moseley GW. 2015. The nucleolar interface of RNA viruses. Cell Microbiol 17:1108–1120. doi: 10.1111/cmi.12465. [DOI] [PubMed] [Google Scholar]
- 4.Fulcher AJ, Jans DA. 2011. Regulation of nucleocytoplasmic trafficking of viral proteins: an integral role in pathogenesis? Biochim Biophys Acta 1813:2176–2190. doi: 10.1016/j.bbamcr.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutten S, Kehlenbach RH. 2007. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol 17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Kutay U, Guttinger S. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Emmott E, Hiscox JA. 2009. Nucleolar targeting: the hub of the matter. EMBO Rep 10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. 2002. Directed proteomic analysis of the human nucleolus. Curr Biol 12:1–11. doi: 10.1016/S0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 9.Scott MS, Boisvert FM, McDowall MD, Lamond AI, Barton GJ. 2010. Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res 38:7388–7399. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam YW, Lamond AI, Mann M, Andersen JS. 2007. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sirri V, Hernandez-Verdun D, Roussel P. 2002. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J Cell Biol 156:969–981. doi: 10.1083/jcb.200201024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiscox JA. 2003. The interaction of animal cytoplasmic RNA viruses with the nucleus to facilitate replication. Virus Res 95:13–22. doi: 10.1016/S0168-1702(03)00160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salvetti A, Greco A. 2014. Viruses and the nucleolus: the fatal attraction. Biochim Biophys Acta 1842:840–847. doi: 10.1016/j.bbadis.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiscox JA, Wurm T, Wilson L, Britton P, Cavanagh D, Brooks G. 2001. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol 75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowland RR, Kervin R, Kuckleburg C, Sperlich A, Benfield DA. 1999. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res 64:1–12. doi: 10.1016/S0168-1702(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 16.Michel MR, Elgizoli M, Dai Y, Jakob R, Koblet H, Arrigo AP. 1990. Karyophilic properties of Semliki Forest virus nucleocapsid protein. J Virol 64:5123–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balinsky CA, Schmeisser H, Ganesan S, Singh K, Pierson TC, Zoon KC. 2013. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J Virol 87:13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SH, Syu WJ, Huang KJ, Lei HY, Yao CW, King CC, Hu ST. 2002. Intracellular localization and determination of a nuclear localization signal of the core protein of dengue virus. J Gen Virol 83:3093–3102. doi: 10.1099/0022-1317-83-12-3093. [DOI] [PubMed] [Google Scholar]
- 19.Hirano M, Kaneko S, Yamashita T, Luo H, Qin W, Shirota Y, Nomura T, Kobayashi K, Murakami S. 2003. Direct interaction between nucleolin and hepatitis C virus NS5B. J Biol Chem 278:5109–5115. doi: 10.1074/jbc.M207629200. [DOI] [PubMed] [Google Scholar]
- 20.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pryor MJ, Rawlinson SM, Butcher RE, Barton CL, Waterhouse TA, Vasudevan SG, Bardin PG, Wright PJ, Jans DA, Davidson AD. 2007. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic 8:795–807. doi: 10.1111/j.1600-0854.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 22.Tay MY, Fraser JE, Chan WK, Moreland NJ, Rathore AP, Wang C, Vasudevan SG, Jans DA. 2013. Nuclear localization of dengue virus (DENV) 1-4 nonstructural protein 5: protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antiviral Res 99:301–306. doi: 10.1016/j.antiviral.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Lindenbach BD, Thiel H, Rice CM. 2007. Flaviviridae: the viruses and their replication, 5th ed Wolters Kluwer/Lippincott/Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 24.Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. 2002. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin β1 and importin alpha/beta-recognized nuclear localization signals. J Biol Chem 277:36399–36407. doi: 10.1074/jbc.M204977200. [DOI] [PubMed] [Google Scholar]
- 25.Forwood JK, Brooks A, Briggs LJ, Xiao CY, Jans DA, Vasudevan SG. 1999. The 37-amino-acid interdomain of dengue virus NS5 protein contains a functional NLS and inhibitory CK2 site. Biochem Biophys Res Commun 257:731–737. doi: 10.1006/bbrc.1999.0370. [DOI] [PubMed] [Google Scholar]
- 26.Johansson M, Brooks AJ, Jans DA, Vasudevan SG. 2001. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J Gen Virol 82:735–745. doi: 10.1099/0022-1317-82-4-735. [DOI] [PubMed] [Google Scholar]
- 27.Rawlinson SM, Pryor MJ, Wright PJ, Jans DA. 2009. CRM1-mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J Biol Chem 284:15589–15597. doi: 10.1074/jbc.M808271200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannemann H, Sung PY, Chiu HC, Yousuf A, Bird J, Lim SP, Davidson AD. 2013. Serotype-specific differences in dengue virus nonstructural protein 5 nuclear localization. J Biol Chem 288:22621–22635. doi: 10.1074/jbc.M113.481382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, Buhler S, Selisko B, Davidson A, Mulder K, Canard B, Miller S, Bartenschlager R. 2013. Nuclear localization of dengue virus nonstructural protein 5 does not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling. J Virol 87:4545–4557. doi: 10.1128/JVI.03083-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser JE, Watanabe S, Wang C, Chan WK, Maher B, Lopez-Denman A, Hick C, Wagstaff KM, Mackenzie JM, Sexton PM, Vasudevan SG, Jans DA. 2014. A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal dengue virus infection. J Infect Dis 210:1780–1791. doi: 10.1093/infdis/jiu319. [DOI] [PubMed] [Google Scholar]
- 31.Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. 2012. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J 443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser JE, Rawlinson SM, Wang C, Jans DA, Wagstaff KM. 2014. Investigating dengue virus nonstructural protein 5 (NS5) nuclear import. Methods Mol Biol 1138:301–328. doi: 10.1007/978-1-4939-0348-1_19. [DOI] [PubMed] [Google Scholar]
- 33.Wang QY, Patel SJ, Vangrevelinghe E, Xu HY, Rao R, Jaber D, Schul W, Gu F, Heudi O, Ma NL, Poh MK, Phong WY, Keller TH, Jacoby E, Vasudevan SG. 2009. A small-molecule dengue virus entry inhibitor. Antimicrob Agents Chemother 53:1823–1831. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Moreland NJ, Tay MY, Lee CC, Swaminathan K, Vasudevan SG. 2014. Identification and molecular characterization of human antibody fragments specific for dengue NS5 protein. Virus Res 179:225–230. doi: 10.1016/j.virusres.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Ghildyal R, Ho A, Wagstaff KM, Dias MM, Barton CL, Jans P, Bardin P, Jans DA. 2005. Nuclear import of the respiratory syncytial virus matrix protein is mediated by importin beta1 independent of importin alpha. Biochemistry 44:12887–12895. doi: 10.1021/bi050701e. [DOI] [PubMed] [Google Scholar]
- 36.Roth DM, Moseley GW, Glover D, Pouton CW, Jans DA. 2007. A microtubule-facilitated nuclear import pathway for cancer regulatory proteins. Traffic 8:673–686. doi: 10.1111/j.1600-0854.2007.00564.x. [DOI] [PubMed] [Google Scholar]
- 37.Dundr M, Misteli T, Olson MO. 2000. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol 150:433–446. doi: 10.1083/jcb.150.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S. 2004. HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol 6:642–647. doi: 10.1038/ncb1144. [DOI] [PubMed] [Google Scholar]
- 39.Oksayan S, Nikolic J, David CT, Blondel D, Jans DA, Moseley GW. 2014. Identification of a role for nucleolin in rabies virus infection. J Virol 89:1939–1943. doi: 10.1128/JVI.03320-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuusisto HV, Wagstaff KM, Alvisi G, Jans DA. 2008. The C-terminus of apoptin represents a unique tumor cell-enhanced nuclear targeting module. Int J Cancer 123:2965–2969. doi: 10.1002/ijc.23884. [DOI] [PubMed] [Google Scholar]
- 41.Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. 2005. Regulation of ubiquitin ligase dynamics by the nucleolus. J Cell Biol 170:733–744. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mekhail K, Rivero-Lopez L, Al-Masri A, Brandon C, Khacho M, Lee S. 2007. Identification of a common subnuclear localization signal. Mol Biol Cell 18:3966–3977. doi: 10.1091/mbc.E07-03-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashour J, Laurent-Rolle M, Shi PY, Garcia-Sastre A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. J Virol 83:5408–5418. doi: 10.1128/JVI.02188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pryor MJ, Carr JM, Hocking H, Davidson AD, Li P, Wright PJ. 2001. Replication of dengue virus type 2 in human monocyte-derived macrophages: comparisons of isolates and recombinant viruses with substitutions at amino acid 390 in the envelope glycoprotein. Am J Trop Med Hyg 65:427–434. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Soh TS, Zheng J, Chan KW, Phoo WW, Lee CC, Tay MY, Swaminathan K, Cornvik TC, Lim SP, Shi PY, Lescar J, Vasudevan SG, Luo D. 2015. A crystal structure of the dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog 11:e1004682. doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutz P, Puvion-Dutilleul F, Lutz Y, Kedinger C. 1996. Nucleoplasmic and nucleolar distribution of the adenovirus IVa2 gene product. J Virol 70:3449–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dundr M, Leno GH, Hammarskjöld ML, Rekosh D, Helga-Maria C, Olson MO. 1995. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J Cell Sci 108(Pt 8):2811–2823. [DOI] [PubMed] [Google Scholar]
- 48.Siomi H, Shida H, Maki M, Hatanaka M. 1990. Effects of a highly basic region of human immunodeficiency virus Tat protein on nucleolar localization. J Virol 64:1803–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falcon V, Acosta-Rivero N, Chinea G, de la Rosa MC, Menendez I, Duenas-Carrera S, Gra B, Rodriguez A, Tsutsumi V, Shibayama M, Luna-Munoz J, Miranda-Sanchez MM, Morales-Grillo J, Kouri J. 2003. Nuclear localization of nucleocapsid-like particles and HCV core protein in hepatocytes of a chronically HCV-infected patient. Biochem Biophys Res Commun 310:54–58. doi: 10.1016/j.bbrc.2003.08.118. [DOI] [PubMed] [Google Scholar]
- 50.James A, Wang Y, Raje H, Rosby R, DiMario P. 2014. Nucleolar stress with and without p53. Nucleus 5:402–426. doi: 10.4161/nucl.32235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacob MD, Audas TE, Uniacke J, Trinkle-Mulcahy L, Lee S. 2013. Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus. Mol Biol Cell 24:2943–2953. doi: 10.1091/mbc.E13-04-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malavige GN, Fernando S, Fernando DJ, Seneviratne SL. 2004. Dengue viral infections. Postgrad Med J 80:588–601. doi: 10.1136/pgmj.2004.019638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buckley A, Gaidamovich S, Turchinskaya A, Gould EA. 1992. Monoclonal antibodies identify the NS5 yellow fever virus nonstructural protein in the nuclei of infected cells. J Gen Virol 73(Pt 5):1125–1130. doi: 10.1099/0022-1317-73-5-1125. [DOI] [PubMed] [Google Scholar]