ABSTRACT
Few studies have evaluated the impact of the viral challenge route on protection against a heterologous simian immunodeficiency virus (SIV) challenge. We vaccinated seven macaques with a live attenuated SIV that differed from SIVmac239Δnef by 24 amino acids, called m3KOΔnef. All animals were protected from an intrarectal SIVmac239 challenge, whereas only four animals were protected from subsequent intravenous SIVmac239 challenge. These data suggest that immune responses elicited by vaccination with live attenuated SIV in an individual animal can confer protection from intrarectal challenge while remaining insufficient for protection from intravenous challenge.
IMPORTANCE Our study is important because we show that vaccinated animals can be protected from a mucosal challenge with a heterologous SIV, but the same animals are not necessarily protected from intravenous challenge with the same virus. This is unique because in most studies, either vaccinated animals are challenged multiple times by the same route or only a single challenge is performed. An individually vaccinated animal is rarely challenged multiple times by different routes, so protection from different challenge routes cannot be measured in the same animal. Our data imply that vaccine-elicited responses in an individual animal may be insufficient for protection from intravenous challenge but may be suitable for protection from a mucosal challenge that better approximates human immunodeficiency virus (HIV) exposure.
INTRODUCTION
Two important variables that can influence the apparent efficacy of preclinical human immunodeficiency virus (HIV)/simian immunodeficiency virus (SIV) vaccines are the route of infection and the sequence of the challenge virus. For example, live attenuated SIV offers effective and consistent protection from intravenous challenge with a homologous virus but incomplete protection from intravenous challenge with a heterologous virus. This incomplete protection from heterologous challenge was observed after vaccination with several live attenuated SIV strains (e.g., SIVmac239Δnef, SIVmac239Δ3, SIVsmE543Δnef, and SIVmacC8), followed by challenge with different pathogenic SIV stocks (e.g., SIVsmE660, SHIV89.6p, SIV239/EnvE543, and SIVmac239) (1–6). Although those studies were each different, incomplete protection from an intravenous heterologous challenge was observed for both rhesus and cynomolgus macaques.
The failure to offer complete protection from an intravenous challenge with a heterologous virus may be because some immune responses elicited by live attenuated SIV are localized to the mucosa and are not mobilized systemically at the time of challenge, making an intravenous challenge too stringent for testing of the efficacy of live attenuated SIV vaccines (7, 8). Perhaps, immune responses elicited by a vaccine in an individual animal are sufficient to protect from mucosal challenge with a high dose of a heterologous virus, even if they cannot protect from intravenous challenge with the same virus.
In this study, we wanted to test the hypothesis that a live attenuated SIV vaccine can offer protection from a high-dose mucosal challenge with a heterologous SIV but not an intravenous challenge with the same virus in an individual animal. If this hypothesis is correct, then challenging vaccinated animals intravenously with SIV may be an inappropriately high bar for testing of preclinical HIV vaccines.
MATERIALS AND METHODS
Animals and virus infections.
The seven experimental animals, animals cy0348, cy0379, cy0381, cy0382, cy0383, cy0384, and cy0385, were used in a previous study (9). Animals cy0379, cy0381, cy0384, and cy0385 were homozygous for the M3 major histocompatibility complex (MHC) haplotype and are referred to as “M3 positive.” Animals cy0348, cy0382, and cy0383 expressed no MHC alleles from the M3 MHC haplotype and are referred to as “M3 negative.” All seven animals were immunized intravenously with 10 ng p27 m3KOΔnef, a strain of live attenuated SIV that differs from SIVmac239Δnef by 24 amino acids.
Twenty weeks after immunization, animals cy0379, cy0381, cy0382, cy0383, cy0384, and cy0385 were challenged intrarectally with 7,000 50% tissue culture infective doses (TCID50) of SIVmac239. Animal cy0348 was immunized with m3KOΔnef 98 days prior to the other six animals. Thus, he was challenged intrarectally with 7,000 TCID50 SIVmac239 20 weeks after m3KOΔnef vaccination and then rechallenged intrarectally with 7,000 TCID50 SIVmac239 at day 98 after the first challenge, alongside the other six animals, so that the timeline for the intrarectal and intravenous challenge studies described in this study would align across all 7 animals. Fourteen weeks after this intrarectal challenge, all seven animals were rechallenged intravenously with 100 TCID50 SIVmac239.
The four control animals, animals cy0684, cy0686, cy0687, and cy0689, were immunized originally with 10 ng p27 SIVmac239Δnef and then challenged intravenously 28 weeks later with 100 TCID50 SIVmac239. All four of these animals were homozygous for the M3 MHC haplotype and are called M3-positive animals here.
All animals used in this study were purchased from Charles River Laboratories or the Bioculture Group and cared for by the Wisconsin National Primate Research Center (WNPRC) according to protocols approved by the University of Wisconsin Graduate School Animal Care and Use Committee.
Plasma viral load analysis.
Viral loads were determined from plasma obtained from venous blood drawn into EDTA tubes. SIV gag loads were determined essentially as previously described (9, 10). Viral RNA (vRNA) was isolated from plasma, reverse transcribed, and amplified with the SuperScript III Platinum one-step quantitative reverse transcription-PCR (qRT-PCR) system (Invitrogen). Quantification of amplified templates was performed on a LightCycler 480 instrument (Roche). Serial dilutions of an SIV gag in vitro transcript were used as an internal standard curve for each run. LightCycler version 1.5 software was used to interpolate values for samples onto the standard curve to determine the copy number. The detection limit of the assay was either 50 or 100 vRNA copy equivalents per ml of plasma, depending on the vRNA isolation protocol used. The limit-of-detection value was reported when the viral load was at or below the limit of detection.
Full-length nef and Δnef viral loads were determined as previously described (11). Briefly, highly specific, real-time RT-PCR assays were used with primers that accurately differentiate viruses containing full-length nef from those that contain nef with a 182-bp deletion, using the methods described above. Serial dilutions of in vitro transcripts for both full-length nef and nef with a 182-bp deletion were used as internal standards for each run. The same machines and software used for the gag viral load assay were used to detect and quantify the nef and Δnef viral loads. The limit of detection was identical to that for the SIV gag viral load assay.
IFN-γ ELISPOT analysis.
Fresh PBMCs (peripheral blood mononuclear cells) were subjected immediately to an gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISPOT) assay, as previously described (12). Briefly, a precoated monkey IFN-γ ELISPOTplus plate (Mabtech, Mariemont, OH) was blocked, and peptides corresponding to each of the wild-type epitope sequences were added to each well at a final concentration of 1 μM. Peptides were tested in duplicate. A total of 105 cells were added to each well, and plates were incubated overnight. Plates were developed according to the manufacturer's protocol. Spots were imaged with an ELISPOT reader (AID Autoimmun Diagnostika Gmbh). Numbers of SFCs (spot-forming cells) per 106 PBMCs were calculated by subtracting the average number of background spots (average of data from four wells not stimulated with any peptide, which served as negative controls) and then multiplying this value by 10. Values for duplicate wells were then averaged and graphed. A response was considered positive if it exceeded our threshold of the no-stimulation average plus 2 times the standard deviation for the nonstimulated wells or if it had 50 SFCs per 106 PBMCs, whichever was greater. Concanavalin A was used as a positive control at 10 μM.
Deep sequencing of SIV.
Genome-wide deep sequencing of replicating virus populations from animals cy0381, cy0348, and cy0382 at necropsy was performed as previously described (12). Briefly, circulating plasma vRNA was isolated with the MinElute virus spin kit (Qiagen). The Superscript III One-Step RT-PCR system with Platinum Taq High Fidelity (Invitrogen) was used to generate viral cDNA and four overlapping amplicons spanning the entire SIV coding sequence. PCR products were purified with the MinElute gel extraction kit (Qiagen) and were then quantified with the Quant-IT dsDNA HS assay kit (Invitrogen). Libraries were generated from 1 ng of pooled amplicons. The libraries were tagged with barcodes by using the Nextera XT kit (Illumina) and then quantified with the Quant-IT dsDNA HS assay kit. An Agilent Bioanalyzer was used to assess the quality of the library preparation. Libraries were pooled and sequenced on an Illumina MiSeq instrument using 2-by-250 kits.
Sequences were analyzed with Geneious (Biomatters, Ltd.). Reads were initially quality trimmed. Paired reads were then mapped to the SIVmac239 sequence (GenBank accession number M33262). Variant nucleotides were called at a threshold of 5%. The variants detected in the analyzed virus populations were then compared to the mutant sites originally incorporated into m3KOΔnef that were 5′ of the 182-bp deletion in nef. The frequencies of variant nucleotides in the virus populations at necropsy that matched those of the original inoculum are reported in Table 2.
TABLE 2.
Frequencies of m3KOΔnef mutations detectable in virus populations at necropsya
| nt position of original mutation | Frequency (%) in animal: |
||
|---|---|---|---|
| cy0381 | cy0348 | cy0382 | |
| 1316 | 99.8 | 95.7 | 96.6 |
| 1481 | 64.4 | <5 | 47.6 |
| 1510 | 99.7 | <5 | 48.3 |
| 2467 | 99.7 | 99.6 | 99.4 |
| 2723 | 99.6 | 84.1 | 99.8 |
| 2850 | 99.8 | 95.5 | 99.7 |
| 2860 | 99.9 | 96.6 | 99.5 |
| 3721 | 99.9 | 99.7 | 99.9 |
| 4260 | 99.8 | 92.6 | 99.8 |
| 4945 | 99.5 | 99.5 | 99.4 |
| 5815 | 100.2 | 99.9 | 99.8 |
| 6199 | 98.9 | 97.0 | 96.5 |
| 6639 | 99.4 | 65.6 | 99.7 |
| 6923 | 97.4 | 11.6 | 98.1 |
| 6925 | 97.4 | 11.6 | 98.1 |
| 7058 | 99.8 | 99.8 | 99.8 |
| 8390 | 98.0 | 11.3 | 97.7 |
| 8750 | 56.4 | 6.8 | 87.4 |
| 8850 | 92.1 | 52.3 | 25.5 |
| 9110 | 99.7 | 99.7 | 99.5 |
| 9176 | 85.0 | 5.2 | 5.3 |
| 9181 | 84.0 | 46.7 | 6.0 |
The nucleotide (nt) positions in the m3KOΔnef inoculum virus that had the original mutations, relative to SIVmac239, are shown. The frequencies of the individual variants, relative to SIVmac239, are shown for each of the virus populations isolated from the three animals. This percentage reflects the frequency that the original mutation from the m3KOΔnef inoculum was still found in the virus population at necropsy.
Nucleotide sequence accession number.
Sequences have been deposited in the Sequence Read Archive (SRA) under accession number SRP073659.
RESULTS
To test our hypothesis, we used seven Mauritian cynomolgus macaques (MCMs) that had been vaccinated with m3KOΔnef, a live attenuated strain of SIV that differs from SIVmac239Δnef by 24 amino acids (9). Four of these amino acid mutations are located in known CD8 T cell epitopes presented by MHC class I molecules expressed in the four M3-positive animals (animals cy0379, cy0381, cy0384, and cy0385) (9). gag viral loads were measured in these seven animals during the first 20 weeks after vaccination with m3KOΔnef (9). Viral loads peaked at 105 virus copy equivalents per ml of blood plasma, or higher, in all 7 animals. We found that the four M3-positive animals had a mean viral load set point of 2,042 copies/ml, whereas the three M3-negative animals (animals cy0348, cy0382, and cy0383) had a mean viral load set point of 39 copies/ml (9).
At least 20 weeks after immunization with m3KOΔnef, all seven animals were challenged intrarectally with 7,000 TCID50 SIVmac239, a dose and challenge route that have been used successfully to infect unvaccinated MCMs (13, 14). To distinguish between replication of SIVmac239 and m3KOΔnef, we used a qRT-PCR assay that uniquely quantifies the number of copies of full-length nef and Δnef (a nef gene with a 182-bp deletion) (11). Even though three of the animals were unable to control the replication of m3KOΔnef (Table 1), we did not detect replication of the full-length nef sequence in the seven animals (Table 1).
TABLE 1.
All seven animals vaccinated with m3KOΔnef are resistant to intrarectal challenge with SIVmac239a
| Challenge virus and animal | Viral load (ceq/ml plasma) |
|||
|---|---|---|---|---|
| Day 0 | Day 7 | Day 14 | Day 21 | |
| ΔNef | ||||
| cy0348 | <50 | <50 | <50 | <50 |
| cy0379 | 82,500 | 21,500 | 29,000 | 102,000 |
| cy0381 | 681 | 178 | 358 | 273 |
| cy0382 | <50 | <50 | <50 | <50 |
| cy0384 | <50 | <50 | <50 | <50 |
| cy0383 | <50 | <50 | <50 | 54 |
| cy0385 | 4,730 | 231 | 1,330 | 7,660 |
| Full-length Nef | ||||
| cy0348 | <50 | <50 | <50 | <50 |
| cy0379 | <50 | <50 | <50 | <50 |
| cy0381 | <50 | <50 | <50 | <50 |
| cy0382 | <50 | <50 | <50 | <50 |
| cy0384 | <50 | <50 | <50 | <50 |
| cy0383 | <50 | <50 | <50 | <50 |
| cy0385 | <50 | <50 | <50 | <50 |
The time points listed indicate the number of days after intrarectal challenge. nef qRT-PCR was performed on plasma samples after SIVmac239 challenge. Viral loads for the 182-bp nef deletion and full-length nef were measured. A viral load of <50 virus copy equivalents (ceq) per ml of blood plasma represents the limit of detection.
We continued to monitor gag viral loads in these seven animals up to ∼100 days after intrarectal challenge with SIVmac239. Of the four M3-positive animals, viral loads ranged from undetectable to >105 copies/ml. For the three M3-negative animals, viral loads remained at <500 copies/ml (data not shown).
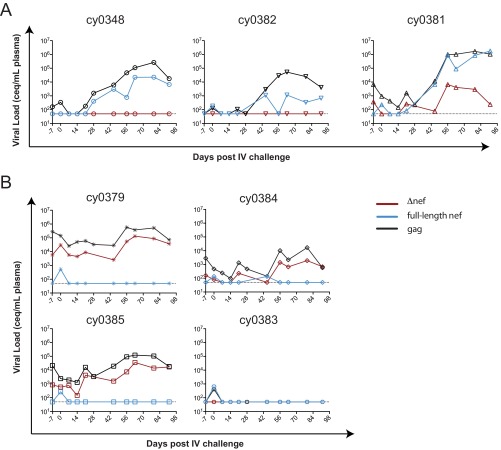
We rechallenged all seven animals intravenously with 100 TCID50 SIVmac239 at ∼14 weeks (∼100 days) after intrarectal challenge. Blood was sampled from these animals immediately after challenge (day 0) (Fig. 1) and then for several weeks. Determinations of gag viral loads were performed on plasma from all animals. In three of the animals (Fig. 1A), the gag viral loads became detectable ∼2 to 3 weeks after infection and then continued to increase for several weeks thereafter (Fig. 1A, black lines). For these three animals, the increase in full-length nef viral loads paralleled the same increases in gag viral loads (Fig. 1A, blue lines). Notably, both M3-positive and M3-negative animals had detectable full-length nef, so we could not ascribe a link between MHC genetics and susceptibility to intravenous SIVmac239 challenge.
FIG 1.
Animals vaccinated with m3KOΔnef are partially susceptible to intravenous (IV) challenge with SIVmac239. gag and nef qRT-PCRs were performed on plasma samples at the indicated times after intravenous SIVmac239 challenge. (A) Three animals with evidence of full-length nef. (B) Four animals without replicating full-length nef. Data for each animal are shown on independent plots. The horizontal dotted line at 50 copies/ml (copy equivalents [ceq] per milliliter) represents the limit of detection of the assay.
This delayed detection of nef is consistent with previous studies suggesting that time is required for live attenuated SIV to recombine with the challenge virus prior to expansion (2). To assess recombination, we deep sequenced virus populations isolated at necropsy. We found that at least 50% of the variants upstream of the nef deletion, and present in m3KOΔnef, were present in the replicating virus population at a frequency of 90% or higher in all three animals (Table 2). This includes a variant present at nucleotide position 9110, which is ∼500 bp upstream of the deletion in nef. Together, these data strongly suggest that the live attenuated SIV recombined with the SIVmac239 challenge virus.
Of the four animals that appeared to control intravenous challenge with SIVmac239 (Fig. 1B), all four of them tested positive for full-length nef in the blood sampled immediately after infection (day 0) (Fig. 1B, blue lines). This detection of full-length nef at day 0 was transient, indicating successful inoculation with SIVmac239 in these four animals that was subsequently controlled.
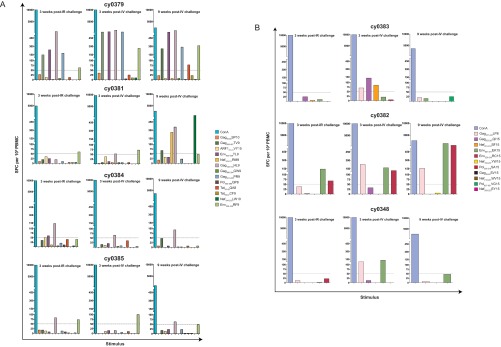
We wanted to determine if the specificity of the CD8 T cell response in these animals was different after intrarectal and intravenous challenges with SIVmac239. We performed IFN-γ ELISPOT assays 3 weeks after intrarectal and intravenous challenges. We used peptides for the epitopes that we predicted could be targeted in the animals with each MHC genotype. We found that there were no overt and consistent differences in the specificity of the T cell response after intrarectal or intravenous challenge (Fig. 2), independent of whether viruses with full-length nef ultimately replicated in the animals. Notably, 9 weeks after intravenous challenge with SIVmac239, we found that the M3-positive animal with replicating full-length nef, animal cy0381, had IFN-γ ELISPOT responses to two epitopes in Nef that are absent in m3KOΔnef but present in full-length Nef.
FIG 2.
Minimal differences in T cell responses elicited following intrarectal and intravenous SIVmac239 challenges. IFN-γ ELISPOT assays were performed by using peptides for epitopes that could be targeted in animals that were M3 positive (A) and M3 negative (B). T cell responses were measured 3 weeks after intrarectal (IR) challenge, 3 weeks after intravenous challenge, and 9 weeks after intravenous challenge, with concanavalin A (ConA) being used as a positive control at all time points. Of note, at the time of the assay, it was unknown that the Nef194–203LW10 peptide contained the Nef196–203HW8 epitope used for Fig. 3B. A response was considered positive if the number of SFCs per 106 PBMCs exceeded our threshold of the no-stimulation average plus 2 times the standard deviation for nonstimulated wells or 50 SFCs per 106 PBMCs, whichever was greater. This threshold of 50 SFCs per 106 PBMCs is represented with a horizontal dotted line. Each bar color represents a different peptide and shows the average of data for two duplicate wells following subtraction of the average for the nonstimulated wells.
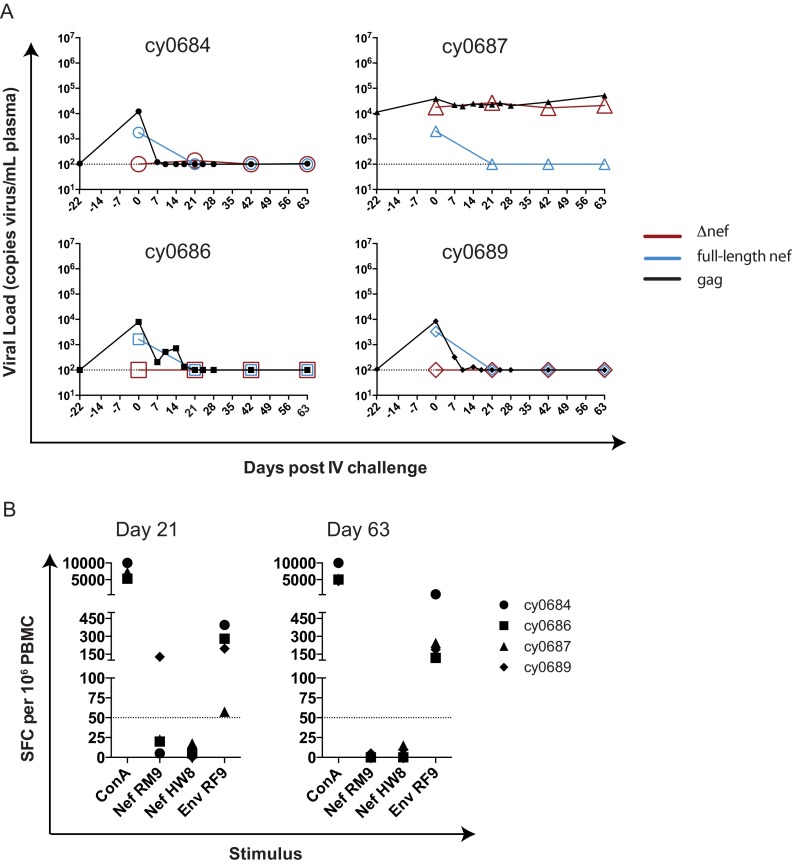
We wanted to confirm that SIVmac239Δnef vaccination protects MCMs from intravenous homologous SIVmac239 challenge, as observed previously in rhesus macaques (4). Four MCMs that were vaccinated with SIVmac239Δnef for 28 weeks were challenged intravenously with 100 TCID50 SIVmac239. Again, blood was sampled from these animals immediately after challenge (day 0) and then for several weeks. gag qRT-PCR was performed to measure total SIV loads prior to and after SIVmac239 challenge (Fig. 3A). We observed minor transient increases in virus replication ∼2 to 3 weeks after SIVmac239 challenge. We then quantified full-length nef and Δnef for select time points, as described above (Table 1 and Fig. 1). All four animals had full-length nef detectable in their blood immediately after challenge (day 0), but this became undetectable at days 21, 42, and 63 (Fig. 3A). This stands in stark contrast to the detection of full-length nef observed in the three m3KOΔnef-immunized animals with replicating full-length nef (Fig. 1A). This control experiment demonstrates that SIVmac239Δnef vaccination protects from intravenous homologous challenge.
FIG 3.
Animals vaccinated with SIVmac239Δnef are resistant to intravenous challenge with SIVmac239. (A) gag qRT-PCR was performed on plasma samples at the indicated times following intravenous SIVmac239 challenge, and data were graphed. Full-length nef and Δnef qRT-PCRs were performed at select time points for each animal. The limit of detection of the assay is shown with a horizontal dotted line. (B) IFN-γ ELISPOT assays were performed with two epitopes (Nef103–111RM9 and Nef196–203HW8) at days 21 and 63 post-intravenous challenge. An additional epitope present in SIVmac239Δnef, Env338–346RF9, is shown as a positive control, as is concanavalin A (ConA). A response was considered positive if the number of SFCs per 106 PBMCs exceeded our threshold of the no-stimulation average plus 2 times the standard deviation of data for nonstimulated wells or 50 SFCs per 106 PBMCs, whichever was greater. This threshold of 50 SFCs per 106 PBMCs is represented with a horizontal dotted line. Each shape represents a different animal and is the average of data for two duplicate wells following subtraction of the average for the nonstimulated wells.
To confirm that there was no full-length Nef antigen in the four SIVmac239Δnef-vaccinated animals after SIVmac239 challenge, we performed IFN-γ ELISPOT assays at days 21 and 63 using two CD8 T cell epitopes that are present in full-length Nef but absent in SIVmac239Δnef: Nef103–111RM9 and Nef196–203HW8. These epitopes are restricted by Mafa-A1*063, an MHC class I molecule expressed in all four animals. CD8 T cell responses to Nef103–111RM9 are routinely detected 2 to 4 weeks after SIVmac239 infection, and responses to Nef196–203HW8 are detected 8 weeks after SIVmac239 infection (13, 15–17). We found that no animals tested positive for a response against either Nef epitope (Fig. 3B), and all animals tested positive for a response against the Env338–346RF9 epitope that is present in SIVmac239Δnef. Animal cy0689 appeared to have an elevated response to Nef103–111RM9 at day 21, but this value did not exceed the positive threshold calculated for this animal at this time point (see Materials and Methods) and is therefore not considered a positive response. These data provide further evidence that there was no full-length nef sequence replicating in these four animals.
DISCUSSION
Unlike other studies of live attenuated SIV vaccines, we challenged seven vaccinated animals both mucosally and intravenously with a high dose of a minimally heterologous virus. Even though there is no absolute definition of a heterologous virus, we found that all seven animals were protected from mucosal challenge, but only four animals were protected from intravenous challenge. These results were independent of the specificity of vaccine-elicited CD8 T cells or host MHC genetics. Similar studies that sequentially challenge vaccinated animals with a high dose of virus are rare (18) because a subset of animals is typically infected during the initial SIV challenge. Nonetheless, challenging animals serially but by different routes can help discern whether vaccine-elicited immune responses can offer protection at one site of exposure but not another one.
It is possible that serial inoculation of the animals in our study affected the differential protection observed with intravenous challenge. Notably, serial exposures are used in models that use rapidly repeated low-dose SIV challenges. While serial challenges may be a concern, this approach is necessary when assessing if host immune responses in a single individual protect from different types of virus exposures.
Unfortunately, there was no clear reason why three of the m3KOΔnef-vaccinated animals had replication of full-length nef after intravenous challenge with SIVmac239, whereas four animals did not. Four of the seven animals were M3 positive and MHC identical, but we found that M3-positive animals were in both outcome groups. Although the numbers of animals are small, this suggests that host MHC genetics did not play a role in the susceptibility of vaccinated MCMs to intravenous challenge with SIVmac239. Furthermore, there were no overt differences in the specificity of the T cell responses 3 weeks after intrarectal and intravenous challenges, as measured by IFN-γ ELISPOT assays (Fig. 2 and data not shown), suggesting that the specificity of T cells in the periphery did not determine whether animals were able to prevent the replication of viruses with full-length nef sequences after intrarectal, but not intravenous, challenge. Together, these observations are consistent with data from previous studies suggesting that control of SIV replication may be independent of the CD8 T cell response, especially among animals that do not have protective MHC alleles (19, 20). It is alternately possible that persistent replication of the m3KOΔnef virus in some animals offered the maintenance of effective host immune responses, but this hypothesis is difficult to test in the small cohort of animals in this study. Future studies will need to address the immunological reasons for this differential outcome.
Overall, it is not that surprising that a single-vaccine approach offers better protection from mucosal challenge than from intravenous challenge. In fact, animals vaccinated with SIVmac239Δnef and then subjected to rapidly repeated low-dose challenges with SIVsmE660 were less susceptible to pathogenic infection than a separate vaccinated cohort that was challenged intravenously with SIVsmE660 (2, 21). Our study is unique, however, for two reasons. First, we challenged the same cohort of animals both mucosally and intravenously with the same virus. Second, our mucosal challenge used a dose of SIV with >10 times the number of copies of virus particles as those used in previous rapidly repeated low-dose challenges (21), yet none of the vaccinated animals in our study became infected with a dose of virus that readily infects unvaccinated animals (14).
Even though the sample size for this study is small, our data suggest that immune responses elicited by strains of live attenuated SIV may protect from a high-dose mucosal challenge with SIV, even if they do not offer similar protection from intravenous challenge. We build upon previous vaccine studies because we show that vaccine-elicited immunity from mucosal exposure does not necessarily protect the same animal from intravenous exposure. Since mucosal transmission accounts for most HIV transmission globally, these results suggest that an intravenous challenge may be an inappropriately high bar for evaluating vaccine performance, and this difference should be a definite point to consider in future vaccine studies.
ACKNOWLEDGMENTS
We thank members of the WNPRC for their support with the animal care and other members of the O'Connor laboratory for critical discussions about the manuscript. We thank Dane Gellerup for deep sequencing of viruses.
We declare that we have no competing interests.
Samples were collected by M.S.S., A.J.B., A.B., C.M.B., and S.L.O. Viral loads were measured by A.M.W. and G.L.-B. as part of virology services at the WNPRC, which is managed by T.C.F. A.B., S.L.O., A.J.B., and M.S.S. performed IFN-γ ELISPOT assays. S.L.O. conceived of the study. M.S.S. and S.L.O. wrote the manuscript.
This work was supported by NIH grant R01 AI108415, an ESI CHAVI/HVTN award, and University of Wisconsin—Madison startup funds to S.L.O. M.S.S. and A.J.B. were supported by NIH grant R01 AI108415. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM081061 to M.S.S. C.M.B. and A.B. were supported by an ESI CHAVI/HVTN award. The research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grants RR15459-01 and RR020141-01. We also thank members of the Wisconsin National Primate Research Center, a facility supported by grants P51RR000167 and P51OD011106.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Funding Statement
C.M.B., A.B., and S.L.O. were supported partially or in full by an ESI CHAVI/HVTN award. S.L.O. was also supported in part by University of Wisconsin—Madison startup funds.
REFERENCES
- 1.Berry N, Ham C, Mee ET, Rose NJ, Mattiuzzo G, Jenkins A, Page M, Elsley W, Robinson M, Smith D, Ferguson D, Towers G, Almond N, Stebbings R. 2011. Early potent protection against heterologous SIVsmE660 challenge following live attenuated SIV vaccination in Mauritian cynomolgus macaques. PLoS One 6:e23092. doi: 10.1371/journal.pone.0023092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reynolds MR, Weiler AM, Weisgrau KL, Piaskowski SM, Furlott JR, Weinfurter JT, Kaizu M, Soma T, Leon EJ, MacNair C, Leaman DP, Zwick MB, Gostick E, Musani SK, Price DA, Friedrich TC, Rakasz EG, Wilson NA, McDermott AB, Boyle R, Allison DB, Burton DR, Koff WC, Watkins DI. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J Exp Med 205:2537–2550. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyand MS, Manson K, Montefiori DC, Lifson JD, Johnson RP, Desrosiers RC. 1999. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J Virol 73:8356–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, McDermott AB, Schultz A, Zamb TJ, Boyle R, Desrosiers RC. 2006. HIV vaccine design: insights from live attenuated SIV vaccines. Nat Immunol 7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- 5.Manrique J, Piatak M, Lauer W, Johnson W, Mansfield K, Lifson J, Desrosiers R. 2013. Influence of mismatch of Env sequences on vaccine protection by live attenuated simian immunodeficiency virus. J Virol 87:7246–7254. doi: 10.1128/JVI.00798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, Mahyari E, Hagen SI, Bae JY, Reyes MD, Swanson T, Legasse AW, Sylwester A, Hansen SG, Smith AT, Stafova P, Shoemaker R, Li Y, Oswald K, Axthelm MK, McDermott A, Ferrari G, Montefiori DC, Edlefsen PT, Piatak MJ, Lifson JD, Sekaly RP, Picker LJ. 2012. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat Med 18:1673–1681. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Zeng M, Duan L, Voss JE, Smith AJ, Pambuccian S, Shang L, Wietgrefe S, Southern PJ, Reilly CS, Skinner PJ, Zupancic ML, Carlis JV, Piatak MJ, Waterman D, Reeves RK, Masek-Hammerman K, Derdeyn CA, Alpert MD, Evans DT, Kohler H, Muller S, Robinson J, Lifson JD, Burton DR, Johnson RP, Haase AT. 2014. Live simian immunodeficiency virus vaccine correlate of protection: local antibody production and concentration on the path of virus entry. J Immunol 193:3113–3125. doi: 10.4049/jimmunol.1400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith AJ, Wietgrefe SW, Shang L, Reilly CS, Southern PJ, Perkey KE, Duan L, Kohler H, Muller S, Robinson J, Carlis JV, Li Q, Johnson RP, Haase AT. 2014. Live simian immunodeficiency virus vaccine correlate of protection: immune complex-inhibitory Fc receptor interactions that reduce target cell availability. J Immunol 193:3126–3133. doi: 10.4049/jimmunol.1400822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris M, Burns CM, Becker EA, Braasch AT, Gostick E, Johnson RC, Broman KW, Price DA, Friedrich TC, O'Connor SL. 2013. Acute-phase CD8 T cell responses that select for escape variants are needed to control live attenuated simian immunodeficiency virus. J Virol 87:9353–9364. doi: 10.1128/JVI.00909-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cline AN, Bess JW, Piatak MJ, Lifson JD. 2005. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol 34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 11.Salisch NC, Kaufmann DE, Awad AS, Reeves RK, Tighe DP, Li Y, Piatak MJ, Lifson JD, Evans DT, Pereyra F, Freeman GJ, Johnson RP. 2010. Inhibitory TCR coreceptor PD-1 is a sensitive indicator of low-level replication of SIV and HIV-1. J Immunol 184:476–487. doi: 10.4049/jimmunol.0902781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gellerup DD, Balgeman AJ, Nelson CW, Ericsen AJ, Scarlotta M, Hughes AL, O'Connor SL. 2016. Conditional immune escape during chronic simian immunodeficiency virus infection. J Virol 90:545–552. doi: 10.1128/JVI.02587-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budde ML, Greene JM, Chin EN, Ericsen AJ, Scarlotta M, Cain BT, Pham NH, Becker EA, Harris M, Weinfurter JT, O'Connor SL, Piatak MJ, Lifson JD, Gostick E, Price DA, Friedrich TC, O'Connor DH. 2012. Specific CD8+ T cell responses correlate with control of simian immunodeficiency virus replication in Mauritian cynomolgus macaques. J Virol 86:7596–7604. doi: 10.1128/JVI.00716-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Connor SL, Becker EA, Weinfurter JT, Chin EN, Budde ML, Gostick E, Correll M, Gleicher M, Hughes AL, Price DA, Friedrich TC, O'Connor DH. 2012. Conditional CD8+ T cell escape during acute simian immunodeficiency virus infection. J Virol 86:605–609. doi: 10.1128/JVI.05511-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bimber BN, Burwitz BJ, O'Connor S, Detmer A, Gostick E, Lank SM, Price DA, Hughes A, O'Connor D. 2009. Ultradeep pyrosequencing detects complex patterns of CD8+ T-lymphocyte escape in simian immunodeficiency virus-infected macaques. J Virol 83:8247–8253. doi: 10.1128/JVI.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burwitz BJ, Pendley CJ, Greene JM, Detmer AM, Lhost JJ, Karl JA, Piaskowski SM, Rudersdorf RA, Wallace LT, Bimber BN, Loffredo JT, Cox DG, Bardet W, Hildebrand W, Wiseman RW, O'Connor SL, O'Connor DH. 2009. Mauritian cynomolgus macaques share two exceptionally common major histocompatibility complex class I alleles that restrict simian immunodeficiency virus-specific CD8+ T cells. J Virol 83:6011–6019. doi: 10.1128/JVI.00199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budde ML, Lhost JJ, Burwitz BJ, Becker EA, Burns CM, O'Connor SL, Karl JA, Wiseman RW, Bimber BN, Zhang GL, Hildebrand W, Brusic V, O'Connor DH. 2011. Transcriptionally abundant major histocompatibility complex class I alleles are fundamental to nonhuman primate simian immunodeficiency virus-specific CD8+ T cell responses. J Virol 85:3250–3261. doi: 10.1128/JVI.02355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu W, Chen S, Lai C, Guo W, Fu L, Andrieu JM. 2012. Induction of CD8+ regulatory T cells protects macaques against SIV challenge. Cell Rep 2:1736–1746. doi: 10.1016/j.celrep.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Bruel T, Hamimi C, Dereuddre-Bosquet N, Cosma A, Shin SY, Corneau A, Versmisse P, Karlsson I, Malleret B, Targat B, Barre-Sinoussi F, Le Grand R, Pancino G, Saez-Cirion A, Vaslin B. 2015. Long-term control of simian immunodeficiency virus (SIV) in cynomolgus macaques not associated with efficient SIV-specific CD8+ T-cell responses. J Virol 89:3542–3556. doi: 10.1128/JVI.03723-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansfield K, Lang SM, Gauduin MC, Sanford HB, Lifson JD, Johnson RP, Desrosiers RC. 2008. Vaccine protection by live, attenuated simian immunodeficiency virus in the absence of high-titer antibody responses and high-frequency cellular immune responses measurable in the periphery. J Virol 82:4135–4148. doi: 10.1128/JVI.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynolds MR, Weiler AM, Piaskowski SM, Kolar HL, Hessell AJ, Weiker M, Weisgrau KL, León EJ, Rogers WE, Makowsky R, McDermott AB, Boyle R, Wilson NA, Allison DB, Burton DR, Koff WC, Watkins DI. 2010. Macaques vaccinated with simian immunodeficiency virus SIVmac239Delta nef delay acquisition and control replication after repeated low-dose heterologous SIV challenge. J Virol 84:9190–9199. doi: 10.1128/JVI.00041-10. [DOI] [PMC free article] [PubMed] [Google Scholar]