Abstract
The transcription-coupled repair pathway (TC-NER) plays a vital role in removing transcription-blocking DNA lesions, particularly UV-induced damage. Clinical symptoms of the two TC-NER-deficiency syndromes, Cockayne syndrome (CS) and UV-hypersensitivity syndrome (UVSS) are dissimilar and the underlying molecular mechanism causing this difference in disease pathology is not yet clearly understood. UV-stimulated scaffold protein A (UVSSA) has been identified recently as a new causal gene for UVSS. Here we describe a functional homolog of the human UVSSA gene in the nematode Caenorhabditis elegans, uvs-1 (UVSSA-like-1). Mutations in uvs-1 render the animals hypersensitive to UV-B irradiation and transcription-blocking lesion-inducing illudin-M, similar to mutations in TC-NER deficient mutants. Moreover, we demonstrate that TC-NER factors including UVS-1 are required for the survival of the adult animals after UV-treatment.
Keywords: Nucleotide excision repair, C. elegans, Cockayne syndrome, UV-hypersensitivity syndrome, Ultraviolet light, DNA damage
1. Introduction
NER is the main repair pathway involved in the repair of bulky DNA lesions that disturb the normal double-helical structure of DNA and is capable of repairing a variety of structurally unrelated lesions. The most important substrates of NER are the ultraviolet (UV) light-induced cyclobutane pyrimidine dimers (CPDs) and pyrimidine 6-4 pyrimidone photoproducts (6-4PPs) [1–3]. Along with the repair of these photoproducts, NER is also involved in the removal of DNA adducts induced by chemotherapeutic drugs like cisplatin [4,5], aromatic hydrocarbons [6], arylamine carcinogens [7], that thermodynamically destabilize the DNA helix [8]. This conserved repair process employs two different mechanisms of DNA damage detection depending on the location of the damage: global genome NER (GG-NER) and transcription-coupled NER (TC-NER). GG-NER scans the entire genome for DNA helix-distorting lesions, whereas, TC-NER is initiated in the transcribed strand of active genes upon the stalling of RNAP II during transcription.
TC-NER deficiencies are associated with two photosensitive syndromes: Cockayne syndrome (CS) and UV-hypersensitivity syndrome (UVSS) [9]. CS is characterized by sun sensitivity, developmental delay, short stature, microcephaly, and severe neurological abnormalities. Most of the CS patients belong to the complementation groups A and B. UVSS patients on the other hand display mild clinical manifestations including sun sensitivity, skin dryness, freckles, pigmentation abnormalities but no developmental or neurological defects [10]. UVSS cells exhibit normal GG-NER but a reduced recovery of RNA synthesis upon UV-B-treatment, similar to TC-NER-deficient cells [11]. The syndrome can be caused by mutations in CSA and CSB and the recently identified UVSSA (previously known as KIAA1530) [11,12]. UVSSA (UV-stimulated scaffold protein A) has been implicated in regulating TC-NER and identified as a causal gene for UVSS in recent studies by various approaches: exome sequencing of UVSS patient derived cells, genetic complementation by chromosome transfer, and SILAC-based proteomic approaches [13–15]. Despite these findings the reasons for the clinical differences between the two TC-NER deficiency syndromes have remained unclear.
Dissecting the roles of the homologs of the mammalian NER genes in a simpler model organism could improve the understanding of the molecular mechanisms underlying the two diseases [16]. C. elegans is being increasingly used for the characterization of various DNA repair pathways since most of the major mammalian repair pathways, including NER, are conserved at the molecular level in this organism [17,18]. C. elegans has a functional NER that repairs UV-induced DNA damage [19,20] with the CSB homolog csb-1 and XPC homolog xpc-1 igniting TC-NER and GG-NER, respectively. Moreover, we recently demonstrated that the ORF D2013.3 encodes the functional CSA homolog in C. elegans [21].
In this study, using the OrthoList database [22], which provides a platform for identifying worm-human orthology, we identified the uncharacterized ORF ZK742.2 as a C. elegans homolog of UVSSA. Consistent with phenotypes observed in UVSS patient cells, ZK742.2 mutant animals are hypersensitive to UV and the transcription-blocking lesion-inducing drug illudin-M. Similar to csa-1 and csb-1 animals, the somatic tissues of ZK742.2 mutant animals are exquisitely sensitive to UV irradiation during development and adulthood. In contrast to GG-NER-deficient xpc-1 mutants, however, the germ cells of ZK742.2 mutants show similar UV resistance as wildtype animals. Furthermore, combined loss of function of ZK742.2 and GG-NER factor xpc-1 increases UV-sensitivity to the levels of complete NER-deficient xpa-1 animals. Taken together, our data describe a function of ZK742.2 in the TC-NER pathway and we therefore name this previously uncharacterized gene uvs-1 for UVSSA-like-1.
2. Materials and methods
2.1. Growth conditions
C. elegans strains were cultured under standard conditions at 20 •C on nematode growth media (NGM) plates with E. coli strain OP50 [23]. Strains used were N2 (Bristol; wildtype), csb-1(ok2335), xpa-1(ok698), xpc-1(tm3886), uvs-1(tm6134), uvs-1(tm6311), uvs-1(tm6311); csb-1(ok2335), uvs-1(tm6311); csb-1(tm3886), uvs-1(tm6311); xpa-1(ok698). uvssa-1 mutant worm strains were obtained from National Bioresource Project and other strains from the Caenorhabditis Genetics Center.
2.2. Somatic UV-sensitivity assay
Synchronized L1 larvae obtained by hypochlorite treatment were plated on NGM-agar plates and UV-B-irradiated (310 nm) with the indicated doses, using a UV6 lamp (Philips) in a Waldmann UV236B device. A minimum of 500 worms was used for each UV-B-dose, and each treatment was conducted in triplicate.
After UV irradiation, worms were washed off with M9 buffer, concentrated by centrifugation and put on NGM plates with a pre-grown OP50 E. coli lawn. Plates were incubated at 20 °C for 48 h or 72 h and analyzed by large particle flow cytometry using a Union Biometrica COPAS Biosort system. Larval stages were determined by measuring ‘time of flight’ (length) and ‘laser extinction’ (optical density) of individual worms using the Biosort 5291 software and confirmed by manual inspection under a Leica M 165C stereomicroscope.
2.3. Germline UV-sensitivity assay
Staged young adults were treated with indicated UV-doses and allowed to recover for 16 h. Following recovery, timed egg-laying was conducted by transferring three animals per UV-dose to a new NGM plate with OP50 for 3 h. Each treatment was conducted in triplicate. The total number of eggs laid was counted and 48 h later the number of viable eggs was determined. To assess UV-sensitivity, the percentage of eggs that were laid after UV-treatment was compared to untreated and the percentage of viable eggs was determined after hatching.
2.4. Adult-survival after UV-treatment
Synchronized day-1 adult animals obtained by hypochlorite treatment were plated on NGM agar plates with OP50 and UV-treated with the indicated doses. 140 animals were used for each dose and the number of dead animals was scored every day until all the animals were dead. During the assay, the live animals were transferred daily to new plates until the end of the egg-laying period and then onwards every second day.
2.5. Illudin-M sensitivity assay
Synchronized L1 larvae were obtained by hypochlorite treatment and were treated with indicated concentrations of illudin-M in liquid S-basal medium with OP50 for 24 h at 20 °C on a shaking platform. A minimum of 50 animals was used for each illudin-M dose and each treatment was conducted in triplicate. Illudin-M sensitivity was measured by determining the percentage of different larval stages in each treatment. Illudin-M was a kind gift from Prof. Dr. Rainer Schobert (Bayreuth).
2.6. Paraquat survival assay
Synchronized day-1 adult worms obtained by hypochlorite treatment were plated on NGM agar plates with 5 mM methyl viologen dichloride hydrate (Paraquat 856177 Sigma-Aldrich) and OP50. 100 animals were used for each dose and the number of dead animals was scored every day until all the animals were dead.
2.7. KBrO3 sensitivity assay
Synchronized L1 larvae were obtained by hypochlorite treatment and were treated with indicated concentrations of KBrO3 (309087 Sigma-Aldrich) in liquid S-basal medium with OP50 for 24 h at 20 °C on a shaking platform. A minimum of 250 animals was used for each KBrO3 dose and each treatment was conducted in triplicate. Larval stages were identified using large particle flow cytometry.
2.8. CPD repair assay
Synchronized L1 larvae were treated with 60 J/m2 UV-B, collected in cold M9 buffer. Half the sample was immediately flash frozen in liquid nitrogen (0h sample) and the other half was allowed to grow on pre-seeded NGM plates for 24 h. 24 h samples were washed 5 times in M9 and flash frozen in liquid nitrogen. Genomic DNA was extracted using Qiagen Puregene kit and quantified.
DNA was denatured for 5 minutes at 95 °C, put directly on ice, and blotted onto Amersham Hybibd-N+ membrane (RPN119B GE Healthcare) using a Whatman 96-well slot blotting device at 300 mbar vacuum. Cross-linking of the DNA was carried out for 2 hours at 80 °C. The membrane was blocked for 30 minutes in 3% milk-PBS-T, incubated with anti-CPD antibody (TDM-2 Cosmo Bio 1:10000) overnight at 4 °C, and peroxidase-conjugated AffiniPure secondary antibody (Jackson Immuno Research 1:10000) for 1 h. After incubation with ECL (RPN2232 GE Healthcare), DNA Lesions were visualized and quantified on the Molecular Imager Gel Doc Imaging System (Bio-Rad) using Image Lab 5.0.
3. Results and discussion
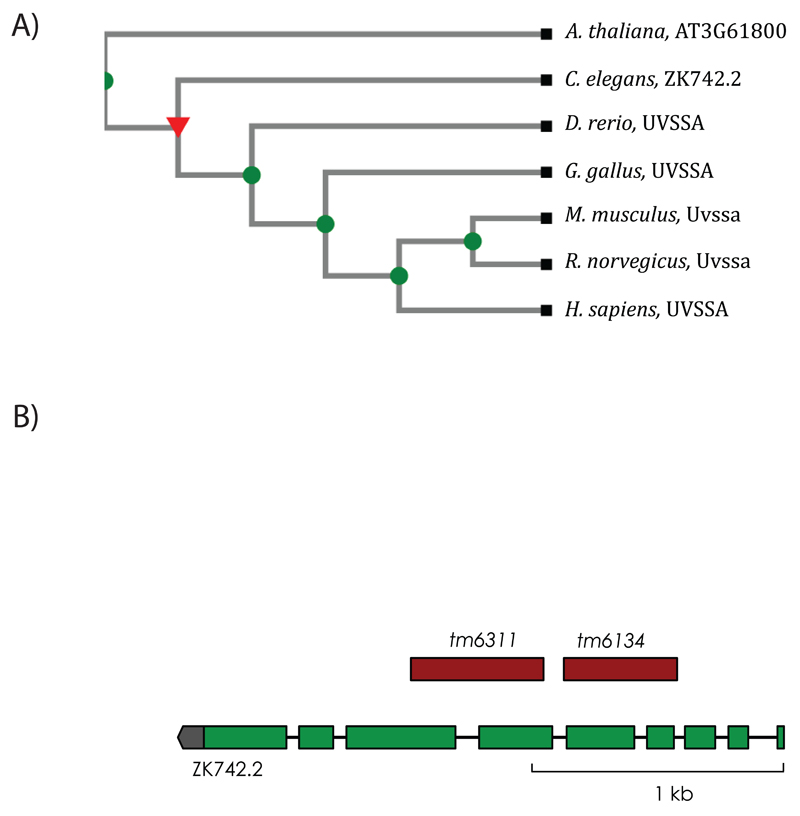
In order to identify the homolog for UVSSA we used the OrthoList database [22], which is a compendium of C. elegans genes with the human orthologs obtained from the meta-analysis of results from four orthology prediction programs. OrthoList identified the uncharacterized ORF ZK742.2 as a C. elegans homolog of UVSSA. A phylogenetic tree created using the TreeFam database [24] placed ZK742.2 with other UVSSA homologs (Figure 1A). The ORF ZK742.2 is 2378 nucleotides long and is organized into nine exons. The coding sequence is 1785 nucleotides long and encodes a 594 aa protein. We obtained two deletion alleles, tm6134 and tm6311, for ZK742.2 from the National BioResource Project [25]. The tm6311 allele contains a complex substitution with a 525 bp deletion and a 2 bp substitution, which deletes parts of exon 6, intron 6 and exon 7 causing a frame shift (Figure 1B). The allele is expected to encode a null mutation by premature termination as it introduces a frameshift. The 6134 allele contains a 441 bp deletion that removes part of intron 3 and exon 4, intron 4 and exon 5, and could lead to a non-functional gene product.
Figure 1. Identification of the C. elegans UVSSA homolog.
(A) Phylogenetic tree displaying the evolutionary relationship between UVSSA homologs in different species constructed using the TreeFam database. Uncharacterized C. elegans ORF ZK742.2 clusters with the UVSSA proteins. (B) Representation of the genomic architecture of ZK742.2. Green boxes represent exons, black lines represent introns, and untranslated region is in gray. The region deleted in the corresponding alleles is represented in red.
3.1. ZK742.2 mutants are UV-hypersensitive during development
In C. elegans, the involvement of the two NER sub pathways differs spatially and temporally [19,20,26]. TC-NER is the major repair pathway in transcriptionally active late embryos and early larval stages, whereas GG-NER takes over the role in early embryos and the germ cells, which are highly proliferative.
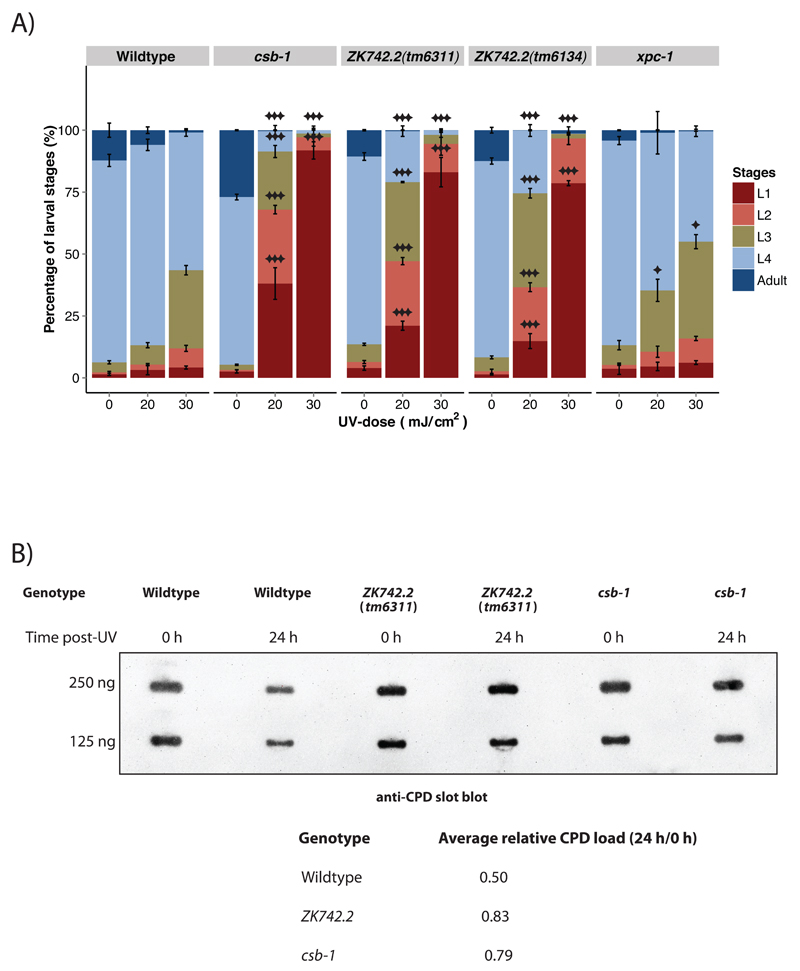
Cells derived from UVSS patients are hypersensitive to UV and exhibit reduced recovery of RNA synthesis (RRS) after UV [13]. To investigate whether ZK742.2 is involved in TC-NER in C. elegans, we subjected synchronized L1 larvae carrying the deletion alleles tm6134 and tm6311 to different UV-B doses and followed the development. 48 h post-UV-B treatment, the untreated animals reached the L4 larval stage, whereas the ZK742.2 mutant animals displayed UV-dependent development arrest in a dose dependent manner, similar to a TC-NER-deficient csb-1 mutant strain (Figure 2A). The GG-NER-deficient xpc-1 animals were only mildly affected.
Figure 2. ZK742.2 is involved in the repair of UV-induced DNA damage.
(A) The percentage of larval stages 48 h after UV-B treatment administered at the L1 larval stage. A minimum of 500 animals were used for each UV-B-dose, and each treatment was conducted in triplicate. Larval stages were determined by flow cytometry. Error bars represent SD, statistical analysis is a two-tailed t-test comparing treatment groups to wildtype, *=p<0.05, **=p<0.01, ***=p<0.001. (B) Slot-blot DNA repair assay using α-CPD antibody. Wildtype, ZK742.2(tm6311) and csb-1 animals were treated with 60 mJ/cm2 UV-B at L1 larval stage and CPD lesions were measured by antibody staining in slot blots of worm extracts directly after treatment and 24 h post-UV. The average relative CPD load were calculated comparing 24 h to 0 h and the values shown are the average of two independent experiments.
To determine if ZK742.2 animals are defective in repairing UV-induced CPDs, DNA from the animals was isolated immediately (0 h) or 24 h after UV-treatment and repair of UV-induced CPDs was assessed with slot-blot assay using a •-CPD antibody (Figure 2B). 24 h post-UV, CPD levels were reduced in wildtype animals demonstrating the effect of functional NER. In contrast, ZK742.2 and the TC-NER deficient csb-1 mutant animals retained higher levels of CPDs indicative of compromised NER. These results demonstrate that ZK742.2 is required for the repair of UV-induced CPDs.
The UV-hypersensitivity as well as reduced CPD repair capacity of the ZK742.2 mutant animals during development supports the bioinformatics identification of ZK742.2 as a UVSSA homolog and its role in UV-induced damage repair.
3.2. ZK742.2 is dispensable for GG-NER in the presence of a functional TC-NER
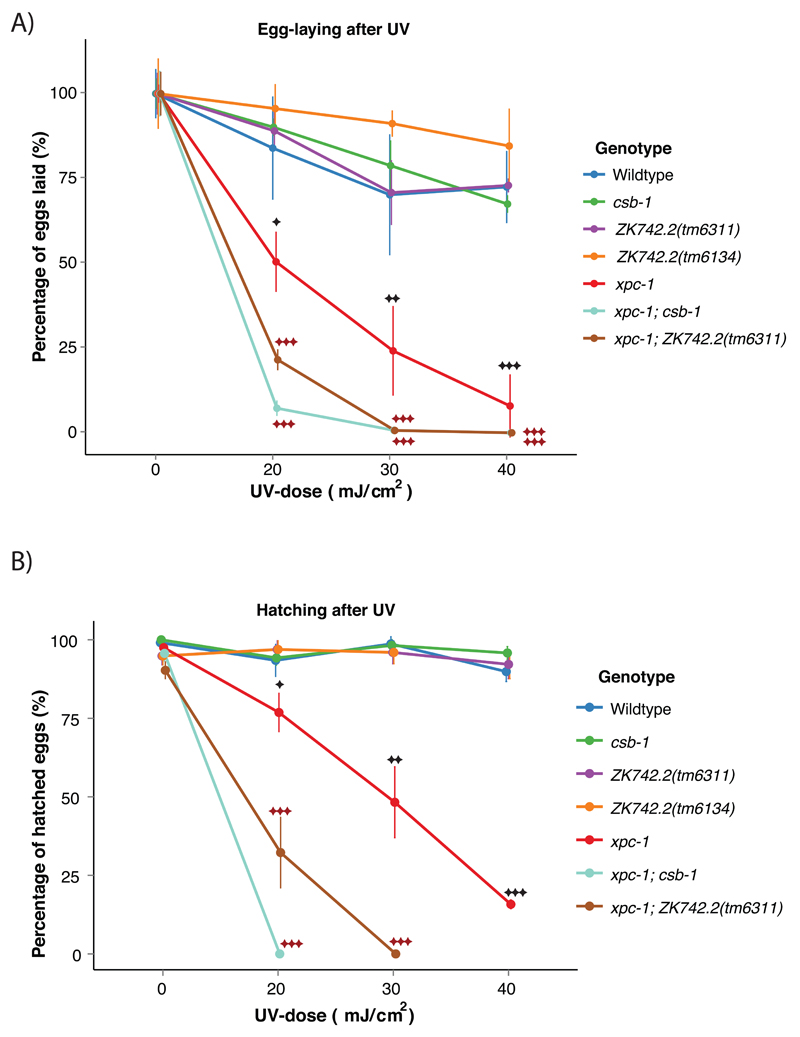
UVSS cells display normal UV-induced DNA repair synthesis (UDS) which is a measure of GG-NER efficiency [15]. In order to further characterize the role of ZK742.2 in UV-response in C. elegans, we examined the UV-sensitivity of the germ cells. The germ cells in C. elegans mainly depend on GG-NER for repairing UV-induced DNA damage and GG-NER-deficient xpc-1 mutant animals exhibit reduced fertility upon UV-treatment, while TC-NER deficient csa-1 and csb-1 mutant worms do not display UV-induced germline defects [21]. The germ cells of strains carrying mutations in ZK742.2 did not show any UV-hypersensitivity compared to wildtype animals as determined by subsequent egg-laying activity (Figure 3A) and hatching efficiency (Figure 3B) after UV-treatment. These results suggest that ZK742.2 is dispensable for GG-NER.
Figure 3. ZK742.2 is dispensable for GG-NER in the presence of a functional TC-NER.
(A) Egg-laying activity of the indicated mutants 16 h after UV-B-treatment at young adult stage. Timed egg-laying was conducted with three animals per UV-dose for 3 h. Each treatment was conducted in triplicate. (B) Percentage of hatched eggs after UV-treatment. Error bars represent SD, statistical analysis is a two-tailed t-test comparing treatment groups to wildtype (black asterisk) or xpc-1 (red asterisk), *=p<0.05, **=p<0.01, ***=p<0.001.
Although germ cells predominantly rely on GG-NER for the repair, TC-NER has a residual role that becomes evident when GG-NER is compromised. Combined loss of xpc-1 and csb-1 lead to an enhanced UV-hypersensitivity compared to the single mutant animals. The same situation is mirrored in the xpc-1; ZK742.2 animals and these results place ZK742.2 in the TC-NER pathway along with csb-1 (Figure 3A, B).
3.3. ZK742.2 is genetically epistatic with csb-1 and specific to transcription-coupled NER
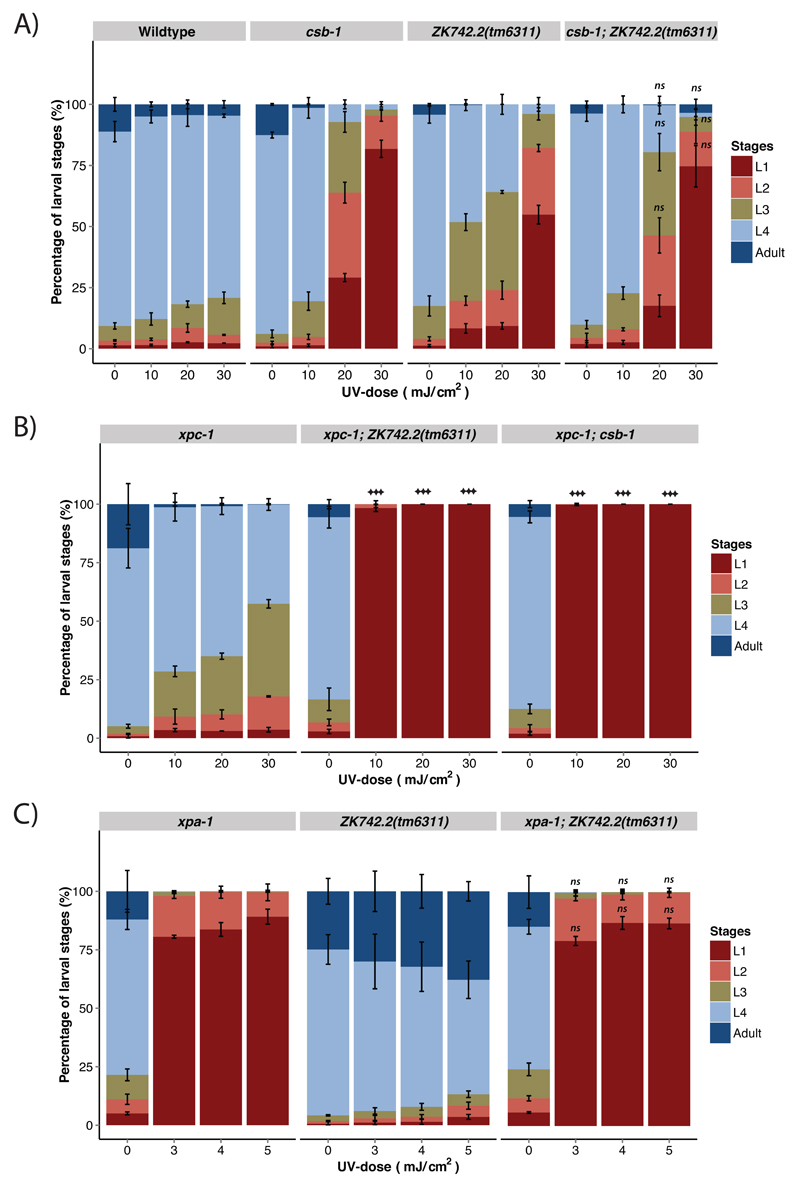
This study further aimed to integrate ZK742.2 into the genetic pathway of NER. To this end, we tested the genetic interaction of ZK742.2 with TC-NER defective csb-1 alleles, GG-NER deficient xpc-1 alleles and total NER deficient xpa-1 alleles (Figure 4). Upon stalling of RNAP II at a transcription-blocking lesion, CSB initiates TC-NER by recruiting CSA, followed by core NER factors including XPA to the site of damage [27]. Upon subjecting synchronized L1 larvae to UV-B, csb-1; ZK742.2 double mutant animals displayed similar UV-sensitivity as the corresponding single mutant animals (Figure 4A). This indicates that CSB-1 and ZK742.2 function epistatically to promote UV-resistance. Mutation of xpa-1 causes a higher UV-sensitivity compared to csb-1, as NER is completely abrogated in this mutant. UV-sensitivity of xpa-1; ZK742.2 was similar to xpa-1 (Figure 4C). Combined loss of xpa-1 and ZK742.2 did not cause UV-sensitivity greater than that of xpa-1 or ZK742.2 alone, suggesting that ZK742.2 acts in the same pathway as csb-1 and upstream of xpa-1. On the contrary, the UV-sensitivity of xpc-1; ZK742.2 double mutant animals was enhanced compared to both single mutant animals, reminiscent of the UV-sensitivity of xpc-1; csb-1 double mutants (Figure 4B). This relationship suggests that in xpc-1; ZK742.2 double mutants, both of the NER subpathways are compromised, as it is in xpc-1; csb-1 double mutant animals. Hence, ZK742.2 is TC-NER specific and, along with csb-1, acts in parallel to xpc-1 to repair UV-B-induced DNA lesions in C. elegans.
Figure 4. ZK742.2 is epistatic with csb-1 and TC-NER specific.
Percentage of larval stages 48 h after UV-B-treatment at the L1 larval stage of the indicated genotypes. A minimum of 200 animals was used for each UV-B-dose, and each treatment was conducted in triplicate. Error bars represent SD, statistical analysis is a two-tailed t-test comparing treatment groups to csb-1 (A), or xpc-1 (B) or xpa-1 (C), *=p<0.05, **=p<0.01, ***=p<0.001, ns = not significant.
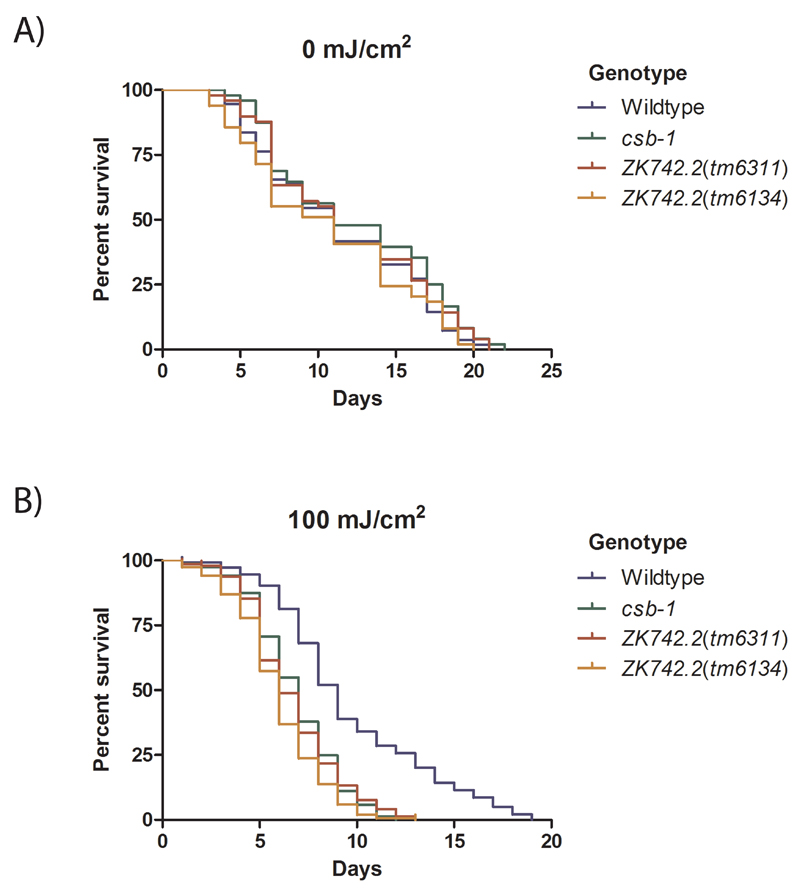
3.4. ZK742.2 animals display reduced lifespan after UV-exposure
TC-NER activity is required for resisting UV-induced DNA lesions during adulthood [20,26,28,29]. We further investigated whether ZK742.2 is involved in the UV-survival of adult worms, which consists almost entirely of post-mitotic cells, except for the germline that undergo mitotic and meiotic cell divisions. Animals were UV-treated on day-1 adulthood and the survival was monitored. csb-1 and ZK742.2 mutant animals have a lifespan similar to the wildtype animals (Figure 5A) but displayed reduced survival upon UV-treatment, confirming a requirement of TC-NER factors for withstanding UV damage during development as well as adulthood (Figure 5B).
Figure 5. ZK742.2 animals display reduced survival upon UV exposure.
The percentage survival after UV-B-treatment administered at the day-1 adult stage. A minimum of 140 animals were tested for each dose. Comparison of the survival curves using the Log-rank (Matel-Cox) test showed that there is no significant difference among the different genotypes in the untreated condition (A). After UV-treatment, the median life span of csb-1 and ZK742.2 animals are significantly reduced (p<0.001) compared to the wildtype (B).
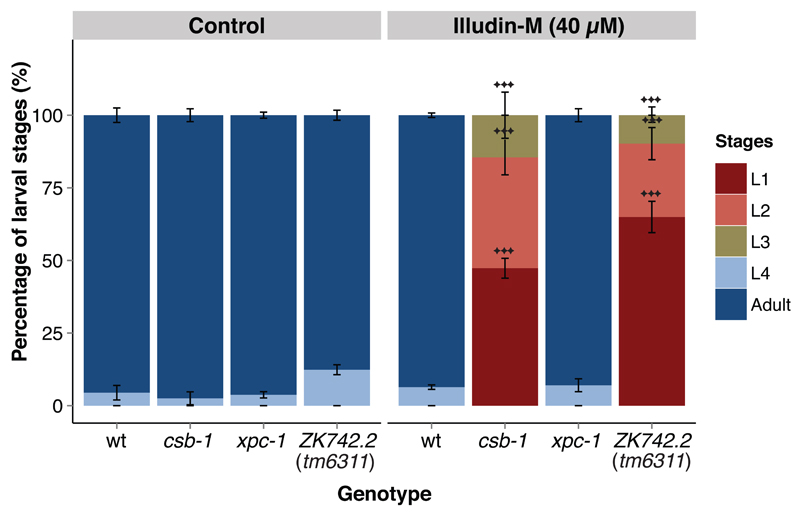
3.5. ZK742.2 animals are hypersensitive to illudin-M
Previous studies have demonstrated that cells derived from UVSS patients are sensitive to illudin-S, similar to cells from CS patients, as the drug sensitizes TC-NER deficient cells [15,30]. We have demonstrated that in C. elegans TC-NER deficient animals are hypersensitive to the closely related illudin-M and undergo a developmental arrest in a dose-dependent manner [21]. Treating ZK742.2 mutant animals with 40 μM illudin-M at the L1 larval stage induced a developmental arrest similar to the UV-induced arrest, indicating a deficiency in repairing illudin-M-induced DNA lesions (Figure 6). Since illudin-induced lesions are specifically repaired by TC-NER, these results further support a TC-NER-specific role for ZK742.2.
Figure 6. ZK742.2 animals are illudin-M hypersensitive.
The percentage of larval stages 72 h after Illudin-M-treatment administered at the L1 larval stage. A minimum of 50 animals were used for each dose, and each treatment was conducted in triplicate. Error bars represent SD, statistical analysis is a two-tailed t-test comparing treatment groups to wildtype, *=p<0.05, **=p<0.01, ***=p<0.001.
3.6. ZK742.2 is dispensable for resistance to oxidative stress
The involvement of TC-NER factors in the repair of oxidative damage has remained subject to debate, as evidence exist for and against such a role [31,32]. To test whether ZK742.2 mutant animals are sensitive to oxidative damage, sensitivity to paraquat was assessed (Supplementary Figure 1). Paraquat is enzymatically reduced to radicals, which then react with oxygen to produce superoxide anions. This reaction gives rise to hydrogen peroxide and thereby induces oxidative stress [33]. Although the survival of wildtype as well as TC-NER mutant animals was reduced on 5 mM paraquat, no hypersensitivity was exhibited by ZK742.2 or csb-1 animals (Supplementary Figure 1). We also tested the effect of another oxidizing agent KBrO3 during development. ZK742.2 mutant animals displayed a similar sensitivity to wildtype and csb-1 mutant animals with the exception of a very mild hypersensitive at a low dose (Supplementary Figure 2). These results suggest that neither csb-1 nor ZK742.2 are required for withstanding oxidative damage.
Taken together, we demonstrate that ZK742.2 is a TC-NER factor in C. elegans and we propose to name it uvs-1.
4. Discussion
Many aspects of NER is not completely elucidated in the mammalian system and C. elegans is a well suited in vivo model for the further characterization and understanding of this repair pathway [16]. In the absence of any helix-distorting DNA lesions, the C. elegans strains with a defective NER are phenotypically wildtype-like [20,26,28,29]. UV-irradiation affects C. elegans germline and leads to embryonic lethality, especially if NER is compromised, as GG-NER is the predominant repair pathway in the C. elegans germline [19,20]. TC-NER plays a partially redundant role in the repair of UV-induced damage in the germline and comes to play only in the absence of a functional GG-NER [20]. UV-irradiation leads to the developmental arrest of somatic cells and contrary to situation in the germline and TC-NER is the predominant repair pathway in the soma [21,26]. Defects in the common NER pathway sensitizes the somatic cells as well as germline to UV-irradiation and induces a UV-hypersensitivity higher than that of GG-NER and TC-NER defective animals.
In this study we have identified the C. elegans ORF ZK742.2 as the homolog for UVSSA and propose the name uvs-1. Mutant animals are hypersensitive to UV-B and illudin in a manner similar to the TC-NER-defective nematode strains. Similar to TC-NER defective animals uvs-1 mutant animals do not exhibit UV-hypersensitivity in the germ cells, unlike GG-NER defective xpc-1 animals. However, combined loss of uvs-1 and xpc-1 enhances the UV-sensitivity of both the single mutants, similar to total NER-deficient csb-1; xpc-1 animals. Moreover, we demonstrate that the TC-NER factors csb-1 and uvs-1 are required for the survival of adult animals after UV-treatment.
One possible hypothesis to explain the differences in the disease phenotypes between CS and UVSS patients is the involvement of CSA and CSB in processes other than their regular role in TC-NER, while UVSSA is not involved in these processes. CS proteins are reported to be involved in the repair of oxidative DNA damage, which is normally repaired by base excision repair [34,35]. Although, CS cells are hypersensitive to oxidative stress, unlike UVSS cells [36], both csb-1 and uvs-1 animals displayed largely similar sensitivity towards oxidative stress inducing agents paraquat and KBrO3. This suggests that TC-NER factors are not essential for the repair of oxidative lesions in C. elegans. Further investigation using this newly identified C. elegans mutant strain will shed light on to the differential roles of the TC-NER components and phenotypic differences between the TC-NER deficiency syndromes.
5. Conclusions
Genetic studies have demonstrated that mammalian NER and NER in C. elegans work in similar fashion, making the nematode an effective model to study the repair pathway. The distinct outcomes of GG-NER and TC-NER defects in C. elegans have already provided new insights into how metazoans respond to unrepaired DNA damage in distinct tissues during development and with aging. This study provides the basis for better understanding the disease-causing gene for UV-hypersensitivity syndrome UVSSA in a simple metazoan model.
Supplementary Material
Supplementary Figure 1. ZK742.2 animals are not hypersensitive to paraquat. The percentage survival after 5 mM paraquat administered at the day-1 adult stage. A minimum of 80 animals were tested for each group. Comparison of the survival curves using the Log-rank (Matel-Cox) test showed that there is no significant difference between the treatment groups.
Supplementary Figure 2. ZK742.2 animals are mildly hypersensitive to KBrO3. The percentage of larval stages 48 h after KBrO3-treatment administered at the L1 larval stage. A minimum of 500 animals were used for each dose, and each treatment was conducted in triplicate. Larval stages were determined by flow cytometry. Error bars represent SD, statistical analysis is a two-tailed t-test comparing treatment groups to wildtype, *=p<0.05, **=p<0.01, ***=p<0.001.
Highlights.
UV-stimulated scaffold protein A (UVSSA) gene, a causal gene of UV-hypersensitivity syndrome, has a C. elegans homolog – uvs-1.
uvs-1 mutant animals are defective in repairing CPD lesions and are hypersensitive to UV during development.
UV-B irradiation reduces the life span of uvs-1 animals.
uvs-1 and csb-1 function epistatically to promote UV-resistance.
uvs-1 mutant animals are hypersensitive to the transcription-blocking DNA lesions induced by illudin-M.
Acknowledgments
We thank Rainer Schobert for providing illudin-M. Worm strains were provided by the National Bioresource Project and the Caenorhabditis Genetics Center (funded by the NIH National Center for Research Resources). VB received an IGSDHD fellowship. BS acknowledges funding from the Deutsche Forschungsgemeinschaft (CECAD, SFB 829, SFB 670, and KFO 286), the European Research Council (ERC Starting grant 260383), Marie Curie (FP7 ITN CodeAge 316354, aDDRess 316390, MARRIAGE 316964, and ERG 239330), the German-Israeli Foundation (GIF, 2213-1935.13/2008 and 1104-68.11/2010), the Deutsche Krebshilfe (109453), and the BMBF (SyBaCol)
Abbreviations
- bp
Base pairs
- 6-4 PP
6-4 photoproduct
- cm
Centimeter
- CPD
Cyclobutane pyrimidine dimer
- CS
Cockayne syndrome
- GG-NER
Global-genome nucleotide excision repair
- mJ
Milli-Joule
- NER
Nucleotide excision repair
- NGM
Nematode growth medium
- nt
Nucleotide
- ORF
Open reading frame
- RNAP II
RNA polymerase II
- RRS
Recovery of RNA synthesis
- TC-NER
Transcription-coupled nucleotide excision repair
- UDS
UV-induced DNA repair synthesis
- UTR
Un-translated region
- UV
Ultraviolet
- UVSS
UV-hypersensitivity syndrome
- XP
Xeroderma pigmentosum
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interests.
References
- [1].Setlow RB. Cyclobutane-type pyrimidine dimers in polynucleotides. Science. 1966;153:379–386. doi: 10.1126/science.153.3734.379. [DOI] [PubMed] [Google Scholar]
- [2].Matsunaga T, Hieda K, Nikaido O. Wavelength dependent formation of thymine dimers and (6-4) photoproducts in DNA by monochromatic ultraviolet light ranging from 150 to 365 nm. Photochem Photobiol. 1991;54:403–410. doi: 10.1111/j.1751-1097.1991.tb02034.x. [DOI] [PubMed] [Google Scholar]
- [3].Douki T, Cadet J. Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry. 2001;40:2495–2501. doi: 10.1021/bi0022543. [DOI] [PubMed] [Google Scholar]
- [4].Fraval HN, Rawlings CJ, Roberts JJ. Increased sensitivity of UV-repair-deficient human cells to DNA bound platinum products which unlike thymine dimers are not recognized by an endonuclease extracted from Micrococcus luteus. Mutat Res. 1978;51:121–132. doi: 10.1016/0027-5107(78)90014-3. [DOI] [PubMed] [Google Scholar]
- [5].Chu G, Berg P. DNA cross-linked by cisplatin: a new probe for the DNA repair defect in xeroderma pigmentosum. Mol Biol Med. 1987;4:277–290. [PubMed] [Google Scholar]
- [6].Gillet LCJ, Schärer OD. Molecular Mechanisms of Mammalian Global Genome Nucleotide Excision Repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- [7].Mu D, Bertrand-Burggraf E, Huang JC, Fuchs RP, Sancar A, Fuchs BP. Human and E.coli excinucleases are affected differently by the sequence context of acetylaminofluorene-guanine adduct. Nucleic Acids Res. 1994;22:4869–4871. doi: 10.1093/nar/22.23.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H. Bipartite substrate discrimination by human nucleotide excision repair. Proc Natl Acad Sci USa. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wolters S, Schumacher B. Genome maintenance and transcription integrity in aging and disease. Front Genet. 2013;4:19. doi: 10.3389/fgene.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Itoh T, Ono T, Yamaizumi M. A new UV-sensitive syndrome not belonging to any complementation groups of xeroderma pigmentosum or Cockayne syndrome: siblings showing biochemical characteristics of Cockayne syndrome without typical clinical manifestations. Mutat Res. 1994;314:233–248. doi: 10.1016/0921-8777(94)90068-x. [DOI] [PubMed] [Google Scholar]
- [11].Nardo T, Oneda R, Spivak G, Vaz B, Mortier L, Thomas P, et al. A UV-sensitive syndrome patient with a specific CSA mutation reveals separable roles for CSA in response to UV and oxidative DNA damage. Proc Natl Acad Sci USa. 2009;106:6209–6214. doi: 10.1073/pnas.0902113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Horibata K, Iwamoto Y, Kuraoka I, Jaspers NGJ, Kurimasa A, Oshimura M, et al. Complete absence of Cockayne syndrome group B gene product gives rise to UV-sensitive syndrome but not Cockayne syndrome. Proc Natl Acad Sci USa. 2004;101:15410–15415. doi: 10.1073/pnas.0404587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nakazawa Y, Sasaki K, Mitsutake N, Matsuse M, Shimada M, Nardo T, et al. Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat Genet. 2012;44:586–592. doi: 10.1038/ng.2229. [DOI] [PubMed] [Google Scholar]
- [14].Zhang X, Horibata K, Saijo M, Ishigami C, Ukai A, Kanno S-I, et al. Mutations in UVSSA cause UV-sensitive syndrome and destabilize ERCC6 in transcription-coupled DNA repair. Nat Genet. 2012;44:593–597. doi: 10.1038/ng.2228. [DOI] [PubMed] [Google Scholar]
- [15].Schwertman P, Lagarou A, Dekkers DHW, Raams A, van der Hoek AC, Laffeber C, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat Genet. 2012;44:598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- [16].Edifizi D, Schumacher B. Genome Instability in Development and Aging: Insights from Nucleotide Excision Repair in Humans, Mice, and Worms. Biomolecules. 2015;5:1855–1869. doi: 10.3390/biom5031855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lans H, Vermeulen W. Nucleotide Excision Repair in Caenorhabditis elegans. Mol Biol Int. 2011;2011:542795. doi: 10.4061/2011/542795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ribezzo F, Shiloh Y, Schumacher B. Systemic DNA damage responses in aging and diseases. Semin Cancer Biol. 2016 doi: 10.1016/j.semcancer.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stergiou L, Doukoumetzidis K, Sendoel A, Hengartner MO. The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ. 2007;14:1129–1138. doi: 10.1038/sj.cdd.4402115. [DOI] [PubMed] [Google Scholar]
- [20].Lans H, Marteijn JA, Schumacher B, Hoeijmakers JHJ, Jansen G, Vermeulen W. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. 2010;6:e1000941. doi: 10.1371/journal.pgen.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Babu V, Hofmann K, Schumacher B. A C. elegans homolog of the Cockayne syndrome complementation group A gene. DNA Repair (Amst.) 2014;24:57–62. doi: 10.1016/j.dnarep.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS ONE. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li H, Coghlan A, Ruan J, Coin LJ, Hériché J-K, Osmotherly L, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–80. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mitani S. Nematode, an experimental animal in the national BioResource project. Exp Anim. 2009;58:351–356. doi: 10.1538/expanim.58.351. [DOI] [PubMed] [Google Scholar]
- [26].Mueller MM, Castells-Roca L, Babu V, Ermolaeva MA, Müller R-U, Frommolt P, et al. DAF-16/FOXO and EGL-27/GATA promote developmental growth in response to persistent somatic DNA damage. Nat Cell Biol. 2014;16:1168–1179. doi: 10.1038/ncb3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LHF. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- [28].Boyd WA, Crocker TL, Rodriguez AM, Leung MCK, Lehmann DW, Freedman JH, et al. Nucleotide excision repair genes are expressed at low levels and are not detectably inducible in Caenorhabditis elegans somatic tissues, but their function is required for normal adult life after UVC exposure. Mutat Res. 2010;683:57–67. doi: 10.1016/j.mrfmmm.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Astin JW, O’Neil NJ, Kuwabara PE. Nucleotide excision repair and the degradation of RNA pol II by the Caenorhabditis elegans XPA and Rsp5 orthologues, RAD-3 and WWP-1. DNA Repair (Amst.) 2008;7:267–280. doi: 10.1016/j.dnarep.2007.10.004. [DOI] [PubMed] [Google Scholar]
- [30].Jaspers NGJ, Raams A, Kelner MJ, Ng JMY, Yamashita YM, Takeda S, et al. Anti-tumour compounds illudin S and Irofulven induce DNA lesions ignored by global repair and exclusively processed by transcription- and replication-coupled repair pathways. DNA Repair (Amst.) 2002;1:1027–1038. doi: 10.1016/s1568-7864(02)00166-0. [DOI] [PubMed] [Google Scholar]
- [31].Boiteux S, le Page F. Repair of 8-oxoguanine and Ogg1-incised apurinic sites in a CHO cell line. Prog Nucleic Acid Res Mol Biol. 2001;68:95–105. doi: 10.1016/s0079-6603(01)68092-9. [DOI] [PubMed] [Google Scholar]
- [32].Thorslund T, Sunesen M, Bohr VA, Stevnsner T. Repair of 8-oxoG is slower in endogenous nuclear genes than in mitochondrial DNA and is without strand bias. DNA Repair (Amst.) 2002;1:261–273. doi: 10.1016/s1568-7864(02)00003-4. [DOI] [PubMed] [Google Scholar]
- [33].Ali S, Jain SK, Abdulla M, Athar M. Paraquat induced DNA damage by reactive oxygen species. Biochem Mol Biol Int. 1996;39:63–67. doi: 10.1080/15216549600201061. [DOI] [PubMed] [Google Scholar]
- [34].Dianov G, Bischoff C, Sunesen M, Bohr VA. Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res. 1999;27:1365–1368. doi: 10.1093/nar/27.5.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. Faseb J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- [36].Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst.) 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.