Abstract
Circadian clocks are fundamental to the biology of most eukaryotes, coordinating behavior and physiology to resonate with the environmental cycle of day and night through complex networks of clock-controlled genes1–3. A fundamental knowledge gap exists however, between circadian gene expression cycles and the biochemical mechanisms that ultimately facilitate circadian regulation of cell biology4,5. Here we report circadian rhythms in the intracellular concentration of magnesium ions, [Mg2+]i, which act as a cell-autonomous timekeeping component to determine key clock properties in both a human cell line and a unicellular alga that diverged from metazoans more than 1 billion years ago6. Given the essential role of Mg2+ as a cofactor for ATP, a functional consequence of [Mg2+]i oscillations is dynamic regulation of cellular energy expenditure over the daily cycle. Mechanistically, we find that these rhythms provide bilateral feedback linking rhythmic metabolism to clock-controlled gene expression. The global regulation of nucleotide triphosphate turnover by intracellular Mg2+ availability has potential to impact upon many of the cell’s >600 MgATP-dependent enzymes7 and every cellular system where MgNTP hydrolysis becomes rate limiting. Indeed, we find that circadian control of translation by mTOR8 is regulated through [Mg2+]i oscillations. It will now be important to identify which additional biological processes are subject to this form of regulation in tissues of multicellular organisms such as plants and humans, in the context of health and disease.
Circadian rhythms occur cell-autonomously and are not restricted to metazoans or multicellular organisms, being found throughout eukaryotes and some prokaryotes9. Although explicit clock gene identities share no similarity across phylogenetic kingdoms, in every case temporal orchestration of gene expression is driven by timekeeping mechanisms that result in rhythmic clock protein transcription factor activity. In human cells, for example, a heterodimeric complex of BMAL1 and CLOCK positively regulates the expression of genes (Period1/2, Cryptochrome1/2) that encode its own repressor complex9, whereas in the marine unicellular alga Ostreococcus tauri, a reduced version of the stereotypical plant-like circadian clock consists of a feedback loop between the morning-expressed MYB-like transcription factor CCA1 and the evening-expressed protein TOC110. Intriguingly, the role of enzymes such as casein kinase 1 and 2 in the post-translational regulation of clock protein activity/stability, and the speed at which biological clocks run, is functionally conserved across eukaryotes11. Also, we recently reported a circadian rhythm in the redox state of peroxiredoxin proteins that is conserved across phylogenetic kingdoms4,12. Critically, this metabolic rhythm persists in the absence of nascent gene expression, both in human cells (anucleate erythrocytes)13 and in Ostreococcus, which ceases mRNA production upon prolonged photosynthetic inactivity under constant darkness14, indicating that circadian regulation of cellular metabolism is not strictly reliant upon rhythmic transcription. These and a number of other observations render it plausible that circadian rhythms observed in diverse eukaryotes incorporate features of a post-translational timing mechanism that was present in the last eukaryotic common ancestor, LECA15.
A rich diversity of evolutionarily conserved membrane transporters and channels mediate uptake of ions and micronutrients essential for cellular biochemistry, with several studies having reported their circadian regulation in a variety of contexts (e.g.16), and with membrane models of the circadian clock actually predating the identification of any clock genes17,18. It is plausible that rhythmic regulation of ion transport may have conferred an adaptive advantage upon early eukaryotes, allowing the global regulation of biochemical equilibria and reaction rates to tune cellular metabolism with environmental cycles. We therefore asked whether circadian regulation of transmembrane ion transport might constitute a fundamental feature of circadian timekeeping in eukaryotic cells. To find evidence for such regulation in modern organisms, we compared two eukaryotic lineages separated by ~1.5 billion years of evolution6: human U2OS cells and Ostreococcus tauri.
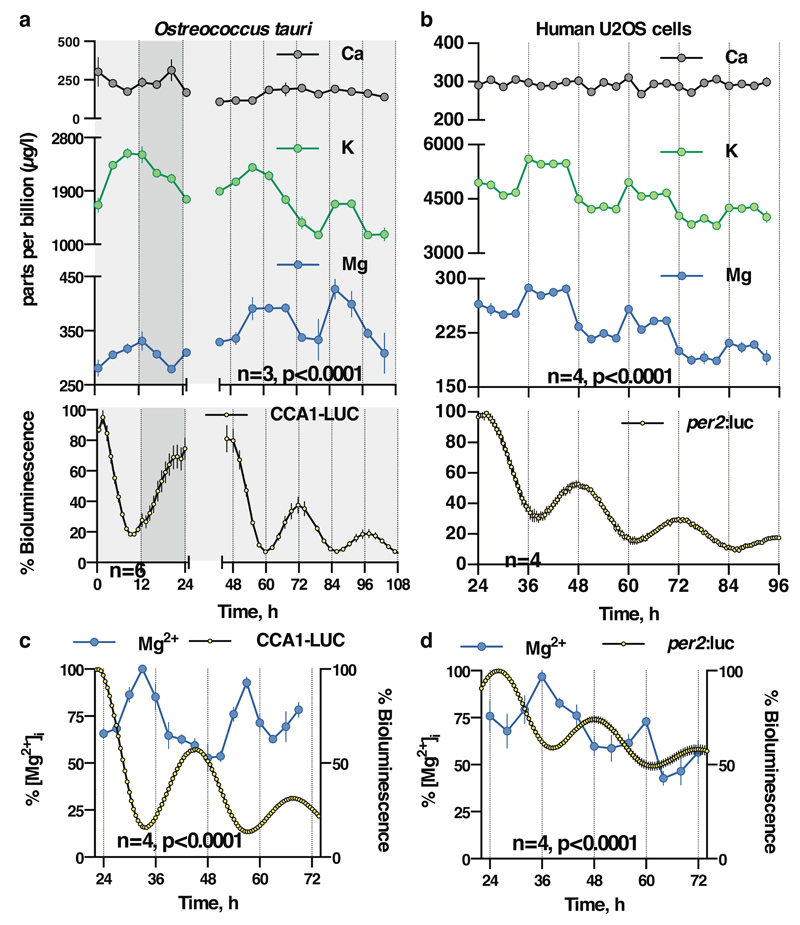
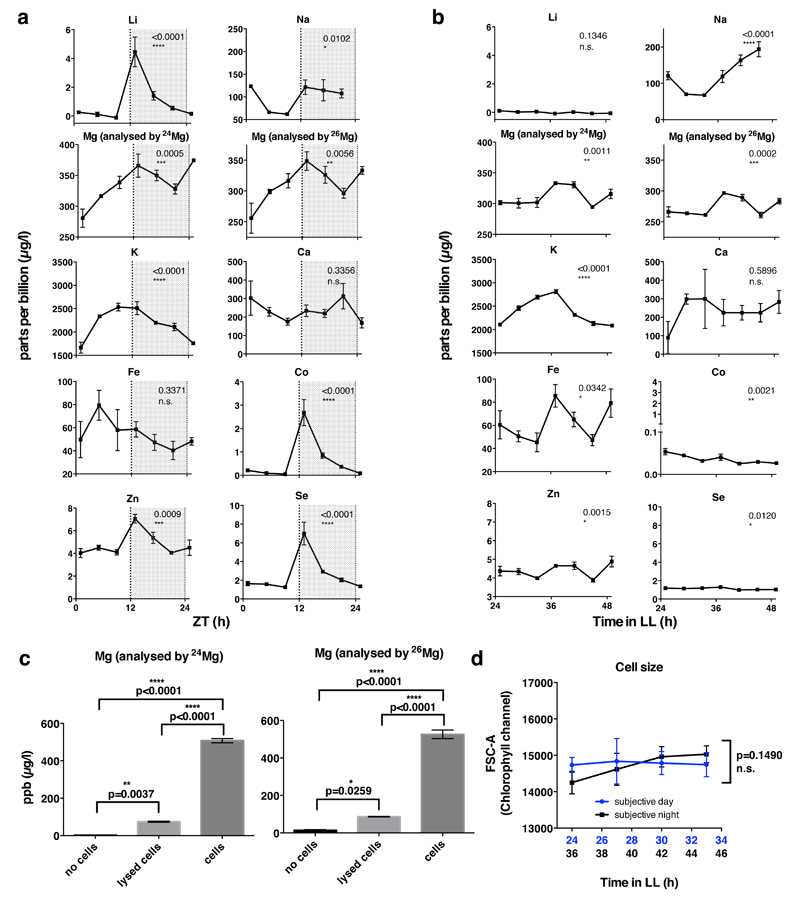
Inductively Coupled Plasma Mass Spectrometry (ICP-MS) was employed to generate an unbiased analysis of the total cellular elemental composition, including all organelles and cellular structures19,20, over circadian time series. In Ostreococcus, clear daily rhythms were detected under natural light/dark cycles for many different ions, including potassium and magnesium (Fig. 1a and Extended Data Fig. 1). Analyses of cells maintained under constant light revealed that, whilst rhythms of some metal ions ceased under these conditions, oscillations in potassium and magnesium persisted, indicating their regulation by cell-autonomous circadian clock mechanisms. Strikingly, circadian rhythms of magnesium and potassium were also observed in non-proliferating human U2OS cells maintained over three days under constant conditions (Fig. 1b and Extended Data Fig. 2). The oscillation in intracellular potassium is likely to be a consequence of rhythmic Na+-dependent pump activity, as circadian regulation of sodium-dependent solute-transport and plasma membrane ATPases has been reported widely (e.g.2,16,21,22). Calcium is largely found in, and released from, intracellular stores and although calcium has a clearly established role in circadian rhythms11, we infer the absence of any obvious cell-autonomous Ca2+ oscillations to mean that its net cellular flux does not vary over the circadian cycle in these cells. We considered the oscillation in Mg2+ to be of particular interest, since Mg2+ is an essential cofactor for (deoxy-) nucleotide triphosphates. Cellular Mg2+ therefore has the potential to regulate many intracellular metabolic reactions, through its requirement for the activities of >600 enzymes7, including those involved in ATP production, as well as DNA, RNA and protein synthesis. Furthermore, free intracellular Mg2+ can act as a second messenger. For example, epidermal growth factor stimulation induces a rapid increase in [Mg2+]i, which acts via highly Mg-sensitive mTOR to activate protein synthesis without any change in total ATP levels23.
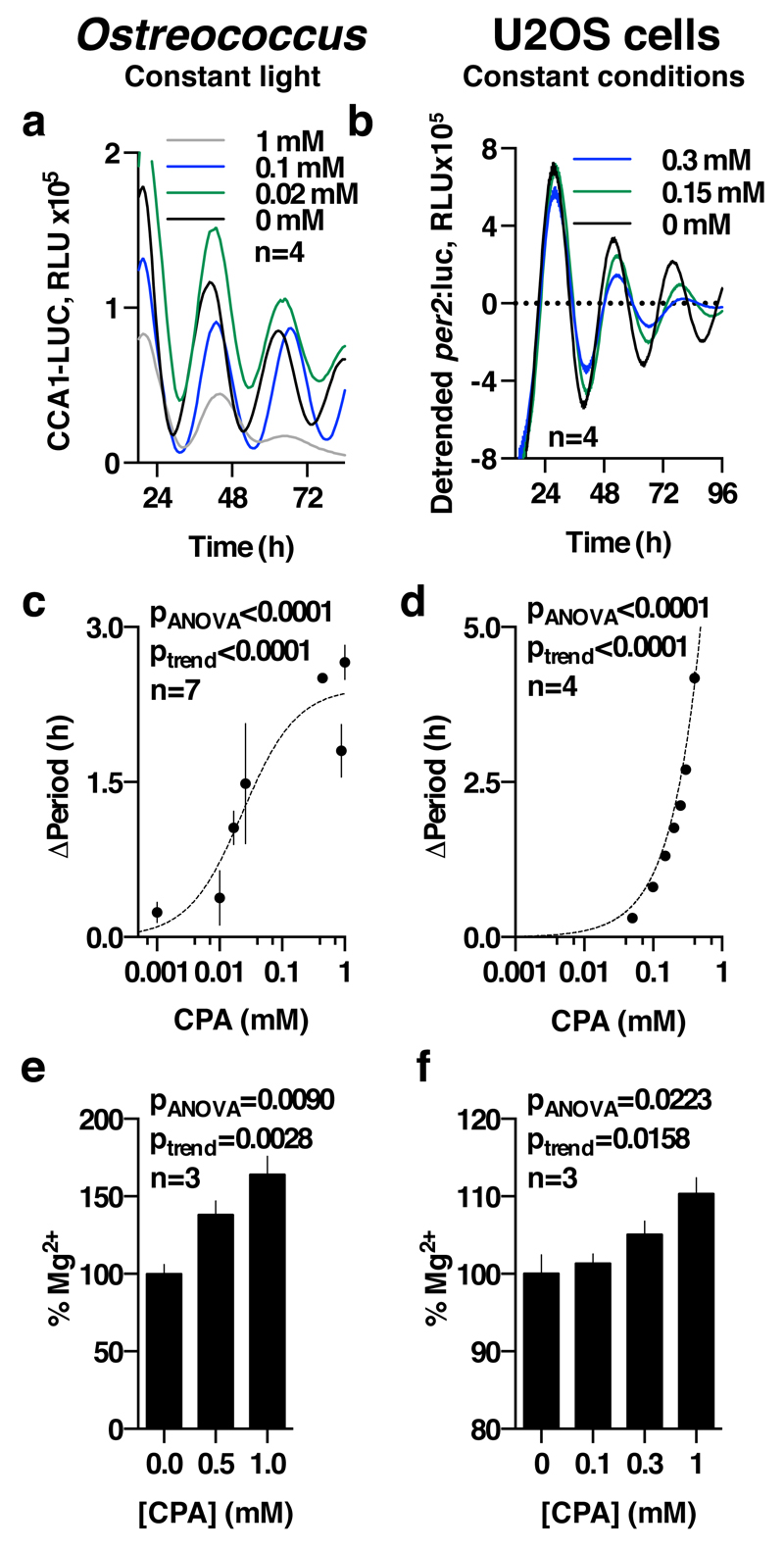
Figure 1. Conserved cellular rhythms in intracellular magnesium concentrations.
Time-series of lysates prepared from Ostreococcus (a, light/dark into constant light) or human U2OS cells (b, constant conditions) were subjected to Inductively Coupled Plasma Mass Spectrometry. Rhythms in magnesium concentration in cell lysates were confirmed with luciferase-based assays (c,d). Bioluminescence reporters for morning-phased clock gene expression were analysed in parallel (CCA1-LUC and Per2:luc) during both assays. All plots show mean±SEM, with replicate numbers (n) indicated. P-values report significance by 2-way ANOVA for time vs. interaction for each element (a,b) or 1-way ANOVA (c,d).
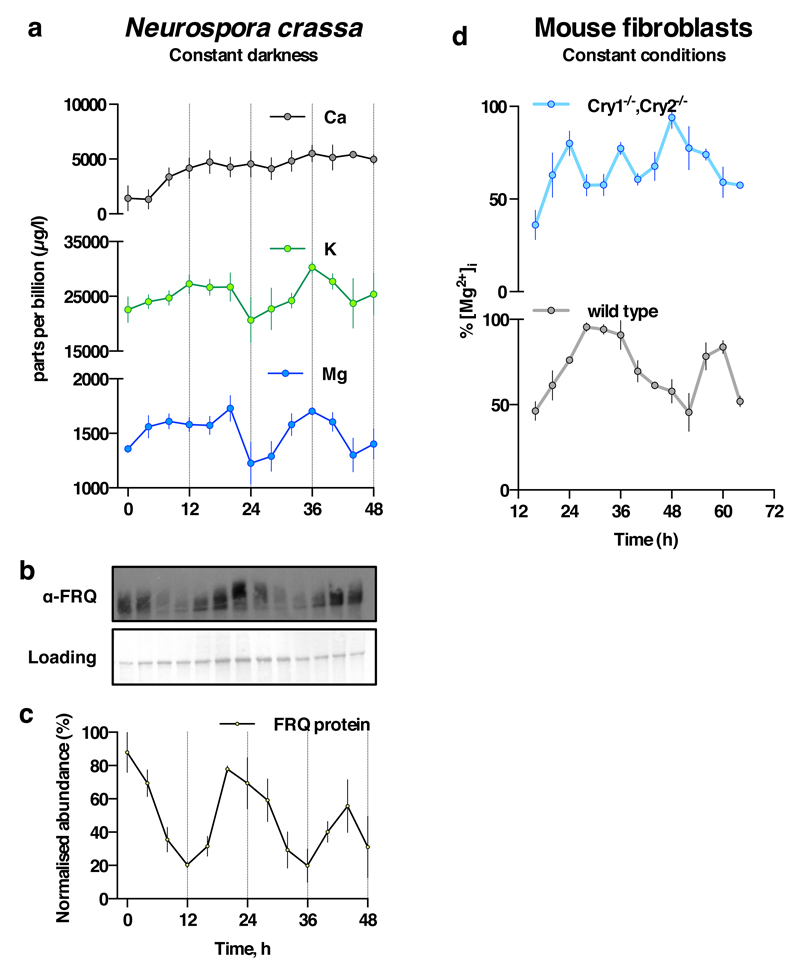
Firstly, we exploited the role of Mg2+ as an ATP cofactor, to measure freely available intracellular magnesium concentrations in cellular extracts using the MgATP-dependent enzyme firefly luciferase (Fig. 1c,d). Our initial ICP-MS observations were confirmed using this assay, and we observed clear rhythms of [Mg2+]i over two days under constant conditions in both cell types. Bioluminescent reporters for clock gene activity, recorded in parallel, confirmed the [Mg2+]i oscillation to occur roughly in antiphase with circadian markers normally expressed around (subjective) dawn (CCA1-LUC, Per2:luc, Fig. 1a,b). Conservation between human and algal cells indicates that rhythmic cation transport might constitute a general feature of cellular rhythmicity. We therefore investigated whether such oscillations were also present in the fungus Neurospora crassa, representing the third eukaryotic kingdom. Similar to our observations in algal and human cells, a circadian rhythm in cellular magnesium content was observed in antiphase with the abundance of the clock protein FRQ9 (Extended Data Fig. 3a-c). We also observed cellular magnesium rhythms in cultured mouse fibroblasts isolated from adult lung, indicating that magnesium rhythms are also present in non-transformed, terminally differentiated mammalian cells (Extended Data Fig. 3d). These rhythms were disrupted in fibroblasts isolated from Cryptochrome1-/-,Cryptochrome2-/- mice, suggesting their dependence upon clock gene activity. Incidentally, we note that approximately 24 h [Mg2+]i rhythms occur cell-autonomously, are temperature-compensated (Extended Data Fig. 4) and entrain to relevant external cues and are therefore circadian by definition24.
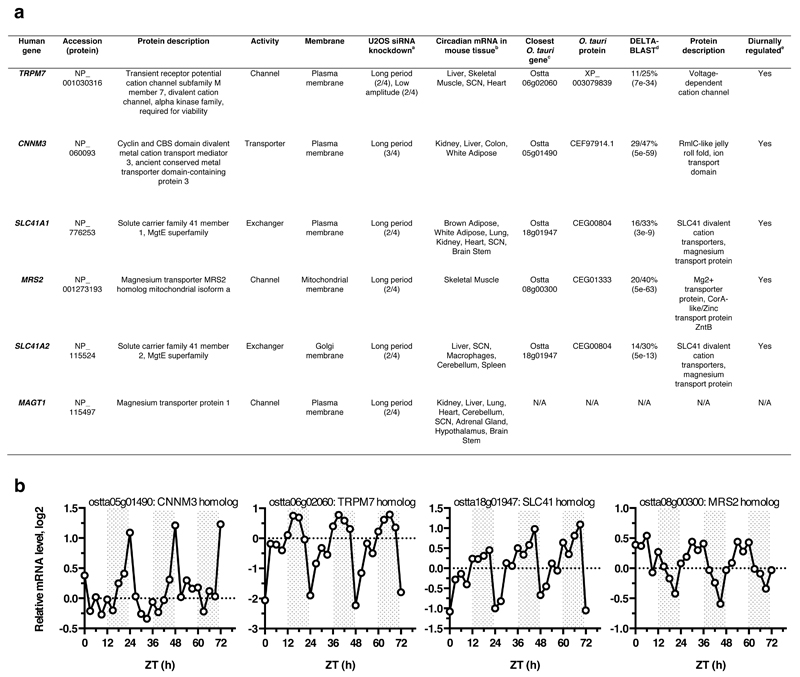
The rhythm in total cellular Mg2+ measured by ICP-MS must result from daily cycles between net cellular Mg2+ influx and efflux, through circadian regulation of plasma membrane Mg2+ channel and transporter activity7. All known magnesium transporting proteins in animals (channels TRPM7, MAGT1, MMGT1 and CNMM3, as well as Mg2+-transporter SLC41) exhibit circadian rhythms at the mRNA level in four or more tissues25 (Extended Data Fig. 5). Ostreococcus encodes homologs of TRPM7, CNMM3 and SLC41, which are also differentially expressed over the daily cycle (Extended Data Fig. 5). Moreover, siRNA-mediated knockdown of each Mg2+-channel/transporter in U2OS cells results in lengthened circadian period26, suggesting that as well as being clock-regulated, [Mg2+]i might also feed back to regulate the cellular clock.
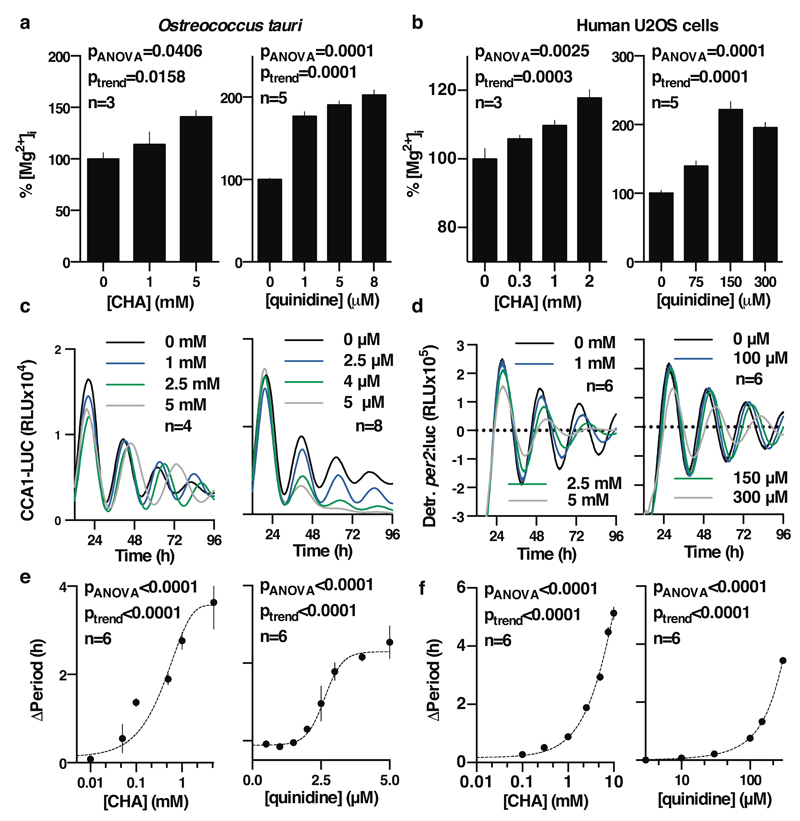
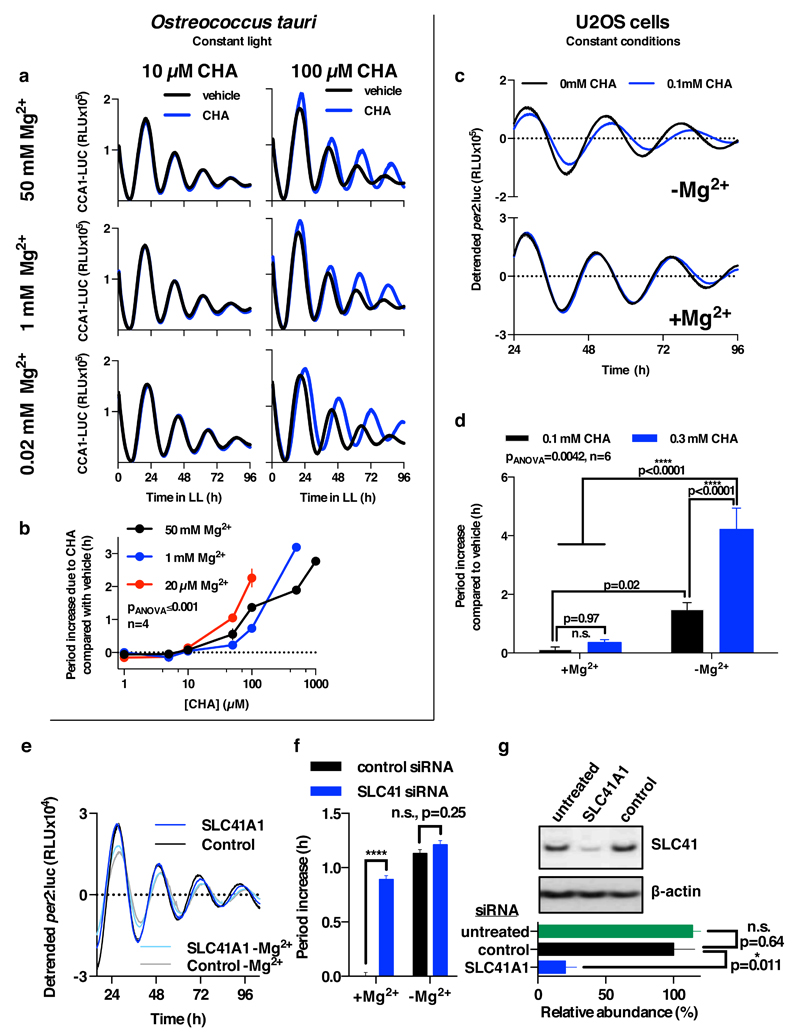
To determine whether [Mg2+]i oscillations are relevant to timekeeping mechanism therefore, we next employed inhibitors of magnesium transport. Cobalt(III)hexammine (Co(NH3)62+, CHA) and cobalt(III)chloro-pentammine (Co(NH3)5Cl2+, CPA) closely resemble a single-solvation shell hydrated Mg2+ ion, and have been shown to block Mg2+ transport through at least two different transporters/channels27,28. We found both compounds to dose-dependently increase [Mg2+]i in both cell types (Fig. 2a,b and Extended Data Fig. 6), indicating that these compounds do act to block Mg2+ transport. Increased [Mg2+]i was associated with clear dose-dependent lengthening of circadian period (Fig. 2c-f). Importantly, the effects of CHA on circadian period were dependent on the concentration of extracellular magnesium (Extended Data Fig. 7a-d), indicating a specific role for magnesium in determining the speed at which both algal and human cellular clocks run. To further substantiate this observation, we used quinidine, an inhibitor that acts on several ion transport activities including the SLC41 Na+/Mg2+ antiporter29. Similarly to CHA and CPA, quinidine led to dose-dependent accumulation of intracellular Mg2+ and lengthening of circadian period in both cell types (Fig. 2). SLC41 constitutes the sole protein known to exhibit sodium-dependent Mg2+-transport activity29 that is conserved between human and Ostreococcus cells and so, to test its specific cellular clock function, we performed siRNA-mediated SLC41 knock-down: observing a clear Mg2+-dependent lengthening of circadian period (Extended Data Fig. 7e-g).
Figure 2. Chronic inhibition of magnesium transport leads to increased [Mg2+]i and long circadian period.
Chronic inhibition of magnesium transport by CHA or quinidine increases [Mg2+]i (a,b) and increases circadian period (c-f), traces and period dose-response of the CCA1-LUC (Ostreococcus) or Per2:luc (U2OS cells) reporters are shown. All plots are mean±SEM, with replicate numbers (n) indicated, p-values report significance by 1-way ANOVA and post-test for linear trend.
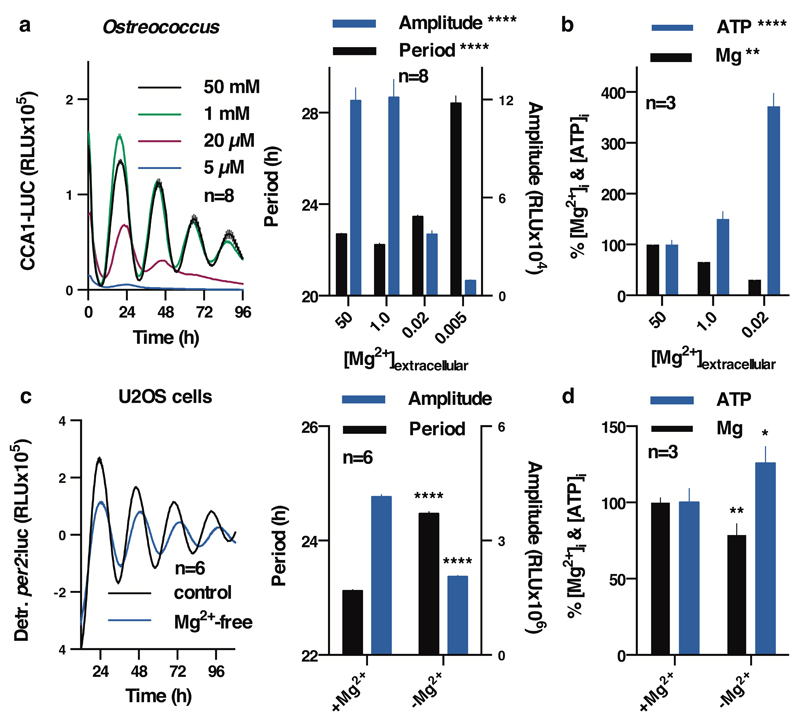
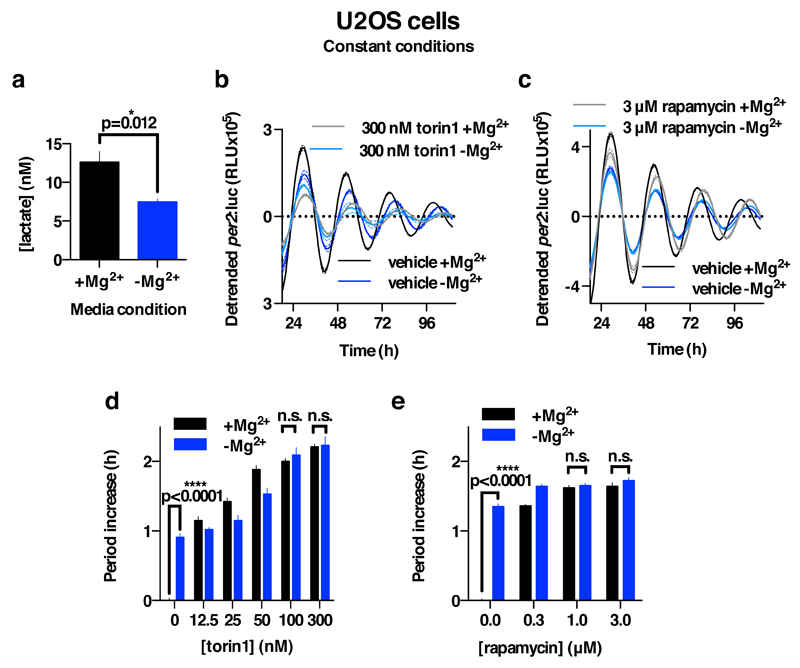
We also observed that depletion of magnesium from the growth medium led to reduced [Mg2+]i, and had dramatic effects on the amplitude and period of the circadian clock in Ostreococcus (Fig. 3a,b). Although prolonged growth in low Mg2+ media had adverse effects on cell viability of the U2OS line, cells that were simply transferred to Mg2+-free media showed reduced [Mg2+]i and exhibited circadian rhythms with increased period and decreased bioluminescence amplitude relative to normal media controls (Fig. 3c,d). In neither case was the effect of [Mg2+]i-depletion attributable to decreased ATP availability, since in both cases cellular ATP levels were significantly increased (Fig. 3b,d).
Figure 3. Reduced [Mg2+]i affects properties of cellular timekeeping and leads to an increase in ATP.
Bioluminescence traces showing reduced extracellular magnesium significantly affects amplitude and period length of circadian reporters in algal (a) and human cells (c). Low extracellular magnesium leads to decreased [Mg2+]i and increased [ATP]i in both cell types (b,d), measured after 4 days. All plots show mean±SEM, with replicate numbers (n) indicated, p-values (****p<0.0001, **p=0.01, *p=0.04) report significance by 1-way ANOVA (Ostreococcus) or t-test (U2OS).
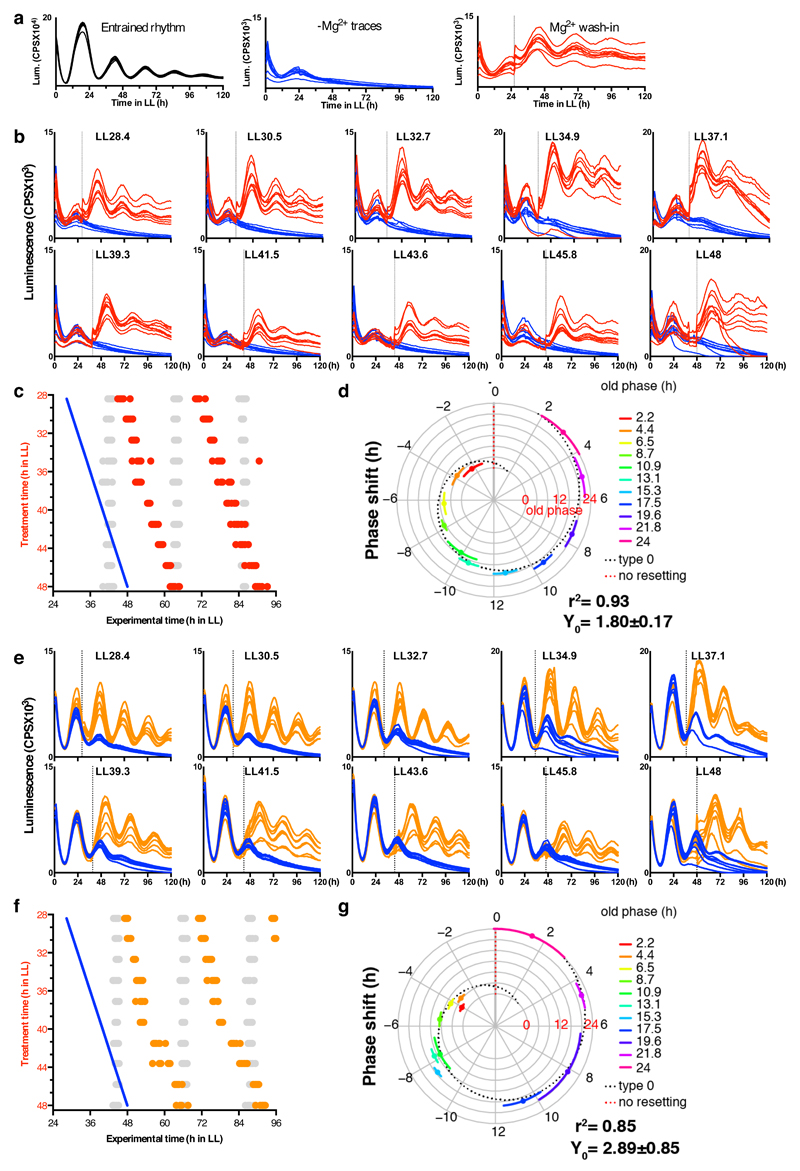
Thus, as observed previously for cAMP signalling30, treatments which constitutively elevate or reduce [Mg2+]i both result in period lengthening of clock gene expression in these cell types, indicating that dynamic circadian regulation of [Mg2+]i might be a cellular clock component. On the other hand however, it remained possible that Mg2+ transport might not contribute to clock mechanism, but instead simply be permissive for cellular timekeeping, analogous to the function of ‘housekeeping’ genes. To distinguish between these two possibilities, we determined whether an enforced transition in [Mg2+]i acts as a state variable for cellular circadian oscillations. Upon introduction of magnesium to Ostreococcus cells starved of magnesium, we observed strict resetting of the subsequent rhythm to subjective dawn, regardless of prior circadian phase, indicating that changes in [Mg2+]i can act as a strong zeitgeber (Extended Data Fig. 8). Therefore, our data indicate that not only does [Mg2+]i exhibit a bona fide circadian rhythm across diverse eukaryotic cells, but also that appropriate manipulation of [Mg2+]i is sufficient to determine the key properties of the oscillation (period, amplitude, and phase), making [Mg2+]i indistinguishable from a core clock component.
We considered that the increased cellular ATP levels we observed during Mg2+ depletion might be attributable to differential sensitivity of MgATP-dependent cytosolic enzymes compared with the organellar ATP synthesis machinery. For example, ATP accumulation was accompanied by a marked reduction in extracellular lactate accumulation in U2OS cultures (Extended Data Fig. 9a), indicative of reduced glycolysis. We therefore considered whether rate-changes in gross cytosolic energy metabolism might be a functional consequence of cell-autonomous circadian [Mg2+]i oscillations. A clear prediction would be that global rates of translation should be limited at circadian phases of low [Mg2+]i, since protein synthesis is one of the most energetically expensive processes that cells undertake.
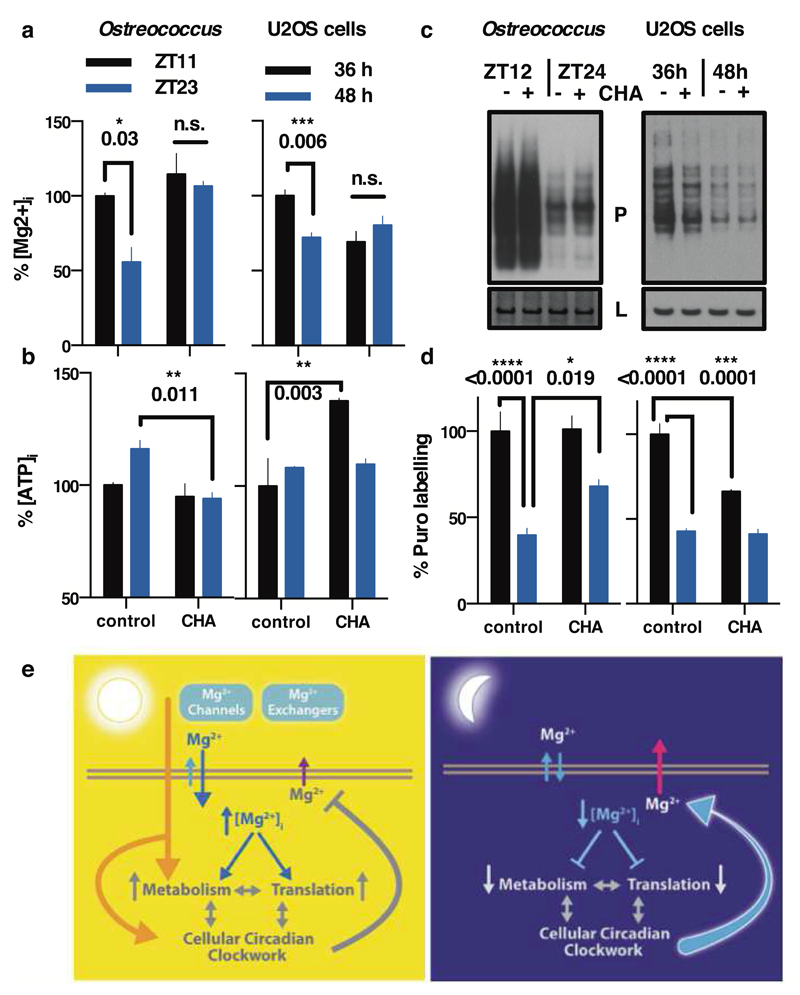
We assayed translation rate by puromycin incorporation8 in both cell types just before (anticipated) biological dusk and dawn; at the phase of lowest and highest [Mg2+]i, respectively. The Ostreococcus experiment was performed under its natural light/dark cycle so as to best model an organism in its natural environment, whereas the U2OS experiment was performed under constant conditions to model innate peripheral cellular clock function. We observed that both Ostreococcus and U2OS cells did exhibit significantly higher translation rates at the peak of [Mg2+]i, as predicted (Fig. 4c,d). In mammalian cells, the highly MgATP-sensitive mTOR pathway was recently shown to mediate circadian control of translation8. We hypothesised that [Mg2+]i oscillations might act via mTOR to effect circadian translational regulation, and tested this using two pharmacologically distinct mTOR inhibitors (torin1 and rapamycin). Both inhibitors lengthened period dose-dependently, and abolished any additional period lengthening due to depletion of extracellular magnesium that was observed in controls. This clear ‘ceiling effect’ strongly suggests changes in [Mg2+]i act through mTOR activity to regulate cellular circadian period (Extended Data Fig. 9b-e). To test the extent to which marked differences in translation rates between dawn and dusk were attributable to cell-autonomous [Mg2+]i oscillations, we incubated cells acutely with CHA to block magnesium transport before assaying overall translational rates. CHA significantly attenuated the difference in [Mg2+]i between dawn and dusk (Fig. 4a), and also phase-dependently affected ATP levels (Fig. 4b). Crucially, differential translation rates were attenuated similarly by CHA treatment (Fig. 4c,d), indicating that in both cell types, circadian regulation of magnesium levels contributes to circadian rhythms in global translation rate.
Figure 4. Magnesium transport phase-dependently affects cellular energy balance.
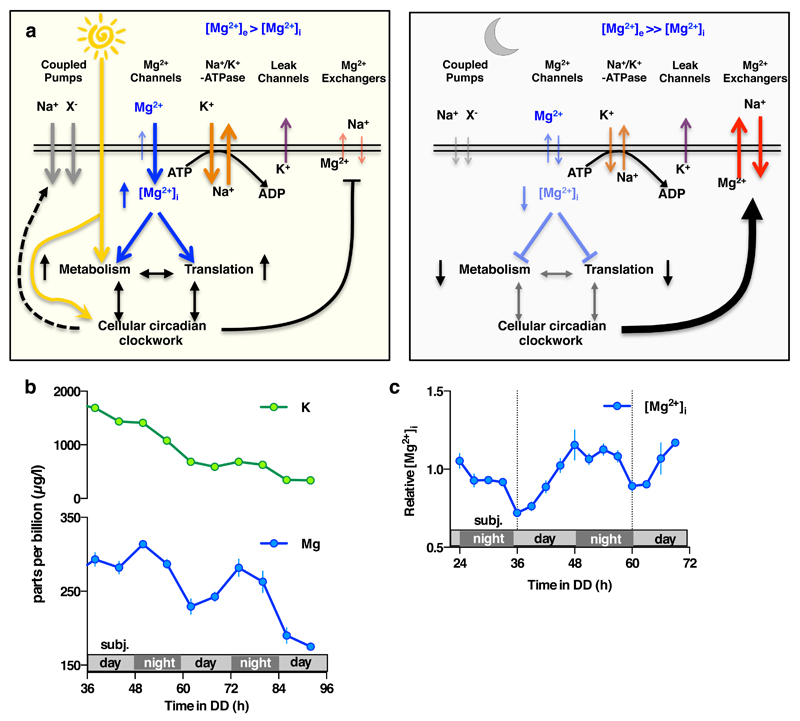
Cells were treated with CHA or vehicle during the second half of the day (black bars) or night (blue bars) to test acute effects on magnesium (a) and ATP (b) levels in both cell types. Incorporation of puromycin was analysed by western blot (c) then quantified to give relative translation rates at the indicated times (d). Mean±SEM are shown, n=3, t-test significance reported (n.s., p>0.18). e, Simplified model for a feedback mechanism between circadian [Mg2+]i rhythms and the clockwork to temporally regulate global cellular metabolism; orange arrow represents extracellular stimuli.
Our data support a model (Fig. 4e, Extended Data Fig. 10a) where the cellular clockwork regulates the expression of plasma membrane Mg2+ channels and transporters to generate rhythmic magnesium fluxes. These rhythms appear to facilitate the higher energetic demands and protein production of human cells during the (biological) day, as well as the global down-regulation of ATP turnover and translation in photosynthetic cells at night. Reciprocally, the [Mg2+]i rhythm feeds back to regulate the period, phase and amplitude of clock gene expression rhythms, acting as a “meta-regulator” to integrate metabolic rhythms with transcriptional feedback models of cellular timekeeping. It is noteworthy however, that the magnesium rhythm observed in Ostreococcus persists under transcriptionally inactive conditions14 of constant darkness (Extended Data Fig. 10b,c), in the absence of transcriptional regulation of membrane transport. It is therefore likely that some aspects of circadian magnesium flux are regulated by, and may contribute to, the same uncharacterised non-transcriptional clock mechanism13,14 that also drive persistent peroxiredoxin overoxidation rhythms in transcriptionally inactive cells across taxa, and which we speculate was present in the LECA.
Cell-autonomous rhythms in [Mg2+]i availability have the potential to impart circadian regulation to any cellular system where MgNTP hydrolysis becomes rate limiting. Although the clinical relevance of [Mg2+]i in various tissues is beginning to garner more attention, the interactions between magnesium transport and human health are poorly understood. Further investigation of the downstream consequences of circadian regulation of [Mg2+]i will therefore be important.
Methods
All materials were purchased from Sigma-Aldrich unless otherwise stated. ICP-MS data are reported as “parts per billion (µg/L)”, Mg2+ measured by luciferase assay are normalised to the highest value and reported as “% [Mg2+]i”.
Ostreococcus tauri
Wild-type cells or cells transgenically expressing a translational fusion of CCA1 to luciferase from the CCA1 promoter (CCA1-LUC) 10 were grown under 12h/12h light/dark cycles in artificial sea water (24 g/l NaCl, 4 g/l Na2SO4, 0.68 g/l KCl, 200 mg/l NaHCO3, 100 mg/l KBr, 25 mg/l H3BO3, 3 mg/l NaF, plus hydrous salts: 50 mM MgCl2*6H2O, 10 mM CaCl2*2H2O, 0.1 µM SrCl*6H20), supplemented with Guillard's F/2 marine enrichment solution and 10 nM H2SeO3. Full medium was adjusted to a salinity of 30 ppt.
Imaging and analysis of luminescent rhythms was performed as described31–33. For resetting experiments, magnesium-free media were removed with a multichannel pipette and replaced with magnesium-containing media. In all luminescent imaging experiments, 8 replicate wells constitute n=8, and presented experiments are representative of 3 or more replicate experiments.
For ICP-MS analyses, 30 ml culture was pelleted, washed three times in 1 M Sorbitol to remove sea water, and digested in 100 µl nitric acid (69%, ARISTAR grade, VWR International) spiked with 345 ppb indium (VWR International) at RT for ~3 hours. Samples were then diluted to a final concentration of 2% v/v nitric acid and 10 ppb Indium prior to analysis on an Agilent 7500ce with octopole reaction system. Serial dilutions of ICP-Multi-element solution IV (Merck, Certipur) was used for calibration of all the metals analysed and to check for instrument drift. A standard reference material SRM1643e (NIST) was analysed to validate the calibration. Indium was used to correct for dilution errors introduced during handling. ICP-MS data reported is based on three replicate flasks, each sampled every timepoint (n=3). Results presented have been verified in a replicate experiment, and outliers were excluded if they were >2 S.D. from the mean.
Cell extracts for luminescent Mg2+ and ATP assays were made from 3 replicates (n=3) of 5 ml cell culture, pelleted and washed with 1 M Sorbitol, and resuspended in 100 µl medium before adding 100 µl 2x extraction buffer (1% Triton X-100, 300 mM NaCl, 100 mM HEPES). 25 µl of extract was boiled and added to 75 µl of assay mix (40 mM HEPES, 1 mM luciferin, 0.05 mg/ml QuantiLum (Promega), and either 1 mM MgCl2 or 10 µM ATP). Luminescence was measured on a TopCount (Packard) plate reader against a standard curve. As Mg2+ ions that remain tightly bound to cellular macromolecules such as membrane components and DNA are not detected by this alternative assay, the relative amplitude of [Mg2+]i changes observed using this assay were substantially larger than measured by ICP-MS. Quinidine, CHA and CPA were made up in medium and added 24 hours prior to cell lysis for chronic treatments. Results were verified in one or more replicate experiments. For puromycin experiments (Fig. 4), cobalt ammines or vehicle were added at ZT6 or ZT18, and 0.5 mg/ml puromycin was added 20 minutes before harvesting cells at ZT11 and ZT23 (required concentration and incubation time determined empirically to reduce expression from a constitutive promoter driving luciferase by ~half). Analysis of incorporation was performed as described for U2OS cells below. Loading control was RbcL (Coomassie).
To identify potential Ostreococcus transporter proteins, mammalian sequences for all classes of SLC and all known magnesium transporters were blasted onto the Ostreococcus proteome using DELTA-BLAST (NCBI), and gene models were then taken from the latest version of the Ostreococcus genome 34 using the Orcae service 35 (Gent University).
Neurospora crassa
Plates with 25 ml of Vogel’s medium containing 2% glucose, 0.5% arginine, 10 ng ml−1 biotin and 0.2% Tween 80 were inoculated with 106 conidia of wild-type ras-1bd strain and incubated under constant light for 48 h at 30 °C. Two 2-cm disks cut from the mycelial pad were placed in a series of 50 ml cultures (Vogel’s medium containing 0.03% glucose, 0.05% arginine and 10 ng ml–1 biotin). These cultures were incubated at 25 °C under constant light before staggered transfers at 4h intervals to constant darkness, shaking at 125 r.p.m. Mycelia were then washed 4 times in a falcon tube with 25ml of sterile water, dried on filtration paper and frozen in liquid N2, and stored at −80°C. Samples were then lyophilised and ground to a powder. These samples were split into two: one for western blot analysis of FRQ oscillations (as described in 36) and one for ICP-MS analysis. ICP-MS was performed as described for Ostreococcus, except that the dissolved tissue was filtered through a 0.22 µM filter before application on ICP-MS.
Mammalian cells
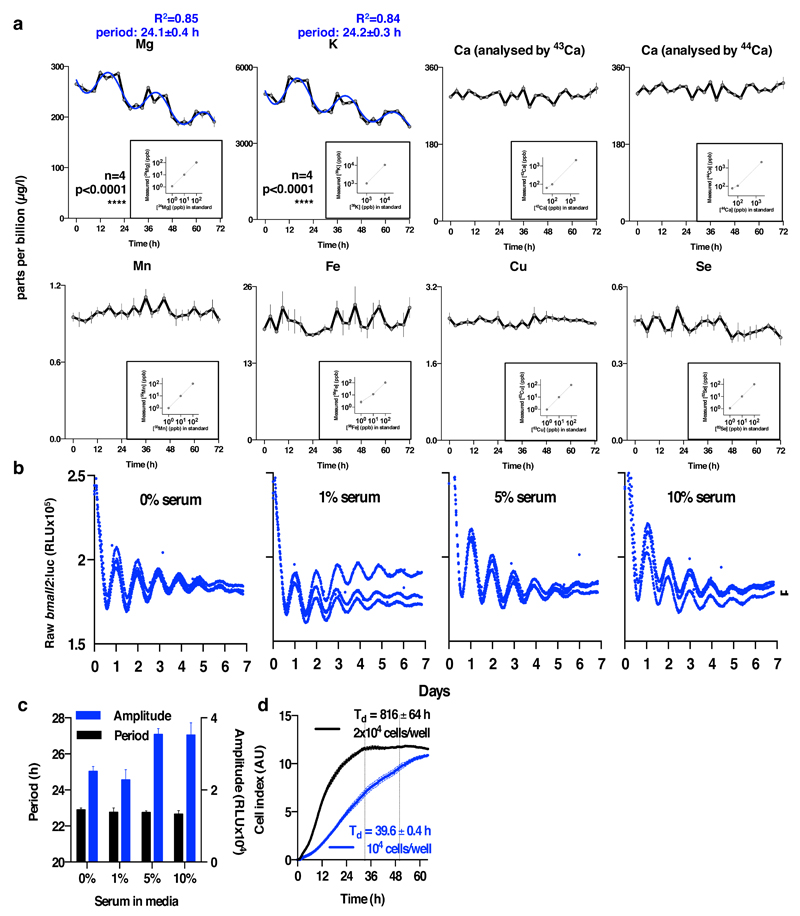
Human U2OS cells, purchased from ATCC, were stably transfected with Per2:luc and cultured between passage numbers 31-51 as described previously37, except that 10% FetalClone™ II serum (HyClone™) was used in place of fetal bovine serum. Cells were mycoplasma-free (Mycoalert, Lonza) and authenticated by Multiplex PCR. Cells were seeded into 6, 24 or 96-well white plates at a density of 105 cells/ml, and incubated in a humidified incubator (37°C, 5% CO2) under 12 h:12 h 32:37°C temperature cycles until confluent for a minimum of 3 days. We confirmed previous reports that U2OS do not proliferate appreciably once confluent and that in the presence of B-27 supplement U2OS circadian period is not affected by the presence of additional serum during bioluminescence recordings (Extended Data Fig. 2b-d). We cannot formally exclude the rhythmic excretion of growth factors. Empirically we determined that upon transfer to constant 37°C Per2:luc bioluminescence peaks around the anticipated transition from 32°C to 37°C, and also 28 h after a media/serum change. To maximize intercellular synchrony therefore, for all U2OS experiments, media was changed at 4 h prior to the warm phase for HEPES-buffered “air medium”38 and then maintained at constant 37°C under a gas impermeable seal. Air medium stock was prepared as described previously38 and supplemented with 2% B-27 (Life Technologies, 50X), 300 µM luciferin (Biosynth AG), 1% glutamax (Life Technologies), 100 units/ml penicillin/100 µg/ml streptomycin, as well as FetalClone™ II serum (HyClone™). Serum was present at 10% for every U2OS experiment except those performed with Mg-free air medium (and controls). Mg-free air medium was prepared from its individual components, replacing magnesium with sodium. The final osmolarity was adjusted to 350 mOsm with NaCl and sterile filtered. Bioluminescence recordings were performed in a lumicycle (Actimetrics), a LB962 plate reader (Berthold technologies) or an Alligator (Cairn Research). Primary fibroblasts were isolated from the lung tissue of adult Cryptochrome1-/- ,Cryptochrome2-/- male mice39 or wild type controls, and cultured as described previously38, then immortalized by serial passage40. All animal work was licensed under the UK Animals (Scientific Procedures) Act of 1986 with local ethical approval. For time courses, confluent cultures were instead synchronized by 2 h incubation with 100 nM dexamethasone, then changed to air medium supplemented as described above, but containing 1% FetalClone™ III (instead of II) serum (HyClone™).
For ICP-MS analyses cells were washed twice in a salt-free isosmotic buffer (300 mM sucrose, 10 mM Tris pH7.4, 350 mOsM) at room temperature to remove ions in the cell media. Cells were then digested in 65 % nitric acid supplemented with 100 ppb cerium for 30 minutes at room temperature and flash frozen. Upon thaw, samples were heated at 90°C for 1 h then centrifuged at 18,000g for 20 minutes to remove any debris, then diluted 1:12 in HPLC-grade water to give a final matrix concentration of 5% HNO3. The ICP-MS time course data shown are representative of three separate time courses all with the indicated number of biological replicates per time point. ICP-MS was performed on a Perkin Elmer Elan DRC II. SPS-SW2 (LGC) was used as a routine standard but linear sensitivity was confirmed for each element (Extended data Fig. 2). Cerium in the HNO3 extraction reagent used to correct for dilution errors introduced during handling. Outliers were excluded if they were >2 S.D. from the mean.
For intracellular ATP and Mg2+ assays, U2OS cells were washed twice with ice-cold PBS + 5 mM EDTA then lysed in buffer containing 30 mM HEPES pH 7.4, 100 mM NaCl, 10% Glycerol, 1% Triton and 5 mM Na3VO4 then flash frozen. Mouse fibroblasts were instead lysed in 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.1% LDS, 1% Triton, 0.5% NaDOC and protease inhibitors. Samples were thawed, and proteins denatured, by heating at 90°C for 10 minutes followed by centrifugation at 18,000 g at 4°C. Assays were performed by diluting samples 1:1 in 2X assay buffer giving final concentrations of 30 mM HEPES (pH 7.4), 1 mM luciferin, 50 nM QuantiLum (Promega) and either 15 mM MgSO4 or 100 µM ATP (dependent on whether ATP or Mg2+ was being assayed). For Mg2+ assays upon mouse fibroblast lysates, 5% bovine serum albumin was also included in the assay buffer to quench LDS. Bioluminescence was measured using a LB962 CentroPRO microplate reader (Berthold technologies) or Spark 10M microplate reader (Tecan). Lactate assay kits (MAK064-1KT) were used in accordance with manufacturer’s instructions and measured on a Spark 10M microplate reader (Tecan). SLC41A1 siRNA (Santa Cruz sc-88707) or control siRNA-A (Santa Cruz sc-37007) was used at 80 pM to 6 µl of transfection agent (Santa Cruz sc-29528) to transfect Per2:luc U2OS cells at 60% confluence in 35 mm2 dishes as per manufacturer’s instructions. Quinidine, rapamycin, torin1, CHA and CPA stock solutions were made up in serum and B-27-free air medium, with chronic treatments lasting for 24 h prior to enzymatic [Mg2+]i assay. For acute treatments, 2 mM CHA (final concentration) was added 3h prior to the peak or trough of Per2:luc bioluminescence and harvested 3h later. Puromycin labeling was performed for 10 minutes at 37°C with 10 µg/ml puromycin before cells were washed twice in ice-cold PBS + 5mM EDTA then lysed on ice for 20 minutes in buffer containing 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% SDS, 1% Triton and 0.5% sodium deoxycholate. Gel electrophoresis, transfer and blotting were performed as described previously4. SLC41A1 rabbit antibody (Abcam ab83701) was diluted by 1:1000, and mouse monoclonal anti-puromycin ascites41 was diluted 1:100 in blocking buffer (w/v 0.25% BSA, 0.25 % dried skimmed milk in Tris-buffered saline/0.05% Tween-20) and incubated overnight with nitrocellulose membranes. Anti-β actin (sc-47778, Santa Cruz) was used as a loading control at 1:5000 dilution. U2OS impedence measurements were performed upon an xCELLigence RTPA DP under our standard bioluminescence recording conditions (air media with supplements, including 10% serum) according to the manufacturer’s instructions.
Analysis
Statistical tests were performed using Graphpad Prism, with all tests two-sided. No significant difference in variance was detected between groups under comparison, Brown-Forsythe’s test (p>0.2). Numbers of biological replicate numbers were chosen, based on preliminary experiments, so that an effect size of at least 10% could be detected between experimental groups for α=0.05 and β=0.9. For phase response curves the circular mean and standard deviation were calculated using the “circular” R package implementing the methods described previously42,43. A least squared fit of the phase response vs phase prior to addition to a linear model where the gradient was constrained to -1 was performed. The Y intercept where X=0 is indicative of the nascent phase relative to the prior phase in hours.
Extended Data
Extended Data Figure 1. Additional ICP-MS data and controls (Ostreococcus).
Inductively Coupled Plasma Mass Spectrometry analyses of cell lysates from 12 h:12 h light/dark cycles (a) or on the second day of constant light (b). P-values report significance by one-way ANOVA (mean±SEM, n=3). c, ICP-MS analyses on cell lysates compared with media control (no cells) and membrane fractions (lysed cells) (mean±SD plotted, n=2), indicating that magnesium signal in panel a and main Fig. 1 comes predominantly from the intracellular space. Groups are significantly different by one-way ANOVA (p<0.0001) Tukey's multiple comparisons p-values are indicated. d. Fluctuations in measured concentrations are not related to fluctuations in cell size over time. No significance of time as source of variation in cell size was observed by FACS analyses (mean±SD plotted, one-way ANOVA p-value is indicated, n=5).
Extended Data Figure 2. Additional ICP-MS data (U2OS cells) and controls.
a. Inductively Coupled Plasma Mass Spectrometry analyses of U2OS cell extracts for several stable isotopes of several biologically relevant ions (mean±SEM, grey/black, n≥4), with insets showing standards that indicate linearity over the observed concentration ranges (mean ± %CV). We compared how well a straight-line + damped sine wave model (adapted from44) fit to each time series compared with a straight-line only (null hypothesis, no rhythm). The null hypothesis was preferred in each case except for Mg and K (analysed by 24Mg and 39K), where the sinusoidal fit with a circadian period was preferred (blue line, R2 and fit period±SEM are reported). b. Bmal1:luc bioluminescence data showing no effect of serum concentration on circadian rhythms in U2OS cells in the presence of B-27 supplement. c. Quantification of period and amplitude for data shown in b, mean±SEM (n=3), one-way ANOVA for period, p=0.79, one-way ANOVA for amplitude, p=0.01. d. Cellular impedance measurements indicate that U2OS cells do not proliferate upon reaching stationary phase under our assay conditions, reported doubling times (Td) were calculated from data collected between the dotted lines.
Extended Data Figure 3. Circadian rhythms of [Mg2+]i in Neurospora crassa and mouse fibroblasts.
a. Circadian regulation of [Mg2+]i detected by ICP-MS in the fungus Neurospora crassa under constant darkness (mean±SEM, n=3). b. Representative (out of 3) FRQ immunoblot sampled in parallel. c. Quantification of FRQ abundance (mean±SEM, n=3). d. Circadian regulation of [Mg2+]i measured by luciferase-based assay is dependent upon CRYPTOCHROME in immortalised adult mouse fibroblasts under constant conditions (mean±SEM, n=3).
Extended Data Figure 4. Rhythms of [Mg2+]i entrain to relevant external cues and are temperature-compensated.
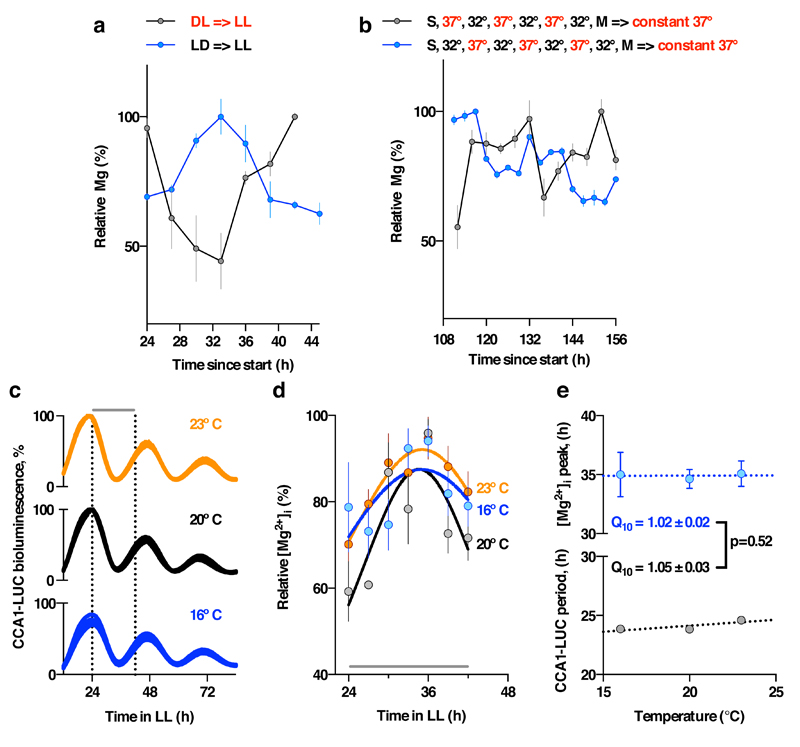
a. Inversion of 12 h : 12 h light/dark entrainment cycles is sufficient to entrain the phase of [Mg2+]i in Ostreococcus cells, measured by luciferase assay under constant light (mean±SEM, n=3). b. From the start of the experiment (S), 3 days of 12 h : 12 h temperature cycles between 32 and 37 ºC, followed a change to air medium (M) is sufficient to entrain the phase of [Mg2+]i in U2OS cells measured by ICP-MS over two circadian cycles under constant conditions (mean±SEM, n=3). c. Ostreococcus bioluminescence recordings (CCA1-LUC) at the indicated temperatures (n=8). Vertical dotted lines indicate sampling window for Mg2+ assays reported in panel d: assays performed during the second cycle under constant conditions show that circadian [Mg2+]i rhythms are temperature compensated (n=4). Each data set was fit with a Lorentzian curve to estimate peak [Mg2+]i. e. No significant difference in temperature compensation (Q10) between CCA1-LUC rhythms and the timing of the second Mg2+i peak; unpaired t-test p-value is reported.
Extended Data Figure 5. Human magnesium transporters and conservation in Ostreococcus tauri.
a. Ubiquitously expressed human proteins with a clearly defined Mg2+ transport activity7 are listed. Note that many additional putative Mg2+-transporters are annotated, with several of these also being circadian-regulated in multiple mouse tissues. b. Expression profiles of Ostreococcus homologs of mammalian Mg2+ channels & transporters listed in (a), mined from publically available microarray data45.
bFrom CircaDB25 with JTK cycle p-value < 0.05.
d% sequence identity/similarity with human protein sequence (E-value). DELTA-BLAST47 performed using default settings.
eFrom micro-array data45 shown in (b).
Extended Data Figure 6. Chronic CPA treatment dose-dependently lengthens period.
Traces (a, b) of the CCA1-LUC (Ostreococcus) or per2:luc (U2OS cells) reporters, showing the effect of inhibition of magnesium transport by Co(NH3)5Cl2+ (CPA) upon period dose-response (c, d) and upon [Mg2+]i (e, f). All plots show mean±SEM, with replicate numbers (n) indicated, p-values report significance by 1-way ANOVA and post-test for linear trend.
Extended Data Figure 7. Period lengthening by CHA and SLC41 knockdown is dependent upon extracellular magnesium.
a. Extracellular magnesium-depletion and CHA act synergistically to lengthen circadian period in Ostreococcus cells (mean±SEM, n=4). b. Quantification of period lengthening by CHA at different concentrations of extracellular magnesium (mean±SEM, n=4), p-value for two-way ANOVA (interaction effect) is reported. c. Extracellular magnesium-depletion and CHA act synergistically to lengthen circadian period in human U2OS cells (mean±SEM, n=6). d. Quantification of period lengthening by CHA in Mg2+-depleted vs. normal media (mean±SEM, n=4), p-values for two-way ANOVA (interaction effect) and Fisher’s exact test are reported. e. Period lengthening due to knockdown of plasma membrane Mg2+/Na+ antiporter SLC41A1 is attenuated by depletion of extracellular magnesium (mean±SEM, n=8). f. Quantification of period lengthening due to knockdown of SLC41A1 in normal vs. Mg2+-depleted media (mean±SEM, n=8); two-way ANOVA interaction effect, p<0.0001, p-values for Sidak’s multiple comparisons test are also reported. g. Quantification of SLC41A1 knockdown efficacy, unpaired t-test p-values are reported, a representative immunoblot (of 3) is shown (mean±SEM, n=3).
Extended Data Figure 8. Bioluminescence data of wedge experiment.
Peak expression phase of the clock protein CCA1 was analysed upon re-introduction of magnesium to cultures in low extracellular magnesium, to test whether the phase of cellular rhythms is dictated by the prior phase of entrainment or by this enforced transition from low to high [Mg2+]i. a, Bioluminescence traces showing that circadian rhythms in Ostreococcus are reversibly attenuated by depletion of extracellular Mg2+, and restored by Mg2+ wash-in. b,e. Bioluminescence traces from cells in low extracellular magnesium (b; 5 µM, e; 20 µM)with rhythms rescued by release into media containing normal physiological concentrations of magnesium at the indicated times (vertical dotted lines) in constant light (LL), compared to their respective controls where no magnesium was added in (blue traces). Data from 7-8 replicate wells are shown in each panel. c,f. Summary graphs where results from b,e are plotted in circadian wedge graphs: peak phases of CCA1-LUC rhythms in untreated control cells (grey dots) are compared with peak phase of rhythms reinstated by introduction of physiological magnesium following depletion to 5 µM (c, red dots) or 20 µM (f, orange dots), revealing that the phase of resulting rhythms is dictated solely by the phase of magnesium reintroduction (blue line). d,g. radial plots of phase shift (mean±SD, circumferential axis) depicted in panels c and f, versus phase prior to addition of Mg2+ to normal levels (old phase, radial axis and colour). The expected phase responses for type 0 resetting (black dotted line) and no resetting (red dotted line) are indicated. The goodness of fit (r2) and Y intercept (Y0) to the type 0 model are shown. Dose-dependent effects of intracellular magnesium on a critical clock parameter are confirmed by the observation that resetting is less strong when magnesium was reintroduced to cells adapted to intermediate levels of extracellular magnesium (e-g) compared to lowest extracellular magnesium (b-d).
Extended Data Figure 9. The effects of magnesium depletion and role of mTOR.
a. Extracellular lactate was measured in U2OS cells after 24 hours in Mg2+-depleted compared with normal media. b,c. Combined action of extracellular magnesium depletion and mTOR inhibition using torin1 (b, n=3) or rapamycin (c, n=6) to lengthen circadian period in U2OS cells is less than additive (mean±SEM). d,e. Quantification of period lengthening due to torin1 (d, n=3) and rapamycin (e, n=6) in Mg2+-depleted compared with normal media (mean±SEM). Note the apparent ‘ceiling effect’ at high concentrations of both drugs, such that Mg2+-depletion elicits no additional lengthening of cellular circadian period. Two-way ANOVA interaction effect: p<0.0001 for both drugs vs. Mg2+, selected p-values for Sidak’s multiple comparisons test are also reported (n.s., p>0.33).
Extended Data Figure 10. Factors potentially contributing to maintenance of membrane electroneutrality in light of [Mg2+]i oscillations.
a. Model indicating potential ion fluxes that might explain how clock-regulated [Mg2+]i oscillations impact on global cellular metabolism whilst membrane electroneutrality is maintained, during the day versus the night. The observed phase dependency of acute CHA was different between Ostreococcus and U2OS cells (Fig. 4a-c), and is consistent with the very different environmental niches inhabited by a marine alga compared with a peripheral human tissue. In Ostreococcus, CHA maintained [Mg2+]i at daytime levels when added prior to the normal trough, resulting in increased nighttime translation and a concomitant reduction in relative ATP levels. This result suggests that Ostreococcus pumps magnesium out of the cell during the dark period, against a large electrochemical potential gradient (magnesium is the second most abundant cation in seawater, at 50 mM in this study) in order to globally down-tune ATP turnover. In U2OS cells, CHA treatment significantly reduced [Mg2+]i accumulation and translation rates as well as significantly increasing ATP levels when added prior to the [Mg2+]i peak. Human cells inhabit an environment where nutrient availability is homeostatically regulated (0.8 mM magnesium in cell culture medium). As such, circadian regulation of increased magnesium transport into the cell during the feeding, active phase of day serves to facilitate higher metabolic rate constants. Please note that light has no direct effect on the clock in human peripheral cells, instead being mediated by systemic cues. b-c. [Mg2+]i oscillations persist in transcriptionally inactive Ostreococcus cells kept in constant darkness, as analysed by both ICP-MS (b) and luciferase assay (c), indicating that circadian regulation of ion transport can occur post-translationally in addition to its transcriptional regulation (mean±SEM, n=3 for ICP-MS data and n=4 for luciferase assays).
Acknowledgements
GvO is supported by a Royal Society University Research Fellowship (UF110173) and research grants (RS120372 and RS140275). JSO is supported by the Medical Research Council (MC_UP_1201/4) and the Wellcome Trust (093734/Z/10/Z). MP is funded by KWF BUIT 2014-6637. LFL and COY are supported by Millennium Nucleus for Fungal Integrative and Synthetic Biology (NC120043), and Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT 1131030). At the MRC LMB, we are grateful to the Biomedical Services Group for animal care; Mick Hastings and Jo Chesham for supplying mouse tissue; Paul Margiotta of MRC-LMB Visual Aids for assistance with figures. The authors would also like to thank Prof. David E. Salt, Prof. Marc Knight, Dr. Ellen Grünewald, Priya Crosby, Dr. Laura Hewitt and Dr. Ben Cross for constructive criticism. The anti-puromycin ascites was a kind gift from Manu Hegde (MRC LMB).
Footnotes
Author contributions GvO and JSO conceived the approach and designed the study. LFL and COY generated the Neurospora result. JD and LE performed ICP analyses. GvO and LLH performed Ostreococcus experiments. Human U2OS cell experiments were performed by KAF. MP performed mouse fibroblast experiments. NPH provided analytical and intellectual contributions. GvO and JSO wrote the manuscript.
Author information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to GvO (Gerben.vanOoijen@ed.ac.uk) or JSO (oneillj@mrc-lmb.cam.ac.uk).
References
- 1.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endo M, Shimizu H, Nohales MA, Araki T, Kay SA. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature. 2014;515:419–22. doi: 10.1038/nature13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar RS, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–64. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–56. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 6.Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 7.de Baaij JH, Hoenderop JG, Bindels RJ. Magnesium in man: implications for health and disease. Physiol Rev. 2015;95:1–46. doi: 10.1152/physrev.00012.2014. [DOI] [PubMed] [Google Scholar]
- 8.Lipton JO, et al. The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell. 2015;161:1138–51. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–90. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 10.Corellou F, et al. Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell. 2009;21:3436–49. doi: 10.1105/tpc.109.068825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastings MH, Maywood ES, O'Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 12.Olmedo M, et al. Circadian regulation of olfaction and an evolutionarily conserved, nontranscriptional marker in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2012;109:20479–84. doi: 10.1073/pnas.1211705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–8. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Ooijen G, Millar AJ. Non-transcriptional oscillators in circadian timekeeping. Trends Biochem Sci. 2012;37:484–92. doi: 10.1016/j.tibs.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Ko GY, Shi L, Ko ML. Circadian regulation of ion channels and their functions. J Neurochem. 2009;110:1150–69. doi: 10.1111/j.1471-4159.2009.06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Njus D, Sulzman FM, Hastings JW. Membrane model for the circadian clock. Nature. 1974;248:116–20. doi: 10.1038/248116a0. [DOI] [PubMed] [Google Scholar]
- 18.Nitabach MN, Holmes TC, Blau J. Membranes, ions, and clocks: testing the Njus-Sulzman-Hastings model of the circadian oscillator. Methods Enzymol. 2005;393:682–93. doi: 10.1016/S0076-6879(05)93036-X. [DOI] [PubMed] [Google Scholar]
- 19.Danku JM, Lahner B, Yakubova E, Salt DE. Large-scale plant ionomics. Methods Mol Biol. 2013;953:255–76. doi: 10.1007/978-1-62703-152-3_17. [DOI] [PubMed] [Google Scholar]
- 20.Danku JMC, Gumaelius L, Baxter I, Salt DE. A high-throughput method for Saccharomyces cerevisiae (yeast) ionomics. J Anal At Spectrom. 2009;24:103–107. [Google Scholar]
- 21.Nishinaga H, et al. Circadian expression of the Na+/H+ exchanger NHE3 in the mouse renal medulla. Biomed Res. 2009;30:87–93. doi: 10.2220/biomedres.30.87. [DOI] [PubMed] [Google Scholar]
- 22.Wang YC, Chen YS, Cheng RC, Huang RC. Role of Na(+)/Ca(2)(+) exchanger in Ca(2)(+) homeostasis in rat suprachiasmatic nucleus neurons. J Neurophysiol. 2015;113:2114–26. doi: 10.1152/jn.00404.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin H. The logic of the Membrane, Magnesium, Mitosis (MMM) model for the regulation of animal cell proliferation. Arch Biochem Biophys. 2007;458:16–23. doi: 10.1016/j.abb.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 24.Pittendrigh CS. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–84. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;41:D1009–13. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang EE, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolisek M, et al. SLC41A1 is a novel mammalian Mg2+ carrier. J Biol Chem. 2008;283:16235–47. doi: 10.1074/jbc.M707276200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kucharski LM, Lubbe WJ, Maguire ME. Cation hexaammines are selective and potent inhibitors of the CorA magnesium transport system. J Biol Chem. 2000;275:16767–73. doi: 10.1074/jbc.M001507200. [DOI] [PubMed] [Google Scholar]
- 29.Kolisek M, Nestler A, Vormann J, Schweigel-Rontgen M. Human gene SLC41A1 encodes for the Na+/Mg(2)+ exchanger. Am J Physiol Cell Physiol. 2012;302:C318–26. doi: 10.1152/ajpcell.00289.2011. [DOI] [PubMed] [Google Scholar]
- 30.O'Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–53. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Ooijen G, Dixon LE, Troein C, Millar AJ. Proteasome function is required for biological timing throughout the twenty-four hour cycle. Curr Biol. 2011;21:869–75. doi: 10.1016/j.cub.2011.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Ooijen G, et al. Functional analysis of Casein Kinase 1 in a minimal circadian system. PLoS One. 2013;8:e70021. doi: 10.1371/journal.pone.0070021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Bihan T, et al. Label-free quantitative analysis of the casein kinase 2-responsive phosphoproteome of the marine minimal model species Ostreococcus tauri. Proteomics. 2015 doi: 10.1002/pmic.201500086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanc-Mathieu R, et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genomics. 2014;15:1103. doi: 10.1186/1471-2164-15-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sterck L, Billiau K, Abeel T, Rouze P, Van de Peer Y. ORCAE: online resource for community annotation of eukaryotes. Nat Methods. 2012;9:1041. doi: 10.1038/nmeth.2242. [DOI] [PubMed] [Google Scholar]
- 36.Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell. 2009;34:354–63. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valekunja UK, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci U S A. 2013;110:1554–9. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Neill JS, Hastings MH. Increased coherence of circadian rhythms in mature fibroblast cultures. J Biol Rhythms. 2008;23:483–8. doi: 10.1177/0748730408326682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–30. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 40.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005;Chapter 28(Unit 28):1. doi: 10.1002/0471142727.mb2801s70. [DOI] [PubMed] [Google Scholar]
- 41.David A, et al. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher NI. Statistical analysis of circular data. xviii. Cambridge University Press, Cambridge England; New York, NY, USA: 1993. p. 277. [Google Scholar]
- 43.Jammalamadaka SR, SenGupta A. Topics in circular statistics. 322 S World Scientific; Singapore: 2001. [Google Scholar]
- 44.Hirota T, et al. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci U S A. 2008;105:20746–51. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monnier A, et al. Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles. BMC Genomics. 2010;11:192. doi: 10.1186/1471-2164-11-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boratyn GM, et al. Domain enhanced lookup time accelerated BLAST. Biol Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]