Abstract
The maintenance of the genome is of pivotal importance for the functional integrity of cells and tissues. The gradual accumulation of DNA damage is thought to contribute to the functional decline of tissues and organs with ageing. Defects in multiple genome maintenance systems cause human disorders characterized by cancer susceptibility, developmental failure, and premature ageing. The complex pathological consequences of genome instability are insufficiently explained by cell-autonomous DNA damage responses (DDR) alone. Quality control pathways play an important role in DNA repair and cellular DDR pathways. Recent years have revealed non-cell autonomous effects of DNA damage that impact the physiological adaptations during ageing. We will discuss the role of quality assurance pathways in cell-autonomous and systemic responses to genome instability.
DNA repair machineries maintain genome integrity
The nuclear genome, with the exception of a few mitochondrial genes, harbours the entire genetic information of a cell. The genomic sequence, once altered or lost, cannot be replaced. However, the genome is constantly attacked by a large variety of genotoxic insults. It has been estimated that in each cell tens of thousands of damaging events occur on a daily basis (De Bont and van Larebeke, 2004). DNA damage can be inflicted by exogenous sources such as the UV irradiation of the sun, ionizing radiation, or chemicals. Also endogenous by-products of the cellular metabolism such as reactive oxygen species (ROS) attack the genome. The types of DNA lesions can vary widely. Single strand breaks are probably the most frequently occurring lesions, followed by spontaneous depurination, alkylations, various oxidative base modifications, and deamination. Even highly cytotoxic lesions such as double strand breaks and interstrand crosslinks that are induced during anti-tumour therapeutic interventions also occur endogenously (De Bont and van Larebeke, 2004; Schärer, 2005). Genotoxins have from the early steps of evolution threatened the maintenance and inheritance of the genetic material and thereby of life itself. Therefore, DNA repair systems are required to remove the damage and maintain genome integrity (Table 1). The first challenge of the DNA repair machinery is the recognition of the altered DNA structure. This might be rather obvious if a strand break occurs or a replication fork stalls at obstructive lesions. However, slight structural alterations require highly specialized recognition molecules that allow distinguishing the damaged DNA from normal structural alterations occurring e.g. during decondensation of the double helix as part of the DNA metabolism. The damage recognition is tightly linked with the most appropriate DNA repair mechanism. For instance, the frequently occurring oxidative lesions are effectively removed by base excision repair (BER) that uses glycosylases to excise the damaged base and short-patch or long-patch repair to refill the gap (Sung and Demple, 2006). Single strand break repair rapidly joins the frequently arising breaks in one of the DNA strands (Caldecott, 2008). UV-induced cyclobutane pyrimidine dimers (CPDs) lead to a slight helix distortion that requires highly sophisticated recognition systems before they are excised by nucleotide excision repair (NER) (Cleaver et al., 2009). Global genome (GG-) NER scans throughout the genome for helix-distorting lesions, while transcription-coupled (TC-) NER activates the repair once RNA polymerase II (RNAPII) stalls at a lesion. While transcription is a relatively slow process, replication forks need to move quickly through the genome to enable timely replication during quick cycles of cell divisions. Therefore, specialized DNA polymerases are able to read through damaged templates at the risk of incorporating a wrong nucleotide (Sale et al., 2012). Translesion synthesis (TLS) thus facilitates speedy replication fork progression at the cost of elevated error rates. DNA double strand breaks (DSBs) form a serious threat to the genomic integrity of the cell, as aneuploidy might result from aberrant chromosome segregation (Chapman et al., 2012). DSBs can be repaired quickly by non-homologous end joining (NHEJ) and yet again speed comes at the expense of accuracy as the break sites are resected prior to end joining. More laborious but error free, homologous recombination (HR) uses the undamaged template that is available during late S-phase and G2 phase. HR is also used to resolve replicative impediments that result in strand breaks. During replication, however, HR can lead to chromosomal aberrations demonstrating that DNA repair systems might also themselves become obstructive at times (Wolters et al., 2014).
Table 1. Overview of DNA repair pathways.
| Repair system | Type of lesions | Accuracy |
|---|---|---|
| Base Excision Repair (BER) | Oxidative lesions | error free |
| Nucleotide Excision Repair (NER) | Helix-distorting lesions | error free |
| Translesion synthesis | Various lesions | error prone |
| Miss Match Repair (MMR) | Replication errors | error free |
| Single Stand Break Repair (SSBR) | Single strand breaks | error free |
| Homologous Recombination (HR) | Double-strand breaks | mostly error free |
| Non-Homologous End Joining (NHEJ) | Double-strand breaks | mostly error prone |
| DNA Interstrand Crosslink Repair Pathway | Interstrand crosslinks | largely error free |
The importance of DNA repair systems for human health has become particularly apparent in a wide variety of rare congenital syndromes that are caused by mutations in genome maintenance genes (Schumacher et al., 2008a). Importantly, DNA repair deficiency syndromes precipitate in three major disease components namely developmental impairment, cancer susceptibility, and accelerated ageing (Wolters and Schumacher, 2013). The most severe types already impair early development. For example, while several glycosylases redundantly excise oxidized bases, a complete lack of BER lead to embryonic lethality in mice (Ludwig et al., 1998). Mutations in NER genes can also give rise to growth and mental retardation. Particularly, mutations in the TC-NER specific components CSA and CSB usually give rise to Cockayne syndrome (CS) that leads to postnatal growth defects, mental retardation, and eventually many signs of premature ageing (Marteijn et al., 2014). Other mutations in the same genes can lead to cerebro-occulo-facio-skeletal syndrome (COFS) with patients developing abnormalities already prenatally (Laugel et al., 2010). Strikingly, other mutations in NER genes lead to skin cancer predisposition and pigmentation abnormalities on sun-exposed areas of the skin in Xeroderma pigmentosum (XP) patients (Cleaver et al., 2009). Depending on the specific mutation in NER, XP patients also suffer from mental retardation. However, the clearest link to cancer predisposition is evident when specifically the damage recognition by GG-NER is impaired leading to elevated mutation rates that then fuel the malignant transformation of the damaged cells. In contrast, the TC-NER deficient CS cells are confronted with persistent stalling of the RNA polymerase II (RNAPII) eventually leading to cell death thus fuelling cell loss and tissue degeneration. TC-NER and GG-NER defects have helped to clarify the distinct contributions of unrepaired DNA lesions to cancer development and accelerated ageing. Most patients suffering from premature ageing, however, also show enhanced susceptibility to develop cancer. For example defects in responding to DSBs in ataxia telangiectasia (AT) or Nijmegen breakage syndrome (NBS) patients confers premature ageing as well as highly elevated lymphoma risk (Shiloh, 1997).
The consequences of DNA damage largely depend on the action of the DDR. It has first been recognized in yeast, that cells respond to DNA damage not only by activating the respective repair machinery, but also by halting cell cycle activity until the damage is repaired (Hartwell and Weinert, 1989; Rowley et al., 1992). The DNA damage checkpoints are conserved from yeast to mammals and are important for preventing damaged cells to transforming into cancer cells (Bartek and Lukas, 2007). The most frequent mutations found in human cancers alter the function of the tumour suppressor p53 (Reinhardt and Schumacher, 2012). The p53 gene has evolved during metazoan evolution and orchestrates the checkpoint response. In the simple nematode C. elegans p53 controls the apoptotic demise of germ cells that carry unrepaired DSBs in late stages of meiotic pachytene (Derry et al., 2001; Schumacher et al., 2001). The meiotic DNA damage checkpoint exemplifies the significance of apoptosis as additional outcome of the DDR as it allows the removal of genomically compromised germ cells before maternal resources are deposited into growing oocytes during diakinesis (Schumacher and Gartner, 2006; Schumacher et al., 2005). In mammals, the apoptotic outcome of p53 activity not only removes damaged germ cells but also compromised somatic cells. In addition, the mammalian DDR also withdraws cells permanently from the cell cycle into cellular senescence. The inactivation of DNA damage checkpoints, for example by p53 mutations, allows cells to continuously divide despite the presence of unrepaired DNA damage. Indeed, karyograms of cancer cells show fusion, losses, and fragmentations of chromosomes. Evidently, in the absence of a checkpoint response, cells not only survive but also actively proliferate. In contrast, constitutive activity of p53 can lead to cell loss and tissue degeneration thus accelerating the ageing process (Maier et al., 2004; Tyner et al., 2002). The consequences of p53 activity exemplifies the two extremes of checkpoint signalling: too little checkpoint activity gives rise to cancerous growth, while checkpoint-mediated apoptosis and cellular senescence can lead to cell loss and impair regeneration of tissues thus promoting the ageing process. DNA damage checkpoint signalling needs, therefore, to balance cancer prevention with tissue maintenance and regeneration. Increased gene dosage of p53 in combination with increased dosage of another checkpoint component, the senescence-driving factor ARF, could indeed raise the cancer protection and still permit enhanced tissue maintenance and regeneration with the final outcome of extended longevity of double transgenic mice beyond normal longevity of wild type counterparts (Matheu et al., 2007).
Protein quality control mechanisms in DNA repair and genome maintenance
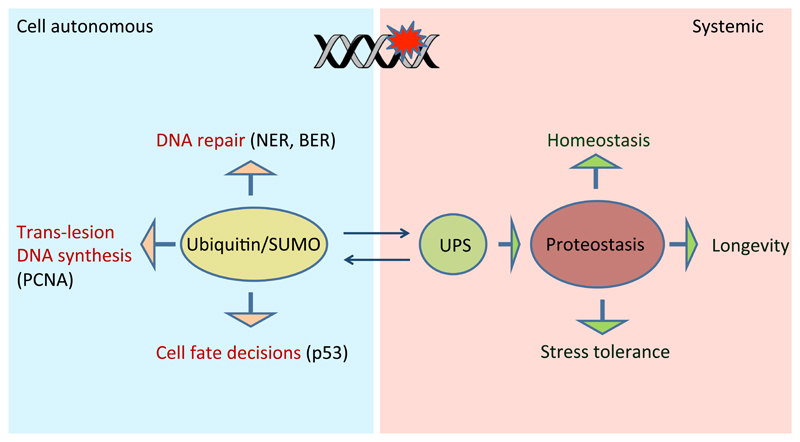
Not only the recognition of DNA lesions and the choice of the most appropriate DNA repair pathway pose a great challenge. Also the actual repair process requires fine-tuning of the action of numerous distinct proteins that are involved in the repair process. The site of the damage needs to be made accessible through remodelling of the chromatin and the chemical reactions to remove the lesions and replace the genetic information need to proceed. A large variety of post-translational modifications are employed throughout the repair process including phosphorylation, acetylation and neddylation of histones as well as dynamic phosphorylation of DNA damage checkpoint and repair factors. Here we will focus on ubiquitinylation and SUMOylation of repair-related proteins as well as on degradation of specific factors through the ubiquitin proteasome system (UPS) (Figure 1). For example, efficient repair of DSBs depends on dynamic K63-linked di- and poly-ubiquitinylation of histone variants H2A and H2AX at the site of damage. The ubiquitinylation is thought to provide critical signals for the assembly and function of the repair complex (Doil et al., 2009; Huen et al., 2007; Mailand et al., 2007). SUMOylation plays an important role in base excision repair where it putatively promotes dissociation of DNA glycosylases (the enzymes initiating repair cascade by removing altered nitrogenous base from the sugar-phosphate backbone of DNA) before the abasic site can be accessed by the subsequent short-patch or long-patch repair systems (Hardeland et al., 2002).
Figure 1. Roles of protein quality control during DNA damage responses.
Protein quality control mechanisms execute important functions in the presence of DNA damage. On the one hand ubiquitinylation, SUMOylation as well as associated degradation of specific repair factors directly facilitate repair of DNA lesions by pathways such as HR, NER, and BER. Specific SUMOylation and ubiqutinylation of the processivity factor PCNA regulates the switching between normal DNA replication and replication-associated DNA repair by acting as a docking platform for translesion polymerases and DNA recombination factors. Global cell fate decisions in the presence of DNA damage are regulated at the level of poly-ubiquitinylation and subsequent degradation versus stabilization of the transcription factor p53. Not only cell-autonomous but also systemic outcomes of DNA damage are linked to protein quality control. For instance it was recently found that enhanced somatic stress tolerance in response to genome instability in the germline of the nematode C. elegans is mediated by enhanced proteostasis and the ubiquitin proteasome system.
Ubiquitinylation and the UPS also play an important role in various steps during the NER reaction. The GG-NER damage recognition factor XPC has a high affinity for UV-induced 6-4PPs, while the most abundant UV-induced lesions, the CPDs are only poorly recognized by XPC (Kusumoto et al., 2001). Therefore, XPC is assisted by a multi-protein complex containing the more effective lesion receptor DDB2 and CUL4 -an E3 ubiquitin ligase belonging to a cullin family (Angers et al., 2006; Groisman et al., 2003). Upon binding to the lesion, the complex mediates polyubiquitinylation of XPC resulting in increased affinity to the DNA. The complex then ubiquitinylates itself thus triggering the dissociation from the damaged DNA and degradation of DDB2 via the proteasome (Sugasawa et al., 2005). Subsequently, the downstream repair factors can assemble and proceed to excise the damaged strand. While DDB2 is degraded, polyubiquitinylated XPC is protected from degradation through binding to RAD23 that contains a ubiquitin-like domain that exerts an inhibitory effect on the proteasome (Ng et al., 2003; Ortolan et al., 2004). Ubiquitinylation is also involved in the TC-NER procedure. The CSB protein initiates TC-NER upon RNAPII stalling at bulky lesions (Fousteri et al., 2006). Once activated, CSB recruits a complex containing the lesion receptor CSA and the CUL4 E3 ligase. The composition of the complex is similar to the one acting in GG-NER, only that instead of DDB2 the CSA protein is recruited (Groisman et al., 2003). CSB is then ubiquitinylated and subsequently degraded, thereby facilitating disassembly of the repair complex and re-start of transcription once the lesion is repaired (Groisman et al., 2006). Also stalled RNAPII is polyubiquitinylated in a dynamic manner (Bregman et al., 1996). It has been postulated that in case of successful and rapid DNA repair ubiquitin chains are fully removed and the RNAPII directly restarts transcription elongation (Daulny and Tansey, 2009). However, when bulky adducts cannot be removed, the polyubiquitinylation targets RNAPII for degradation via the proteasome (Ratner et al., 1998).
Switching between normal DNA replication and distinct types of replication-associated DNA repair is another example of how protein quality control-related mechanisms contribute to genome integrity. The switching is established via specific ubiquitinylation and SUMOylation of PCNA -a ring like homotrimeric clamp that acts as a processivity factor for DNA polymerases (Ulrich, 2009). PCNA is loaded onto DNA at the start of replication in an ATP-dependent manner. In yeast, DNA loading triggers SUMOylation of PCNA on lysine residues 164 and 127 by SUMO-specific E2 conjugating enzyme Ubc9 and the SUMO ligase Siz1. SUMOylated Lys164 creates a high affinity binding site for Srs2, a helicase that displaces the Rad51 recombinase from the replication fork, thus preventing aberrant recombination, which is otherwise likely to occur due to close proximity of sister chromatids during normal replication (Hoege et al., 2002; Pfander et al., 2005). SUMOylated Lys127 functions similar to Lys164 by inhibiting interaction between PCNA and Eco1, a factor responsible for sister chromatid cohesion (Moldovan et al., 2006). Once the replication fork stalls at a DNA lesion, single stranded DNA intermediates are formed as helicases continue to unwind DNA despite polymerase stalling. The ssDNA is rapidly coated with RPA proteins, which in turn recruits the Rad18-Rad6 ubiquitin ligase to the site of damage. The Rad18-Rad6 complex outcompetes SUMOylation at Lys164 of PCNA and establishes monoubiquitinylation of this residue (Hoege et al., 2002). Monoubiquitinylated Lys164 creates a binding surface for low-fidelity TLS polymerases of the Y-family that contain ubiquitin-binding domains (Bienko et al., 2005). The Y-family polymerases lack requirement for Watson-Crick base pairing and can accommodate bulky DNA adducts in their active site, thereby facilitating DNA synthesis across lesions in an error-prone manner (Yang, 2003). Once replication is re-established, ubiquitin is removed from Lys164 by the de-ubiquitinylating enzyme Usp1 followed by dissociation of TLS polymerases from the replication fork and re-start of high fidelity DNA synthesis (Zhuang et al., 2008). If TLS is unsuccessful, stalled replication forks persist attracting additional factors including Ubc13-MMS2 ubiquitin-conjugating enzyme and Rad5 ubiquitin ligase. The Ubc13-MMS2-Rad5 complex extends monoubiquitinylation of Lys164 of PCNA to K63-linked polyubiquitin chains causing dissociation of TLS polymerases from the stalled replication fork followed by fork regression and switching to nascent lagging DNA strand as a template for error-free DNA synthesis (Blastyák et al., 2007; Parker and Ulrich, 2009; Ulrich and Jentsch, 2000).
Finally, the processing of inter-strand crosslinks (ICLs) is a remarkable example of how defects in ubiquitinylation related to DNA repair may lead to severe human disease. ICLs are typically inflicted by chemicals reacting with DNA such as cisplatin and mitomycin C and represent a highly toxic lesion type that interferes with replication and transcription and in addition promotes DSB generation (Deans and West, 2011). The inability to repair ICLs in humans is linked to the rare genetic disorder Fanconi anemia (FA), which is characterized by bone marrow failure, developmental abnormalities, and increased risk of cancer. Mutations in thirteen different genes were found to cause FA (Wang, 2007). Eight of these genes (FA proteins A, B, C, E, F, G, L and M) are involved in the formation of a single large protein complex called “FA core complex” (de Winter et al., 2000; Medhurst et al., 2001). The only known enzymatic activity of this complex is to act as a ubiquitin ligase for the so-called “ID complex” consisting of FA proteins I and D2 (Sims et al., 2007; Smogorzewska et al., 2007). The ID complex is mono-ubiquitinylated by FA core complex in a checkpoint-dependent manner leading to the translocation of the ID complex to the damage site to initiate the repair process.
It has been speculated that decreased quality control with aging might impact the capacity of DNA repair systems to maintain genome integrity (Grillari et al., 2006). Senescent cells accumulate ubiquitinylated proteins while the pool of free ubiquitin declines (Merker et al., 2000) and in various tissues in mammals proteasomal activity progressively declines with aging (Carrard et al., 2002). It is thus conceivable that an accumulation of ubiquitinylated substrates and shortage of free ubiquitin negatively influence events that involve de-novo ubiquitinylation of DNA repair proteins. Similarly to ubiquitinylation, the levels of SUMOylated proteins in various tissues in mammals have been reported to increase with aging (Li et al., 2008).
Systemic activation of protein quality control and multi-tissue responses by nuclear DNA damage
Not only the various DNA repair processes rely on protein quality control but also the cell-fate outcomes of genome instability are regulated by ubiquitinylation and degradation of specific factors. Probably the most intensely studied example of such regulation is DNA-damage dependent stabilization of p53 protein that leads to cell cycle arrest and/or apoptosis depending on levels of genome instability (Reinhardt and Schumacher, 2012). In unperturbed cells p53 is rapidly ubiquitinylated by the E3 ligase MDM2 resulting in the degradation of p53 by the proteasome. The constant cycle of p53 synthesis and MDM2-controlled degradation ensures the availability of the p53-mediated checkpoint surveillance while preventing unnecessary cell cycle arrest or even death of healthy cells. In response to DNA damage, MDM2 is phosphorylated by the DNA damage checkpoint kinase ATM thus disrupting oligomerisation of MDM2 and its specific ability to modify p53 (while other targets can still be ubiquitinylated). Consequently, p53 accumulates in the nucleus and executes the DNA damage response (Manfredi, 2010). Interestingly, the proapoptotic activity of p53 in response to genome instability can also be regulated at the systemic level. It has been demonstrated that enhanced stabilization of HIF-1 (hypoxia induced factor 1) in specific sensory neurons of C. elegans leads to production of the tyrosinase TYR-2 that acts in the germline to inhibit the nematode p53 homolog CEP-1 through a yet to be determined mechanism. The alterations result in reduction of CEP-1 dependent apoptosis of germ cells in response to genotoxic stress (Sendoel et al., 2010). The HIF-1-mediated CEP-1 inhibition was suggested to protect the germ cells under conditions when food shortage forces the worms to migrate into deeper soil layers where oxygen levels are strongly reduced.
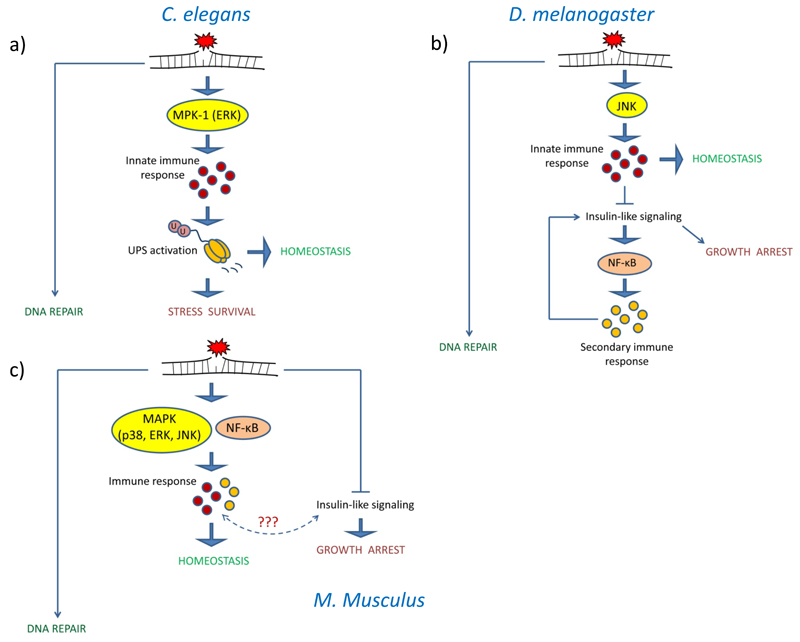
While non-cell-autonomous signalling alterations could influence cell-autonomous DNA damage responses, a number of systemic physiological events could also be triggered by local nuclear DNA damage. Again, the nematode was used as model system of choice as only germ cells proliferate and evoke DNA damage checkpoint signalling upon genotoxic stress while somatic tissues are entirely postmitotic and highly radio-resistant. Various types of DNA damage including hydroxyurea treatment that leads to genome instability specifically during replication and meiotic DSBs that only occur in germ cells led to systemic resistance of somatic tissues to a number of stressors including heat and oxidative stress, a process coined “germline DNA damage-induced systemic stress resistance” (GDISR) (Figure 2). The ability to resist stress conditions is a typical hallmark of lifespan extension across species. Elevated stress survival upon DNA damage was shown to depend on MPK-1/ERK signalling in affected germ cells and on the functionality of the UPS in somatic tissues (Ermolaeva et al., 2013). Genome instability in germ cells triggers MPK-1 dependent expression of genes associated with innate immune defence of the nematode (Ermolaeva and Schumacher, 2014a). Interestingly, many of these genes encode peptides of putative secreted nature leading to their hypothesized function as systemic mediators of DNA damage response and inducers of non-cell-autonomous protein quality control (Ermolaeva and Schumacher, 2014b). Consistent with this hypothesis, MAP kinase (in this case the p38 homolog PMK-1) dependent innate immune priming of C. elegans by immunogenic bacteria also results in activation of UPS and systemic resistance to stress. In contrast, inhibition of innate immune signalling abolishes DNA-damage induced stress tolerance. Interestingly, recent experimental evidence has established several examples that link innate immunity and protein quality control pathways in C. elegans. It was demonstrated that nematodes lacking unfolded protein response in the endoplasmic reticulum (UPRER) fail to develop on immunogenic P. aeruginosa. Of note, worms lacking UPRER and at the same time are unable to mount an innate immune response due to a mutation in the p38 MAPK pmk-1 develop normally under the same conditions suggesting that massive protein synthesis during immune response is likely to apply additional pressure on the proteostatic machinery (Richardson et al., 2010). Under these circumstances it is plausible that activation of the UPS during innate immune response represents an additional mechanism to support cellular proteostasis when large amounts of innate immune peptides are synthesized and processed for secretion. Autophagy as well as activity of specific E3 ubiquitin ligases was recently shown to be induced during immune response in C. elegans (Bakowski et al., 2014; Visvikis et al., 2014). These quality control cascades were suggested to play a role in immune defence against viral and bacterial pathogens by supporting direct degradation of pathogenic material within infected cells. The effect of such activity on cell-specific substrates during an immune response was, however, not investigated. It is, therefore, highly interesting to study the relationship between the different types of protein quality control cascades, genome instability in the germline, DNA-damage induced systemic immune responses and stress tolerance. Another important question is the biological significance of GDISR. The DNA damage checkpoint arrests germ cells of C. elegans thus halting the production of progeny until the repair has been completed. Interestingly, comparison of progeny generation by control and UV-treated nematodes revealed early reduction of viable offspring in the UV-irradiated population that was, however, compensated by enhanced offspring generation at the stage when untreated animals had already passed beyond their reproductively active period (Ermolaeva et al., 2013). GDISR might thus act as a systemic DNA damage checkpoint to support somatic endurance during the time needed to repair DNA lesions in germ cells and resume generation of healthy offspring once genome stability is reinstated. In combination with the previously discussed role of neuronal HIF-1 in preventing apoptosis of germ cells, it appears that both the soma can provide cyto-protective signals to the germline and the germline can support survival of the soma during stress, thus eventually supporting successful passage of genetic material to the next generation. It will be highly interesting to better understand the intriguing systemic interactions between reproductive and somatic compartments under the influence of genome instability.
Figure 2. Systemic DNA damage responses through evolutionary conserved pathways.
Organismal quality control pathways are involved in systemic DNA damage responses that slow down systemic growth and support tissue homeostasis to allow time for appropriate DNA repair. (a) In C. elegans it was recently demonstrated that enhanced tissue homeostasis and elevated stress survival upon innate immune induction (by DNA damage or other means) is executed via activation of the ubiquitin proteasome system (UPS). DNA damage in C. elegans is linked to innate immune response through the MAP kinase (MPK-1/ERK) signaling. (b) In D. melanogaster UV-induced lesions in the epidermis lead to MAPK-dependent induction of cytokines. Cytokines act systemically to inhibit insulin signaling (IIS) via the central nervous system. The inhibition of IIS mediates growth arrest and activates NF-kB/Relish transcription factor in the fat body of the fly leading to the secondary immune response that, among other effects, re-activates insulin signaling leading to eventual resumption of growth. (c) In mammals DNA damage leads to the induction of immune response (both innate and adaptive) as well as to reduced insulin signaling and growth arrest. Both MAP kinases and NF-kB participate in the triggering of immune responses. The cytokines taking part in the innate immune response have been implicated in tissue maintenance and regeneration in both Drosophila and mammals.
Responding to DNA damage through the activation of the innate immune system is by no means specific to C. elegans. For instance, it was demonstrated that UV-induced DNA damage in the larval epidermis of D. melanogaster causes innate immune induction and secretion of cytokines (Karpac et al., 2011) (Figure 2). Consistent with the systemic DNA damage responses in C. elegans, the initial steps of this process are regulated by MAP kinase signaling (in this case by the JNK pathway). The local cytokine induction is amplified by hemocytes - the immune cells of the fly, in a JAK/STAT dependent manner leading to a systemic innate immune response. The JAK/STAT pathway is an evolutionary conserved signaling cascade implicated in innate immunity and cytokine signaling in various species including mammals. Janus tyrosine kinases (JAKs) associate with respective receptors such as IL-6R, IL-10R or type I interferon receptors and are activated upon ligand binding. Activated JAKs phosphorylate specific tyrosine residues of the receptor leading to the recruitment and phosphorylation of STAT (Signal Transducer and Activator of Transcription) transcription factors. Phosphorylated STATs dimerize, translocate into the nucleus and bind to specific promoters thus inducing transcription of genes responsive to a given STAT combination (STATs can form both homo- and heterodimers). Among genes regulated by STATs there are multiple molecules involved in activation and regulation of immune response turning the JAK/STAT module into a critical signaling hub of immunity (Murray, 2007; Stark and Darnell, 2012). Interestingly, in Drosophila DNA damage-induced cytokine production causes systemic inhibition of insulin signaling and larval growth arrest by limiting the secretion of insulin-like peptides by the central nervous system. Among other effects, reduced insulin signaling activates the NF-κB/Relish transcription factor in the fat body of the larvae in a FOXO-dependent manner leading to induction of a secondary non-destructive immune response (Karpac et al., 2011). The components of the secondary immune response, in addition to their direct immune function, also re-establish systemic production of insulin-like peptides thereby completing the tightly regulated interaction between DNA damage, innate immune response, and somatic growth in the Drosophila larvae. It is, therefore, plausible that the temporary growth attenuation acts in this case as a systemic quality control checkpoint to allow time for DNA repair and tissue regeneration before further developmental growth can be resumed.
In mammals, inflammatory responses to DNA damage are particularly well documented in UV-irradiated skin, where DNA lesions trigger MAP kinase and NF-κB dependent secretion of inflammatory cytokines in keratinocytes, thus triggering programmed death of skin cells and attracting immune cells to the skin (Lee et al., 2013). In addition to local inflammation, UV irradiation of the skin leads to systemic immunosuppression. One important molecule to trigger the immunosuppression is the RANK ligand that is produced by keratinocytes. RANKL binds to its receptor on the surface Langerhans cells, the immune cells of the skin. Conditioned Langerhans cells then migrate into lymph nodes where they induce peripheral expansion of regulatory T-lymphocytes thereby conferring systemic immunosuppression (Loser and Beissert, 2009). Additional evidence of mammalian non-cell-autonomous responses to nuclear genome instability comes from human cells that are kept in senescence as a result of persistent DNA damage. These cells halt mitotic divisions and develop a “senescence associated secretory phenotype” (SASP) characterized by a constant production of pro-inflammatory cytokines that might impact the proliferation of surrounding cells and modulate tissue remodeling (Rodier et al., 2009).
Due to higher organismal complexity of Drosophila and mammals when compared to C. elegans, it might be more difficult to establish direct mechanistic links between DNA damage, innate immune response, and protective quality control in these systems. Nonetheless, the involved pathways -namely MAPK-induced innate immune responses- are highly conserved throughout evolution.
Moreover, the requirement of innate immunity for tissue remodeling and regeneration was described in insects and mammals. In Drosophila JAK/STAT dependent production of cytokines was shown to be essential for the regeneration of the intestine upon infection and/or injury (Jiang et al., 2009). Similar involvement of cytokines in maintenance of infected and injured intestine was demonstrated in mice (Iizuka and Konno, 2011). Another example of components of the innate immune system playing a role in mammalian tissue maintenance is the involvement of immune cells and pro-inflammatory mediators in the wound healing response of the skin (Eming et al., 2014). The exact molecular events orchestrating recovery of normal cell homeostasis in all described cases are still incompletely understood. It is, however, plausible that protein quality control activation downstream of the innate immune response contributes to the preservation of tissue functioning similarly to its consequences in C. elegans. Given the conservation of the connection between DNA damage and innate immunity among species it is tempting to hypothesize that systemic quality control induction via pro-inflammatory factors as well as temporary systemic attenuation of somatic growth are conserved protective responses to local genome instability.
Indeed, the systemic growth attenuation has been observed in distinct DNA repair compromised mouse models that showed impaired postnatal growth and premature ageing. Mice with defects in Sirt6 or the NER components Csb and Xpc or Ercc1 showed reduced levels of IGF-1 (Mostoslavsky et al., 2006; Niedernhofer et al., 2006; van der Pluijm et al., 2006). Mechanistically, cells respond with somatotropic attenuation specifically to the persistence of transcription-blocking lesions (Garinis et al., 2009). Based on the prominent role of transcription-blocking lesions, RNAPII might survey the integrity of genes even in cell types that are postmitotic (Wolters and Schumacher, 2013). Transcription-coupled genome surveillance might thus orchestrate endocrine adjustments to DNA damage accumulation with ageing (Garinis and Schumacher, 2009; Schumacher, 2009). Systemic attenuation of the IGF-1/GH-mediated somatotropic axis in mammals might thus enhance maintenance of postmitotic tissues, however, possibly at the expense of tissue renewal that depends on the availability of endocrine growth factors (Behrens et al., 2014). Intriguingly, a recent study in C. elegans, which carries highly conserved TC-NER genes (Babu et al., 2014), has revealed that transcription-blocking DNA lesions lead to the activation of the FOXO transcription factor DAF-16 (Mueller et al., 2014). DAF-16 is negatively regulated by IGF-1 signaling and thus active when inhibitory IGF-1 signaling is attenuated. In response to DNA damage, DAF-16 could enhances the threshold of DNA damage-induced functional decline of tissues, suggesting that longevity assurance factors -such as DAF-16- might extend lifespan by increasing the DNA damage tolerance (Mueller et al., 2014).
Enhanced innate immune responses and systemic attenuation of the GH/IGF-mediated somatic growth axis similar to the prematurely aged DNA repair deficient animals has also been observed during natural aging (Schumacher et al., 2008b). It was hypothesized that the growth axis is attenuated in an attempt to antagonize the consequences of genome instability that otherwise might lead to cancer development (Schumacher et al., 2008a). However, the inflammatory responses to DNA damage might become detrimental and contribute to the tissue damage in the mice. Indeed, inhibition of pro-inflammatory signaling could alleviate various symptoms of premature aging in Hutchinson-Gilford-Progeria-Syndrome mice as well as in Ercc1 mutants (Osorio et al., 2012; Tilstra et al., 2012). Anti-inflammatory interventions might therefore offer useful treatment options for progeroid syndromes and aging-associated diseases.
Concluding remarks
The maintenance of the nuclear genome integrity is essential for the organism’s development and the maintenance of homeostasis throughout the lifespan. Given how frequently DNA damaging events occur, genome maintenance becomes a particular challenge during the process of aging where accumulation of DNA lesions is thought to contribute to the functional decline of cells and tissues. The critical role of genome instability in age-related deterioration of tissues has become particularly evident in various human progeroid syndromes and mice carrying mutations in various DNA repair genes. DNA repair machineries rely on precise temporal and spatial regulation, to which the cellular protein quality control mechanisms contribute. Failure to repair genomic lesions can result in cellular senescence and apoptosis of affected cells causing degeneration of tissues and functional decline. Cell-autonomous responses to DNA damage, however, insufficiently explain the complex pathological outcomes of genome instability for instance in progeroid syndromes. Growing evidence obtained from different animal models has established intriguing links between nuclear DNA damage and systemic adaptations that affect multiple organs. The innate immune responses that are mounted upon genotoxic stress have emerged as central mediators of non-cell-autonomous DNA damage responses. Evolutionary conserved MAP kinase pathways appear to play an important role in the secretion of cytokines in response to DNA damage. Direct connection between innate immunity and systemic induction of stress tolerance via activation of cellular protein quality control has recently been revealed in C. elegans. The molecular events underlying systemic adaptations to genome instability in higher organisms are as of yet less well understood. However, the involvement of cytokine secretion and its requirement for tissue maintenance and regeneration upon damage has become evident also in higher animals. Another important component of the systemic DNA damage response is the attenuation of IGF/insulin signaling (IIS), which inhibits further growth of affected organisms. The DNA damage-induced inhibition of IIS is mediated by cytokines interacting with the central nervous systems at least in Drosophila. The attenuation of growth in response to genotoxic stress may serve as a systemic checkpoint mechanism that halts metabolic and developmental processes to allow time for DNA repair and damage removal before resuming full tissue functionality. More studies using different animal models will be instrumental in further dissecting molecular events underlying systemic adaptation to genome instability. Given the complexity of metabolic signaling and immune responses in mammals, simple model organisms like Drosophila and C. elegans become particularly important. Drosophila provides the opportunity to study interactions between dynamically regenerating tissues during systemic response to DNA damage, while the systemic adaptation to genotoxic stress in postmitotic tissues is particularly well accessible in the nematode. Further studies should integrate information obtained from simple organisms with mammalian models of pre-mature as well as normal aging and tumorigenesis thus deepening the understanding of organismal adaptations to genome instability impacting cancer, immunity, the biology of ageing, and mechanisms of age-related diseases.
Acknowledgements
ME received the EMBO long-term fellowship, BS acknowledges funding from the Deutsche Forschungsgemeinschaft (CECAD, SFB 829, SFB 670, and KFO 286), the European Research Council (ERC Starting grant 260383), Marie Curie (FP7 ITN CodeAge 316354, aDDRess 316390, MARRIAGE 316964), the German-Israeli Foundation (GIF 1104-68.11/2010), the Deutsche Krebshilfe (109453), and the Bundesministerium für Forschung und Bildung (Sybacol FKZ0315893A-B).
References
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Babu V, Hofmann K, Schumacher B. A C. elegans homolog of the Cockayne syndrome complementation group A gene. DNA Repair (Amst.) 2014;24:57–62. doi: 10.1016/j.dnarep.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TA, Lopez-Moyado IF, Rifkin SA, Cuomo CA, Troemel ER. Ubiquitin-Mediated Response to Microsporidia and Virus Infection in C. elegans. PLoS Pathog. 2014;10:e1004200. doi: 10.1371/journal.ppat.1004200.s019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Behrens A, van Deursen JM, Rudolph KL, Schumacher B. Impact of genomic damage and ageing on stem cell function. Nat Cell Biol. 2014;16:201–207. doi: 10.1038/ncb2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Blastyák A, Pintér L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Molecular Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Carrard G, Bulteau A-L, Petropoulos I, Friguet B. Impairment of proteasome structure and function in aging. Int J Biochem Cell Biol. 2002;34:1461–1474. doi: 10.1016/s1357-2725(02)00085-7. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Taylor MRG, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- Daulny A, Tansey WP. Damage control: DNA repair, transcription, and the ubiquitin-proteasome system. DNA Repair (Amst.) 2009;8:444–448. doi: 10.1016/j.dnarep.2009.01.017. [DOI] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- de Winter JP, van der Weel L, de Groot J, Stone S, Waisfisz Q, Arwert F, Scheper RJ, Kruyt FA, Hoatlin ME, Joenje H. The Fanconi anemia protein FANCF forms a nuclear complex with FANCA, FANCC and FANCG. Hum Mol Genet. 2000;9:2665–2674. doi: 10.1093/hmg/9.18.2665. [DOI] [PubMed] [Google Scholar]
- Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry WB, Putzke AP, Rothman JH. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science. 2001;294:591–595. doi: 10.1126/science.1065486. [DOI] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: Mechanisms, signaling, and translation. Sci Transl Med. 2014;6:265sr6. doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Schumacher B. Insights from the worm: The C. elegans model for innate immunity. Seminars in Immunology. 2014a doi: 10.1016/j.smim.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Schumacher B. Systemic DNA damage responses: organismal adaptations to genome instability. Trends Genet. 2014b;30:95–102. doi: 10.1016/j.tig.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva MA, Segref A, Dakhovnik A, Ou H-L, Schneider JI, Utermöhlen O, Hoppe T, Schumacher B. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature. 2013;501:416–420. doi: 10.1038/nature12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol Cell. 2006;23:471–482. doi: 10.1016/j.molcel.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Garinis GA, Schumacher B. Transcription-blocking DNA damage in aging and longevity. Cell Cycle. 2009;8:2134–2135. [PubMed] [Google Scholar]
- Garinis GA, Uittenboogaard LM, Stachelscheid H, Fousteri M, van Ijcken W, Breit TM, van Steeg H, Mullenders LHF, van der Horst GTJ, Brüning JC, Niessen CM, et al. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol. 2009;11:604–615. doi: 10.1038/ncb1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillari J, Katinger H, Voglauer R. Aging and the ubiquitinome: Traditional and non-traditional functions of ubiquitin in aging cells and tissues. Exp Gerontol. 2006;41:1067–1079. doi: 10.1016/j.exger.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Groisman R, Kuraoka I, Chevallier O, Gaye N, Magnaldo T, Tanaka K, Kisselev AF, Harel-Bellan A, Nakatani Y. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 2006;20:1429–1434. doi: 10.1101/gad.378206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J-I, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–367. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- Hardeland U, Steinacher R, Jiricny J, Schär P. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Huen MSY, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka M, Konno S. Wound healing of intestinal epithelial cells. World J Gastroenterol. 2011;17:2161–2171. doi: 10.3748/wjg.v17.i17.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpac J, Younger A, Jasper H. Dynamic coordination of innate immune signaling and insulin signaling regulates systemic responses to localized DNA damage. Dev Cell. 2011;20:841–854. doi: 10.1016/j.devcel.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto R, Masutani C, Sugasawa K, Iwai S, Araki M, Uchida A, Mizukoshi T, Hanaoka F. Diversity of the damage recognition step in the global genomic nucleotide excision repair in vitro. Mutat Res. 2001;485:219–227. doi: 10.1016/s0921-8777(00)00082-3. [DOI] [PubMed] [Google Scholar]
- Laugel V, Dalloz C, Durand M, Sauvanaud F, Kristensen U, Vincent MC, Pasquier L, Odent S, Cormier-Daire V, Gener B, Tobias ES, et al. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31:113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- Lee C-H, Wu S-B, Hong C-H, Yu H-S, Wei Y-H. Molecular Mechanisms of UV-Induced Apoptosis and Its Effects on Skin Residential Cells: The Implication in UV-Based Phototherapy. Int J Mol Sci. 2013;14:6414–6435. doi: 10.3390/ijms14036414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhang L, Craddock J, Bruce-Keller AJ, Dasuri K, Nguyen A, Keller JN. Aging and dietary restriction effects on ubiquitination, sumoylation, and the proteasome in the heart. Mech Ageing Dev. 2008;129:515–521. doi: 10.1016/j.mad.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loser K, Beissert S. Regulation of cutaneous immunity by the environment: an important role for UV irradiation and vitamin D. Int Immunopharmacol. 2009;9:587–589. doi: 10.1016/j.intimp.2009.01.024. [DOI] [PubMed] [Google Scholar]
- Ludwig DL, MacInnes MA, Takiguchi Y, Purtymun PE, Henrie M, Flannery M, Meneses J, Pedersen RA, Chen DJ. A murine AP-endonuclease gene-targeted deficiency with post-implantation embryonic progression and ionizing radiation sensitivity. Mutat Res. 1998;409:17–29. doi: 10.1016/s0921-8777(98)00039-1. [DOI] [PubMed] [Google Scholar]
- Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, Sutherland A, Thorner M, Scrable H. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes \& development. 2010;24:1580–1589. doi: 10.1101/gad.1941710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JHJ. Understanding nucleotide excisionrepair and its roles in cancer andageing. Nat Rev Mol Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- Medhurst AL, Huber PA, Waisfisz Q, de Winter JP, Mathew CG. Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum. Mol Genet. 2001;10:423–429. doi: 10.1093/hmg/10.4.423. [DOI] [PubMed] [Google Scholar]
- Merker K, Sitte N, Grune T. Hydrogen Peroxide-Mediated Protein Oxidation in Young and Old Human MRC-5 Fibroblasts. Archives of Biochemistry and Biophysics. 2000;375:50–54. doi: 10.1006/abbi.1999.1657. [DOI] [PubMed] [Google Scholar]
- Moldovan G-L, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Molecular Cell. 2006;23:723–732. doi: 10.1016/j.molcel.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Mueller MM, Castells-Roca L, Babu V, Ermolaeva MA, Müller R-U, Frommolt P, Williams AB, Greiss S, Schneider JI, Benzing T, Schermer B, et al. DAF-16/FOXO and EGL-27/GATA promote developmental growth in response to persistent somatic DNA damage. Nat Cell Biol. 2014;16:1168–1179. doi: 10.1038/ncb3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ. The JAK-STAT Signaling Pathway: Input and Output Integration. The Journal of Immunology. 2007;178:2623–2629. doi: 10.4049/jimmunol.178.5.2623. [DOI] [PubMed] [Google Scholar]
- Ng JMY, Vermeulen W, van der Horst GTJ, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JHJ. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev. 2003;17:1630–1645. doi: 10.1101/gad.260003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–1043. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- Ortolan TG, Chen L, Tongaonkar P, Madura K. Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway. Nucleic Acids Res. 2004;32:6490–6500. doi: 10.1093/nar/gkh987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio FG, Bárcena C, Soria-Valles C, Ramsay AJ, de Carlos F, Cobo J, Fueyo A, Freije JMP, López-Otín C. Nuclear lamina defects cause ATM-dependent NF-κB activation and link accelerated aging to a systemic inflammatory response. Genes \& development. 2012;26:2311–2324. doi: 10.1101/gad.197954.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Ulrich HD. Mechanistic analysis of PCNA poly-ubiquitylation by the ubiquitin protein ligases Rad18 and Rad5. EMBO J. 2009;28:3657–3666. doi: 10.1038/emboj.2009.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Schumacher B. The p53 network: cellular and systemic DNA damage responses in aging and cancer. Trends Genet. 2012;28:128–136. doi: 10.1016/j.tig.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley R, Hudson J, Young PG. The wee1 protein kinase is required for radiation-induced mitotic delay. Nature. 1992;356:353–355. doi: 10.1038/356353a0. [DOI] [PubMed] [Google Scholar]
- Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer OD. DNA Interstrand Crosslinks: Natural and Drug-Induced DNA Adducts that Induce Unique Cellular Responses. ChemBioChem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- Schumacher B. Transcription-blocking DNA damage in aging: a mechanism for hormesis. Bioessays. 2009;31:1347–1356. doi: 10.1002/bies.200900107. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Garinis GA, Hoeijmakers JHJ. Age to survive: DNA damage and aging. Trends Genet. 2008a;24:77–85. doi: 10.1016/j.tig.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Gartner A. Translational regulation of p53 as a potential tumor therapy target. Future Oncol. 2006;2:145–153. doi: 10.2217/14796694.2.1.145. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee M-H, Nayak S, Volkmann K, Hofmann ER, Hofmann R, Hengartner M, Schedl T, Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hofmann K, Boulton S, Gartner A. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr Biol. 2001;11:1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- Schumacher B, van der Pluijm I, Moorhouse MJ, Kosteas T, Robinson AR, Suh Y, Breit TM, van Steeg H, Niedernhofer LJ, van Ijcken W, Bartke A, et al. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008b;4:e1000161. doi: 10.1371/journal.pgen.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Kohler I, Fellmann C, Lowe SW, Hengartner MO. HIF-1 antagonizes p53-mediated apoptosis through a secreted neuronal tyrosinase. Nature. 2010;465:577–583. doi: 10.1038/nature09141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- Sims AE, Spiteri E, Sims RJ, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, Auerbach AD, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, Elledge SJ. Identification of the FANCI Protein, a Monoubiquitinated FANCD2 Paralog Required for DNA Repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Darnell JE., Jr The JAK-STAT Pathway at Twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- Sung J-S, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006;273:1620–1629. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- Tilstra JS, Robinson AR, Wang J, Gregg SQ, Clauson CL, Reay DP, Nasto LA, St Croix CM, Usas A, Vo N, Huard J, et al. NF-κB inhibition delays DNA damage-induced senescence and aging in mice. J Clin Invest. 2012;122:2601–2612. doi: 10.1172/JCI45785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Hee PS, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Ulrich HD. Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair (Amst) 2009;8:461–469. doi: 10.1016/j.dnarep.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pluijm I, Garinis GA, Brandt RM, Gorgels TG, Wijnhoven SW, Diderich KE, de Wit J, Mitchell JR, van Oostrom C, Beems R, Niedernhofer LJ, et al. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2006;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves A-MF, Wollenberg AC, Stuart LM, Stormo GD, Irazoqui JE. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014;40:896–909. doi: 10.1016/j.immuni.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Wolters S, Ermolaeva MA, Bickel JS, Fingerhut JM, Khanikar J, Chan RC, Schumacher B. Loss of Caenorhabditis elegans BRCA1 promotes genome stability during replication in smc-5 mutants. Genetics. 2014;196:985–999. doi: 10.1534/genetics.113.158295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters S, Schumacher B. Genome maintenance and transcription integrity in aging and disease. Front Genet. 2013;4:19. doi: 10.3389/fgene.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Damage repair DNA polymerases Y. Curr Opin Struct Biol. 2003;13:23–30. doi: 10.1016/s0959-440x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- Zhuang Z, Johnson RE, Haracska L, Prakash L, Prakash S, Benkovic SJ. Regulation of polymerase exchange between Poleta and Poldelta by monoubiquitination of PCNA and the movement of DNA polymerase holoenzyme. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5361–5366. doi: 10.1073/pnas.0801310105. [DOI] [PMC free article] [PubMed] [Google Scholar]