Abstract
Primitive streak formation in the chick embryo involves large scale highly coordinated flows of over 100.000 cells in the epiblast. These large scale tissue flows and deformations can be correlated with specific anisotropic cell behaviours in the forming mesendoderm through a combined light-sheet microscopy and computational analysis. Relevant behaviours include apical contraction, elongation along the apical-basal axis followed by ingression as well as asynchronous directional cell intercalation of small groups of mesendoderm cells. Cell intercalation is associated with sequential, directional contraction of apical junctions, the onset, localisation and direction of which correlate strongly with the appearance of active Myosin II cables in aligned apical junctions in neighbouring cells. Use of a class specific Myosin inhibitors and gene specific knockdowns show that apical contraction and intercalation are Myosin II dependent and also reveal critical roles for Myosin I and Myosin V family members in the assembly of junctional Myosin II cables.
Gastrulation is a key event in the development of higher organisms. In amniotes this process is characterised by the formation of the primitive streak, a structure through which the mesendoderm cells ingress to form the deeper layers of the embryo 1. Before streak formation the embryo consists of a sheet of epithelial cells with a well-developed apical-basal polarity. Cells are connected by apical tight and adherens junctions, while at the basal side a developing basal membrane separates the cells from the forming hypoblast2–5. In chick embryos streak formation involves large scale vortex-like tissue flows that transport the mesendoderm precursors located in the epiblast at the interface between the extra embryonic area opaca and the embryonic area pellucida into the central midline of the embryo 6–9 (Fig.1d). There has been considerable speculation about the cellular mechanisms driving these large scale tissue flows 10. Experiments so far have relied on labelling a small subset of cells and following their movements during streak formation 7, 9, 11. Based on these observations several hypotheses, oriented cell divisions, intercalation of cells in the streak region, chemotaxis of subpopulations of cells, movement of the extracellular matrix, localised ingression of cells into the hypoblast, have been put forward to explain tissue flows during streak formation 7–9, 12–15. Progress has been impeded by lack of a detailed description of the epiblast cell behaviours underlying streak formation, due to absence of methods to investigate the behaviour of the >100,000 cells in the 4 mm diameter epiblast disk at cellular resolution and good methods to identify all cells.
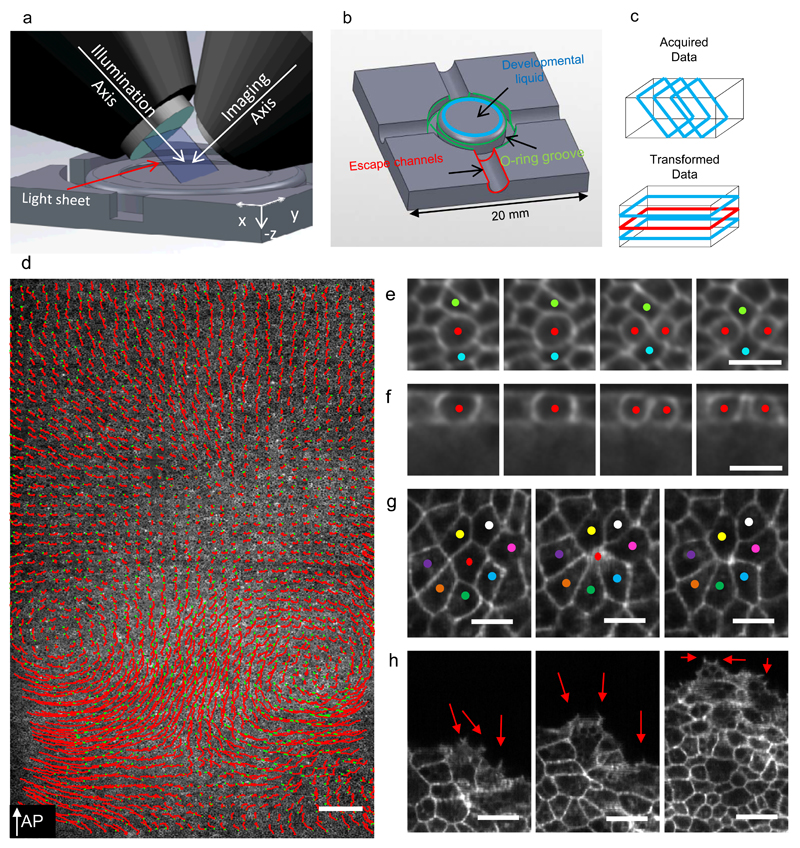
Figure 1. Light-sheet microscopy setup to study gastrulation in chicken embryos.
a-) Schematics of the light-sheet microscope for large flat samples, the illumination and imaging objectives positioned at 45° to the embryo’s surface. Successive 45° cross-sections are collected by moving the embryo through the light-sheet. b-) Sample plate designed to keep the early embryo flat and isolated from external environment. c-) Schematic representations of acquisition geometry (images marked with blue rectangles) and the transformed data for analysis. d-) Single Z plane (red rectangle in C) overlaid with the cell tracks of 5% of all cells over a 180 minute time interval shown as red lines with green dots indicating their final positions (7950×2560 pixels, 5.17mm×1.66mm) (Supplementary Video 1). e-) Four frames 5 minutes apart showing a dividing cell (red dots) and its local neighbourhood shown at full recorded resolution (blue dots) (Supplementary Videos 2,3). f-) Cross-section through the dividing cell seen in E. g-) Three frames 10 minutes apart showing an ingressing cell (red dot, Supplementary Video 4). h-) Three frames 20 minutes apart showing embryo expansion driven by Area Opaca boundary cells making active protrusions (red arrows). The AP arrow in D indicates the direction of the anterior posterior axis, the white scale bar in D is 200 µm. The white scale bars in E, F, G and H are 25 µm in length.
In order to address these problems we have developed a transgenic chick line in which the cell membranes of all cells in the embryonic and extra embryonic tissues are labelled with a green fluorescent protein tag (myr-EGFP), allowing a detailed characterisation of cell behaviours. We have furthermore built a dedicated Light Sheet Microscope (LSM) especially designed to image these large fragile flat live tissue samples 16–18. We also have developed new methods that allow us to culture the early chick embryos in liquid with the epiblast side up, conditions required to take advantage of the high resolution long working distance immersion optics of the lightsheet microscope. We have developed and implemented computational methods to characterise the large scale tissue flows and automated segmentation and tracking methods to characterise cell behaviours streak formation, allowing us to correlate tissue and cell behaviour on the scale of the embryo 19, 20.
Results
LSM imaging of streak formation
To image the majority of cells in the early chick embryo we have designed and constructed a LSM (fig.1a), devised a dedicated liquid culture method (Fig. 1b) optimised for recording the early development of chick embryos at cellular resolution at 2-3 minute time-intervals (Fig. 1d,e,g,h). A key feature of the LSM is an automatic height adjustment that continuously calculates the position of the surface of the embryo and through dynamic feedback to the microscope stage keeps it the focus of the light sheet during the scanning process, resulting in images with optimal resolution. The LSM enables us to see all the cells in a 1.66 mm stripe across the embryo as a 3D data stack (typically ~8000×2560×500 voxels) at excellent cellular resolution (Fig. 1d, Supplementary Video 1, 2). Image series at full spatial and temporal resolution are available at DOI 10.15132/10000100 to be viewed via an Omero dataviewer 21. A major challenge is to analyse these large data sets. We analyse tissue behaviour using Particle Image Velocimetry (PIV)7, 11, 22, 23 and automated segmentation and tracking algorithms to analyse detailed cell behaviours of epiblast cells. PIV provides information about the local velocity of small approximately cell sized tissue regions (10 × 10 µm) and allows calculation of detailed local spatiotemporal behaviours of these domains, such as movement and deformation over time.
We have used the LSM to obtain a detailed characterisation of cell and tissue behaviours during primitive streak formation in Myr GFP embryos. From the start of development the behaviour of cells in the epiblast is highly dynamic and characterised by frequent cell divisions, cell shape changes and ingressions (Fig. 1e,f,g,h, Supplementary Videos 3,4). Cells at the Area Opaca boundary extend active lamelopodia and filopodia while actively moving outward over the vitelline membrane, keeping the growing embryo under tension (Fig. 1h) 24. Tracking of epiblast cells over time confirms the existence of large scale vortex cell flows during streak formation (Fig. 1d, 2c) 7–9. We have used PIV to fate map the tissue regions that give rise to the streak, by marking the streak outline at the extended streak stage and following the tissue backward in time to the start of the experiment (Fig. 2a, Supplementary Video 5, Supplementary Fig. 1A). This procedure identified a sickle shaped area in the posterior area pellucida epiblast as the precursor region to the streak, a domain essentially congruent with known expression domains of mesoderm specific genes 25, 26.
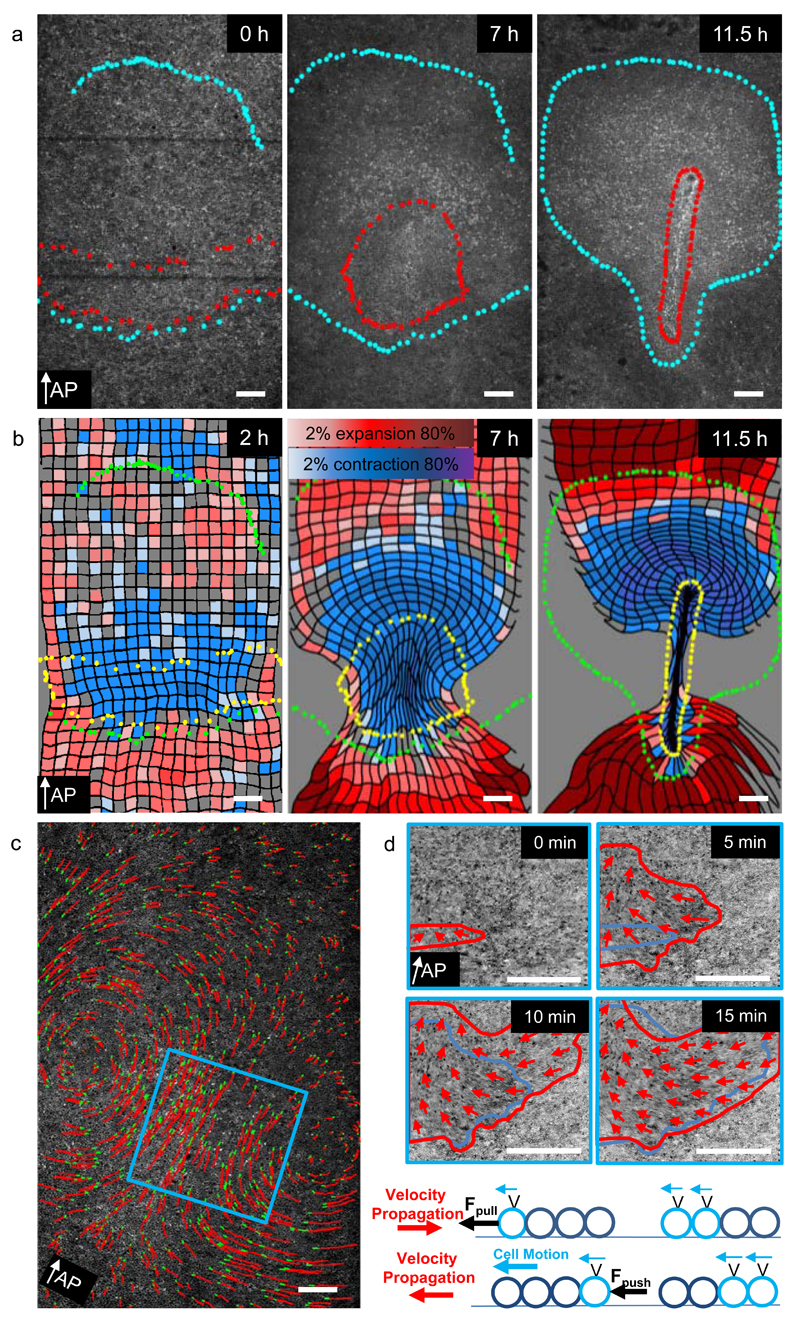
Figure 2. Tissue dynamics using PIV analysis.
a-) Three time points illustrating formation of the primitive streak. Cells contained within red and blue dotted boundary were marked at the streak stage (11.5 h) and tracked backwards to reveal their origin at 0h. White arrow indicates direction of AP axis from posterior to anterior. b-) Three panels illustrating the contraction expansion fate map calculated using the velocity fields as described in Methods. c-) Development of tissue motion pattern over time. Left panel shows the cell flow pattern midway during streak formation. Red lines indicate cell tracks of 5% of randomly selected cells over a 150 minute time interval with green dots indicating their final positions. The blue square indicates the location used to analyse the onset of motion. Each image in the right panel is a maximum intensity projection of 5 consecutive frames (10 min) 5 minutes apart highlighting actively moving cells. Red arrows indicate direction of the tissue flow. Red lines mark the area of moving cells at the current time, while the blue line shows the area moving cells at the previous time point. The area of actively moving cells expands from medial to lateral. The schematic drawing illustrates possible scenarios for cell motion and velocity propagation depending on the type of active force. In case of a pulling force (upper schematic) the onset of motion propagates in a direction opposite to the direction of motion, while in case of a pushing force (lower schematic) onset of cell motion and movement are oriented in the same direction. The white scale bars are 200 µm in length
Tissue deformations during streak formation
To analyse the deformations of the mesendoderm tissue over time we have used the PIV derived velocity fields to calculate the changes in shape of initially square tissue domains. Contracting domains are coloured blue and expanding domains are coloured red (Fig 2b, Supplementary Fig 1b, Supplementary Videos 6, 7). Contrary to expectation the mesendoderm is strongly contracting (Fig 2b, Supplementary Fig 1b), while undergoing a characteristic biphasic deformation during streak formation. Domains located in front of the extending streak, are also seen to contract, while elongating perpendicular to the direction of streak extension (Fig. 2b). Averaging of the deformations of 9 embryos shows the consistency of these deformation patterns (Supplementary Fig. 2, Supplementary Video 8). The onset of tissue motion reveals that it starts in the central mesendoderm region and then rapidly spreads lateral, while the motion of the tissue (red arrows) is in the opposite direction, i.e. towards the streak (Fig. 2d), implying that the mesendoderm tissue is pulled from the centre, rather than being pushed from the lateral sides (Fig. 2d).
To further characterise the mesendoderm deformation we have calculated spatiotemporal distribution of the isotropic and anisotropic strain rate components from the PIV derived velocity fields. The isotropic strain rate component describes uniform tissue expansion or contraction rate, the anisotropic strain rate component describes the shearing rate. At the onset of the tissue motion the mesendoderm starts to contract (blue circles) and shear strain rate in the direction of contraction increases in the direction towards the midline (blue lines). Both processes increase during development (Fig 3a, Supplementary Fig1C, Supplementary Videos 7, 9).
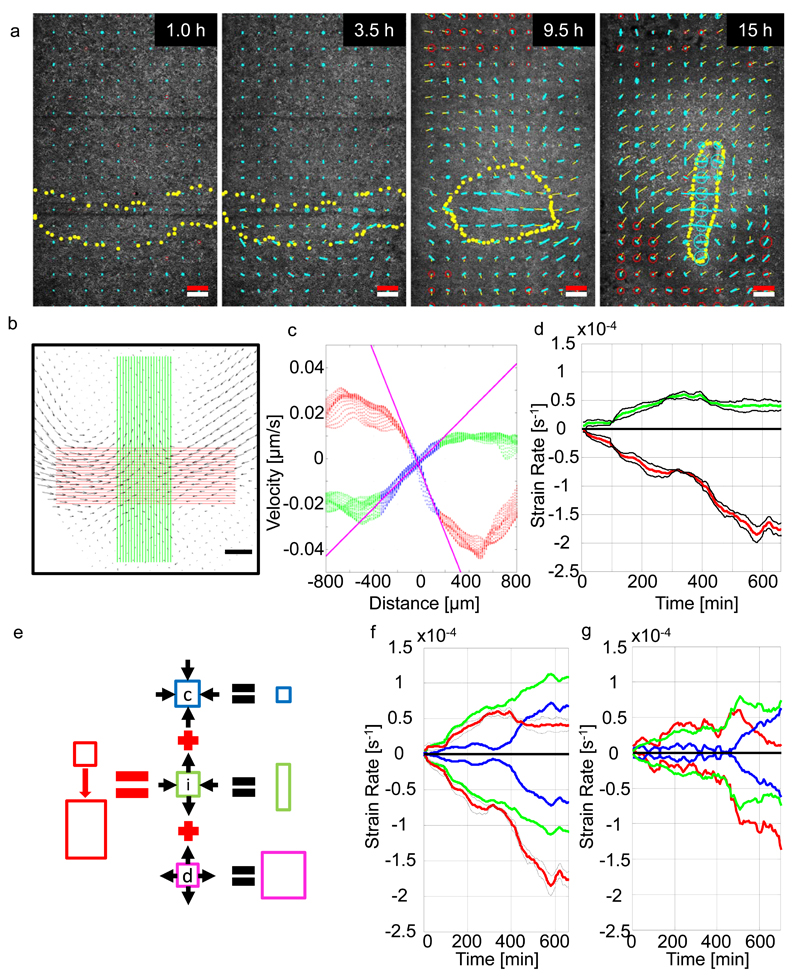
Figure 3. Analysis of strain rates during streak formation.
a-) Evolution of the expansion/compression strain rate of the strain rate tensor and the shear strain rate of the strain rate tensor calculated as described in methods during streak formation. The expansion contraction rates of the strain tensor are shown as circles; blue indicates contraction, red expansion. The anisotropic part indicating the shear strain rate is shown as a line blue line in the direction of contraction. b-) Velocity field at t = 600 min. Green and red lines indicate location of the velocity field vectors used for analysis. Black scale bar is 200 µm. c-) Velocity components as a function of distance along green and red lines in A. Red and green dots indicate velocity components from red and green areas. Blue dots mark fitting range used to determine the strain rate (spatial velocity gradients). Slopes of fitted magenta lines are tissue strain rates. d-) Mean tissue strain rates and standard errors as a function of developmental time of 4 experiments. Red and green line indicate tissue strain rate perpendicular and parallel to the streak respectively. e-) Cellular events driving deformation of epithelial tissue. Contraction/ingression (blue square), cell intercalation (green) and cell growth (magenta). Arrows indicate direction of the tissue flows generated by these processes. Panels E-G show tissue strain rate (red lines) for: f-) wild type embryos, g-) an embryo treated with 50 µM H1152. The tissue strain rates are decomposed into the isotropic part (apical contraction/ingression and cell growth, blue lines) and anisotropic part (intercalation, green lines). The white scale bar in panel a is a 200 µm size marker, the red scale bar in panel a indicates a strain rate of 10-4/sec and a tissue domain velocity of 4 µm/min.
To analyse this behaviour in several embryos we calculated the average strain rate in 4 embryos in the direction perpendicular to the streak axis and along the streak as a function of time (Fig. 3b, c). The strain rate perpendicular to the streak (red curve) is always larger than the strain rate along the streak (green curve) (Fig 3d). Figure 3e explains how isotropic processes, such as uniform tissue contraction, uniform expansion and anisotropic processes contribute to the overall deformation of the tissue 20, 27. Separation of the strain rate in its isotropic and anisotropic components shows that the anisotropic strain rate increases continuously over time, while the isotropic strain rate increases only slowly at first, but then increases more rapidly at around ~6 hours of development, the onset of streak extension (Fig. 3f). The isotropic contraction processes accelerate the movement in the direction of the streak and counteract the expansion of the streak (Fig. 3b, d, f).
Cellular behaviours driving tissue deformations
To visualise local cell behaviours, the images are segmented and specific groups of cells are visualised in a moving frame of reference, allowing investigation of their behaviours for extended periods of time. Manual verification of the automatic segmentation and tracking data, in four selected regions in the embryo, has provided us with gold standard data (Fig. 4a,b,c). The regions analysed include a region initially close to the centre of the mesendoderm (blue)(Fig 4b), a region in the lateral mesendoderm (black) moving towards the streak, a region in the posterior area opaca (red) and a region in front of the streak (green) (Fig. 4a). To understand which cell behaviours contribute to the observed tissue flows we have analysed the following cell behaviours:
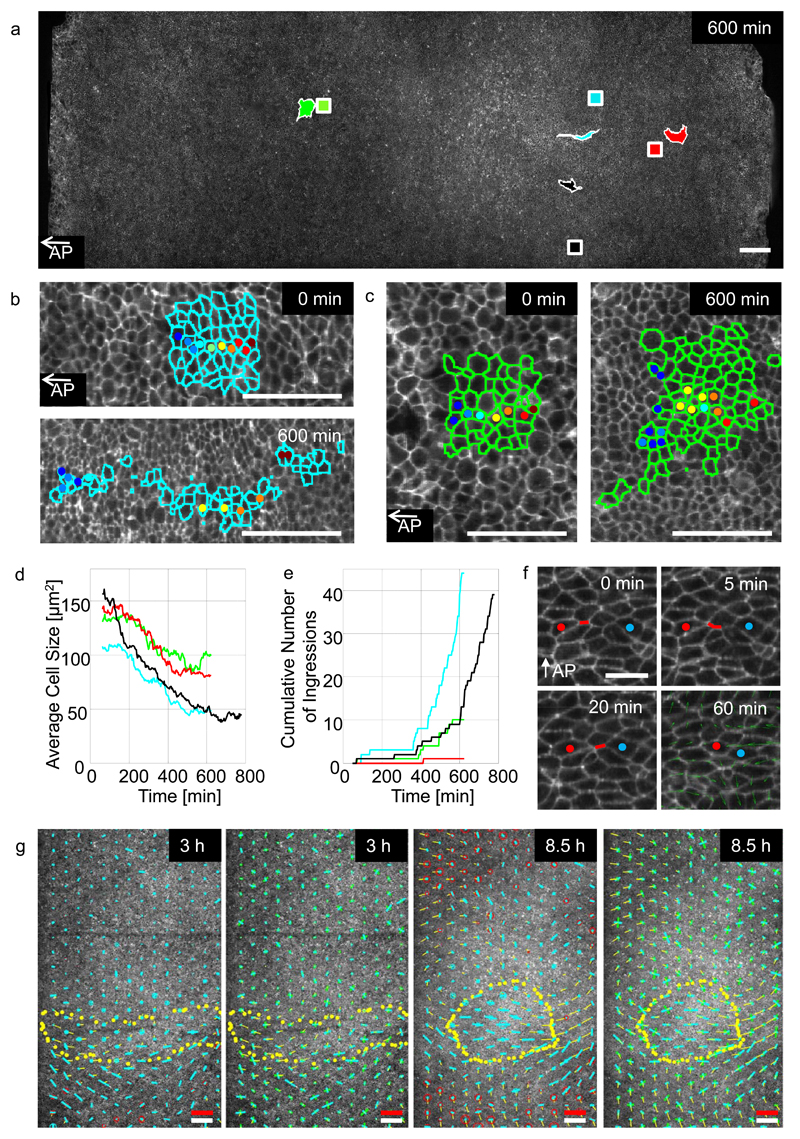
Figure 4. Analysis of cell behaviours controlling gastrulation.
a-) Analysis of cell behaviour in four distinct regions marked with different colour squares: the middle (blue) and lateral (black) sickle region, the area opaca (red) and a region in front of the sickle (green). The initial position of the domains are shown as squares, the same coloured irregular shaped domains indicate their positions and shape after 600 minutes. Scale bar 200 µm. b-) Shows outlines of cells manually tracked in the middle sickle domain (blue square) at the start 0 and after 600 minutes respectively. Scale bar 100 µm. c-) Cells tracked in the anterior domain (green) at 0 and 600 minutes respectively. Scale bar 100 µm. d-) Average cell size in the four domains (colours as in A). e-) Ingression of cells in the same four regions. f-) Example of sequential junctional contraction event in sickle region. The successive contractions generate a pulling force perpendicular to the streak axis and a pushing force along AP axis as shown by the velocity field (green arrows, averaged over 60 min). Scale bar 25 µm. g-) Quantitative analysis of the contraction expansion (red/blue circles) rate and shear rate (blue bar in the direction of contraction) of the total tissue strain rate at 3 and 8.5 hours of development (first and third panel). The strain rates are calculated from the changes in the lengths and direction of the vectors linking the centroid positions of all cells with their immediate neighbours as described in methods. The strain rate contribution of the rate of cell shape change (green bars) and the rate of cell rearrangement (blue bars) shown in the direction of contraction (second and fourth panel), are shown in the second and 4th panel. The dotted yellow line indicates the outline of the deforming endoderm. The thin yellow lines indicate the instantaneous velocities of the tissue at the time the strain rate tensors were calculated. The white scale bar in is 200 µm size marker, the red scale bar indicates a strain rate of 10-4/sec and a tissue velocity of 4 µm/min.
Apical contraction and ingression
The apical cross-sectional area of cells in all areas decreased significantly over time (40-65% depending on position) (Fig. 4b,d, Supplementary Fig. 3a), however measurement of the radii of the mitotically rounded cells showed that cell volumes changed only slightly (<10%) during streak formation (Supplementary Fig.3b,c). This implies that mesendoderm cells elongate along their apical-basal axis, a process that continues until they start to ingress in the streak a process starting at around 6-7 hours after the start of the experiment (Fig. 4e).
Division
Cells divide on average every 6 hours everywhere in the embryo (Supplementary Fig. 4a), but the cell division rates are not constant in time (Supplementary Fig.4b). We note a rise in the number of cell divisions at the time of the initiation of streak elongation in all experiments, the significance of which is presently unknown (Supplementary Fig. 4a,b).
Intercalation
Cells in different domains showed marked differences in their local neighbourhood dynamics. Cells in the streak forming region showed extensive directional rearrangements (Fig. 4b), while cells in outside the mesendoderm did not (Fig. 4c). Within the sickle region cells rearrange by directional sequential contraction of aligned apical junctions involving 2 - 6 cells (Fig. 4f, Supplementary Videos 10, 11). These junctional contraction events result in local cell rearrangements and are aligned towards the midline, thereby contributing to the initial mesendoderm tissue contraction and expansion of the forming streak (fig 4g).
Alignment
A marked alignment of mesendoderm cells in the direction of motion is already detectable before the onset of tissue motion. This alignment increases during the early stages of sickle contraction but then dissipates (Supplementary Fig 5a,b). Cell divisions are also seen to align in the mesendoderm in the direction of motion especially in the early phases of streak formation (Supplementary Fig. 4c).
Statistical analysis of cell behaviours
We have used recently developed statistical techniques to quantify the different cell behaviour changes over time 20, 28. Calculation of the cell based analogue of the isotropic and anisoptropic tissue strain rate components (Fig. 4g, panel 1&3, Supplementary Video 12) show them to be in excellent quantitative agreement with the PIV based strain rates (Fig. 3a, Supplementary Video 9). These calculations also show that the onset of motion of the mesendoderm is accompanied by an increasing contraction rate of cells in the anterior mesendoderm (Fig. 4g, blue circles in first and third panel, Supplementary Fig 5b) as well as an strong increase in magnitude and alignment of the shear strain rate in the direction of contraction of the mesendoderm (Fig. 4g, blue bars in first and third panel). The anisotropic strain rate can be decomposed into the contributions made by the cell deformation rate (shape change) and the cell rearrangement rate (intercalation) (Fig. 4g, panel 2&4, Supplementary Video 12). Before the onset of motion the cell shape changes (Fig. 4g green bars, second and fourth panel) and directional cell rearrangements (Fig. 4g blue bars, second and fourth panel) are of similar magnitude and arranged in perpendicular orientation, thereby cancelling each other’s effects on local tissue rearrangement. At around 3.5 hr the directional cell rearrangements become more prominent in the mesendoderm resulting in the onset of motion and contraction of the mesendoderm (Fig. 4g panel 2&4, Supplementary Video 12). From around 9 hours the time the streak start to extend it can be seen that the tissue in front of the streak shows increasing intercalation along the direction of streak extension (Fig. 4g panel 2&4, Supplementary Video 12) explaining the observed compression and lateral extension of the tissue in this area at this time of development (Fig. 2b).
The role of Myosins in controlling cell behaviours
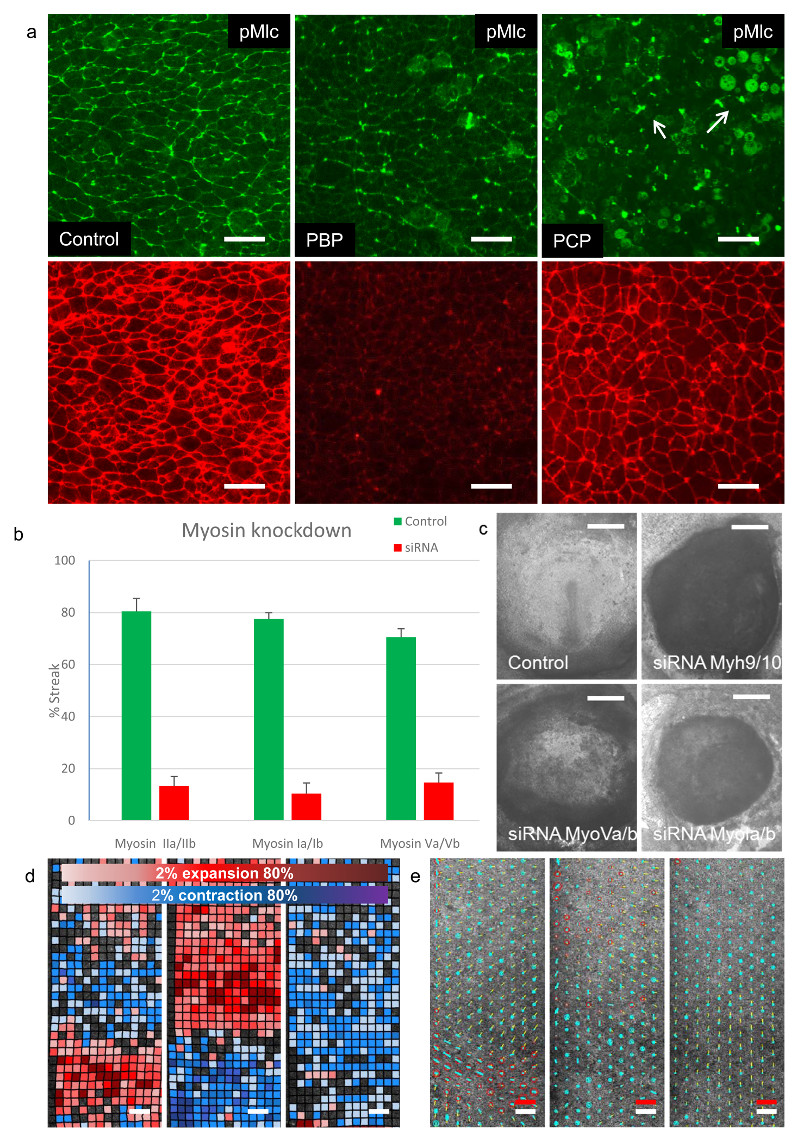
During Drosophila gastrulation apical junctional contraction and cell intercalation are mediated by Myosin II 29–32. This prompted us to investigate the patterns of Myosin II activation via detection of the phosphorylation of the Myosin light chain (Mlc)23, 29. In the posterior embryo we detected active Myosin II in apical cell junctions, organised in supercellular patterns of Myosin cables spanning 2 -8 cells (Fig. 5a,b, Supplementary Fig 6a). These Myosin cables appear at the time mesendoderm starts to contract and are aligned along the axis of contraction (Fig. 5b). Application of Blebbistatin a specific Myosin II inhibitor, 7, 33 showed neglible effects on streak formation at concentrations up to 50µM and little effect on Mlc phosphorylation (Supplementary Fig. 6a,b) 7. In Drosophila the asymmetric apical accumulation of Myosin II is dependent on the segment polarity genes and controlled by interactions with the PAR3 system in a RhoGef2 and Rho kinase dependent manner34–36. We found no detectable asymmetry in apical junctional localisation of Par3 or the PDZ RhoGef (AHRGef11), the RhoGef2 homologue in the chick embryo (Supplementary Fig. 7a,b,c). The Rho kinase inhibitor H1152 37 reduced Mlc phosphorylation in apical cell junctions and partially impaired tissue flows resulting in embryos with short fat streaks (Supplementary Fig. 6a,b). Quantitative analysis showed that H1152 partially inhibited both the anisotropic as well as the isotropic strain rate (Fig. 3g). These results could suggest the involvement of other signalling pathways or other Myosins. A global analysis of Myosin expression in an RNA seq experiment showed that several members of the Myosin I, Myosin II and Myosin V families were expressed at detectable levels during early development (Table 1). To investigate the possible involvement of members of the Myosin I, Myosin II and Myosin V families in streak formation we used two potent Inhibitors, Pentachloro- and Pentabromo-pseudilin. Pentabromopseudilin (PCB) inhibits Myosin V family members and to a lesser extent Myosin II family members (IC50 1.2 µM and 28 µM respectively) 38 39, while Pentachloropseudilin (PCP) very specifically inhibits members of the Myosin I family (IC50 1-5 µM, IC50 for class II and class 5 Myosins >95 µM)40. PCB at 5 µM showed a very strong inhibitory effect on streak formation, and resulted in a significant decrease in Myosin II light chain phosphorylation and cable formation (Fig. 6a). PCP showed an immediate inhibition of tissue motion as well as very strong inhibition of Mlc phosphorylation (Fig. 6a, Supplementary Video 13). It strongly inhibited the association of Myosin Ib with apical junctions, but had only little effect on the association of Myosin Va with apical junctions in the mesendoderm (Supplementary Fig 6c,d). PCP completely blocked apical contraction, intercalation and streak formation and resulted in a complete block of tissue flow (Fig. 6d, e, Supplementary Videos 13, 14, 15).
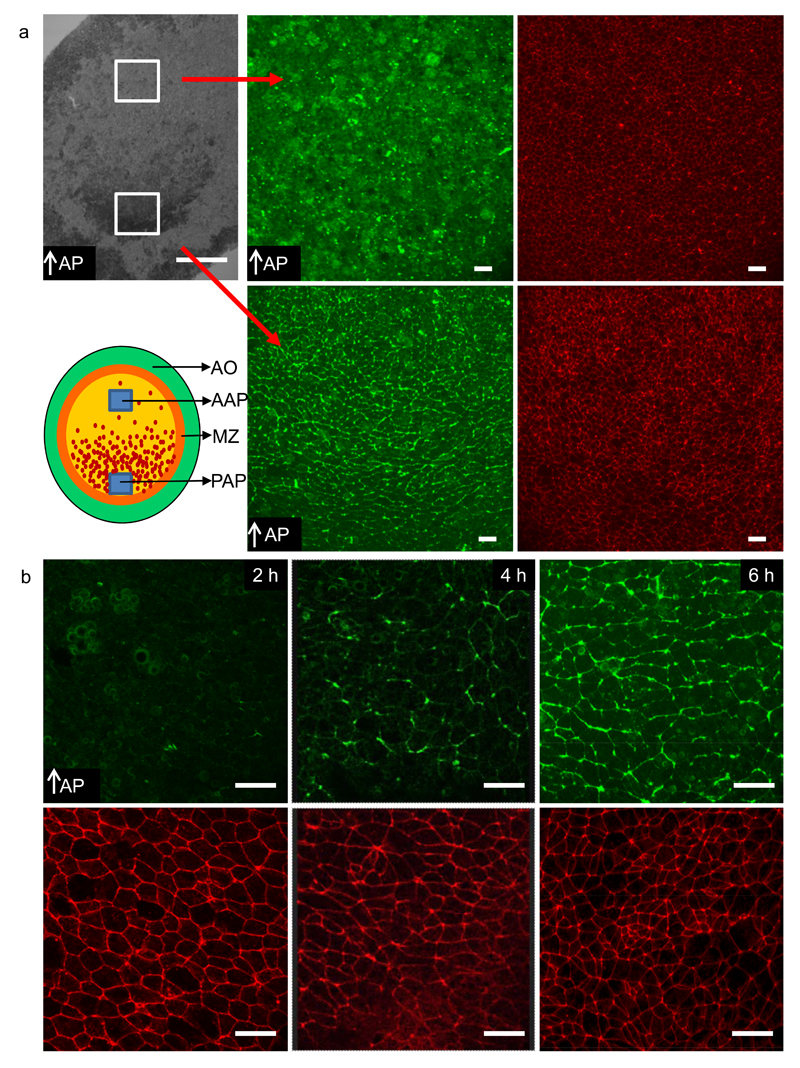
Figure. 5. Spatiotemporal localisation of phosphorylated Myosin light chain cables.
a-) Phosphorylated Myosin light chain (pMlc) (green) and actin (red) staining reveals the existence of supercellular active Myosin cables in the posterior are pellucida (sickle region) aligned perpendicular to the axis of streak elongation. pMlc cables are absent in the anterior area pellucida. The red arrows link the areas in the overview image of the embryo with the two high magnification images of pMlc staining in the right hand side panels of a. b-) pMlc staining (green) and actin staining (red) in the posterior area pellucida for embryos at different stages of development (2h, 4h and 6h). Appearance of the pMlc cables coincides with initiation of cell motion. The data shown are representative for results obtained in 3 experiments with 12- 20 embryos in each experiment. Scale bar in first panel is 1 mm, the white scale bars in all other panels are 25 µm.
Table 1. Analysis of RNA expression of Myosin family members.
Table shows the relative expression of all detected Myosin heavy chain family members as well as MyosinII light chains at various times of development in a RNA seq experiment performed as detailed in methods. 0 hrs represents freshly laid unincubated eggs, 5 hrs represents stage EGXIII, 10 hrs is stage HH2, 15 hrs stage HH3+, 20 hrs stage HH4. RKPM is denotes relative expression level (reads per kilobase per million reads).
| Feature ID | Description | RKPM | RKPM | RKPM | RKPM | RKPM |
|---|---|---|---|---|---|---|
| 0 hrs | 5hrs | 10 hrs | 15 hrs | 20hrs | ||
| ENSGALT00000020472 | MYH9 (Myosin IIa) | 98.21 | 256.26 | 294.09 | 143.99 | 202.84 |
| ENSGALT00000034774 | MYL9 (Myosin Light Chain 2) | 28.31 | 156.29 | 110.16 | 26.43 | 67.04 |
| ENSGALT00000008744 | MYL3 (Myosin Light Chain 1) | 37.48 | 86.09 | 118.94 | 11.54 | 29.19 |
| ENSGALT00000012496 | MYO1B | 36.61 | 67.18 | 51.03 | 35.19 | 37.8 |
| ENSGALT00000007538 | MYO1A | 9.69 | 31.77 | 45.52 | 3.98 | 7.59 |
| ENSGALT00000023397 | MYH10 (Myosin IIb) | 9.82 | 20.91 | 17.52 | 21.13 | 16.61 |
| ENSGALT00000030933 | MyL12A (Myosin Light chain, smooth muscle isoform) | 4.46 | 10.07 | 4.77 | 1.93 | 2.09 |
| ENSGALT00000008228 | MYO1C | 2.36 | 3.64 | 1.92 | 3.5 | 2.51 |
| ENSGALT00000039066 | MYO5A | 1.9 | 2.98 | 2.44 | 2.46 | 1.82 |
| ENSGALT00000007393 | MYO5C | 0.89 | 0.89 | 1.1 | 0.29 | 0.36 |
| ENSGALT00000031656 | MYO6 | 0.03 | 0.15 | 0.12 | 0.05 | 0.03 |
| ENSGALT00000009051 | MYO1G | 0.11 | 0.13 | 0.24 | 0.07 | 0.08 |
| ENSGALT00000022380 | MYH7 | 0.04 | 0.06 | 0.05 | 0.04 | 0 |
| ENSGALT00000001184 | MYH13 | 0.02 | 0.06 | 0.07 | 0.02 | 0.05 |
| ENSGALT00000010535 | MYH11 | 0 | 0.02 | 0 | 0.02 | 0 |
| ENSGALT00000000816 | Myosin light chain, embryonic (L23) | 0.24 | 0 | 0.11 | 0 | 0 |
Figure 6. Effect of inhibition of convention and non-conventional Myosins on streak formation.
a-) Phosphorylated Myosin light chain (green) and actin staining (red) for a control embryo at 8hrs of development (left panel), an embryo developed for 6hrs in EC culture followed by a treatment with 5µM pentabromopseudilin (PCB) for 2 hours showing partial inhibition of Myosin light chain phosphorylation (middle panel), an embryo developed for 6hrs in EC culture followed by a treatment with 5µM pentachloropseudilin (PCP) for 2 hours showing complete inhibition of Myosin light chain phosphorylation (right panel). White arrows indicate pMLC localisation in contraction furrow of dividing cells in the PCP treated embryo. The results shown are representative for 3-5 experiments with n> 20 embryos in control and treatment. All embryos were visually inspected by fluorescence microscopy and at least 1 control and 1 inhibitor treated embryo was analysed in detail by confocal microscopy. b-) Effects of knockdown of different classes of Myosins on streak formation. Results shown are means and standard deviations of Myosin IIa/IIb (control n=95 embryos/7 independent experiments, siRNA n=59 embryos/7 independent experiments,), Myosin Ia/Ia (control n=111 embryos/10 independent experiments, siRNA knockdown n=90 embryos/10 independent experiments) and Myosin Va/Vb (control n=31 embryos/3 independent experiments, siRNA n=41 embryos/3 independent experiments) on streak formation 18rs after transfection with the respective siRNA’s. c-) Images of typical embryos 18hrs after transfection with control or specific siRNA’s. The controls mostly develop streaks, the Myosin knockdowns typically do not as indicated in b. Knockdown of Myosin IIa/IIb and Myosin Ia/Ib results in very contracted embryos, knockdown of Myosin Va/Vb results in embryos that expand but the majority do not develop streaks. Scale bar 1 mm. d-) Contraction/expansion map of an embryo before and after treatment with 5µM pcp. First panel 5 hrs of development before treatment. Middle panel 30 minutes after pcp addition, right panel 5hrs after PCP addition. Note the immediate relaxation of the epiblast after PCP addition followed by a later contraction of the embryo. e-) Analysis of the isotropic and anisotropic shear strain rate of the embryo shown in d. Analysis was performed as described for the embryo shown in Fig. 3A. Note the expansion of the tissue and the loss of shear strain after pcp addition, followed by contraction 5hrs after addition of PCP. The white scale bar represent 200µm, the red scale bar indicates a strain rate of 10-4/s and an instantaneous tissue speed of 4 µm/min.
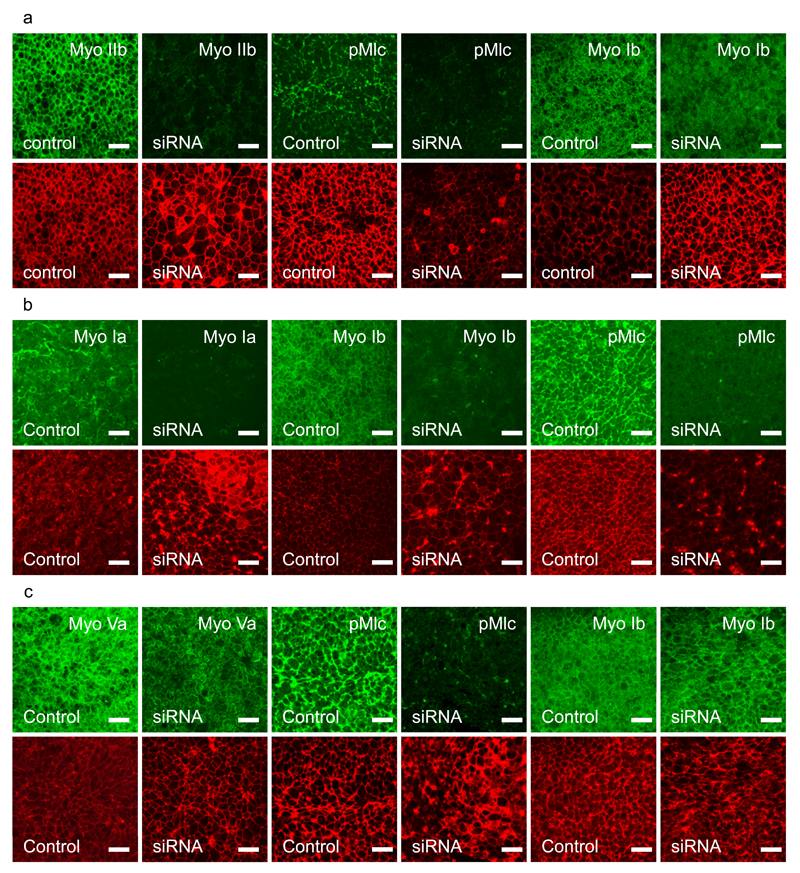
We down regulated the expressed Myosins using a directed siRNA approach to independently verify their effects on streak formation (Fig. 7). Simultaneous downregulation of Myosin IIa/IIb blocked streak formation and led to a strong reduction of Mlc phosphorylation in apical junctions in the mesoderm cells without a significant effect on Myosin I localisation (Fig. 7a). Myosin IIA and/or IIBIIB are evidently key mediators of streak formation. Observation of cells behaviours in the LSM showed that 5-10 hours after siRNA transfection the tissue flows stopped and the embryos started to contract (Supplementary Video 16). Simultaneous downregulation of Myosin Ia/Ib also strongly inhibited streak formation and resulted in strong inhibition Mlc phosphorylation (Fig. 7b). Knockdown of Myosin Ia/Ib resulted in the formation of large actin rich protrusions and hair-like structures especially at the cell boundaries (Fig. 7B). Knockdown of Myosin Va/b was less effective as judged by Myosin Va antibody staining, but still resulted a significant loss of Mlc phosphorylation, while having little effect on Myosin Ib localisation (Fig. 7c). Altogether the results of the knockdown of Myosin Ia/Ib and Myosin Va/Vb are in line with the observations obtained using their chemical inhibitors PCP and PCB respectively. The experiments highlight unexpected roles for unconventional Myosins in controlling cell behaviours resulting in streak formation and suggest that these unconventional Myosins most likely act through effects on Mlc phosphorylation.
Figure 7. Effects of siRNA mediated Myosin I, II and V downregulation.
a-) Effect of simultaneous Myosin IIa and Myosin IIb knockdown. Upper panels show from left to right, expression of Myosin IIb in control embryos, after Myosin IIa/IIb knockdown. pMlc phosphorylation in control and after Myosin IIa/IIb knockdown. Myosin Ib expression in control and after Myosin IIa/IIb knockdown. The lower panels show phalloidin staining for the same samples directly above. There is strong knockdown of Myosin IIb and pMlc, but little effect on Myosin Ib staining. Embryos were incubated for 3hrs, put in EC culture and transfected with specific or control siRNA’s. All samples were fixed 18 hrs after transfection. The controls had reached the primitive streak stages (HH3-4), while the transfected embryos did not develop into streaks, Scalebar, 25µm.
b-) Effect of simultaneous Myosin Ia and Myosin Ib knockdown. Upper panels show from left to right; expression of Myosin Ia in control and after Myosin Ia/Ib double knockdown, Myosin Ib expression in control and after Myosin Ia/Ib double knockdown. pMlc expression in a control and after Myosin Ia/Ib knockdown. The red panels show Phalloidin staining for the same samples positioned directly above. There is strong knockdown of Myosin Ia, Myosin Ib and pMlc, note the effect on the changes in actin distribution. Other conditions as in A.
c-) Effects of simultaneous Myosin Va and Myosin Vb knockdown. Upper row from left to right, Myosin Va expression in control and after Myosin Va/Vb knockdown; pMLC expression in a control embryo and after Myosin Va/Vb knockdown; Myosin Ib expression in control and after Myosin Va/Vb double knockdown. The lower panels show Phalloidin staining for the same samples directly above. There is a noticeable loss of Myosin Va membrane staining in the knockdown samples and a great loss of pMlc expression, but little effect on Myosin Ib expression. Other conditions as in A.
The data shown in this figure are representative for outcomes of the siRNA knockdown experiments shown in Fig 6b. All samples (control and siRNA of each experiment were stained with the relevant antibodies and inspected by fluorescence microscopy, at least 1 control embryo and 1 siRNA treated embryo of each experiment was investigated in detail by confocal microscopy).
Discussion
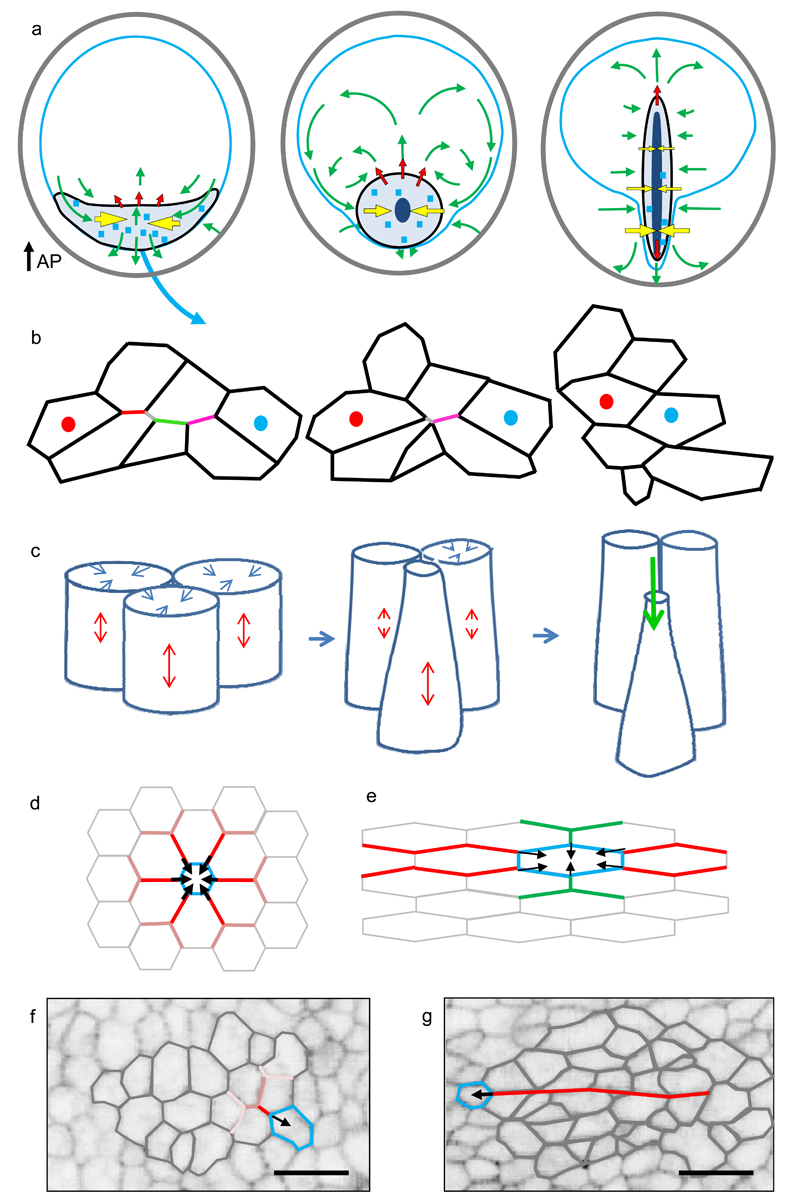
The development of the light sheet microscope enabled us to visualise the large scale tissue flows during streak formation in the chick embryo, while at the same time providing enough resolution to quantitate individual cell behaviours underlying these flows. The streak forms in an essentially biphasic process, the mesendoderm initially contracts in the direction of midline and then extends along the anterior posterior axis (Fig. 2b, 4d,f, Supplementary Videos 1, 5). The apical contraction of aligned asymmetric mesendoderm cells contributes to the directional shortening of the mesendoderm as does directional cell intercalation via shortening of aligned junctions (Fig 8a,b,c). The alignment of asymmetrically shaped cells favours the formation of long junctional “cables” spanning 2-8 cells, which favour asymmetric force propagation along these cables, contrary to cases where alignment does not occur (Fig. 8 d,e,f,g). Therefore the initial alignment of cells before the onset of motion (Supplementary Fig 5) is likely a key factor in triggering the directional contraction of the mesendoderm (Fig. 2a, b, 8a). The phase of streak extension is associated with increased ingression of cells in the streak and further strong directional cell intercalation perpendicular to the direction of streak elongation mediated by directional shortening of junctions (Fig 2a,b, 3a, 4g, Supplementary Videos 1, 9, 12). The ingression of cells helps puling mesendoderm cells towards the streak, but counteracts its extension (Fig. 3,e,f).
Figure 8. Model of the forces and cell behaviours controlling streak formation.
a-) Diagrams depicting forces generating the tissue flows during streak formation. The active pulling forces - yellow arrows, the passive pushing forces - red arrows, the direction of tissue flows - green arrows. The sickle region is indicated in black, the Area Pellucida outline in blue. Light blue squares indicate scattered events of junctional contraction, while dark blue shapes indicate regions of ingression. b-) Schematic of sequential junctional contraction (in a region marked by the blue squares in A). The sequentially contracting junctions are indicated with different (red grey, green, magenta) colours. c-) Schematic of cells showing apical contraction (blue arrows, coupled to elongation (red arrows) along the apical axis followed by ingression (green arrow). d-) Propagation of the force generated by contracting/ingressing cell between symmetrically and e-) asymmetrically shaped cells. In case of symmetrical cells the magnitude of the force decreases strongly and symmetrically at every successive junction bifurcation. However, for asymmetric cells, the broken symmetry favours force transmission along the aligned junctions (red lines), while damping transmission in perpendicular directions (green lines). f-) Image of randomly oriented cells outside the sickle region, showing lack of junctional alignment. g-) Image of cells inside the sickle, showing many aligned junctions in neighbouring cells forming long chains (red line). The alignment of asymmetrically shaped cells inside the sickle region (Fig. S5) enables anisotropic force propagation by apical contraction and directional junctional shortening resulting in large scale directed motion. Scale bar in F and G, 25 µm.
The deformation of tissue in the forming neural plate is likely caused by the pressure exerted by the extending streak. It results in directional cell intercalation in the direction of streak extension (Fig 4g) and also coincides with ingression of isolated epiblast cells into the deeper parts of the embryo (Fig. 2a, Supplementary Videos 1, 9, 12). These cells could either be squeezed out from the epiblast 41 or could ingress actively. The role of ingression has been independently highlighted in a recently published study, where it was argued there that the ingression of cells is cooperative and coordinated via a Nodal dependent community effect 42. How Nodal signals mediate ingression remains unresolved.
Junctional contraction is coincident with patterns of Myosin light chain phosphorylation, organised in supercellular cable-like structures, spanning 2-8 cells that are aligned in the direction of motion towards the streak (Fig. 5). Inhibition of Myosin II in aligned cell junctions and a strong inhibition of streak formation, showing that Myosin II is a key motor in junctional contraction (Fig. 6a,b,c).
In several systems cortical MyosinII assembly has been shown to be tension sensitive 43, 44 Our working hypothesis is that an active tension dependent mechanism of assembly of active myosin in cell junctions underlies its supercellular alignment. Contraction of a given junction results in an increase in tension of neighbouring aligned junctions. The junctions rapidly assemble more myosin to counteract the experienced increase in tension. This Myosin accumulation then in turn can result in contraction of those aligned junctions, leading to the observed sequential junctional contractions. The level of MLC phosphorylation is effectively a read-out of the tension on cell junctions. Surprisingly inhibition of Myosin V and Myosin I activity by chemical inhibitors and knockdown strongly inhibit formation of active Myosin II cables as well as streak formation (Fig 6c,d,e,7b). Myosin I members have been shown to be able to act as tension sensors at the membrane-cytoskeleton interface 45, possibly explaining their key role in apical Myosin II cable formation.
In the mouse the role of cell intercalation appears to be less important and it has been proposed that the streak forms essentially by in situ by ingression of cells 46. This difference could be due to the much larger size and faster development of the chick embryo requiring coordination of behaviours of around hundred thousand cells by a robust highly self-organising mechanism 47
The basic cellular mechanisms that drive gastrulation in the chick embryo appear very similar to those observed during Drosophila, gastrulation and germband extension, where epiblast cells move towards the cleavage furrow by pulling forces generated by apical contraction of ingressing cells and cell intercalations driven by Myosin II mediated directional junctional contraction 29, 30, 48, while the MyosinV homologue Dachs, has been shown to control of junctional remodelling during epithelial morphogenesis in the dorsal thorax in Drosophila 20. The main difference is that in the chick embryo the junctional contractions appear to be sequential, while in Drosophila they often appear to occur simultaneous giving rise to clear rosette structures 30. In the chick embryo the rosette structures observed are mainly caused by dividing or ingressing cells 15, 26, 49. The mechanism of intercalation appears to be different from the sliding intercalation mechanism proposed to drive convergent extension of the mesoderm during gastrulation in the frog embryo 50, 51.
Another key question is why do myosin cables assemble preferentially in the forming mesendoderm? In Drosophila myosin assembly requires folded gastrulation (Fog) a ligand secreted by mesoderm cells 52 which through a serpentine transmembrane receptor, a heterotrimeric G protein, a RhoGef, Rho and Rho kinase results in Myosin II activation and apical junction contraction52–54. Given the high mechanistic similarity at the cellular level it would appear likely that a similar signalling mechanism is required to initiate and coordinate cell behaviours during amniote apical contraction in a larger region during gastrulation.
Online Methods
Production of the membrane-localised GFP transgenic chicken line
Gateway technology (InVitrogen) was used to generate a lentiviral expression vector for ubiquitous expression of myr-EGFP. Myr-Flag-pEGFP-N3 plasmid containing the N-terminal myristoylation/palmitoylation sequence MGCGCSSHPED of LCK was digested using XhoI and NotI and the myr-EGFP fragment was inserted into pENTR2B (Invitrogen), which had been linearised by NotI/SalI digestion. The CAG promoter/enhancer (CMV-IE enhancer fused to the chicken β-actin promoter/1st intron 55 was cloned between two EcoRI sites in pENTR5’-TOPO vector (InVitrogen) from which the blasticidin resistance gene had been deleted. The three plasmids were recombined using LR ClonaseII enzyme mix to generate the myr-EGFP lentiviral expression vector. HIV cPPT and the modified RNA regulatory element oPRE 56 were then inserted 5’ upstream of CAG promoter and 3’ downstream of myr-EGFP gene respectively, to enhance viral titre and transgene expression.
Packaged viral vector stocks were generated by FuGENE6 (Roche, Lewes, UK) cotransfection of HEK 293T cells with the vector plasmid, gag/pol plasmid (psPAX2, Addgene) and VSV-G plasmid (pLP/VSV-G, Invitrogen). Viral particles were concentrated from cell culture medium by centrifugation and resuspended in a final volume of ~60-80µl. The virus preparation had a titre of approximately 109 TU/ml. A ~2 μl volume of concentrated virus was injected into the subgerminal cavity of newly laid eggs and the embryos incubated in host shells to hatch 57. A total of 38 eggs were injected from which 23 (60.5%) chicks hatched. G1 birds were bred from one cockerel (MCG12-14), by screening for GFP fluorescence in newly hatched chicks. Nine G1 chicks were identified and analysis of genomic DNA by Southern blot indicated that each chick had a single integrated copy of the myr-GFP expression vector and that all 9 chicks carried the same genomic site insertion of the lentiviral vector. A transgenic line was established from one of these chicks. Fertile eggs used in the experiments described here were hemizygous for the transgene. All experiments, animal breeding and care procedures were carried out under license from the UK Home Office.
Culture conditions
Early stage chicken embryos can develop in vitro- up to the beating heart stage in EC culture. In EC and New’s culture method embryos younger than stage HH2 cannot be grown with their dorsal-side up 58 59. Contact with a large volume of gel based or liquid substrata impairs development, most likely since factors secreted by the embryo get diluted too much to sustain development. It is of great interest however to grow young stage embryos not only dorsal-side up but also in liquid environment to take advantage of high NA immersion lenses as in Lightsheet microscopy systems. To overcome these limitations we have designed special culture plates that allow cultivating un-incubated chicken embryos dorsal-side up and also in liquid environment (Fig. 1B). Our method is an extension to EC culture method 58. The embryos were cultured on a specially designed plate 2x2cm wide and 2.5 mm high as indicated in figure 1B. The central depression is 1cm in diameter and 0.3mm deep (volume of ~25 ul) and filled with 30 ul of heavy silicone oil (Poly dimethylsiloxane-co-methylphenylsiloxane, density 1.066g/ml,125cSt viscosity, Aldrich, 378488). Embryos are collected using circular 30 mm Whatman 1 filter paper carriers with a ~2.2 cm internal hole. The embryo is transferred on the culture plate with the epiblast side up. The embryo is placed on the plate over the central depression and the vitelline membrane is fixed using a tight fitting rubber O-ring, sealing the hypoblast side of the embryo off from the external environment. The silicon oil filled well under the embryo supports the embryo and helps to sustain development allowing the embryo to condition the small amount of fluid surrounding the hypoblast cells with essential signalling and growth factors. The embryos mounted epiblast side up on these plates are then fixed in a silicone mould at the bottom of a plastic container (7x3x1.4 cm) and immersed in 10 ml pure albumin and imaged in the Light Sheet Microscope at 37.5 °C. To prevent evaporation of liquid from the preparation the sample is overlaid with 2ml of light silicon oil (density 0.93g/ml, 10cSt viscosity, Aldrich 378321). Embryos can be cultured from stage EGXII up to HH10 60, 61 using this method. The culture method allows development of early embryos from the un-incubated egg stage to stage HH8 with timing and morphology indistinguishable from the commonly used EC culture58. A step-by-step protocol describing the mounting of the embryo on the culture plat as well as a technical drawing of the culture plate culture plate can be found at Nature Protocol Exchange 62.
Antibodies and Reagents
Antibodies used were from Cell Signalling Technologies: Phospho Myosin Light Chain2 (Thr18/Ser19, #3674), MyosinVa (#3402S), Phospho-Ezrin(#9967), Developmental Hybridoma Bank: Myosin IIb (CM23 and CM25), Santa Cruz Biotech Myosin Ia (sc-32698), Sigma: Myosin Ib (HPA013607), AHRGef11 (HPA011026), Zymed: Par3 (36-2301). All these antibodies were used in 1:100 dilutions in PBT (PBS containing 2% BSA, 0.1% TritonX100). Phalloidin-Alexa 595 and Alexa 488 and Alexa 594 secondary antibodies were obtained from InVitrogen. Phalloidin was dissolved in ethanol and used following the manufacturers protocol at 1 unit/ml. The secondary antibodies were used at 1:200 dilution in PBT (PBS containing 2% BSA, 0.1% TritonX100, 0.1%tween 20). The inhibitors pentabromopseudilin and pentachloropseudilin were synthesised by optimisation of a procedure reported in the literature 63.
Inhibitor experiments
Fertile white leghorn eggs (Winter Farm) were incubated at 37oC for 6hrs. Embryos were isolated and put in EC culture according to published procedures 58. The plates either contained no additives (control) or 5µM pentachloropseudilin or 5µM pentabromopseudilin prepared from 10mM stock solutions in DMSO. The embryos were incubated over night for live LSM microscopy or fixed after 2hrs and 4hrs for immunocytochemistry. Embryos were fixed for 3hrs in 4% para-formaldehyde/PBS on ice, followed by three washes times in PBS, followed by antibody detection using published procedures 64 The embryos were mounted in and investigated by confocal microscopy using a Leica TCS SP2 confocal microscope at 40x and 100x magnifications.
RNA Seq experiment
Total RNA was prepared from batches of 10-20 embryos of mixed sex incubated for 0, 5, 10, 15 and 20hrs. The area pellucida was cut out with fine tungsten needles and collected in ice cold PBS. RNA was isolated with a Qiagen RNA Easy kit according to the manufacturer’s instructions. The RNA sequencing libraries were prepared using the Illumina mRNA seq 8 sample prep kit, following the manufacturer’s protocol. Briefly, high quality total RNA (RIN >7) is used as input to mRNA capture by Poly A. The mRNA is then fragmented and randomly primed for reverse transcription followed by second-strand synthesis to create double-stranded cDNA fragments. Ends are repaired to produce blunt ends and an ‘A’- base is added to the blunt ends followed by ligation to Illumina Paired-End Sequencing adapters. The library is enriched by a limited number of cycles of PCR amplification. The Libraries were assessed using Agilent Bioanalyser DNA 100 chips and quantified by qPCR using the Kapa Biosystems Kapa Illumina Library quantification kit. The concentration of the sequencing libraries were normalised to 10 nM before denaturation and dilution for loading at 4.75 pM concentration onto an Illumina paired end flow cell using the Illumina Paired End cluster kit and cluster station instrument. The samples were loaded at one sample per lane. After the clustering the flow cell was immediately placed on the Illumina GAIIX sequencer, previously primed with Illumina SBS reagents sufficient for 36 cycle paired end sequencing, and the sequencing run initiated. Sequencing was carried out by Edinburgh Genomics, The University of Edinburgh. Sequence analysis was performed using CLC genomics work bench software.
siRNA knockdown experiments
Transfections: embryos were incubated for 3hrs followed by preparation in EC culture. Embryos were transfected using a dedicated electroporation apparatus. Embryos were placed epiblast apical side down on a 3 mm diameter round electrode, covered with 20ul of 0.6% saline epiblast side down. 1ul of siRNA (1 ug/ml) dissolved in stabilisation buffer was pipetted on basal side of the epiblast. Another 20µl 0.6% NaCl was pipetted on the top, forming a small drop in which the second circular electrode was positioned 2mm from the bottom electrode. The embryos were electroporated by 3 consecutive 50msec pulses at a field strength of 17.5 V/cm. After electroporation the embryos were immediately transferred to EC culture and allowed to develop for 18hrs after which they were fixed with 4%paraformadehyde in PBS for 4hrs and processed for immuno-fluorescence staining with a variety of antibodies using standard procedures 64. Experiments were started with batches of 36 fertilised eggs. Typically 20-25 embryos were selected for transfection experiments, where half were controls and the other half were transfected with a mixture of Myosin class specific siRNA’s. All experiments were repeated at least three times with different batches of eggs. All fixed and stained embryos were investigated by epifluorescence microscopy and samples of 2-3 embryos for control and treatment were chosen for more detailed analysis and recording of results by confocal microscopy (Leica SP2) using standardised settings at 100x magnification.
We used the following Dharmacon siRNA’s pairs for silencing of specific Myosin genes. The oligo’s were designed and checked for optimal specificity using BLAST searches against the chick genome data base.
MYH9: GAGAAGAAGCUGAAAGAUAUU & GGGAAGAAGGUGAAAGUAAUU
MYH10: GGAAGAACCUGGAGAAACAUU & AGGAAGAUCCCGAGAGAUAUU
MYOIA: CUGAGAAGGUGGAGGAGUAUU &ACGUGAAGGGCGAGAGGAAUU
MYOIB: GGAAGGAUUUGUUGAUAAAUU & GGAAUUGAGUGGACACAUAUU
MYOVA: AGGAUGAACUGGACAGAAAUU & GGGCAAGGACCUAGAAUAUUU
MYOVB: CCGGAAUGCUGGAGAGUGAUU & CCAAGUAUGCCAUGAGAUAUU
Microscopy setup
To image chick embryos development on the scale of the whole organism and with cellular resolution we have designed and constructed a designated light-sheet fluorescent microscope (LSFM). Light-sheet microscopy offers high-speed high-resolution three dimensional time lapse imaging with very low levels of photo-bleaching and photo-toxicity. Our setup is based on Stelzer and Keller LSFM 65 design, however, to image flat chick embryos we have placed imaging and illumination arms above the sample at 45° to the worktop surface. The system is equipped with a high speed scientific CMOS camera (PCO.Edge), a GSI 2-axis galvo scanner and a Physik Instrumente – 3 axis motorized stage combination, which allows for imaging at rates exceeding 50 frames per second. Illumination and imaging are performed using water dipping 10× Nikon objectives with a working distance of 3.5 mm and 0.3 numerical aperture. During the experiment the sample is kept in a heated chamber at 37°C. Imaging is performed by moving the sample through the light-sheet along the x-axis and collecting subsequent cross sections, which after acquisition are transformed into “confocal” stacks (Fig. 1A). Currently imaging of a 4mm diameter embryo at 1.88 um step takes 2 minutes. Additionally, to accommodate for sample shape and its changes over time we have implemented active surface contour detection algorithm, which corrects the sample position along the z-axis for each scan to keep the top of the sample in the focal point of the light-sheet, necessary for assuring high and consistent data quality. Typically 3000 images 2560x400 pixels are collected per time point resulting in 1.5 TB of data per 15h experiment.
Data analysis
We have divided data processing pipeline into 3 steps, each of which not only generates input for the following ones but also provides information about embryo behaviour at the given level of analysis.
(i). 2D surface projection
In the first stage of data processing the embryo surface shape is determined and a flat Z stack is constructed from sections conformal to the surface. The acquired 3D-stack is transformed into rectangular coordinates. Next, each Z plane is divided into square interrogation areas (64×64 pixels) for which a FFT power-spectrum is evaluated. Along the z axis the square that shows the highest number of frequencies above certain threshold in the FFT spectrum is selected as the surface tile. This process is repeated for all squares in the stack. A new, flat 3D-stack is constructed by shifting tiles along z-axis so that the surface tiles are occupying the sample z-plane. This method allows not only correcting for the large scale height differences between different parts of the sample but also removes small surface undulations. The apical plane is selected for further steps of analysis
(ii). PIV analysis
Tissue velocity is calculated by image cross-correlation using standard particle image velocimetry algorithm (PIVLab vs1.32 for Matlab 66). Results of this calculation are used to study local tissue deformation, tissue fate mapping and as an input for automatic cell tracking algorithm. We found that setting the interrogation window to 64×64 pixels for the first pass and 32×32 pixels for the second pass with 50% overlap at each pass gave the best results.
To understand the way tissue deforms we have derived a new velocity field averaging scheme that focuses on tissue level dynamics of developing chick embryo and helps to reduce noise by removing small fluctuations of vector fields. The embryo is divided into a square grid with a lattice constant of 125 pixels and 10 points per square side. Typically the grid contains 60 x 25 squares. Each lattice point is displaced in time according to the local velocity field. For points located in between the PIV computational grid, their velocity vectors are linearly interpolated from the nearest points using the MATLAB built-in function interp2. Initial squares deform into polygons and their area is calculated at each time point using the MATLAB built-in function polyarea. The areas are colour coded on a dynamic grid as total change in area of the squares (Fig. 2b, Supplementary Fig. 1) to identify and select active tissue regions for further analysis at the cellular level.
Averaging of velocity fields from different experiments
To average the expansion contraction behaviour from several embryos we needed to align their developmental axis. This was achieved using points from the anterior and posterior end of primitive streak, for each time point in each experiment. We also estimated time of onset of motion from each experiment. The annotated points were used to translate centre point of primitive streak into the origin of generalised coordinate system for each embryo and for each time point. The spatial alignment, rotation and temporal alignment was used to align their corresponding velocity fields which were then averaged. The averaged velocity field over the nine experiments was then used evaluate the grid deformation over the whole time sequence.
Calculation and Decomposition of Tissue Strain Rate
The strain rate describes how much the material (tissue) contracts or expands based on the differences in velocity between neighbouring points/areas. To compare the contraction rate along the left-right axis and the expansion rate along the posterior-anterior axis we have calculated stain rates for those two directions as shown in Figure 3b. To reliably estimate values we have taken 25 cuts for each direction at the resolution of PIV calculations and fitted a linear slope to linear parts of the curves (blue points, Fig. 3c). By analysing tissue flows generated by contraction/ingression, cell growth and intercalation (Fig. 3d) it is evident that ingression generates anisotropic effects (expansion in one axis and contraction in the other), while growth and contraction leads to isotropic effects expansion and contraction in both axis respectively. This allows us to decompose tissue strain rate into isotropic part (growth + apical contraction/ingression) and anisotropic part (intercalation) (Fig. 3e, f):
Alternatively we have calculated the strain rate for the whole image. The velocity tensor L 67
can be decomposed in a symmetric part, which defines the strain rate tensor ξ as follows ξ = (L+L ⊤)/2.
This can be written as
The strain rate tensor can then be further decomposed in a term reflecting the expansion/contraction strain rate and a term reflecting the shear strain rate as follows.
We have plotted the isotropic contraction/expansion term as a circle, red for expanding and blue for contraction and the anisotropic shear strain rate tensor as a blue bar in the direction of contraction (the direction of the negative eigenvalue). Velocity field gradients of the matrix L were estimated using central differences over intervals of 32 pixels. For visualisation the computed strain rate fields were averaged over 10 time points (~ 30 min) and over 13 spatial points (~200 pixels) along both spatial dimensions.
Segmentation
Prior segmentation in order to smooth images and equalise intensities images are filtered with a FFT based band-pass filter using standard Image J and Matlab routines. A Watershed segmentation algorithm is used to find outlines of cells in membrane GFP images 68. Local intensity minima are used as seed points for the algorithm. As a result of the segmentation each cell in an image is presented as a collection of labelled segments.
(iii). Automatic tracking
Cells are tracked forward in time starting from the first time point. An initial segmentation is obtained using watershed segmentation 68. To track cells between time points, the locations of cells in future time points are estimated from their previous locations using the tissue velocity fields calculated by PIV between these time points fields. These locations are used to constrain the seedpoints of the watershed algorithm. Ingression, division events are detected utilising the characteristic changes in cell size and shape of the segmented cells. The automatic tracking method results in the tracking of 70% of the >100.000 cells correctly for between >100 successive time points. For the cell based statistical analysis described below it is only required to track cells confidently over ~20 successive time points. The routines segmentation routines are written in Matlab.
(iv). Manual tracking
To improve the automatic tracking a manual curation option for smaller selected regions was introduced. The manual tracking scheme is similar to that used in the automatic tracking using algorithms coded in Matlab. The initial image is segmented and checked by visual inspection. The locations of local minima in each image are adjusted manually if needed to achieve correct segmentation results. In addition, ingressions, divisions and emerging cells are marked manually if necessary.
(v). Cell orientation measurements
To measure orientation of cells from segmented images, an ellipse is fitted into each segment utilising the normalised second order central moments of pixels of the segment 69. Orientation of a cell is taken to be the orientation of the major axis of the fitted ellipse. Orientation of a cell is taken to be the orientation of the major axis of the fitted ellipse. In this study we consider a cell to be polarised if the eccentricity of the fitted ellipse exceeds 0.87.
(VI). Calculation of the statistical strain rate from the cell segments and tracks
Methods have been derived to calculate the discrete counterparts of the isotropic and anisotropic tissue deformations based on the dynamics of the spatial links connecting the centroids of cells 70. For the calculation of the cellular deformation based on the positions of the cell centroids we calculate the texture tensor M which provides information on the size, shape and alignment of the cells at a given time point in a particular region 70.
The links l are defined as vectors connecting the centroids of neighbouring cells. The length and orientation of the links are captured in the so called link matrices which are averaged over the domain of interest. We calculate these averages based on the links between all neighbouring cells in circular domains of 65 µm radius that cover the embryo.
We can use the dynamic changes in link length and orientation to calculate the statistical symmetrized velocity gradient V, the discrete analogue of the strain rate tensor. It is calculated as follows 70:
Here Nc is the number of links present at both ends points of time interval Δt and Ntot is number of all links of cells that have their centroid within the circular domain at the time t. ⊗ represents the tensor outer product. M is calculated as previously shown. The tensor variation was evaluated between successive time intervals, typically 2.5-3 min.
We also calculate the variation of statistical internal strain rate dU/dt. This tensor quantifies how cell size and shape change contribute to the total tissue deformation. The tensor is defined as follows 70 :
Here links are chosen from end points of the same time interval (Δt) as for evaluation the tensor V.
Finally, we calculate the statistical topological rearrangement rate P. This tensor measures contribution of cell intercalation to the total tissue deformation 70.
Where, ma are links that exist in the end the time interval Δt but not in the beginning and md are links that exist in the beginning of the time interval but not in the end. Na is number of appearing links and Nd is number of disappearing links. In the figures in the results we show the traceless parts of the dU/dt and P tensor in the direction of negative eigenvalues. These signify the rate of the change in cell shape and the rate for cell rearrangement respectively. For visualisation, the computed tensor fields were displayed as a moving average over 10 time points (~ 30 min).
Code availability: The code used for all data processing and analyses described above has been written in Matlab. The scripts will be made available upon request.
Reproducibility of experiments
Fig 1 d,e,f,g,h, the images presented are representative for over 150 successfully recorded embryos acquired over a 3 year period. Fig 2a, the results shown were replicated in over 20 embryos, another example is shown in Sup Fig 1. Fig 2c, these results are representative for analyses in over 20 embryos. Fig.2d, these results are representative for observations in 4 embryos. Fig. 3a this detailed analysis has been performed in 3 embryos. Fig 4a,b,c,d,e, this analysis has been performed in 2 embryos with comparable results. Fig 4G this analysis has been performed in 3 embryos. Fig 6d, e, this analysis has been performed in 2 embryos out of 11 embryos recorded. Supplementary Fig 3, this analysis has been performed in 4 embryos with similar results. Supplementary Fig 4, this analysis has been performed in 4 embryos with similar results Supplementary Fig 5, this analysis has been performed in 3 embryos with similar results. For other results details of the statistical analyses and reproducibility is provided in the figure legends.
Data Access
The RNA seq have been submitted to the European Nucleotide Archive (ENA) and can be accessed at http://www.ebi.ac.uk/ena/data/view/PRJEB8414
Supplementary Material
Acknowledgements
We thank E. Stelzer and P. Keller for advice on the construction of LSMs, and A. Sherman, F. Thomson, M. Hutchison and R. Mitchell for support in generation and breeding of the transgenic chick line and supplying the fertilized transgenic eggs. This work was supported by BBSRC (BB/E011276/1) to H.M.S. and C.J.W., (BB/G015082/1) to C.J.W. and M.P.M., (BB/K00204X/1) to C.J.W., Institute Strategic Grant funding to H.M.S., Wellcome Trust (094131/Z/10/Z) to C.J.W. RNA sequencing was carried out by Edinburgh Genomics, The University of Edinburgh. Edinburgh Genomics is partly supported through core grants from NERC (R8/H10/56), MRC (MR/K001744/1) and BBSRC (BB/J004243/1).
Footnotes
Contributions
ER built the LSM hardware and software, performed experiments and PIV based data analysis, MC performed the biological and Myosin perturbation experiments. AIK developed the cell based image analysis software and analysed experiments. FS and HS developed the Myr-GFP embryos, RM and HJK developed and produced the Myosin Inhibitors. CJW and MPM conceived the design and use of the LSM in the investigation of chick development.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Stern CD. Gastrulation, from cells to embryo′s. Cold Spring Harbor Laboratory Press; New York: 2004. [Google Scholar]
- 2.Weinberger C, Brick I. Primary hypoblast development in the chick II. The role of cell division. Wilhelm Roux' Arch. Entwicklungsmechan.Org. 1982;191:127–133. doi: 10.1007/BF00848450. [DOI] [PubMed] [Google Scholar]
- 3.Stern CD, Downs KM. The hypoblast (visceral endoderm): an evo-devo perspective. Development. 2012;139:1059–1069. doi: 10.1242/dev.070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakaya Y, Sheng G. Epithelial to mesenchymal transition during gastrulation: an embryological view. Dev Growth Differ. 2008;50:755–766. doi: 10.1111/j.1440-169X.2008.01070.x. [DOI] [PubMed] [Google Scholar]
- 5.Nakaya Y, Sukowati EW, Wu Y, Sheng G. RhoA and microtubule dynamics control cell-basement membrane interaction in EMT during gastrulation. Nat Cell Biol. 2008;10:765–775. doi: 10.1038/ncb1739. [DOI] [PubMed] [Google Scholar]
- 6.Graeper L. Die Primitiventwicklung des Huehnchens nach stereokinematographischen Untersuchungen kontrolliert durch vitale Farbmarkierung und verglichen mit der Entwicklung anderer Wirbeltiere. Wilhelm Roux' Arch. Entwicklungsmechan.Org. 1929;116:382–429. doi: 10.1007/BF02145235. [DOI] [PubMed] [Google Scholar]
- 7.Chuai M, et al. Cell movement during chick primitive streak formation. Dev Biol. 2006;296:137–149. doi: 10.1016/j.ydbio.2006.04.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamir EA, Rongish BJ, Little CD. The ECM moves during primitive streak formation--computation of ECM versus cellular motion. PLoS Biol. 2008;6:e247. doi: 10.1371/journal.pbio.0060247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voiculescu O, Bertocchini F, Wolpert L, Keller RE, Stern CD. The amniote primitive streak is defined by epithelial cell intercalation before gastrulation. Nature. 2007;449:1049–1052. doi: 10.1038/nature06211. [DOI] [PubMed] [Google Scholar]
- 10.Chuai M, Hughes D, Weijer CJ. Collective epithelial and mesenchymal cell migration during gastrulation. Current genomics. 2012;13:267–277. doi: 10.2174/138920212800793357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui C, Yang X, Chuai M, Glazier JA, Weijer CJ. Analysis of tissue flow patterns during primitive streak formation in the chick embryo. Dev Biol. 2005;284:37–47. doi: 10.1016/j.ydbio.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 12.Sandersius SA, Chuai M, Weijer CJ, Newman TJ. A 'chemotactic dipole' mechanism for large-scale vortex motion during primitive streak formation in the chick embryo. Phys Biol. 2011;8:045008. doi: 10.1088/1478-3975/8/4/045008. [DOI] [PubMed] [Google Scholar]
- 13.Wei Y, Mikawa T. Formation of the avian primitive streak from spatially restricted blastoderm: evidence for polarized cell division in the elongating streak. Development. 2000;127:87–96. doi: 10.1242/dev.127.1.87. [DOI] [PubMed] [Google Scholar]
- 14.Alev C, Wu Y, Nakaya Y, Sheng G. Decoupling of amniote gastrulation and streak formation reveals a morphogenetic unity in vertebrate mesoderm induction. Development. 2013;140:2691–2696. doi: 10.1242/dev.094318. [DOI] [PubMed] [Google Scholar]
- 15.Yanagawa N, Sakabe M, Sakata H, Yamagishi T, Nakajima Y. Nodal signal is required for morphogenetic movements of epiblast layer in the pre-streak chick blastoderm. Dev Growth Differ. 2011;53:366–377. doi: 10.1111/j.1440-169X.2010.01244.x. [DOI] [PubMed] [Google Scholar]
- 16.Verveer PJ, et al. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat Methods. 2007;4:311–313. doi: 10.1038/nmeth1017. [DOI] [PubMed] [Google Scholar]
- 17.Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EH. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 18.Tomer R, Khairy K, Keller PJ. Shedding light on the system: studying embryonic development with light sheet microscopy. Curr Opin Genet Dev. 2011;21:558–565. doi: 10.1016/j.gde.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Blanchard GB, et al. Tissue tectonics: morphogenetic strain rates, cell shape change and intercalation. Nat Methods. 2009;6:458–464. doi: 10.1038/nmeth.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosveld F, et al. Mechanical control of morphogenesis by Fat/Dachsous/Four-jointed planar cell polarity pathway. Science. 2012;336:724–727. doi: 10.1126/science.1221071. [DOI] [PubMed] [Google Scholar]
- 21.Allan C, et al. OMERO: flexible, model-driven data management for experimental biology. Nat Meth. 2012;9:245–253. doi: 10.1038/nmeth.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czirok A, Rongish BJ, Little CD. Extracellular matrix dynamics during vertebrate axis formation. Dev Biol. 2004;268:111–122. doi: 10.1016/j.ydbio.2003.09.040. [DOI] [PubMed] [Google Scholar]
- 23.Amat F, Myers EW, Keller PJ. Fast and robust optical flow for time-lapse microscopy using super-voxels. Bioinformatics. 2013;29:373–380. doi: 10.1093/bioinformatics/bts706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Downie JR. The mechanism of chick blastoderm expansion. J Embryol Exp Morphol. 1976;35:559–575. [PubMed] [Google Scholar]
- 25.Chapman SC, Schubert FR, Schoenwolf GC, Lumsden A. Analysis of spatial and temporal gene expression patterns in blastula and gastrula stage chick embryos. Dev Biol. 2002;245:187–199. doi: 10.1006/dbio.2002.0641. [DOI] [PubMed] [Google Scholar]
- 26.Chuai M, Weijer CJ. The mechanisms underlying primitive streak formation in the chick embryo. Curr Top Dev Biol. 2008;81:135–156. doi: 10.1016/S0070-2153(07)81004-0. [DOI] [PubMed] [Google Scholar]
- 27.Butler LC, et al. Cell shape changes indicate a role for extrinsic tensile forces in Drosophila germ-band extension. Nat Cell Biol. 2009;11:859–864. doi: 10.1038/ncb1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bardet PL, et al. PTEN controls junction lengthening and stability during cell rearrangement in epithelial tissue. Dev Cell. 2013;25:534–546. doi: 10.1016/j.devcel.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 29.Bertet C, Sulak L, Lecuit T. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429:667–671. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- 30.Blankenship JT, Backovic ST, Sanny JS, Weitz O, Zallen JA. Multicellular rosette formation links planar cell polarity to tissue morphogenesis. Dev Cell. 2006;11:459–470. doi: 10.1016/j.devcel.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF. Integration of contractile forces during tissue invagination. J Cell Biol. 2010;188:735–749. doi: 10.1083/jcb.200910099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zallen JA, Wieschaus E. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev Cell. 2004;6:343–355. doi: 10.1016/s1534-5807(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 33.Limouze J, Straight AF, Mitchison T, Sellers JR. Specificity of blebbistatin, an inhibitor of myosin II. J Muscle Res Cell Motil. 2004;25:337–341. doi: 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- 34.Simoes Sde M, et al. Rho-kinase directs Bazooka/Par-3 planar polarity during Drosophila axis elongation. Dev Cell. 2010;19:377–388. doi: 10.1016/j.devcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levayer R, Pelissier-Monier A, Lecuit T. Spatial regulation of Dia and Myosin-II by RhoGEF2 controls initiation of E-cadherin endocytosis during epithelial morphogenesis. Nat Cell Biol. 2011;13:529–540. doi: 10.1038/ncb2224. [DOI] [PubMed] [Google Scholar]
- 36.Warrington SJ, Strutt H, Strutt D. The Frizzled-dependent planar polarity pathway locally promotes E-cadherin turnover via recruitment of RhoGEF2. Development. 2013;140:1045–1054. doi: 10.1242/dev.088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura M, et al. Development of specific Rho-kinase inhibitors and their clinical application. Biochim Biophys Acta. 2005;1754:245–252. doi: 10.1016/j.bbapap.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 38.Fedorov R, et al. The mechanism of pentabromopseudilin inhibition of myosin motor activity. Nat Struct Mol Biol. 2009;16:80–88. doi: 10.1038/nsmb.1542. [DOI] [PubMed] [Google Scholar]
- 39.Bond LM, Tumbarello DA, Kendrick-Jones J, Buss F. Small-molecule inhibitors of myosin proteins. Future medicinal chemistry. 2013;5:41–52. doi: 10.4155/fmc.12.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chinthalapudi K, et al. Mechanism and specificity of pentachloropseudilin-mediated inhibition of myosin motor activity. J Biol Chem. 2011;286:29700–29708. doi: 10.1074/jbc.M111.239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marinari E, et al. Live-cell delamination counterbalances epithelial growth to limit tissue overcrowding. Nature. 2012;484:542–545. doi: 10.1038/nature10984. [DOI] [PubMed] [Google Scholar]
- 42.Voiculescu O, Bodenstein L, Lau IJ, Stern CD. Local cell interactions and self-amplifying individual cell ingression drive amniote gastrulation. eLife. 2014;3:e01817. doi: 10.7554/eLife.01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernandez-Gonzalez R, Simoes Sde M, Roper JC, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev Cell. 2009;17:736–743. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ren Y, et al. Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin I. Curr Biol. 2009;19:1421–1428. doi: 10.1016/j.cub.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I can act as a molecular force sensor. Science. 2008;321:133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams M, Burdsal C, Periasamy A, Lewandoski M, Sutherland A. Mouse primitive streak forms in situ by initiation of epithelial to mesenchymal transition without migration of a cell population. Dev Dyn. 2012;241:270–283. doi: 10.1002/dvdy.23711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pouille PA, Ahmadi P, Brunet AC, Farge E. Mechanical signals trigger Myosin II redistribution and mesoderm invagination in Drosophila embryos. Sci Signal. 2009;2:ra16. doi: 10.1126/scisignal.2000098. [DOI] [PubMed] [Google Scholar]
- 48.Zallen JA, Blankenship JT. Multicellular dynamics during epithelial elongation. Semin Cell Dev Biol. 2008;19:263–270. doi: 10.1016/j.semcdb.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagstaff LJ, Bellett G, Mogensen MM, Munsterberg A. Multicellular rosette formation during cell ingression in the avian primitive streak. Dev Dyn. 2008;237:91–96. doi: 10.1002/dvdy.21390. [DOI] [PubMed] [Google Scholar]
- 50.Keller R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 2002;298:1950–1954. doi: 10.1126/science.1079478. [DOI] [PubMed] [Google Scholar]
- 51.Honda H, Nagai T, Tanemura M. Two different mechanisms of planar cell intercalation leading to tissue elongation. Dev Dyn. 2008;237:1826–1836. doi: 10.1002/dvdy.21609. [DOI] [PubMed] [Google Scholar]
- 52.Dawes-Hoang RE, et al. folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132:4165–4178. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- 53.Kolsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315:384–386. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- 54.Manning AJ, Peters KA, Peifer M, Rogers SL. Regulation of epithelial morphogenesis by the g protein-coupled receptor mist and its ligand fog. Sci Signal. 2013;6:ra98. doi: 10.1126/scisignal.2004427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 56.Schambach A, et al. Woodchuck hepatitis virus post-transcriptional regulatory element deleted from X protein and promoter sequences enhances retroviral vector titer and expression. Gene Ther. 2006;13:641–645. doi: 10.1038/sj.gt.3302698. [DOI] [PubMed] [Google Scholar]
- 57.McGrew MJ, et al. Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep. 2004;5:728–733. doi: 10.1038/sj.embor.7400171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chapman SC, Collignon J, Schoenwolf GC, Lumsden A. Improved method for chick whole-embryo culture using a filter paper carrier. Dev Dyn. 2001;220:284–289. doi: 10.1002/1097-0177(20010301)220:3<284::AID-DVDY1102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 59.New D. A new technique for the cultivation of the Chick embryo in vitro. J Embryol Exp Morphol. 1955;3:320–331. [Google Scholar]
- 60.Eyal-Giladi H, Kochav S. From cleavage to primitive streak formation: a complementary normal table and a new look at the first stages of the development of the chick. I. General morphology. Dev Biol. 1976;49:321–337. doi: 10.1016/0012-1606(76)90178-0. [DOI] [PubMed] [Google Scholar]
- 61.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 62.Rozbicki EMC, CJ W. Liquid Culture Technique for Early Chick Embryos Suitable for Long Term Live Imaging. Nature Protocol Exchange. 2015 doi: 10.1038/protex.2015.012. [DOI] [Google Scholar]
- 63.Martin R, et al. Total synthesis of pentabromo- and pentachloropseudilin, and synthetic analogues--allosteric inhibitors of myosin ATPase. Angew Chem Int Ed Engl. 2009;48:8042–8046. doi: 10.1002/anie.200903743. [DOI] [PubMed] [Google Scholar]
- 64.Leslie NR, Yang X, Downes CP, Weijer CJ. PtdIns(3,4,5)P(3)-dependent and -independent roles for PTEN in the control of cell migration. Curr Biol. 2007;17:115–125. doi: 10.1016/j.cub.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 66.Thielicke W, Stamhuis EJ. PIVlab - Time-Resolved Digital Particle Image Velocimetry Tool for MATLAB [Google Scholar]
- 67.Heller E, Kumar KV, Grill SW, Fuchs E. Forces generated by cell intercalation tow epidermal sheets in mammalian tissue morphogenesis. Dev Cell. 2014;28:617–632. doi: 10.1016/j.devcel.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meyer F. Topographic distance and watershed lines. Signal Processing. 1994;38:113–125. [Google Scholar]
- 69.Mulchrone KF, Choudhury KR. Fitting an ellipse to an arbitrary shape: implications for strain analysis. Journal of Structural Geology. 2004;26:143–153. [Google Scholar]
- 70.Graner F, Dollet B, Raufaste C, Marmottant P. Discrete rearranging disordered patterns, part I: robust statistical tools in two or three dimensions. The European physical journal. E Soft matter. 2008;25:349–369. doi: 10.1140/epje/i2007-10298-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.