Abstract
In the recent decades, the evolution of omics technologies has led to advances in all biological fields, creating a demand for effective storage, management and exchange of rapidly generated data and research discoveries. To address this need, the development of databases of experimental outputs has become a common part of scientific practice in order to serve as knowledge sources and data-sharing platforms, providing information about genes, transcripts, proteins or metabolites. In this review, we present omics databases available currently, with a special focus on their application in kidney research and possibly in clinical practice. Databases are divided into two categories: general databases with a broad information scope and kidney-specific databases distinctively concentrated on kidney pathologies. In research, databases can be used as a rich source of information about pathophysiological mechanisms and molecular targets. In the future, databases will support clinicians with their decisions, providing better and faster diagnoses and setting the direction towards more preventive, personalized medicine. We also provide a test case demonstrating the potential of biological databases in comparing multi-omics datasets and generating new hypotheses to answer a critical and common diagnostic problem in nephrology practice. In the future, employment of databases combined with data integration and data mining should provide powerful insights into unlocking the mysteries of kidney disease, leading to a potential impact on pharmacological intervention and therapeutic disease management.
Keywords: bioinformatics, data integration, kidney disease, omics databases, system biology
Introduction
In recent decades, major advances in the field of omics analyses have led to an exponential increase in available experimental data. Omics platforms offer high-throughput, detailed exploration of the genome, transcriptome, proteome and metabolome, analysed using a variety of techniques including mRNA and miRNA arrays, next-generation sequencing and mass spectrometry. In the light of technological improvements, generation of large amounts of high-quality data is no longer the main challenge in all fields of research, as the bottleneck has now shifted to the handling and extensive analysis of these omics datasets. Thus, the huge volume of data produced by each platform is still largely underexplored and leads in the majority of cases to the study of a restricted number of molecules, focusing on those that are of direct interest to the researchers and thus ignoring (and maybe wasting) the vast majority of the remaining information. Moreover, the increased number of omics-based publications hinders the thorough and effective reuse of data; the task becomes laborious and significantly prolongs the research process. Hence, efficient data storage and fast information retrieval based on data and literature mining becomes vital in order to confirm the findings from omic studies, avoiding incorrect data interpretation and ultimately elucidating novel hypotheses, which can be beneficial in answering everyday clinical problems. For these reasons, the creation, management and utilization of omics databases is particularly important. In this context, the field of nephrology is no exception.
Kidney disease can be defined as any disorder that affects renal structure and function, thus high-throughput methods have become essential to capture the complex molecular signatures underlying this broad spectrum of distinct pathologies. Therefore, in chronic and acute kidney diseases, which are highly variable and multifactorial, a large number of differentially expressed molecules will vary with regards to cause of pathology, severity and rate of progression [1]. Moreover, renal function decline is commonly accompanied by comorbidities such as diabetes, cardiovascular disease, mineral and bone disorder or anaemia, adding further layers of complexity to the picture of the dysregulated molecular networks associated with the disease. The wealth of information produced by omics experiments has allowed parallel analysis of modifications of large numbers of molecules in different parts of the kidney, across different diseases and sometimes from rare samples such as kidney biopsies.
Omics databases are invaluable tools in nephrology research and greatly facilitate the work of scientists. For each omics platform, available data repositories contain information about molecules of interest coming from experimental datasets, computational annotations or manually curated literature searches. Most of the database resources are of high quality, publicly accessible and frequently updated. They provide up-to-date information about the function, the localization and the expression of the molecules and can help with study design. They also store information about similar experiments that have already been conducted, thus easing literature searches and facilitating fast results validation and confirmation of findings.
Yet how can the omics databases be helpful for clinical practice? It is a long way from data collection to clinical usage, but the databases are the core of this process, being the central storage of the raw data. Databases currently utilized by clinicians are mostly used for administrative purposes—electronic patient records (EPR) store clinical and health information, allowing treatment of the patients to be more effective [2]. Nevertheless, systems biology approaches and high-throughput technologies push the revolution of medicine from reactive towards proactive and preventive. This so-called P4 medicine is defined by four features—predictive, preventive, personalized and participatory—and is fuelled by systems approaches to disease, emerging technologies and novel analytical tools [3]. Omics sciences provide a wealth of information that, with the use of powerful computational methods, can be used for patient screening, diagnosis, monitoring and prevention. The development of omics diagnostic tools is ongoing [4], and it is expected that they will be gradually introduced in clinical practice within the next decade [5]. Moreover, integrative methods of data analysis can lead to the discovery of new biomarkers, correlation of molecular changes with disease outcome and ultimately elucidation of mechanisms of various diseases [6]. Pharmacogenomic companies benefit the most from the use of these databases by creating models for possible therapies, which later can be proven essential for treatment during clinical practice [7]. Additionally, databases may work as an encyclopaedia for clinicians by offering a collective list of the molecules of interest with fast retrieval of information and possibly ‘connecting the dots’ in cases without prior logical links. To conclude, in the future, omics databases will be proven to be vital tools for the treatment of the patients, as personalized medicine is coming of age [8].
In this review, we focus on and describe the application of the main resources that can be useful in kidney disease studies: the general omics databases that cover a wide range of information on molecules and pathologies (Table 1), as well as the specific databases that target information explicitly connected to kidney diseases and the urinary tract (Table 2). Furthermore, we will introduce basic technical aspects and showcase the use of these databases in an attempt to solve an everyday problem in clinical practice, distinguishing diabetic nephropathy from other causes of glomerular disease, such as IgA nephropathy.
Table 1.
General omics databases
| Name | Description | Main features |
|---|---|---|
| Genomics | ||
| GeneCards (http://www.genecards.org/) | Detailed information on all annotated and predicted human genes | Contains >152 000 GeneCards genes |
| Gene-centric data from >100 Web sources from all kind of omics | ||
| Very detailed description of genes (aliases, compounds, proteins, domains, expression, related publications, transcripts, pathways) | ||
| Online Mendelian Inheritance in Man (http://www.omim.org/) | Comprehensive, authoritative compendium of human genes and genetic phenotypes | >15 000 genes |
| Information on all known Mendelian disorders | ||
| Relationship between phenotype and genotype | ||
| Transcriptomics | ||
| Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/gds) | Repository for gene expression datasets supplied by researchers | 3848 array and sequence-based datasets |
| Common data submission procedures ensure good data quality | ||
| Tools for data analysis and visualization are provided | ||
| ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) | Functional genomics archive | 60 054 high-throughput experiments |
| Stores both processed and raw data | ||
| Standardized data submission process, frequent updates, connected with GEO | ||
| Expression Atlas (https://www.ebi.ac.uk/gxa/) | Similar to GEO but with fewer datasets that are more focused on baseline experiments | 1572 datasets |
| Two components: Baseline Atlas for expression in ‘normal’ conditions and differential Atlas for experimental expression data | ||
| miRBase (http://www.mirbase.org) | Detailed information on all published and annotated miRNAs | 28 645 entries of miRNAs from 223 species |
| DIANA tools (http://diana.imis.athena-innovation.gr) | Web tool dedicated to miRNA studies | miRNA target identification, and pathway analysis |
| Published validated miRNA–gene interactions | ||
| Automated pipelines to analyse user data | ||
| miRNA-related publication search | ||
| Proteomics | ||
| PRoteomics IDEntifications (PRIDE) (https://www.ebi.ac.uk/pride/archive/) | Proteomics data repository | Stores 3342 projects on protein/peptide identifications and post-translational modifications and supporting spectral evidence |
| Additional annotation of datasets for better organization | ||
| Human Protein Atlas (http://www.proteinatlas.org/) | Protein expression and localization in different tissues and organs (immunochemistry) | Additional information regarding genes, annotations and organs Nice graphical interface |
| Metabolomics | ||
| Human Metabolome Database (http://www.hmdb.ca/) | Detailed information on metabolites (chemical, clinical and molecular biology/biochemistry levels) | Contains thousands of metabolites |
| Search in 17 different biofluids and 617 diseases | ||
| Connections with pathways, proteins and reactions | ||
| Multi-omics | ||
| Multi-Omics Profiling Expression Database (www.proteinspire.org/MOPED/) | Processed multi-omics data | Interactive visualization tools |
Table 2.
Kidney-specific databases
| Name | Description | Main features |
|---|---|---|
| Transcriptomics | ||
| Nephroseq (https://www.nephroseq.org/) | Gene expression in renal disease; integration with clinical data | 26 datasets (1989 samples) |
| Analysis and visualization tools (differential expression, co-expression, outlier, etc.) | ||
| Upload and export tools | ||
| Renal Gene Expression Database (http://rged.wall-eva.net/) | Gene expression in renal disease | 88 research papers analysed |
| Easy-to-use interface | ||
| Proteomics | ||
| Human Kidney and Urine Proteome Project (HKUPP) (http://www.hkupp.org/) | Protein expression in normal urine and normal or diseased kidney | Search for proteins in kidney structures (glomerulus, human medulla) and urine |
| Enables viewing two-dimensional gels and query fractions | ||
| Urinary Protein Biomarker Database (http://122.70.220.102/biomarker) | Candidate protein biomarkers in urine | >400 reports on human and animals |
| 819 human biomarkers, 33 animal biomarkers | ||
| Peptidomics | ||
| Urinary Peptidomics and Peak-maps (http://www.padb.org/updb) | Urinary peptides modified in disease | Search by detection methodology and disease |
| Multi-omics | ||
| Kidney and Urinary Pathway Knowledge Base (KUPKB) (http://www.kupkb.org/) | Collection of publically available omics data related to renal disease | >220 experiments |
| Easy and fast interface | ||
| Pathway visualization with KUPKB Network Visualizer | ||
| Chronic Kidney Disease database (CKDdb) (http://www.padb.org/ckddb) | Collection of publically available omics data related to chronic kidney disease | 366 datasets |
| Search by study, sample, tissue, disease and molecule type | ||
General databases
Functional genomics and transcriptomics are considered the most advanced omics technologies, due to early (compared to other omics traits) major technological improvements and progress in data analysis [9]. One of the universal resources in the field of genomics, the Online Mendelian Inheritance in Man (OMIM) [10] represents a publicly available, daily updated source of information about human genes. The OMIM catalogues >15 000 gene entries focusing mainly on the molecular relationships between genetic and phenotypic variations, with a special emphasis on, but not restricted to, human genetic disorders. Observations on animal models are also available. By obtaining knowledge from the OMIM on genetic disorders, a researcher can acquire some clues about particular genes, observe the phenotyping changes and further evaluate putative differences at the proteome level, setting the OMIM ultimately as an all-around omics database. Due to this universality, the OMIM can be used as a source of information to connect genes and renal phenotypes, as was presented by Parsa et al. [11]. In this publication, the authors systematically compiled the 258 OMIM genes described to be responsible for diseases associated with kidney phenotypes such as renal hypoplasia, dysplasia or agenesis, end-stage renal disease and proteinuria and excluding those causing renal malignancy. Using the previously published genome-wide association study (GWAS) meta-analysis data of the CKDGen Consortium [12], they further studied the potential association of common variants within these genes and kidney function in the general population. Although the authors did not find any new gene variants that could be linked to kidney function decline, this study highlights the value of the OMIM as a source of information to connect genes and renal phenotypes [11].
Besides providing information about the function of genes and/or proteins, databases can be a useful source of information about their localization within the kidney or other organs. One of the largest resources on spatial protein tissue expression is the Human Protein Atlas (HPA) [13]. The HPA database regroups millions of immunohistochemistry-based high-resolution images, presenting the spatial distribution of proteins in human tissues (44 tissue types), cancer types (20 types) and human cell lines (46 types). In the context of renal pathophysiology, the HPA has been used mostly as a downstream resource to validate the expression of newly identified kidney or urinary proteins, following transcriptomics or proteomics experiments, in the different renal compartments [14–18]. Regarding the very limited access to kidney tissue, especially from healthy individuals, the HPA database is an important resource to consider when searching for information regarding kidney protein expression and localization.
Other general databases are listed non-exhaustively in Table 1. These resources include the genomics data repository GeneCards [19] for storing annotated and predicted human genes, databases about microRNAs and their targets such as miRBase [20], miRDB [21] and microCosm (http://www.ebi.ac.uk/enright-srv/microcosm) and the Human Metabolome Database [22] for small molecule metabolites. A number of databases at the European Bioinformatics Institute website (http://www.ebi.ac.uk) are also worth mentioning since they host a number of popular and frequently used omics databases, including Expression Atlas, PRIDE and ArrayExpress [23]. All these databases integrate clinical, functional and molecular annotation and enable fast information retrieval with the use of advanced search engines. Moreover, they are often cross-linked with large biological data repositories (e.g. UniProt, GenBank, KEGG, etc.) and are equipped with various tools for data analysis and visualization. Altogether, although not kidney specific, these general databases provide valuable information about newly identified molecular pathways, drug targets or potential biomarkers associated with kidney disease.
Kidney-specific: focus on omics data originating from urine, kidney and kidney disease
Apart from the various general databases, some resources specifically focus on omics data related to kidney and/or kidney diseases and provide knowledge and support in nephrology research.
Proteomics
The Human Kidney and Urine Proteome Project (HKUPP) [24] database was created in 2006 and hosts experimental data from the human kidney and urine proteome using mass spectrometry. The goal of the HKUPP is to facilitate the analysis of proteomes in normal and disease conditions, provide open-source access to experimental data and help with identification of novel putative biomarkers and drug targets for kidney disease. The database consists of proteomic profiles of kidney structures (glomerulus, renal medulla) as well as urine (normal, proteinuric, exosomes) obtained from the experiments performed in the context of the HKUPP consortium. In addition to human datasets, the HKUPP database also stores profiling data from rat inner medullary collecting ducts (IMCDs) [25]. Therefore, it is feasible to search and compare human and rat data, correlate findings between the two species and thus enrich the proteomic knowledge of kidney development and pathology. Besides these advantages, some limitations have to be mentioned. First, despite being of high quality, the entries are limited to results generated by the HKUPP consortium, while many other urinary or kidney proteomics experiments have been published. Moreover, although some new features have been added recently, such as the ‘Human Renal Medulla Identified Proteome and Mass and Antibody-based Proteomics databases, the resource is neither updated nor upgraded regularly and the data are not confirmed with another technology or with other publications. Finally, proteins have been attributed accession numbers based on the international protein index (IPI), an outdated and obsolete protein database that was closed in 2011 and replaced by Uniprot identifiers, making it difficult to search for information about a protein. Despite these shortcomings, in 2012, Simonson et al. [26] successfully used the HKUPP database as part of an analytical workflow designed to identify novel non-invasive biomarkers of early renal damage associated with type 2 diabetic nephropathy. Using transcriptomics, 36 mRNAs coding for secreted proteins were differentially expressed in the kidney of diabetic db/db mice compared with non-diabetic db/m mice. The HKUPP was used to verify whether some or all of the corresponding proteins were known to be present in human urine. Six of these potential urinary biomarkers of diabetic kidney disease [endothelin-1, lipocalin-2, transforming growth factor β (TGF-β), growth and differentiation factor-15 (GDF-15), interleukin-6 (IL-6) and macrophage chemoattractant protein-1] were further validated in a cohort of 56 patients with type 2 diabetes using ELISA. Urinary protein levels of all six candidates were elevated in type 2 diabetic patients with renal function decline and three of them (endothelin-1, GDF-15 and IL-6) were also associated with proximal tubular damage [26]. This study demonstrates that the HKUPP database, as a resource for reference proteomics datasets, can be easily integrated in basic and clinical research workflows to facilitate studies to understand kidney biology and disease.
The Urinary Protein Biomarker Database (UPBD) [27] contains approximately 1500 urinary protein entries, extracted manually from the literature from >500 reports associated with different human and animal disorders, including a wide range of acute and chronic kidney diseases (CKDs) such as acute renal allograft rejection, diabetic nephropathy or polycystic kidney disease. The proteins of interest can be searched individually, but the database also allows all the proteins modified in one specific condition (disease and/or species) to be found. An interesting feature of the database is the possibility to download the entries, as well as to contribute to the database by submitting third-party study results. In contrast to the HKUPP database, which is based on the presence or absence of a given protein in normal urine (or renal tissue), the UPBD can be used to examine if and when the level of the protein of interest was modified in urine in human or animal models of kidney disease [28–30]. Such an application was described in the publication from Jia et al. [30]. In this work the authors sought to identify new potential urinary biomarkers of kidney disease by targeting specifically the proteins of kidney origin in urine. Indeed, determination of quantitative changes in those protein levels in urine may have a greater potential for detecting modifications of renal function at early stages than proteins deriving from plasma and/or other organs. In order to exclude proteins present in urine as the result of plasma ultrafiltration and selectively collecting proteins from kidney tissue, a model of isolated rat kidney perfusion was used. A total of 1400 proteins were identified in the isolated rat kidney perfusion-driven urine, including 990 proteins expressed in the human kidney. After comparison with the UPBD, 923 of these 990 potential human kidney origin proteins in urine were found to have never been studied as candidate biomarkers of kidney diseases and, hence, could be examined using direct targeted proteomics studies in the discovery phase for new kidney disease biomarkers [30]. The UPBD is an interesting resource compiling modifications of urinary proteins in the context of renal disease. The term ‘biomarker’, however, should be considered with caution. In the past decade, an increasingly large number of papers have been published on novel so-called biomarkers of kidney disease. Yet, their translation to clinical practice is limited. This might be due to problems associated with the design of discovery and validation studies and/or the lack of real clinical benefit of these proteins, which could be associated with the disease pathophysiological mechanisms without any diagnosis or prognosis value [31]. Therefore, most of the proteins present in the database might be considered preferably as candidate biomarkers awaiting validation instead of as biomarkers per se.
Transcriptomics
Nephroseq [32, 33] combines 26 publicly available renal gene expression profiles from renal disease studies and related disorders in humans and mice with a sophisticated analysis engine and an application for data mining and visualization of gene expression data. The power of this database is that the transcriptomics data are reanalysed and associated with detailed clinical information, which allows for more thorough analysis. The interface integrates different analysis modules for differential expression, cluster, outliers and concept analysis. Analysis results can be visualized in the form of summary tables, block plots and co-expression matrices. The success of Nephroseq can be assessed by the number of publications that used the database as part of their analysis workflow [34–40]. It is interesting to note that although Nephroseq compiles transcriptomics data from different kidney diseases such as IgA nephropathy, aging, focal and segmental glomerulosclerosis, transplantation, hypertension or lupus, so far the majority (if not all) of the publications that have used the resource were interested in the datasets associated with diabetic nephropathy. For example, in a recent report, McKay et al. [34] combined two bioinformatics databases, Nephroseq and the Jaspar database (a database containing a set of transcription factor binding sites), in order to predict in silico the involvement of key transcription factors regulating the development of diabetic nephropathy, identifying the transcription factor AP-2 alpha (TFAP2A), myeloid zinc finger 1 (MZF1) and specificity protein 1 (SP1) [34]. Literature mining relieved that TFAP2A and MZF1 are involved in epithelial-to-mesenchymal transition and SP1 regulates TGF-β signalling and fibrogenesis, three mechanisms related to diabetic nephropathy pathogenesis. Using Nephroseq, the authors further confirmed that gene expression of MZF1 was increased in diabetic nephropathy and gene expression of TFAP2A was decreased in an in vitro model of tubular fibrosis (HK2 cells treated with TGF-β). This publication is an example of how the use of databases and bioinformatics tools may offer potential therapeutic targets for the treatment of renal disorders.
Another kidney-specific database of renal gene expression profiles is the Renal Gene Expression Database [41]. This resource consolidates information on gene expression from next-generation sequencing and microarrays from renal cell lines, human kidney tissue and animal models related to various kidney diseases. A user interface enables querying of the database by gene or disease and plots gene expression within the samples. Additionally, a Similarity Analysis Tool Box allows the comparison of selected gene expression with other gene sets from Biocarta, KEGG and Reactome Pathways. It is a new and promising database with a goal to link genes of interest with their expression in different kidney diseases and pathways involved, providing possibilities to drive researchers towards biomarker and drug discovery.
Multi-omics
The Kidney and Urinary Pathway Knowledge Base (KUPKB) is a web tool that includes data from different omics datasets on kidney diseases and facilitates rapid hypothesis generation in the context of renal pathophysiology in a simple environment [42]. The KUPKB offers the network visualization tool KUPNetViz, which enables users to integrate multilayered experimental data over different species, renal locations and renal diseases and protein–protein interaction networks and identify associations with biological functions, biochemical pathways and other functional elements such as miRNAs [43]. In a recent publication, Sanchez-Nino et al. [44] examined the role of the brain abundant signal protein 1 (BASP1) in albumin-induced tubular cell death and its correlation with CKD. BASP1 was increased in apoptotic tubular cells in response to puromycin aminonucleoside–induced nephritic syndrome in rats and in human renal biopsies of proteinuric nephropathy. In vitro, albumin-induced BASP1 expression in tubular cells and inhibition of BASP1 protected from albumin-induced apoptosis. Using the KUPKB, the authors also found that the BASP1 protein was increased in urine from patients with type 2 diabetes with microalbuminuria as compared with those with normoalbuminuria, despite both groups displaying high glucose levels. These results suggest that tubular apoptosis observed in diabetic nephropathy cannot be solely attributed to high glucose levels but also to albumin-induced toxicity [44].
The Chronic Kidney Disease database (CKDdb) [45] is an integrated and clustered information resource that covers multi-omic studies (microRNA, genomics, peptidomics, proteomics and metabolomics) of CKD. The resource was developed by mining the existing literature on the featured topic followed by manual curation of the assembled data. In order to deal with the high heterogeneity and diversity of the gathered data, a specific ontology was applied to tie together and harmonise multilevel omic studies based on gene and protein clusters and mapping of orthologous genes across species. This database is primarily aimed at allowing disease pathway analysis by a data-driven system approach. A workflow utilizing the design of interlinked kidney disease–themed databases, including CKDdb (http://www.padb.org/), was recently published by Husi et al. [46]. It consisted of combined top-down and bottom-up systems biology methods to find novel disease pathways in diabetic arteriopathy. Proteomics analysis was used to identify statistically significant proteins further integrated using GO terms and cluster analysis into interactomes of metabolic and signalling pathways. Additionally, OMIM disease clustering combined with literature mining found the link to human disease pathways and identified decreased glycolysis and fatty acid metabolism as the molecular processes perturbed in diabetic arteriopathy.
Technical note on data integration: from databases to model development
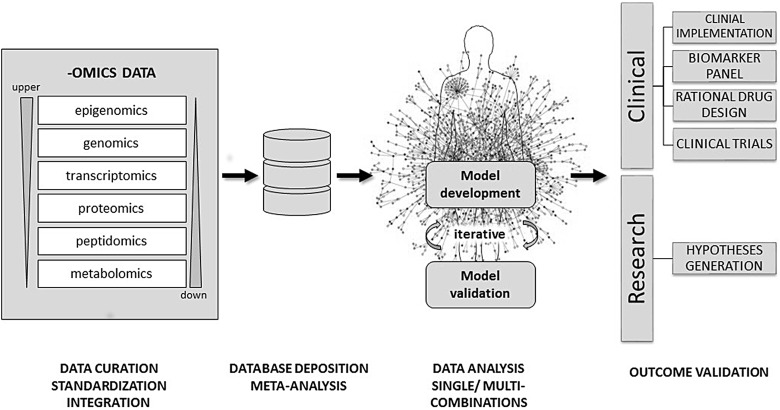
Combinations of different omics data sources obtained from various biological levels and/or closely related conditions should allow for better understanding of complex biological systems and elucidation of progression mechanisms, hence enabling the discovery of novel biomarkers and therapeutic targets [47]. However, integration and analysis of heterogeneous omics data is difficult and, until recently, was not a common approach in data analysis workflows. A detailed discussion on the bioinformatic tools for data integration and system biology approaches in CKD can be found in the recent review by Cisek et al. [48]. After data acquisition via different high-throughput omics platforms, pre-processing steps are required to obtain normalized, standardized, high-quality and statistically significant data. Such data can be later used for initial meta-analysis and to perform further iterative, single and multicombinatorial analysis to elucidate existing inter- and intradependencies in the form of interaction pathways, statistical models and others [49]. Database systems have found their position as the tools to bridge this gap, as they can provide data in a common form, both in structure and content, that facilitates data analysis, comparison and integration on a greater scale [50]. Analysis outcomes can provide novel insights that need to be further validated in either clinical or research environments to ultimately be used in the clinic or to pursue new research hypotheses [51]. In Figure 1 we describe this approach from omics data generation to the creation and validation of a model via the use of databases.
Fig. 1.
Proposed workflow for a data-driven approach in multi-omics data integration. After data acquisition via different high-throughput omic platforms, raw data can be stored locally in the owner's database and be pre-processed (data cleaning, filtering, normalization, reduction, etc.). After the pre-processing steps the data are matched with current reference repositories (data curation). The latter metadata can be then deposited in a different database that only displays statistically relevant features, which is much more amenable for collaborative use for researchers in a common project. Single and/or multiple combinations can be used in order to integrate data coming from up- and downstream levels and then used to develop models that together try to mimic the cell environment and represent their own interactome. The state-of-art model would consider simultaneously any network topology, molecular interaction and statistical relevance in order to provide the most robust representation of the cell dynamics when undergoing disease. Every new model requires confirmation by validating some selected molecular features using in vivo or in vitro experiments (immunohistochemistry, qRT-PCR, ELISA, etc.) This is an iterative step, in which to obtain a final model, it could involve several cycles of incrementing new data and testing its validity until an optimal phase is reached where the model is considered suitable for scientific scrutiny.
Showcasing the use of different databases in kidney research
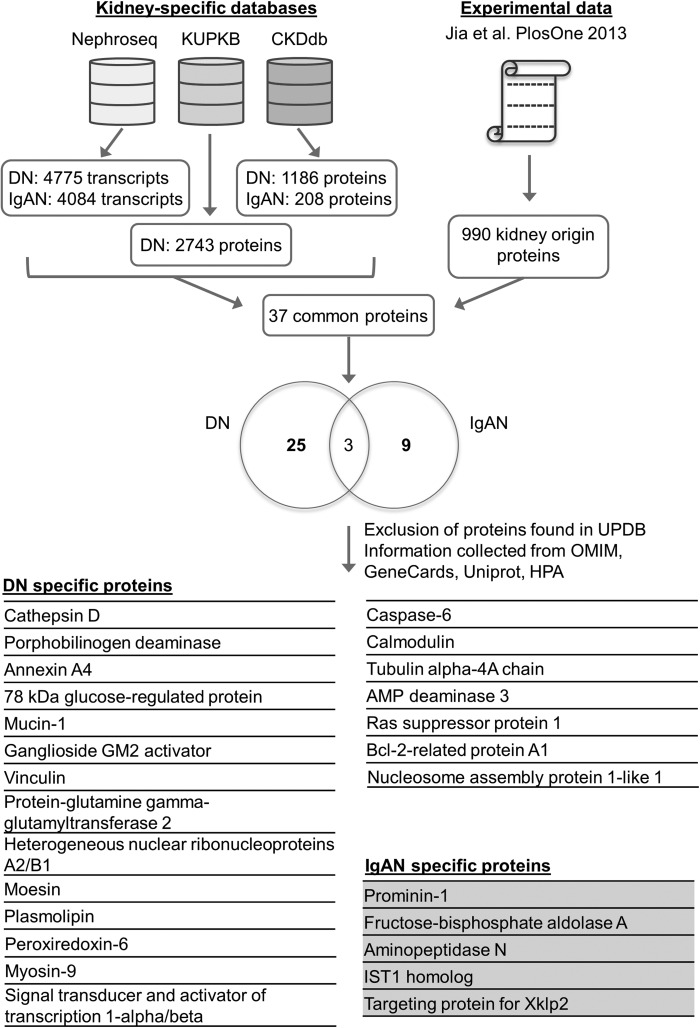
In order to showcase the potential utility of combining the existing kidney disease databases, we analysed kidney transcriptomic and proteomic datasets originating from diabetic nephropathy and IgA nephropathy human studies with the aim to propose markers of IgA nephropathy in diabetic individuals. In most patients with type 2 diabetes, renal disease is due to diabetic nephropathy. However, the prevalence of other, non-diabetic nephropathies such as IgA nephropathy, isolated or superimposed on a diabetic nephropathy, can range from 6 to 64% [52]. Therefore, in everyday clinical practice, there is a critical need to identify specific biomarkers to exclude the possibility of a non-diabetic, potentially treatable glomerulonephritis in diabetic patients [53]. The aim of this analysis was to identify new potential biomarkers and/or mechanistic targets that could help differentiate diabetic and/or IgA nephropathy in diabetic patients. To do so we chose to further analyse the study from Jia et al. [30], where the authors investigated the proteome from perfused rat kidneys and identified 990 proteins as kidney proteins with human orthologs. By comparing this dataset with the UPBD, 67 proteins were already described as candidate biomarkers for human kidney diseases, leaving 923 proteins with unknown potential association to nephropathy. In the test case, we compared the 990 proteins with Nephroseq, the KUPKB and the CKDdb, focusing on identifying proteins for IgA nephropathy and diabetic nephropathy. Comparison of the 107 IgA nephropathy proteins and 212 diabetic nephropathy proteins extracted from the kidney-specific databases and the 990 proteins from Jia et al. yielded 5 proteins specific for IgA nephropathy and 21 proteins specific for diabetic nephropathy that were not present in the UPBD. Analysis workflow is depicted in Figure 2. In addition to the kidney-specific databases, we also used the general databases OMIM and Human Protein Atlas in order to gather further knowledge about these proteins. Some of these proteins are already known to be potentially involved in diabetic and IgA nephropathies, such as aminopeptidase N [54], vinculin [55], plasmolipin [56, 57], transglutaminase-2 [58, 59] and signal transducer and activator of transcription 1-α/β [60, 61]. Some proteins, however, although not yet found to be directly involved, showed potential interest, as they were associated with mechanisms related to diabetic nephropathy. For example, 78-kDa glucose-regulated protein is a molecular chaperone involved in endoplasmic reticulum stress, a mechanism mediating cell death in diabetic nephropathy [62, 63]. Another example is ganglioside GM2 activator, an enzyme involved in lipid metabolism, which could be of significant interest as a biomarker regarding the pathogenic potential of gangliosides in the development of diabetic nephropathy [64, 65].
Fig. 2.
The test case pipeline. The comparison of the transcriptomics and proteomics datasets for Diabetic Nephropathy (DN) and IgA nephropathy (IgAN) from the CKDdb, the KUPKB and Nephroseq with the 990 proteins identified in urine in the Jia et al. [30] study yielded 37 common proteins, 25 DN-specific proteins, 9 IgAN-specific and 3 for both cases (Venn diagram). The proteins found as possible biomarker candidates in Jia et al. through analysis in the Urinary Proteome Biomarker Database were excluded, resulting to 21 proteins specific for DN and 5 proteins specific for IgAN.
In conclusion, we show with this example that the combination of different general and kidney-specific databases can lead to the generation of new hypotheses and to new candidate molecules for further in-depth research for mechanistic, drug and/or biomarker discovery to help answer some frequent problems that clinicians face, such as the precise diagnosis of diabetic nephropathy versus IgA nephropathy in diabetic individuals.
Conclusion
Kidney-specific databases are becoming more mature. Technological progress, additional knowledge and information on kidney pathology and physiology and the plurality of information found in the different kidney-specific and general databases produces confidence in the presented results. Web resources such as Nephroseq, the KUPKB and the CKDdb attempt to unify all available information from different sample origins and omics levels and provide nephrology researchers with tools for more comprehensive research and analysis. Conversely, in scientific fields like biology and molecular biology that are advancing at a rapid pace, the limited rate of updates in these resources may lead to a large number of false discoveries. Users must be careful when selecting a database to support their research. They should take into consideration the source of the experimental data (e.g. Nephroseq for genes or UPBD for urinary proteins). Most importantly, users must keep in mind that the databases provide a static picture which might be true only until proven otherwise at a later point in time. Nevertheless, the development and use of omics databases represents a major step forward in aiding the fast confirmation of findings, but also to ultimately elucidate novel hypotheses in the context of renal diseases. These databases at the current form do not yet change everyday life of the clinicians. Nevertheless, it is likely that in the near future this large amount of available data, combined with powerful in silico analysis tools and user friendly interfaces, will provide significant help to shed light on the pathophysiology of the kidney and improve future diagnosis and treatment options in clinical nephrology.
Conflict of interest statement
K.C. is an employee in Mosaiques Diagnostics. All other authors declare no conflict of interest.
Acknowledgements
This work was supported by ‘Clinical and system -omics for the identification of the Molecular Determinants of established Chronic Kidney Disease’ (iMODE-CKD; PEOPLE-ITN-GA-2013-608332). This work was supported by the Fondation du Rein sous égide de la Fondation pour la Recherche Médicale et ses partenaires, grant number GENZYME 2014 FDR-SdN/FRM, to JK.
References
- 1.Jiang S, Chuang PY, Liu ZH et al. The primary glomerulonephritides: a systems biology approach. Nat Rev Nephrol 2013; 9: 500–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellazzi R. Big data and biomedical informatics: a challenging opportunity. Yearb Med Inform 2014; 9: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hood L, Flores M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. N Biotechnol 2012; 29: 613–624 [DOI] [PubMed] [Google Scholar]
- 4.Loupy A, Lefaucheur C, Vernerey D et al. Molecular microscope strategy to improve risk stratification in early antibody-mediated kidney allograft rejection. J Am Soc Nephrol 2014; 25: 2267–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Micheel CM, Nass SJ, Omenn GS (eds). Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington, DC: National Academies Press, 2012 [Google Scholar]
- 6.Bebek G, Koyuturk M, Price ND et al. Network biology methods integrating biological data for translational science. Brief Bioinform 2012; 13: 446–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynn DJ, Lloyd AT, O'Farrelly C. Bioinformatics: implications for medical research and clinical practice. Clin Invest Med 2003; 26: 70–74 [PubMed] [Google Scholar]
- 8.Hood L. Systems biology and p4 medicine: past, present, and future. Rambam Maimonides Med J 2013; 4: e0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlaanderen J, Moore LE, Smith MT et al. Application of OMICS technologies in occupational and environmental health research; current status and projections. Occup Environ Med 2010; 67: 136–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamosh A, Scott AF, Amberger JS et al. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005; 33(Database issue): D514–D517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsa A, Fuchsberger C, Kottgen A et al. Common variants in Mendelian kidney disease genes and their association with renal function. J Am Soc Nephrol 2013; 24: 2105–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pattaro C, Kottgen A, Teumer A et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet 2012; 8: e1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhlen M, Fagerberg L, Hallstrom BM et al. Proteomics. Tissue-based map of the human proteome. Science 2015; 347: 1260419. [DOI] [PubMed] [Google Scholar]
- 14.Cui Z, Yoshida Y, Xu B et al. Profiling and annotation of human kidney glomerulus proteome. Proteome Sci 2013; 11: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lennon R, Byron A, Humphries JD et al. Global analysis reveals the complexity of the human glomerular extracellular matrix. J Am Soc Nephrol 2014; 25: 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neiman M, Hedberg JJ, Donnes PR et al. Plasma profiling reveals human fibulin-1 as candidate marker for renal impairment. J Proteome Res 2011; 10: 4925–4934 [DOI] [PubMed] [Google Scholar]
- 17.Hansson J, Hultenby K, Cramnert C et al. Evidence for a morphologically distinct and functionally robust cell type in the proximal tubules of human kidney. Hum Pathol 2014; 45: 382–393 [DOI] [PubMed] [Google Scholar]
- 18.Habuka M, Fagerberg L, Hallstrom BM et al. The kidney transcriptome and proteome defined by transcriptomics and antibody-based profiling. PLoS One 2014; 9: e116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang AD, Dai SX, Huang JF. Reconstruction and analysis of human kidney-specific metabolic network based on omics data. Biomed Res Int 2013; 2013: 187509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014; 42(Database issue): D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015; 43(Database issue): D146–D152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart DS, Jewison T, Guo AC et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res 2013; 41(Database issue): D801–D807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooksbank C, Bergman MT, Apweiler R et al. The European Bioinformatics Institute's data resources 2014. Nucleic Acids Res 2014; 42(Database issue): D18–D25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto T, Langham RG, Ronco P et al. Towards standard protocols and guidelines for urine proteomics: a report on the Human Kidney and Urine Proteome Project (HKUPP) symposium and workshop, 6 October 2007, Seoul, Korea and 1 November 2007, San Francisco, CA, USA. Proteomics 2008; 8: 2156–2159 [DOI] [PubMed] [Google Scholar]
- 25.Pisitkun T, Bieniek J, Tchapyjnikov D et al. High-throughput identification of IMCD proteins using LC-MS/MS. Physiol Genomics 2006; 25: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simonson MS, Tiktin M, Debanne SM et al. The renal transcriptome of db/db mice identifies putative urinary biomarker proteins in patients with type 2 diabetes: a pilot study. Am J Physiol Renal Physiol 2012; 302: F820–F829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao C, Li M, Li X et al. A tool for biomarker discovery in the urinary proteome: a manually curated human and animal urine protein biomarker database. Mol Cell Proteomics 2011; 10: M111.010975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao M, Li M, Li X et al. Dynamic changes of urinary proteins in a focal segmental glomerulosclerosis rat model. Proteome Sci 2014; 12: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Y, Zhang F, Wu J et al. Urinary candidate biomarker discovery in a rat unilateral ureteral obstruction model. Sci Rep 2015; 5: 9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia L, Li X, Shao C et al. Using an isolated rat kidney model to identify kidney origin proteins in urine. PLoS One 2013; 8: e66911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mischak H, Delles C, Vlahou A et al. Proteomic biomarkers in kidney disease: issues in development and implementation. Nat Rev Nephrol 2015; 11: 221–232 [DOI] [PubMed] [Google Scholar]
- 32.Athey BD, Cavalcoli JD, Jagadish HV et al. The NIH National Center for Integrative Biomedical Informatics (NCIBI). J Am Med Inform Assoc 2012; 19: 166–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martini S, Eichinger F, Nair V et al. Defining human diabetic nephropathy on the molecular level: integration of transcriptomic profiles with biological knowledge. Rev Endocr Metab Disord 2008; 9: 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKay GJ, Kavanagh DH, Crean JK et al. Bioinformatic evaluation of transcriptional regulation of WNT pathway genes with reference to diabetic nephropathy. J Diabetes Res 2016; 2016: 7684038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderberg RJ, Meek RL, Hudkins KL et al. Serum amyloid A and inflammation in diabetic kidney disease and podocytes. Lab Invest 2015; 95: 250–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chitra PS, Swathi T, Sahay R et al. Growth hormone induces transforming growth factor-beta-induced protein in podocytes: implications for podocyte depletion and proteinuria. J Cell Biochem 2015; 116: 1947–1956 [DOI] [PubMed] [Google Scholar]
- 37.Wanic K, Krolewski B, Ju W et al. Transcriptome analysis of proximal tubular cells (HK-2) exposed to urines of type 1 diabetes patients at risk of early progressive renal function decline. PLoS One 2013; 8: e57751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Donovan HC, Hickey F, Brazil DP et al. Connective tissue growth factor antagonizes transforming growth factor-beta1/Smad signalling in renal mesangial cells. Biochem J 2012; 441: 499–510 [DOI] [PubMed] [Google Scholar]
- 39.Brennan EP, Morine MJ, Walsh DW et al. Next-generation sequencing identifies TGF-beta1-associated gene expression profiles in renal epithelial cells reiterated in human diabetic nephropathy. Biochim Biophys Acta 2012; 1822: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brosius FC III, Alpers CE. New targets for treatment of diabetic nephropathy: what we have learned from animal models. Curr Opin Nephrol Hypertens 2013; 22: 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, Yang B, Chen X et al. Renal Gene Expression Database (RGED): a relational database of gene expression profiles in kidney disease. Database (Oxford) 2014; 2014: bau092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein J, Jupp S, Moulos P et al. The KUPKB: a novel Web application to access multiomics data on kidney disease. FASEB J 2012; 26: 2145–2153 [DOI] [PubMed] [Google Scholar]
- 43.Moulos P, Klein J, Jupp S et al. The KUPNetViz: a biological network viewer for multiple -omics datasets in kidney diseases. BMC Bioinformatics 2013; 14: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez-Nino MD, Fernandez-Fernandez B, Perez-Gomez MV et al. Albumin-induced apoptosis of tubular cells is modulated by BASP1. Cell Death Dis 2015; 6: e1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandes M, Husi H. FP222 The Chronic Kidney Disease Database (CKDdb). Nephrol Dial Transplant 2015; 30(Suppl 3): iii141 [Google Scholar]
- 46.Husi H, Van Agtmael T, Mullen W et al. Proteome-based systems biology analysis of the diabetic mouse aorta reveals major changes in fatty acid biosynthesis as potential hallmark in diabetes mellitus-associated vascular disease. Circ Cardiovasc Genet 2014; 7: 161–170 [DOI] [PubMed] [Google Scholar]
- 47.Hu ZZ, Huang H, Wu CH et al. Omics-based molecular target and biomarker identification. Methods Mol Biol 2011; 719: 547–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cisek K, Krochmal M, Klein J et al. The application of multi-omics and systems biology to identify therapeutic targets in chronic kidney disease. Nephrol Dial Transplant 2015; doi:10.1093/ndt/gfv364 [DOI] [PubMed] [Google Scholar]
- 49.Meng C, Kuster B, Culhane AC et al. A multivariate approach to the integration of multi-omics datasets. BMC Bioinformatics 2014; 15: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou D, Ma L, Yu J et al. Biological databases for human research. Genomics Proteomics Bioinformatics 2015; 13: 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayers D, Day PJ. Systems medicine: the application of systems biology approaches for modern medical research and drug development. Mol Biol Int 2015; 2015: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanhille P. The diabetic patient with renal insufficiency. Diabetes Metab 2000; 26(Suppl 4): 67–72 [PubMed] [Google Scholar]
- 53.Kamal L, Salvatore S, Hartono C et al. Concomitance of IgA nephropathy and diabetic nephropathy in a kidney allograft: case report and review of the literature. Transplant Proc 2014; 46: 2396–2399 [DOI] [PubMed] [Google Scholar]
- 54.Moon PG, Lee JE, You S et al. Proteomic analysis of urinary exosomes from patients of early IgA nephropathy and thin basement membrane nephropathy. Proteomics 2011; 11: 2459–2475 [DOI] [PubMed] [Google Scholar]
- 55.Millioni R, Iori E, Puricelli L et al. Abnormal cytoskeletal protein expression in cultured skin fibroblasts from type 1 diabetes mellitus patients with nephropathy: a proteomic approach. Proteomics Clin Appl 2008; 2: 492–503 [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez-Fraticelli AE, Bagwell J, Bosch-Fortea M et al. Developmental regulation of apical endocytosis controls epithelial patterning in vertebrate tubular organs. Nat Cell Biol 2015; 17: 241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu XD, Zhang LY, Zhu TC et al. Overexpression of miR-34c inhibits high glucose-induced apoptosis in podocytes by targeting Notch signaling pathways. Int J Clin Exp Pathol 2015; 8: 4525–4534 [PMC free article] [PubMed] [Google Scholar]
- 58.Huang L, Haylor JL, Fisher M et al. Do changes in transglutaminase activity alter latent transforming growth factor beta activation in experimental diabetic nephropathy? Nephrol Dial Transplant 2010; 25: 3897–3910 [DOI] [PubMed] [Google Scholar]
- 59.Berthelot L, Papista C, Maciel TT et al. Transglutaminase is essential for IgA nephropathy development acting through IgA receptors. J Exp Med 2012; 209: 793–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei Q, Dong Z. HDAC4 blocks autophagy to trigger podocyte injury: non-epigenetic action in diabetic nephropathy. Kidney Int 2014; 86: 666–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox SN, Sallustio F, Serino G et al. Activated innate immunity and the involvement of CX3CR1-fractalkine in promoting hematuria in patients with IgA nephropathy. Kidney Int 2012; 82: 548–560 [DOI] [PubMed] [Google Scholar]
- 62.Liu G, Sun Y, Li Z et al. Apoptosis induced by endoplasmic reticulum stress involved in diabetic kidney disease. Biochem Biophys Res Commun 2008; 370: 651–656 [DOI] [PubMed] [Google Scholar]
- 63.Sanchez-Nino MD, Benito-Martin A, Ortiz A. New paradigms in cell death in human diabetic nephropathy. Kidney Int 2010; 78: 737–744 [DOI] [PubMed] [Google Scholar]
- 64.Grove KJ, Voziyan PA, Spraggins JM et al. Diabetic nephropathy induces alterations in the glomerular and tubule lipid profiles. J Lipid Res 2014; 55: 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vukovic I, Bozic J, Markotic A et al. The missing link – likely pathogenetic role of GM3 and other gangliosides in the development of diabetic nephropathy. Kidney Blood Press Res 2015; 40: 306–314 [DOI] [PubMed] [Google Scholar]