Abstract
Protein–protein interactions (PPIs) underlie most biological processes. An increasing interest to investigate the unexplored potential of PPIs in drug discovery is driven by the need to find novel therapeutic targets for a whole range of diseases with a high unmet medical need. To date, PPI inhibition with small molecules is the mechanism that has most often been explored, resulting in significant progress towards drug development. However, also PPI stabilization is gradually gaining ground. In this review, we provide a focused overview of a number of PPIs that control critical regulatory pathways and constitute targets for the design of novel therapeutics. We discuss PPI-modulating small molecules that are already pursued in clinical trials. In addition, we review a number of PPIs that are still under preclinical investigation but for which preliminary data support their use as therapeutic targets.
Introduction
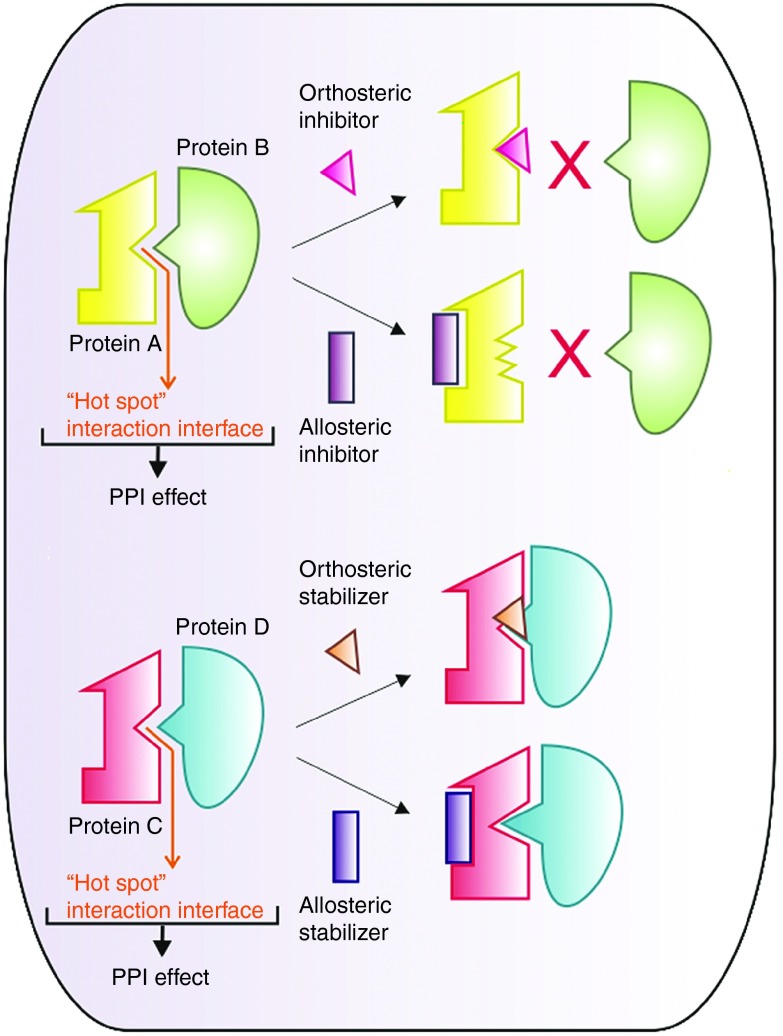
The human interactome has been estimated to cover ~400,000 protein–protein interactions (PPIs), indicating an area of high complexity and organization, which may hide answers to many unsolved questions in biology. In addition, PPIs provide a wealth of opportunities for therapeutic intervention in a broad range of disease conditions. For long, the typical large and flat nature of protein interaction surfaces, often missing clear features (such as pockets, grooves, or clefts) that could act as potential docking sites for small molecule inhibitors, has withheld researchers from exploiting PPIs as drug targets.1 In those cases where such features are present, the structural complexity of the interface often poses an additional challenge; the binding epitopes of PPI surfaces are often created by secondary and tertiary protein structures, precluding the use of a linear peptide sequence as a template for modeling a new therapeutic molecule, e.g., a small molecule peptide mimetic.2 Moreover, the lack of natural small molecule ligands that could serve as an alternative starting point for drug design has been perceived as another major obstacle.1 With the discovery of so-called “hot spots” in PPI interfaces, well-defined regions that contribute most of the binding energy, it became feasible to target a broader range of PPIs with small molecule drugs.3 The identification of hot spots has enabled researchers to identify molecules that interact at these sites, thus interfering with PPIs and the downstream pathways they mediate. Small molecule compounds that modulate PPIs can directly target the protein interaction interface, resulting in its disruption or stabilization (orthosteric PPI inhibitors or stabilizers). Alternatively, PPI-modulating compounds can bind to a neighboring site on one of the interacting proteins and inhibit or enhance the PPI by changing its conformation (allosteric PPI inhibitors or stabilizers) (Figure 1).4,5
Figure 1.
The “hot spot” concept and the rationale for designing PPI modulators. Upper panel: an orthosteric small molecule inhibitor binds to the interaction interface of two proteins, thereby preventing their interaction. An allosteric inhibitor binds to the interacting protein A outside of the PPI surface, inducing a conformational change that inhibits its association with protein B. Lower panel: similarly, an orthosteric small molecule stabilizer binds to the PPI surface and stabilizes the interaction between proteins B and C, whereas an allosteric stabilizer changes the conformation of protein C so that protein D can bind with higher affinity. PPI, protein–protein interaction.
Technological progress has played a key role in the identification of small molecules modulators of PPIs. Indeed, sensitive screening approaches are required to detect the typically low affinity interaction with a protein interaction interface of initial small molecule hits. High-throughput screening-compatible assays that have yielded useful starting points for chemical optimization include fluorescence resonance energy transfer, amplified luminescent proximity homogeneous assay screen (AlphaScreen; PerkinElmer), surface plasmon resonance, and fluorescence polarization.1,6 Alternatively, PPI inhibitor discovery programs driven by a structure-based approach have proven successful.7 Structural information about the PPI interface—obtained through X-ray crystallography, nuclear magnetic resonance or homology modeling—enables in silico screening of virtual compound libraries. In a next stage, promising hits are synthesized and tested in an appropriate protein binding or interaction assay.
Applying this highly diverse set of discovery tools, potent PPI modulators are being developed for a broad spectrum of protein complexes and several of these have already progressed into clinical trials. In this review, we provide an overview of the application range of PPI modulators and present a selection of promising compounds that are currently making their way through (pre-)clinical development.
The “Yin” Face of PPIs – Inhibition of PPIs in Drug Design
Interactions involved in the cell cycle pathway as possible therapeutic targets for cancer
MDM2/p53. One of the best-studied PPIs in cancer research is the interaction of murine double minute 2 (MDM2) with p53. The transcription factor p53 plays a crucial role in cell cycle regulation, apoptosis, DNA repair, senescence, angiogenesis, and innate immunity.8,9 p53 is a potent tumor suppressor and in 50% of human cancers, its antitumor activity is impaired due to mutations within the p53 gene.10 In most other human cancers, p53 retains its wild-type status but its function as a tumor suppressor is compromised by multiple intracellular mechanisms. MDM2 or HDM2 in human is the major inhibitor of p53. MDM2 binds directly to p53, resulting in a repressed p53 transactivation activity, enhanced nuclear export of p53, and degradation of p53 by ubiquitination through its E3 ligase activity (Figure 2).11,12,13 Additionally, overexpression of MDM2 in human tumors correlates with poor clinical prognosis and poor treatment response to current cancer treatments. Amplification of MDM2 was found in 7% of human cancers following an analysis of 28 different cancer types, while amplification of MDM2 and mutations in the p53 gene are mutually exclusive.14 For these reasons, it became clear that interference with the MDM2/p53 interaction could lead to an improved antitumor action of p53 and more efficient anticancer treatments. MDM2 and p53 interact via their N-terminal domains,15,16 more specifically via a hydrophobic surface groove in MDM2 and three key hydrophobic residues in p53, Phe19, Trp23, and Leu26. These residues make up the “hot spot” which was targeted by researchers in an attempt to identify molecules that can interrupt this specific interaction.17 Although still an area of active research, seven MDM2-p53 inhibitors have progressed to clinical trials with impressive results.
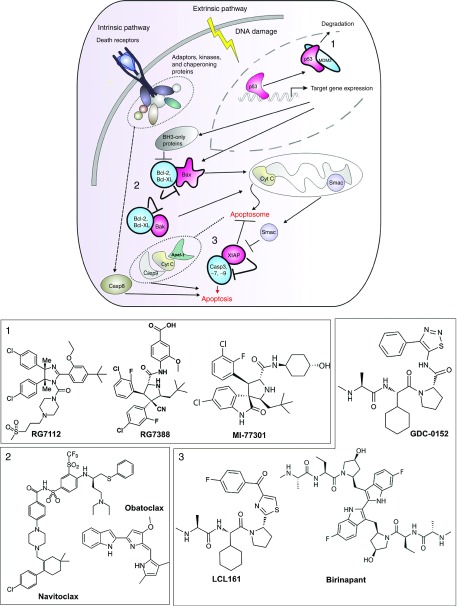
Figure 2.
Small molecule inhibitors in clinical and preclinical stage interfere with PPIs involved in the apoptosis pathway as an anticancer treatment. This scheme illustrates three clinically important PPIs: 1) MDM2/p53; 2) Bcl2, Bcl-XL/Bak, Bax; and 3) IAP/caspases, and their role in the apoptosis cascade. In blue, proteins are depicted that are targeted for inhibition by small molecules and in pink their interaction partners that promote apoptosis. The panels below depict the chemical structures of representative small molecule inhibitors that interfere with the respective PPIs. IAP, inhibitor of apoptosis protein; MDM2, murine double minute 2; PPI, protein–protein interaction.
In 2004, researchers at Roche (Basel, Switzerland) identified the nutlins, a first class of specific and orally active, imidazoline-containing compounds that bind to MDM2 by mimicking the structure of the p53 peptide and the accompanying in vitro data supported cell growth inhibition. Nutlins were identified by screening a small molecule diversity library using an surface plasmon resonance-based p53-MDM2 competition assay.18 Further chemical optimization, aimed at enhancing binding and pharmacokinetics, yielded RG7112 a compound that entered clinical trials for sarcoma, myelogenous leukemia, neoplasm, and hematologic neoplasm.19 The first promising data with RG7112 in clinical trials emerged in 2012. The results from patients with an MDM2-amplified liposarcoma showed clear evidence of p53 reactivation and cell growth inhibition. Unfortunately, its long-term administration is correlated with hematologic cytotoxicity, including neutropenia and thrombocytopenia.20 The same company synthesized the pyrrolidine-containing compound RG7388, a RG7112 analogue.21 This molecule has better pharmacological properties and can activate p53 more potently than RG7112. Upon oral administration, it achieves tumor regression in the SJSA-1 osteosarcoma xenograft model in mice. Currently, RG7388 is ready to enter phase 2 clinical trials for the treatment of patients with acute myelogenous leukemia, solid tumors, or advanced malignancies either as a single agent or in combination with chemotherapeutics such as cytarabine, although hematologic adverse effects remain dose-limiting.22 Sanofi's MI-77301 (SAR405838; Sanofi, Paris, France) is a spirooxindole-containing compound that entered phase 1 clinical trials to assess its safety, pharmacokinetics, and biological activity in patients with advanced tumors.23 Identified by using structure-based approaches to mimic the three key binding residues of p53, MI-77301 binds to MDM2 with kinetics in the nanomolar range.24 Still more molecules are being developed by different pharmaceutical companies to target this specific interaction (Table 1).25,26 MDM2 inhibitors are used as single agents or in combination with traditional chemotherapeutic agents. Combined treatment is needed because the MDM2 inhibitors are highly selective for MDM2 but not for MDMX, which also interacts directly with p53 and represses its action. Conventional chemotherapeutics such as irinotecan and doxorubicin can effectively downregulate the levels of MDMX and consequently their combination with MDM2 inhibitors effectively treats human cancers characterized by high expression of both MDM2 and MDMX. Efforts towards the identification of inhibitors of MDMX led to the discovery of additional molecules such as RO-5963,27 which has high binding affinities to MDM2 as well as MDMX.
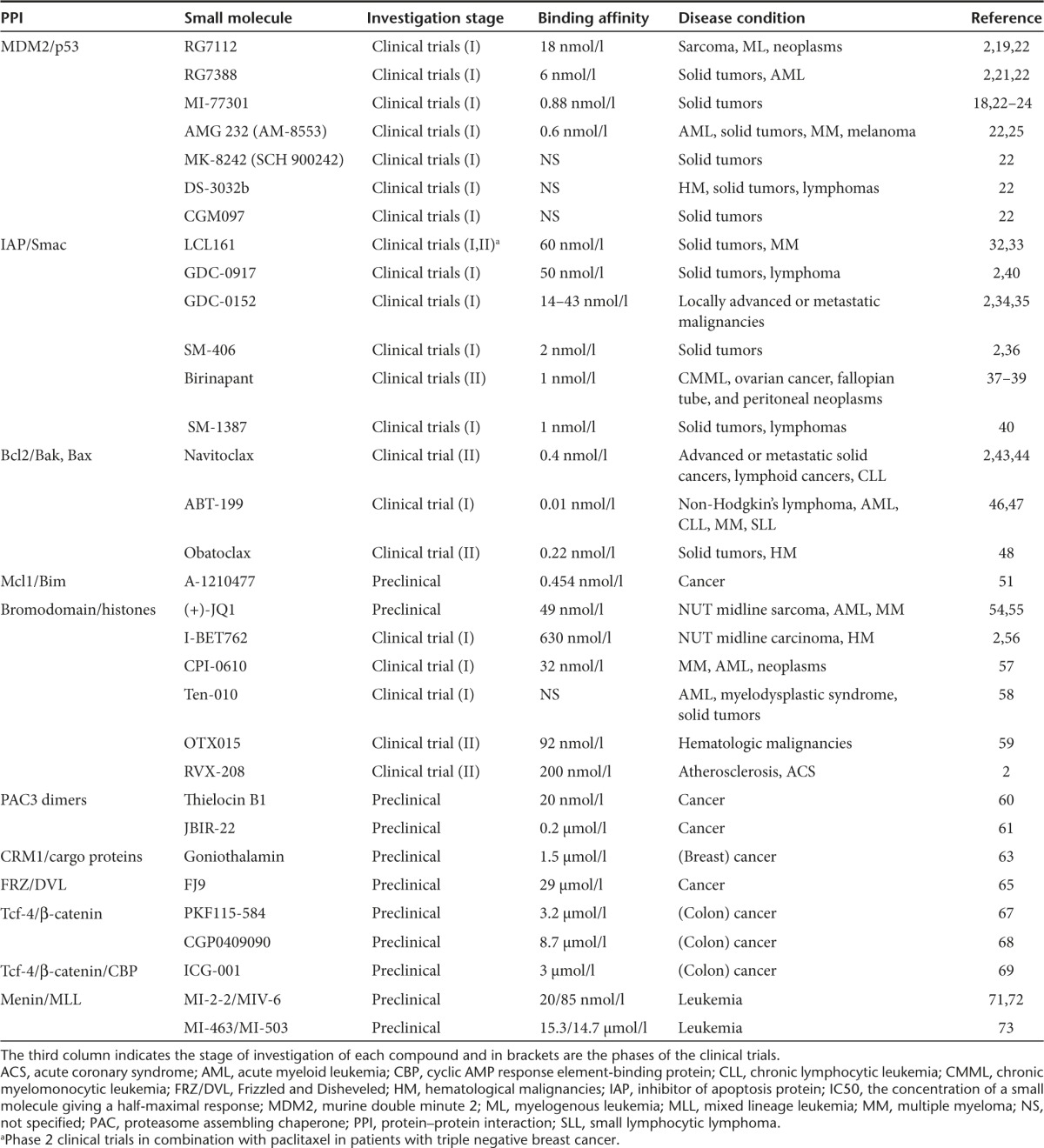
Table 1. Modulation of PPIs involved in cancer by small molecule inhibitors.
The caspase 9, XIAP/BIR3, SMAC system. Apoptosis, or programmed cell death, is mediated mainly through two different pathways, yet these intrinsic and extrinsic pathways culminate in the activation of caspases. Caspase-9 is an initiator caspase in the intrinsic pathway that dimerizes into a catalytically active form able to cleave and activate procaspase-3 and procaspase-7.28 The inhibitor of apoptosis proteins (IAPs) are overexpressed or constitutively activated in tumor cells, resulting in evasion of programmed cell death. The XIAP (X-linked IAP) is the most potent caspase inhibitor among the IAP protein family.29 XIAP contains three baculoviral inhibitory repeat (BIR) domains and a ring domain. This protein interacts with initiator caspase-9 through its BIR3 domain and with caspases 3 and 7 through its BIR1/2 domains.30 BIR3 inhibits caspase-9 by preventing dimerization, which is required for its catalytic activity. The search for new compounds that are able to disrupt the XIAP–caspase interaction has attracted attention of the scientific community as a promising strategy for cancer treatment.
The natural protein inhibitor of XIAP, SMAC/DIABLO (second mitochondrial activator of caspases/direct IAP-binding protein with Low PI), is released from the mitochondria into the cytosol in response to apoptotic stimuli (Figure 2). SMAC competes with caspase binding to BIR domains through its interaction with the AVPI tetrapeptide (Ala-Val-Pro-Ile) present in the N-terminal part of SMAC.31 Since the discovery of the SMAC protein in 2000, there has been an enormous interest by academic laboratories and pharmaceutical companies to design small molecule SMAC mimetics.32 Seven SMAC mimetics have reached clinical trials and five molecules remain in clinical development. Current IAP inhibitors belong to two distinct classes, i.e., the monovalent and the bivalent inhibitors. The monovalent inhibitors, like LCL161 (Novartis, Basel, Switzerland),29,33 GDC-0917/CUDC-427, GDC-0152 (RG7419) (Genentech, San Francisco, CA/Curis, Lexington, MA),34,35 and SM-406/AT-406 (Wang lab/Ascenta Therapeutics, Malvern, PA),36 were designed based on the AVPI peptide and have IC50 values in the nanomolar range. The bivalent inhibitors, such as Birinapant (TL-32711) (Tetralogic Pharmaceuticals, Malvern, PA)37,38,39 and SM-1387 (APG1387),40 are dimerized SMAC mimetics, which as homodimers bind simultaneously both XIAP's BIR1/2 and BIR3 domains.
Bcl2 family. The B-cell lymphoma 2 (BCL2) family of proteins is composed of more than 20 members. Some members are antiapoptotic, like Bcl2, Bcl-XL, Bcl2l2, Mcl-1, whereas some others are proapoptotic such as Bax, Bak1, Bid, and Bcl2l11. The members of this family can engage in PPIs to modulate the intrinsic apoptotic pathway.41 The antiapoptotic Bcl2 members protect the cells against apoptosis by inhibiting the actions of the proapoptotic members (Figure 2). In some cancer types, the antiapoptotic members are overexpressed and the discovery of molecules that can bind to their hydrophobic grooves are predicted to induce apoptosis in cancer cells by antagonizing their protective effect. In 2005, researchers from the Abbott Laboratories (Chicago, IL), having utilized nuclear magnetic resonance-based screening and structure-based design, identified ABT-737, a potent small molecule inhibitor of Bcl2, Bcl-XL, and Bcl2l2.42 Further optimization of ABT-737 in terms of pharmacokinetics and efficacy via a fragment-based approach launched Navitoclax (ABT-263) as a potent antiapoptotic Bcl2 inhibitor, but its administration led to thrombocytopenia due to suppression of Bcl-xL.43,44,45 Later on, a Bcl2-specific version was designed, i.e., ABT-199 (RG7601). This compound is in phase 1 trials for chronic lymphocytic lymphoma or small lymphocytic lymphoma with an encouraging response rate of 84%.46,47 Another auspicious Bcl2 inhibitor is Obatoclax (GX015-070) from Gemin X Pharmaceuticals (Montreal, Quebec, Canada), an indole bipyrrole-containing drug, which is currently assessed in multiple phase 2 clinical trials. The attractive safety profile of Obatoclax offers the opportunity to treat many forms of cancer both as a single agent and in combination with current treatments. Advantageously, it is well tolerated, without any evidence of immuno- or myelosuppression.48
Overexpression of Mcl-1 in cancer cells results in the sequestration of the proapoptotic Bak, Bax, Bad, and Bim, thus Mcl-1 has also been subjected to inhibitor screens. Apart from natural compounds reported to inhibit this interaction in fluorescence resonance energy transfer assays,49 Varadarajan et al. introduced the small molecule TW-37 as a specific Mcl-1 inhibitor and as a lead compound for further synthetic programs.50 Recently, a series of indole-2-carboxylic acids have been described to bind Mcl-1 selectively at nanomolar concentrations and to efficiently disrupt Mcl-1/Bim complexes in living cells. A-1210477, one of the most potent binders, induced apoptosis in Mcl-1-dependent cancer cell and showed synergistic effects when combined with Navitoclax.51 A variety of other promising Mcl-1 inhibitor compounds that are currently being evaluated, including A*STAR compounds, MIM1, and Maritoclax, have been recently reviewed by Belmar and Fesik.52
Bromodomains. PPIs contributing to the formation of dynamic transcription complexes may determine chromatin modifications such as acetylation, hence controlling the transcription fate of a specific gene locus. Bromodomains are epigenetic readers that recognize acetylated lysines (Kacs) on histones and mediate transcription complexes to switch on genes. They share a conserved structure comprised by a left-handed bundle of four α-helices linked by diverse loop regions of variable charge and length. A hydrophobic pocket including a conserved asparagine and five water molecules recognizes the acetylated lysines. The interaction of this pocket with synthetic molecules has been explored in order to control gene transcription.53 Accordingly, structure-based molecular modeling performed by Bradner's laboratory at the Dana Farber Cancer Institute revealed the thienodiazepine (+)-JQ1 compound which is specific for bromodomain-containing protein 4 (BRD4) and has been used as an anticancer agent.54,55 Other BRD4 inhibitors based on the (+)-JQ1 structure that are in clinical trials for cancer treatment include I-BET762 (GSK525762) (Glaxosmithkline, Middlesex, UK),56 CPI-0610 (Constellation Pharmaceutical, Cambridge, MA),57 Ten-010 (Tensha Therapeutics, Cambridge, MA),58 and OTX015 (OncoEthix, Lausanne, Switzerland).59 Additionally, RVX-208 is a quinazoline specific for BRD3 that is now in phase 2 clinical trials for atherosclerosis.2
Oncology-related PPI targets in the preclinical stage. Proteasome assembling chaperone (PAC) 3 acts as a homodimer and plays an important role in proteasome formation. The fungal metabolite Thielocin 1 (TB1) was identified through a fragment complementation assay to inhibit the dimerization of PAC3 and suppresses the growth of cancer cells.60 Another compound, named JBIR-22 and isolated from Verticillus sp., had a specific inhibitory activity on PAC3 homodimerization, hereby inhibiting the assembly of functional proteasomes.61
Protein shuttling between cytoplasmic and nuclear compartments is critical for the accurate processing of signaling cascades. The transport of proteins from the nucleus to the cytoplasm by the exportin CRM1, which recognizes cargo proteins through a leucine-rich nuclear export signal, has also been targeted for inhibition as an antitumor strategy.62 In the quest for analogous compounds to anguinomycins, which are potent anticancer agents belonging to the leptomycin family, the natural product goniothalamin was identified from plants of the genus Goniothalamus. Goniothalamin has been reported to induce cytotoxicity in breast cancer cells through disruption of the PPI between CRM1 and cargo proteins, leading to inhibition of nuclear export.63
Since the identification of Wnt1 as a proto-oncogene in a model of mouse breast cancer, the knowledge about this important pathway has expanded. Perturbation of Wnt signaling is associated with stimulation of proliferation and with prevention of apoptosis in a number of human cancers, which is reflected by an elevated transcriptional activity of β-catenin.64 A few PPIs involved in the Wnt pathway have been targeted for inhibition in order to limit the negative effect in cancer progression. The interaction between Frizzled (FRZ) and Disheveled (DVL) is one of the first steps in this pathway. Particularly promising for drug design is the observation that Wnt signaling associates with oncogenesis via the FRZ-7 receptor and both DVL and FRZ-7 are reported to be overexpressed in tumor cell lines. Indeed, inhibiting the FRZ-7/DVL interaction by the small molecule FJ9 induced apoptosis in human cancer cell lines and inhibited tumor growth in mouse xenograft (H460) models in vivo.65 The interaction of β-catenin with Tcf-4, further downstream in the Wnt pathway, has also been targeted in PPI inhibitor screens. About 7,000 natural compounds were tested in a high-throughput ELISA screen for their ability to inhibit the β-catenin/Tcf-4 interaction in the context of colorectal cancer.66 Among them, two fungal compounds, PKF115-584 and CGP0409090, are interaction-specific inhibitors able to inhibit growth of colon cancer and adrenocortical67 and hepatocellular carcinomas.68 For the transcriptional activation of the β-catenin/Tcf-4 complex, the coactivator cyclic AMP response element-binding protein (CBP) is required. The small molecule ICG-001 binds specifically to CBP, leading to reduced transcriptional activity of the complex. ICG-001 induces apoptosis in transformed colon cells and in mouse xenograft model of colon cancer.69
Inhibition of PPIs has also been a strategy for the management of leukemias. Menin functions as a critical oncogenic cofactor of mixed lineage leukemia (MLL) fusion proteins in the development of acute leukemias, and inhibition of the menin interaction with MLL fusion proteins represents a very promising strategy to reverse their oncogenic activity. In an effort to identify small molecule inhibitors of the menin–MLL interaction, Grembecka et al. screened 49,000 small molecules using a fluorescence polarization assay and identified MI-2.70 Based on the crystal structures of menin with MI-2, more potent second-generation inhibitors were generated, namely MI-2-271 and MIV-6,72 that efficiently mimic the MLL peptide hot spots. Both compounds have binding activity in the nanomolar range and the preliminary in vitro data provided proof-of-concept for the development of PPI inhibitors to fight leukemia. In addition, two highly potent and orally bioavailable menin–MLL inhibitors (MI-463 and MI-503) were recently described that show profound effects in MLL leukemia cells and provide substantial survival benefit in vivo in mouse models of MLL leukemia.73
Targeting PPIs to combat infections by pathogens
The majority of viruses invade hosts by taking advantage of the cellular machinery to accomplish integration, replication, and survival. PPIs between viral and host proteins or among viral proteins which are imperative for their maintenance in the host represent important clinical targets.
The homotetrameric retroviral integrase (IN) is an enzyme produced by retroviruses such as the human immunodeficiency virus, which integrates its genetic material into the DNA of the infected cell by catalyzing 3′-processing and strand transfer reactions. The human protein lens epithelium-derived growth factor (LEDGF/p75) is a cellular cofactor of IN that promotes viral integration by tethering the preintegration complex to the chromatin and protects IN from proteolytic degradation.74,75 Intensive drug discovery efforts over the past years have validated the LEDGF-IN interaction as a druggable target for antiviral therapy and, through the use of structure-based approaches, have resulted in the design and synthesis of small molecule inhibitors, the so-called LEDGINs. These molecules not only disrupt the interaction but also allosterically inhibit the catalytic function of IN.76 The most potent LEDGF-IN inhibitors are tert-Butoxy-(4-phenyl-quinolin-3-yl)-acetic acid (tBPQA) derivatives, including the clinical compound BI 224436, which was similarly identified through structure-based drug design (Boehringer-Ingelheim, Ingelheim, Germany/Gilead, Foster City, CA).77,78,79 tBPQAs inhibit both early and late steps of the viral replication cycle, warranting their further clinical development.
A different strategy has been used for the inhibition of human papilloma virus replication, where the target is a PPI among viral proteins rather than between a viral and a host protein. Once entered into the host, human papilloma virus-11 requires the replication initiation factor E1 helicase to bind to the E2 transcription factor at specific DNA sites, and the small molecule compound BILH434 was identified to interrupt this interaction.80
Also PPIs involved in bacterial infections have been addressed as therapeutic targets. For example, FtsZ (a homologue of eukaryotic tubulin) and ZipA (a membrane-anchored protein) interact to form the septal ring that mediates cell division and this PPI has been validated as a potential target in strategies to limit infection by Gram-negative bacteria. Compounds sharing the indolo[2,3-a]quinolizin-7-one structure have been shown to inhibit this interaction in in vitro assays.81 Additionally, an nuclear magnetic resonance-based fragment screening approach revealed a hit series able to inhibit the FtsZ/ZipA interaction through binding the C-terminal domain of ZipA.82
PPIs involved in neuronal diseases
Amyloid β (Aβ), and specifically Aβ40 and Aβ42, constitutes the main component of the amyloid plaques found in the brains of Alzheimer patients.83 The essential factors generating of Aβ are β- and γ-secretase, which are primary amyloidogenic proteases. The initial cleavage of amyloid precursor protein (APP) is mediated by β-secretase and results in two products, an amino-terminal fragment of APP, sAPPb and a membrane embedded carboxy-terminal fragment, C99. C99 is the immediate substrate for γ-secretase, resulting in the generation of Aβ40 and Aβ42, which contribute to the progression of Alzheimer's disease.84 Inhibition of the interaction between γ-secretase and the APPs can lead to new therapeutic opportunities for the treatment of Alzheimer's disease. Different approaches were followed to design γ-secretase inhibitors, including transition state analogues, α-helical peptide-based inhibitors and nontransition state analogues.85 The small molecule compound LY450139 or Semagacestat is a benzolactam γ-secretase inhibitor, which entered clinical trials in 2005. Unfortunately, the results from phase 3 clinical trials showed that Semagacestat was associated with impaired lymphocyte differentiation and an increased risk for skin cancer, so it was withdrawn.86 As secretase activity is still in the center of research interest, control of β-secretase gradually gains attention for the same therapeutic purpose.87
Parkinson's disease is the second most common neurodegenerative disorder in most Western countries.88 Experimental data suggest that an important factor driving this pathology is the misfolding and oligomerization of the protein α-synuclein, which consequently forms a series of self-associating β-pleated sheets that spontaneously form aggregates called “Lewy bodies.”89,90 Therefore, inhibitors of the aggregation of α-synuclein are in the center of scientific interest and several inhibitors have been identified.91,92 An intriguing finding is that catecholamines are capable of inhibiting α-synuclein aggregation93 while still constitutes an area of active research for the development of novel therapeutics in Parkinson's disease.
Modulation of PPIs of liganded receptors
Activated receptors control an array of physiological functions upon binding of their respective ligands. Numerous pathological conditions have been attributed to the deregulation of liganded receptor-dependent signaling pathways. This deregulation can be mediated partially by PPIs that alter the receptor-dependent signaling cascades; hence their control has attracted scientific attention. A few small molecules have been reported for their in vitro ability to interfere with activated receptors. Cushing's disease is a neuroendocrine condition caused by partially glucocorticoid-resistant corticotroph adenomas leading to hypercortisolism.94 The effects of glucocorticoids are mediated by the glucocorticoid receptor. In its unliganded form, glucocorticoid receptor exists in a complex with chaperoning proteins, like Hsp90, which play a significant role in the proper conformation of the receptor. Silibinin binds to the C-terminal part of Hsp90, inhibiting its interaction with glucocorticoid receptor. In an allograft mouse model, administration of silibinin alleviated symptoms of Cushing's syndrome, indicating that a reduced response to glucocorticoids can be overcome pharmacologically with selective Hsp90 inhibitors.95
The interaction between interleukin-17 (IL-17) and its receptor has also been under investigation for inhibition since IL-17 is a potent proinflammatory cytokine involved in the pathogenesis of multiple inflammatory diseases such as psoriasis, rheumatoid arthritis, and Crohn's disease.96 Ensemble Therapeutics has identified a series of unique small molecule macrocycles, or Ensemblins that are antagonists of IL-17. In 2012, this company announced positive preclinical oral efficacy data with its first-in-class small molecule IL-17 antagonists. Similarly, an nuclear magnetic resonance-based approach yielded Ro26-455, a competitive inhibitor of IL-2 for binding to its receptor IL-2Ra, with an IC50 of 3 mM.97 Further evolvement of the Ro26-4550 scaffold by fragment-based methods into a more potent and drug-like inhibitor of IL-2:IL-2Ra resulted in the generation of SP4206.98 However, more functional studies are needed to evaluate the significance and efficiency of this molecule in interfering with IL-2 signaling.99 An analogous strategy was followed for targeting tumor necrosis factor signaling. Some molecules that interfere with the tumor necrosis factor/tumor necrosis factor receptor interaction are under preclinical investigation.100,101 Among the scientific advances for immunoregulation, the PPI inhibitor lifitegrast (SAR1118) plays a predominant role. Lifitegrast inhibits the interaction between LFA-1 (Cd11a/α2, CD18/β2) and ICAM1. LFA-1 is a β2 integrin receptor found on leukocytes and involved in T-cell activation through binding to its ligand ICAM1.102 Lifitegrast successfully passed clinical trials for treatment of inflammatory dry eye syndrome,103 resulting in a drug application being filed for this new inhibitor in the beginning of 2015 (Figure 3a).
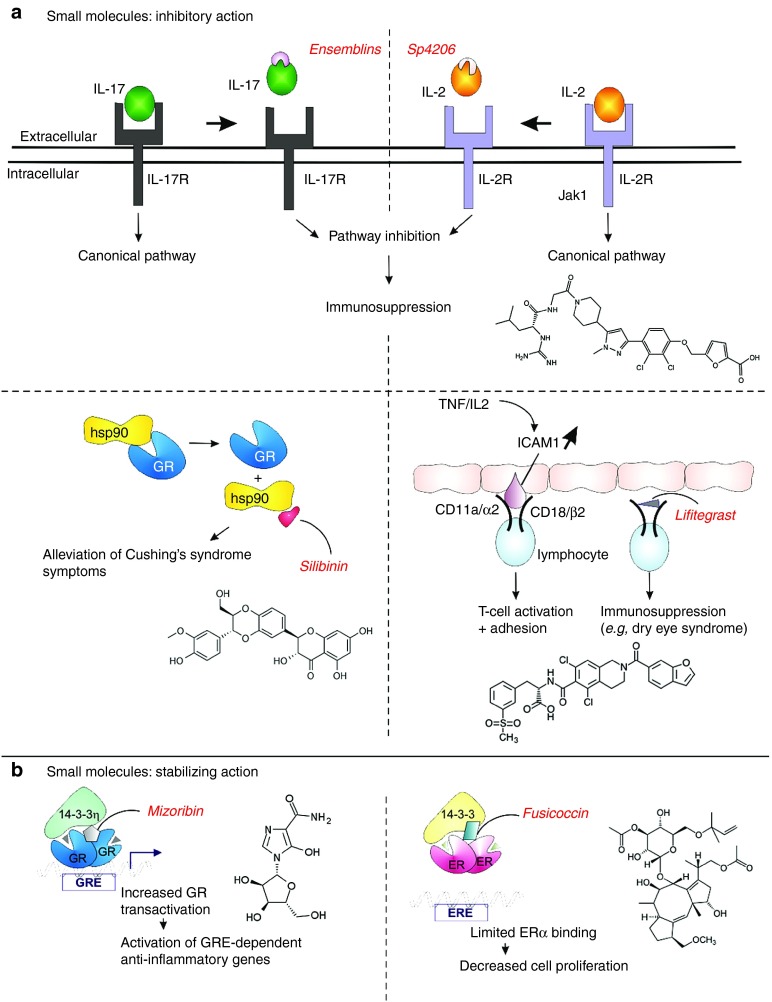
Figure 3.
Modulation of liganded receptors with small molecules. (a) PPI inhibitors of liganded receptors. Small molecules designed to inhibit cytokine signaling of IL-17 and IL-2 through their respective receptors IL-17R and IL-2R. The GR/Hsp90 inhibition by silibinin is involved in Cushing's syndrome and the interaction of ICAM1 with LFA-1 is targeted by lifitegrast to obtain immunosuppression. (b) Beneficial effects of stabilization of 14-3-3 protein interaction with GR or ER. The chemical structures of the depicted molecules are provided except for the structure of Ensemblins, which is not publicly available. ER, estrogen receptor; GR, glucocorticoid receptor; IL, interleukin; PPI, protein–protein interaction.
The “Yang” Face of PPIs – Stabilization of Beneficial PPIs
The “other side” of PPI control is stabilization. Small molecule PPI stabilizers act through two distinct mechanisms of action. First, allosteric stabilizers interact with one of the interaction partners of the complex, increasing the mutual affinity binding of the interacting proteins. Second, direct stabilizers may interact within the interfacial surface of a protein complex, creating contacts with the participating partners, similarly leading to an increased binding affinity (Table 2).4
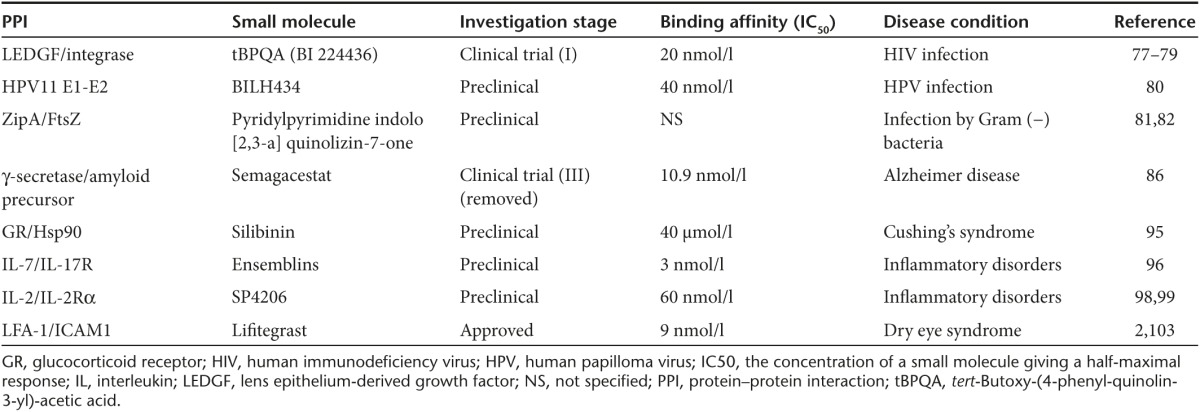
Table 2. PPI inhibitors for different pathological conditions.
PPI stabilization as an anticancer treatment
One of the most common PPI stabilizers widely used in the clinic as an anticancer agent is the Taxus brevifolia-derived paclitaxel, which interferes with the normal breakdown of microtubules during cell division.104 Paclitaxel and other compounds of this category induce cell cycle arrest by modulating microtubules' polymerization status.105 Microtubules consist of α- and β-tubulin and paclitaxel binds with high affinity to a hydrophobic pocket located on β-tubulin, thereby stabilizing polymerized microtubule structures in an allosteric fashion (Figure 4a).106
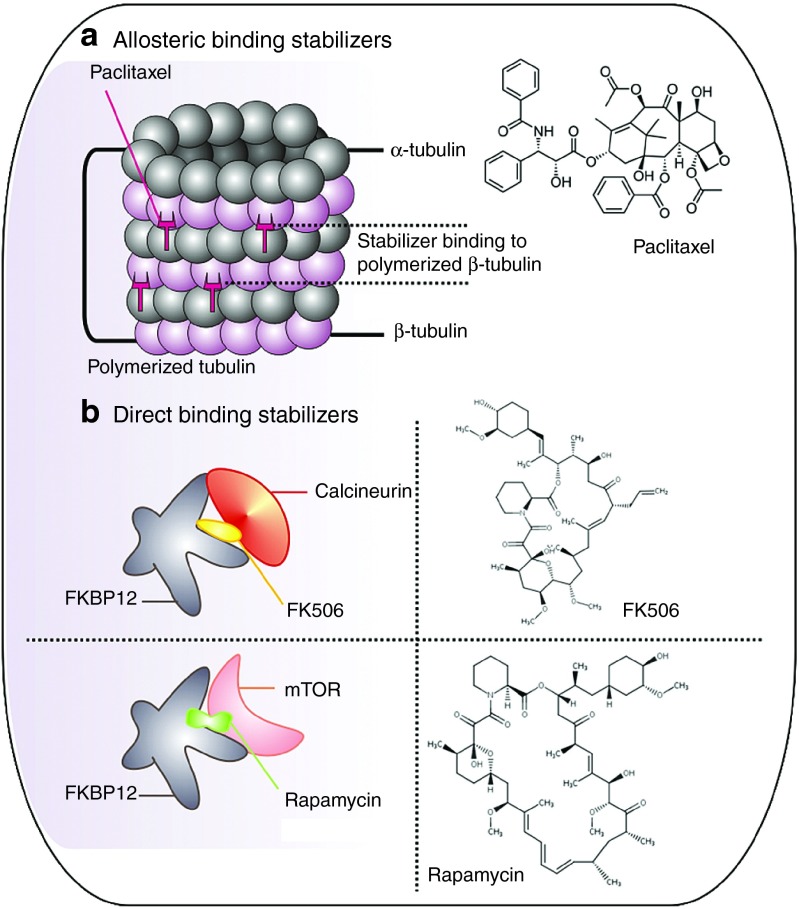
Figure 4.
PPI stabilizers in cancer treatment and in the modulation of immunosuppression. (a) Paclitaxel is a representative allosteric PPI stabilizer that preserves tubulin formation. (b) FK506 and rapamycin are potent immunosuppressants acting as direct stabilizers of protein interactions with FKBP12. PPI, protein–protein interaction.
14-3-3 proteins have also been described to participate in diverse cancers, neurodegenerative diseases, or virulence of human pathogenic organisms.107,108,109 Due to their versatile mode of action, these proteins constitute a novel target class for pharmacological intervention by either stabilizing or inhibiting 14-3-3 PPIs. In a number of cases, 14-3-3 proteins have been shown to support the stability and bioavailability of their interaction partners such as TASK3.110 Dysregulation of TASK3 has been linked to cancer, inflammation, and epilepsy,111 hence stabilization of the 14-3-3 (β and ɛ isoforms)/TASK3 interaction could prove therapeutically promising. Furthermore, the stabilization of estrogen receptor α (ERα) binding to 14-3-3β has been reported to have anticancer effects.112 Fusicoccin directly binds to the interface rim of the 14-3-3σ/ERα and stabilizes this interaction (Figure 3b). Fusicoccin treatment leads to diminished estradiol-mediated ERα dimerization, limitation of ERα binding to chromatin, and downstream gene activation as well as decreased cell proliferation.113 Current breast cancer treatments are based on the suppression of the transcriptional potency of ERα by aromatase inhibitors or antiestrogens. Due to the onset of resistance of patients to these treatments, there is an urgent need for alternative therapeutics. Inhibition of ERα activity through stabilization of its interaction with 14-3-3 represents a new strategy for drug development in the field of breast cancer (Table 3).
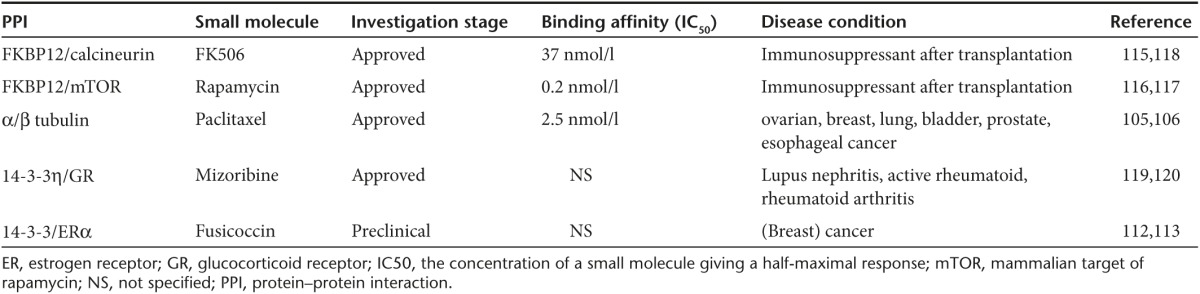
Table 3. PPI stabilizers for the control of immunosuppression and cancer progression.
Stabilizers of PPIs acting as immunosuppressants
Two directly stabilizing molecules, rapamycin (Sirolimus) and FK506 (Tacrolimus), are well-known immunosuppressants in the clinic. Although they have considerably different structures, they share a remarkably common mechanism of action. FK506 and rapamycin stabilize the interactions between FKBP12/protein phosphatase calcineurin and FKBP12/mTOR (mammalian target of rapamycin), respectively. Interestingly, initially FK506 and rapamycin bind with high affinity to FKBP12, which is an immunophilin.114 In a next step, FK506/FKBP12 and rapamycin/FKBP12 bind to calcineurin and mTOR, respectively, via the newly formed interface. This results in suppression of the catalytic activity of these enzymes (Figure 4b). Of note, in the absence of FK506 and rapamycin, FKBP12 is not able to interact with calcineurin115 or with mTOR.116 Rapamycin117 and FK506118 have been investigated as immunosuppressive agents for treatment of transplant patients in different clinical trials.
Mizoribine is an imidazole nucleoside with immunosuppressive activity. Mizoribine has been approved in Japan for combinatorial therapy with glucocorticoids in lupus nephritis, rheumatoid arthritis, or after renal transplantation.119 A possible mechanism of mizoribine is the enhancement of the interaction of 14-3-3η with the glucocorticoid receptor leading to enhanced activity of the receptor and subsequent increased immunosuppression120 (Figure 3b).
Conclusions and Future Perspectives
Natural products like taxanes and rapamycin, which were discovered in the late 90s as potent stabilizers of PPIs, raised initial enthusiasm for small molecule modulation of PPIs as a therapeutic rationale. During the next decade, the advance of “omics” technologies, greatly expanding our knowledge of genes, proteins, and their interactions, highlighted the central importance of PPI networks both towards enhancing our basic understanding of cellular processes and as a vast source of potential drug targets, further increasing interest in PPI-targeted drug discovery. Yet, at the same time high-resolution structures revealed that PPI interfaces are often made up of large shallow surfaces which were thought to be difficult, if not impossible, to interfere with, significantly lowering confidence in the approach.
The cases of inhibitors and stabilizers described in this review however clearly illustrate the potential of PPIs in drug development. Novel small molecules targeting specific PPIs have entered clinical trials, and in some cases already resulted in new therapeutics or optimized treatments.
Looking at the technologies that lead to these successful programs, it is striking to note the variety of discovery approaches. The huge diversity in PPIs and in the characteristics of their interfaces clearly precludes a one-fits-all approach. The reported progress in PPI drug discovery should be attributed at least partly to the growing availability of a varied and complementary set of both in vitro and in silico screening approaches from which can be drawn depending on the nature of the PPI target.121 Also at the compound side, there should be a proper match with the nature of the target. An often-cited caveat is the fact that classic small molecule libraries applied in high-throughput screening campaigns consist mainly of small, simple, and flat structures, whereas successful disruption of a PPI interface generally requires larger and more complex molecules. Studies aimed at determining common features among successful PPI inhibitors yielded a number of rational design principles that can be used to compile PPI-specific compound collections which should increase hit rates in PPI inhibitor screens.122
In addition to small molecules, also peptides were shown to be promising tools for targeting PPIs. Yet, despite the exciting preliminary in vitro data, the use of peptides as therapeutics has been hampered by fast renal clearance, poor metabolic stability, and biodegradability. Nevertheless, different strategies were applied to improve plasma half-lives of these therapeutic peptides, resulting in potent PPI modulators, for instance for Bcl2, caspases, and ERα.123 Particularly encouraging is the case of the “stapled” peptides that reactivate the p53 pathway by binding and inhibiting HDM2 and HDMX. These entered clinical trials in 2014.124
Traditionally, drug design has been directed towards targets containing well-defined binding pockets such as enzymes, nuclear receptors, and ion channels. However, our increased understanding of PPIs, their interfaces, and how to interfere with these open up new horizons for drug development. Resolving PPI modulation currently constitutes an area of intense research and numerous protein complexes await further investigation as potential new therapeutic agents.
References
- Arkin, MR and Wells, JA (2004). Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov 3: 301–317. [DOI] [PubMed] [Google Scholar]
- Arkin, MR, Tang, Y and Wells, JA (2014). Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol 21: 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W, Wisniewski, JA and Ji, H (2014). Hot spot-based design of small-molecule inhibitors for protein-protein interactions. Bioorg Med Chem Lett 24: 2546–2554. [DOI] [PubMed] [Google Scholar]
- Thiel, P, Kaiser, M and Ottmann, C (2012). Small-molecule stabilization of protein-protein interactions: an underestimated concept in drug discovery? Angew Chem Int Ed Engl 51: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Turnbull, AP, Boyd, SM and Walse, B (2014). Fragment-based drug discovery and protein-protein interactions. Res Rep Biochem 4: 13–26. [Google Scholar]
- Monroy, CA, Mackie, DI and Roman, DL (2013). A high throughput screen for RGS proteins using steady state monitoring of free phosphate formation. PLoS One 8: e62247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounnas, V, Ritschel, T, Kelder, J, McGuire, R, Bywater, RP and Foloppe, N (2013). Current progress in Structure-Based Rational Drug Design marks a new mindset in drug discovery. Comput Struct Biotechnol J 5: e201302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden, KH and Lu, X (2002). Live or let die: the cell's response to p53. Nat Rev Cancer 2: 594–604. [DOI] [PubMed] [Google Scholar]
- Brown, CJ, Lain, S, Verma, CS, Fersht, AR and Lane, DP (2009). Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 9: 862–873. [DOI] [PubMed] [Google Scholar]
- Feki, A and Irminger-Finger, I (2004). Mutational spectrum of p53 mutations in primary breast and ovarian tumors. Crit Rev Oncol Hematol 52: 103–116. [DOI] [PubMed] [Google Scholar]
- Wu, X, Bayle, JH, Olson, D and Levine, AJ (1993). The p53-mdm-2 autoregulatory feedback loop. Genes Dev 7(7A): 1126–1132. [DOI] [PubMed] [Google Scholar]
- Freedman, DA, Wu, L and Levine, AJ (1999). Functions of the MDM2 oncoprotein. Cell Mol Life Sci 55: 96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon, T and Oren, M (1999). Mdm2: the ups and downs. Mol Med 5: 71–83. [PMC free article] [PubMed] [Google Scholar]
- Momand, J, Jung, D, Wilczynski, S and Niland, J (1998). The MDM2 gene amplification database. Nucleic Acids Res 26: 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoulade, C, Bressac-de Paillerets, B, Lefrère, I, Ronsin, M, Feunteun, J, Tursz, T et al. (1998). Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt's lymphoma cells. Oncogene 16: 1603–1610. [DOI] [PubMed] [Google Scholar]
- Momand, J, Wu, HH and Dasgupta, G (2000). MDM2–master regulator of the p53 tumor suppressor protein. Gene 242: 15–29. [DOI] [PubMed] [Google Scholar]
- Kussie, PH, Gorina, S, Marechal, V, Elenbaas, B, Moreau, J, Levine, AJ et al. (1996). Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274: 948–953. [DOI] [PubMed] [Google Scholar]
- Vassilev, LT, Vu, BT, Graves, B, Carvajal, D, Podlaski, F, Filipovic, Z et al. (2004). In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848. [DOI] [PubMed] [Google Scholar]
- Vu, B, Wovkulich, P, Pizzolato, G, Lovey, A, Ding, Q, Jiang, N et al. (2013). Discovery of RG7112: a small-molecule MDM2 inhibitor in clinical development. ACS Med Chem Lett 4: 466–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray-Coquard, I, Blay, JY, Italiano, A, Le Cesne, A, Penel, N, Zhi, J et al. (2012). Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol 13: 1133–1140. [DOI] [PubMed] [Google Scholar]
- Ding, Q, Zhang, Z, Liu, JJ, Jiang, N, Zhang, J, Ross, TM et al. (2013). Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem 56: 5979–5983. [DOI] [PubMed] [Google Scholar]
- Zhao, Y, Aguilar, A, Bernard, D and Wang, S (2015). Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 inhibitors) in clinical trials for cancer treatment. J Med Chem 58: 1038–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- clinicaltrials.gov/SAR405838 (2015). <https://clinicaltrials.gov/ct2/results?term=SAR405838&Search=Search>.
- Wang, S, Sun, W, Zhao, Y, McEachern, D, Meaux, I, Barrière, C et al. (2014). SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression. Cancer Res 74: 5855–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D, Li, Z, Rew, Y, Gribble, M, Bartberger, MD, Beck, HP et al. (2014). Discovery of AMG 232, a potent, selective, and orally bioavailable MDM2-p53 inhibitor in clinical development. J Med Chem 57: 1454–1472. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno, P, Brambilla, E, Thomas, R and Soria, JC (2012). Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin Cancer Res 18: 2443–2451. [DOI] [PubMed] [Google Scholar]
- Graves, B, Thompson, T, Xia, M, Janson, C, Lukacs, C, Deo, D et al. (2012). Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci USA 109: 11788–11793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, CD, Wu, H, Bolaños, B, Bergqvist, S, Brooun, A, Pauly, T et al. (2009). Structural and biophysical characterization of XIAP BIR3 G306E mutant: insights in protein dynamics and application for fragment-based drug design. Chem Biol Drug Des 74: 212–223. [DOI] [PubMed] [Google Scholar]
- Dubrez, L, Berthelet, J and Glorian, V (2013). IAP proteins as targets for drug development in oncology. Onco Targets Ther 9: 1285–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber, EC, Schoeffler, AJ, Lingel, A, Elliott, JM, Fedorova, AV, Giannetti, AM et al. (2011). Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science 334: 376–380. [DOI] [PubMed] [Google Scholar]
- Crisóstomo, FR, Feng, Y, Zhu, X, Welsh, K, An, J, Reed, JC et al. (2009). Design and synthesis of a simplified inhibitor for XIAP-BIR3 domain. Bioorg Med Chem Lett 19: 6413–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H, Nikolovska-Coleska, Z, Yang, CY, Qian, D, Lu, J, Qiu, S et al. (2008). Design of small-molecule peptidic and nonpeptidic Smac mimetics. Acc Chem Res 41: 1264–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- clinicaltrials.gov/LCL161 (2014). <https://clinicaltrials.gov/ct2/show/NCT01617668?term=LCL161&rank=6>.
- Flygare, JA, Beresini, M, Budha, N, Chan, H, Chan, IT, Cheeti, S et al. (2012). Discovery of a potent small-molecule antagonist of inhibitor of apoptosis (IAP) proteins and clinical candidate for the treatment of cancer (GDC-0152). J Med Chem 55: 4101–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- clinicaltrials.gov/GDC0152 (2010). <https://clinicaltrials.gov/ct2/show/NCT00977067?term=GDC-0152&rank=1>.
- Cai, Q, Sun, H, Peng, Y, Lu, J, Nikolovska-Coleska, Z, McEachern, D et al. (2011). A potent and orally active antagonist (SM-406/AT-406) of multiple inhibitor of apoptosis proteins (IAPs) in clinical development for cancer treatment. J Med Chem 54: 2714–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon, SM, Mitsuuchi, Y, Deng, Y, LaPorte, MG, Rippin, SR, Haimowitz, T et al. (2014). Birinapant, a smac-mimetic with improved tolerability for the treatment of solid tumors and hematological malignancies. J Med Chem 57: 3666–3677. [DOI] [PubMed] [Google Scholar]
- Benetatos, CA, Mitsuuchi, Y, Burns, JM, Neiman, EM, Condon, SM, Yu, G et al. (2014). Birinapant (TL32711), a bivalent SMAC mimetic, targets TRAF2-associated cIAPs, abrogates TNF-induced NF-κB activation, and is active in patient-derived xenograft models. Mol Cancer Ther 13: 867–879. [DOI] [PubMed] [Google Scholar]
- clinicaltrials.gov/Birinapant (2015). <https://clinicaltrials.gov/ct2/show/NCT01681368?term=Birinapant&rank=6>.
- Bai, L, Smith, DC and Wang, S (2014). Small-molecule SMAC mimetics as new cancer therapeutics. Pharmacol Ther 144: 82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, JC (1998). Bcl-2 family proteins. Oncogene 17: 3225–3236. [DOI] [PubMed] [Google Scholar]
- Oltersdorf, T, Elmore, SW, Shoemaker, AR, Armstrong, RC, Augeri, DJ, Belli, BA et al. (2005). An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435: 677–681. [DOI] [PubMed] [Google Scholar]
- Tse, C, Shoemaker, AR, Adickes, J, Anderson, MG, Chen, J, Jin, S et al. (2008). ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res 68: 3421–3428. [DOI] [PubMed] [Google Scholar]
- clinicaltrials.gov/Navitoclax (2014). <https://clinicaltrials.gov/ct2/show/NCT01557777?term=Navitoclax&rank=9>.
- Park, CM, Bruncko, M, Adickes, J, Bauch, J, Ding, H, Kunzer, A et al. (2008). Discovery of an orally bioavailable small molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J Med Chem 51: 6902–6915. [DOI] [PubMed] [Google Scholar]
- Cancer Discovery (2014). <http://cancerdiscovery.aacrjournals.org/content/4/2/OF5.full>. [DOI] [PubMed]
- clinicaltrials.gov/ABT-199 (2015). <https://clinicaltrials.gov/ct2/results?term=ABT-199&Search=Search>.
- clinicaltrials.gov/Obatoclax (2015). <https://clinicaltrials.gov/ct2/results?term=obatoclax&Search=Search>.
- Calcul, L, Chow, R, Oliver, AG, Tenney, K, White, KN, Wood, AW et al. (2009). NMR strategy for unraveling structures of bioactive sponge-derived oxy-polyhalogenated diphenyl ethers. J Nat Prod 72: 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan, S, Vogler, M, Butterworth, M, Dinsdale, D, Walensky, LD and Cohen, GM (2013). Evaluation and critical assessment of putative MCL-1 inhibitors. Cell Death Differ 20: 1475–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson, JD, Zhang, H, Chen, J, Tahir, SK, Phillips, DC, Xue, J et al. (2015). Potent and selective small-molecule MCL-1 inhibitors demonstrate on-target cancer cell killing activity as single agents and in combination with ABT-263 (navitoclax). Cell Death Dis 6: e1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmar, J and Fesik, SW (2015). Small molecule Mcl-1 inhibitors for the treatment of cancer. Pharmacol Ther 145: 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos, P and Knapp, S (2014). Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 13: 337–356. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos, P, Qi, J, Picaud, S, Shen, Y, Smith, WB, Fedorov, O et al. (2010). Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkina, AC and Denis, GV (2012). BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 12: 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirguet, O, Gosmini, R, Toum, J, Clément, CA, Barnathan, M, Brusq, JM et al. (2013). Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem 56: 7501–7515. [DOI] [PubMed] [Google Scholar]
- Gehling, VS, Hewitt, MC, Vaswani, RG, Leblanc, Y, Côté, A, Nasveschuk, CG et al. (2013). Discovery, design, and optimization of Isoxazole Azepine BET inhibitors. ACS Med Chem Lett 4: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- clinicaltrials.gov/Ten-010 (2015). <https://clinicaltrials.gov/ct2/results?term=Ten-010+&Search=Search>.
- clinicaltrials.gov/OTX015 (2015). <https://www.clinicaltrials.gov/ct2/results?term=OTX015&Search=Search>.
- Hashimoto, J, Watanabe, T, Seki, T, Karasawa, S, Izumikawa, M, Seki, T et al. (2009). Novel in vitro protein fragment complementation assay applicable to high-throughput screening in a 1536-well format. J Biomol Screen 14: 970–979. [DOI] [PubMed] [Google Scholar]
- Izumikawa, M, Hashimoto, J, Hirokawa, T, Sugimoto, S, Kato, T, Takagi, M et al. (2010). JBIR-22, an inhibitor for protein-protein interaction of the homodimer of proteasome assembly factor 3. J Nat Prod 73: 628–631. [DOI] [PubMed] [Google Scholar]
- Gademann, K (2011). Controlling protein transport by small molecules. Curr Drug Targets 12: 1574–1580. [DOI] [PubMed] [Google Scholar]
- Wach, JY, Güttinger, S, Kutay, U and Gademann, K (2010). The cytotoxic styryl lactone goniothalamin is an inhibitor of nucleocytoplasmic transport. Bioorg Med Chem Lett 20: 2843–2846. [DOI] [PubMed] [Google Scholar]
- Barker, N and Clevers, H (2006). Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov 5: 997–1014. [DOI] [PubMed] [Google Scholar]
- Fujii, N, You, L, Xu, Z, Uematsu, K, Shan, J, He, B et al. (2007). An antagonist of dishevelled protein-protein interaction suppresses beta-catenin-dependent tumor cell growth. Cancer Res 67: 573–579. [DOI] [PubMed] [Google Scholar]
- Lepourcelet, M, Chen, YN, France, DS, Wang, H, Crews, P, Petersen, F et al. (2004). Small-molecule antagonists of the oncogenic Tcf/beta-catenin protein complex. Cancer Cell 5: 91–102. [DOI] [PubMed] [Google Scholar]
- Doghman, M, Cazareth, J and Lalli, E (2008). The T cell factor/beta-catenin antagonist PKF115-584 inhibits proliferation of adrenocortical carcinoma cells. J Clin Endocrinol Metab 93: 3222–3225. [DOI] [PubMed] [Google Scholar]
- Wei, W, Chua, MS, Grepper, S and So, S (2010). Small molecule antagonists of Tcf4/beta-catenin complex inhibit the growth of HCC cells in vitro and in vivo. Int J Cancer 126: 2426–2436. [DOI] [PubMed] [Google Scholar]
- Emami, KH, Nguyen, C, Ma, H, Kim, DH, Jeong, KW, Eguchi, M et al. (2004). A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected]. Proc Natl Acad Sci USA 101: 12682–12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grembecka, J, He, S, Shi, A, Purohit, T, Muntean, AG, Sorenson, RJ et al. (2012). Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol 8: 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, A, Murai, MJ, He, S, Lund, G, Hartley, T, Purohit, T et al. (2012). Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia. Blood 120: 4461–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, S, Senter, TJ, Pollock, J, Han, C, Upadhyay, SK, Purohit, T et al. (2014). High-affinity small-molecule inhibitors of the menin-mixed lineage leukemia (MLL) interaction closely mimic a natural protein-protein interaction. J Med Chem 57: 1543–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkin, D, He, S, Miao, H, Kempinska, K, Pollock, J, Chase, J et al. (2015). Pharmacologic inhibition of the Menin-MLL interaction blocks progression of MLL leukemia in vivo. Cancer Cell 27: 589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov, P, Maertens, G, Proost, P, Devreese, B, Van Beeumen, J, Engelborghs, Y et al. (2003). HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278: 372–381. [DOI] [PubMed] [Google Scholar]
- Maertens, G, Cherepanov, P, Pluymers, W, Busschots, K, De Clercq, E, Debyser, Z et al. (2003). LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem 278: 33528–33539. [DOI] [PubMed] [Google Scholar]
- Christ, F and Debyser, Z (2013). The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy. Virology 435: 102–109. [DOI] [PubMed] [Google Scholar]
- clinicaltrials.gov/BI-224436 (2014). <https://clinicaltrials.gov/ct2/show/NCT02183662?term=BI-224436&rank=2>.
- Fader, LD, Malenfant, E, Parisien, M, Carson, R, Bilodeau, F, Landry, S et al. (2014). Discovery of BI 224436, a Noncatalytic Site Integrase Inhibitor (NCINI) of HIV-1. ACS Med Chem Lett 5: 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick, C, Amad, M, Bailey, MD, Bethell, R, Bös, M, Bonneau, P et al. (2014). Preclinical profile of BI 224436, a novel HIV-1 non-catalytic-site integrase inhibitor. Antimicrob Agents Chemother 58: 3233–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y, Coulombe, R, Cameron, DR, Thauvette, L, Massariol, MJ, Amon, LM et al. (2004). Crystal structure of the E2 transactivation domain of human papillomavirus type 11 bound to a protein interaction inhibitor. J Biol Chem 279: 6976–6985. [DOI] [PubMed] [Google Scholar]
- Jennings, LD, Foreman, KW, Rush, TS 3rd, Tsao, DH, Mosyak, L, Li, Y et al. (2004). Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction. Bioorg Med Chem Lett 14: 1427–1431. [DOI] [PubMed] [Google Scholar]
- Tsao, DH, Sutherland, AG, Jennings, LD, Li, Y, Rush, TS 3rd, Alvarez, JC et al. (2006). Discovery of novel inhibitors of the ZipA/FtsZ complex by NMR fragment screening coupled with structure-based design. Bioorg Med Chem 14: 7953–7961. [DOI] [PubMed] [Google Scholar]
- Murphy, MP and LeVine, H 3rd (2010). Alzheimer's disease and the amyloid-beta peptide. J Alzheimers Dis 19: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman, MS, Beher, D, Clarke, EE, Lewis, HD, Harrison, T, Hunt, P et al. (2000). L-685,458, an aspartyl protease transition state mimic, is a potent inhibitor of amyloid beta-protein precursor gamma-secretase activity. Biochemistry 39: 8698–8704. [DOI] [PubMed] [Google Scholar]
- Churcher, I, Williams, S, Kerrad, S, Harrison, T, Castro, JL, Shearman, MS et al. (2003). Design and synthesis of highly potent benzodiazepine gamma-secretase inhibitors: preparation of (2S,3R)-3-(3,4-difluorophenyl)-2-(4-fluorophenyl)-4- hydroxy-N-((3S)-1-methyl-2-oxo-5- phenyl-2,3-dihydro-1H-benzo[e][1,4]-diazepin-3-yl)butyramide by use of an asymmetric Ireland-Claisen rearrangement. J Med Chem 46: 2275–2278. [DOI] [PubMed] [Google Scholar]
- Fan, LY and Chiu, MJ (2010). Pharmacological treatment for Alzheimer's disease: current approaches and future strategies. Acta Neurol Taiwan 19: 228–245. [PubMed] [Google Scholar]
- Nie, Q, Du, XG and Geng, MY (2011). Small molecule inhibitors of amyloid β peptide aggregation as a potential therapeutic strategy for Alzheimer's disease. Acta Pharmacol Sin 32: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub, D, Comella, CL and Horn, S (2008). Parkinson's disease–Part 1: Pathophysiology, symptoms, burden, diagnosis, and assessment. Am J Manag Care 14 (suppl. 2): S40–S48. [PubMed] [Google Scholar]
- Duda, JE, Lee, VM and Trojanowski, JQ (2000). Neuropathology of synuclein aggregates. J Neurosci Res 61: 121–127. [DOI] [PubMed] [Google Scholar]
- Norris, EH, Giasson, BI and Lee, VM (2004). Alpha-synuclein: normal function and role in neurodegenerative diseases. Curr Top Dev Biol 60: 17–54. [DOI] [PubMed] [Google Scholar]
- Norris, EH, Giasson, BI, Hodara, R, Xu, S, Trojanowski, JQ, Ischiropoulos, H et al. (2005). Reversible inhibition of alpha-synuclein fibrillization by dopaminochrome-mediated conformational alterations. J Biol Chem 280: 21212–21219. [DOI] [PubMed] [Google Scholar]
- Skovronsky, DM, Lee, VM and Trojanowski, JQ (2006). Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu Rev Pathol 1: 151–170. [DOI] [PubMed] [Google Scholar]
- Conway, KA, Rochet, JC, Bieganski, RM and Lansbury, PT Jr. (2001). Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science 294: 1346–1349. [DOI] [PubMed] [Google Scholar]
- Sulentic, P, Morris, DG and Grossman, A (2000). Cushing's disease [updated 18 August 2014]. In: De Groot, LJ, Beck Peccoz, P, Chrousos, G, Dungan, K, Grossman, A, Hershman, JM, et al. (eds). Endotext. South Dartmouth, MA. <http://www.ncbi.nlm.nih.gov/books/NBK279088/>. [Google Scholar]
- Riebold, M, Kozany, C, Freiburger, L, Sattler, M, Buchfelder, M, Hausch, F et al. (2015). A C-terminal HSP90 inhibitor restores glucocorticoid sensitivity and relieves a mouse allograft model of Cushing disease. Nat Med 21: 276–280. [DOI] [PubMed] [Google Scholar]
- Kirkham, BW, Kavanaugh, A and Reich, K (2014). Interleukin-17A: a unique pathway in immune-mediated diseases: psoriasis, psoriatic arthritis and rheumatoid arthritis. Immunology 141: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, SD, Palermo, R, Liu, CM, Tilley, JW, Chen, L, Danho, W et al. (2003). NMR characterization of interleukin-2 in complexes with the IL-2Ralpha receptor component, and with low molecular weight compounds that inhibit the IL-2/IL-Ralpha interaction. Protein Sci 12: 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimundo, BC, Oslob, JD, Braisted, AC, Hyde, J, McDowell, RS, Randal, M et al. (2004). Integrating fragment assembly and biophysical methods in the chemical advancement of small-molecule antagonists of IL-2: an approach for inhibiting protein-protein interactions. J Med Chem 47: 3111–3130. [DOI] [PubMed] [Google Scholar]
- Wilson, CG and Arkin, MR (2011). Small-molecule inhibitors of IL-2/IL-2R: lessons learned and applied. Curr Top Microbiol Immunol 348: 25–59. [DOI] [PubMed] [Google Scholar]
- Ma, L, Gong, H, Zhu, H, Ji, Q, Su, P, Liu, P et al. (2014). A novel small-molecule tumor necrosis factor α inhibitor attenuates inflammation in a hepatitis mouse model. J Biol Chem 289: 12457–12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, MM, Smith, AS, Oslob, JD, Flanagan, WM, Braisted, AC, Whitty, A et al. (2005). Small-molecule inhibition of TNF-alpha. Science 310: 1022–1025. [DOI] [PubMed] [Google Scholar]
- Long, EO (2011). ICAM-1: getting a grip on leukocyte adhesion. J Immunol 186: 5021–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard, JD, Torkildsen, GL, Lonsdale, JD, D'Ambrosio, FA Jr., McLaurin, EB, Eiferman, RA et al.; OPUS-1 Study Group. (2014). Lifitegrast ophthalmic solution 5.0% for treatment of dry eye disease: results of the OPUS-1 phase 3 study. Ophthalmology 121: 475–483. [DOI] [PubMed] [Google Scholar]
- Downing, KH (2000). Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol 16: 89–111. [DOI] [PubMed] [Google Scholar]
- Jordan, MA (2002). Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr Med Chem Anticancer Agents 2: 1–17. [DOI] [PubMed] [Google Scholar]
- Nogales, E (2001). Structural insight into microtubule function. Annu Rev Biophys Biomol Struct 30: 397–420. [DOI] [PubMed] [Google Scholar]
- Rose, R, Rose, M and Ottmann, C (2012). Identification and structural characterization of two 14-3-3 binding sites in the human peptidylarginine deiminase type VI. J Struct Biol 180: 65–72. [DOI] [PubMed] [Google Scholar]
- Berg, D, Holzmann, C and Riess, O (2003). 14-3-3 proteins in the nervous system. Nat Rev Neurosci 4: 752–762. [DOI] [PubMed] [Google Scholar]
- Fu, H, Coburn, J and Collier, RJ (1993). The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci USA 90: 2320–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan, S, Preisig-Müller, R, Wischmeyer, E, Nehring, R, Hanley, PJ, Renigunta, V et al. (2002). Interaction with 14-3-3 proteins promotes functional expression of the potassium channels TASK-1 and TASK-3. J Physiol 545(Pt 1): 13–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner, S, Budde, T, Wiendl, H and Meuth, SG (2010). From the background to the spotlight: TASK channels in pathological conditions. Brain Pathol 20: 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y, Kim, H, Jang, SW and Ko, J (2011). The role of 14-3-3β in transcriptional activation of estrogen receptor α and its involvement in proliferation of breast cancer cells. Biochem Biophys Res Commun 414: 199–204. [DOI] [PubMed] [Google Scholar]
- De Vries-van Leeuwen, IJ, da Costa Pereira, D, Flach, KD, Piersma, SR, Haase, C, Bier, D et al. (2013). Interaction of 14-3-3 proteins with the estrogen receptor alpha F domain provides a drug target interface. Proc Natl Acad Sci USA 110: 8894–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordanetto, F, Schäfer, A and Ottmann, C (2014). Stabilization of protein-protein interactions by small molecules. Drug Discov Today 19: 1812–1821. [DOI] [PubMed] [Google Scholar]
- Harding, MW, Galat, A, Uehling, DE and Schreiber, SL (1989). A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 341: 758–760. [DOI] [PubMed] [Google Scholar]
- Griffith, JP, Kim, JL, Kim, EE, Sintchak, MD, Thomson, JA, Fitzgibbon, MJ et al. (1995). X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12-FK506 complex. Cell 82: 507–522. [DOI] [PubMed] [Google Scholar]
- clinicaltrials.gov/Rapamycin (2015). <https://clinicaltrials.gov/ct2/results?term=rapamycin&Search=Search>.
- clinicaltrials.gov/FK506 (2015). <https://clinicaltrials.gov/ct2/results?term=FK506&Search=Search>.
- clinicaltrials.gov/Mizoribine (2015). <https://clinicaltrials.gov/ct2/results?term=mizoribine&Search=Search>.
- Takahashi, S, Wakui, H, Gustafsson, JA, Zilliacus, J and Itoh, H (2000). Functional interaction of the immunosuppressant mizoribine with the 14-3-3 protein. Biochem Biophys Res Commun 274: 87–92. [DOI] [PubMed] [Google Scholar]
- Higueruelo, AP, Jubb, H and Blundell, TL (2013). Protein-protein interactions as druggable targets: recent technological advances. Curr Opin Pharmacol 13: 791–796. [DOI] [PubMed] [Google Scholar]
- Kuenemann, MA, Sperandio, O, Labbé, CM, Lagorce, D, Miteva, MA and Villoutreix, BO (2015). In silico design of low molecular weight protein-protein interaction inhibitors: overall concept and recent advances. Prog Biophys Mol Biol 119: 20–32. [DOI] [PubMed] [Google Scholar]
- Verdine, GL and Hilinski, GJ (2012). Stapled peptides for intracellular drug targets. Methods Enzymol 503: 3–33. [DOI] [PubMed] [Google Scholar]
- Aileron Therapeutics (2015). <http://www.aileronrx.com/programs_p53.php>.