Abstract
Spread of oncolytic viruses through tumor tissue is essential to effective virotherapy. Interstitial matrix is thought to be a significant barrier to virus particle convection between “islands” of tumor cells. One way to address this is to encode matrix-degrading enzymes within oncolytic viruses, for secretion from infected cells. To test the hypothesis that extracellular DNA provides an important barrier, we assessed the ability of DNase to promote virus spread. Nonreplicating Ad5 vectors expressing actin-resistant DNase (aDNAse I), proteinase K (PK), hyaluronidase (rhPH20), and chondroitinase ABC (CABC) were injected into established DLD human colorectal adenocarcinoma xenografts, transcomplemented with a replicating Ad5 virus. Each enzyme improved oncolysis by the replicating adenovirus, with no evidence of tumor cells being shed into the bloodstream. aDNAse I and rhPH20 hyaluronidase were then cloned into conditionally-replicating group B adenovirus, Enadenotucirev (EnAd). EnAd encoding each enzyme showed significantly better antitumor efficacy than the parental virus, with the aDNAse I-expressing virus showing improved spread. Both DNase and hyaluronidase activity was still measurable 32 days postinfection. This is the first time that extracellular DNA has been implicated as a barrier for interstitial virus spread, and suggests that oncolytic viruses expressing aDNAse I may be promising candidates for clinical translation.
Introduction
Oncolytic viruses have been engineered for tumor-selective replication, leaving normal cells virtually unharmed while specifically lysing cancer cells.1 The ability to replicate within tumor cells before spreading to infect adjacent cells provides a renewable supply of virus within the tumor and should endow a high therapeutic index. Adenoviruses have been widely developed as oncolytic agents, however while clinical trials show little toxicity, anticancer efficacy is also usually limited, particularly after intravenous administration.2,3,4 This is thought largely to reflect the difficulties of delivery, both in the bloodstream and within the tumor itself. Recently clinical studies using the group B oncolytic adenovirus Enadenotucirev (EnAd) have shown good progress with successful systemic delivery5,6 and here, we will focus on strategies to improve virus delivery within tumor deposits.
One of the major barriers for oncolytic viruses is the challenge of particulate spread through solid tumors. Possible contributory factors include the high interstitial fluid pressure (IFP) that restricts convection, coupled with phagocytosis of virus particles by immune cells. However, the dominant inhibitory effect is thought to be the dense extracellular matrix (ECM) that physically interferes with the movement of macromolecules and particles.7,8,9,10 Tumor ECM is a complex, multicomponent structure and several components may interfere with spread of virus particles.
Previous studies have shown that pretreatment of tumors with free enzymes to degrade the ECM could improve tumor interstitial convection. These included proteases11,12 to degrade extracellular proteins and hyaluronidase13,14 (e.g., rhPH20) to degrade hyaluronan, a viscous glycosaminoglycan. In exploiting these findings to improve the performance of therapeutic viruses, oncolytic adenoviruses have been “armed” to express ECM-degrading enzymes including relaxin, a peptide hormone that reduces expression of collagen and increases matrix-metalloproteinases, showing ECM degradation, improved virus spread and better antitumor efficacy.15,16 Oncolytic adenoviruses expressing hyaluronidase have also shown improved interstitial spread and are now undergoing clinical evaluation.17 Similarly chondroitin sulfate, usually found as a proteoglycan, interacts with multiple constituents in the tumor tissue reducing fluid convection and drug permeation.18 Chondroitinase ABC (CABC)19,20 can degrade proteoglycans by removing glycosaminoglycan side chains from the protein core. This has been exploited in an oncolytic herpes virus expressing CABC which also leads to both better intratumoral spread and improved anticancer efficacy.21
Cell death within tumors is likely to include nonapoptosis mechanisms such as ischemic death (oncosis),22 and this may release large fragments of genomic DNA into the extracellular space.23,24 Since DNA is very hydrophilic and viscous, we postulated it might also inhibit interstitial spread of oncolytic virus particles. DNAse I25,26 is an endonuclease that cleaves both double-stranded and single-stranded DNA producing a mixture of 5'-phosphate mononucleotides and oligonucleotides. DNAse I is an important treatment for cystic fibrosis where the enzyme greatly reduces the viscosity of cystic fibrosis sputum.
In this study, we therefore explored the use of actin-resistant recombinant human DNAse I (aDNAse I) as a means to improve interstitial virus convection. To provide context, we included the use of hyaluronidase (PH20), CABC and proteinase K in parallel experiments. Proteinase K (PK) is a serine protease with broad substrate specificity.27 We engineered it to carry a human signal peptide and introduced a furin cleavage site between the propeptide and mature protein to allow activation by proteolytic cleavage at the cell surface.
We constructed E1,E3-deleted type 5 adenovirus vectors expressing aDNAse I, rhPH20, CABC and PK, under the control of a CMV promoter, using the AdZ system developed by Prof G. Wilkinson.28 Constructs were characterized for enzyme production in vitro and effects of the ECM-degrading enzymes on oncolytic activity was assessed in vivo. Expression of all of the enzymes, including aDNAse I, led to a strong reduction in tumor growth. We then compared conditionally-replicating group B chimeric adenovirus, EnAd, expressing aDNAse I and rhPH20. Both “armed” oncolytic viruses showed greater replication in vivo than parental EnAd, and both also showed significantly enhanced anticancer efficacy.
Results
Production and characterization of type 5 adenovirus vectors expressing aDNAse I, rhPH20, CABC, and PK
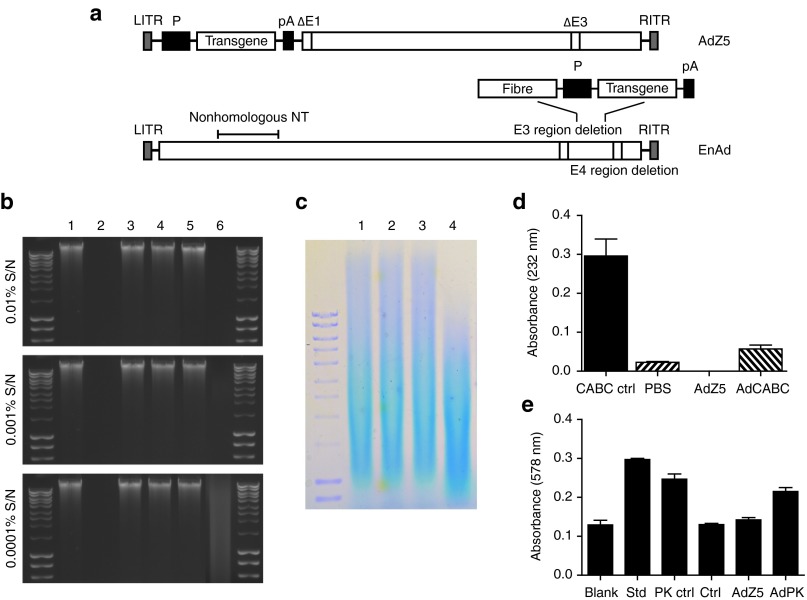
We constructed four replication-incompetent E1,E3-deleted type 5 adenoviruses expressing either human aDNAse I, rhPH20, CABC, or PK under control of the CMV promoter, using the AdZ5 recombination system (Figure 1a).28 In addition, an AdZ5 control virus was engineered which had the same backbone as the other constructs but lacked a transgene.
Figure 1.
Adenovirus construct and enzymatic activity of the transgenes. (a) Schematic representation of the adenoviruses used in this study. Each virus was engineered to express the transgene immediately downstream of a CMV promoter (P) followed by a polyadenylation sequence (pA) using either the AdZ5 recombination system or EnAd. AdZ5 is a replication deficient recombinant adenovirus with E1/E3 deletions. EnAd is a conditionally-replicating chimeric group B adenovirus that contains frequent nonhomologous nucleotide (non-homologous NT) substitutions of Ad3 for Ad11p. In addition, it has a nearly complete E3 deletion and a smaller E4 deletion mapped to E4orf4. Concentrated supernatants of HEK293 cells 48 hours after infection with enzyme-expressing AdZ5 viruses were each tested for their ability to degrade their corresponding substrate. (b) aDNAse I degradation of calf thymus DNA was assessed by agarose gel electrophoresis. (1) Undigested DNA, (2) DNA digested with recombinant DNAse, (3) DNA digested with supernatant (S/N) from uninfected cells, (4) DNA digested with supernatant from AdZ5-infected cells, (5) DNA digested with supernatant from AdPH20 infected cells, (6) DNA digested with supernatant from AdDNAse-infected cells. (c) Agarose gel electrophoresis-stained patterns of hyaluronidase degrading hyaluronic acid (HA). (1) Undigested HA, (2) HA digested with supernatant from uninfected cells, (3) HA digested with supernatant from AdZ5 infected cells, (4) HA digested with supernatant from AdPH20-infected cells. (d) Degradation of chondroitin sulfate by chondroitinase ABC measured by UV absorbance. Mean ± SD is plotted (n = 3). (e) Liberation of Folin-positive amino acids as a result of proteinase K hydrolysis of hemoglobin denatured with urea; tyrosine standard (Std), proteinase K (PK ctrl), uninfected cells (Ctrl). Mean ± SD is plotted (n = 3).
Analysis of enzyme activity in the supernatants of HEK293 cells infected with each of the four viruses indicated each virus expressed a soluble enzyme that was capable of degrading its corresponding substrate (Figure 1b–e). As expected, no activity was detected with any of the substrates in the supernatants of HEK293 cells infected with the AdZ5 control virus.
Enzyme expression does not affect virus replication and production
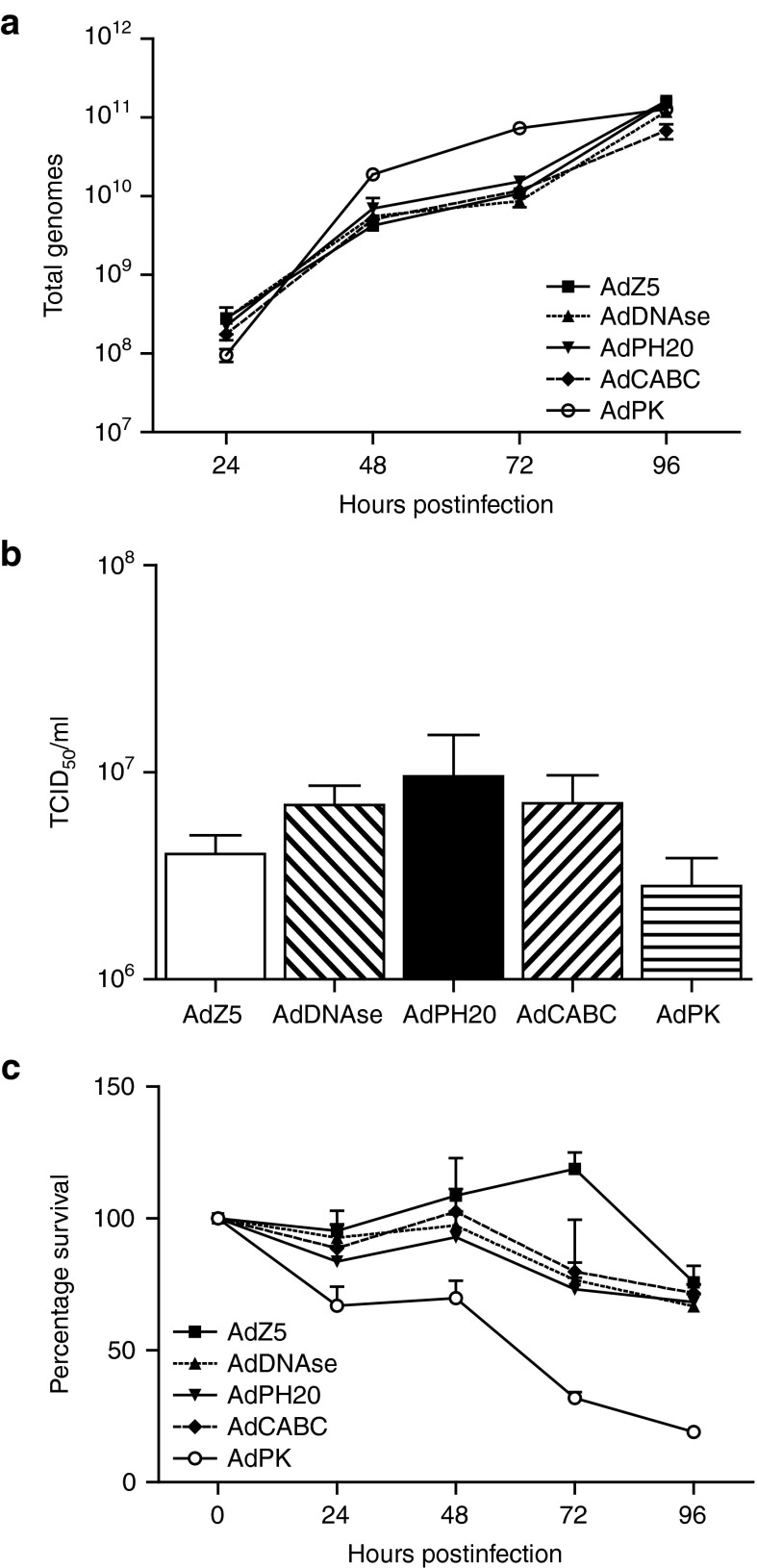
To analyze the impact of incorporating the transgenes on virus replication, HEK293 cells were infected separately with each virus. As shown in Figure 2a, all viruses produced very similar amounts of viral genomes over the period 24–96 hours. To ensure these genomes were packaged into viable infectious particles, viruses from the supernatants of infected HEK293 cells were harvested on day 4 and titrated on HEK293 Trex cells (to inhibit transgene expression by inhibiting tet operators in the CMV promoters) allowing direct measurement and comparison of infectivity (Figure 2b). There was no significant difference in viral titers between the adenoviruses encoding the enzymes and the control AdZ5 virus. Collectively, these results indicate a similar level of viral replication for all viruses and the secreted enzymes had no direct effect on virus infection or replication.
Figure 2.
Enzyme expression does not affect virus replication and production. (a) HEK293 cells were infected separately with each replication-deficient virus to assess genome production ensuring all viruses were replicating efficiently. (b) Supernatants from day 4 infected HEK293 cells were subsequently titrated on HEK293 Trex cells to measure infectivity directly. (c) MTS assay of each virus to measure impact of transgene expression on cell viability. In all experiments, mean ± SD is plotted (n = 3).
Next, we assessed the impact of the transgenes on viability of infected cells. HEK293 cells were infected separately with each Ad5 virus and cell viability was measured every 24 hours using an MTS assay. The data were normalised to uninfected control cells. As shown in Figure 2c, all viruses showed slightly greater cytotoxicity compared to AdZ5, although AdPK showed significantly greater cytotoxicity than the other viruses (P ≤ 0.001), most likely reflecting nonspecific cytotoxicity of the secreted protease.
Ad5 viruses display antitumor activity after intratumoral administration in vivo
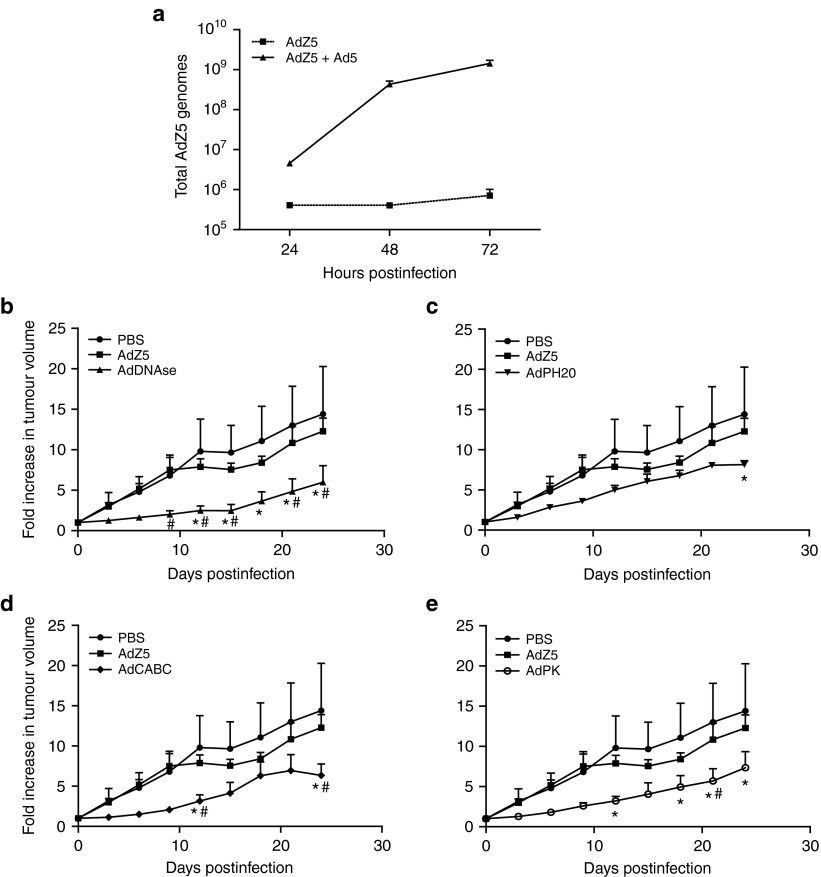
Given that the enzyme-expressing Ad5 viruses are nonreplicating, assessment of their ability to potentiate virotherapy in vivo is easiest to address by transcomplementation with a conditionally-replicating or replication-competent virus. For this purpose, we used a modified wild-type Ad5 with luciferase fused to the 3' terminal of E1A (“E1A-AdLuc”), providing both transcomplementation and luminescence imaging potential.29 To confirm suitability of E1A-AdLuc for transcomplementation, A549 cells were infected with AdZ5 alone or AdZ5 with E1A-AdLuc. Genomes of AdZ5 were measured by quantitative polymerase chain reaction (QPCR) using primers and probe designed to the CMV promoter. As shown in Figure 3a, over a 3-day period, increasing numbers of AdZ5 genomes were detected in cells coinfected with both viruses while there was no increase in genome number in cells infected with AdZ5 only. This suggests that there is sufficient E1A produced by the E1A-AdLuc to allow transcomplementation and replication of the engineered Ad5 viruses.
Figure 3.
In vivo antitumor activity after intratumoral administration. (a) Coadministration of a conditionally replicating virus (AdZ5) with E1A-Adluc allows in-trans complementation of E1A. Genomes of AdZ5 were measured by quantitative polymerase chain reaction (QPCR) using primers and probe designed toward the CMV promoter. Mean ± SD is plotted (n = 3). (b) CD1 nude mice bearing DLD tumor xenografts were treated with a single intratumoral injection of PBS, or E1A-AdLuc plus either AdZ5, AdDNAse, AdPH20, AdCABC, or AdPK (1 × 109 total vp/tumor). Tumor volume was measured every 3 days. * Significant (P ≤ 0.05) compared with tumors treated with PBS; # Significant (P ≤ 0.05) compared with tumors treated with AdZ5 (two-way analysis of variance, Bonferroni post-tests). Tumor volume ± SEM is plotted (n = 5).
Next, we analyzed the antitumor activity of all the enzyme-expressing Ad5 viruses (coadministered with E1A-AdLuc) in subcutaneous colon carcinoma tumors. SCID mice bearing DLD tumor xenografts were treated with a single intratumoral injection of PBS, or E1A-AdLuc (5 × 108 vp/tumor) plus AdZ5, AdDNAse, AdCABC, or AdPK (5 × 108 vp/tumor) (Figure 3b). Tumors were allowed to develop to 100 mm3 prior to injection, and sizes were subsequently measured every 3 days. There was no significant difference in tumor growth observed between the two control groups of PBS-treated and E1A-AdLuc/AdZ5-treated mice. Treatment with AdPH20, AdCABC, and AdPK (combined with E1A-AdLuc) all inhibited tumor growth, for example, at day 24, tumors were significantly smaller compared to the control groups in each case (P ≤ 0.05). Interestingly, treatment with AdDNAse (combined with E1A-AdLuc) inhibited tumor growth with tumor sizes significantly smaller than PBS alone (P ≤ 0.05) or AdZ5 (P ≤ 0.05) from day 9 onwards.
Expression of exogenous enzymes within tumors often raises questions about possible enhancement of metastasis. To investigate whether these treatments would promote spontaneous metastasis, we assessed the level of human DNA present within the bloodstream using human-specific QPCR. This would allow detection of circulating tumor cells that have been shed from the primary tumor after oncolytic treatment, with a sensitivity of less than 80 pg DNA/sample (approximately 14 cells). QPCR was performed using human-specific primers and probe, targeting the PTGER2 gene, against a mouse background.30 There was no detection of human-specific DNA in blood samples from any mice treated with oncolytic adenovirus expressing aDNAse I, rhPH20, CABC, or PK, suggesting none of the treatments mobilized appreciable numbers of tumor cells (Supplementary Figure S1).
The ability of DNase to improve oncolytic virus activity has not been previously reported. Coupled with published reports of the antimetastatic activity of this enzyme, we prioritized oncolytic viruses expressing aDNAse I for further development. Hyaluronidase (rhPH20) was also selected given that a hyaluronidase-expressing adenovirus is already in clinical study.
Engineering replicating group B adenoviruses, EnAdDNAse, and EnAdPH20
EnAd is an oncolytic group B adenovirus (previously known as ColoAd1), currently undergoing several early phase clinical trials for treatment of cancer. The virus combines good systemic kinetics and promising clinical activity31,32 with the possibility to encode and express transgenes. Accordingly, we constructed EnAd expressing aDNAse I and rhPH20 hyaluronidase. These enzymes, under the control of a CMV promoter, were inserted into the EnAd backbone, downstream of the fiber gene (Figure 1a).
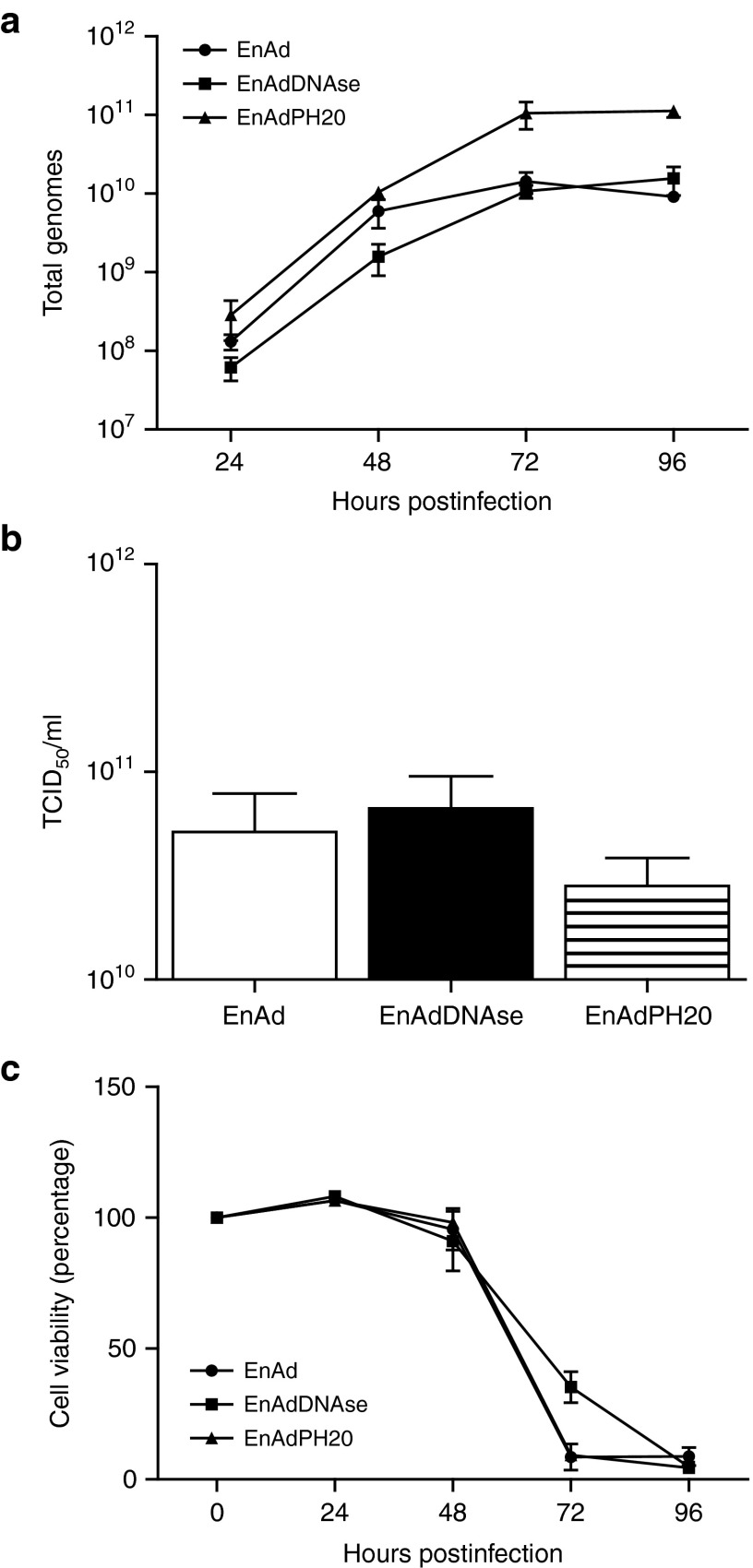
Analysis of aDNAse I and rhPH20 activity in the supernatant of A549 cells infected with “armed” EnAd confirmed the enzymes were secreted and capable of digesting exogenous DNA and hyaluronic acid respectively (data not shown). To analyze the effect of transgene expression on virus replication, the viruses were tested for their replication kinetics by measuring effects on cell viability, virus quantification, and genome production. While there were slightly more virus genomes produced in EnAdPH20-infected cells compared to EnAdDNAse and EnAd (measured by QPCR), there was no significant difference in the number of infectious particles produced, nor in cell kill (Figure 4).
Figure 4.
Enzyme expression does not affect virus replication and production. (a) A549 cells were infected separately with each EnAd virus to assess genome production ensuring all viruses were replicating efficiently. (b) Purified virus was titrated in 10-fold dilutions on A549 cells to measure infectivity directly. (c) MTS assay of each virus to measure impact of transgene expression on cell viability. In all experiments, mean ± SD is plotted (n = 3).
EnAd expressing aDNAse or PH20 exhibit superior antitumor activity in vivo compared to unmodified virus
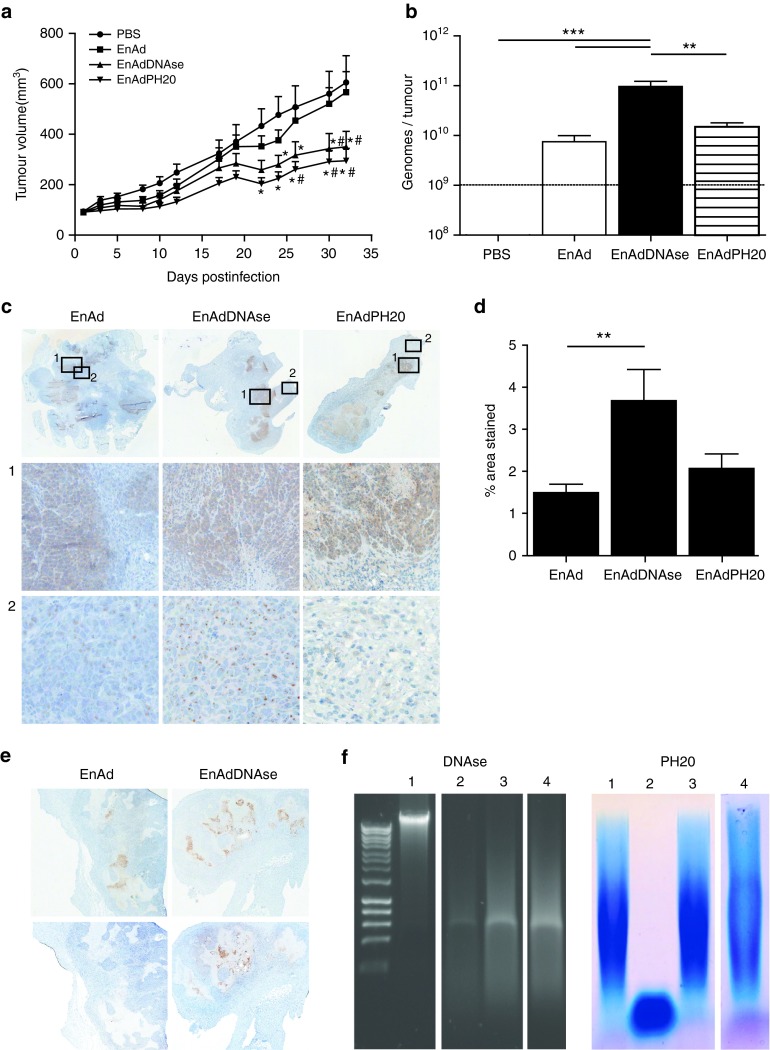
Next, we analyzed the antitumor activity of the enzyme-expressing EnAd viruses after a single intratumoral injection of a relatively low virus dose (1 × 109 vp/tumor) into pre-established DLD human colon carcinoma xenografts. This low dose and protocol was adopted to mimic a poor delivery scenario, and to maximize the potential for improved spread to enhance oncolytic activity. Both engineered viruses mediated considerable inhibition of tumor growth in comparison mice treated with PBS and unmodified EnAd (Figure 5a). At 32 days, animals were killed and the tumors removed for measurement of virus genomes by QPCR (Figure 5b) and immunohistochemical analysis. Tumors treated with EnAdDNAse showed significantly higher numbers of virus genomes present compared to both EnAd and EnAdPH20 treated tumors (12.9- and 6.5-fold respectively).
Figure 5.
EnAdDNAse and EnAdPH20 exhibit antitumor efficacy. (a) CD1 nude mice bearing DLD tumors were treated with a single intratumoral injection of PBS, EnAd, EnAdDNAse, or EnAdPH20 (1 × 109 total vp/tumor). Tumor volume was measured every 3 days. * Significant (P ≤ 0.05) compared with tumors treated with PBS; # Significant (P ≤ 0.05) compared with tumors treated with EnAd (two-way analysis of variance (ANOVA), Bonferroni post-tests). Tumor volume ± SEM is plotted (n = 10). (b) Total genomes per tumor were measured by QPCR using primers and probe against the hexon gene of EnAd; ** Significant (P ≤ 0.01) and *** Significant (P ≤ 0.001) (one-way ANOVA, Bonferroni post-tests). Mean ± SD is plotted (n = 5). (c) Immunohistochemical analysis of adenovirus distribution in colorectal tumors treated with either EnAd, EnAdDNAse or EnAdPH20. Upper panel: original magnification ×20. Lower panels: original magnification ×400. (d) Hexon staining (which is indicative of virus replication) was quantified using ImageJ software. Eight random fields in viable tissue zones were selected for each tumor (n = 5) and quantified as a percentage of stained area. ** Significant (P ≤ 0.01) compared with EnAd-treated tumors. (e) Upper panel: sections stained with antihexon antibody and counterstained with hematoxylin; original magnification ×20. Lower panel: sequential sections stained with anti-DNAse antibody and counterstained with hematoxylin; original magnification ×20. (f) Assessment of enzyme activity 32 days postinfection. Degradation of calf thymus DNA was assessed by agarose gel electrophoresis. (1) Undigested DNA, (2) DNA digested with lysate from tumors infected with EnAdDNAse, (3) DNA digested with lysate from tumors infected with EnAd, (4) DNA digested with lysate from tumors infected with EnAdPH20. Agarose gel electrophoresis-stained patterns of hyaluronic acid (HA) degradation. (1) Undigested HA, (2) HA digested with lysate from tumors infected with EnAdPH20, (3) HA digested with lysate from tumors infected with EnAd, (4) DNA digested with lysate from tumors infected with EnAdDNAse.
To assess whether enhanced efficacy observed in mice treated with either EnAdDNAse or EnAdPH20 was as a result of improved distribution of virus, immunohistochemical analysis was performed on tumors 32 days postinfection (Figure 5c–e). All of the DLD tumors treated with either EnAdDNAse or EnAdPH20 showed evidence of active virus infection (5/5 tumors for each enzyme), with many cells showing strong intranuclear hexon stain, indicative of virus packaging. It was noticeable that large patches of infection were spread throughout the tumor mass in each case. In the case of EnAdDNAse, the patches of hexon expression were coincident with strong expression of DNAse activity (Figure 5e). In contrast, only two out of five tumors in the group treated with unmodified EnAd showed signs of ongoing virus infection at day 32 (one of these is shown in Figure 5c), and no expression of DNAse (Figure 5e). Tumors treated with PBS showed no adenoviral staining (data not shown). Intriguingly, tumors treated with EnAdDNAse showed particularly wide-spread staining pattern, with many individual cells staining strongly for hexon even in regions spatially separated from the main areas of infection. This effect was not observed for the other viruses, except very close to areas of strong infection, and suggests that the DNAse-expressing virus is capable of better intratumoral spread (Figure 5c-2). This was further supported by quantifying the percentage of stained areas. EnAdDNAse-treated tumors showed significantly more positive staining compared to tumors treated with EnAd alone (Figure 5d).
Enzyme activity can be detected in tumors 32 days postinfection
To determine whether enzymes encoded within viruses remained active 32 days after intratumoral administration, tumor xenografts were resected and homogenates were assayed for substrate-specific degradation. Homogenates of tumors injected with unmodified EnAd showed a significant ability to degrade calf thymus DNA, reflecting endogenous cell-associated DNAse activity, although EnAdDNAse injected tumors showed much stronger DNA degradation due to the encoded aDNAse I activity (Figure 5f). Similarly, EnAdPH20-injected tumors showed powerful ability to digest high molecular weight hyaluronic acid, and in this case there was no evidence of any endogenous hyaluronic acid-degrading activity in the control groups. These data support the possibility that the increased intratumoral spread of EnAdDNAse, and the improved anticancer activity of both EnAdDNAse and EnAdPH20 (compared to the unmodified parental virus) is associated with persistent expression and activity of the encoded enzymes.
Discussion
Limited spread of adenovirus particles through solid tumor tissue is thought to have played a significant role in limiting the success of early clinical trials.2,3,9,33 The dense ECM restricts the intercellular percolation of macromolecules and particles,10,34,35,36 and degradation of the ECM by injection of enzymes has already been shown to enhance therapeutic activity of oncolytic viruses.12,17 However, the need for multiple reinjections of enzyme was found to limit the utility of this approach,37 paving the way for ECM-degrading enzymes to be encoded within the virus, with enzymes consequently produced and secreted from infected tumor cells throughout the duration of virus infection.
Tumor ECM is a complex mixture including proteins, carbohydrates, nucleic acids and lipids, all of which components may impact on virus particle convection. Here, we have explored the use of several ECM-degrading enzymes, encoded within adenoviruses as a strategy to enhance oncolytic virus spread through tumors. Hyaluronidase has previously been shown to enhance anticancer activity when encoded in replicating adenovirus.17 Chondroitinase ABC21 and proteases such as metalloproteinases11 and protease-inducers such as relaxin15,16 have also shown promising activity, and the wide substrate specificity of proteinase K, coupled with its high activity at a broad pH and temperature range, makes it a promising candidate. We also explored the usefulness of DNAse, reasoning that the ongoing cell death within tumors, particularly in areas of ischemia/necrosis, might release genomic DNA that could form a viscous interstitial barrier. This hypothesis was inspired by the known DNA content of cystic fibrotic mucous, shed from dying bronchiolar epithelial cells,25,38 and the fact that many pathogenic bacteria produce extracellular DNAse as a virulence factor, most likely to assist their cellular infection and spread despite the presence of extracellular DNA.
In vitro, all viruses were analyzed for their expression and secretion of soluble enzymes and the activity of each enzyme was confirmed by substrate-specific assays. It was also observed, while each enzyme did not impair virus particle production and replication, they had varying effects on cell viability with AdPK being the most cytotoxic. In DLD colorectal cancer xenografts, complemented by replication competent E1A-AdLuc, expression of proteinase K, chondroitinase ABC, hyaluronidase and actin-resistant DNase all showed the ability to improve oncolytic adenovirus activity compared to the unarmed control AdZ5. This anticancer effect was achieved after a single low dose intratumoral injection and at the time of treatment, tumors were a significant burden. It is possible that the encoded enzymes may exert direct anticancer activity, as well as improving spread of the oncolytic virus. For example, hyaluronidase is known to exert anticancer activity, perhaps by decreasing hyaluronan signaling via CD44.14 Chondroitinase activity has also been linked with inhibiting proliferation and invasion of melanoma cells, suggesting chondroitin sulfates may play a role in metastasis.39 In addition, DNase activity has been widely shown to inhibit metastasis, most likely by degrading extracellular DNA. This is discussed in more detail below. In contrast, protease activity is associated predominantly with tumor spread and progression, and we could find no reports that proteinase K has direct anticancer activity. Given that the ability of DNase to potentiate virotherapy has not been previously reported, coupled with its reported ability to inhibit metastasis, an oncolytic virus encoding DNase was explored in greater depth. An equivalent oncolytic virus, expressing rhPH20 hyaluronidase, was also engineered for comparison.
Incorporation of aDNAse I and rhPH20 into the replicating group B adenovirus, EnAd, had little effect on virus replication kinetics, and in both cases a single intratumoral dose produced significant anticancer activities. At the end of the experiment (32 days after injection), high levels of DNAse I and hyaluronidase activity were present in tumor lysates, indicating the viruses were still viable. Immunohistochemical analysis of treated tumors showed both enzymes appeared to increase the spread of virus infection within the tumor, although when quantified only DNase mediated a significant increase in spread. However, Guedan et al.17 reported both improved intratumoral spread and improved anticancer activity for an oncolytic type 5 adenovirus expressing PH20, hence it seems most likely that the improved activities of both enzyme-expressing EnAd viruses is predominantly due to improved spread of the oncolytic activity.
The powerful activity of the aDNAse-expressing EnAd supports our hypothesis that nonapoptotic cell death within tumors (whether caused by virus infection or simply as a result of cell turnover) leads to the deposition of considerable extracellular DNA. Clinically, it is well established that many tumors shed DNA into the circulation that can be used for diagnosis, much more than is shed from normal tissues. Indeed, a recent study documented that over 50% of tumors actively shed free DNA into the bloodstream including colorectal, gastroesophageal, pancreatic, and breast cancer.40 This fits with the concept of macromolecular DNA being shed from dying tumor cells in the clinical setting, as well as in experimental models.40,41,42,43,44 Expressing DNase from oncolytic viruses should also have the fortuitous consequence of helping to degrade contaminating genomic DNA during manufacture (a contaminant which is normally removed by expensive treatment with benzonase), simplifying and accelerating the manufacturing process.
It is important to consider whether expression of these degradatory enzymes might increase toxicity effects of oncolytic viruses, for example by enhancing tumor metastasis. Previous studies have shown that, when used in combination or encoded within oncolytic viruses, neither heparanase, hyaluronidase nor decorin increased the incidence of metastasis, and our studies also failed to detect any circulating tumor cells.37,45,46 The inability of DNAse to degrade rigid protein components of the ECM, combined with the absence of significant extracellular DNA in normal tissues suggests the enzyme is unlikely to enhance the spread of tumor cells into normal tissues. Indeed, it has been observed that the presence of extracellular DNA actively promotes cell invasion and metastasis47 and the addition of DNAse I systemically, prevents the occurrence of metastases in both pancreatic and liver cancers.47,48,49 Hence it is possible that the DNase-expressing oncolytic virus developed here may exert both an enhanced oncolytic anticancer activity and a potential to decrease background tumor metastasis.
To conclude, our results indicate that degrading several components of the ECM can augment virus particle spread and oncolytic activity. Degradation of extracellular DNA is surprisingly effective and may improve interstitial fluid convection without disrupting the protein matrix. We believe that oncolytic adenoviruses expressing actin-resistant DNAse are highly promising for clinical development and have the potential to be used alone or in combination with other chemotherapeutic agents.
Materials and Methods
Cell lines. HEK293, HEK293 Trex cells, A549 lung adenocarcinoma cells, and DLD colorectal adenocarcinoma cells were obtained from ATCC (Manassas, VA). All cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum.
Molecular engineering and virus production. A schematic diagram outlining the structure of the viruses is shown in Figure 1a. All enzymes were synthesized (Life Technologies) and optimized for human codon usage. aDNAse I was engineered to be actin resistant by including an A114R point mutation.25,26 rhPH20 was synthesized to exclude the sequence encoding the GPI membrane attachment motif. The 24 aa signal peptide from chondroitinase ABC was replaced with the 22 aa signal peptide from aDNAse I. This was also the case for the 15 aa signal peptide of proteinase K. In addition, since PK is expressed as a propeptide, a furin cleavage site was introduced before the mature protein to allow activation upon cleavage. All genes were flanked by regions of homology to the E1 and E3-deleted AdZ5 vector and subjected to recombination as described by Stanton et al.28 Correct sequences of each vector were confirmed by DNA sequencing. Successful recombinants were then transfected into HEK293 cells using Lipofectamine (Invitrogen, Carlsbad, CA). Resultant viruses were double-purified by cesium chloride gradient centrifugation and viral DNA concentrations determined by PicoGreen assay (PicoGreen, dsDNA quantitation reagent, Invitrogen). Infectious particles were measured by TCID50 on HEK293 cells at day 7 following fixation with formaldehyde-crystal violet. The P/I ratio (particle/infectious particles) of AdZ5 and ColoAd1 viruses are from 91–97 and 35–41, respectively.
Construction of plasmids. aDNAse I and rhPH20 was introduced into the genome of the serotype B Ad3/Ad11 chimeric adenovirus, EnAd,50 by directed ligation into the plasmid pColoAd2.4. The pColoAd2.4 plasmid contains a p15A origin of replication, a kanamycin resistance cassette, and the EnAd genome with introduced unique SgfI and SbfI restriction sites inserted after the late gene, L5.
The plasmid pColoAd2.4 was obtained by homologous recombination between a shuttle vector, pColoAd2.4 Shuttle, and the EnAd genome.
The detailed construction of the pColoAd2.4 plasmid was as follows. A ~12 kb shuttle plasmid, pEnAd Shuttle, was initially constructed in order that unique restriction sites could be introduced in the late gene, L5, region of the EnAd genome. The 5′ (nt 1-4632) and 3′ (nt 27837–32326) ends of EnAd were amplified from the EnAd genome by PCR using the primer 5′-TTGGCGGCGCGCCTATCTATATAATATACC-3′ and primers 5′-AATGCAAATCTGTGAGGGG-3′ or 5′-CTTAGTGGTGTTGTGGTATTGG-3′ respectively. The 5′ arm PCR product contained a 5′ introduced AscI site and 3′ PspOMI site that corresponds to the PspOMI site at nt 4626 in the EnAd genome. The 3′ arm PCR product contained a 5′ PspOMI site that corresponds to the PspOMI site at nt 27837 in the EnAd genome and a introduced 3′ AscI site. The PCR products were restriction digested with AscI/PspOMI and ligated in a one-step three-way ligation into an AscI linearised plasmid that contained a p15A origin of replication and a kanamycin resistance cassette. This generated the pEnAd Shuttle plasmid.
A DNA fragment corresponding to the region of the EnAd genome that is flanked by PspOMI and AclI restriction sites and contains the late gene, L5, (nt 27837–30060) was synthesized with an added region of 19 bp 5′-GCGATCGCTACCCTGCAGG-3′ inserted at position corresponding to EnAd nt 29355 (Mwg-Eurofins, Ebersberg, Germany). This additional region included restriction sites for two enzymes that are not present in the EnAd genome, SgfI and SbfI. The synthesized DNA fragment was restriction digested with the enzymes PspOMI and AclI and cloned into the corresponding region in the PspOMI/AclI digested pEnAd shuttle plasmid to create the plasmid, pColoAd2.4 shuttle. To obtain the pColoAd2.4 plasmid by homologous recombination, the pColoAd2.4 shuttle plasmid was linearised by restriction digest with the enzyme PspOMI and treated with alkaline phosphatase to remove 5' phosphates. The linearised plasmid and the EnAd genome were cotransfected into BJ5183 cells by electroporation according to the manufacturer's protocol and the generation of the pColoAd2.4 plasmid by homologous recombination was determined by restriction digest.
Correct construction of all plasmids was confirmed by DNA sequencing (MWG-Eurofins)
Assay for DNAse activity. HEK293 cells were grown to 80% confluence and infected with AdDNAse at 50 vppc (virus particles per cell). After 24 hours, infection medium was removed and replaced with serum-free medium. After 24 hours, medium was collected and concentrated by filtration using Amicon Ultra-4 columns (Millipore, Billerica, MA). DNAse activity was assessed by incubating calf thymus DNA (Sigma, Poole, UK) at a concentration of 1 mg/ml with the supernatants in the presence of 5 mmol/l MgCl2 for 1 hour at 37 °C. DNA fragment sizes after digestion with samples were analyzed by DNA agarose gel electrophoresis.
Assay for hyaluronidase activity. Hyaluronidase containing supernatants were collected as above by infecting cells with AdPH20. Hyaluronidase activity was assessed by mixing the supernatants with a 5 mg/ml HA solution containing 50 mmol/l HEPES, 0.15M NaCl, and 1 mmol/l MgCl2, pH 7.4. These were incubated for 24 hours at 37 °C before being run on a 1% agarose gel. The gel was then stained using Stains-All (0.005% in 50% ethanol) for 18 hours in the dark with gentle agitation. After incubation, the gel was washed with H20 and subjected to white light from a transilluminator for ~30 minutes until the background became colorless.
Assay for chondroitinase ABC activity. Chondroitinase ABC containing supernatants were collected as above by infecting cells with AdCABC. Chondroitinase ABC activity was assessed using the protocol described at http://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Enzyme_Assay/c2905enz.pdf.
Assay for proteinase K activity. Proteinase K containing supernatants were collected as above by infecting cells with AdPK. Proteinase K activity was assessed using the protocol described at http://www.worthington-biochem.com/PROK/assay.html.
Real-time QPCR. Real-time QPCR was used to detect and quantify adenoviral DNA from extracted DNA samples using an ABI 7000 Step One Plus Sequence Detection System and software. Primers and probe were designed to an 84 base pair fragment of the adenovirus type 5 fiber gene and amplification carried out as outlined by Bachtarzi et al.51 Amplification of the CMV promoter was carried out using primers, probe and conditions as outlined by Moulay et al.52 EnAd DNA was quantified using primers and probe targeting Hexon.53 The reaction mixture (total volume, 25 μl) contained qPCR BIO probe mix HI ROX (PCR Biosystems), 5 μl of DNA, 1 μmol/l forward/reverse primers and 100 nmol/l probe. Amplification parameters were as described in Bachtarzi et al.51 Amplification of the human-specific PTGER2 gene was carried out using primers, probe and conditions as outlined by Alcoser et al.30
In vivo studies. All animal experiments were performed in accordance with the terms of UK Home Office guidelines and the UKCCCR Guidelines for the Welfare of Animals. For in vivo coadministration studies, 6-week-old female SCID mice (Charles River, Margate, UK) were injected subcutaneously with 5 × 106 DLD cells. Once xenografts developed to ~100 mm3, mice were randomized and tumors treated with a single intratumoral injection of 10 μl of PBS, or a combination of E1A-AdLuc (5 × 108 vp/tumor) plus either AdZ5, AdDNAse, AdPH20, AdCABC, or AdPK (5 × 108 vp/tumor).
Tumor progression and morbidity status were monitored every 3 days. At the desired time points, mice were euthanised and xenografts removed. Tumors were homogenized in lysis buffer (Luciferase Assay System; Promega, Fitchburg, WI) using a motorized homogenizer (Ultra Turrax IKA T18 Basic; Fisher Scientific, Loughborough, UK) to obtain a 150 mg/ml homogenate.
Immunohistochemistry. 4-μm-thick sections were cut from paraffin embedded tumor blocks and stained using the Envision G/2 Doublestain System kit (Dako) according to the manufacturer's instructions. For Adenovirus hexon staining, the slides were incubated with 1 μg/ml of anti-adenovirus antibody (Ab8251; Abcam, Cambridge, UK) overnight at 4 °C. Sections were stained for DNAse using the anti-DNAse antibody (sc-19267; Santa Cruz, Dallas, TX) at a concentration of 2 μg/ml overnight at 4 °C. Images of sections were obtained on an Aperio Scanscope CS2 (Oxford, UK). Quantification of percentage of stained areas was performed using ImageJ software.
SUPPLEMENTARY MATERIAL Figure S1. Expression of soluble enzymes from oncolytic adenoviruses does not increase the likelihood of metastases.
Acknowledgments
The authors would like to gratefully acknowledge support from Cancer Research UK, programme grant number C552/A17720 and EPSRC grant EP/L024012/1.
Supplementary Material
Expression of soluble enzymes from oncolytic adenoviruses does not increase the likelihood of metastases.
References
- Russell, SJ, Peng, KW and Bell, JC (2012). Oncolytic virotherapy. Nat Biotechnol 30: 658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn, D (2001). Clinical research results with dl1520 (Onyx-015), a replication-selective adenovirus for the treatment of cancer: what have we learned? Gene Ther 8: 89–98. [DOI] [PubMed] [Google Scholar]
- Liu, TC, Galanis, E and Kirn, D (2007). Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nat Clin Pract Oncol 4: 101–117. [DOI] [PubMed] [Google Scholar]
- Nemunaitis, J, Senzer, N, Sarmiento, S, Zhang, YA, Arzaga, R, Sands, B et al. (2007). A phase I trial of intravenous infusion of ONYX-015 and enbrel in solid tumor patients. Cancer Gene Ther 14: 885–893. [DOI] [PubMed] [Google Scholar]
- Boni, V, De La Portilla, F, Cubillo, A, Gil-Martin, M, Calvo, E, Salazar, R et al. (2014). A phase 1 mechanism of action study of intra-tumoural (IT) or intravenous (IV) administration of enadenotucirev, an oncolytic Ad11/Ad3 chimeric group B adenovirus. Ann Oncol 25: iv361–iv372. [Google Scholar]
- Calvo, E, Gil-Martin, M, Machiels, JP, Rottey, S, Cubillo, A, Salazar, R et al. (2014). A first-in-class, first-in-human phase I study of enadenotucirev, an oncolytic Ad11/Ad3 chimeric group B adenovirus, administered intravenously in patients with metastatic epithelial tumors. J Clinical Oncol 32:5s: Abstract 3103. [Google Scholar]
- Netti, PA, Berk, DA, Swartz, MA, Grodzinsky, AJ and Jain, RK (2000). Role of extracellular matrix assembly in interstitial transport in solid tumors. Cancer Res 60: 2497–2503. [PubMed] [Google Scholar]
- Sauthoff, H, Hu, J, Maca, C, Goldman, M, Heitner, S, Yee, H et al. (2003). Intratumoral spread of wild-type adenovirus is limited after local injection of human xenograft tumors: virus persists and spreads systemically at late time points. Hum Gene Ther 14: 425–433. [DOI] [PubMed] [Google Scholar]
- Swiderek, MS, and Mannuzza, FJ (1997). Effects of ECM Proteins on Barrier Formation in Caco-2 Cells. Becton Dickinson Technical Bulletin 421: 1–4. [Google Scholar]
- Mok, W, Stylianopoulos, T, Boucher, Y and Jain, RK (2009). Mathematical modeling of herpes simplex virus distribution in solid tumors: implications for cancer gene therapy. Clin Cancer Res 15: 2352–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J, Sauthoff, H, Huang, Y, Kutler, DI, Bajwa, S, Rom, WN et al. (2007). Human matrix metalloproteinase-8 gene delivery increases the oncolytic activity of a replicating adenovirus. Mol Ther 15: 1982–1990. [DOI] [PubMed] [Google Scholar]
- Kuriyama, N, Kuriyama, H, Julin, CM, Lamborn, K and Israel, MA (2000). Pretreatment with protease is a useful experimental strategy for enhancing adenovirus-mediated cancer gene therapy. Hum Gene Ther 11: 2219–2230. [DOI] [PubMed] [Google Scholar]
- Brekken, C, Bruland, ØS and de Lange Davies, C (2000). Interstitial fluid pressure in human osteosarcoma xenografts: significance of implantation site and the response to intratumoral injection of hyaluronidase. Anticancer Res 20(5B): 3503–3512. [PubMed] [Google Scholar]
- Shuster, S, Frost, GI, Csoka, AB, Formby, B and Stern, R (2002). Hyaluronidase reduces human breast cancer xenografts in SCID mice. Int J Cancer 102: 192–197. [DOI] [PubMed] [Google Scholar]
- Ganesh, S, Gonzalez Edick, M, Idamakanti, N, Abramova, M, Vanroey, M, Robinson, M et al. (2007). Relaxin-expressing, fiber chimeric oncolytic adenovirus prolongs survival of tumor-bearing mice. Cancer Res 67: 4399–4407. [DOI] [PubMed] [Google Scholar]
- Kim, JH, Lee, YS, Kim, H, Huang, JH, Yoon, AR and Yun, CO (2006). Relaxin expression from tumor-targeting adenoviruses and its intratumoral spread, apoptosis induction, and efficacy. J Natl Cancer Inst 98: 1482–1493. [DOI] [PubMed] [Google Scholar]
- Guedan, S, Rojas, JJ, Gros, A, Mercade, E, Cascallo, M and Alemany, R (2010). Hyaluronidase expression by an oncolytic adenovirus enhances its intratumoral spread and suppresses tumor growth. Mol Ther 18: 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, KE and Fawcett, JW (2004). Chondroitin sulphate proteoglycans: preventing plasticity or protecting the CNS? J Anat 204: 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo, D, Asher, RA, Lin, R, Rhodes, KE and Fawcett, JW (2007). How does chondroitinase promote functional recovery in the damaged CNS? Exp Neurol 206: 159–171. [DOI] [PubMed] [Google Scholar]
- Prabhakar, V, Capila, I, Bosques, CJ, Pojasek, K and Sasisekharan, R (2005). Chondroitinase ABC I from Proteus vulgaris: cloning, recombinant expression and active site identification. Biochem J 386(Pt 1): 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitrieva, N, Yu, L, Viapiano, M, Cripe, TP, Chiocca, EA, Glorioso, JC et al. (2011). Chondroitinase ABC I-mediated enhancement of oncolytic virus spread and antitumor efficacy. Clin Cancer Res 17: 1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer, G, Galluzzi, L, Kepp, O and Zitvogel, L (2013). Immunogenic cell death in cancer therapy. Annu Rev Immunol 31: 51–72. [DOI] [PubMed] [Google Scholar]
- Weerasinghe, P and Buja, LM (2012). Oncosis: an important non-apoptotic mode of cell death. Exp Mol Pathol 93: 302–308. [DOI] [PubMed] [Google Scholar]
- Escobar, ML, Vazquez-Nin, G., Echeverria, O. (2011). Oncosis. In: Vazquez-Nin G (ed.). Cell Death in Mammalian Ovary. Springer Science and Business Media, Berlin, Germany. pp. 103–110. [Google Scholar]
- Shak, S, Capon, DJ, Hellmiss, R, Marsters, SA and Baker, CL (1990). Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc Natl Acad Sci USA 87: 9188–9192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer, JS, Herzka, A, Toy, KJ, Baker, DL, Dodge, AH, Sinicropi, D et al. (1996). Engineering actin-resistant human DNase I for treatment of cystic fibrosis. Proc Natl Acad Sci USA 93: 8225–8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunkel, FA and Gassen, HG (1989). Proteinase K from Tritirachium album Limber. Characterization of the chromosomal gene and expression of the cDNA in Escherichia coli. Eur J Biochem 179: 185–194 [DOI] [PubMed] [Google Scholar]
- Stanton, RJ, McSharry, BP, Armstrong, M, Tomasec, P and Wilkinson, GW (2008). Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function. Biotechniques 45: 659–62, 664. [DOI] [PubMed] [Google Scholar]
- Cawood, R, Chen, HH, Carroll, F, Bazan-Peregrino, M, van Rooijen, N and Seymour, LW (2009). Use of tissue-specific microRNA to control pathology of wild-type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcoser, SY, Kimmel, DJ, Borgel, SD, Carter, JP, Dougherty, KM and Hollingshead, MG (2011). Real-time PCR-based assay to quantify the relative amount of human and mouse tissue present in tumor xenografts. BMC Biotechnol 11: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo, E, Machiels, J-PH, Rottey, S, Cubillo, A, Salazar, R, Mardjuadi, FI et al. (2014). A first-in-class, first-in-human phase I study of enadenotucirev, an oncolytic Ad11/Ad3 chimeric group B adenovirus, administered intravenously in patients with metastatic epithelial tumors. J Clin Oncol 32 (15, suppl.) abstract 3103. [Google Scholar]
- Calvo, E, Martín, MG, Cubillo, A, Machiels, J, Rottey, S, Mardjuadi, F et al. (2014). A phase 1 study of enadenotucirev, an oncolytic ad11/ad3 chimeric group b adenovirus, administered intravenously-analysis of dose expansion and repeat cycle cohorts in patients with metastatic colorectal cancer (mCRC). Ann Oncol 25: iv367–iv367. [Google Scholar]
- Parato, KA, Senger, D, Forsyth, PA and Bell, JC (2005). Recent progress in the battle between oncolytic viruses and tumours. Nat Rev Cancer 5: 965–976. [DOI] [PubMed] [Google Scholar]
- Pipiya, T, Sauthoff, H, Huang, YQ, Chang, B, Cheng, J, Heitner, S et al. (2005). Hypoxia reduces adenoviral replication in cancer cells by downregulation of viral protein expression. Gene Ther 12: 911–917. [DOI] [PubMed] [Google Scholar]
- Shen, BH and Hermiston, TW (2005). Effect of hypoxia on Ad5 infection, transgene expression and replication. Gene Ther 12: 902–910. [DOI] [PubMed] [Google Scholar]
- Smith, E, Breznik, J and Lichty, BD (2011). Strategies to enhance viral penetration of solid tumors. Hum Gene Ther 22: 1053–1060. [DOI] [PubMed] [Google Scholar]
- Ganesh, S, Gonzalez-Edick, M, Gibbons, D, Van Roey, M and Jooss, K (2008). Intratumoral coadministration of hyaluronidase enzyme and oncolytic adenoviruses enhances virus potency in metastatic tumor models. Clin Cancer Res 14: 3933–3941. [DOI] [PubMed] [Google Scholar]
- Jones, A, Wallis, C, and Kearney, C (2003). Dornase alfa for cystic fibrosis. Cochrane Database Syst Rev 3: 1–63. [DOI] [PubMed] [Google Scholar]
- Denholm, EM, Lin, YQ and Silver, PJ (2001). Anti-tumor activities of chondroitinase AC and chondroitinase B: inhibition of angiogenesis, proliferation and invasion. Eur J Pharmacol 416: 213–221. [DOI] [PubMed] [Google Scholar]
- Bettegowda, C, Sausen, M, Leary, RJ, Kinde, I, Wang, Y, Agrawal, N et al. (2014). Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6: 224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier, N, Izui, S, Rose, L, Lambert, P, and Miescher, P (1981). The presence of circulating deoxyribonucleic-acid (DNA) in patients with acute or chronic leukemia-relation to serum anti-DNA antibodies and clq binding-activity. Human Lymphocyte Differentiation 1: 93–104. [Google Scholar]
- Shapiro, B, Chakrabarty, M, Cohn, EM and Leon, SA (1983). Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer 51: 2116–2120. [DOI] [PubMed] [Google Scholar]
- Stroun, M, Anker, P, Lyautey, J, Lederrey, C and Maurice, PA (1987). Isolation and characterization of DNA from the plasma of cancer patients. Eur J Cancer Clin Oncol 23: 707–712. [DOI] [PubMed] [Google Scholar]
- Stroun, M, Anker, P, Maurice, P, Lyautey, J, Lederrey, C and Beljanski, M (1989). Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology 46: 318–322. [DOI] [PubMed] [Google Scholar]
- Choi, IK, Lee, YS, Yoo, JY, Yoon, AR, Kim, H, Kim, DS et al. (2010). Effect of decorin on overcoming the extracellular matrix barrier for oncolytic virotherapy. Gene Ther 17: 190–201. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y, Kojima, T, Kagawa, S, Uno, F, Hashimoto, Y, Kyo, S et al. (2010). A novel translational approach for human malignant pleural mesothelioma: heparanase-assisted dual virotherapy. Oncogene 29: 1145–1154. [DOI] [PubMed] [Google Scholar]
- Wen, F, Shen, A, Choi, A, Gerner, EW and Shi, J (2013). Extracellular DNA in pancreatic cancer promotes cell invasion and metastasis. Cancer Res 73: 4256–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patutina, O, Mironova, N, Ryabchikova, E, Popova, N, Nikolin, V, Kaledin, V et al. (2011). Inhibition of metastasis development by daily administration of ultralow doses of RNase A and DNase I. Biochimie 93: 689–696. [DOI] [PubMed] [Google Scholar]
- Sugihara, S, Yamamoto, T, Tanaka, H, Kambara, T, Hiraoka, T and Miyauchi, Y (1993). Deoxyribonuclease treatment prevents blood-borne liver metastasis of cutaneously transplanted tumour cells in mice. Br J Cancer 67: 66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, I, Harden, P, Bauzon, M, Chartier, C, Nye, J, Thorne, S et al. (2008). Directed evolution generates a novel oncolytic virus for the treatment of colon cancer. PLoS One 3: e2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtarzi, H, Stevenson, M, Šubr, V, Ulbrich, K, Seymour, LW and Fisher, KD (2011). Targeting adenovirus gene delivery to activated tumour-associated vasculature via endothelial selectins. J Control Release 150: 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulay, G, Boutin, S, Masurier, C, Scherman, D and Kichler, A (2010). Polymers for improving the in vivo transduction efficiency of AAV2 vectors. PLoS One 5: e15576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, BH, Bauzon, M and Hermiston, TW (2006). The effect of hypoxia on the uptake, replication and lytic potential of group B adenovirus type 3 (Ad3) and type 11p (Ad11p). Gene Ther 13: 986–990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of soluble enzymes from oncolytic adenoviruses does not increase the likelihood of metastases.