Abstract
Inhibition of vascular smooth muscle cell (VSMC) proliferation by drug eluting stents has markedly reduced intimal hyperplasia and subsequent in-stent restenosis. However, the effects of antiproliferative drugs on endothelial cells (EC) contribute to delayed re-endothelialization and late stent thrombosis. Cell-targeted therapies to inhibit VSMC remodeling while maintaining EC health are necessary to allow vascular healing while preventing restenosis. We describe an RNA aptamer (Apt 14) that functions as a smart drug by preferentially targeting VSMCs as compared to ECs and other myocytes. Furthermore, Apt 14 inhibits phosphatidylinositol 3-kinase/protein kinase-B (PI3K/Akt) and VSMC migration in response to multiple agonists by a mechanism that involves inhibition of platelet-derived growth factor receptor (PDGFR)-β phosphorylation. In a murine model of carotid injury, treatment of vessels with Apt 14 reduces neointimal formation to levels similar to those observed with paclitaxel. Importantly, we confirm that Apt 14 cross-reacts with rodent and human VSMCs, exhibits a half-life of ~300 hours in human serum, and does not elicit immune activation of human peripheral blood mononuclear cells. We describe a VSMC-targeted RNA aptamer that blocks cell migration and inhibits intimal formation. These findings provide the foundation for the translation of cell-targeted RNA therapeutics to vascular disease.
Cardiovascular disease represents the primary cause of death and morbidity in the western world and is projected to be the number one killer globally by 2020 (ref. 1) with associated annual costs exceeding $1 trillion.2 A key pathologic process in vascular disorders, including arteriosclerosis, restenosis, vein-bypass graft disease, and allograft arteriopathy, involves vascular smooth muscle cell (VSMC) migration and proliferation. Currently, the only available therapy to directly prevent VSMC activation is the local delivery of antiproliferative/antimigratory drugs (e.g., paclitaxel3,4) by percutaneous drug-eluting stents (DES).5 The development of DES more than 15 years ago revolutionized the treatment of obstructive coronary artery disease by reducing in-stent restenosis, the primary limitation of bare metal stents. Although achieving their primary goal, the safety of DES is limited by delayed re-endothelialization, in part due to the drug's antiproliferative effect on endothelial cells (EC), contributing to late stent thrombosis and the need for prolonged dual platelet therapy.6,7 Subsequent advances in stent technology (e.g., second-generation DES) have focused mostly on stent architecture using struts comprised of thinner cobalt-chromium rather than thicker stainless steel, and using polymer based controlled-release drug delivery strategies.8,9 Unfortunately, these new generations of stents have not significantly improved patient outcomes.10,11 In contrast to changes in stent-delivery platform, little progress has been made in optimizing the therapeutic drug cargo. The changes in drugs eluted by DES have been limited to derivatives of rapamycin (e.g., zotarolimus, everolimus) that include minor changes in serum half-life (e.g., zotarolimun 9.4 hours versus rapamycin 14 hours) and immune-suppressive properties. While these second-generation drugs have resulted in improved efficacy compared to paclitaxel,12 these agents still lack cell specificity. The ideal therapy of hyperproliferative vascular disease would specifically target VSMCs without impairing endothelial cell healing.13 A VSMC-specific drug (smart drug) would allow for more rapid re-endothelialization and the associated antithrombotic, anti-inflammatory, and antiproliferative effects on the vessel wall.
The goal of this study was to identify the first VSMC-targeted smart drug for the treatment of vascular disease. We characterize an RNA aptamer (Apt 14) that selectively binds to VSMCs.14 RNA aptamers are single-stranded oligonucleotides whose binding properties depend on their sequence and structure.15 Aptamers typically have binding affinities and specificities comparable to those of antibody/antigen interactions.16,17 Since their discovery 25 years ago, aptamers have been developed for a variety of technical and therapeutic applications.16,17,18 RNA aptamers generated for therapeutic applications are typically modified with fluoro or O-methyl groups at the 2′ position of their sugar moiety, which renders them resistant to degradation by serum nucleases and reduces their immunogenicity. An important advantage of aptamers over other biologics is the feasibility of generating cross-species reactive aptamers that enable testing the same reagent in preclinical animal models and subsequently, in human clinical trials. Importantly, the clinical potential of aptamers is highlighted by the Food and Drug Administration approval of an aptamer drug for macular degeneration and by clinical trials that demonstrate the safety and efficacy of similar systemically-administered RNA drugs.19,20,21,22
We show that Apt 14 is a potent inhibitor of VSMC migration in response to multiple agonists by a mechanism that involves interruption of PDGFR-β activation and PI3K/Akt-dependent signaling in VSMC. In addition, Apt 14 attenuates neointimal formation in a murine model of vascular injury. These findings, together with the long half-life in human serum, cross-reactivity with human VSMCs, and safety profile with human cells, justify additional preclinical testing of Apt 14 as a novel inhibitor of hyperproliferative vascular disease.
Results
Specificity of RNA aptamer for VSMCs
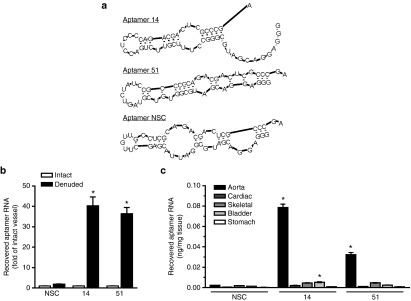
We have previously described the identification of VSMC-specific RNA aptamers using an in vitro cell-based systematic evolution of ligand by exponential enrichment (SELEX) selection process.14 Two of these aptamers (Apt 14 and Apt 51, Figure 1a) were further characterized in this study for their effects on VSMC function. Because these RNA aptamers were selected against cultured rat VSMCs (Supplementary Figure S1), we first confirmed selectivity of Apt 14 and Apt 51 to VSMCs within the arterial wall. Following intraluminal incubation for 30 minutes, VSMC-selected aptamers (Apt 14 and Apt 51), but not a nonselected control RNA aptamer (NSC), were recovered from rat femoral arteries subjected to endothelial denudation (Figure 1b). In contrast, there was no recovery of aptamers incubated with endothelium-intact femoral arteries. These data confirm that the RNA aptamers selected in vitro retain specificity for VSMCs in vivo. Next, we assessed specificity of the aptamers for vascular smooth muscle as compared to other muscle types (cardiac and skeletal) and to other organs containing smooth muscle (bladder and stomach) (Figure 1c). These data indicate high selectivity of the aptamers for vascular-derived VSMCs and highlight the potential for using these reagents to develop cell-targeted therapy to the blood vessel.
Figure 1.
Specificity of RNA aptamers for vascular smooth muscle cells (VSMCs). (a) Secondary structure predictions of selected RNA aptamers (14 and 51) and of a nonselected, control aptamer (NSC). (b) VSMC-specific aptamers (150 nmol/l; 14 and 51), but not control aptamer (150 nmol/l; NSC), bind to denuded femoral arteries but not endothelium-intact arteries. Binding was measured using RT-qPCR in vessels of equal length derived from the same animal and normalized to NSC intact. *P < 0.05 versus intact for same aptamer, n = 3. (c) Binding selectivity of RNA aptamers (150 nmol/l) to various muscle types. *P < 0.05 versus NSC for same tissue, n = 3. RT-qPCR, reverse-transcription quantitative PCR.
Apt 14 inhibits VSMC migration
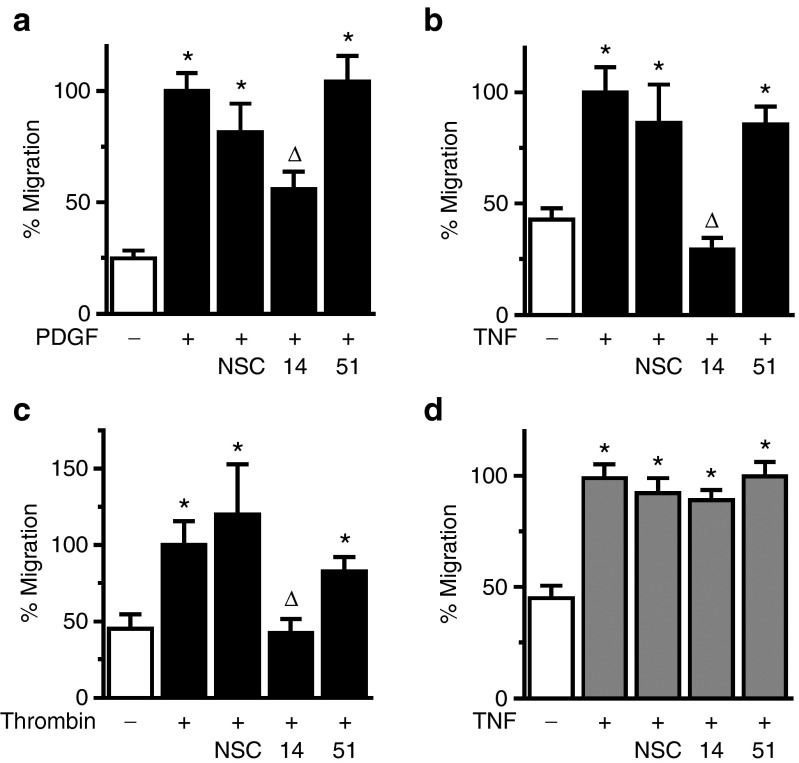
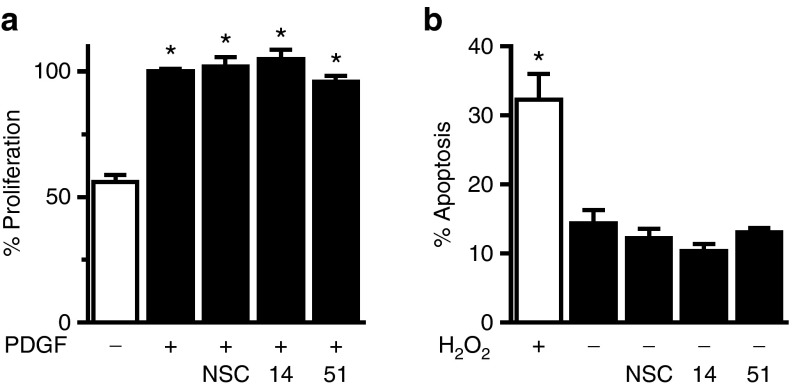
To assess for potential effects of the RNA aptamers on VSMC function, we first evaluated their effect on cell migration in response to multiple agonists. Apt 14, but not Apt 51 or control aptamer (NSC), inhibited migration to PDGF-BB (PDGF) (Figure 2a), TNF-α (TNF) (Figure 2b), and thrombin (Figure 2c). Apt 14 also prevented the basal migration observed in nonstimulated VSMCs (Supplementary Figure S2). In contrast, the aptamers had no effect on the migration of EC under the same experimental conditions (Figure 2d). Despite its ability to inhibit VSMC migration, Apt 14 had no effect on VSMC proliferation (Figure 3a) or apoptosis (Figure 3b). Together, these data suggest that Apt 14 interrupts a converging signaling pathway, shared by multiple agonists, involved in VSMC migration.
Figure 2.
Apt 14 is a selective inhibitor of vascular smooth muscle cell (VSMC) migration. The effect of aptamers (150 nmol/l) on the migration of VSMC (a–c) or ECs (d) in the presence of PDGF-BB (10 ng/ml), TNF-α (20 ng/ml), or (c) thrombin (4 U/ml). *P < 0.05 versus no agonist; ΔP < 0.05 versus NSC. n = 4–9.
Figure 3.
Apt 14 has no effect on vascular smooth muscle cell proliferation or apoptosis. (a) Effect of RNA aptamers (150 nmol/l) on cell proliferation to PDGF-BB (10 ng/ml). *P < 0.05 versus no agonist, n = 5–8. (b) Effect of aptamers on apoptosis. H2O2 (50 μmol/l) served as a positive control for apoptosis. *P < 0.05 versus no RNA control, n = 4.
Apt 14 inhibits neointimal formation in vivo
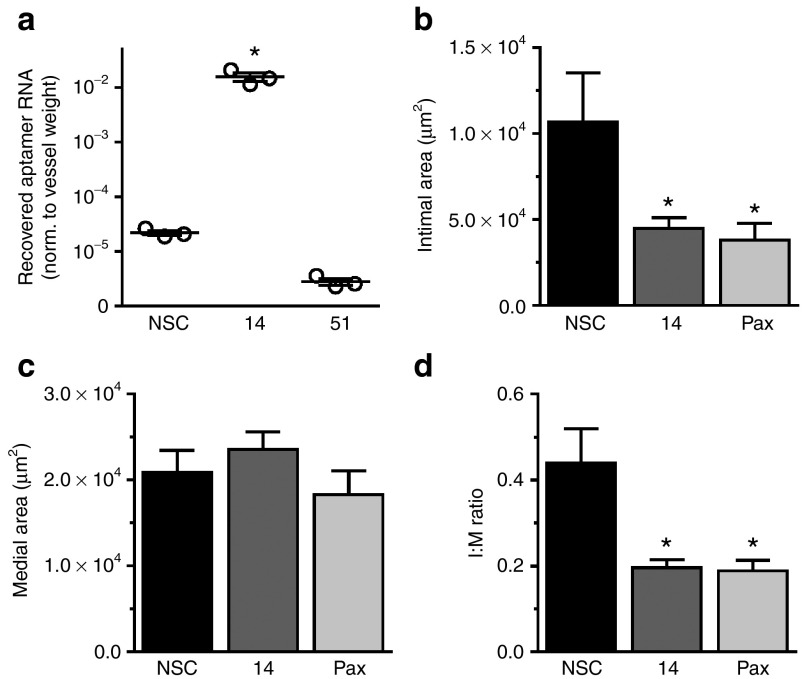
In response to vascular injury, the development of neointima is in part dependent on the migration of VSMCs from the media to the intima of injured blood vessels.23 To evaluate the effect of Apt 14 on neointimal formation, we treated carotid arteries with Apt 14 or control aptamer (NSC) while using the ligation model of vascular injury.24 We first confirmed that the pluronic gel delivery system25 was effective at delivering Apt 14 to the vessel wall. We found that Apt 14, but not NSC or Apt 51, could be recovered from the carotid artery 3 days following ligation (Figure 4a). Interestingly, although Apt 51 binds to rat VSMCs14 and to denuded rat femoral artery (Figure 1b), it did not target murine-derived VSMCs in vitro (data not shown). After confirming delivery of Apt 14 to the ligated vessel, we next assessed efficacy of in the aptamer in preventing neointimal formation. Of note, the periadventitial application of Pluronic gel (containing no RNA) appeared to accelerate intimal formation as compared to the ligated artery with no Pluronic gel control (Supplementary Figure S3), although this difference was not statistically significant. The carotid artery was treated by periadventitial delivery of NSC, Apt 14 or paclitaxel in Pluronic gel immediately following ligation. Twenty-nine days later, Apt 14-treated arteries had less neointima as compared with those treated with control aptamer (NSC), with no effect on the medial area (Figure 4b,c, Supplementary Figure S4). The efficacy of Apt 14 to reduce the intimal:media ratio by >50% was similar to that of paclitaxel, which has been used on drug eluting stents (Figure 4c, Supplementary Figure S4). Together, these data suggest that Apt 14 selectively prevents VSMC activation thereby attenuating neointimal formation.
Figure 4.
Apt 14 inhibits intimal formation in vivo. (a) Recovery of Apt 14 (150 nmol/l) from mouse carotids as measured by RT-qPCR. *P < 0.05 versus nonselected control (NSC), n = 3. (b) Intimal areas and c, medial areas of mouse carotids 29 days following carotid ligation. (d) Ratio of the intima:medial (I:M) areas. The carotid artery was treated with NSC, Apt 14, or paclitaxel (Pax) at the time of ligation. *P < 0.05 versus NSC, NSC (n = 10), Apt 14 (n = 16), and Pax (n = 13). RT-qPCR, reverse-transcription quantitative PCR.
Apt 14 inhibits PDGFR-β signaling
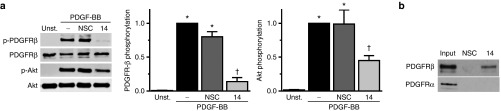
To gain insight into the mechanism(s) by which Apt 14 inhibits VSMC migration, we initially performed target identification analysis using a phospho-array (ProteomeProfiler Antibody Array, R&D Systems). These data suggested that Apt 14 inhibited PDGFR-β receptor phosphorylation (Supplementary Figure S5a). Based on these results, we established that Apt 14 prevents phosphorylation of PDGFR-β receptor by PDGF-BB (Figure 5a). We next confirmed that Apt 14 directly binds to PDGFR-β with a pull-down assay using biotinylated RNA aptamers as affinity capture agents (Figure 5b), whereas no binding was observed between Apt 14 and PDGFR-α. Direct binding to PDGFR-β was further confirmed using recombinant purified protein (Supplementary Figure S5b).
Figure 5.
Apt 14 inhibits PDGFR-β signaling. Immunoblots of PDGFR-β and PI3K/Akt activation in vascular smooth muscle cell (VSMC) treated in the presence of aptamer (400 nmol/l; 14 or nonselected control (NSC)) following a, PDGF-BB (PDGF; 50 ng/ml). *P < 0.05 versus control. †P < 0.05 versus PDGF. Unst = unstimulated. (b) Immunoblots of PDGFR-α and PDGFR-β following pull-down with biotinylated aptamers (200 nmol/l; 14 or NSC) to VSMCs. Input refers to total lysate.
Our observation that Apt 14 also inhibited migration to TNF-α and thrombin (Figure 2b,c) suggested an effect on converging signaling pathways. Recent studies have described cross-talk between TNF-α, thrombin and PDGFR signaling, resulting in the activation of PI3K/Akt.26,27 Apt 14 inhibited PI3K/Akt activation following stimulation with PDGF-BB (Figure 5a), but not TNF-α or Thrombin (Supplementary Figure S6a). Furthermore, Apt 14 did not inhibit NF-κB activation in response to TNF-α (Supplementary Figure S6b). These data suggest that Apt 14 is acting through PDGFR-β to inhibit a converging signaling pathway that is independent of PI3K/Akt or NF-κB activation.
Preclinical assessment of Apt 14
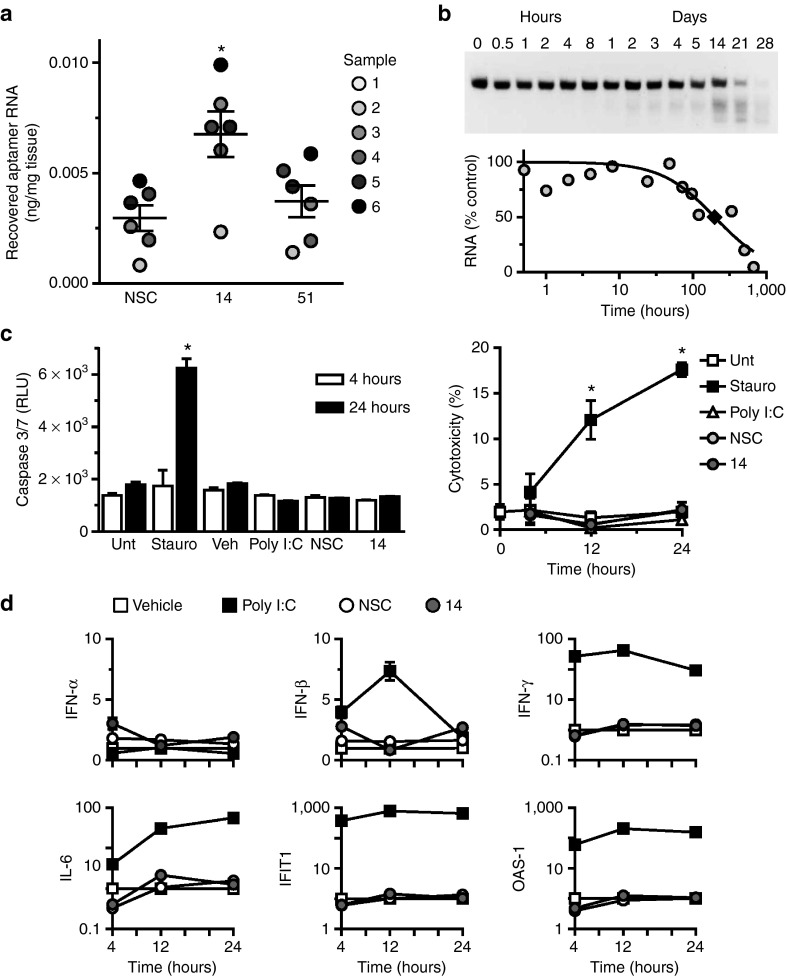
Aptamers produced against nonhuman targets require evaluation for cross-reactivity against human targets prior to consideration for clinical translation.16,17 We confirmed preferential binding of Apt 14 to cultured primary human coronary VSMCs (hVSMCs) over human endothelial cells (hEC) (Supplementary Figure S7). Next, we established recovery of Apt 14 following incubation with human pulmonary artery segments (Figure 6a, Supplementary Table S1). We next assessed stability and safety studies in human blood.17 The serum stability of RNA drugs is limited by endogenous serum nucleases. However, our VSMC aptamer selection was conducted with an aptamer library where every pyrimidine in the RNA sequence was modified at the 2′ position of the ribose with a fluoro (2′-F-pyrimidines).14 This modification has been shown to reduce endonuclease cleavage by serum RNAses.28 All aptamers identified from the VSMC aptamer selection, including Apt 14, are thus expected to be resistant to nuclease activity and have significant stability in serum. To assess the stability of Apt 14 in human serum, we incubated the aptamer RNA in 95% human serum over the course of weeks. RNA/serum samples were then resolved by denaturing polyacrylamide gel electrophoresis (Figure 6b). Based on these data, Apt 14 is stable in human serum with a predicted t1/2 of ~300 hours.
Figure 6.
Human cross-species reactivity, serum stability, and safety of aptamers. (a) Recovery of aptamers after incubation (150 nmol/l; 14, 51 or nonselected control (NSC)) with segments of human pulmonary artery, measured by RT-qPCR. Sample numbers refer to different patients as described in Supplementary Table S1. *P < 0.05 versus NSC. (b) Stability of Apt 14 (5 μmol/l) in 95% human serum. T1/2 = ~10 days (♦). (c) Assessment of aptamer (150 nmol/l; 14 or NSC) toxicity in human peripheral blood mononuclear cells. Measurement of Caspase 3/7 activation (left panel), measurement of lactate dehydrogenase (LDH) release (right panel). Percent cytotoxicity was determined by measuring LDH release as compared to maximum LDH release at each time point. *P < 0.05 versus untreated (Unt) for same time point. (d) Assessment of immunostimulatory effect of aptamers (150 nmol/l; 14 or NSC). Staurosporine: positive control for apoptosis/cytotoxicity; Poly I:C: positive control for immune stimulation. OAS-1, 2'-5' oligoadenylate synthetase 1A; INF-α, interferon-α; IFN-β, interferon-β1; IFN-γ: interferon-γ; IL-6, interleukin-6; IFIT1, interferon-induced protein with tetratricopeptide repeats 1. RT-qPCR, reverse-transcription quantitative PCR.
We next assessed potential cell cytotoxicity and activation of the innate immune system by Apt 14. Treatment of human peripheral blood mononuclear cells (PBMCs) with Apt 14 did not induce activation of Caspase 3 or 7 (markers of apoptosis) (Figure 6c; left panel) or the release of lactate dehydrogenase (Figure 6c; right panel) over a 24-hour incubation. In contrast, cytotoxicity was observed following treatment of PBMCs with staurosporine, a positive control for activation of caspase-dependent and independent apoptotic pathways29 (Figure 6c). Finally, as a measure of immune stimulation, treatment of human PBMCs with Apt 14 for 24 hours did not increase inflammatory cytokines (Il-6), type I (IFN-β) and type II (IFN-γ) interferons, or viral RNA recognition genes (2′-5′-oligoadenylate synthetase 1, OAS-1; interferon-induced protein with tetratricopeptide repeats, IFIT1) (Figure 6d). Together, these studies suggest that Apt 14 is stable in human serum and has an acceptable safety profile.
Discussion
This is the first report of a specific VSMC-targeted drug for the potential treatment of vascular disease. In this study, we show that the RNA aptamer Apt 14 (i) cross-reacts with multiple species, including human, by preferentially targeting VSMCs over ECs and nonvascular myocytes, in vitro and ex vivo; (ii) is a potent inhibitor of VSMC migration in response to multiple agonists by a mechanism that involves interruption of PDGFR-β and PI3K/Akt signaling pathways; (iii) reduces neointimal formation to levels comparable to that of paclitaxel in a murine model of vascular injury; and (iv) has a long t1/2 in human serum and it does not elicit cytotoxicity or immune activation in human PBMCs.
Cell-targeted therapies have long been considered the Holy Grail for treating diseases such as cancer.30 These smart drugs are more effective and have better safety profiles as compared to conventional nontargeted drugs.30 Despite success with this strategy of selectively controlling pathologic cell activation while limiting off-target effects on normal cells, many diseases, including cardiovascular diseases, are not currently being treated with smart drug technology. Here, we describe a VSMC-targeted smart drug for inhibiting neointimal hyperplasia.
Our findings suggest that Apt 14 inhibits VSMC activation and neointimal formation via a mechanism that interferes with PDGFR-β phosphorylation (Figure 5a) and are consistent with several studies showing that prevention of PDGFR-β activation reduces neointimal development.31,32,33 Although PDGF-BB is both mitogenic and chemotactic through its activation of PDGFR-β, PDGF-BB acts predominantly as a chemotactic factor on VSMC. For example, injection of PDGF-BB increases VSMC migration in injured arteries up to 20-fold, but proliferation by no more than 2-fold.23 Furthermore, in the presence of an antibody to PDGFR-β, VSMC migration to PDGF-BB was inhibited whereas proliferation occurred through PDGFR-α signaling.34 These observations are consistent with our finding that Apt 14 inhibited migration (Figure 2a) but not proliferation (Figure 3a) of VSMC to PDGF-BB.
We found that Apt 14 blocks migration not only to PDGF-BB, but also inhibits migration to TNF-α and thrombin (Figure 2b,c), suggesting that the effect of Apt 14 is not primarily the result of interference with ligand binding. Our data suggest that Apt 14 inhibits AKT activation by PDGF-BB (Figure 5a). The PI3K/Akt pathway has been implicated in the regulation of cell proliferation, differentiation, cellular metabolism, and cytoskeletal reorganization. Furthermore, the activation of PI3K/Akt by TNF-α and thrombin involves Src-dependent transactivation of PDGFR.26 Although, Apt 14 did not affect TNF-α and thrombin mediated-activation of PI3K/Akt (Supplementary Figure S6a), it is possible that it is acting downstream of Akt. Consistent with our finding that Apt 14 did not prevent TNF-α activation of NF-κB (Supplementary Figure S6b), inhibition of the PI3K/Akt pathway has no effect on NF-κB translocation in TNF-α-stimulated human tracheal SMCs.27 Taken together, our data suggest that Apt 14 prevents agonist-mediated activation of the c-Src/PDGFR/PI3K/Akt-dependent signaling cascade in VSMC downstream of Akt. See Supplementary Figure S8 for a proposed mechanism of action of Apt 14.
The ability of Apt 14 to selectively target VSMC over EC by binding to PDGFR-β is consistent with previous observations regarding the vascular expression of this receptor under normal and diseased conditions. Expression of PDGFR-β is low in normal human arteries and high in atherosclerotic plaques.35,36 Following vascular injury, PDGFR-β increases in those VSMC localized at or near the luminal surface and PDGFR-β mRNA levels are three to fivefold higher in the neointima than in the media.37 Expression of PDGFR-β is increased In human coronary arteries after angioplasty, and protein expression decreases as these VSMC develop a more differentiated phenotype.38 In contrast to VSMC, PDGFR-β is not expressed, or is expressed only at relatively low levels in ECs35,39 Together, these findings describe dynamic changes in VSMC expression of PDGFR-β in response to vascular injury during the time these cells are involved in vascular repair and neointimal development. The relative absence of PDGFR-β expression on ECs identifies Apt 14 as an ideal “smart drug” to target VSMC in neointimal formation.
In comparison to our description of an RNA aptamer that targets PDGFR-β, a DNA aptamer to the PDGF-B chain was previously shown to inhibit proliferation of VSMC to PDGF-BB in vitro.40 This DNA aptamer attenuated neointimal hyperplasia when measured at 2 weeks but this effect was lost by six weeks after injury.41 In contrast to this DNA aptamer, our RNA aptamer has the advantage of (i) specifically targeting the activation of VSMC signaling rather than targeting a ligand, (ii) preventing the migration to multiple agonists in addition to only PDGF-BB, and (iii) the potential delivery of other therapeutics or small modulating RNAs to VSMCs.
Given the potential of aptamers as smart drugs, considerable effort over the past decade has focused on developing and optimizing aptamers for cell-targeted therapies.17,42 Cell-targeted aptamers can be developed to (i) inhibit the function of target proteins and to control pathological cell growth and survival of the diseased cell,16,17,43 (ii) deliver therapeutic siRNAs/miRNAs44,45,46 or antiproliferative drugs directly to activated-VSMCs, (iii) allow noninvasive tracking of the RNA aptamer drug in vivo,47 (iv) identify regions of endothelial injury, (v) identify vascular segments at high risk for acute thrombotic events, and (vi) identify the presence of subclinical disease. Finally, VSMC- or EC-specific RNA aptamers could be complexed to biodegradable polymers or directly coated onto stents to allow for the local release of the RNA smart drug directly to the vessel wall.
In summary, we describe a novel RNA aptamer that preferentially targets VSMCs and can be developed as a novel therapeutic option with improved efficacy and safety profile for treating neointimal formation. Used alone, the RNA aptamer prevents agonist-mediated VSMC migration. However, this smart drug has the potential to be conjugated to existing drugs (e.g., paclitaxel) or small RNAs for the development of a cell-targeted, one-component, combination therapy. As such, this bioreagent would significantly expand the repertoire of drugs for patients with hyperplastic vascular disease, including the treatment of arteriosclerosis, restenosis, vein-bypass graft disease and allograft arteriopathy, and provide a foundation for the translation of cell-targeted RNA therapeutics to vascular disease.
Materials and Methods
Cell culture. Cell lines and culturing media used are as follows: A7r5 (ATCC, CRL-1444) cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, 11965) media containing 10% fetal bovine serum (FBS) (Atlanta biologicals, S11550); YPEN-1 (ATCC, CRL-2222) cultured in MEM (Gibco, 11095) containing 5% FBS (Atlanta biologicals, S11550), 1.5 g/l Na bicarbonate (Gibco, 25080), 0.1 mmol/l Minimum Essential Medium Non-essential Amino Acids (MEM-NEAA) (Gibco,11140), 1.0 mmol/l Na pyruvate (Gibco, 11360), and 0.03 mg/ml heparin (Sigma, H4784); primary human aortic smooth muscle cells (LifeLine, FC-0015) cultured with VascuLife Smooth Muscle Cell Medium Complete Kit (LifeLine, LL-0014); primary human vascular endothelial cells (ATCC, PCS-100-011) cultured in Vascular Cell Basal Medium (ATCC, PCS-0100-30) with the Endothelial Cell Growth Kit (ATCC, PCS-100-040). Each cell line was cultured under serum-free media conditions for 24–48 hours for experiments that required synchronized cells. Agonists were added to cell culture media included PDGF-BB (Sigma, SRP3229), TNF-α (Sigma, T7539), and thrombin (Sigma, T6634). Other media and supplements used included Opti-MEM (Life Technologies, 31985), tRNA (Invitrogen, 15401-029), penicillin/streptomycin (Life Technologies, 15140-122). All cell lines were cultured at 37 °C under 5% CO2.
RNA aptamers. RNA aptamers were either chemically synthesized (5′ C12-NH2, 5′ biotin C12-NH) by TriLink Biotechnologies (San Diego, CA) or generated by in vitro transcription with 2′fluoro pyrimidines and 2′-OH purines. The in vitro transcription was performed using the Y639F mutant T7 RNAP (obtained from Dr. Rui Sousa, University of Texas, San Antonio) on dsDNA from extended ssDNA template oligos as previously reported.14 The ssDNA templates were synthesized by IDT (Coralville, IA) at 25 nmol/l scale with desalting purification and no modifications as follows:
Aptamer 14 5′-TCGGGCGAGTCGTCTGGGGAGGTCAGAACGAAAGGCCCGCATCGTCCTCCC-3′,
Aptamer 51 5′-TCGGGCGAGTCGTCTGCGGGCGGATAGACTACGACACCGCATCGTCCTCCC-3′
Aptamer NSC 5′-TCGGGCGAGTCGTCTGCGAGGCAACAAGCTCGTAATCCGCATCGTCCTCCC-3′.
Synthetic and in vitro transcribed RNA aptamers were folded at 3 µmol/l concentration using the following protocol: 5 minutes 95 °C; 15 minutes 65 °C, and 20–30 minutes 37 °C. The folded aptamer RNA was diluted to 150 nmol/l in full media.
In vivo perfusion of rat femoral artery experiments. Animal experiments were performed according to the protocol approved by the University of Iowa Institutional Animal Care and Use Committee. Studies conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Adult male Sprague Dawley rats (Harlan) 10–12 weeks old were anesthetized with a mix of 80 mg/kg Ketamine (Fort Dodge Animal Health, Ketastet) and 10 mg/kg xylazine (Shenandoah, AnaSed Injection) and placed on a warming pad. An incision along the leg was made and the femoral artery was exposed by blunt dissection. The iliac artery and deep femoral were clamped and epigastric artery was ligated. Access to the femoral was gained by a small incision proximal to the clamped deep femoral artery. The artery was either mechanically denuded by insertion of a wire or the endothelium was left intact. The artery was then cannulated and washed with Opti-MEM containing 100 µg/ml tRNA. The cannula was anchored using a suture and 150 nmol/l aptamer RNA was introduced into the artery for 30 minutes. Following aptamer/RNA incubation the artery was washed with ice cold phosphate-buffered solution (PBS) and 0.5M NaCl ice cold PBS. The segment of femoral artery was excised and placed into a pre-weighed tube to determine the mass of the vessel segment. The artery segment was washed once with ice cold PBS, once with 0.5M NaCl ice cold PBS, once with a 5 minute 4 °C incubation in 0.5M NaCl PBS and once with ice cold PBS. After the final washes, tissue segments were placed in a 100 µg/ml proteinase K (Qiagen, 19131) lysis buffer (100 nmol/l NaCl, 10 mmol/l Tris HCl pH 8.0, 0.5% SDS) and incubated overnight at 55 °C. RNA aptamer was recovered by TRIzol (Life Technologies, 5596018) extraction of the proteinase K lysed samples. Recovered aptamer RNA was measured by reverse-transcription quantitative PCR (RT-qPCR) and normalized to the previously recorded mass of each femoral arterial segment.
Ex vivo muscle specificity. Rats were euthanized with pentobarbital (150 mg/kg IP) and the aorta, bladder, heart, thigh skeletal muscle, and stomach were collected. Each tissue was washed with Opti-MEM containing penicillin and streptomycin. To remove the endothelial cell layer, each aorta segment was mechanically denuded by rolling each segment with a pair of tweezer inserted into the lumen. Each tissue was sectioned into approximately equal-sized pieces and placed into preweighed tubes containing Opti-MEM to determine the mass of each tissue segment. Segments were then incubated with 150 nmol/l aptamer RNA with 100 µg/ml tRNA for 30 minutes at room temperature (RT), with each tube inverted gently every 10 minutes during the incubation. Following incubation with RNA aptamers, each tissue segment was washed once with ice cold PBS, once with 0.5M NaCl ice cold PBS, once with a 5 minute 4 °C incubation in 0.5M NaCl PBS, and once with ice cold PBS. After the final washes, tissue segments were placed in a 400 µg/proteinase K (Qiagen, 19131) lysis buffer (100 nmol/l NaCl, 10 mmol/l Tris HCl pH 8.0, 0.5% SDS) and incubated overnight at 55 °C. RNA aptamer was recovered by TRIzol (Life Technologies, 5596018) extraction of the proteinase K lysed samples. All samples of each tissue type were pooled and recovered aptamer RNA was measured by RT-qPCR. Recovered RNA was normalized to the average recorded mass of all segments of each tissue type.
Migration assays. A7r5 and YPEN-1 cells were synchronized for 48 hours in 0.5% FBS media. Cells were detached from the cell culture plate with 0.25% trypsin with low ethylenediaminetetraacetic acid, trypsin was inhibited with soybean trypsin inhibitor. Cells were then pelleted by centrifugation, resuspended in 0.5% FBS DMEM, and counted by hemocytometer. 5 × 104 cells were added to the inner chamber of a transwell (Costar, 3422) in a 100 µl volume that included 150 nmol/l aptamer RNA and 100 µg/ml tRNA. The outer chamber of the transwell was filled with 600 α 0.5% FBS DMEM containing either 10 ng/ml PDGF-BB, or 20 ng/ml TNF-α, or 4 units/ml thrombin or vehicle. Cells were allowed to migrate for 5 hours. The inner chamber of the transwell was then scrapped to remove any cells that did not migrate. The inner transwell chamber membrane was washed with PBS, fixed with 4% paraformaldehyde, cut from the transwell and mounted with Vectashield containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, H-1200). Migrated cells were counted at 10× magnification from five different fields of view.
Proliferation assays. 5 × 104 A7r5 cells/well were plated in 12-well plates and synchronized for 48 hours in 0.5% FBS DMEM. Cells were stimulated with either 10 ng/ml PDGF-BB or vehicle with 0.5% FBS media containing 0.05 µCi/ml [methyl-3H] thymidine (Amersham, TRK-120) and either 150 nmol/l aptamer RNA or no RNA. Cells are cultured for 4 hours under 5% CO2 at 37 °C. The media is removed; cells are washed twice with PBS and incubated with 10% tetrachloro acetic acid for 20 minutes at RT. Cells are lysed with 0.75 ml 1N NaOH for 20 minutes. Following lysis, 0.75 ml HCl is added and 1 ml of the NaOH/HCl cell lysate mixture is transferred to a scintillation vial containing 10 ml scintillation cocktail (Research Products International, Bio-Safe II 111195). The 3H DPMs were then measured by a scintillation counter and data were calculated as a percent of the PDGF-BB treated no RNA positive control.
Apoptosis assays. 2.5 × 104 A7r5 cells/well were plated in six-well plates and synchronized for 48 hours in 0.5% FBS DMEM. The A7r5 cells were treated with 150 nmol/l aptamer RNA or vehicle control with 100 µg/ml tRNA for 24 hours. Cells were collected and stained for Annexin V and with Hoechst. Staining was quantified by flow cytometry. Cells were segregated to four quadrants based upon measured Annexin V and Hoechst staining to separate live cells from early/late apoptotic cells and dead cells. The % apoptosis consists of all early/late apoptotic and dead cells as a percentage of all cells measured. Vehicle control cells were treated briefly with H2O2 as a positive control for apoptosis.
Mouse carotid ligation. C57BL/6 mice (Jackson Labs) at an average of 17.2 (range of 15.6–18.9 weeks) weeks of age were anesthetized with 40 mg/kg ketamine (Fort Dodge Animal Health, Ketastet) and 5 mg/kg xylazine (Shenandoah, AnaSed Injection) mixture and placed onto a warming pad. At the time of anesthetic induction, 2 mg/kg Meloxicam (Boehringer Ingelheim Vetmedica, Metacam) was given subcutaneously as an analgesic. Hair at the site of incision was removed using a depilatory cream, rinsed, then swabbed with betadine. An incision along the midline was made and the left common carotid artery was isolated by blunt dissection using a dissection microscope. A 8-0 nylon suture (Ethicon) was passed under the carotid artery just proximal to the bifurcation of the external and internal carotid branches, and tied to totally ligate the artery. The wound was rinsed with sterile saline. A Pluronic F-127 aptamer RNA gel was applied around the carotid artery and allowed to solidify and the site of incision was sutured. The F-127 Pluronic gel was made by diluting folded 3 µmol/l aptamer RNA to 150 nmol/l in 25% Pluronic F-127 (Sigma, P2443) with ice cold DMEM. Pluronic gel with paclitaxel was made by diluting 5 mmol/l paclitaxel in dimethyl sulfoxide to 100 µmol/l in 25% Pluronic F-127 (Sigma, P2443). The Pluronic F-127 RNA aptamer mixture was rotated overnight at 4 °C to fully dissolve the Pluronic F-127. At either 3 or 28 days following surgery, the mice were euthanized by IP overdose with either 150 mg/kg pentobarbital (Akorn, Nembutal) or 0.22 ml/kg Euthasol (Virbac Animal Health, 200-071).
Carotid arteries collected 3 days postsurgery were excised, washed with PBS, and weighed. Each carotid was then placed in a 100 µg/ml proteinase K (Qiagen, 19131) lysis buffer (100 nmol/l NaCl, 10 mmol/l Tris HCl pH 8.0, 0.5% SDS and incubated overnight at 55 °C. Aptamer RNA was recovered by TRIzol (Life Technologies, 5596018) extraction of the proteinase K lysed samples. Recovered aptamer RNA was measured by RT-qPCR and normalized to the previously recorded mass of each carotid segment.
Carotid arteries collected approximately 29 days postsurgery were excised following whole animal perfusion with 20 units/ml heparin (Fresenius Kabi USA, LLC, 63323-540-11) PBS and 4% paraformaldehyde. The carotid vessels were paraffin-embedded (Central Microscopy, University of Iowa) and sectioned 500–1,000 μm proximal to the ligation and stained with Verhoeff-van Gieson and counterstained with eosin (Central Microscopy, University of Iowa). Sections were imaged by light microscopy at 20× magnification. Imaged sections were analyzed using ImageJ by measuring the area of open lumen, the area contained within the internal elastic lamina and the area contained within the external elastic lamina. The intimal to medial ratio (I:M) was calculated by dividing the intimal area (the internal elastic lamina area subtracted by luminal area) by the medial area (the external elastic lamina area subtracted by the internal elastic lamina area).
Immunoblots. Human VSMCs were cultured in six-well plate to 80% confluence and then serum starved overnight. Aptamer RNA at 400 nmol/l was incubated with the VSMCs for 2 hours in serum-free media. The cells were then stimulated with PDGF-BB (50 ng/ml) for 10 minutes in media containing 400 nmol/l aptamer RNA. Cells were washed briefly, lysed (50 mmol/l 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid pH 7.5; 150 mmol/l NaCl; 1% Glycerol; 1% Triton X-100; 1.5 mmol/l MgCl2; 5 mmol/l ethylene glycol tetraacetic acid (EGTA); 1 mmol/l Na3VO4; mammalian protease inhibitors) and the protein concentration of the lysate was determined. Samples were diluted with Laemmli buffer, separated by SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) and immuno-blotted for the following proteins; PDGFR-β (Cell Signaling, 3169), phospho-Tyr771 PDGFR-β (Cell Signaling, 3173), PKB/Akt (Cell Signaling, 9272), and phospho-PKB/Akt (Cell Signaling, 9271).
Biotinylated aptamer pull-down. Human VMSCs (hVSMCs) were cultured in 10 cm dishes until 80% confluent. The hVSMCs were incubated with 200 nmol/l biotinylated aptamer RNA (Apt 14, NSC) in serum-free media for 30 minutes. Cells were washed briefly with PBS and lysed (10 mmol/l Tris-HCl pH 7.5; 200 mmol/l NaCl; 5 mmol/l ethylenediaminetetraacetic acid; 0.1% Triton X-100; 1 mmol/l Na3VO4; mammalian protease inhibitors). Protein concentration was determined and 150 µg of protein lysate was incubated with streptavidin beads (Pierce, 20349) at RT for 2 hours while rotating. The streptavidin beads were washed with lysis buffer. The streptavidin samples along with 20 µg of total protein were resuspended in Laemmli buffer boiled. Samples were separated by SDS-PAGE, transferred to PVDF and immune-blotted for either PDGFR-β (Cell Signaling, 3169) or PDGFR-α (Cell Signaling, 3164).
Ex vivo human pulmonary artery experiments. Pulmonary artery segments were obtained from the University of Iowa Hospitals at the time of lung transplantation using a protocol approved by the Institutional Review Board of the University of Iowa. Pulmonary artery tissue was placed in a sterile nylon filter strainer (BD Falcon, 352350). The strainer with tissue was positioned onto a 50 ml conical and the pulmonary artery tissue was washed with 40 ml 10% FBS VSMC media containing 1× penicillin/streptomycin. Further washing was achieved by rocking the tissue gently in 50 ml conical with 20 ml of VSMC media containing 1× penicillin/streptomycin and plasmocure (InvivoGen, 100 mg/ml). Tissue was then cultured in 10% FBS VSMC media containing 1× penicillin/streptomycin plasmocure for 24 hours. Culture media was collected and tested for presence of bacterial nuclease activity. The pulmonary artery endothelium was denuded and the remaining tissue was sectioned into approximately 25 mg segments. Individual segments were added to preweighed microfuge tube containing Opti-MEM and these tubes were reweighed to determine the mass of each arterial segment. Each segment was washed with Opti-MEM containing plasmocure and 100 µg/ml tRNA. Segments were then incubated with 150 nmol/l aptamer RNA with 100 µg/ml tRNA for 30 minutes at 37 °C, with each tube inverted gently every 10 minutes during the incubation. Following incubation with RNA aptamers, each arterial segment was washed once ice cold PBS, once with 0.5M NaCl ice cold PBS, once with a 5 minute 4 °C incubation in 0.5M NaCl PBS and once with ice cold PBS. After the final washes, arterial segments were placed in a 400 µg/ml proteinase K (Qiagen, 19131) lysis buffer (100 nmol/l NaCl, 10 mmol/l Tris HCl pH 8.0, 0.5% SDS) and incubated overnight at 55 °C. RNA aptamer was recovered by TRIzol (Life Technologies, 5596018) extraction of the proteinase K lysed samples. Recovered RNA aptamer was measured by RT-qPCR. Recovered RNA was normalized to the recorded mass of each pulmonary arterial segment.
Aptamer stability in human serum. Chemically synthesized Apt 14 was incubated at 5 µmol/l in 95% human serum from 30 minutes to 28 days at 37 °C under 5% CO2. The human serum was obtained from male type AB clotted blood (Sigma, H6194). At each time point, 2 µl (10 pmol aptamer RNA) was withdrawn and added to 8 µl 1× TBE and 2 µl 10× Urea RNA loading dye (Invitrogen, LC6876). Each sample was then heated to 95 °C for 5 minutes and stored at −20 °C. All time point samples (10 µl/lane = 5 pmol aptamer RNA) and controls (aptamer RNA alone and human serum alone) were loaded into a 8M urea 10% acrylamide gel and separated by electrophoresis. Size of the RNA was verified using a 100 bp DNA ladder. The gel was stained with a 1:10,000 dilution of SYBR Gold (Life Technologies, S-11494) in PBS for 30 minutes and visualized by UV exposure.
Human PBMC isolation. Heparinized venous blood was obtained from healthy adult volunteers, with donor informed consent, using a protocol approved by the Institutional Review Board of the University of Iowa. PBMCs were separated from each sample by Ficoll-Hypaque density gradient centrifugation. The PBMCs were washed twice with RPMI 1640 with L-Glutamine medium supplemented with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Lonza, 12-702F). Nonheparinized whole blood was incubated at 37 °C for 30 minutes followed by 30 minutes on ice to obtain human serum. The clotted blood was manually disrupted, centrifuged at 1,100×g for 15 minutes at 4 °C and the serum was sterile filtered. Human serum was prepared by pooling from a minimum of 14 different donors as indicated.
Human PBMC cytotoxicity assay, apoptosis assay, and innate immune stimulation. 2 × 106 PBMCs were diluted in 1 ml RPMI with 2.5% human serum. The PBMCs were divided into twelve 75 mm polypropylene round-bottom tubes (BD Biosciences) and incubated at 37 °C under 5% CO2 for 2 hours. A nonstimulated PBMC sample was processed to represent time 0. The remaining PBMC samples were treated with either 150 nmol/l aptamer RNA, vehicle control, 100 µg/ml poly(I:C) or 1 mmol/l staurosporine. Aliquots of 10 µl were collected from each PBMC sample at 4, 12, and 24 hours poststimulation. Each aliquot was mixed with Caspase-Glo 3/7 Reagent (Promega, 8091) according to the manufacturer's protocol. The remaining PBMCs from each sample were pelleted by centrifugation at 900× g for 15 minutes at 4 °C. Supernatants were collected and cytotoxicity was determined by measuring lactate dehydrogenase release with the CytoTox-ONE Homogeneous Membrane Integrity Assay (Promega, G7890). The % cytotoxicity was calculated using the maximum lactate dehydrogenase release obtained from a positive control PBMC sample treated with 9% Triton X-100 at each time point. The cell pellets were resuspended in 1 ml of TRIzol for total RNA isolation using the PureLink RNA Mini Kit (Ambion, 12183018A) and isolated total RNA was assessed for concentration and purity by a NanoDrop 2000c (Thermo Scientific). The expression levels of innate immune responsive genes were determined by RT-qPCR in triplicate and normalized to glyceraldehyde-3-phosphate dehydrogenase expression. The human specific primers used are as follows: glyceraldehyde-3-phosphate dehydrogenase (Forward: AGCCACATCGCTCAGACAC; Reverse: GCCCAATACGACCAAATCC) OAS-1 (Forward: GGTGGAGTTCGATGTGCTG; Reverse: AGGTTTATAGCCGCCAGTCA) IFN-β (Forward: TGCTCTCCTGTTGTGCTTCTCCAC; Reverse: ATAGATGGTCAATGCGGCGTCC), IFN-γ (Forward: CCAACGCAAAGCAATACATGA; Reverse: CCTTTTTCGCTTCCCTGTTTTA), IL-6 (Forward: GGTACATCCTCGACGGCATCT; Reverse: GTGCCTCTTTGCTGCTTTCAC), IFIT1 (Forward: GCCTCCTTGGGTTCGTCTACAA; Reverse: CTCAGGGCCCGCTCATAGTA).
Statistics. Results are expressed as mean ± SEM. Statistical comparisons were performed by a two-tailed t-test, or one-way or two-way analysis of variance with appropriate post-hoc analysis. A P value of < 0.05 was considered significant.
Software. Aptamer structure predictions were made by RNAstructure 5.3. Data analysis and statistics were performed with Microsoft Excel 2010, and Prism v 6.05 for Windows.
SUPPLEMENTARY MATERIAL Figure S1. Binding of selected RNA aptamers to vascular smooth muscle cells in culture. Figure S2. Apt 14 inhibits cell migration in the absence of agonist. Figure S3. Effect of Pluronic gel on neointimal formation. Figure S4. Effect of Apt 14 and paclitaxel (Pax) on intimal formation. Figure S5. PDGFR-β is the target of Apt 14. Figure S6. Apt 14 inhibits PDGFR-β signaling. Figure S7. Internalization of RNA aptamers to human vascular smooth muscle cells. Figure S8. Proposed mechanism of action of Apt 14. Table S1. Patient information of pulmonary artery segments obtained at time of lung transplantation.
Disclaimer
The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Acknowledgments
The authors thank Rui Sousa (University of Texas, San Antonio) for his gift of the mutant (Y639F) T7 RNAP. The authors thank James O McNamara (University of Iowa) and Lee-Ann Allen (University of Iowa) for helpful discussions. The authors also thank Xiu-Ying (Lisa) Liu for technical assistance with RNA transcription and purification. W.H.T., C.L.E., D.D.D., J.P.D., M.E.L., J.A., J.S., B.S., M.T., and K.S.F. designed and conducted all aspects of the experiments. J.K.T. provided the human arterial samples. Vd.F. contributed to the experimental design of the mechanistic studies. F.J.M. and P.H.G. oversaw the overall project, designed experiments, were responsible for funding and drafted and edited the manuscript. W.H.T. was supported by the American Heart Association (11POST7620018, 13POST17070101, 14SDG18850071). D.D.D. was supported by a postdoctoral training grant from the National Institutes of Health (T32 HL07344). J.P.D. was supported by a postdoctoral training grant from the National Institutes of Health (T32HL07344). This work was supported by grants to P.H.G. from the National Institutes of Health (R01CA138503 and R21DE019953), Mary Kay Foundation (9033-12 and 001-09), Elsa U Pardee Foundation (E2766) and the Roy J Carver Charitable Trust (RJCCT 01-224) and to FJM from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Program (Merit Review Award #1BX001729) and the National Institutes of Health (HL081750). Research reported in this publication was also supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA086862. Disclosure: None.
Supplementary Material
References
- Lopez, AD, Mathers, CD, Ezzati, M, Jamison, DT and Murray, CJ (2006). Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet 367: 1747–1757. [DOI] [PubMed] [Google Scholar]
- Heidenreich, PA, Trogdon, JG, Khavjou, OA, Butler, J, Dracup, K, Ezekowitz, MD et al.; American Heart Association Advocacy Coordinating Committee; Stroke Council; Council on Cardiovascular Radiology and Intervention; Council on Clinical Cardiology; Council on Epidemiology and Prevention; Council on Arteriosclerosis; Thrombosis and Vascular Biology; Council on Cardiopulmonary; Critical Care; Perioperative and Resuscitation; Council on Cardiovascular Nursing; Council on the Kidney in Cardiovascular Disease; Council on Cardiovascular Surgery and Anesthesia, and Interdisciplinary Council on Quality of Care and Outcomes Research. (2011). Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation 123: 933–944. [DOI] [PubMed] [Google Scholar]
- Sollott, SJ, Cheng, L, Pauly, RR, Jenkins, GM, Monticone, RE, Kuzuya, M et al. (1995). Taxol inhibits neointimal smooth muscle cell accumulation after angioplasty in the rat. J Clin Invest 95: 1869–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel, DI, Kunert, W, Göggelmann, C, Oberhoff, M, Herdeg, C, Küttner, A et al. (1997). Paclitaxel inhibits arterial smooth muscle cell proliferation and migration in vitro and in vivo using local drug delivery. Circulation 96: 636–645. [DOI] [PubMed] [Google Scholar]
- Fattori, R and Piva, T (2003). Drug-eluting stents in vascular intervention. Lancet 361: 247–249. [DOI] [PubMed] [Google Scholar]
- Curcio, A, Torella, D and Indolfi, C (2011). Mechanisms of smooth muscle cell proliferation and endothelial regeneration after vascular injury and stenting: approach to therapy. Circ J 75: 1287–1296. [DOI] [PubMed] [Google Scholar]
- Centemero, MP and Stadler, JR (2012). Stent thrombosis: an overview. Expert Rev Cardiovasc Ther 10: 599–615. [DOI] [PubMed] [Google Scholar]
- Kedia, G and Lee, MS (2007). Stent thrombosis with drug-eluting stents: a re-examination of the evidence. Catheter Cardiovasc Interv 69: 782–789. [DOI] [PubMed] [Google Scholar]
- Garg, S, Bourantas, C and Serruys, PW (2013). New concepts in the design of drug-eluting coronary stents. Nat Rev Cardiol 10: 248–260. [DOI] [PubMed] [Google Scholar]
- Corti, R (2012). Long-term patient benefit with biodegradable polymer biolimus eluting stent. Minerva Cardioangiol 60: 629–636. [PubMed] [Google Scholar]
- Lupi, A, Rognoni, A, Secco, GG, Lazzero, M, Nardi, F, Fattori, R et al. (2014). Biodegradable versus durable polymer drug eluting stents in coronary artery disease: insights from a meta-analysis of 5,834 patients. Eur J Prev Cardiol 21: 411–424. [DOI] [PubMed] [Google Scholar]
- Kaul, U, Bangalore, S, Seth, A, Arambam, P, Abhaychand, RK, Patel, TM et al.; TUXEDO–India Investigators. (2015). Paclitaxel-Eluting versus Everolimus-Eluting Coronary Stents in Diabetes. N Engl J Med 373: 1709–1719. [DOI] [PubMed] [Google Scholar]
- Tang, R and Chen, SY (2014). Smooth muscle-specific drug targets for next-generation drug-eluting stent. Expert Rev Cardiovasc Ther 12: 21–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, WH, Bair, T, Peek, AS, Liu, X, Dassie, J, Stockdale, KR et al. (2012). Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS One 7: e43836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington, AD and Szostak, JW (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Thiel, KW and Giangrande, PH (2009). Therapeutic applications of DNA and RNA aptamers. Oligonucleotides 19: 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe, AD, Pai, S and Ellington, A (2010). Aptamers as therapeutics. Nat Rev Drug Discov 9: 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram, P, Kurniawan, H, Byrne, ME and Wower, J (2013). Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci 48: 259–271. [DOI] [PubMed] [Google Scholar]
- Sundaram, P, Kurniawan, H, Byrne, ME and Wower, J (2013). Therapeutic RNA aptamers in clinical trials. Eur J Pharm Sci 48: 259–271. [DOI] [PubMed] [Google Scholar]
- Esposito, CL, Passaro, D, Longobardo, I, Condorelli, G, Marotta, P, Affuso, A et al. (2011). A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS One 6: e24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povsic, TJ, Vavalle, JP, Aberle, LH, Kasprzak, JD, Cohen, MG, Mehran, R et al.; RADAR Investigators. (2013). A Phase 2, randomized, partially blinded, active-controlled study assessing the efficacy and safety of variable anticoagulation reversal using the REG1 system in patients with acute coronary syndromes: results of the RADAR trial. Eur Heart J 34: 2481–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, MY, Rusconi, CP, Alexander, JH, Tonkens, RM, Harrington, RA and Becker, RC (2008). A randomized, repeat-dose, pharmacodynamic and safety study of an antidote-controlled factor IXa inhibitor. J Thromb Haemost 6: 789–796. [DOI] [PubMed] [Google Scholar]
- Jawien, A, Bowen-Pope, DF, Lindner, V, Schwartz, SM and Clowes, AW (1992). Platelet-derived growth factor promotes smooth muscle migration and intimal thickening in a rat model of balloon angioplasty. J Clin Invest 89: 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A and Lindner, V (1997). Remodeling with neointima formation in the mouse carotid artery after cessation of blood flow. Arterioscler Thromb Vasc Biol 17: 2238–2244. [DOI] [PubMed] [Google Scholar]
- Oshiro, A, da Silva, DC, de Mello, JC, de Moraes, VW, Cavalcanti, LP, Franco, MK et al. (2014). Pluronics f-127/l-81 binary hydrogels as drug-delivery systems: influence of physicochemical aspects on release kinetics and cytotoxicity. Langmuir 30: 13689–13698. [DOI] [PubMed] [Google Scholar]
- Cheng, SE, Lee, IT, Lin, CC, Hsiao, LD and Yang, CM (2014). Thrombin induces ICAM-1 expression in human lung epithelial cells via c-Src/PDGFR/PI3K/Akt-dependent NF-κB/p300 activation. Clin Sci (Lond) 127: 171–183. [DOI] [PubMed] [Google Scholar]
- Lee, CW, Lin, CC, Lin, WN, Liang, KC, Luo, SF, Wu, CB et al. (2007). TNF-alpha induces MMP-9 expression via activation of Src/EGFR, PDGFR/PI3K/Akt cascade and promotion of NF-kappaB/p300 binding in human tracheal smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 292: L799–L812. [DOI] [PubMed] [Google Scholar]
- Lok, CN, Viazovkina, E, Min, KL, Nagy, E, Wilds, CJ, Damha, MJ et al. (2002). Potent gene-specific inhibitory properties of mixed-backbone antisense oligonucleotides comprised of 2'-deoxy-2'-fluoro-D-arabinose and 2'-deoxyribose nucleotides. Biochemistry 41: 3457–3467. [DOI] [PubMed] [Google Scholar]
- Zhang, XD, Gillespie, SK and Hersey, P (2004). Staurosporine induces apoptosis of melanoma by both caspase-dependent and -independent apoptotic pathways. Mol Cancer Ther 3: 187–197. [PubMed] [Google Scholar]
- Tarner, IH, Müller-Ladner, U and Fathman, CG (2004). Targeted gene therapy: frontiers in the development of ‘smart drugs'. Trends Biotechnol 22: 304–310. [DOI] [PubMed] [Google Scholar]
- Hart, CE, Kraiss, LW, Vergel, S, Gilbertson, D, Kenagy, R, Kirkman, T et al. (1999). PDGFbeta receptor blockade inhibits intimal hyperplasia in the baboon. Circulation 99: 564–569. [DOI] [PubMed] [Google Scholar]
- Sirois, MG, Simons, M and Edelman, ER (1997). Antisense oligonucleotide inhibition of PDGFR-beta receptor subunit expression directs suppression of intimal thickening. Circulation 95: 669–676. [DOI] [PubMed] [Google Scholar]
- Caglayan, E, Vantler, M, Leppänen, O, Gerhardt, F, Mustafov, L, Ten Freyhaus, H et al. (2011). Disruption of platelet-derived growth factor-dependent phosphatidylinositol 3-kinase and phospholipase Cγ 1 activity abolishes vascular smooth muscle cell proliferation and migration and attenuates neointima formation in vivo. J Am Coll Cardiol 57: 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama, N, Hart, CE and Clowes, AW (1994). Different functions of the platelet-derived growth factor-alpha and -beta receptors for the migration and proliferation of cultured baboon smooth muscle cells. Circ Res 75: 682–691. [DOI] [PubMed] [Google Scholar]
- Karvinen, H, Rutanen, J, Leppänen, O, Lach, R, Levonen, AL, Eriksson, U et al. (2009). PDGF-C and -D and their receptors PDGFR-alpha and PDGFR-beta in atherosclerotic human arteries. Eur J Clin Invest 39: 320–327. [DOI] [PubMed] [Google Scholar]
- Rubin, K, Tingström, A, Hansson, GK, Larsson, E, Rönnstrand, L, Klareskog, L et al. (1988). Induction of B-type receptors for platelet-derived growth factor in vascular inflammation: possible implications for development of vascular proliferative lesions. Lancet 1: 1353–1356. [DOI] [PubMed] [Google Scholar]
- Majesky, MW, Reidy, MA, Bowen-Pope, DF, Hart, CE, Wilcox, JN and Schwartz, SM (1990). PDGF ligand and receptor gene expression during repair of arterial injury. J Cell Biol 111(5 Pt 1): 2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa, S, Ueda, M, van der Loos, CM, van der Wal, AC and Becker, AE (1996). Expression of platelet derived growth factor B chain and beta receptor in human coronary arteries after percutaneous transluminal coronary angioplasty: an immunohistochemical study. Heart 75: 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, R, Raines, EW and Bowen-Pope, DF (1986). The biology of platelet-derived growth factor. Cell 46: 155–169. [DOI] [PubMed] [Google Scholar]
- Green, LS, Jellinek, D, Jenison, R, Ostman, A, Heldin, CH and Janjic, N (1996). Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry 35: 14413–14424. [DOI] [PubMed] [Google Scholar]
- Leppänen, O, Janjic, N, Carlsson, MA, Pietras, K, Levin, M, Vargeese, C et al. (2000). Intimal hyperplasia recurs after removal of PDGF-AB and -BB inhibition in the rat carotid artery injury model. Arterioscler Thromb Vasc Biol 20: E89–E95. [DOI] [PubMed] [Google Scholar]
- Cerchia, L, Giangrande, PH, McNamara, JO and de Franciscis, V (2009). Cell-specific aptamers for targeted therapies. Methods Mol Biol 535: 59–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, P, Viles, KD, Soule, EE and Woodruff, RS (2013). Application of aptamers for targeted therapeutics. Arch Immunol Ther Exp (Warsz) 61: 255–271. [DOI] [PubMed] [Google Scholar]
- McNamara, JO 2nd, Andrechek, ER, Wang, Y, Viles, KD, Rempel, RE, Gilboa, E et al. (2006). Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 24: 1005–1015. [DOI] [PubMed] [Google Scholar]
- Dassie, JP, Liu, XY, Thomas, GS, Whitaker, RM, Thiel, KW, Stockdale, KR et al. (2009). Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol 27: 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, KW, Hernandez, LI, Dassie, JP, Thiel, WH, Liu, X, Stockdale, KR et al. (2012). Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res 40: 6319–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassie, JP, Hernandez, LI, Thomas, GS, Long, ME, Rockey, WM, Howell, CA et al. (2014). Targeted inhibition of prostate cancer metastases with an RNA aptamer to prostate-specific membrane antigen. Mol Ther 22: 1910–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.