Abstract
An effective human immunodeficiency virus type 1 (HIV-1) vaccine is the best solution for halting the acquired immune deficiency syndrome epidemic. Here, we describe the design and preclinical immunogenicity of T-cell vaccine expressing novel immunogens tHIVconsvX, vectored by DNA, simian (chimpanzee) adenovirus, and poxvirus modified vaccinia virus Ankara (MVA), a combination highly immunogenic in humans. The tHIVconsvX immunogens combine the three leading strategies for elicitation of effective CD8+ T cells: use of regions of HIV-1 proteins functionally conserved across all M group viruses (to make HIV-1 escape costly on viral fitness), inclusion of bivalent complementary mosaic immunogens (to maximize global epitope matching and breadth of responses, and block common escape paths), and inclusion of epitopes known to be associated with low viral load in infected untreated people (to induce field-proven protective responses). tHIVconsvX was highly immunogenic in two strains of mice. Furthermore, the magnitude and breadth of CD8+ T-cell responses to tHIVconsvX-derived peptides in treatment-naive HIV-1+ patients significantly correlated with high CD4+ T-cell count and low viral load. Overall, the tHIVconsvX design, combining the mosaic and conserved-region approaches, provides an indisputably better coverage of global HIV-1 variants than previous T-cell vaccines. These immunogens delivered in a highly immunogenic framework of adenovirus prime and MVA boost are ready for clinical development.
Traditional vaccine approaches using killed or live attenuated virus cannot be used for human immunodeficiency virus type 1 (HIV-1), and so subunit vaccines are favored.1,2 They employ HIV-1-derived immunogens, which determine the specificity of the vaccine-elicited responses, and effective responses must contend with HIV-1 variability.3 Combinations of vaccine modalities are used to present the immunogens to the immune system, influencing the magnitude, type, location and durability of the elicited effector functions, and subsequent immunological memory. The quality of both the immunogens and their delivery is critical to achieve protective immunity: suboptimal design of either may cause vaccine failure.
The best HIV-1 vaccine strategy will likely involve induction of both broadly neutralizing antibodies and effective CD8+ T cells. The path to each requires different approaches, therefore most studies focus on developing T cell- and broadly neutralizing antibody-vaccine strategies separately, before ultimately combining into one field vaccine. Meanwhile, either type of vaccine alone, if effective, could decrease HIV-1 spread and benefit infected individuals. In humans, the protective role of T cells is supported indirectly by detection of HIV-1-specific CD8+ T cells in HIV-1-exposed seronegative subjects,4 the kinetics of early partial control of viremia as the first CD8+ T-cell responses appear, extensive virus escape in targeted epitopes5,6, and the protective effects of certain human leukocyte antigen (HLA) class I allotypes.7 Model vaccine and simian immunodeficiency virus-challenge studies in rhesus macaques have provided direct evidence that CD8+ T cell-vaccine responses can improve viral control and outcome in infected animals in a traditional vaccine setting8 and, remarkably, vaccine-elicited CD8+ T cells can both protect9 and clear10 simian immunodeficiency virus infection when elicited using a particular molecular clone of cytomegalovirus vector.
Currently, there are three leading strategies for antigen design for induction of effective anti-HIV-1 CD8+ T-cell responses. In the first strategy, in silico-designed multivalent-mosaic proteins efficiently capture the most common epitope variants in the Los Alamos National Laboratory HIV Sequence Database (LANL-HSD, www.hiv.lanl.gov).11 In the nonhuman primate model, full-length protein mosaic vaccines induced CD8+ and CD4+ T-cell responses of higher magnitude, greater breadth and cross-reactivity than either consensus-based or natural-protein vaccines against peptides representing diverse natural strains.12,13,14 Nevertheless, the use of full-length proteins only partially addresses HIV-1 diversity and escape, because efficacy is likely to be compromised by the inclusion of variable immunodominant and less protective regions.1 In the second strategy, we and Mullins et al.1,15,16,17 hypothesized that focusing vaccine-elicited T cells on the most conserved regions of HIV-1 proteins would be beneficial because mutations in these regions are more likely to cause replicative fitness loss,18,19,20,21 and conserved epitopes are common to most HIV-1 variants, both within- and between clades, offering the potential for broad protection against diverse natural strains and global deployment. We pioneered the approach using 14 highly conserved consensus regions of HIV-1 proteins assembled into a chimeric immunogen HIVconsv.15 When delivered in combinations of plasmid DNA, nonreplicating simian (chimpanzee) adenovirus (ChAdV) serotype 63, and nonreplicating poxvirus modified vaccinia virus Ankara (MVA), HIVconsv induced high frequencies of oligofunctional HIV-1-specific T cells capable of inhibiting HIV-1 replication in autologous CD4+ cells in vitro.22,23 The third strategy selects beneficial CD8+ T-cell epitopes that are associated with low viral load, based on data from ~1,000 HIV-1 clade-B and C-infected individuals.24 Beneficial epitopes assembled into a vaccine immunogen have induced broad CD8+ T-cell responses in preclinical models.25
Our first-generation conserved-region vaccine15 provided the proof of concept that taking subdominant conserved regions out of the context of full-length proteins/HIV-1 and delivering them by a potent ChAdV-MVA regimen can induce robust human immune responses (re)focused on conserved epitopes22,23 (and T.H., unpublished). Furthermore, the elicited T cells could recognize and kill autologous HIV-1-infected cells, where the epitopes were expressed in the context of natural HIV-1 proteins.22,23 Encouraged by this foundational work, here we designed a second-generation vaccine also focusing on functionally conserved regions, but improved by incorporating advances made in the intervening years since our original conserved vaccines design in 2005. Thus, the main improvement is in combining the three above-mentioned T-cell approaches into one strategy as conserved bivalent-mosaic immunogens, designated tHIVconsvX, which have a superior coverage of global HIV-1 variants to any previous vaccine approach. This second-generation design also benefited from the information gained by the clinical testing of the original HIVconsv vaccine as well as by natural-infection studies defining epitopes that are associated with viral control. The tHIVconsvX vaccines are vectored by plasmid DNA, ChAdOx126, and MVA. Collectively, the very high immunogenicity of the first-generation vaccine in humans23 (and T.H., unpublished), the highly rational in silico immunogen design, the strong preclinical overall immunogenicity in mice, and the demonstration that tHIVconsvX-specific CD8+ T cells correlated with high CD4+ T-cell count in untreated patients strongly argue for timely clinical development of this novel, global T-cell vaccine approach.
Results
Design of the tHIVconsvX immunogens
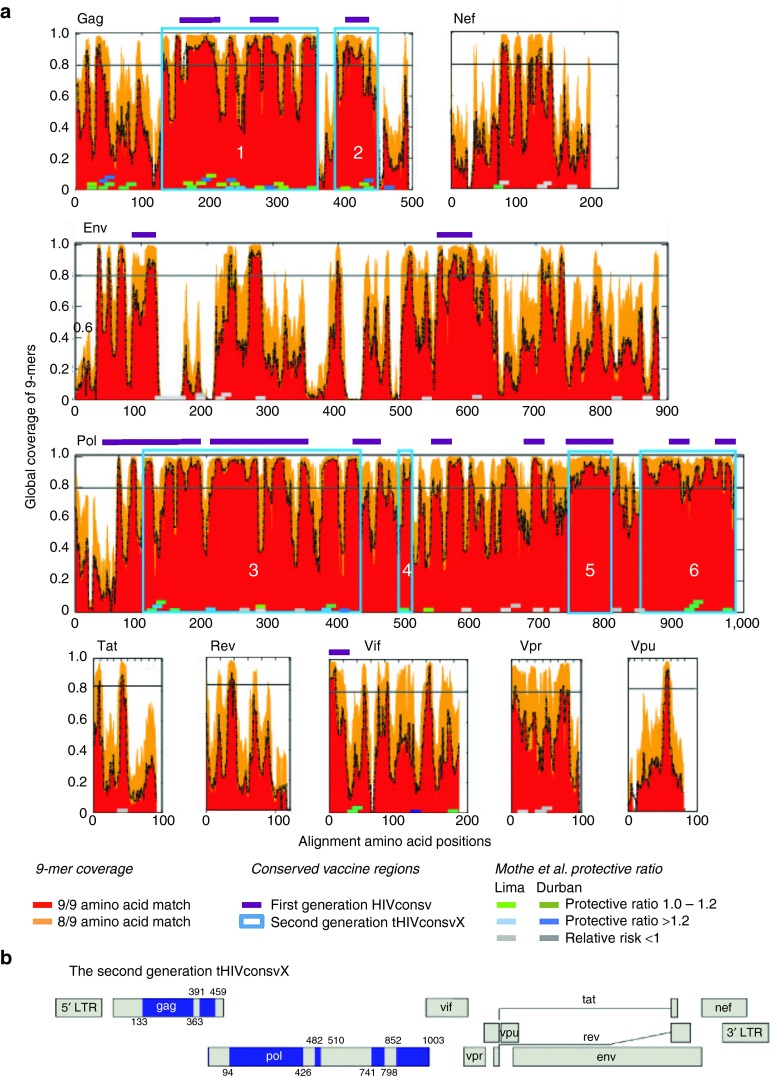
Open-reading frames (ORFs) of up to ~2.7 kbp can be easily inserted into commonly used vaccine vectors and support high protein expression. Thus, we aimed to include conserved regions totaling ~900 amino acids (aa), ~28% of the HIV-1 proteome, while minimizing the number of junctional regions and maximizing the number of beneficial epitopes.24 Our final second-generation vaccine was 872 aa in length, well within the range of the vector capacities, and contained only six fragments. In contrast, the first generation was 778 aa long and contained 14 fragments, and thus had many more junctional domains. The LANL-HSD curated global-HIV-protein alignments, circa September 2013, were used as the baseline data to define conserved regions. These alignments included only one sequence per individual, and only sequences spanning full-length proteins. The alignments were “cleaned” to exclude all proteins with frame-shifting mutations or uncertain aa calls, leaving 1,800–3,600 sequences per protein. Cooptimized, complementary pairs of two mosaic proteins were designed,11 spanning each HIV-1 protein (Figure 1a). Conservation was defined by the capacity of the two mosaic proteins to have a perfect 9/9-aa match to at least 80% of the potential T-cell epitope (PTE) variants found among the diverse HIV-1 strains in the LANL-HSD alignments; the 80% cutoff translated into a design of six fragments from 29 to 333 aa long. Next, both beneficial and bad epitopes as defined by Mothe et al.24 were overlaid on vaccine epitope coverage maps for the final selection of regions for the second-generation conserved vaccine based on all criteria above, and we included only regions that were both conserved and enriched for beneficial epitopes. Thus, our second-generation vaccine contained 33 of the 48 beneficial epitopes defined by Mothe et al.24 (69%)—the missing 15 beneficial epitopes were in variable regions in p17 and Vif; in contrast, our original design only contained only 14 (29%) beneficial epitopes. The six regions we used included the whole of Gag p24, one region in Gag p15 and four regions of Pol overlapping with protease, polymerase, and integrase (Figure 1a,b). As a trade-off between fragment length and coverage, very short stretches of modestly increased 9-mer diversity were tolerated. In contrast, the original vaccine matched only 66% of the PTE regions spanned by the vaccine regions (Figure 2a; see Supplementary Figures S1 and S2 for more detail). The improvement in PTE coverage in the second generation was due to two factors. First, the second-generation vaccine spanned more conserved regions than our original vaccine (Figure 2b). Second, the inclusion of two mosaic sequences, which provides the two most common forms of each 9-mer as the protein region traversed, enables much greater coverage of natural diversity by the vaccine (Supplementary Figures S1 and S2). Supplementary Figure S3 provides a simple summary of diversity and coverage of three representative epitopes with different levels of diversity, to provide a more intuitive view of the implications of including bivalent vaccine in terms of improved coverage of HIV-1 diversity.
Figure 1.
Definition of the second-generation conserved regions employed in tHIVconsvX. (a) Global HIV-1 protein alignments taken from the LANL-HSD were used to design complementary bivalent mosaics for each protein spanning the full proteome, as described previously.11 The epitope coverage plots show, on the y-axis, the fraction of PTEs in the alignment that are matched by one of the two 9-mers in the mosaic pair; the fractions with a 9/9 and 8/9 matches are shown in red and orange, respectively. The x-axis tracks along the protein alignment, such that column that contains the first nine positions in an alignment (positions 1–9) is used to provide the coverage data shown in the left-most point, then the second 9-mer column that contains positions 2–10 provides the data for the next point, etc. The black dashed line shows the sum of the frequencies of the two most common 9-mers in each position. The purple bars above the graphs and the blue boxes illustrate the location of the 14 regions included, which are used in the first-generation conserved HIVconsv immunogen and the 6 regions in the second-generation tHIVconsvX immunogens, respectively. The small squares show the beneficial (green, modest reduction in viral loads, and blue more dramatic reductions in viral loads) and bad (grey) epitopes.24 The shades of blue and green indicate the cohort, in which the associations were found: Spain, Peru, and South Africa. (b) Six regions (blue) of HIV-1 Gag and Pol chosen to compose the tHIVconsvX immunogens with aa positions indicated above and below.
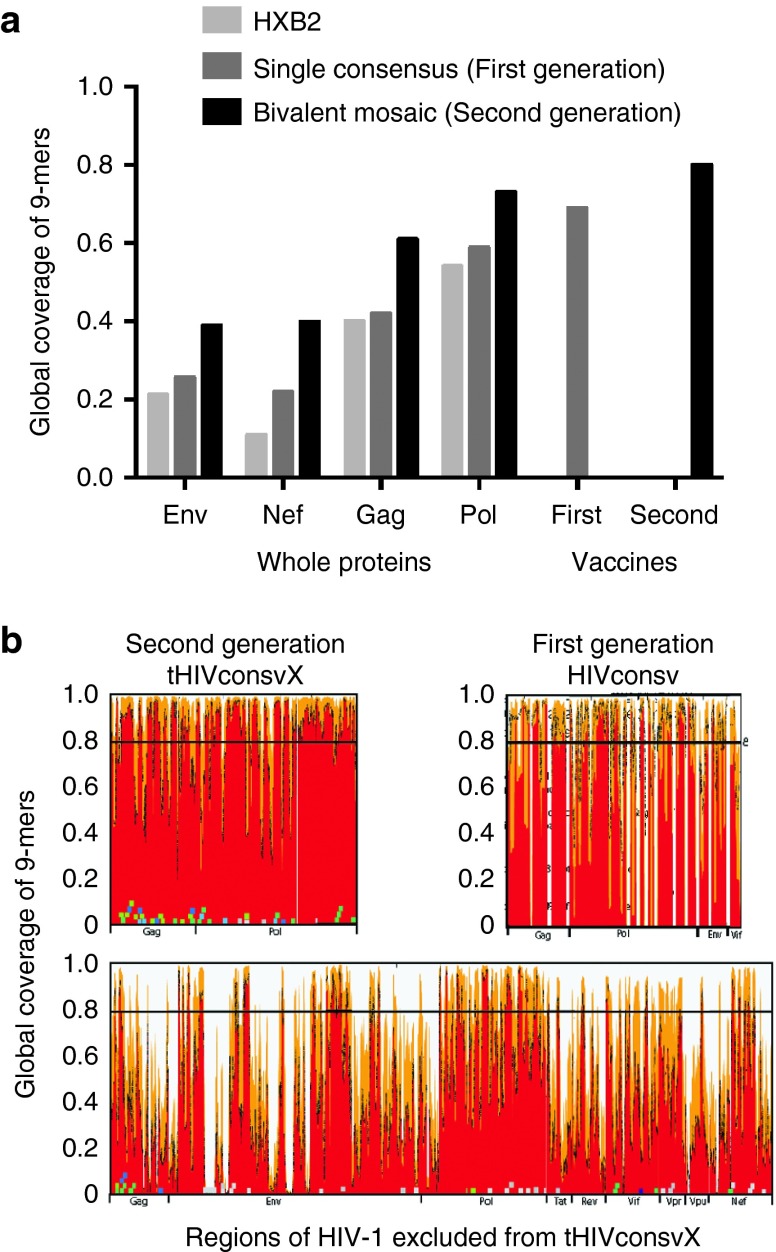
Figure 2.
Comparisons of epitope coverage using different vaccine strategies. Graph (a) shows a comparison of the global PTE coverage with 9/9 aa match, contrasting the coverage provided by a natural virus isolate HXB2 (light grey), a single consensus3 (dark grey), and bivalent mosaics (black). Four whole HIV-1 proteins that are commonly included in vaccines are compared with the regions included in the HIVconsv (first-generation) and tHIVconsvX (second-generation) immunogens. This calculation is not alignment based—all 9-mers in each protein in the original alignment are extracted, gaps are excluded and the fraction of 9-mers that are covered (i.e., perfectly matched) by the vaccine candidate is calculated. This number is equivalent to the average number of epitopes per natural strain that are matched by a given vaccine candidate. (b) These graphs show the match with group M 9-mer PTE variant sequences for the regions of the second-generation tHIVconsvX bivalent mosaic (top left), those of the first-generation HIVconsv consensus amino acid sequences (top right) and the rest of the HIV-1 proteome after removing the second-generation regions (bottom). The white vertical gaps in the conserved-region vaccines figures indicate the boundaries of each of the included regions. The top left and bottom figures are essentially a reordering of the data shown in Figure 1a, to enable a visual comparison of epitope coverage in the second-generation vaccine relative to the rest of the proteome.
Env was deliberately excluded from the second-generation design; it has a short conserved stretch, but carries no beneficial epitopes.24 Often after vaccination, Env specificity has dominated anti-HIV-1 T-cell responses, yet Env-specific T cells have never been associated with good virus control.24,27,28,29,30 Also excluded were several highly conserved regions that contained only unfavourable epitopes, and regions that densely populated with beneficial epitopes but were variable, e.g., Gag p17 and Vif (Figure 1a). Finally, while the boundaries of our conserved regions were defined based on 9-mer coverage, the precise boundaries were confirmed by per-position Shannon entropy scores.31 We also confirmed that each conserved region was richly populated with validated human CD8+ and CD4+ T-cell epitopes (Supplementary Figure S4). Part of our motivation in maximizing the immunogen length potential of the vaccine is that an effective vaccine will not only have to contend with HIV-1 diversity, but will need to accommodate global human HLA class I diversity as well, hence we focused on epitope-dense regions. Based on the LANL epitope database, as of June 2015, 752 distinct epitopes have been experimentally defined in the literature within the boundaries of six conserved regions we included. These epitopes utilized 84 different HLA class I presenting molecules, a consequence of including some of the most epitope-rich regions of the viruses. While these numbers, based on the database summary of the literature, emphasize the rich immunogenic potential of the conserved regions we selected, we fully expect these regions will contain many as yet undescribed epitopes.17 The aa sequences of the tHIVconsvX mosaic 1 and mosaic 2 differ in about 10% aa (Supplementary Figure S5).
Minimizing responses to possible junctional epitopes by reordering regions
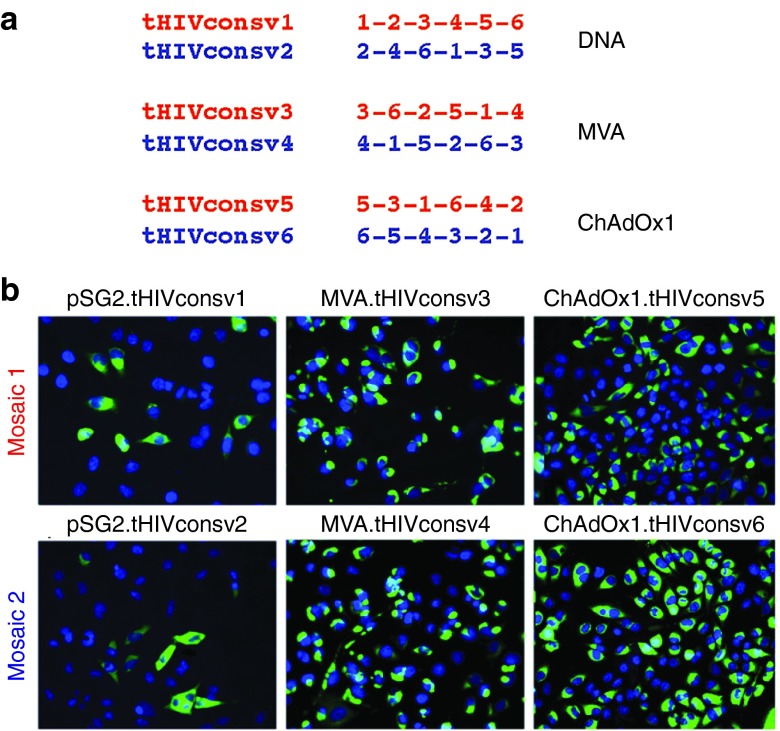
Unnatural junctions between adjacent regions are an unavoidable feature of chimeric proteins. Up to 20% of HIVconsv-specific T cells induced by the first-generation immunogen recognized junctional epitopes.23 This is not a major concern as long as sufficiently broad anti-HIV-1 responses are also induced (note that many MVA and ChAdV vector-specific responses are elicited, too), and there are no significant matches to proteins in the human proteome. Nevertheless, we aimed to minimize induction of irrelevant junction-specific responses, and so included 6 larger fragments that spanned a few variable positions, rather than 14 small conserved regions. We also scrambled the conserved regions in different orders into six immunogens designated from tHIVconsv1 to tHIVconsv6 (collectively called tHIVconsvX), such that no single junction is present more than once (Figure 3a). Two distinctly ordered mosaics are paired for the same vector modality (DNA, ChAdOx1, or MVA), thus the dose of each junction is halved for priming and boosting. Moreover, heterologous prime-boost regimens never immunize twice with the same junction.
Figure 3.
Arrangement of regions and expressions of the tHIVconsvX immunogens. (a) Serial organization (scramble) of the six regions used to minimize induction T-cell responses by potentially newly formed junctional epitopes. These are irrelevant for protection, because they are not present in natural HIV-1 proteins. The delivery vectors, into which the individual immunogens were inserted for presentation to the immune system, are also indicated. Note that mosaic 1 and mosaic 2 are always used together in the same vaccine dose. (b) Expression of individual tHIVconsvX proteins from the plasmid DNA, ChAdOx1, and MVA vaccine vectors. HeLa cells were either transfected with 2.5 µg of pSG2.tHIVconsv1 and pSG2.tHIVconsv2 for 2 days, infected with MVA.tHIVconsv3 and MVA.tHIVconsv4 MOI of 5 for 1 day, or infected with ChAdOx1.tHIVconsv5 and ChAdOx1.tHIVconsv6 at MOI of 10 for 1 day. Recombinant proteins were detected using primary mAb ID (91-5) followed by FITC-conjugated goat-antihuman IgG secondary antibody (green). DAPI stain shows the positions of cell nuclei (blue).
Comparable preclinical immunogenicity of the first- and second-generation vaccines
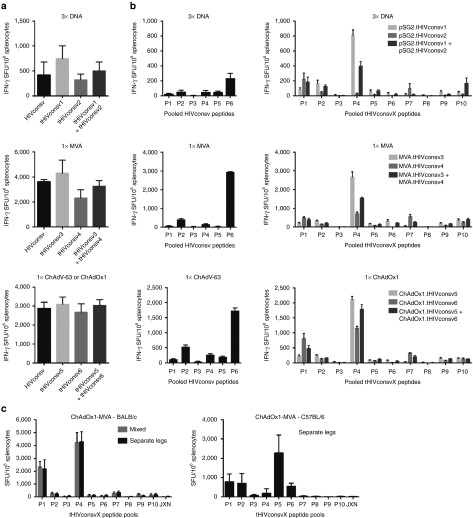
DNA fragments coding for the six tHIVconsvX immunogens were inserted into pSG2 plasmid DNA, ChAdOx126 (note that the first generation used ChAdV-63), and MVA. Because each is scrambled differently, forming a unique protein (Figure 3a), expression of all was confirmed in HeLa cells and protein levels were comparable (Figure 3b). Next, BALB/c mice were immunized with HIVconsv, or tHIVconsvX mosaic 1 or mosaic 2 vaccines alone, or with combined half doses of both with the aim of assessing overall immunogenicity. Separate comparisons were carried out for three administrations of plasmid DNAs and one administration of either recombinant MVAs or ChAdVs alone. Vaccine-elicited, HIV-1-specific splenocytes were enumerated in an interferon (IFN)-γ ELISPOT assay using pools of 15-mer peptides overlapping by 11 aa (15/11) (Supplementary Figure S8). The HIVconsv peptides were arranged into six pools P1–P6 23; P6 contained an immunodominant H-2d-restricted epitope32 coupled to the C terminus of HIVconsv to facilitate preclinical development. The two sets of peptides matching the two mosaics were organized into 10 pools, P1–P10, with variant peptide pairs combined into the same pools, so that individual pool frequencies can be added to assess the total magnitude without counting responses to the same epitope twice. Induction of T cells by the second- and first-generation vaccines was high and comparable, no matter in which order the conserved regions were scrambled in the tHIVconsvX proteins (Figure 4a). In tHIVconsvX pools P1 and P4, the likely immunodominant epitopes were mapped previously33 and the highest frequencies induced by the two stronger variants AMQMLKETI (mosaic 2) and LVGPTPVNI (mosaic 1) over their weaker variants AMQMLKDTI (mosaic 1) and LIGPTPVNI (mosaic 2) concurred with their presence in the particular mosaic. Notably, coadministration of mosaic 1 and 2 resulted in frequencies halfway between those induced by individual mosaics alone (Figure 4a). Thus, each mosaic on its own would induce one strong and one (relatively) weak response, but used together, the mosaic pair can respond strongly to both epitope variants.
Figure 4.
Comparable immunogenicity of the first- and second-generation of conserved-region vaccines in mice. Groups of six BALB/c mice were immunized with either the first- or second-generation immunogens as indicated below the graphs. The immunogens were vectored by DNA, MVA, or one of the chimpanzee adenoviruses ChAdV-63 (first generation) or ChAdOx1 (second generation) as shown above the graphs. Vaccine-elicited, conserved-region-specific T cells were enumerated in a fresh unexpanded IFN-γ ELISPOT assay using 15-mer peptides overlapping by 11 amino acids assembled into 6 pools P1–P6 or 10 pools P1–P10 of over 30 peptides each corresponding to the first-(HIVconsv) and second (tHIVconsvX)-generation vaccines, respectively. Graphs show (a) the totals (sum of all pools) of net T-cell frequencies as spot-forming units (SFU) per million of splenocytes for each vaccine modality indicated above the graphs and (b) the net frequencies of T cells recognizing individual peptide pools. (c) BALB/c and C57BL/6 mice were vaccinated using the rChAdV prime-rMVA boost regimens using mixed or anatomically separated deliveries of the two mosaic immunogens. JXN indicates a pool of junctional peptides. Frequencies are shown as mean ± SEM (n = 6).
We also tested immunogenicity of the combined rChAdOx1 prime-rMVA boost regimen, assessed the effect of mixed versus anatomically separate administration of the two mosaics and immunized C57BL/6 mice, which lack the Th2 bias of BALB/c mice. Enhanced responses to tHIVconsvX in heterologous prime-boost regimen relative to the single-vector delivery were consistently induced in both inbred strains (Figure 4b). The mixed and separate administrations of the two mosaic variants were not statistically separable (Figure 4c, left). Both CD8+ and CD4+ T cells were oligofunctional for production of IFN-γ, TNF-α, IL-2, and degranulation (CD107a) upon specific antigen restimulation (Supplementary Figure S6). Importantly, no responses were detected to the pooled peptides spanning junctional regions even using the potent rChAdOx1-rMVA regimen (Figure 4c, JXN).
Magnitude and breadth of responses to tHIVconsvX epitopes correlate with low viral load and high CD4+ cell count in treatment-naive patients
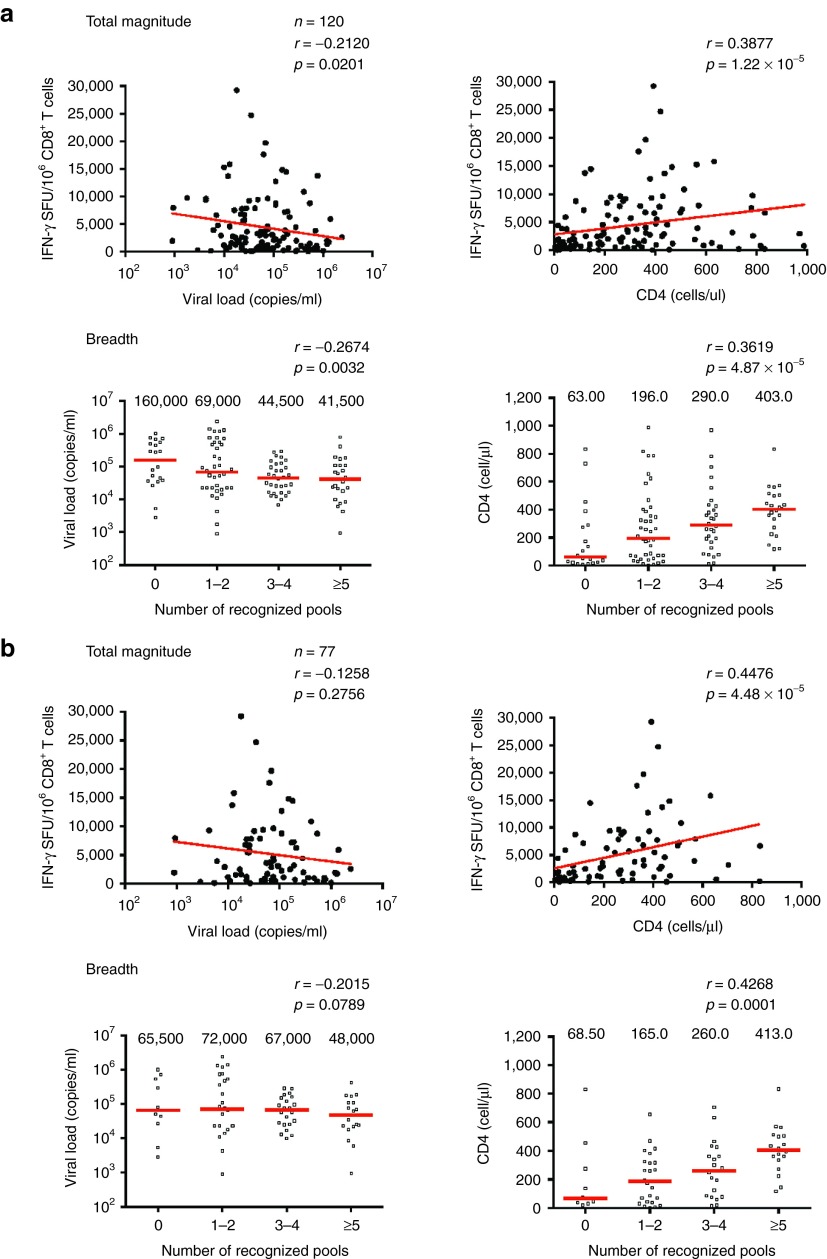
A key rationale for the conserved-region T-cell vaccine approach is the prediction that HIV-1+ patients whose CD8+ T cells recognize the tHIVconsvX immunogen will have lower virus loads and their immune system will be more preserved than those lacking these responses. This is based on the argument that virus control is determined early in infection rather than during the chronic phase, because progressors do not spontaneously change into controllers during the chronic stage. To test this hypothesis, PBMC samples from 120 treatment-naive Japanese individuals with chronic HIV-1 clade-B infection (Table 1) were tested for their ability to recognize tHIVconsvX peptide pools P1–P10. The median frequencies of CD8+ T cells recognizing each of the ten tHIVconsvX peptide pools were determined using IFN-γ ELISPOT and flow cytometry. The total magnitude, or the sum of T-cell frequencies responding to individual peptides pools, showed a significant inverse correlation with plasma viral load (P = 0.0201) and a direct correlation with the CD4+ T-cell counts (P = 1.22 × 10-5) (Figure 5a). Correlations were also found for responses to individual peptide pools P3 and P9 for viral loads (Supplementary Figure S7a) and P1, P2, P3, P6, P8, and P9 for CD4+ cell counts (Supplementary Figure S7b). Furthermore, the protective potential of tHIVconsvX-specific T cells in treatment-naive patients was supported by statistically significant correlations between the number of peptide pools (out of 10) that the patients' CD8+ T cells recognized and their viral load (P = 0.0032), and CD4+ cell counts (P = 4.87 × 10−5) (Figure 5a). Even after excluding patients with protective HLA B*52:01 and B*67:01 alleles34 (n = 77), the magnitude and breadth of T-cell responses still significantly correlated with the CD4+ T-cell counts (P = 4.48 × 10-5 and P = 0.0001, respectively), but not with the plasma viral load (Figure 5b). Thus, the specificity of the anti-HIV-1 CD8+ T cells, in particular the magnitude and breadth of CD8+ T cells recognizing conserved epitopes included in tHIVconsvX, determine the suppression of chronic virus replication and impact the preservation of the patients' immune system.
Table 1. Characteristics of the treatment-naive HIV-1 clade B-positive patient cohort.
Figure 5.
tHIVconsvX-specific CD8+ T-cell responses are beneficial in chronically infected treatment-naive patients. PBMCs responsive to tHIVconsvX peptide pools P1–P10 from treatment-naive patients in Japan chronically infected with HIV-1 clade B were enumerated in an IFN-γ ELISPOT assay and the frequencies of CD8+ and CD4+ T cells were determined by flow cytometry. Correlations between the magnitude (top) or breadth (bottom) and the viral load (left) or CD4+ cell counts (right) were statistically analyzed using Spearman's rank test. The P values are given above individual graphs. (a) Correlations in all patients (n = 120) and (b) in a subgroup of patients without the two known protective HLA alleles B*52:01 and B*67:01 (n = 77).
Discussion
Here, we describe the design and construction of novel candidate T-cell vaccines against HIV-1. The major novelty in the design is the combination of conserved regions with the mosaic design such that both the computer-optimized vaccine immunogens and their vector delivery maximize effective targeting of global HIV-1 variants by vaccine-elicited CD8+ T cells. This is shown by the very high level of perfect sequence matching between the vaccine and actual and potential epitope sequences across all the major clades of HIV-1. Thus, the conserved nature of the epitopes included in the vaccine makes immunogen responses more likely to be initially cross-reactive and less likely to be escaped. We confirmed expression of all six differentially scrambled immunogen proteins in human cells and demonstrated high T-cell immunogenicity of the tHIVconsvX regions in the absence of any junctional responses in two inbred mouse strains. This predicts that they will be as immunogenic in humans as was the first-generation HIVconsv vaccine, which generated uniquely strong T-cell responses in phase 1 vaccine trials23 (and T.H., unpublished). Furthermore, the advantages of the mosaic design shown previously in rhesus monkeys13,14 and transgenic mice with HLA-A*02:01 presentation12 should enhance the magnitude, breadth, and depth of the induced CD8+ T-cell responses in human subjects, too. This will need to be tested in clinical trials in humans, who are the only relevant species for this level of detailed analysis. It is very encouraging that we found a correlation between the magnitude and breadth of CD8+ T-cell responses to the tHIVconsvX-conserved regions and low viral load and preservation of CD4+ T-cell counts in treatment-naive HIV-1+ patients.
Although substantial experimental progress was made with our first-generation conserved design, supporting the general concept22,23,35 (and T.H., unpublished), the improvements in the second-generation design are substantial enough to merit changing over. In summary, there is a clearly improved coverage of known group M variants by the second relative to the first-generation regions and certainly compared to the rest of the HIV-1 proteome (Figures 1 and 2). The first-generation design was based on a single consensus for each region, while the second generation utilizes complementing bivalent-mosaic proteins intended to be used together. While this increases the cost, there are several advantages. First, this improves the potential epitope coverage at the population level, as even the most conserved regions of HIV-1 are somewhat variable reflecting the enormous overall HIV-1 global variation. Also, eliciting T-cell responses that can recognize common epitope variants may block relatively fit immune escape routes that otherwise may be used by the virus in vivo.11 The second-generation vaccine is 12% longer than the first and so contains more potential epitopes (872 aa versus 778). Despite the additional length, the second generation spans regions that are more consistently conserved than the first (Figure 2). The second generation contains fewer segments (6 versus 14), hence fewer unnatural junctional domains that could misdirect the immune response. The first generation contained only 15 protective epitopes compared to 25 that have been associated with high viral load in large patient cohorts in South Africa, Peru, and Spain.24 In contrast, the second generation contains 33 protective epitopes and only 3 associated with high viral load, and all the beneficial epitopes included in the vaccine are highly conserved (Figure 2a). Our selection of these regions is supported by a recent study of 401 treatment-naive HIV-1+ patients in Japan, infected with B-clade virus. In that cohort, CD8+ T-cell responses to 13 epitopes were significantly associated with low viral load and high CD4+ cell counts,36 10 of these are present in tHIVconsvX as perfect 100% aa matches (Supplementary Table S1). Together, the matching to all M group viruses and the presence of protective epitopes from four geographically separated cohorts support the global relevance of this second-generation design.
HIV-1-induced CD8+ T cells can be compromised by virus escape and ultimately limited in their effectiveness,6,37,38 and past T-cell vaccine strategies have failed to induce protective immunity.39,40 Thus, inducing “more of the same” CTL by vaccines will not advance the vaccine development. The second generation of T-cell vaccines described here should avoid induction of ineffective responses, focusing on those that are less likely to select escape and therefore control virus control better24,36 (and this study), targeting HIV-1's “Achilles heel”. In natural infection, ineffective T-cell responses often dominate and may compete with the protective ones, reducing their relative magnitude and therefore impact.41 The tHIVconsvX-conserved mosaic proteins will focus T-cell responses toward conserved subdominant epitopes, by excluding the hypervariable regions of full-length HIV-1 proteins that are often immunodominant both in natural infection41 and in responses stimulated by previous vaccines.38,40 Further insights into definition of HIV-1 vulnerability recently suggested that association of high fitness cost with changes in functionally conserved regions may be further increased by considerations of structure stability.42,43,44 Nonetheless, the contribution of structural constraints on the top and above conservation will be a relatively small fraction of the benefits gained already by focusing on conserved regions, which resulted from structural/functional requirements. In any case, conserved mosaic is currently the most developmentally and clinically advanced approach to tackle HIV-1 diversity and escape.
Immunogen delivery is also critical for induction of protective responses.1 While the design of the tHIVconsvX immunogens has a strong rationale, their presentation to the immune system has to induce potent CD8+ T-cell responses and memory, that will not be superseded by less protective primary T-cell responses after exposure to HIV-1. The heterologous regimen of ChAdV and MVA (±DNA) currently stimulates the strongest CD8+ T cells of any nonreplicating subunit vaccines in humans.23,45,46 ChAdOx1 has low seroprevalence in humans,26 and recombinant ChAdVs have been safely used in over 1,500 volunteers including African infants (Adrian Hill, University of Oxford, personal communication). The comparable immunogenicity of the first and second generations of conserved vaccines in mice suggests that in humans, the second generation will be as potent as the first generation was,23 with all the advantages of second-generation design.
There is a strong rationale for using the conserved mosaic tHIVconsvX vaccines in both prophylactic and therapeutic settings. For prevention, tHIVconsvX vaccines by matching transmitted/founder viruses will aim to decrease HIV-1 acquisition, eradicate HIV-1 early in infection or establish very good virus control, and will complement any broadly neutralizing antibodies-eliciting strategies.2 Therapeutic vaccines aim to either control HIV-1 replication without the need of antiretroviral drugs or eradicate HIV-1 from the body completely following reactivation of latent HIV-1 genomes.20,47 The tHIVconsvX strategy is particularly suited for the therapeutic use, because refocusing cytotoxic T-cell effectors on the conserved regions of HIV-1 is greatly preferable to reactivating T cells that have already failed to control the virus and selected for escape mutations retained in provirus reservoirs.20,47 Therapeutic approaches may not require coinduction of broadly neutralizing antibodies.
Vaccine-induced CD8+ T cells will have to suppress transmitted/founder HIV-1 or reactivated HIV-1 replication,2 acting efficiently from the onset of infection/reactivation.5,6 The correlation in treatment-naive patients of the magnitude and breadth of tHIVconsvX-specific CD8+ T cells with high CD4+ T-cell counts and with low virus loads indicates that these responses are beneficial.24,28,36,37 High CD4+ counts, the best indicator of slowed disease progression, remained significantly correlated even after excluding patients with known protective HLA alleles. Thus, benefits of tHIVconsvX CD8+ T-cell responses could reach the whole population.
This improved approach to targeting T-cell responses on biologically highly sensitive regions of HIV-1 proteins, common to majority of global variants, early for prophylaxis or refocusing for treatment, merits clinical evaluation and could lead to development of a truly global HIV-1 vaccine.
Materials and Methods
Synthetic genes for tHIVconsvX. Six DNA fragments carrying the six tHIVconsvX open-reading frame were synthesized (Life Technologies) using humanized codons and were preceded by a consensus Kozak sequence at −6 nucleotides to maximize protein expression.
Construction of plasmid pSG2.tHIVconsv1 and pSG2.tHIVconsv2 vaccines. For plasmid DNA, the backbone of pSG2 was used with the enhancer/intron A/human cytomegalovirus promoter immediate early promoter cassette, bovine growth hormone polyadenylation site and genes tHIVconsv1 and tHIVconsv2 were inserted to construct vaccine components pSG2.tHIVconsv1 and pSG2.tHIVconsv2. The plasmid DNAs for immunizations were prepared using the Endo-Free Gigaprep (Qiagen) and stored at −20 °C until use.
Construction of MVA.tHIVconsv3 and MVA.tHIVconsv4 vaccines. The parental nonreplicating MVA originates directly from Professor Anton Mayr, passage 575 dated 14 December 1983. The tHIVconsv3 and tHIVconsv4 genes were cloned into transfer plasmid p856MVA-GFP-TD-mH5 under control of the modified H5 promoter. Through homologous recombination, the expression cassettes were directed into the thymidine kinase locus on the MVA genome. The initially coinserted green fluorescent protein marker was removed by subsequent transdominant recombination to generate markerless vaccine components MVA.tHIVconsv3 and MVA.tHIVconsv4. Recombinant MVAs were made as described elsewhere. Briefly, chicken embryo fibroblast cells grown in Dulbeco's Modified Eagle's Medium supplemented with 10% FBS, penicillin/streptomycin and glutamine (DMEM 10) were infected with parental MVA at multiplicity of infection (MOI) 1 and transfected using Superfectin (Qiagen) with 3 µg of p4016MVA-GFP-TD-mH5.tHIVconsv3 or p4065MVA-GFP-TD-mH5.tHIVconsv4. The cell lysate from this recombination was harvested and used to infect chicken embryo fibroblast. These cells were MoFlo single-cell sorted into 96-well plates and these were used to culture recombinant virus upon addition of fresh chicken embryo fibroblast. Those wells containing suitably infected cells were harvested and screened by PCR to confirm identity and test purity. Plaque picking was performed until the culture was free of parental virus, as determined by PCR. The virus was then prepared in bulk and purified on a 36% sucrose cushion, titred and stored at –80 °C until use.
Construction of ChAdOx1.tHIVconsv5 and ChAdOx1.tHIVconsv6 vaccines. ChAdOx1 is derived from ChAdV isolate Y25 of group E adenoviruses and preexisting antibodies to group E are rare in human populations. Its genome modifications include removal of the E1, E3, and a substitution of simian region E4 with the human adenovirus 5 E4 orf4 and orf6/7 genes. For the generation of recombinant ChAdOx1s, the tHIVconsv5 and tHIVconsv6 genes were subcloned under the control of the human cytomegalovirus immediate early promoter into plasmid pENTR4_Mono and inserted at the E1 locus of the ChAdOx1 genome by GalK recombineering. Recombinant ChAdOx1 vaccines were rescued by transfection of HEK293A T-Rex cells using linearized plasmid and grown in suspension culture of HEK 293 cells. The presence of the transgene and absence of contaminating empty parental adenovirus was confirmed by PCR. The virus was titred to obtain infectious units per milliliter, assayed by spectrophotometry to quantify the number of virus particles per milliliter and stored at –80 °C until use.
Immunofluorescent staining for tHIVconsvX vaccines. First, DNA transfection was used to identify a primary anti-Gag p24 mAb, which recognized both mosaic variants. This was then used to confirm expression of the other four immunogen proteins expressed in recombinant virus-infected human cells. Briefly, HeLa cells were transfected with 2.5 µg of pSG2.tHIVconsv1 and pSG2.tHIVconsv1 for 48 hours, using Lipofectamine 3000 (Invitrogen), according to the manufacturer's specifications. For MVA.tHIVconsv3 and MVA.tHIVconsv4, HeLa cells (ATCC, mycoplasmafree) were infected at MOI of 5 for 2 hours at 37 °C, 5% CO2 in serum-free media. Following infection, the cells were washed with phosphate buffered saline (PBS) and incubated in 3 ml of complete DMEM media for 24 hours. Infection with ChAdOx1.tHIVconsv5 and ChAdOx1.tHIVconsv6 were conducted in a similar way, except that a MOI of 10 was used. At the end of overnight incubation, the cells were washed twice with ice cold PBS and fixed for 10 minutes on ice with 10% neutral buffered formalin solution containing 4% formaldehyde (Sigma). The fixed cells were incubated at room temperature for 20 minutes, then washed three times with PBS and permeabilized with 0.2% Triton (TX-100) for 5 minutes at room temperature. Cells were washed again, blocked with 1% bovine serum albumin in PBS for 30 minutes and incubated overnight with a 1:200 dilution of the primary antibody (NIH reagent mAb ID (91-5), catalogue #1238). The cells were washed three times for 15 minutes each, using PBS and stained with 1:200 dilution of FITC-conjugated goat-antihuman IgG secondary antibody (Millipore) for 2 hours at room temperature. The cells were washed three times as before and the coverslips mounted on microscope slides using Vectashield DAPI nuclear stain mounting media (Vector laboratories). The slides were examined on a fluorescence microscope (Leica DMI 3000B) and images were analyzed with the ImageJ program.
Mice and immunization regimens. Six-week-old female BALB/c or C57BL/6 mice were purchased from Harlan Laboratories (UK) and housed at the Functional Genomics Facility, University of Oxford. Groups of six animals are a standard group size for vaccine initial vaccine experiments using inbred mouse strains. Mice were immunized intramuscularly under general anaesthesia either with 100 μg of plasmid DNA, 108 infectious units of rChAdOx1s, 5 × 106 plaque-forming units of rMVAs. In the combined regimens of mosaics 1 and 2, half a dose for each vaccine type was used injecting mosaic 1 and 2 into the left and right hind quadriceps, respectively. Mice were sacrificed 2 weeks after the last rChAdOx1 and 1 week after the last rMVA or DNA. All procedures and care were approved by the local Research Ethics Committee, University of Oxford and conformed strictly to the United Kingdom Home Office Guidelines under the Animals (Scientific Procedures) Act 1986. Experiments were conducted under Project License 30/2833 held by T.H.
Peptides. Over 90% pure 15-mer peptides overlapping by 11 aa (15/11) spanning the entire HIVconsv, mosaic 1, and mosaic 2 six regions were used in IFN-γ ELISPOT assay to assess the vaccine immunogenicity. One hundred and ninety-nine HIVconsv-derived peptides (Ana Spec, USA) were assembled into six pools P1–P6. Two hundred and 201 peptides corresponding to the six regions of mosaic 1 and mosaic 2, respectively, were assembled into 10 pools P1–P10 of between 34 and 47 peptides in a way that variant peptides were always present in the same pool. Thirty one 16-mer peptides corresponding to 30 junctions of tHIVconsvX and the tPA leader as 22-mer were assembled into pool JXN. Supplementary Figure S8 illustrates the peptide derivation. Individual peptides were dissolved in DMSO at a concentration of 20 mg/ml and stored at −80 °C. Working stocks of 4 mg/ml were prepared by diluting 20 mg/ml stocks with PBS. Peptides were used in assays at a final concentration of 2 μg/ml.
Murine IFN-γ ELISPOT assay. The ELISPOT assay was performed using the Mouse IFN-γ ELISpot kit (Mabtech) according to the manufacturer's instructions. Immune splenocytes were collected and tested separately from individual mice. Spots were visualized using sequential applications of a biotin-conjugated secondary anti-IFN-γ mAb (R4-6A2, Rat IgG1), an alkaline phosphatase and a chromogenic substrate (Bio-Rad) and counted using the AID ELISpot Reader System (Autoimmun Diagnostika).
HIV-1+ subjects. One hundred and twenty treatment-naive Japanese individuals with chronic HIV-1 clade-B infection were enrolled in the National Center for Global Health and Medicine from 2011 to 2012. This study was approved by the ethics committees of the National Center for Global Health and Medicine and Kumamoto University. The study was conducted according to the principles of the Declaration of Helsinki (2008) and complied with the International Conference on Harmonization Good Clinical Practice guidelines.
Human IFN-γ ELISPOT assay. Peptide pools P1–P10 at a concentration of 1.5 μg/ml of each peptide and PBMCs separated from whole blood at 1 × 105 cells/well were added to 96-well polyvinylidene plates (Millipore, Bedford, MA) that had been precoated with 5 mg/mL anti-IFN-γ mAb 1-D1K (Mabtech, Stockholm, Sweden). The plates were incubated at 37 °C in 5% CO2 for 16 hours and washed with PBS before the addition of biotinylated anti-IFN-γ Mab (Mabtech) at 1 mg/ml at room temperature for 90 minutes. The plates were then washed with PBS, incubated with streptavidin-conjugated alkaline phosphatase (Mabtech) at room temperature for 60 minutes, washed with PBS, and individual cytokine-producing units were detected as dark spots after a 20-minute reaction with 5-bromo-4-chloro-3-idolyl phosphate and nitro blue tetrazolium using an alkaline phosphatase-conjugate substrate (Bio-Rad, Richmond, CA, USA). Spot-forming units were counted with an Eliphoto-Counter (Minerva Teck, Tokyo, Japan). The frequencies of responding cell were expressed as a number of spot-forming units/106 CD8+ T cells by measuring frequency of CD8+ T cells using a flow cytometry. A mean ± 2 SD of the spot number of samples from 12 HIV-1-naive individuals for these peptides was <176 spots/106 CD8+ T cells. Therefore, we defined >200 spot-forming units/106 CD8+ T cells as positive responses. Ninety six volunteers were used based on availability and provided sufficient power to detect statistically significant differences for a proportion of samples.
Statistical analysis. Statistical analyses were performed using Graph Pad Prism version 5. Simple comparisons were performed using two-way Student's t-test. Multiple comparisons were performed using the Kruskal–Wallis test with Dunn's multiple comparison posttest for nonparametric data. Correlations between the breadths or the magnitudes and pVL or CD4 counts were statistically analyzed using Spearman's rank test. For comparison of two groups, two-tailed Mann–Whitney's U-tests were performed. A P value <0.05 was considered significant.
SUPPLEMENTARY MATERIAL Figure S1. Comparing coverage by Gag conserved regions of the first- and second-generation vaccines. Figure S2. Comparing coverage of the second generation (a) and original (b) conserved region vaccines. Figure S3. Examples of the diversity of three experimentally defined epitopes. Figure S4. Known epitopes in the LANL-HSD recognized by human CD8+ and CD4+ T cells in individual HIV-1 proteins as of 2013. Figure S5. Amino acid sequences of the two complementing mosaic regions and their amino acid differences. Figure S6. Functionality of tHIVconsvX-specific CD8+ and CD4+ T cells induced by the rChAdOx1-prime rMVA-boost regimen. Figure S7. Correlation between magnitude of responses in IFN-γ ELISPOT assay to individual tHIVconsvX peptide pools and plasma viral load or CD4+ T cell count in treatment-naïve HIV-1+ patients (n = 120). Figure S8. Peptide pools used for immune monitoring. Table S1. Protective Japanese epitopes in the tHIVconsvX vaccines.
Acknowledgments
The authors would like to thank Jo Cox, Jill Gilmour, Eddy Sayeed, Jan De Bont, Pat Fast, Wayne Koff, and Bart Haynes for useful discussions. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 p24 Gag Monoclonal (#24-2) from Michael H. Malim; mAbs toHIV-1 p24 (specificity clone, 71-31, 91-5) from Susan Zola-Pazner. The work is jointly funded by the UK Medical Research Council (MRC G1001757) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreements, the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (UM1 AI100645) and AIDS International Collaborative Project Grant in Center for AIDS Research Kumamoto University. T.H. and A.J.McM. are the Jenner Institute Investigators. B.O. was funded in part by the International Vaccine Initiative and made possible by the support of the United States Agency for International Development (USAID) and other donors. The full list of IAVI donors is available at http://www.iavi.org. The authors have no competing interests other than T.H., B.K., and A.J.McM., the inventors on PCT Application No. PCT/US2014/058422.
Supplementary Material
References
- Hanke, T (2014). Conserved immunogens in prime-boost strategies for the next-generation HIV-1 vaccines. Expert Opin Biol Ther 14: 601–616. [DOI] [PubMed] [Google Scholar]
- McMichael, AJ and Haynes, BF (2012). Lessons learned from HIV-1 vaccine trials: new priorities and directions. Nat Immunol 13: 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen, B, Taylor, J, Yusim, K, Foley, B, Gao, F, Lang, D et al. (2002). Diversity considerations in HIV-1 vaccine selection. Science 296: 2354–2360. [DOI] [PubMed] [Google Scholar]
- Kuebler, PJ, Mehrotra, ML, McConnell, JJ, Holditch, SJ, Shaw, BI, Tarosso, LF et al. (2015). Cellular immune correlates analysis of an HIV-1 preexposure prophylaxis trial. Proc Natl Acad Sci USA 112: 8379–8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freel, SA, Picking, RA, Ferrari, G, Ding, H, Ochsenbauer, C, Kappes, JC et al. (2012). Initial HIV-1 antigen-specific CD8+ T cells in acute HIV-1 infection inhibit transmitted/founder virus replication. J Virol 86: 6835–6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonetilleke, N, Liu, MK, Salazar-Gonzalez, JF, Ferrari, G, Giorgi, E, Ganusov, VV et al.; CHAVI Clinical Core B (2009). The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 206: 1253–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay, J, Shianna, KV, Ge, D, Colombo, S, Ledergerber, B, Weale, M et al. (2007). A whole-genome association study of major determinants for host control of HIV-1. Science 317: 944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer, M, Keele, BF, Schmidt, SD, Mason, RD, Welles, HC, Fischer, W et al. (2014). Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505: 502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, SG, Ford, JC, Lewis, MS, Ventura, AB, Hughes, CM, Coyne-Johnson, L et al. (2011). Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473: 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, SG, Piatak, M Jr, Ventura, AB, Hughes, CM, Gilbride, RM, Ford, JC et al. (2013). Immune clearance of highly pathogenic SIV infection. Nature 502: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, W, Perkins, S, Theiler, J, Bhattacharya, T, Yusim, K, Funkhouser, R et al. (2007). Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med 13: 100–106. [DOI] [PubMed] [Google Scholar]
- Abdul-Jawad, S, Ondondo, B, van Hateren, A, Gardner, A, Elliott, T, Korber, B et al. (2015). Increased valency of conserved-mosaic vaccines enhances the breadth and depth of epitope recognition. Mol Ther (epub ahead of print). [DOI] [PMC free article] [PubMed]
- Barouch, DH, O'Brien, KL, Simmons, NL, King, SL, Abbink, P, Maxfield, LF et al. (2010). Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat Med 16: 319–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra, S, Liao, HX, Zhang, R, Muldoon, M, Watson, S, Fischer, W et al. (2010). Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat Med 16: 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Létourneau, S, Im, EJ, Mashishi, T, Brereton, C, Bridgeman, A, Yang, H et al. (2007). Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2: e984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, M, Nickle, DC and Mullins, JI (2007). HIV-1 group M conserved elements vaccine. PLoS Pathog 3: e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, OO, Ali, A, Kasahara, N, Faure-Kumar, E, Bae, JY, Picker, LJ et al. (2015). Short conserved sequences of HIV-1 are highly immunogenic and shift immunodominance. J Virol 89: 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, JM, Schaefer, M, Monaco, DC, Batorsky, R, Claiborne, DT, Prince, J et al. (2014). HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science 345: 1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claiborne, DT, Prince, JL, Scully, E, Macharia, G, Micci, L, Lawson, B et al. (2015). Replicative fitness of transmitted HIV-1 drives acute immune activation, proviral load in memory CD4+ T cells, and disease progression. Proc Natl Acad Sci USA 112: E1480–E1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, K, Pertea, M, Rongvaux, A, Wang, L, Durand, CM, Ghiaur, G et al. (2015). Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, AL, Mann, JK, Omarjee, S, Ndung'u, T, Walker, BD and Chakraborty, AK (2013). Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity 38: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, T et al. (2016).Control of HIV-1 replication by vaccine-induced CD8+ T cells in humans through conserved subdominant non-Gag epitopes. Vaccine (epub ahead of print). [DOI] [PMC free article] [PubMed]
- Borthwick, N, Ahmed, T, Ondondo, B, Hayes, P, Rose, A, Ebrahimsa, U et al. (2014). Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol Ther 22: 464–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe, B, Llano, A, Ibarrondo, J, Daniels, M, Miranda, C, Zamarreño, J et al. (2011). Definition of the viral targets of protective HIV-1-specific T cell responses. J Transl Med 9: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe, B, Hu, X, Llano, A, Rosati, M, Olvera, A, Kulkarni, V et al. (2015). A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques. J Transl Med 13: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicks, MD, Spencer, AJ, Edwards, NJ, Wadell, G, Bojang, K, Gilbert, SC et al. (2012). A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One 7: e40385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela, P, Ngumbela, K, Thobakgale, C, Ramduth, D, Honeyborne, I, Moodley, E et al. (2007). CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med 13: 46–53. [DOI] [PubMed] [Google Scholar]
- Masemola, A, Mashishi, T, Khoury, G, Mohube, P, Mokgotho, P, Vardas, E et al.; HIVNET 028 Study Team. (2004). Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J Virol 78: 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, M, Heckerman, D, Deng, W, Rousseau, CM, Coovadia, H, Bishop, K et al. (2008). Broad and Gag-biased HIV-1 epitope repertoires are associated with lower viral loads. PLoS One 3: e1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuñiga, R, Lucchetti, A, Galvan, P, Sanchez, S, Sanchez, C, Hernandez, A et al. (2006). Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J Virol 80: 3122–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber, BT, Farber, RM, Wolpert, DH and Lapedes, AS (1993). Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc Natl Acad Sci USA 90: 7176–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H, Germain, RN, Moss, B and Berzofsky, JA (1990). An immunodominant class I-restricted cytotoxic T lymphocyte determinant of human immunodeficiency virus type 1 induces CD4 class II-restricted help for itself. J Exp Med 171: 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondondo, B, Abdul-Jawad, S, Bridgeman, A and Hanke, T (2014). Characterization of T-cell responses to conserved regions of the HIV-1 proteome in BALB/c mice. Clin Vaccine Immunol 21: 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruto, T, Gatanaga, H, Nelson, G, Sakai, K, Carrington, M, Oka, S et al. (2012). HLA class I-mediated control of HIV-1 in the Japanese population, in which the protective HLA-B*57 and HLA-B*27 alleles are absent. J Virol 86: 10870–10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternette, N, Block, PD, Sánchez-Bernabéu, Á, Borthwick, N, Pappalardo, E, Abdul-Jawad, S et al. (2015). Early kinetics of the HLA class I-associated peptidome of MVA.HIVconsv-infected cells. J Virol 89: 5760–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi, H, Akahoshi, T, Koyanagi, M, Chikata, T, Naruto, T, Maruyama, R et al. (2015). Clinical control of HIV-1 by cytotoxic T cells specific for multiple conserved epitopes. J Virol 89: 5330–5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, G, Yang, H, Yorke, E, Wainwright, E, Bourne, V, Frisbee, A et al. (2015). Identification of effective subdominant anti-HIV-1 CD8+ T cells within entire post-infection and post-vaccination immune responses. PLoS Pathog 11: e1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F, Finnefrock, AC, Dubey, SA, Korber, BT, Szinger, J, Cole, S et al. (2011). Mapping HIV-1 vaccine induced T-cell responses: bias towards less-conserved regions and potential impact on vaccine efficacy in the Step study. PLoS One 6: e20479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchbinder, SP, Mehrotra, DV, Duerr, A, Fitzgerald, DW, Mogg, R, Li, D et al.; Step Study Protocol Team (2008). Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372: 1881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, SM, Sobieszczyk, ME, Janes, H, Karuna, ST, Mulligan, MJ, Grove, D et al. (2013) Efficacy trial of a DNA/rAd5 HIV-1 preventive vaccine. N Engl J Med 369: 2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, MK, Hawkins, N, Ritchie, AJ, Ganusov, VV, Whale, V, Brackenridge, S et al.; CHAVI Core B (2013). Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J Clin Invest 123: 380–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manocheewa, S, Lanxon-Cookson, EC, Liu, Y, Swain, JV, McClure, J, Rao, U et al. (2015) Pairwise growth competition assay for determining the replication fitness of human immunodeficiency viruses. J Vis Exp 4: e52610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manocheewa, S, Mittler, JE, Samudrala, R and Mullins, JI (2015). Composite sequence-structure stability models as screening tools for identifying vulnerable targets for HIV drug and vaccine development. Viruses 7: 5718–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland, M, Manocheewa, S, Swain, JV, Lanxon-Cookson, EC, Kim, M, Westfall, DH et al. (2013). HIV-1 conserved-element vaccines: relationship between sequence conservation and replicative capacity. J Virol 87: 5461–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer, KJ, O'Hara, GA, Duncan, CJ, Collins, KA, Sheehy, SH, Reyes-Sandoval, A et al. (2013). Protective CD8+ T-cell immunity to human malaria induced by chimpanzee adenovirus-MVA immunisation. Nat Commun 4: 2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy, SH, Duncan, CJ, Elias, SC, Collins, KA, Ewer, KJ, Spencer, AJ et al. (2011). Phase Ia clinical evaluation of the Plasmodium falciparum blood-stage antigen MSP1 in ChAd63 and MVA vaccine vectors. Mol Ther 19: 2269–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis, DM and Hazuda, DJ (2013). Combined approaches for HIV cure. Curr Opin HIV AIDS 8: 230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.