Abstract
Limb girdle muscular dystrophies types 2B (LGMD2B) and 2D (LGMD2D) are degenerative muscle diseases caused by mutations in the dysferlin and alpha-sarcoglycan genes, respectively. Using patient-derived induced pluripotent stem cells (iPSC), we corrected the dysferlin nonsense mutation c.5713C>T; p.R1905X and the most common alpha-sarcoglycan mutation, missense c.229C>T; p.R77C, by single-stranded oligonucleotide-mediated gene editing, using the CRISPR/Cas9 gene-editing system to enhance the frequency of homology-directed repair. We demonstrated seamless, allele-specific correction at efficiencies of 0.7–1.5%. As an alternative, we also carried out precise gene addition strategies for correction of the LGMD2B iPSC by integration of wild-type dysferlin cDNA into the H11 safe harbor locus on chromosome 22, using dual integrase cassette exchange (DICE) or TALEN-assisted homologous recombination for insertion precise (THRIP). These methods employed TALENs and homologous recombination, and DICE also utilized site-specific recombinases. With DICE and THRIP, we obtained targeting efficiencies after selection of ~20%. We purified iPSC corrected by all methods and verified rescue of appropriate levels of dysferlin and alpha-sarcoglycan protein expression and correct localization, as shown by immunoblot and immunocytochemistry. In summary, we demonstrate for the first time precise correction of LGMD iPSC and validation of expression, opening the possibility of cell therapy utilizing these corrected iPSC.
Introduction
The discovery of human induced pluripotent stem cells1 (hiPSC) opened up new therapeutic possibilities for genetic diseases such as muscular dystrophies, including combined gene and cell therapy. For example, iPSC can be generated from patient cells, genetically engineered to correct the mutation, differentiated to the appropriate cell type, and transplanted into the patient. The ability to genetically engineer iPSC and expand them indefinitely is attractive as a means to generate corrected regenerative cells for large tissues like muscle. Several studies have demonstrated that iPSC can be differentiated in vitro to generate muscle precursor cells that can be engrafted into mouse models of muscular dystrophy.2,3 These studies suggest a pathway for generation of cells that could be therapeutic for muscular dystrophy, if disease mutations in hiPSC can be corrected in an accurate and efficient manner.
Fortunately, precise gene correction strategies now exist, including both correction of mutations at their endogenous chromosomal loci and site-specific integration of wild-type cDNA at safe heterologous locations. Genetic information can be corrected in situ by recombination using single-stranded oligonucleotides (ssODNs) as a source of the correct sequence. Such ssODNs can be used for precise gene editing of hiPSC via the homology-directed repair (HDR) pathway.4,5 Addition of wild-type sequences at safe harbor sites can be achieved by homologous recombination (HR), through flanking the therapeutic gene with homology arms corresponding to the desired target site. However, both incorporation of oligonucleotides by HDR and site-specific integration of plasmids by HR are inefficient processes.6,7 Designer nucleases have been found to stimulate gene correction frequencies greatly for these approaches by generating double-strand DNA breaks in the area where recombination is desired.8,9 To date, several distinct classes of sequence-specific nucleases have been described, including zinc finger nucleases (ZFN), meganucleases, transcription activator-like effector nucleases (TALEN), and CRISPR/Cas910. In addition, site-specific recombinases such as phage integrases carry out precise and efficient recombination in human cells.11,12 We apply several of these gene-editing tools here to achieve precise correction of iPSC derived from two autosomal recessive forms of limb girdle muscular dystrophy (LGMD).
LGMDs are a genetically heterogeneous group of muscle disorders. Disease onset can range from childhood to early adulthood and includes variable progression and distribution of muscle weakness and wasting. Specific parts of the hip, pelvic, upper arm, and shoulder girdle muscles are affected, resulting in walking disabilities.13 Most LGMD2B patients show partial to complete deficiency of dysferlin, a 220 kD type II transmembrane protein located at the sarcolemma and involved in skeletal muscle membrane repair.14,15 The dysferlin gene (DYSF) is located on chromosome 2p13 (ref. 16). LGMD 2D leads to a more severe phenotype. It can be diagnosed by missense, nonsense, and frameshift mutations in the alpha-sarcoglycan (SGCA) gene located on chromosome 17q21 (refs. 17,18). Absence of alpha-sarcoglycan destabilizes the dystrophin-associated protein complex, leading to progressive muscle wasting and fibrosis.19 While no cure currently exists for LGMD2B or 2D, the precise correction reported here of iPSC derived from these diseases opens up the possibility of developing cell therapies for these diseases.
Results
Gene-editing strategies to correct point mutations in patient-derived iPSC with programmable SpCas9 nucleases and oligonucleotides
We developed strategies to reverse the missense mutation c.229C>T (R77C) of SGCA and the stop codon mutation c.5713C>T (R1905X) in human dysferlin (DYSF). Both strategies were optimized in HEK293 cells and then applied to patient-derived iPSC. To allow more efficient gene editing, we made use of the CRISPR/Cas9 system, which included the Streptococcus pyogenes Cas9 (SpCas9) nuclease and a 20-nt long single guide RNA (sgRNA) (Figure 1a). Expression of both components from the plasmid pX330 allowed the sgRNA to form a complex with SpCas9 and guide the nuclease to a 20-nt complementary genomic region of either DYSF or SGCA and introduce a sequence-specific, blunt-ended, double-strand break that enhanced the frequency of gene editing. Together with the CRISPR/Cas9 plasmid, ssODN was introduced into the cells. The antisense ssODN sequences were designed individually for DYSF and SGCA and contained the corrected nucleotide information to reverse the point mutations. The ssODN was used as a template for HDR and also made it possible to insert several other useful nucleotide changes in the vicinity7 (Figure 1a).
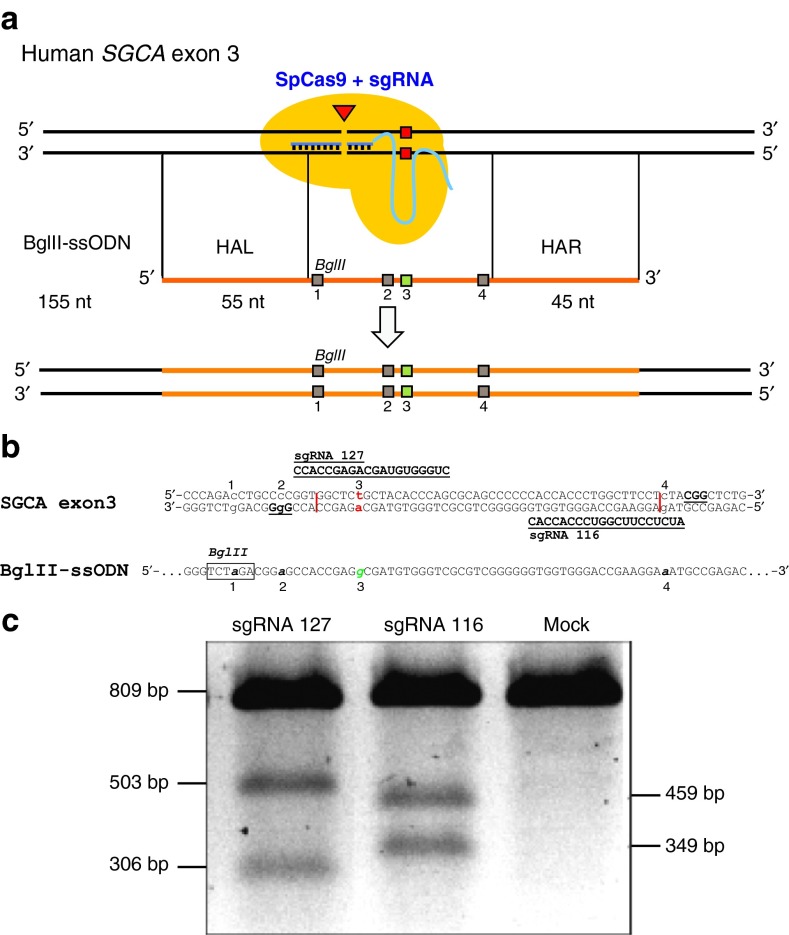
Figure 1.
Targeting strategy for c.229C>T point mutation in exon 3 of human SGCA. (a) Single guide RNA (sgRNA) 127 or sgRNA 116 directs Streptococcus pyogenes (SpCas9) close to the point mutation c.229C>T on human SGCA exon 3 and introduces a blunt double strand break (DSB). BglII-ssODN is 155 nucleotides (nt) in antisense orientation with a homology arm left (HAL) of 55 nt and a homology arm right (HAR) of 45 nt length. Homology arms flank a sequence that harbors four silent mutations. Mutation 1 creates a BglII restriction site. Mutations 2 and 4 change the PAM sequence of sgRNA 127 or the PAM proximal seed sequence of sgRNA 116, in order to protect the ssODN from SpCas9-mediated degradation. Mutation 3, changes T229 to C229 resulting in change of amino acid C77 to R77. (b) Sequence comparison of human SGCA exon 3 with BglII-ssODN. c.229C>T point mutation is labeled 3. Correcting guanosine base is located on BglII-ssODN and labeled as 3. sgRNA 127 and sgRNA 116 sequences and their corresponding PAM are shown bold and underlined. Proposed cutting sites are shown as a vertical line. All four introduced point mutations are numbered below. (c) Characterization of the sgRNA 127 and sgRNA 116 by T7 Endonuclease assay in HEK293 cells. Both sgRNA/SpCas9 complexes showed similar activity. As predicted, Cas9 generates indels, which can be detected by T7 endonuclease. The PCR band (809 bp) is cut by T7 endonuclease into two bands (503 and 306 bp for sgRNA 127 and 459 and 349 bp for sgRNA 116). Mock = HEK293 cells nucleofected with equal amount of pMaxgfp plasmid.
ssODN-mediated gene editing of SGCA exon 3 in patient-derived hiPSC
To correct the c.229C>T (R77C) missense mutation in exon 3 of human SGCA, we designed sgRNAs 127 and 116 (Figure 1b). These sgRNAs were designed to enable SpCas9 to cut close to the c.229C>T point mutation. The T7 endonuclease 1 (T7E1) assay20 revealed cutting activity for both sgRNAs in HEK293 cells (Figure 1c). We designed a first generation ssODN (EcoRI-ssODN) for SGCA exon 3 and tested it in HEK293 cells (Supplementary Figure S1a,b). After co-nucleofection of sgRNA and EcoRI-ssODN, we detected ssODN-mediated HDR by genotyping PCR and restriction fragment length polymorphism (RFLP) (Supplementary Figure S1c–e). In HEK293 cells, we saw higher HDR efficiencies by applying both sgRNAs simultaneously, compared to using only one sgRNA (Supplementary Figure S1f). For use in LGMD2D iPSC, we modified the ssODN by deleting the EcoRI site and adding a silent mutation to create a BglII restriction site to enable detection of RFLPs (Figure 1a,b).
We utilized a characterized hiPSC line referred to as B3 from a LGMD2D patient. The patient had two different autosomal recessive mutations, one at each SGCA allele. He carried a c.229C>T (R77C) missense mutation on the C229T allele and a c.409G>A missense mutation on the G409A allele. Because R77C is the most prevalent mutation (32%) of all LGMD2D-associated mutations,21 we chose to target and correct this mutation. Simultaneous application of two sgRNAs had been reported to enhance the efficiency of gene replacement by HR in hiPSC.22 Since we had seen a similar result in HEK293 cells (Supplementary Figure S1f), we were interested in testing the double sgRNA method for oligonucleotide-mediated gene editing in hiPSC. Figure 2 depicts the experimental scheme we employed for gene editing of hiPSC, encompassing nucleofection, picking, subcloning, and analysis. We compared two editing strategies. In strategy 1 (S1), we used sgRNA 116 and sgRNA 127, while in strategy 2 (S2), only sgRNA 127 was used (Figure 3a). Experiment S1 yielded two RFLP-positive clones (1.4%) (Figure 3b–d, Supplementary Figure S2a,b), one (0.7%) of which was found to be precisely edited at the C229T allele (Supplementary Figure S3a,b). In experiment S2, we found five clones positive for RFLP (3.5%) (Figure 3b–d, Supplementary Figure S2c,d) of which one (0.7%) was corrected seamlessly at the C229T allele, (Supplementary Figure S3c,d). To separate edited from nonedited clones, we subcloned the hiPSC lines once or twice until they were ~90% pure (Supplementary Figure S3a–d).
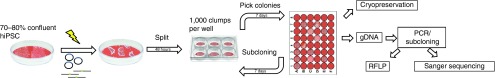
Figure 2.
Experimental scheme for gene editing of hiPSC. 70–80% confluent hiPSC were harvested and nucleofected with 5 µg plasmid pX330 that expressed SpCas9 and the sgRNA of choice. Furthermore, 0.2 nmol of a single-stranded oligonucleotide was added to the reaction. Subsequently, cells were treated with mTesR1 (Stem Cell Technologies, Vancouver, Canada) medium containing 10 µmol/l Rho-kinase (ROCK) inhibitor Y-27632 (Tocris, Minneapolis, MN) for 24 hours. After 48 hours, cell clumps were plated at 1,000 clumps per well. Small clones were picked and transferred to a 48-well plate. Emerging colonies were split 1:2. Half of the split was used to generate genomic DNA (gDNA) for PCR- and restriction fragment length polymorphism (RFLP)-based screening purposes. The other half was transferred into a new well of a 48-well plate for cryopreservation. PCR amplicons of positive clones were subcloned and sequenced to assess sequence information of individual alleles and degree of clonal purification. If sequences revealed correction of the c.229C>T point mutation in B3 iPSC or the c.5713C>T in JF25 iPSC at the right allele, candidate hiPSC were subcloned repeatedly until the gene edited clone was 90% pure.
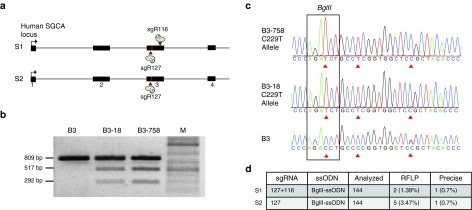
Figure 3.
CRISPR/Cas9 assisted ssODN-mediated gene correction at SGCA exon 3 in B3 hiPSC. (a) In strategy 1 (S1), two SpCas9 nucleases sgRNA 127 and sgRNA 116 target the point mutation c.229C>T (illustrated as a vertical line) in exon 3 of SGCA simultaneously. Arrows depict the cutting sites of SpCas9. For strategy 2 (S2), only sgRNA 127 was used. (b) RFLP analysis of candidate clones. Genomic sequences flanking the putative BglII site were amplified to yield an 809 bp PCR product. The BglII-digested PCR product of the B3 hiPSC clone showed only the 809 bp amplicon, whereas clones B3-18 and B3-758 revealed two bands of 517 and 292 bp size in addition to the 809 bp band, indicating BglII-ssODN-mediated incorporation of the BglII site. M = DNA ladder. (c) 809 bp PCR product was subcloned into a subcloning vector to allow individual Sanger sequencing analysis of both alleles. Top and middle sequences are two chromatogram graphics of the C229T allele of clone B3-758 and B3-18 with three integrated silent mutations depicted by the triangular arrow. Bottom: chromatogram of naive B3 hiPSC created by direct sequencing of the PCR product is shown for comparison. (d) Summary and comparison of S1 with S2. For S1, 144 clones were analyzed. Two of them were found positive for RFLP (1.39%). One clone (0.7%) named B3-18 underwent precise editing of the disease allele (C229T). S2 revealed 5 out of 144 clones (3.47%) positive for RFLP. One of the clones (0.7%) B3-758 was targeted at the C229T allele precisely.
CRISPR/Cas9-mediated gene editing in iPSC from a LGMD2B patient
To target DYSF exon 51 in HEK293 cells, we designed and tested novel sgRNAs 3 and 24 (Supplementary Figure S4a–c). We verified precise gene editing with a DYSF-specific ssODN (HindIII-ssODN) (Supplementary Figure S4a,c) by PCR and RFLP (Supplementary Figure S4d,e). Similar to our previous results (Supplementary Figure S1f), combined use of sgRNA 3 and sgRNA 24 enhanced HDR efficiencies up to 2.2% in HEK 293 cells (Supplementary Figure S4f). However, we were not able to reproduce these results in LGMD2B patient-derived hiPSC using a slightly different setup with two sgRNAs (Supplementary Figure S5a–d). We concluded that the use of two sgRNAs simultaneously might have more deleterious effects in hiPSC than in HEK 293 cells. Consequently, for editing LGMD2B hiPSC, we used just one sgRNA. Since we had found poor allele-specific targeting with sgRNA 3T, we also designed another allele-specific sgRNA called 150, together with an adapted BauI-ssODN (Figure 4a–c).
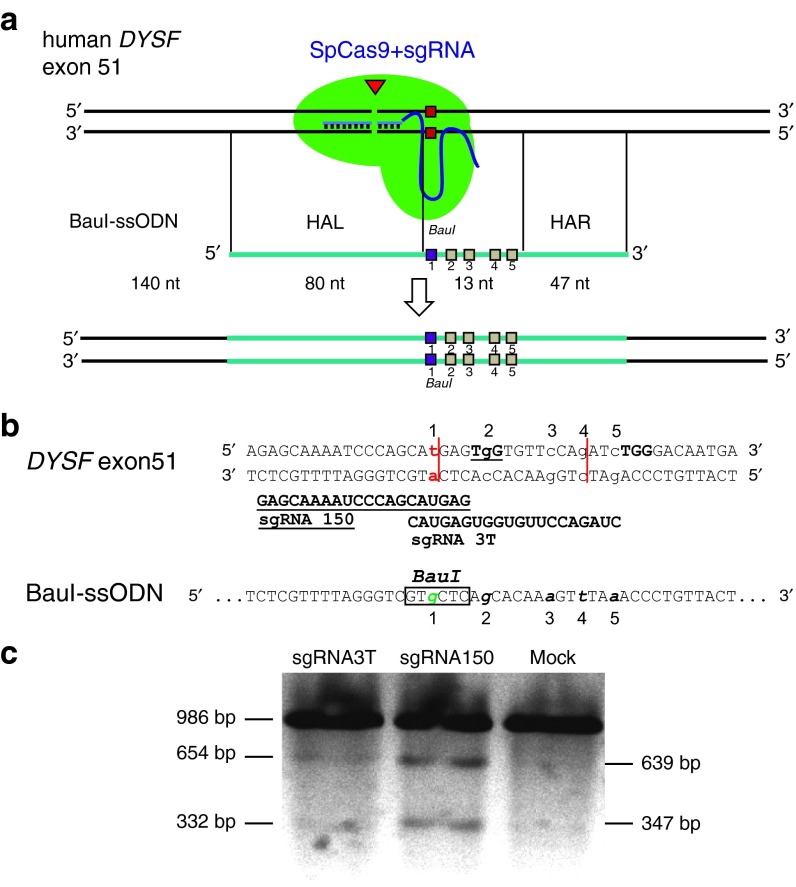
Figure 4.
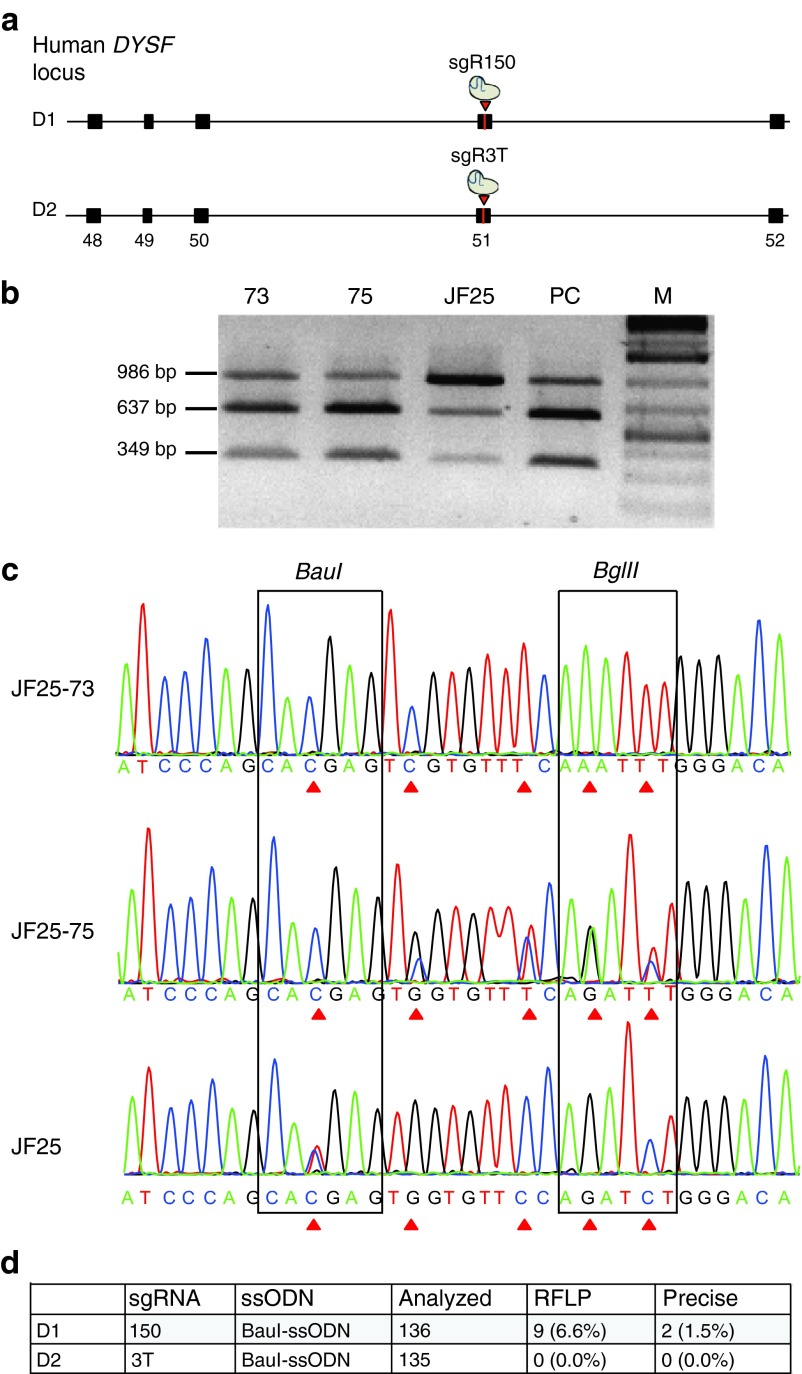
Targeting strategy for c.5713C>T point mutation in exon 51 of human DYSF. (a) sgRNA 3T or sgRNA 150 lead SpCas9 to the proximity of the point mutation c.C5713C>T in human dysferlin (DYSF) exon 51 and introduce a blunt double-strand break (DSB) depicted by the triangle. Co-delivery of a BauI-ssODN homologous to this region of exon 51 allows homology-directed repair (HDR) of the DSB. The BauI-ssODN is 140 nt and in antisense orientation, with homology arm left (HAL) of 80 nt and homology arm right (HAR) of 47 nt length. The homology arms flank five silent mutations over a 13 nt sequence area depicted as squares. Mutation 1 changes T5713 to C5713 and introduces a BauI restriction site. Mutation 2 inactivates the PAM motif of sgRNA 150 from TGG to TCG, mutations 3–5 change the PAM proximal seed sequence of sgRNA 3T. (b) DNA sequence of human dysferlin exon 51 compared to BauI-ssODN. The c.5713C>T point mutation in DYSF exon 51 and C5713 on the ssODN are numbered as 1. sgRNA 150 target sequence is shown bold, underlined and sgRNA 3T target sequence is shown bold. The genomic PAM sequence for each sgRNA is depicted as the target sequence. The proposed cutting sites in exon 51 are shown as vertical lines. All five introduced point mutations on the ssODN are lower case, italic and numbered. (c) Characterization of sgRNA 3T and 150 activity by the T7 endonuclease assay in JF25 hiPSC line. Predicted cutting products from 986 bp PCR amplicon are 654 and 332 bp for sgRNA 3T and 639 and 347 bp for sgRNA 150. sgRNA 150 shows superior cutting activity compared to sgRNA 3T. Mock = JF25 hiPSC nucleofected with equal amount of pMaxgfp plasmid.
We obtained the characterized JF25 hiPSC line derived from a patient with LGMD2B. On each allele, the patient carried a distinct autosomal recessive mutation in the coding region of DYSF. The first mutation was a duplication of a thymidine nucleotide in exon 32 (c.3517dupT), designated to be located in allele 3517TT. This mutation caused a frameshift, leading to formation of a premature stop codon (S1173FfsX2). The second mutation was a C to T transition in exon 51 (c.5713C>T) on allele C5713T. It caused the change of a CGA triplet encoding an arginine residue to a TGA triplet coding for a premature stop codon (R1905X). Both mutations were anticipated to cause nonsense-mediated mRNA decay, with dysferlin protein expected to be reduced or absent. We chose to correct the point mutation c.5713C>T in order to restore dysferlin protein expression (Figure 4a). Allele-specific targeting of the C5713T allele was necessary, since allele 3517TT was wild type for exon 51. For targeting, we used C5713T allele-specific sgRNA variants 3T or 150 and the BauI-ssODN (Figure 4b). We nucleofected JF25 cells with either the BauI-ssODN and plasmid pX330-sgRNA 150 alone (D1) or pX330-sgRNA 3T alone (D2) (Figure 5a). We checked if gene editing occurred by BauI-RFLP and sequencing (Figure 5b,c). No RFLP-positive clones were found with the combination sgRNA 3T/BauI-ssODN (D2). However, we found 9 out of 136 clones positive for RFLP with the combination sgRNA 150/BauI-ssODN (D1) (Figure 5d, Supplementary Figure S5e–h). We identified two seamlessly edited clones, JF25-73 and JF25-75, targeting the C5713T allele (1.5%) (Figure 5b,c). These two clones were purified by two rounds of subcloning until the level of purity was more than 90% (Supplementary Figure S6a,b).
Figure 5.
CRISPR/Cas9-assisted ssODN-mediated gene correction at DYSF exon 51 in JF25 hiPSC. (a) Schematic diagram of exons 48–52 of the human dysferlin locus. In D1, SpCas9 sgRNA 150 can target and cut (arrow) close to the point mutation c.5713C>T (vertical line) in exon 51. In D2, sgRNA 3T was used instead of sgRNA 150. (b) RFLP analysis of representative candidate clones. Exon 51 flanking sequences from naive JF25 and gene-edited clone JF25-73 (73) and JF25-75 (75) were PCR amplified. Amplicons were digested with BauI. Agarose gel analysis of the naive JF25 hiPSC clone showed the 986 bp band and two bands 637 and 349 bp. The edited clones 73 and 75 showed a fainter 986 bp band and the two smaller bands with stronger intensity, indicating a more complete digest of the PCR product similar to the positive control JF10 (PC), which was wild-type at DYSF exon 51. (c) PCR products from (b) naive JF25, clone 73, and clone 75 were directly sequenced. Top: chromatogram of JF25-73. All five mutations were introduced on both alleles as indicated by the triangular arrows. Middle bar shows sequence of JF25-75. Only the C5713T allele was targeted by the BauI-ssODN. As predicted, all five mutations were introduced on this allele. Bottom: chromatogram of naive JF25 for comparison. Sequences of restriction sites BauI and BglII are framed. After homology-directed repair, BauI site was established on gene-edited clones, whereas the BglII site was destroyed. Triangular arrows show the nucleotides that were changed in the gene-edited clones. (d) Summary of both experiments. D1: sgRNA 150 and BauI-ssODN were nucleofected into JF25, and 9 out of 136 (6.6%) clones were positive for allele-specific RFLP. JF25-73 and JF25-75 (1.5%) edited the C5713T allele precisely. D2: sgRNA 3T and BauI-ssODN were nucleofected into JF25 and 135 clones were analyzed, but none of these were RFLP positive.
Targeted insertion of dysferlin cDNA into the H11 safe harbor by dual integrase cassette exchange and TALEN-assisted homologous recombination insertion precise
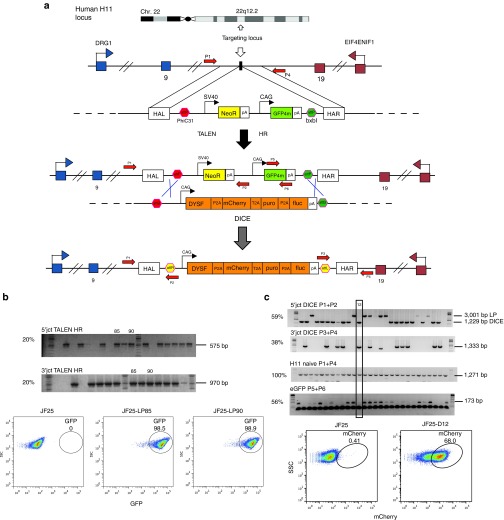
In addition to oligonucleotide-based in situ gene editing, we corrected the LGMD2B mutations by site-specific cDNA addition at a safe harbor sequence. The H11 safe harbor site was characterized in mouse,23 and the cognate site was identified in human cells12 (Figure 6a). H11 is intergenic, universally transcribed, recombinogenic, and distant from known oncogenes. Our lab established a method called dual integrase cassette exchange (DICE) for gene addition at the human H11 safe harbor locus in human embryonic stem cells and hiPSC.12 DICE is a two-step procedure that first requires insertion of a landing pad (LP) into H11 (Figure 6a). The second step is a cassette exchange mediated by concerted action of the serine integrases phiC31 and BxbI (Figure 6a). We took advantage of this method to integrate wild-type DYSF cDNA driven by the CAG promoter into H11. We generated a JF25-LP line as described.12 G418-resistant clones with moderate GFP fluorescence were picked, and ~20% of them had undergone HR-mediated integration of the LP at H11 (Figure 6b). To bring about DICE at the integrated LP, we employed a donor plasmid that carried phiC31 and a BxbI attB sites flanking an 11.8 kb CAG-DYSF containing insert (Figure 6a). To initiate DICE, we nucleofected line JF25-LP85 with the DYSF donor and plasmids expressing phiC31 and Bxb1 integrases. Ten of 32 puromycin-resistant clones (31%) were positive for both junction PCRs (Figure 6c). In order to exclude the possible formation of intermediates during DICE, we performed a PCR detecting eGFP. All junction PCR-positive clones were negative for eGFP, verifying replacement of the LP sequence during the DICE reaction. We found 68% expression of the mCherry donor marker by flow cytometry in representative clone 12, verifying that the cassette exchange was successful (Figure 6c).
Figure 6.
Dual integrase cassette exchange (DICE) to add DYSF cDNA to the H11 site. (a) DICE is a two-step method. First, a TALEN-assisted homologous recombination (HR) step allows integration of a landing pad (LP) plasmid12 in a specific genomic location, in this case the H11 locus. The LP contained a neomycin resistance gene and a GFP4m color marker for selection and screening purposes. PhiC31 and BxbI attP sites flanked this cassette. After integration by TALEN-assisted HR, a donor plasmid was used to achieve cassette exchange. Our donor contained the human dysferlin cDNA and the mCherry, puromycin resistance, and firefly luciferase genes for selection and screening purposes. To allow DICE, PhiC31, and BxbI attB sites flank this cassette. DICE occurred when the PhiC31 and BxbI integrases and the donor plasmid were cointroduced into the cell. The product of DICE is the irreversible exchange of the LP with the donor cassette. (b) Verification of site-specific integration of the LP into H11. After co-nucleofection of the LP and TALEN plasmids, JF25 hiPSCs were selected, and genomic DNA from the resistant clones was extracted for junction PCR. Site-specific integration of the LP into H11 was verified in about 20% of the clones by amplification of the 5′ and 3′ junctions of the LP and genomic region. Clones 85 and 90 (PCR positive for both junctions) showed high levels of GFP4m expression (98.5 and 98.8%) by flow cytometric analysis and were chosen for further DICE experiments. (c) Top: Verification of DICE in JF25 hiPSC. We co-nucleofected clone 85 with the DICE donor and phiC31 and BxbI expressing plasmids and screened puromycin-resistant clones by junction PCRs. We screened for the 5′ junction with primer combination P1+P2 (59% positive) and 3′ junction with primer combination P3+P4 (37.5% positive). To check if both H11 alleles were targeted, we conducted an H11 PCR with primer P1+P4 to amplify the naive H11 locus (100%). To check for residual presence of the LP after DICE, we checked for presence of GFP4m with primer set P5+P6. We found that 56.3% of all puromycin-resistant clones were positive for the GFP PCR. We found clone 12 to be positive for DICE and negative for random integration events. Bottom: Flow cytometric analysis of clone 12 (JF25-D12). We found 68% of the cells positive for mCherry, while 0.4% of the starting JF25 cells were positive.
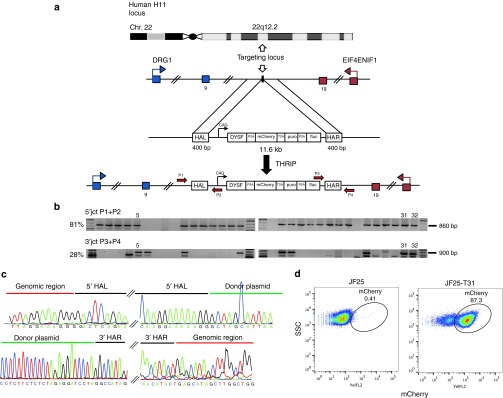
We also tested whether a single-step knock-in mechanism based solely on TALEN-assisted HR was feasible at H11. First, we added 400-bp homology arms identical to those used for the H11-LP to the DYSF cDNA donor (Figure 7a). We nucleofected the homology-flanked DYSF donor with TALENs L2 and R2 to initiate a “TALEN-assisted homologous recombination insertion precise” (THRIP) reaction. Eight of 32 screened clones (25%) were positive for both junctions (Figure 7b). We sequenced junction PCR products and verified seamless insert knock-in by HR (Figure 7c). Flow cytometric analysis of representative clone JF25-T31 confirmed stable mCherry expression, verifying insertion of the DYSF donor cassette (Figure 7d).
Figure 7.
TALEN-assisted homologous recombination for insertion precise (THRIP) targeting strategy at the human H11 locus. (a) A TALEN pair was designed and evaluated to cut in H11 (ref. 12). The donor contained two 400-bp homology arms, left (HAL) and right (HAR), flanking a CAG promoter-driven human dysferlin (DYSF) cDNA. DYSF was connected to mCherry, selection marker puromycin, and firefly luciferase using skipping peptides P2A and T2A. After co-nucleofection of donor and TALEN plasmids into hiPSC line JF25, HR-mediated integration of the donor took place. Recovery of recombinants was enhanced by 6 days of puromycin selection. (b) 5′ junction PCR verified correct integration of the donor into H11. Thirty-two puromycin-resistant clones were picked and genomic DNA was prepared. Primer pair P1+P2 was used to amplify the 5′ junction, yielding an 860-bp amplicon. Twenty-six of 32 (81%) clones showed a positive signal. 3′ junction PCR: primer pair P3+P4 were used to amplify the 3′ junction of the donor–H11 junction sequence. Nine of 32 (28%) clones revealed a positive signal. (c) PCR products of (b) were subcloned and sequenced. Top: sequence junction between genomic H11 region and HAL is shown. On the right, junction between HAL and donor plasmid is shown. Bottom: sequence junction between donor plasmid and HAR is shown. Junction between HAR and genomic sequence is shown on the bottom right. (d) Flow cytometry analysis of mCherry expression of clone JF25-T31 (87.3%) compared with naive JF25 (0.41%) ~30 days after nucleofection showed stable expression of the inserted cassette.
Correction of the R77C mutation restores SGCA at the cell membrane of differentiated hiPSC
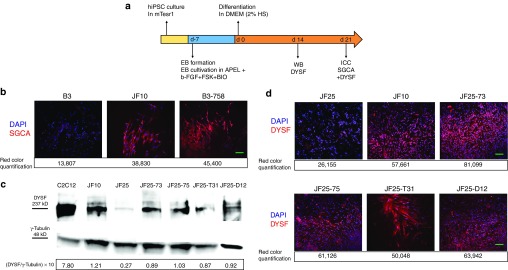
To determine whether functional SGCA protein was expressed in gene-edited clones, we first differentiated B3-18 and B3-758 hiPSC toward the muscle lineage, since the SGCA protein was not expressed in iPSC. The corrected iPSC clones were differentiated for 21 days using a previously published protocol24 (Figure 8a). We noted increased proliferation and differentiation capacity of B3-18 and B3-758 compared to uncorrected B3 (data not shown). The misfolded R77C mutant protein is synthesized but is retained in the endoplasmic reticulum and does not reach the cell membrane.25 To allow staining of only membrane-localized SGCA, a detergent-free protocol was used for immunocytochemistry.25 We quantified the red signal of each image to elucidate the differences between JF10, B3, and the B3-758 corrected clone. See Supplementary Figure S7 for further information about quantification of SGCA signal in corrected and control samples. We observed strong SGCA-positive staining in JF10 and corrected clone B3-758 (Figure 8b), but little in uncorrected B3 cells (Figure 8b). These results demonstrated that the R77C phenotype was rescued by gene editing.
Figure 8.
Rescue of SGCA and DYSF expression by CRISPR/Cas9-assisted HDR, DICE, and THRIP. (a) Scheme for differentiation of hiPSC into muscle progenitor cells. hiPSC were cultured and expanded. After embryoid body (EB) formation, EBs were cultivated in APEL medium with three small molecules, b-FGF, forskolin, and BIO, for 7 days. Terminal muscle differentiation was induced in Dulbecco's Modified Eagle Medium (DMEM) containing 2% horse serum for 21 days. Protein lysate from differentiated cells for immunoblot detection of dysferlin protein was generated after 14 days of terminal differentiation. Immunocytochemistry (ICC) for SGCA and DYSF was prepared after 21 days of differentiation. (b) ICC for SGCA in non-cell membrane penetrating conditions (without Tween 20) allowed staining of the membrane-localized SGCA only. Corrected B3-758 clone expressed SGCA (red) after differentiation. B3 and JF10, respectively, were used as negative and positive controls. Numbers below each picture refer to the red color quantification of the respective image. 4′,6-diamidino-2-phenylindole (DAPI) is in blue. Bar = 100 µm. (c) Top: Immunoblot for staining of DYSF. JF-iPSC lines and corrected hiPSC clones were differentiated toward muscle lineage (a). DYSF protein (237 kD) was detected in ssODN-corrected lines JF25-73 and JF25-75. Furthermore, DYSF protein was detected in THRIP clone T31 and DICE clone D12. Background levels of DYSF protein were detected in naive JF25. Mouse myoblasts C2C12 and JF10 were positive controls. Loading control was 48 kD γ-tubulin. Bottom: numbers show DYSF/γ-tubulin ratios × 10 of each sample. (d) ICC to detect dysferlin. Twenty-one days differentiated iPSC lines were plated on chamber slides. Dysferlin (red) is located in cells from CRISPR/Cas9-corrected and THRIP- and DICE-corrected clones. The red signal of each image was quantified and is shown numerically below each image. In general, JF25 clones showed fewer differentiated cells, and only very few cells stained in dim red. Bar = 100 µm.
Rescue of DYSF expression by CRISPR/Cas9 gene editing, DICE, and THRIP
We were also interested in determining whether correction of the c.5713C>T mutation by ssODN-mediated HDR rescued dysferlin protein expression. Furthermore, we wanted to compare DYSF expression levels between clones that underwent CRISPR/Cas9-mediated correction, DICE, or THRIP. All corrected iPSC clones were differentiated for 21 days toward the muscle lineage using the same protocol24 (Figure 8a). We detected dysferlin protein by western blot in both ssODN-corrected clones JF25-73 and JF25-75 at levels comparable to the amount of dysferlin present in iPSC line JF10, derived from the patient's healthy father (Figure 8c). As expected, little dysferlin was detected in the JF25 patient line. Dysferlin expression was also detected in the THRIP-corrected clone JF25-T31 and the DICE-corrected clone JF25-D12. We calculated the ratios of dysferlin/γ-tubulin for each sample (see Supplementary Materials and Methods), which revealed differences between JF25 (0.27) and the other samples. The ratios of the corrected clones (0.87–1.03) were comparable to the JF10-positive control (1.21) (Figure 8d). To provide additional evidence for restoration of dysferlin, we stained differentiated cells for dysferlin by immunocytochemistry. We found cells that stained positively for dysferlin in all the corrected clones and the JF10-positive control, while little dysferlin staining was observed in differentiated cells derived from the JF25 patient iPSC (Figure 8d). We quantified the red signal of each image to further elucidate the difference between JF25 and JF10 and the corrected clones (Figure 8d, Supplementary Figure S7). As we had noticed during differentiation of corrected SGCA clones, the proliferation and differentiation capacity of clones corrected for dysferlin was greater than that of uncorrected clones (data not shown).
sgRNA 127, sgRNA 116, and sgRNA 150 had limited detectable off-target activity and did not change the karyotype of corrected hiPSC
We were interested in determining whether the guide RNAs used in conjunction with SpCas9 for targeting of DYSF and SGCA had any significant activity at proposed off-target sites. We tested the top five scored off-target sites for SGCA sgRNA 127, sgRNA 116, and DYSF sgRNA 150 for off-target activity by the T7E1 assay. For sgRNA 127 and sgRNA 150, we found no apparent off-target activity (Supplementary Figure S8a). For sgRNA 116, we detected signs of off-target activity only at locus OT5 (Supplementary Figure S8a). We examined the karyotypes of CRISPR/Cas9 gene-edited LGMD2D clone B3-758 and LGMD2B clone JF25-75 and found them to be normal (Supplementary Figure S8b).
Discussion
We report for the first time allele-specific, seamless correction of single point mutations from two patients diagnosed with LGMD2B and LGMD2D with CRISPR/Cas9-assisted ssODN-mediated HDR in hiPSC. Furthermore, as an alternative, we also used the DICE12 and novel THRIP gene-targeting strategies to deliver the wild-type DYSF coding sequence precisely to the H11 safe harbor in the LGMD2B iPSC. We differentiated the corrected LGMD2B and LGMD2D iPSC lines in vitro into muscle progenitor cells and demonstrated rescue of protein expression for dysferlin and relocation of corrected alpha-sarcoglycan protein to the cell membrane, providing appropriate levels of correctly localized protein in all cases.
Our in situ gene correction method relied on gene editing with ssODNs.4 SSODNs can be used for subtle HDR-mediated gene correction in pluripotent stem cells, as well as in muscle stem cells.4,26 With the CRISPR/Cas9 system, almost any genomic information can be targeted. Introduction of double-strand breaks with SpCas9 is limited only by the need for a NGG protospacer adjacent motif (PAM) sequence.27 Fortunately, in our case, we were able to find suitable PAM sequences in the desired regions. For example, sgRNA 150 produced Cas9 cutting precisely at the c.5713C>T mutation in DYSF, and sgRNA 127 led to Cas9 cutting 5-bp upstream of the c.229C>T mutation in SGCA. We noticed increased HDR efficiencies with sgRNAs that bound and cut closer to the targeted mutations. Cutting directly at the mutation with sgRNA 150 proved to be the most efficient sgRNA for targeting of DYSF exon 51 (Figure 5d). Others have also noted this trend.9,28,29 In order to increase the ability to target any genomic sequence, Cas9 variants derived from other species with different PAM requirements have been characterized.30 Furthermore, directed evolution of SpCas9 can be used to change its PAM specificity.31 Further developments in this area would make ssODN-mediated targeting feasible for all mutations that are associated with LGMD2B32 and LGMD2D.17
Since most LGMD2B mutations are single events on one allele, we were interested in achieving allele-specific gene targeting. However, even with allele-specific sgRNA variants such as sgRNA 150 and sgRNA 3T, we found a significant number of clones in which the undesired 3517TT allele was edited by the ssODN (Supplementary Figure S5d,g). This result was in contrast to another study, which reported robust allele specificity with sgRNA variants that had been changed in one nucleotide.33 Other studies have reported gene correction efficiencies of 0.47–2% in hESC and hiPSC using ZFN, TALENs, or CRISPR/Cas99,34,35,36,37, and the efficiencies we observed were similar. To target the disease mutations DYSF c.5173C>T and SGCA c.229C>T, we used either one sgRNA or a combination of two sgRNAs. We tested the use of two sgRNAs based on a report22 that described higher HR efficiencies with two sgRNAs in a hiPSC clone, as well as our own observation of higher efficiencies in HEK293 cells targeted with two sgRNAs (Supplementary Figures S1f and S4f). Surprisingly, in our hiPSC, we found that the yield of RFLP-positive clones with sgRNA 150 (6.6%) and sgRNA 127 (3.5%) used alone were higher than using sgRNA 127 + sgRNA 116 in combination (1.39%; Figures 3d and 5d). Also, we saw evidence of imprecise HDR when two gRNAs were used, with insertions of parts of the ssODN in the vicinity of SGCA exon 3 or DYSF exon 51 in several instances (Supplementary Figures S2b and S5d). CRISPR/Cas9 can induce significant off-target activity.20 However, when tested with the T7E1 assay, we observed little or no detectable off-target activity at the most highly ranked off-target loci for sgRNA 127 and sgRNA 116 for SGCA and sgRNA 150 for DYSF in B3 or JF25 hiPSC (Supplementary Figure S8a).
An alternative method to correct hiPSC to CRISPR/Cas9-assisted HDR is site-specific addition of cDNA expression cassettes at the H11 safe harbor locus. The H11 locus is attractive as a gene addition site, since it is intergenic, universally transcribed, and distant from known oncogenes.12,23 We compared the two-step DICE method previously reported by our lab12 with the novel one-step THRIP method developed here. For DICE, the LP from our earlier study was inserted at the H11 locus of JF25 by TALEN-assisted HR. This LP carried two phage integrase recognition sites flanking selection and screening markers. We confirmed targeted integration of the LP at postselection efficiencies of about 20% (Figure 6b). The subsequent dual integrase-mediated cassette exchange reaction correctly inserted the DYSF expression cassette at efficiencies of 31% of drug-resistant clones. The THRIP method for precise gene addition is based simply on TALEN-assisted HR and is thus similar to step one of the DICE reaction. THRIP had a surprisingly high efficiency after selection (25%) (Figure 7b), especially considering that it relied on HR of a relatively large 11.6-kb expression cassette, and it is known that insert size limits HR efficiencies.38 This high efficiency highlights the recombinogenic nature of the H11 locus, a further benefit of this safe harbor site.12 Given that the THRIP method involves only one step, providing an overall higher efficiency, THRIP may be preferable if a recombination will only be done once. On the other hand, if a number of different expression cassettes will be inserted at the same target site, the DICE method may be advantageous, since once a LP is correctly inserted, the cassette exchange step proceeds at a high efficiency (>30%). Another potential advantage of the DICE method is for insertion of large cassettes. While the homologous recombination involved in the THRIP method is known to be affected by insert size, the phage integrase-mediated recombination involved in the cassette exchange reaction of DICE has no known size limit. Both methods should be useful for the rapid and safe generation of transgenic hiPSC cell lines for gene correction, lineage tracking, and disease modeling.
Because alpha-sarcoglycan and dysferlin are preferentially expressed in skeletal muscle and were not well expressed in undifferentiated iPSC, we differentiated the iPSC lines corrected by CRISPR/Cas9/ssODN, DICE, and THRIP into myogenic cells using an in vitro differentiation protocol24 (Figure 8a) to allow phenotypic characterization. We detected dysferlin protein by western blot analysis and immunocytochemistry in the differentiated, corrected JF25 lines at levels roughly comparable to the JF10-positive control line, while only trace levels were seen in the differentiated uncorrected JF25 iPSC (Figure 8c,d). Dysferlin expression was observed in the corrected iPSC clones, regardless of whether they were corrected by CRISPR/Cas9/ssODN, DICE, or THRIP. The human R77C missense mutation in SGCA produces a protein that is arrested in the endoplasmic reticulum and does not localize to the cell membrane.39,40 Consequently, to show that SGCA was absent at the membrane in uncorrected 2D-B3 cells, we used a cell membrane nonpermeablizing form of immunocytochemistry that omitted the use of Tween (Figure 8b). Cell membrane nonpermeabilizing immunocytochemistry had been shown in earlier studies to reveal the absence of R77C-SGCA at the membrane of a reporter cell line.25,40 This approach clearly revealed the presence of membrane-localized SGCA protein in the corrected cells and its absence in the uncorrected patient iPSC.
The observation that we obtained roughly similar levels of corrected gene products whether we applied the CRISPR/Cas9 method of in situ gene correction or pursued precise addition of the therapeutic cDNA sequence to a safe, well-expressed heterologous site suggests that either of these approaches may be an effective means to correct iPSC. The potential advantage of the CRISPR/Cas9 strategy is that one can anticipate an appropriate level of gene expression, due to presence of the endogenous gene control elements. On the other hand, addition of a cDNA to a heterologous site was approximately an order of magnitude more efficient by DICE and THRIP compared to CRISPR/Cas9, so that finding correctly targeted clones required screening of fewer colonies. This feature produced a significant savings of time, labor, and materials. To ensure the appropriate level of expression of the therapeutic gene, the gene addition strategy can encompass any desired transcriptional control elements, including the endogenous sequences if desired. Furthermore, the gene addition approach produces a general strategy that would work for any patient, regardless of the mutation involved. This feature may be particularly important in the case of larger, more complex mutations and in situations, such as LGMD2B, where there is no large mutational hotspot.32
In conclusion, we carried out allele-specific, precise gene editing of two point mutations with the CRISPR/Cas9/ssODN system in LGMD2B and LGMD2D patient-derived hiPSC and also demonstrated site-specific cDNA addition in LGMD2B iPSC using DICE and THRIP. The gene correction evidenced by rescue of appropriate levels of DYSF and SGCA proteins to the cell membrane suggests that these corrected iPSC could form part of a cell therapy strategy for these muscular dystrophies.41,42,43,44 The ability to carry out sophisticated genome editing methods on iPSC, as demonstrated by this work, represents one of the strengths of using iPSC in a cell therapy strategy. By contrast, it would be difficult to generate clonal corrected lines using primary cells. On the other hand, iPSC possess several features that make their use in a clinical strategy challenging. For example, the iPSC would have to be expanded extensively without introduction of an unacceptable level of mutations. iPSC would then have to be differentiated in a manner that would assure that they could engraft in muscle, without generation of tumors. Then, as with any cell therapy, methods would need to be devised to enable adequate delivery of cells to the large muscle target involved in most forms of muscular dystrophy. The work presented here demonstrated that the first steps in this process are feasible, while the subsequent challenges of an iPSC cell therapy strategy remain to be solved.
Materials and Methods
iPSC lines. The JF10 and JF25 iPSC lines were generated at Cellular Dynamics from patient blood cells using Epstein–Barr virus nonintegrating reprogramming vectors and characterized there for pluripotentcy and karyotype. The B3 iPSC line was generated at the Stanford Stem Cell Core from patient fibroblasts using Sendai virus nonintegrating reprogramming vectors (CytoTune 2.0-iPS Reprogramming Kit, Life Technologies, South San Francisco, CA) and was characterized there for pluripotency. The iPSC line referred to in this study as JF10 is officially designated as Jain Foundation line JF010i-DYSFHZ1 and CDI#01460.101.10. The iPSC line referred to as JF25 in this study is officially designated as Jain Foundation line JFNY1 or CDI#01456.103.10. All tissue donations were obtained under informed consent.
sgRNA and ssODN design and cloning. All single guide RNAs (sgRNA) for human dysferlin (DYSF) exon 51 and human alpha-sarcoglycan (SGCA) exon 3 were designed with the web tool CRISPR Design (http://crispr.mit.edu). The oligonucleotides containing the sgRNA sequence (Supplementary Table S1; Integrated DNA Technologies; IDT, Coralville, IA) were cloned into the pX330 backbone (Addgene plasmid #42230) according to the protocol from the Zhang lab (https://www.addgene.org/crispr/zhang/). Candidate clones were confirmed by sequencing (Sequetech, Mountain View, CA). All plasmids for nucleofection were isolated using the Nucleobond Midi Plus EF kit (Macherey-Nagel, Bethlehem, PA). Single-stranded oligodeoxynucleotide (ssODN) templates (Supplementary Table S2) were ordered from IDT as Ultramers. A list of all plasmids used can be found in Supplementary Table S7.
Cloning of DICE and THRIP donor plasmids. Detailed descriptions of cloning of the DICE and THRIP DYSF donor plasmids are found in Supplementary Materials and Methods. A list of all plasmids used can be found in Supplementary Table S7.
Generation of corrected B3 and JF25 hiPSC by CRISPR/Cas9-assisted HDR-mediated ssODN integration. To generate ssODN-corrected B3 iPSC, we nucleofected 1.2 × 106 B3 hiPSCs with 5 µg of pX330-sgRNA 127 and 0.2 nmol of BglII-ssODN (S2; Figure 3a) or 2.5 µg of pX330-sgRNA 116 and pX330-sgRNA 127 each and 0.2 nmol of BglII-ssODN (S1; Figure 3a). To generate ssODN-corrected JF25 iPSC, we nucleofected 1.2 × 106 JF25 iPSCs with 5 µg of pX330-sgRNA 150 and 0.2 nmol of BauI-ssODN (D1; Figure 5a) or 5 µg of pX330-sgRNA 3T and 0.2 nmol of BauI-ssODN (D2; Figure 5a). We always generated one control sample that was nucleofected with 5 µg of plasmid pMaxGFP (Lonza, Basel, Switzerland). If more than 80% of the living cells were eGFP positive, the experiment was continued. As illustrated in Figure 2, hiPSCs were split with Versene (Thermo Fisher Scientific, Waltham, MA) 48 hours after nucleofection. Around 1,000 clumps were distributed in a well of a six-well plate. We picked small-sized hiPSC colonies 7–10 days after plating using an EVOS XL Core microscope (Thermo Fisher Scientific) with a P200 Gilson pipette under the flow hood. For each experiment, 135–144 colonies were transferred into 48-well plates. Finally, we split clones that reached confluency with Versene and used one half of the split to generate genomic DNA (gDNA) using QuickExtract DNA Extraction Solution (Epicentre, Madison, WI) according to the manufacturer's instructions. The other half was transferred to another 48-well plate for downstream applications such as freezing or expanding. In order to screen for gene-edited hiPSC clones, we performed a RFLP. First, we PCR-amplified the target locus of SGCA exon 3 or DYSF exon 51 with flanking primers (Supplementary Table S3). The PCR conditions for both PCRs were as follows: initial denaturation at 98 °C for 30 seconds; denaturation at 98 °C for 10 seconds; annealing for 30 seconds at 67 °C for primer P118/P119 and at 68 °C for primer P114/P115; extension at 72 °C for 20 seconds. The PCR had 35 cycles in total with a final denaturation for 2 minutes. The expected PCR amplicon had a length of 809 bp (SGCA) or 986 bp (DYSF). We excised and purified the PCR products with the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and digested 200 ng of the amplicon DNA with BglII for experiments in which a BglII-ssODN was used to correct the c.229C>T mutation of SGCA exon 3. We used BauI for experiments in which the BauI-ssODN was used to correct the c.5713C>T mutation of DYSF exon 51. We detected positive gene-editing events by 517 and 292 bp bands for SGCA and intense 637 and 349 bp fragments for DYSF. Allelic discrimination of gene-edited clones is described in the Supplementary Materials and Methods.
Integration of the LP at the H11 locus of JF25 hiPSC. Three plasmids were co-nucleofected into hiPSC JF25. In detail, 3 µg of LP plasmid p2attng H11 short was mixed with 1 µg MR015-H11-L2-TALEN and 1 µg of MR015-H11-R2-TALEN. Forty-eight to 72 hours after nucleofection we started selection with 25 µg/ml G418 (Invitrogen, Carlsbad, CA) for 2 weeks. We picked up to 24 G418 resistant clones manually and transferred them to a 24-well plate. We generated gDNA from cell splits of the resistant cells with DNeasy Blood & Tissue Kit (Qiagen). The rest of the cells were further analyzed by flow cytometry analyses and cryopreserved. Characterization of G418-resistant clones included 5′H11-LP and 3′LP-H11 junction PCR. Primers and PCR protocols for these reactions will be made available upon request.
DICE reaction to integrate DYSF cDNA into H11. We nucleofected JF25-LP85 hiPSCs with 3 µg of donor pKLD-CAG-Dysf-CPL that contained a CAG promoter-driven human DYSF cDNA connected to a CPL marker gene expression cassette (see Supplementary Materials and Methods for details). For DICE, 1 µg of phiC31 integrase-expressing plasmid pCS-kI and 1 µg of Bxb1 integrase-expressing plasmid pCS-Bxb1 were added to the reaction. Forty-eight to 72 hours after nucleofection, 0.5 µg/ml puromycin (Life Technologies) was added to mTesr1 medium for 5 days. We picked puromycin-resistant colonies and transferred them into a new 24-well plate. Seven days later, growing cell colonies were split, and some of them were used for gDNA generation with the QuickExtract DNA Extraction Solution (Epicentre, Madison, WI). Genomic DNA was used for PCR-mediated characterization of the clones. Primer for all reactions can be found in Supplementary Table S4 and S6. The following PCR protocols were used: 5′ junction PCR: 98 °C for 30 seconds, 98 °C for 7 seconds, 66 °C for 13 seconds, 72 °C for 1 minute, 50 cycles, 72 °C for 2 minutes, giving an amplicon size of 1,106 bp. 3′ junction PCR: 98 °C for 30 seconds, 98 °C for 7 seconds, 68 °C for 13 seconds, 72 °C for 1 minute, 50 cycles, 72 °C for 2 minutes, giving an amplicon size of 1,333 bp. H11 PCR: same protocol as for 3′ junction PCR with an amplicon size of 1,271 bp. GFP PCR: 98 °C for 30 seconds, 98 °C for 7 seconds, 66 °C for 13 seconds, 72 °C for 20 seconds, 72 °C for 2 minutes, with an amplicon size of 173 bp. Another portion of the cells was analyzed by flow cytometry. Detailed information about flow cytometry experiments conducted in this study can be found in the Supplementary Materials and Methods.
THRIP reaction to integrate DYSF into H11 via homologous recombination. For the THRIP reaction, we co-nucleofected 3 µg of DYSF donor plasmid pKLD-C-5HA-hdysf-CPL-3HA with 1 µg of TALEN MR015-H11-L2 and 1 µg of MR015-H11-R2. Puromycin selection, picking, handling of clones, and gDNA generation were performed as described for insertion of the LP. For characterization (see Supplementary Figure S6 for primers), we used the following PCR protocols: 5′ junction PCR: 98 °C for 30 seconds, 98 °C for 7 seconds, 68 °C for 13 seconds, 72 °C for 40 seconds, 50 cycles, 72 °C for 2 minutes, giving an amplicon size of 868 bp and 3′ junction PCR: 98 °C for 30 seconds, 98 °C for 7 seconds, 68 °C for 13 seconds, 72 °C for 60 seconds, 50 cycles, giving an amplicon size of 900 bp.
In vitro differentiation. For in vitro differentiation of hiPSC into muscle precursors, we used an optimized protocol that had been originally published by the Zon group.24 Details of this protocol can be found in the Supplementary Materials and Methods.
Immunoblot. For detection of human dysferlin, we used 1:100 dilution of primary antibody NCL Hamlet (Leica Biosystems, Buffalo Grove, IL) and 1:500 dilution of goat antimouse IgG horseradish peroxidase secondary antibody (Thermo Fisher Scientific). A more detailed protocol can be found in the Supplementary Materials and Methods.
Immunocytochemistry. Differentiated cells were stained for SGCA or dysferlin using immunocytochemistry. For detection of membrane-bound SGCA, we used a protocol without the use of Tween 20 as described earlier.25 Primary antibody against SGCA was NCL-L-a-SARC (Leica Biosystems) and NCL-Hamlet against DYSF. More details of these protocols can be found in the Supplementary Materials and Methods.
Off-target analysis. For each sgRNA used, the five highest scoring predicted off-targets of CRISPR design (http://crispr.mit.edu) were selected (see Supplementary Table S5 for primer). A detailed protocol can be found in the Supplementary Materials and Methods.
Additional methods are described in the Supplementary Materials and Methods.
SUPPLEMENTARY MATERIAL Figure S1. ssODN-mediated HDR at SGCA exon 3 in HEK 293 cells. Figure S2. Gene editing of B3 hiPSC with sgRNA/Cas9 and ssODN. Figure S3. Subcloning of RFLP-positive clones B3-18 and B3-758. Figure S4. ssODN-mediated HDR at dysferlin exon 51 in HEK 293 cells. Figure S5. ssODN-mediated HDR at dysferlin (DYSF) exon 51 in JF25 hiPSC was feasible with sgRNA 150. Figure S6. Subcloning of RFLP-positive clones JF25-73 and JF25-75. Figure S7. Immunocytochemistry supplementary information. Figure S8. CRISPR/Cas off-target analysis of sgRNA 127, sgRNA 116 and sgRNA 150. Table S1. Primer sequences for cloning of sgRNAs into pX330. Table S2. ssODN sequences 5′-3′. Table S3. Primers for RFLP, T7E1 assays, and allele discrimination. Table S4. Primers for genotyping PCR. Table S5. Primers for off target analysis. Table S6. Primers for genotyping PCR DICE/THRIP. Table S7. Plasmids used in this study. Materials and Methods
Acknowledgments
We thank the following members of the Calos lab for assistance: Jon Geisinger for discussions and comments during the project, Michael Wilkinson for help on optimizing the protocols for the genotyping PCR and T7 endonuclease assay, Christophe Pichavant for critical reading of the manuscript. We thank the Jain Foundation and Cellular Dynamics for generation, characterization, and provision of JF25 and JF10 hiPSC and the Stem Cell Core of the Department of Genetics at Stanford University and Gavin Wang for help with generation and characterization of B3 iPSC. We are grateful to the patients and their fathers for tissue donations used for iPSC generation. ST was supported by postdoctoral fellowship TU418/1-1 from the Deutsche Forschungsgemeinschaft. This work was supported by grant TR4-06711 from the California Institute for Regenerative Medicine to MPC and a gift fund donated to the Calos lab by James Kanagy. MPC is an inventor on Stanford-owned patents covering phiC31 integrase, and MPC and APF are inventors on a Stanford-owned patent application covering DICE.
Supplementary Material
References
- Takahashi, K, Tanabe, K, Ohnuki, M, Narita, M, Ichisaka, T, Tomoda, K et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. [DOI] [PubMed] [Google Scholar]
- Awaya, T, Kato, T, Mizuno, Y, Chang, H, Niwa, A, Umeda, K et al. (2012). Selective development of myogenic mesenchymal cells from human embryonic and induced pluripotent stem cells. PLoS One 7: e51638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chal, J, Oginuma, M, Al Tanoury, Z, Gobert, B, Sumara, O, Hick, A et al. (2015). Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat Biotechnol 33: 962–969. [DOI] [PubMed] [Google Scholar]
- Aarts, M and te Riele, H (2011). Progress and prospects: oligonucleotide-directed gene modification in mouse embryonic stem cells: a route to therapeutic application. Gene Ther 18: 213–219. [DOI] [PubMed] [Google Scholar]
- Ding, Q, Lee, YK, Schaefer, EA, Peters, DT, Veres, A, Kim, K et al. (2013). A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 12: 238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, C and Capecchi, MR (1992). Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol 12: 3365–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igoucheva, O, Alexeev, V and Yoon, K (2001). Targeted gene correction by small single-stranded oligonucleotides in mammalian cells. Gene Ther 8: 391–399. [DOI] [PubMed] [Google Scholar]
- Urnov, FD, Miller, JC, Lee, YL, Beausejour, CM, Rock, JM, Augustus, S et al. (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651. [DOI] [PubMed] [Google Scholar]
- Yang, L, Guell, M, Byrne, S, Yang, JL, De Los Angeles, A, Mali, P et al. (2013). Optimization of scarless human stem cell genome editing. Nucleic Acids Res 41: 9049–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, D (2014). Genome engineering with targetable nucleases. Annu Rev Biochem 83: 409–439. [DOI] [PubMed] [Google Scholar]
- Zhao, C, Farruggio, AP, Bjornson, CR, Chavez, CL, Geisinger, JM, Neal, TL et al. (2014). Recombinase-mediated reprogramming and dystrophin gene addition in mdx mouse induced pluripotent stem cells. PLoS One 9: e96279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, F, Gamboa, M, Farruggio, AP, Hippenmeyer, S, Tasic, B, Schüle, B et al. (2014). DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res 42: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi, S and Kang, PB (2012). Update on the genetics of limb girdle muscular dystrophy. Semin Pediatr Neurol 19: 211–218. [DOI] [PubMed] [Google Scholar]
- Piccolo, F, Moore, SA, Ford, GC and Campbell, KP (2000). Intracellular accumulation and reduced sarcolemmal expression of dysferlin in limb–girdle muscular dystrophies. Ann Neurol 48: 902–912. [PubMed] [Google Scholar]
- Bansal, D, Miyake, K, Vogel, SS, Groh, S, Chen, CC, Williamson, R et al. (2003). Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature 423: 168–172. [DOI] [PubMed] [Google Scholar]
- Liu, J, Aoki, M, Illa, I, Wu, C, Fardeau, M, Angelini, C et al. (1998). Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat Genet 20: 31–36. [DOI] [PubMed] [Google Scholar]
- Piccolo, F, Roberds, SL, Jeanpierre, M, Leturcq, F, Azibi, K, Beldjord, C et al. (1995). Primary adhalinopathy: a common cause of autosomal recessive muscular dystrophy of variable severity. Nat Genet 10: 243–245. [DOI] [PubMed] [Google Scholar]
- Kawai, H, Akaike, M, Endo, T, Adachi, K, Inui, T, Mitsui, T et al. (1995). Adhalin gene mutations in patients with autosomal recessive childhood onset muscular dystrophy with adhalin deficiency. J Clin Invest 96: 1202–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fougerousse, F, Bartoli, M, Poupiot, J, Arandel, L, Durand, M, Guerchet, N et al. (2007). Phenotypic correction of alpha-sarcoglycan deficiency by intra-arterial injection of a muscle-specific serotype 1 rAAV vector. Mol Ther 15: 53–61. [DOI] [PubMed] [Google Scholar]
- Cradick, TJ, Fine, EJ, Antico, CJ and Bao, G (2013). CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res 41: 9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrié, A, Piccolo, F, Leturcq, F, de Toma, C, Azibi, K, Beldjord, C et al. (1997). Mutational diversity and hot spots in the alpha-sarcoglycan gene in autosomal recessive muscular dystrophy (LGMD2D). J Med Genet 34: 470–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, SM, Ortiz, L, Mali, P, Aach, J and Church, GM (2015). Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res 43: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic, B, Hippenmeyer, S, Wang, C, Gamboa, M, Zong, H, Chen-Tsai, Y et al. (2011). Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proc Natl Acad Sci USA 108: 7902–7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C, Tabebordbar, M, Iovino, S, Ciarlo, C, Liu, J, Castiglioni, A et al. (2013). A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell 155: 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soheili, T, Gicquel, E, Poupiot, J, N'Guyen, L, Le Roy, F, Bartoli, M et al. (2012). Rescue of sarcoglycan mutations by inhibition of endoplasmic reticulum quality control is associated with minimal structural modifications. Hum Mutat 33: 429–439. [DOI] [PubMed] [Google Scholar]
- Nik-Ahd, F and Bertoni, C (2014). Ex vivo gene editing of the dystrophin gene in muscle stem cells mediated by peptide nucleic acid single stranded oligodeoxynucleotides induces stable expression of dystrophin in a mouse model for Duchenne muscular dystrophy. Stem Cells 32: 1817–1830. [DOI] [PubMed] [Google Scholar]
- Jinek, M, Chylinski, K, Fonfara, I, Hauer, M, Doudna, JA and Charpentier, E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialk, P, Rivera-Torres, N, Strouse, B and Kmiec, EB (2015). Regulation of gene editing activity directed by single-stranded oligonucleotides and CRISPR/Cas9 systems. PLoS One 10: e0129308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F, Pruett-Miller, SM, Huang, Y, Gjoka, M, Duda, K, Taunton, J et al. (2011). High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods 8: 753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, FA, Cong, L, Yan, WX, Scott, DA, Gootenberg, JS, Kriz, AJ et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver, BP, Prew, MS, Tsai, SQ, Topkar, VV, Nguyen, NT, Zheng, Z et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, K, Bassez, G, Bernard, R, Krahn, M, Labelle, V, Figarella-Branger, D et al. (2005). Dysferlin mutations in LGMD2B, Miyoshi myopathy, and atypical dysferlinopathies. Hum Mutat 26: 165. [DOI] [PubMed] [Google Scholar]
- Smith, C, Abalde-Atristain, L, He, C, Brodsky, BR, Braunstein, EM, Chaudhari, P et al. (2015). Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol Ther 23: 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner, F, Laganière, J, Cheng, AW, Hockemeyer, D, Gao, Q, Alagappan, R et al. (2011). Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell 146: 318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, PD, Scott, DA, Weinstein, JA, Ran, FA, Konermann, S, Agarwala, V et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X, Wang, Y, Huang, H, Chen, B, Chen, X, Hu, J et al. (2014). Precise gene modification mediated by TALEN and single-stranded oligodeoxynucleotides in human cells. PLoS One 9: e93575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkert, S, Wunderlich, S, Bednarski, C, Beier, J, Haase, A, Dreyer, AK et al. (2014). Efficient designer nuclease-based homologous recombination enables direct PCR screening for footprintless targeted human pluripotent stem cells. Stem Cell Reports 2: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle, EA, Rock, JM, Lee, YL et al. (2007). Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci USA 104: 3055–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam, RA, Wang, B, Shand, SH, Xiao, X and Watkins, SC (2006). Alpha-sarcoglycan is recycled from the plasma membrane in the absence of sarcoglycan complex assembly. Traffic 7: 793–810. [DOI] [PubMed] [Google Scholar]
- Bartoli, M, Gicquel, E, Barrault, L, Soheili, T, Malissen, M, Malissen, B et al. (2008). Mannosidase I inhibition rescues the human alpha-sarcoglycan R77C recurrent mutation. Hum Mol Genet 17: 1214–1221. [DOI] [PubMed] [Google Scholar]
- Darabi, R, Arpke, RW, Irion, S, Dimos, JT, Grskovic, M, Kyba, M et al. (2012). Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell 10: 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudenege, S, Lebel, C, Huot, NB, Dufour, C, Fujii, I, Gekas, J et al. (2012). Myoblasts derived from normal hESCs and dystrophic hiPSCs efficiently fuse with existing muscle fibers following transplantation. Mol Ther 20: 2153–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco, FS, Gerli, MF, Perani, L, Benedetti, S, Ungaro, F, Cassano, M et al. (2012). Transplantation of genetically corrected human iPSC-derived progenitors in mice with limb-girdle muscular dystrophy. Sci Transl Med 4: 140ra89. [DOI] [PubMed] [Google Scholar]
- Tanaka, A, Woltjen, K, Miyake, K, Hotta, A, Ikeya, M, Yamamoto, T et al. (2013). Efficient and reproducible myogenic differentiation from human iPS cells: prospects for modeling Miyoshi Myopathy in vitro. PLoS One 8: e61540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.