Adeno-associated virus (AAV) continues to be a promising viral vector for delivery of therapeutic genes in human gene therapy. In 2012, the European Commission approved Glybera (alipogene tiparvovec),1 an AAV vector carrying the human lipoprotein lipase gene for commercial use in treatment of lipoprotein lipase deficiency. In addition, within the past five years there have been numerous successes in preclinical, early, and late-stage clinical trials of AAV for treatment of a variety of diseases, including muscular dystrophy, hemophilia, Leber's congenital amaurosis retinopathy, age-related macular degeneration, cardiac disorders, cancer, and neurodegenerative disorders, such as Parkinson's disease and Canavan disease. However, despite this progress, the infectious pathway of AAV vectors is an evolving story. In a study recently reported in Nature, Pillay and colleagues2 employed a powerful genome-wide haploid genetic screen to gain a better understanding of the AAV infectious entry pathway. Among other factors consistent with the known pathways of entry and infection, the authors identified a new cellular factor that is required for infection of multiple AAV serotypes, which they term AAV receptor (AAVR).
At least 12 serotypes of AAV from human and primate sources have been identified. Many bind to distinct receptors on the surface of host cells, with several of the serotypes reported to bind to a primary cell surface receptor followed by interaction with a secondary receptor that is capable of facilitating viral entry. Primary cell surface attachment receptors identified to date include heparan sulfate proteoglycans for AAV serotypes 2, 3, and 6; N-terminal galactose for AAV9; and specific N- or O-linked sialic acid moieties for AAV1, 4, 5, and 6 (ref. 3). Secondary receptors include fibroblast growth factor receptor (FGFR) and integrin (which has been disputed) for AAV2; hepatocyte growth factor receptor (c-Met) for AAV2 and 3, and platelet-derived growth factor for AAV5, which is also modified by sialic acid.4,5 However, no single receptor common to all the serotypes has previously been identified.
Haploid genetic screens have been used to identify both extracellular and intracellular receptors that impact the infectious entry pathway of several viruses.6,7,8 A cholesterol transporter that is localized in the late endosome/lysosome, Niemann-Pick C1 (NPC1), was identified as a critical intracellular host receptor for Ebola and later for other filoviruses, and is speculated to trigger membrane fusion. Additionally, cell surface heparan sulfate proteoglycan and cell surface sialic acid moieties were recently identified as attachment receptors for Rift Valley fever virus and enterovirus D68, respectively.
In the new study, Pillay et al.2 generated a library of mutagenized haploid HAP1 cells by using a retroviral gene-trap vector to create knockouts of almost all nonessential genes in the human genome. They exposed this knockout library to recombinant AAV2 expressing red fluorescent protein and sorted cells that were resistant to infection. When they mapped the location of gene-trap insertions in this sorted population, the authors identified 46 significant hits, including genes involved in heparan sulfate proteoglycan biosynthesis, and genes known to be important for endocytic trafficking and retrograde transport from endosomes to the Golgi. The high number of hits in these pathways is reassuring, given that we know they play critical roles in AAV2 attachment and subcellular trafficking. In addition to increasing our confidence in the screen, these hits offer opportunities for further exploration in that many of these genes have not been shown to play a direct role in AAV infection. Unexpectedly, the most significantly enriched gene the authors identified was KIAA0319L, which encodes a type I transmembrane protein containing a MANSC domain, five polycystic kidney disease (PKD) domains, and a C6 region near the N terminus.9 Little is known about the gene encoding AAVR, although it has been linked to dyslexia and is implicated in neuronal migration and axon guidance.9,10
Pillay et al.2 evaluated the ability of AAV to infect AAVR knockout (AAVRKO) cell lines created using CRISPR-Cas9 technology. They also generated knockout cell lines of the reported secondary receptors for AAV2, METKO, and FGFRKO. Interestingly, METKO and FGFRKO had little to no effect on AAV infection, whereas AAVRKO rendered cells highly resistant to AAV infection using 20,000 viral genomes per cell. AAVRKO cells were still poorly infected by AAV2 at 100,000 viral genomes per cell. The importance of AAVR in AAV infection was validated by demonstrating that expression of recombinant AAVR in the AAVRKO cells rescued infection. Intriguingly, infection by distinct serotypes, including AAV1, AAV2, AAV3b, AAV5, AAV6, AAV8, and AAV9, in AAVRKO cells was also dependent upon the presence of AAVR.
The identification of AAVR as an important receptor for a diverse cast of AAV serotypes is a significant observation that raises many questions. For example, what is the universal function of AAVR? Additionally, with respect to AAV biology, is there an evolutionarily conserved epitope or structure common to all AAV serotypes that interacts with AAVR? When considering mechanism, is a direct or indirect interaction between AAV and AAVR necessary? On an even more basic level, at what stage of the infectious pathway is AAVR important?
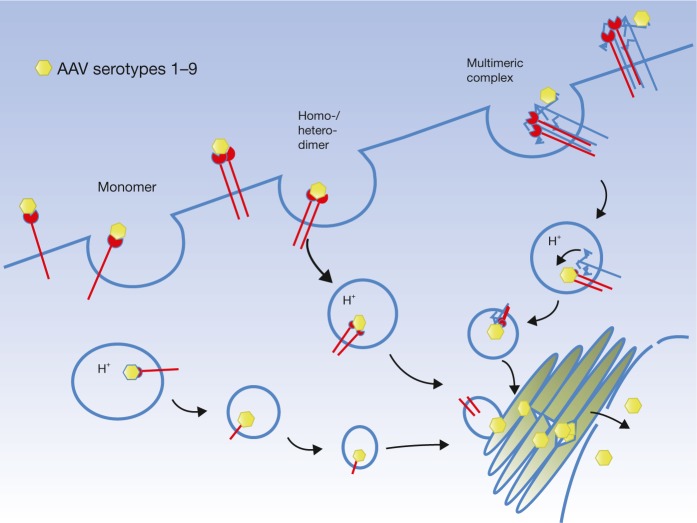
At first blush, the data appear to suggest that the interaction between AAV and AAVR occurs at the cell surface and that AAVR serves as a cell surface receptor. Soluble AAVR (with PKD domains 1–5) bound directly to AAV2 particles in an enzyme-linked immunosorbent assay, and soluble AAVR as well as AAVR-specific antibody inhibited AAV2 infection in a concentration-dependent manner. In addition, deleting the C terminus of AAVR, which encodes a motif required for endocytic sorting, prevented internalization of AAVR and was not able to rescue AAV2 infection when expressed in AAVRKO cells. Intriguingly, AAVR fusions with different C-terminal domains could rescue infection. Although these data may support a direct interaction at the cell surface, the authors concede that they cannot rule out the possibility that AAV interacts with AAVR after entry into the host cell. Going forward, it will be important to tease apart this distinction, i.e., if AAVR directly interacts with AAV at the cell surface, or is part of a multimeric receptor complex at the cell surface that is important for uptake (Figure 1) or later steps in entry, or rather if AAVR interacts with AAV after internalization of the virus and promotes subsequent steps in the life cycle, such as escape into the cytoplasm (Figure 2). Perhaps all are true. A logical next step would be to interrogate AAV binding, uptake, and subcellular trafficking in AAVRKO cells. If cells deficient in AAVR still efficiently facilitate AAV attachment and endocytosis, it would indicate AAVR as a cellular factor mediating later steps in entry.
Figure 1.
Potential scenarios for AAV cellular uptake mediated by AAVR. AAV serotypes 1–9 may exhibit a direct interaction with AAVR by binding to a monomer, a dimer, or even a heterodimer of AAVR, followed by subsequent endocytosis. Alternatively, AAVR may form a multimeric complex with other AAV receptors (e.g., glycans or other receptors) necessary for endocytosis, and a direct interaction with AAVR at the cell surface is not required.
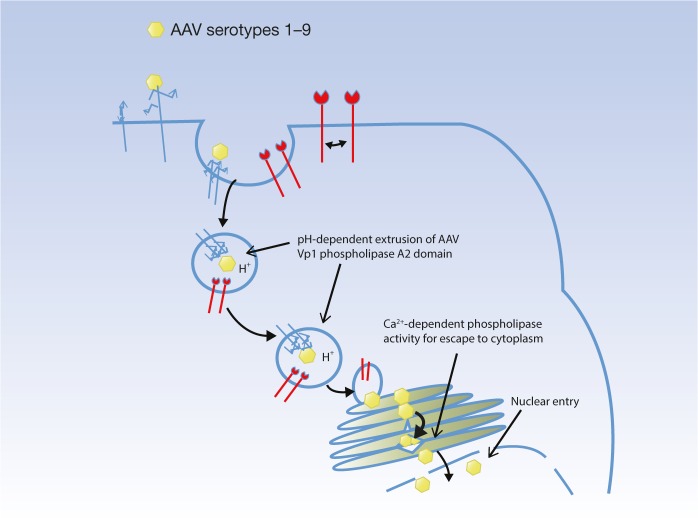
Figure 2.
The role of AAVR may not be virus uptake. The AAV serotypes 1–9 may internalize into the cell using distinct surface receptors, and the function of AAVR may lie within the endosome, e.g., aiding in a conformational change for VP1 extrusion, or trafficking the different serotypes to the trans-Golgi network, where Ca2+ levels may be optimal for VP1 PLA2 activity leading to efficient escape to the cytoplasm and subsequent nuclear transport.22 This is not inconsistent with the data of Pillay et al.2 The soluble AAVR and polyclonal antibody used by Pillay et al. may disrupt an interaction of AAVR with another plasma or endosomal membrane protein (e.g., disrupt a homo- or hetero- interaction) that is necessary for function of AAVR. The function of AAVR may be aiding in the conformational change of AAV necessary for infection, or the function may be for trafficking of AAVR to the TGN. It will be important to look at the effect of soluble receptor and antibody on the steps of binding, internalization, and trafficking of AAV to gain a better understanding of the stage at which AAVR is actually functioning.
We note that Pillay et al.2 report the binding affinity of AAV2 for AAVR with a Kd of 150 nm, a much lower affinity than that observed for the AAV2 heparan sulfate interaction, which has been estimated to range from 0.1 to 3.7 nm Kd.11,12 AAVR has also been described to form homodimeric interactions,13 and thus one alternative explanation of the data is that the soluble AAVR, and the polyclonal AAVR-specific antibody are inhibiting infection by disrupting a multimeric receptor interaction important for AAV entry (Figure 1) or infection (Figure 2), and that a direct interaction of AAVR with AAV at the cell surface is not necessary.
Variable transduction efficiencies observed in different tissues for AAV serotypes have been attributed, in part, to their binding to different cell surface receptors. However, there are many potential stages in the infectious entry pathway in which the efficiency of AAV transduction can be affected, including, e.g., the rate of endocytic uptake, endosomal trafficking, endosomal escape, nuclear translocation, virus uncoating, and genome conversion/or expression. Pillay et al. demonstrated that AAVR associates with a cis-medial marker (giantin) and co-localizes with a trans-Golgi network (TGN) marker, TGN46, at steady state. The authors were able to label a pool of AAVR at the plasma membrane by cooling cells to 4°C and showed that AAVR rapidly migrated from the plasma membrane to the Golgi once cells were warmed to initiate endocytosis. Although the authors remark that this trafficking pattern is similar to what is known for AAV2 infection,14 this study does not directly address whether AAVR plays a role in virus uptake or trafficking. Curiously, Pillay et al. also demonstrated that AAVR fusion mutants carrying the C-terminal domain of low-density lipoprotein receptor and poliovirus receptor, which are known to internalize and traffic through endosomal pathways that are not associated with the Golgi, were able to partially rescue AAV2 infection in AAVRKO cells. Trafficking through the Golgi may be the preferred route, but this finding supports the idea that AAV can use multiple distinct endocytic pathways and that trafficking through the Golgi may not be an absolute requirement for infection. Indeed, evidence for AAV exploiting multiple endocytic pathways can be found in the literature, including clathrin-mediated endocytosis, non-clathrin-coated pathways, e.g., CLIG/GEEC, and macropinocytosis.4,14,15,16 The ability of AAV to use multiple independent pathways is an example of virus flexibility and has long been thought to be driven by pressures specific to certain cell types or environmental conditions, making the discovery of AAVR seem all the more impressive, as it appears to be universally required for all serotypes and cell targets tested.
Exposure to acidic pH in endosomes appears to be essential for virus infection, as it is linked with conformational changes in the AAV capsid that permit escape into the cytoplasm. The N terminus of capsid protein VP1, which normally is tucked away in the AAV interior, becomes extruded through a capsid pore during this conformational change, thereby presenting a phospholipase domain and nuclear localization signals that are required for escape into the cytoplasm and nuclear trafficking, respectively.17 When considering that AAV vectors carrying mutations in these VP1 domains have been shown to accumulate at the Golgi,18 it may not be a coincidence that AAVR migrates to this location as well. If a direct interaction of AAV and AAVR is required for infection of all AAV serotypes either at the surface or after internalization, then an evolutionarily conserved amino acid epitope or structure must be involved. Amino acid sequence identity between serotypes can be as little as 45%,19 with the loop regions showing the least conservation. Of note, regions in VP1 spanning the catalytic residues of the phospholipase domain are highly conserved, even among other parvoviruses, such as minute virus of mice, canine parvovirus, and porcine parvovirus. Regardless of whether a connection can be made between AAVR and VP1 function, it would be interesting to discover if the importance of AAVR during infection extends to viruses outside the dependovirus family.
Pillay et al.2 convincingly demonstrate an in vivo requirement for AAVR, as AAVRKO mice are resistant to infection with AAV9. Interestingly, AAVR has been reported to have little comparative expression in skeletal muscle tissue and the liver,13 which are two tissues that are known to have enhanced transduction with AAV9 as compared to other AAV serotypes.20,21 This might seem inconsistent, but RNA-seq data from both the Genotype-Tissue Expression (GTEx) project and the Illumina Body Map indicate AAVR message is expressed in skeletal muscle and liver. Additionally, the AAVR gene is predicted to have multiple splice variants that could complicate its detection by traditional approaches. In the years ahead, new studies will surely explore AAVR splice variants, tissue distribution, and expression levels to see how they impact not only AAV9, but other serotypes in vivo, particularly if the interaction between the capsid and AAVR is shown to be rate-limiting for AAV infection. It will also be important to understand whether the putative glycosylated isoforms of AAVR affect AAV binding of the various serotypes.13
This exciting study highlights the power of unbiased genetic screens in unraveling the mysteries of biology, as it is staggering to consider that AAV has been studied for more than 50 years, and yet until now, no universal receptor has been found that impacts the entry of all AAV serotypes. Of course, the potential to harness the AAV–AAVR interaction to improve transduction of AAV vectors for gene therapy will be of strong interest.
References
- Moran, N (2012). First gene therapy approved. Nat Biotechnol 30: 1153. [DOI] [PubMed] [Google Scholar]
- Pillay, S, Meyer, NL, Puschnik, AS, Davulcu, O, Diep, JH, Ishikawa, Yet al. (2016). An essential receptor for adeno-associated virus infection. Nature 530: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murlidharan, G, Samulski, RJ and Asokan, A (2014). Biology of adeno-associated viral vectors in the central nervous system. Front Mol Neurosci 7: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, W, Zhang, L, Yan, Z and Engelhardt, JF (2005). Intracellular trafficking of adeno-associated viral vectors. Gene Ther 12: 873–880. [DOI] [PubMed] [Google Scholar]
- Kashiwakura Y, Tamayose, K, Iwabuchi, K, Hirai, Y, Shimada, T, Matsumoto, Ket al. (2005). Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol 79: 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riblett, AM, Blomen, VA, Jae, LT, Altamura, LA, Doms, RW, Brummelkamp, TR et al. (2015). A haploid genetic screen identifies heparan sulfate proteoglycans supporting Rift Valley fever virus infection. J Virol 90: 1414–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay, S and Carette JE (2015). Hunting viral receptors using haploid cells. Annu Rev Virol 2: 219–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggen, J, Thibaut, HJ, Staring, J, Jae, LT, Liu, Y, Guo, Het al. (2016). Enterovirus D68 receptor requirements unveiled by haploid genetics. Proc Natl Acad Sci USA 113: 1399–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, MW, Tsang, WH, Chan, SO, Li, HM, Ng, HK and Waye, MM (2011). Dyslexia-associated kiaa0319-like protein interacts with axon guidance receptor Nogo Receptor 1. Cell Mol Neurobiol 31: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelmans, G, Buitelaar, JK, Pauls, DL and Franke, B (2011). A theoretical molecular network for dyslexia: integrating available genetic findings. Mol Psychiatry 16: 365–382. [DOI] [PubMed] [Google Scholar]
- Zhang, F, Aguilera, J, Beaudet, JM, Xie, Q, Lerch, TF, Davulcu, Oet al. (2013). Characterization of interactions between heparin/glycosaminoglycan and adeno-associated virus. Biochemistry 52: 6275–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi, A, Chen, J, McCarty, DM, Samulski, RJ, Liu, J, Superfine, R (2004). Analysis of the interaction between adeno-associated virus and heparan sulfate using atomic force microscopy. Glycobiology 14: 969–977. [DOI] [PubMed] [Google Scholar]
- Poon, M-W, Chan, H-L, Lim, K-P and Waye, MMY (2011). The dyslexia candidate gene Kiaa0319L encodes N-glycosylated isoforms that form homodimers. J Biochem Mol Biol Post Genomic Era 1: 65–78. [Google Scholar]
- Nonnenmacher, M and Weber, T (2011). Adeno-associated virus 2 infection requires endocytosis through the CLIC/GEEC pathway. Cell Host Microbe 10: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg, MS, Nicolson, S, Bhatt, AP, McLendon, M, Li, C and Samulski RJ (2014). Recombinant adeno-associated virus utilizes cell-specific infectious entry mechanisms. J Virol 88: 12472–12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher, M and Weber, T (2012). Intracellular transport of recombinant adeno-associated virus vectors. Gene Ther 19: 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samulski, RJ and Muzyczka, N (2014). AAV-mediated gene therapy for research and therapeutic purposes. Annu Rev Virol 1: 427–451. [DOI] [PubMed] [Google Scholar]
- Johnson, JS, Li, C, DiPrimio, N, Weinberg, MS, McCown, TJ and Samulski, RJ (2010). Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. J Virol 84: 8888–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli, C, Soltys, S, Rengo, G and Rabinowitz, JE (2008). Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 16: 1073–1080. [DOI] [PubMed] [Google Scholar]
- Bish, LT Morine, K, Sleeper, MM, Sanmiguel, J, Wu, D, Gao, G,et al. (2008). Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum Gene Ther 19: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattia, MA, Nam, HJ, Van Vliet, K, Mitchell, M, Bennett, A, Gurda, BLet al. (2012). Structural insight into the unique properties of adeno-associated virus serotype 9. J Virol 86, 6947–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher, M, Cintrat, J, Gliiet, D and Weber, T (2015). Syntaxin 5-dependent retrograde transport to the trans-Golgi network is required for adeno-associated virus transduction. J Virol 89: 1673–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]