Abstract
Coxiella burnetii and Coxiella-like bacteria (CLB) are genetically and ecologically distinct despite some genetic similarities. Furthermore, CLB are exceptionally diverse and widespread in ticks, but rarely detected in domestic animals. Since Coxiella bacteria can be transmitted from infected horses by inhalation or by coming in contact with ticks during activities such as horseback riding, it is necessary to study their prevalence. To the best of our knowledge, this is the first large-scale nationwide investigation of the prevalence of C. burnetii and CLB among horses reared in South Korea. Of 816 blood samples collected between 2007 and 2013, 11 (1.3%) were identified as C. burnetii by ELISA, and six (0.7%) as CLB by 16S rRNA sequencing. While a sequence from Jeju Island was similar (97.9–100%) to those within clade B, five sequences obtained from the northern region were categorized into a new clade, indicating the sequence diversity of the genus Coxiella. Studies until date had detected CLB only in ticks; here, we describe their detection in mammals. Given their zoonotic potential, strategic monitoring and appropriate control programs for Coxiella species need to be established.
Introduction
Coxiella burnetii, an obligate intracellular gram-negative bacterium, is a zoonotic pathogen that causes Q fever or coxiellosis. C. burnetii has been detected in species across the animal kingdom, including many domestic and wild mammals, birds, and arthropods such as ticks [1]. C. burnetii was originally named as Rickettsia burnetii on the basis of its similarity to Rickettsia species, but 16S rRNA analysis-based phylogeny placed this species in the genus Coxiella, which belongs to the gamma subdivision of the phylum Proteobacteria [2]. C. burnetii is currently the only species in this genus [3]; however, another species, C. cheraxi, presumed to belong to this genus, has been detected in crayfish [4]. Moreover, the fact that ticks transmit both C. burnetii and Coxiella-like bacteria (CLB) emphasizes the need to accurately discriminate between them [5]. In addition, questions about these bacteria remain, including the potential role of CLB in the population dynamics of ticks, and the possibility of CLB conversion leading to the emergence of Q fever [6].
Infertility and abortion caused by C. burnetii have been reported in numerous animals, but it is often difficult to identify the infection owing to its asymptomatic nature [7]. Stillbirth, abortion, and neonatal death caused by C. burnetii lead to economic loss in the horse industry [8]. To date, the role of horses as a reservoir for C. burnetii has not been extensively studied. C. burnetii is a well-known cause of abortion in ruminants; however, several recent studies have examined this characteristic in horses, too. C. burnetii DNA was detected in the aborted fetuses of horses, indicating the abortogenic nature of C. burnetii in horses [9,10]. C. burnetii DNA was also detected in the placenta of horses without any known abortion history in the Netherlands. Seven recent studies had detected C. burnetii in horse samples, in particular, in aborted fetuses, while another 34 studies determined the seroprevalence of C. burnetii in horses [7]. Therefore, horses should be considered as a reservoir for C. burnetii [11].
Although several studies have investigated C. burnetii infection in dairy cattle, goats, and water deer in South Korea [12–14], no studies have examined the occurrence of C. burnetii in horses. Recently, there has been a boom in the horse industry in South Korea, as the international trade of horses has increased. The potential risk of transmission of Coxiella species to humans may increase after exposure to infected horses or ticks during horseback riding. Therefore, the objective of this study was to detect and survey the current epidemiological prevalence and distribution of C. burnetii in horses reared in South Korea, by using ELISA and PCR.
Materials and Methods
Ethics statement
This study did not receive approval from the Institutional Animal Care and Use Committee (IACUC) at Kyungpook National University (KNU) in 2007, as the IACUC at KNU evaluates laboratory animals maintained in indoor facilities, not outdoor animals. Equine veterinarians collected blood samples at horse farms after receiving consent from the horse owners.
Sample size determination and sample collection
The total number of horses reared in South Korea in 2014 was recorded at 25,819 [15]. The sample size for this study was determined using the following formula, with an expected disease prevalence of 50%, accepted absolute error of 5%, and a confidence level of 99% by using a simple random sampling design [16]:
where n = required sample size, pexp = expected prevalence, and d = desired absolute precision
According to the formula, a minimum of 664 samples was required. In this study, 816 horses were randomly selected from multiple regions in South Korea, between 2007 and 2013 (Fig 1). Following blood collection from the jugular vein, whole blood was used for PCR, and serum samples were used for serology. Age, sex, breed, and region were recorded for data analysis, and missing information was recorded as “unknown” (Table 1). The mean age of the study animals was 7.2 years, with a standard deviation of 4.6 years.
Fig 1. A map of South Korea showing the four different study regions where blood samples were collected from horses to detect Coxiella species.
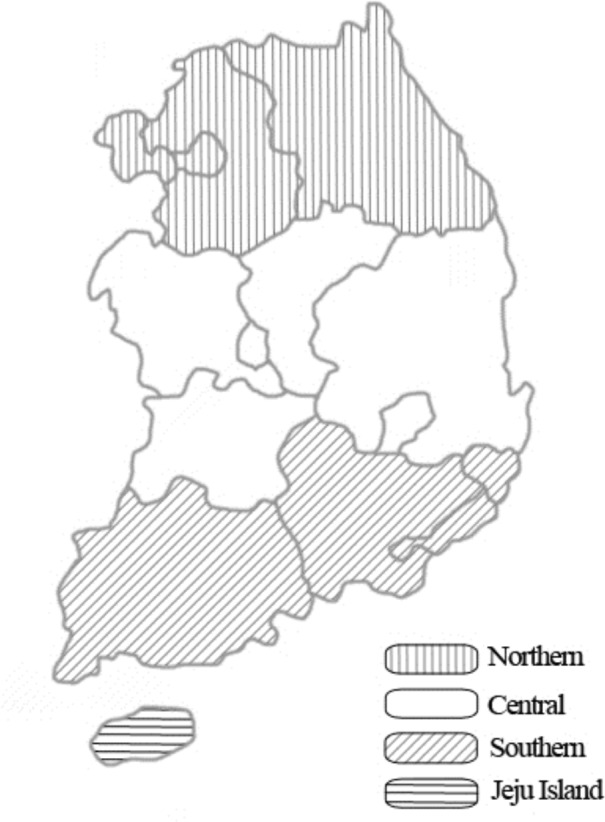
Table 1. Detection of Coxiella infection among horses raised in Korea between 2007 and 2013.
| Group | No. tested | No. (%) of horses | ||||
|---|---|---|---|---|---|---|
| ELISA | PCR | |||||
| Positive | 95% CI† | Positive | 95% CI† | |||
| Region | Northern | 295 | 7 (2.4) | 0.6–4.1 | 5 (1.7) | 0.2–3.2 |
| Central | 184 | 2 (1.1) | 0–2.6 | 0 | 0 | |
| Southern | 243 | 2 (0.8) | 0–2.0 | 0 | 0 | |
| Jeju Island | 94 | 0 | 0 | 1 (1.1) | 0–3.1 | |
| Breed | Thoroughbred | 566 | 11 (1.9) | 0.8–3.1 | 5 (0.9) | 0.1–1.7 |
| Native Korean pony | 109 | 0 | 0 | 1 (0.9) | 0–2.7 | |
| Warm blood | 61 | 0 | 0 | 0 | 0 | |
| Mixed | 80 | 0 | 0 | 0 | 0 | |
| Sex | Male | 159 | 3 (1.9) | 0–4.0 | 1 (0.6) | 0–1.9 |
| Female | 283 | 5 (1.8) | 0.2–3.3 | 3 (1.1) | 0–2.3 | |
| Castrated | 280 | 3 (1.1) | 0–2.3 | 1 (0.4) | 0–1.1 | |
| Unknown | 94 | 0 | 0 | 1 (1.1) | 0–3.1 | |
| Age | <5 | 271 | 5 (1.8) | 0.2–3.5 | 5 (1.8)* | 0.2–3.5 |
| 5–10 | 249 | 4 (1.6) | 0.1–3.2 | 0 | 0 | |
| >10 | 202 | 2 (1) | 0–2.4 | 0 | 0 | |
| Unknown | 94 | 0 | 0 | 1 (1.1) | 0–3.1 | |
| Total | 816 | 11 (1.3) | 0.6–2.1 | 6 (0.7) | 0.2–1.3 |
*Significantly different, p < 0.05
†CI = confidence interval.
Serology
Serum samples were checked for the presence of antibodies against C. burnetii by ELISA with the ID Screen Q Fever Indirect Multi-species Kit (IDvet, Montpellier, France), in accordance with the manufacturer’s instructions. The microwells were coated with C. burnetii phases I and II. The optical density ratio of the sample and the positive control (S/P) was calculated for each sample as follows:
Samples with an S/P value greater than 50% were considered positive; values between 40% and 50% were deemed doubtful, and those less than 40% were determined to be negative. Doubtful results were considered negative.
DNA extraction and PCR
Genomic DNA was extracted from whole blood, using the commercial DNeasy Blood and Tissue Kit (Qiagen, Melbourne, Australia) according to the manufacturer’s instructions. The extracted DNA was stored at −20°C until use. The commercial AccuPower HotStart PCR Premix Kit (Bioneer, Daejeon, South Korea) was used for PCR amplification. Multiple primer sets were used to amplify 16S rRNA of the genus Coxiella. Coxiella was first screened by nested PCR (nPCR), as previously described [6,17]. First-round PCR was performed with the primers Cox16SF1 (5′-CGTAGGAATCTACCTTRTAGWGG-3′) and Cox16SR2 (5′-GCCTACCCGCTTCTGGTACAATT-3′), which produced amplicons with 1,321–1,429 bp. Then, nPCR was performed using the primers Cox16SF2 (5′-TGAGAACTAGCTGTTGGRRAGT-3′) and Cox16SR2, which produced amplicons with 624–627 bp. Samples yielding amplicons of the expected size were sequenced using the primers Cox16SF1 and Cox16SR1 (5′-ACTYYCCAACAGCTAGTTCTCA-3′), which produced amplicons with 719–826 bp. All PCR amplifications were performed using the Mastercycler Pro (Eppendorf, Hamburg, Germany), with a pre-denaturation cycle at 93°C for 3 min, followed by 30 cycles of denaturation at 93°C for 30 s, annealing at 56°C for 30 s, and polymerization at 72°C for 1 min, with a final post-polymerization cycle at 72°C for 5 min. PCR products of the second round of amplification were evaluated by electrophoresis, using 10 μl of the reaction mixture and a 100 bp DNA ladder (Bioneer) in 1.5% agarose gel for 30 min at 100 V, and visualized using UV transillumination, after ethidium bromide staining.
DNA sequencing and phylogenetic analysis
Purified amplicons, obtained from nPCR using the primers Cox16SF1 and Cox16SR1, were sent to Solgent (Daejeon, South Korea) for nucleotide sequencing. The sequences were analyzed using the multiple sequence alignment program CLUSTAL Omega (ver. 1.2.1). Alignment results were corrected using BioEdit (ver. 7.2.5). Phylogenetic analysis was performed using MEGA (ver. 6.0) and the aligned sequences of Coxiella 16S rRNA were compared to determine homology. Stability of the trees obtained was estimated by bootstrap analysis with 1,000 replicates.
Statistical analysis
Chi-square test was used to analyze significant differences among the groups. Data for the “unknown” group were disregarded in the chi-square test. A p value of < 0.05 was considered statistically significant. The analytical software package GraphPad Prism version 5.04 (GraphPad Software Inc., La Jolla, CA, USA) was used for statistical analysis. A confidence interval (CI) of 95% was calculated for all estimates.
Results
Serological and molecular analyses
As shown in Table 1, the sera of 11 horses (1.3%, 95% CI: 0.6–2.1) tested positive for C. burnetii by ELISA. In addition, the sera of six horses (0.7%, 95% CI: 0.2–1.3) tested positive for CLB by 16S rRNA sequencing. With respect to region, sex, and breed, no statistically significant differences were observed. However, prevalence was relatively high (2.4% by ELISA and 1.7% by PCR) in the northern region compared to other regions. In Jeju Island, none of the 94 tested samples were positive for C. burnetii, but one (1.1%, 95% CI: 0–3.1), a native Korean pony, tested positive for CLB by PCR. Although some thoroughbreds were positive by either ELISA or PCR, none tested positive by both assays. When PCR data were analyzed by age, prevalence was observed to be significantly higher (p < 0.05) in horses less than 5 years of age (1.8%, 95% CI: 0.2–3.5).
Prevalence based on ELISA and PCR data
None of the samples tested positive by both assays. Six (0.7%) samples were PCR+/ELISA−, 11 (1.3%) samples were PCR−/ELISA+, and 799 (97.9%) samples were PCR−/ELISA− (Table 2).
Table 2. Comparison of Coxiella detection by ELISA and PCR.
| PCR | Total | |||
|---|---|---|---|---|
| No. positive | No. negative | |||
| ELISA | No. positive | 0 | 11 | 11 |
| No. negative | 6 | 799 | 805 | |
| Total | 6 | 810 | 816 |
DNA sequencing and phylogenetic analysis
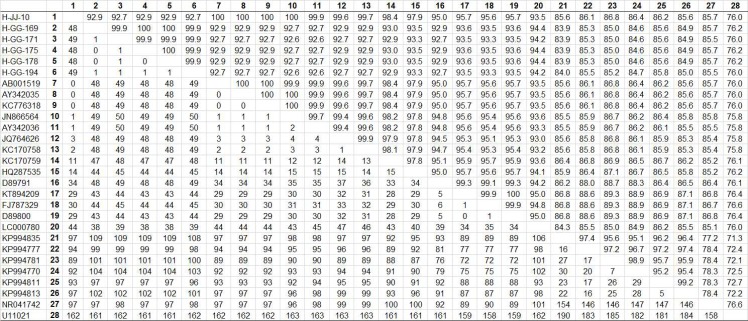
Among the six samples producing an amplicon of the expected size from Coxiella 16S rRNA, one sequence (H-JJ-10) from Jeju Island and five sequences (H-GG-169, 171, 175, 178, and 194) from the northern region of South Korea were studied. This analysis revealed that these sequences shared 92.7–100% similarity. The sequences of Coxiella 16S rRNA obtained from six horses were deposited in GenBank (accession nos. KT835658–KT835661, KU324470–KU324471). Comparative analysis of the 16S rRNA nucleotide sequences from the Korean samples with the 22 Coxiella isolates included in the GenBank database is shown in Fig 2.
Fig 2. Comparison of Coxiella 16S rRNA nucleotide sequences.
The upper matrix shows percent identity between the partial sequences of the Coxiella 16S rRNA gene. The lower matrix presents the number of differences in nucleotide bases.
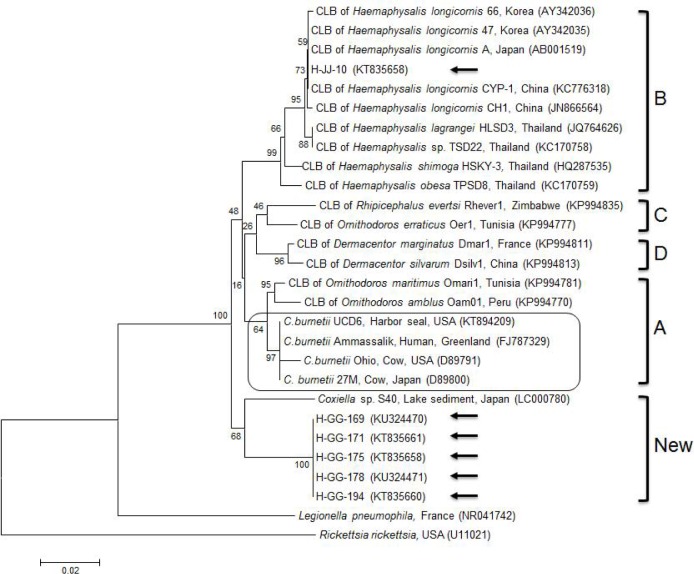
Coxiella 16S rRNA sequences were compared to published sequences (available in GenBank). This phylogenetic analysis (Kimura/neighbor joining) showed that one sequence (H-JJ-10) from Jeju Island shared high similarity (97.9–100%) with those of nine CLB strains, within clade B, isolated from Haemaphysalis ticks, in South Korea (AY342035, AY342036), Japan (AB001519), China (KC776318, JN866564), and Thailand (JQ764626, KC170758, KC170759, and HQ287535) (Figs 2 and 3). In contrast, five sequences (H-GG-169, 171, 175, 178, and 194) from the northern region were clustered into a new clade, showing 94.2–94.4% similarity to Coxiella sp. S40 from Japan (LC000780), again within the same new clade (Figs 2 and 3).
Fig 3. Phylogenetic tree constructed using Kimura/neighbor-joining methods based on 16S rRNA sequences of Coxiella.
The four clades (A–D) of Coxiella are categorized. The sequences of Coxiella-like bacteria obtained in this study are marked using arrows. The C. burnetii group is found within clade A. The accession numbers of sequences obtained from GenBank are shown with the sequence names and countries of origin. Numbers on the branches indicate bootstrap support (1,000 replicates). Scale bar indicates a phylogenetic distance of 0.02 nucleotide substitution per position. CLB = Coxiella-like-bacteria.
Discussion
In 1997, the development of 16S rRNA sequencing led to the first instance of identification of CLB in three species of ticks [18]. The sequences of CLB 16S rRNA were closely related to those of C. burnetii, which indicated diversity within the genus Coxiella, a previously overlooked aspect [19]. C. burnetii and CLB are genetically and ecologically distinct, despite genetic similarities. Furthermore, CLB are exceptionally diverse and widespread in ticks, but rarely described in domestic animals; however, CLB have recently been reported to be a leading cause of lethal systematic infections in domestic birds [20–22]. There is an important risk of misidentification, given that the current protocols for detecting C. burnetii in ticks depend on PCR-based detection of a single gene, without subsequent confirmation by sequencing [23].
An increase in the seroprevalence of C. burnetii in ruminant herds is a useful index for studying their occurrence in humans [24]. This could also be extrapolated to C. burnetii infections in horses. The primary route of transmission to humans is via nasal inhalation, and the rate of tick-borne transmission of Q fever in humans is considered low. However, some cases of possible tick-borne transmission have been reported [25, 26]. Those studies described patients with serological and clinical evidence of a tick-borne disease and subsequent or concomitant Q fever after tick bites. Furthermore, a case report indicates that C. burnetii could be spread by various ticks during horseback riding [27]. This might be a different approach of transmission involving horses. Our study confirms the results of a previous study indicating that humans can be exposed to C. burnetii from infected horses through contact with ticks during horseback riding; contaminated stable materials could also expose humans to polluted air or dust from the environment. In South Korea, the popularity of horseback riding has surpassed that of horseracing.
Recently, a meta-analysis of studies performed over an extended period across countries showed that the mean seroprevalence of C. burnetii in horses was expected to be 15.8% (95% CI: 9.6–23%) [7]. However, seroprevalence rates differ with the geographical area, test design, population, cut-off value used, year, diagnostic method, and sensitivity and specificity of the assay [7]. In the present study, the prevalence of C. burnetii was found to be 1.3% by ELISA and 0.7% by PCR among the 816 horses tested. Although we initially expected to detect C. burnetii in horses by PCR, the sequencing data indicate that only CLB were detected in horses. Because CLB are known to exist in the salivary glands of ticks, they could be transmitted to humans and vertebrates during blood sucking. Potential tick-to-vertebrate transmission of CLB is likely because ticks occur worldwide and feed on various hosts [5]. Because CLB detection has been restricted to ticks, CLB might pose a much lesser threat to vertebrates than C. burnetii [23]. However, the risk of vertebrate infection by CLB is unknown, as these bacteria have not been detected in vertebrates or associated with clinical symptoms [6]. Thus, to the best of our knowledge, the present study is the first to report the detection of CLB in a mammalian species, namely, the horse. Further studies should be performed to determine whether CLB infection manifests the clinical signs of the disease in mammals, including humans.
This study found relatively low positive yield of Coxiella, compared to other studies on ruminants in South Korea. A previous study using ELISA showed a detection rate of 24.2% (119/492) in dairy cattle and 54% (175/324) in bulk milk tanks in the southern regions [12]. Other studies also showed high infection rates in native Korean goats by ELISA (19.1%; 114/597) and PCR (9.5%; 57/597) in the central and southern regions [13], and wild Korean water deer by ELISA (9.2%; 18/196) and real-time PCR (6.6%; 13/196) in the northern, central, and southern regions [14]. Horse studies performed in other countries showed relatively low positive rates in aborted fetuses (0%; 0/122) by using PCR in Italy [7], blood (0%; 0/105) by complement fixation test in Denmark [28], aborted fetuses (1.5%; 6/407) by real-time PCR in France [10], and aborted fetuses (4.3%; 1/23) by real-time PCR in Germany [9]. However, much higher positivity was reported in aborted pregnancies (42.2%; 19/45) by PCR in Croatia [29], blood (22.2%; 4/18) by loop-mediated isothermal amplification in China [30], aborted or non-aborted placenta (7.7%; 3/39) by real-time PCR in the Netherlands [11], and blood (12.5%; 14/112) and urine (7.1%; 1/14) by real-time PCR in Australia [31].
The health status of horses that yielded positive results for C. burnetii could provide better insight into the pathogenesis of C. burnetii; however, this was not recorded in this study. While the data indicated no regional variations, the northern region did have a higher prevalence, as only one horse from Jeju Island was determined to be positive by PCR. This result was unexpected, as infectivity is likely to be observed in regions with a warm and wet climate, such as Jeju Island. However, the one native Korean pony infected with CLB was raised on Jeju Island. All horses of Jeju Island used in this experiment were native Korean ponies. Therefore, additional investigation is required for other breeds of horses reared on Jeju Island. The recent climate change has probably contributed to the widespread distribution of ticks [32] and an increase in the period of activity. With respect to breed, prevalence was mostly detected in thoroughbred horses, except one native Korean pony. Higher prevalence of Coxiella in thoroughbreds was expected based on the geographical distribution of thoroughbreds (46.7%, 12,066 horses were raised in South Korea) [15]. No significant difference was observed with respect to sex. Younger horses (< 5 years) showed significantly higher prevalence of CLB by PCR. This result was also unexpected because older animals are likely to have had more opportunities for exposure than younger animals [12,13].
In this study, 11 (1.3%) and six (0.7%) of the 816 horse blood samples tested positive for Coxiella by ELISA and PCR, respectively. None of the samples tested positive by both assays. The PCR assay employed detects genomic DNA common to all Coxiella species, including C. burnetii and CLB, while the ELISA is based on the specific detection of serum antibodies against only C. burnetii. Hence, one plausible explanation for the lack of consistency observed is that only CLB, but not C. burnetii, were present in the PCR-positive samples. Another possible explanation for this inconsistency may be the detection limit of ELISA, and the fact that this assay can detect antibodies from both active and prior infections. PCR can solely detect active infections [33]. The ID screen ELISA kit is adapted to mammalian IgG antibodies, and it was originally developed to react with cattle, goats, and sheep. However, it has also been used to react with antibodies against C. burnetii in blood samples from other mammalian species, including cats, foxes, and rodents [34].
Genotypically and phenotypically different features of CLB and C. burnetii could also conceivably lead to different rates of positivity. The widespread genetic variability in CLB strains compared to C. burnetii strains has led to a clear sub-classification of this genus into four largely divergent clades (A–D) [5]. The clustering of all C. burnetii strains within clade A indicates that the progenitor of C. burnetii was a tick-related bacterium that succeeded in infecting vertebrates. Based on the phylogenetic analysis, H-JJ-10 on Jeju Island is closely related to the CLB in Haemaphysalis ticks belonging to clade B. Therefore, further studies on CLB in ticks associated with horses are required, especially on Jeju Island. The H-JJ-10 clustered together with the CLB in Haemaphysalis ticks from South Korea, China, Japan, and Thailand, which implies a close epidemiological connection between these isolates. The five isolates from the northern region were placed into a new distinct clade. Because of the geographical differences between the northern region of the mainland and Jeju Island, the origin of Coxiella may differ, contributing to the diversity of this species in South Korea. Further molecular studies are needed to fully understand the diversity of the genus Coxiella.
To the best of our knowledge, this is the first report of a large-scale nationwide study to report the serological and molecular detection of the genus Coxiella in mammals, namely, horses. Nevertheless, further studies will be required to describe these CLB isolates, characterize their genetic relationship, and evaluate their potential to cause infections in vertebrates [5]. Considering the zoonotic potential of the genus Coxiella and climate change, which affects the widespread distribution of ticks and increases their period of activity, it is necessary to establish strategic monitoring, epidemiological insight, and appropriate control programs for Coxiella and other tick-borne diseases. Future research on possible cross-reactivity between C. burnetii and CLB will be essential to better evaluate the specificity of diagnostic assays and screening tools now used in vertebrates [5]. In addition, further epidemiological studies on Coxiella species in healthy horse herds, epizootics in horses, and positive cases of horse abortion are significant for better understanding C. burnetii or CLB infections.
Conclusions
C. burnetii can be transmitted to humans following exposure to ticks during horseback riding. Global warming is expected to cause an increase in the number and distribution of ticks. To the best of our knowledge, this is the first large-scale nationwide investigation to study the prevalence of C. burnetii and CLB among 816 horses in South Korea. Eleven (1.3%) and six (0.7%) samples tested positive by ELISA and PCR, respectively. This study describes the first instance of CLB detection (previously restricted to ticks) in mammals, thereby providing evidence about the potential for transmission of Q fever-causing bacteria to humans from infected horses during activities such as horseback riding.
Data Availability
All relevant data are within the paper.
Funding Statement
This research was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (Grant No. NRF-2013R1A1A2013102). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The Smile Equine Clinic provided support in the form of salary for author EC, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of this author are articulated in the "author contributions" section.
References
- 1.Babudieri B. Q fever: a zoonosis. Adv Vet Sci. 1959; 5:81–181. [Google Scholar]
- 2.Maurin M, Raoult D. Q fever. Clin Microbiol Rev. 1999; 12:518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980; 30:225–420. [PubMed] [Google Scholar]
- 4.Tan CK, Owens L. Infectivity, transmission and 16S rRNA sequencing of a rickettsia, Coxiella cheraxi sp. nov., from the freshwater crayfish Cherax quadricarinatus. Dis Aquat Organ. 2000; 41:115–122. [DOI] [PubMed] [Google Scholar]
- 5.Duron O, Sidi-Boumedine K, Rousset E, Moutailler S, Jourdain E. The importance of ticks in Q fever transmission: What has (and has not) been demonstrated? Trends Parasitol. 2015; 31:536–552. 10.1016/j.pt.2015.06.014 [DOI] [PubMed] [Google Scholar]
- 6.Duron O, Noël V, McCoy KD, Bonazzi M, Sidi-Boumedine K, Morel O, et al. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog. 2015; 11:e1004892 10.1371/journal.ppat.1004892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marenzoni ML, Stefanetti V, Papa P, Casagrande Proietti P, Bietta A, Coletti M, et al. Is the horse a reservoir or an indicator of Coxiella burnetii infection? Systematic review and biomolecular investigation. Vet Microbiol. 2013; 167:662–669. 10.1016/j.vetmic.2013.09.027 [DOI] [PubMed] [Google Scholar]
- 8.Acland HM. Abortion in mares In: McKinnon AO, Voss JL. Equine, reproduction. Philadelphia: Lea & Febiger; 1993. pp. 554–562. [Google Scholar]
- 9.Runge M, Hilbert A, Henning K. Contribution to the occurrence of Coxiella burnetii-infection in horses. Praktische Tierarzt. 2012; 93:220–222. [Google Scholar]
- 10.Leon A, Richard E, Fortier C, Laugier C, Fortier G, Pronost S. Molecular detection of Coxiella burnetii and Neospora caninum in equine aborted foetuses and neonates. Prev Vet Med. 2012; 104:179–183. 10.1016/j.prevetmed.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 11.Roest HI, van Solt CB, Tilburg JJ, Klaassen CH, Hovius EK, Roest FT, et al. Search for possible additional reservoirs for human Q fever, the Netherlands. Emerg Infect Dis. 2013; 19:834–835. 10.3201/eid1905.121489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouh IO, Seo MG, Do JC, Kim IK, Cho MH, Kwak DM. Seroprevalence of Coxiella burnetii in bulk-tank milk and dairy cattle in Gyeongbuk province, Korea. Korean J Vet Serv. 2013; 36:243–248. [Google Scholar]
- 13.Jung BY, Seo MG, Lee SH, Byun JW, Oem JK, Kwak D. Molecular and serologic detection of Coxiella burnetii in native Korean goats (Capra hircus coreanae). Vet Microbiol. 2014; 173:152–155. 10.1016/j.vetmic.2014.06.029 [DOI] [PubMed] [Google Scholar]
- 14.Shin GW, Kim EJ, Lee HB, Cho HS. The prevalence of Coxiella burnetii infection in wild Korean water deer, Korea. J Vet Med Sci. 2014; 76:1069–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ministry of Agriculture, Food and Rural Affairs, South Korea. Report of “A fact finding survey of horse industry in South Korea during 2014”. 2015.
- 16.Thrusfield M. Veterinary epidemiology, 3rd ed. Oxford: Wiley Blackwell; 2005. pp. 228–330. [Google Scholar]
- 17.Duron O, Jourdain E, McCoy KD. Diversity and global distribution of the Coxiella intracellular bacterium in seabird ticks. Ticks Tick Borne Dis. 2014; 5:557–563. 10.1016/j.ttbdis.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 18.Noda H, Munderloh UG, Kurtti TJ. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol. 1997; 63:3926–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong J. Coxiella-like endosymbionts In: Toman R. et al. Coxiella burnetii: Recent advances and new perspectives in research of the Q fever bacterium. New York: Springer; 2012. pp. 365–379. [Google Scholar]
- 20.Shivaprasad HL, Cadenas MB, Diab SS, Nordhausen R, Bradway D, Crespo R, et al. Coxiella-like infection in psittacines and a toucan. Avian Dis. 2008; 52:426–432. [DOI] [PubMed] [Google Scholar]
- 21.Vapniarsky N, Barr BC, Murphy B. Systemic Coxiella-like infection with myocarditis and hepatitis in an eclectus parrot (Eclectus roratus). Vet Pathol. 2012; 49:717–722. 10.1177/0300985811409251 [DOI] [PubMed] [Google Scholar]
- 22.Woc-Colburn AM, Garner MM, Bradway D, West G, D'Agostino J, Trupkiewicz J, et al. Fatal coxiellosis in swainson's blue mountain rainbow lorikeets (Trichoglossus haematodus moluccanus). Vet Pathol. 2008; 45:247–254. 10.1354/vp.45-2-247 [DOI] [PubMed] [Google Scholar]
- 23.Smith TA, Driscoll T, Gillespie JJ, Raghavan R. A Coxiella-like endosymbiont is a potential vitamin source for the Lone Star tick. Genome Biol Evol. 2015; 7:831–838. 10.1093/gbe/evv016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgiev M, Afonso A, Neubauer H, Needham H, Thiery R, Rodolakis A, et al. Q fever in humans and farm animals in four European countries, 1982 to 2010. Euro Surveill. 2013; 18:pii = 20407 [PubMed] [Google Scholar]
- 25.Eklund CM, Parker RR, Lackman DB. A case of Q fever probably contracted by exposure to ticks in nature. Public Health Rep. 1947; 62:1413–1416. [PubMed] [Google Scholar]
- 26.Rolain JM, Gouriet F, Brouqui P, Larrey D, Janbon F, Vene S, et al. Concomitant or consecutive infection with Coxiella burnetii and tickborne diseases. Clin Infect Dis. 2005; 40:82–88. [DOI] [PubMed] [Google Scholar]
- 27.Nett RJ, Book E, Anderson AD. Q Fever with unusual exposure history: a classic presentation of a commonly misdiagnosed disease. Case Rep Infect Dis. 2012; 916142 10.1155/2012/916142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agerholm JS, Svejstrup CA, Christoffersen AB, Agger JFG. Apparent seroprevalence for Coxiella burnetii in the Danish horse population. Dansk Veterinaertidsskrift. 2015; 98:26–29. [Google Scholar]
- 29.Račić I, Spičić S, Galov A, Duvnjak S, Zdelar-Tuk M, Vujnović A, et al. Identification of Coxiella burnetii genotypes in Croatia using multi-locus VNTR analysis. Vet Microbiol. 2014; 173:340–347. 10.1016/j.vetmic.2014.08.016 [DOI] [PubMed] [Google Scholar]
- 30.Pan L, Zhang L, Fan D, Zhang X, Liu H, Lu Q, et al. Rapid, simple and sensitive detection of Q fever by loop-mediated isothermal amplification of the htpAB gene. PLoS Negl Trop Dis. 2013; 7:e2231 10.1371/journal.pntd.0002231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tozer SJ, Lambert SB, Strong CL, Field HE, Sloots TP, Nissen MD. Potential animal and environmental sources of Q fever infection for humans in Queensland. Zoonoses Public Health. 2013; 61:105–112. 10.1111/zph.12051 [DOI] [PubMed] [Google Scholar]
- 32.Dantas-Torres F. Climate change, biodiversity, ticks and tick-borne diseases: The butterfly effect. Int J Parasitol Parasites Wildl. 2015; 4:452–461. 10.1016/j.ijppaw.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirkan S, Kaya O, Tekbiyik S, Parin U. Detection of Coxiella burnetii in cattle by PCR. Turk. J. Vet. Anim. Sci. 2008; 32:215–220. [Google Scholar]
- 34.Meredith AL, Cleaveland SC, Denwood MJ, Brown JK, Shaw DJ. Coxiella burnetii (Q-fever) seroprevalence in prey and predators in the United Kingdom: evaluation of infection in wild rodents, foxes and domestic cats using a modified ELISA. Transbound. Emerg. Dis. 2015; 62:639–649. 10.1111/tbed.12211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.