Abstract
Several key transcription factors regulate cell growth, survival, and differentiation during neural crest and melanoblast development in the embryo, and these same pathways may be reactivated in tumors arising from the progenitors of these cells. The transcription factors PAX3 and FOXD3 have essential roles in melanoblasts and melanoma. In this study, we define a regulatory pathway where FOXD3 promotes the expression of PAX3. Both factors are expressed in melanoma cells and there is a positive correlation between the transcript levels of PAX3 and FOXD3. The PAX3 gene contains two FOX binding motifs within highly conserved enhancer regulatory elements that are essential for neural crest development. FOXD3 binds to both of these motifs in vitro but only one of these sites is preferentially utilized in melanoma cells. Overexpression of FOXD3 upregulates PAX3 levels while inhibition of FOXD3 function does not alter PAX3 protein levels, supporting that FOXD3 is sufficient but not necessary to drive PAX3 expression in melanoma cells. Here, we identify a molecular pathway where FOXD3 upregulates PAX3 expression and therefore contributes to melanoma progression.
Keywords: Melanoma, Transcription factors, DNA-protein interactions, Gene regulation, PAX3, FOXD3
INTRODUCTION
PAX3 (Paired box protein 3) is a transcription factor essential during melanocyte development and is aberrantly expressed in melanoma [Plummer et al., 2008]. In normal melanocyte development, PAX3 is down-regulated upon terminal differentiation but its expression is maintained in melanoma cells. PAX3 acts as an oncogene in melanoma by driving the expression of genes promoting cellular migration, invasion, and metastasis [Bonvin et al., 2012]. While the mechanism responsible for promoting and maintaining PAX3 expression in melanoma is unknown, some insight may be gained from developmental studies. During development, PAX3 expression is maintained by TEAD2, PBX, and POU factors in the neural crest, a population of cells that ultimately gives rise to melanocytes [Chang et al., 2008; Milewski et al., 2004; Pruitt et al., 2004; Smit et al., 2000]. STAT3 and SMADs regulate PAX3 expression in normal melanocytes [Dong et al., 2012; Yang et al., 2008], and the POU proteins OCT1, BRN1 and BRN2 bind to the PAX3 promoter in murine B16 melanoma cells [Zhu and Pruitt, 2005]. Factors that drive or inhibit expression of PAX3 through upstream genetic elements within human melanoma cells are unknown. Inhibition of upstream factors driving PAX3 expression may lead to the down-regulation of PAX3 and, therefore, a loss of oncogenic function within melanoma cells.
The transcription factor FOXD3 is a member of the Forkhead Box (FOX) protein family, a group of factors responsible for lineage decisions during embryonic development [Clark et al., 1993]. FOXD3 expression in the developing neural crest retains a population of uncommitted daughter cells, while loss of FOXD3 leads to a concurrent premature differentiation and reduction of these neural crest-derived cells [Dottori et al., 2001; Teng et al., 2008]. As the neural crest undergoes commitment to lineage specificity, FOXD3 blocks cells from undergoing melanocyte differentiation [Kos et al., 2001; Thomas and Erickson, 2009]. FOXD3 is also expressed in melanoma, regulated by B-RAF, and over-expressed as a possible adaptive response during BRAF-inhibitor drug resistance [Abel and Aplin, 2010; Basile et al., 2012; Katiyar and Aplin, 2011]. The function of FOXD3 in melanoma is not well understood, but it is thought to parallel its developmental role as a transcriptional regulator of genes promoting stem cell-like qualities while simultaneously blocking terminal differentiation.
In this report, we determine that FOXD3 directly binds to enhancers in the PAX3 locus and promotes the expression of this protein. Examination of the PAX3 promoter identified two FOX consensus motifs, both of which are sufficient to activate the PAX3 promoter in 293T cells. Melanoma cells, however, preferentially utilize only one of these motifs for PAX3 promoter activation. We find that FOXD3 overexpression causes an increase in PAX3 levels but inhibition of the transcription factor activity of FOXD3 is not sufficient to reduce PAX3 levels. These data support that FOXD3 is sufficient but not necessary for PAX3 expression in melanoma cells. Here, we discover a molecular pathway where the transcription factor FOXD3 promotes PAX3 expression, thereby promoting melanoma advancement.
MATERIALS AND METHODS
CELL CULTURE AND TRANSFECTION
Human melanoma lines (A375, mel-537, mel-624, mel-888, SKMEL-5, SKMEL-23, and SKMEL-28), 293T, HELA and 3T3 cells (ATCC and University of Chicago Comprehensive Cancer Center Core Facilities) were cultured in DMEM/10% FBS (Sigma). Morphology and melanoma-marker testing verified melanoma cell identity. All cells were negative for the presence of mycoplasma. Cells were transfected with Lipofectamine 2000 (Life Technologies) reagent following manufacturer’s protocols.
WESTERN BLOTS
Cells were lysed in RIPA buffer and 50μg total protein was separated on 7.5% Bis-Tris gels, transferred to nitrocellulose membranes (Bio-Rad), then probed with 1:200 PAX3 (University of Iowa Hybridoma Bank), 1:500 FOXD3 (BioLegend), or 1:1,000,000 vinculin antibody (Sigma).
QUANTITATIVE REAL-TIME POLYMERASE CHAIN REACTION (QPCR)
The expression of FOXD3, PAX3 and GAPDH transcripts was measured utilizing iScript cDNA Synthesis kit and the iTaq Universal SYBR Green Supermix and CFX Connect Real-Time System (Bio-Rad). Each reaction was performed in triplicate. Primer sequences are as follows: PAX3 forward TGA GCG AGC GAG CCT CAG, PAX3 reverse TCT GAC ACA GCT TGT GGA AT, hGAPDH forward ACA TCA TCC CTG CCT CTA CT, hGAPDH reverse CTC TCT TCC TCT TGT GCT CTT G, FOXD3 forward GCT GGT CGA GCA AAC TCA CCG C, and FOXD3 reverse CAA AGA ATT TCC CTC CCA TCC C. Values were normalized to GAPDH mRNA levels.
EXPRESSION AND REPORTER DNA VECTORS
The PAX3 reporter construct (pGL2-PAX3pm) contains a 5’ 1.6Kb region upstream of the murine pax3 start codon [Li et al., 1999] cloned into a pGL2 reporter vector. Mutation of putative FOX sites within pGL2-PAX3pm were created by site directed mutagenesis, by changing sites (F1 and F2, as shown in Fig. 4) to NotI restriction sites. The PAX3 expression construct was created as described [Lang and Epstein, 2003]. The pcS2-mFoxD3 expression construct, containing 75 bp of 5’ UTR, 600 bp of 3’ UTR, and a complete cDNA for mouse FOXD3, was kindly provided by P. Labosky, Vanderbilt University [Labosky and Kaestner, 1998]. For the FOXD3 dominant negative construct (FOXD3-DN), the C-terminal end of FOXD3 (amino acids 232-369) was cloned into pcDNA3 using primers CAG GAT CCA TGC GGC GCC GGA AAC GCT TCA A and CAG AAT TCC TAT TGC GCC GGC CAT TTG GCT T.
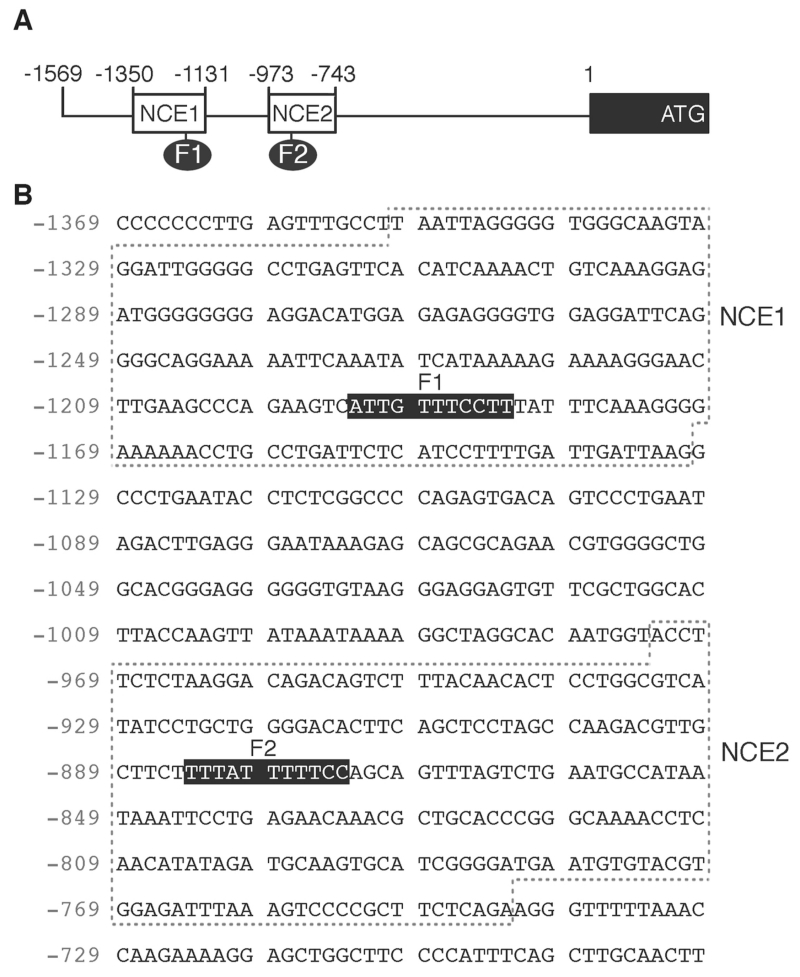
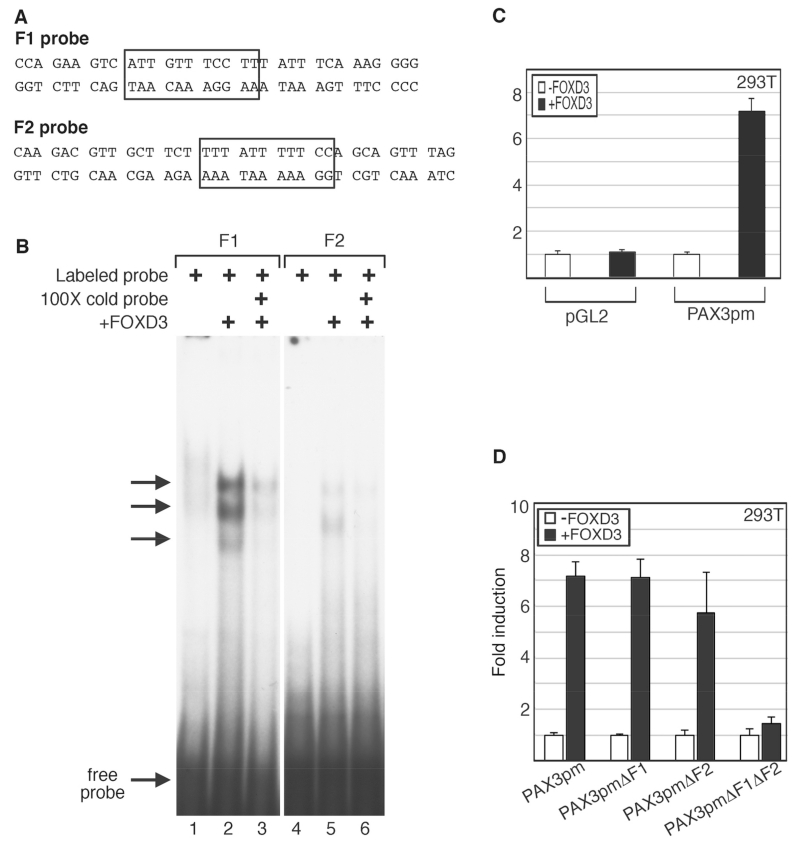
Fig. 4.
The PAX3 promoter contains two highly conserved FOXD3 sites, F1 and F2. (A) Schematic of the PAX3 promoter, with the locations of the ATG, CDS start, and conserved elements NCE1 and NCE2. Two putative FOXD3 sites, F1 and F2, are indicated. (B) Sequence of the NCE1 and NCE2 enhancer islands within the PAX3 promoter. The sequence numbering is in relation to the translational start site, which is numbered +1. The dotted boxes indicate the NCE1 and NCE2 islands of sequence homology between human and mouse sequences. Putative FOX long sites are shown by black boxes.
ELECTROPHORETIC MOBILITY SHIFT ASSAY (EMSA)
Protein lysate samples were prepared from 293T cells transfected with FOXD3. The reaction was performed in 50 mM Tris, HCl pH 7.5, 50 mM NaCl, 4% glycerol, 1mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 50μg/ml poly (dI-dC) (USB) and 500μg/ml BSA. For probes, primer sequences for the PAX3 promoter are CCA GAA GTC ATT GTT TCC TTT ATT TCA AAG GGG and CAA GAC GTT GCT TCT TTT ATT TTT CCA GCA GTT TAG for the FOXD3 F1 and F2 sites, respectively. Annealed probes were labeled by end filing with T4 polynucleotide kinase (New England Biolabs) and purified on microspin G-50 columns (GE healthcare Biosciences). Nuclear extracts were incubated for 30 minutes on ice with labeled probe. Electrophoresis was performed on 7% native gels. Gels were dried and exposed to Kodak Biomax MS film (Carestream Health).
LUCIFERASE ASSAYS
Cells were transfected with pGL2-PAX3pm reporter constructs, pCMV-beta-galactosidase (Clontech) as an internal control, with or without PAX3 and/or FOXD3 expression constructs. The total amount of DNA transfected was kept constant by the addition of empty vector. Cells were transfected, incubated for 48 hours, then luciferase and beta-galactosidase levels were measured (Promega). Luciferase activity was normalized to the internal control beta-galactosidase with all experiments performed in triplicate.
CHROMATIN IMUNOPRECIPITATION (CHIP) ASSAYS
Melanoma cells (SKMEL-28 and A375) were fixed in 1% formaldehyde and quenched in 0.125 M glycine then processed according to manufacturer’s protocol (EMD Millipore). For immunoprecipitation of FOXD3 DNA complexes, 1μl of FOXD3 polyclonal antibody (Cell Signaling) was added per experimental reaction. Normal IgG (Sigma) was used as a negative control against non-specific DNA precipitation by an antibody. Nested PCR was performed with primers to the PAX3 enhancer, first with TTG TGG ACT GCT GAG CGG CTA TAC C (F) and TAT TCA GGG ACT GTC ACT CCG G (R) then a second PCR reaction with primers CAT CTT GCC TTC TGA GTT TGC C (F) and AAA AGG ATG AGA ATC GGG CAG G (R). All ChIP samples were tested for false positive PCR amplification using primers that amplify a sequence from within the fourth large coding exon of the beta-tubulin gene to control against genomic DNA contamination. The nested primer set for this control was AAA GGC CAC TAC ACA GAG GG (F) and TAC CAA CTG ATG GAC GGA GAG G (R) for the first PCR reaction, then TTG ATT CTG TCC TGG ATG TGG (F) and TCA GAC ACT TTG GGT GAA GGC(R) for the second PCR round.
STATISTICAL ANALYSIS
Correlation coefficients between FOXD3 and PAX3 were determined using Spearman rank tests. Significance of the differences between variable conditions was determined by Graphpad Prism statistical software (version 5.0) utilizing the Student’s t-test and Chi-square analysis with a confidence interval of 95%. All values stated as significant have p values of less than or equal to 0.05 unless indicated.
RESULTS
MELANOMA CELL LINES EXPRESS FOXD3 AND PAX3
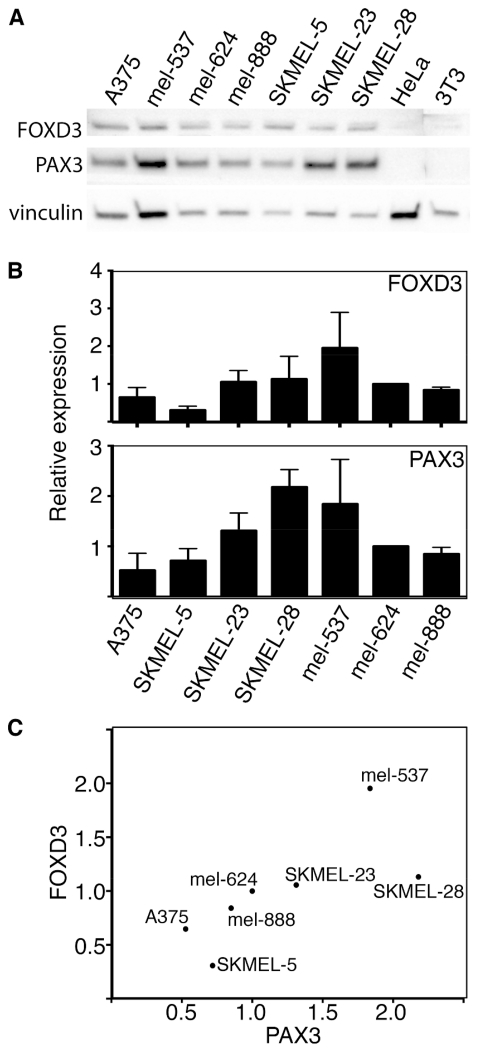
FOXD3 and PAX3 protein and transcript expression levels were determined in seven melanoma cell lines by western analysis and Real-time PCR (qPCR). Both proteins are expressed in all lines tested to variant degrees (Fig. 1A). Cell lines HELA and 3T3 were included as negative controls for PAX3 and FOXD3. Transcript levels of FOXD3 and PAX3 are positively correlated to each other when compared using Spearman rank correlation tests (p=0.0067)(Fig. 1B,C).
Fig. 1.
Melanoma cells express FOXD3 and PAX3 with a positive correlation between the two transcripts. (A) Western analysis for FOXD3 and PAX3 protein levels in a panel of melanoma cell lines. HELA and NIH 3T3 cell lysates are included as negative controls, and vinculin levels are measured as a loading control. (B) FOXD3 and PAX3 transcript levels in melanoma cell lines, as measured by Real-Time PCR analysis. Levels for the cell line mel-624 was arbitrarily assigned as 1, and the other bars represent relative expression levels as compared to mel-624 transcripts. All levels were normalized to GAPDH. (C) Graph demonstrating the correlation between PAX3 (x-axis) and FOXD3 (y-axis) transcript levels in melanoma cell lines.
FOXD3 OVEREXPRESSION INCREASES PAX3 PROTEIN LEVELS
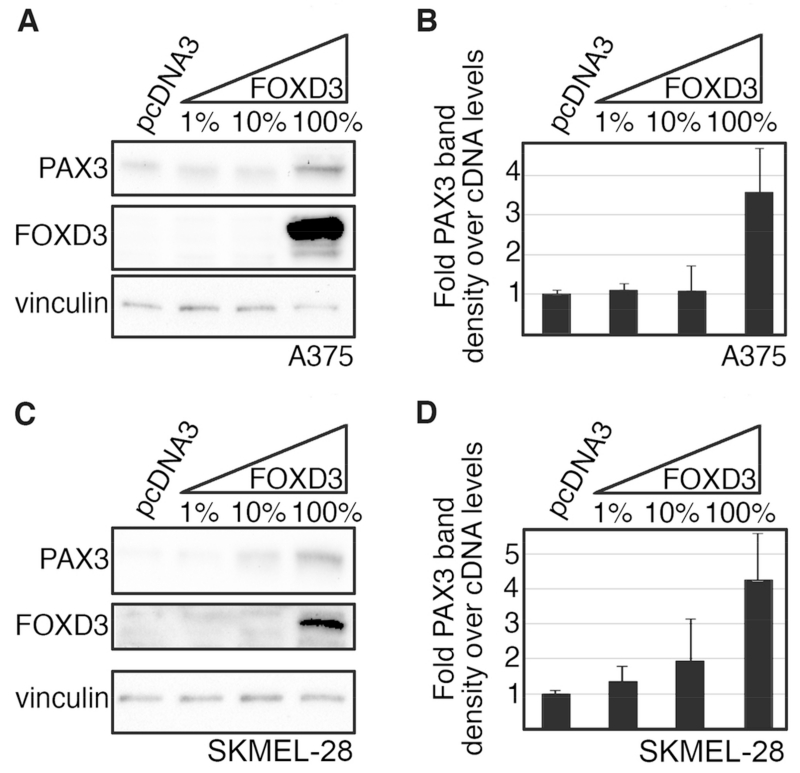
To test if FOXD3 increases PAX3 protein levels in melanoma cells, the FOXD3 construct was transfected into A375 and SKMEL-28 cells at increasing amounts. While the DNA levels were constant for each sample, the ratio of FOXD3 construct to empty vector pcDNA3 was altered, where FOXD3 consisted of 0%, 1%, 10%, and 100% of the total DNA content. Exogenous FOXD3 protein expression increased PAX3 levels in A375 and SKMEL-28 melanoma cells (Fig. 2A,C). Densitometry on these western blots show that FOXD3 increased PAX3 levels 3.6-fold ±1.1-fold and 4.2-fold ±1.3-fold in A375 and SKMEL-28 cells, respectively (Fig. 2B,D). These data support that FOXD3 is upstream of PAX3 and is sufficient to drive PAX3 expression in melanoma.
Fig. 2.
FOXD3 is sufficient to drive PAX3 expression in melanoma cells. (A-D) Increasing levels of exogenous FOXD3 leads to an increase in PAX3 in A375 (A,B) and SKMEL-28 (C,D) melanoma cells. Protein levels of PAX3, FOXD3 and vinculin were measured by western analysis in cells transfected with DNA containing increasing levels (1%, 10%, 100%) of FOXD3 expression construct (A,C). Corresponding densitometric analysis for each western blot is shown (B,D). Due to high levels of exogenous expression, endogenous levels of proteins may not be visible in some samples.
INHIBITION OF FOXD3 FUNCTION DOES NOT REDUCE PAX3 EXPRESSION IN MELANOMA CELLS
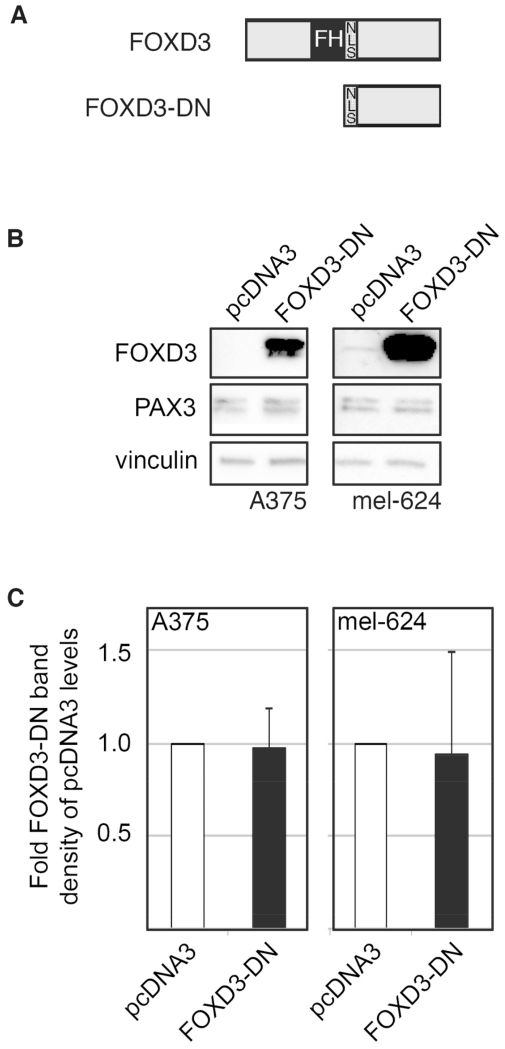
In order to determine if FOXD3 is necessary for PAX3 expression in melanoma cells, a FOXD3 dominant negative (FOXD3-DN) construct was made by cloning the C-terminal half of the protein starting with an endogenous nuclear localization signal (NLS) through the end of the protein (Fig. 3A). Transfection of FOXD3-DN into A375 and mel-624 melanoma cell lines did not reduce PAX3 protein levels (Fig. 3B,C). Similar results were seen when siRNA against FOXD3 were transfected into cells (data not shown). These data support that while FOXD3 is sufficient to drive PAX3 expression in melanoma, it is not necessary.
Fig. 3.
FOXD3 is not necessary for PAX3 expression in melanoma cells. (A) Schematics of the FOXD3 protein (top) and the portion of FOXD3 used in the FOXD3 dominant negative construct (FOXD3-DN, bottom). The C-terminal half of the FOXD3 protein starting from the nuclear localization signal (NLS) through the end of the protein was cloned into pcDNA3 to create FOXD3-DN. (B) Inhibition of FOXD3 activity does not alter PAX3 protein levels in melanoma levels. Western blots were performed on A375 and mel-624 cells transfected with the FOXD3-DN construct or pcDNA3 (as a negative control) and probed for FOXD3, PAX3, and vinculin (loading control). (C) Densitometric measurements of PAX3 levels in western blots.
THE PAX3 PROMOTER CONTAINS TWO FOX BINDING SITES CONTAINED IN TWO CONSERVED ENHANCERS
Since PAX3 levels increased in the presence of FOXD3, the PAX3 promoter was examined for FOX consensus motifs to determine if FOXD3 directly regulates the PAX3 gene. Through phylogenic footprinting and in silico promoter analysis, two highly conserved islands of homology within the proximal 1.6 Kb PAX3 promoter were previously identified, referred to as either NCE1 and NCE2 ([Milewski et al., 2004], shown schematically and by sequence in Fig. 4A,B) or sites I and II [Natoli et al., 1997]. These regions are sufficient to drive pax3 expression during murine neural crest development, where NCE2 initiates induction and dorsal restriction in the embryo and NCE1 is involved in anterior expression [Chang et al., 2008; Li et al., 2000; Li et al., 1999; Milewski et al., 2004; Pruitt et al., 2004; Sanchez-Ferras et al., 2012]. The PAX3 promoter sequence was analyzed for transcriptional factor binding motifs, including FOXD3 long high-affinity sites (A/T)(A/T)T(A/G)TTTN(T/C) N(T/C) and short core FOX DNA recognition sites T(A/G)TT(T/G)(A/G) (T/C) [Overdier et al., 1994; Sutton et al., 1996]. Two FOX long sites were identified in the PAX3 promoter, one in NCE1 (F1) and another in NCE2 (F2). To determine if FOXD3 directly binds to the predicted cis regulatory elements, EMSAs were performed (Fig. 5A,B). Nuclear extracts containing exogenous FOXD3 protein produced slower migrating bands with radioactive probes containing the F1 and F2 sites, and these bands were reduced upon competition with unlabeled probe. Here, we identify two FOX binding sites within the PAX3 promoter, and determine that FOXD3 directly binds these sites in vitro.
Fig. 5.
FOXD3 binds to the F1 and F2 elements in the PAX3 promoter and can drive expression from these elements in cells. (A) Sequences for the probes used for the EMSA analysis, with the putative FOX binding sites indicated by boxes. (B) EMSA analysis for FOXD3 binding of PAX3 promoter sequences in vitro. Labeled probe containing the PAX3 promoter F1 site (lanes 1-3) or F2 site (lanes 4-6) were mixed with 293T lysate without (lanes 1, 4) or with (lanes 2, 3, 5, 6) exogenous FOXD3 protein. Binding of the labeled probe was inhibited with cold probe competition (lanes 3, 6). (C) FOXD3 promotes expression from the PAX3 promoter. Empty vector (pGL2) or PAX3pm reporter constructs were transfected into 293T cells without (white bars) or with (black bars) FOXD3 expression constructs. (D) FOXD3 drives PAX3 promoter activity in 293T cells through the F1 and F2 sites. Reporter constructs containing the PAX3 genomic region (shown in Fig. 3A and B), driving the expression of luciferase without mutations in putative FOX sites (PAX3pm) or with mutations in F1 (PAX3pmΔF1), F2 (PAX3pmΔF2), or both (PAX3pmΔF1μΔF2), were transfected into 293T cells without (white bars) or with (black bars) a FOXD3 expression construct. For luciferase assays shown in C and D, fold induction was calculated for each sample by the measurement of arbitrary light units produced divided by light units obtained from samples transfected with vector without FOXD3. Each bar is n=9, with standard error of the mean as shown.
THE F1 AND F2 SITES WITHIN THE PAX3 PROMOTER ARE ACTIVE IN CELLS
To determine if the PAX3 promoter was active in cultured cells and responsive to FOXD3, a PAX3 promoter reporter construct (PAX3pm) was transfected into 293T cells with or without the FOXD3 expression construct. The PAX3pm reporter construct contained the 1.6Kb proximal PAX3 promoter sequence in the pGL2 luciferase vector. Exogenous FOXD3 drove reporter expression from the PAX3 promoter at significant levels, 7.2 fold ±0.5 fold over reporter alone (Fig. 5C). PAX3pm constructs with or without mutated F1 and/or F2 sites were also transfected into 293T cells in the presence or absence of a FOXD3 expression vector (Fig. 5D). FOXD3 was able to drive reporter expression at significant levels when either or both sites were present in comparison to promoter vector alone. However, when both sites were mutated, the ability of FOXD3 to drive this construct was attenuated.
THE 5’ PROXIMAL PAX3 PROMOTER IS ACTIVE IN MELANOMA CELLS
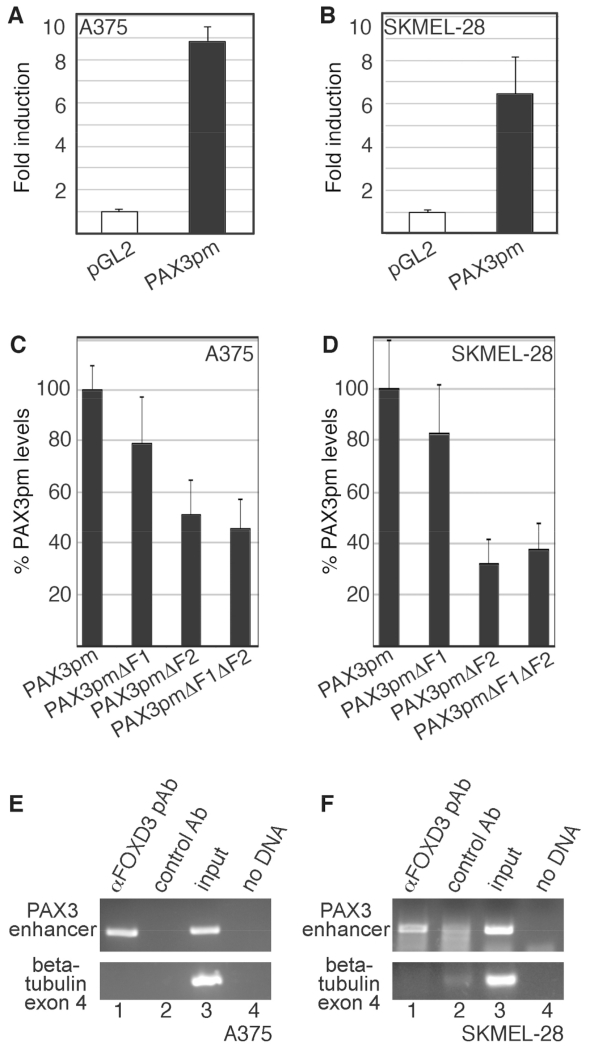
To test if FOXD3 acts on the PAX3 promoter in melanoma cells, it was first determined if the PAX3 proximal promoter element drives expression in melanoma cells. The PAX3 promoter reporter construct (PAX3pm) was transfected into A375 and SKMEL-28 melanoma cells (Fig. 6A,B). The PAX3pm produced significant levels of luciferase reporter activity in A375 and SKMEL-28 melanoma cells over empty pGL2 reporter vector (Fig. 6A,B, black bars, 8.85 ±0.65 and 6.44 ±1.72-fold induction over pGL2 vector, respectively). In 293T cells, FOXD3 was able to drive reporter expression through either of the two conserved FOXD3 binding motifs (Fig. 5C,D). To determine if these FOXD3 sites are active in melanoma cells, PAX3pm constructs containing intact or mutated F1 and F2 sites were transfected into A375 and SKMEL-28 melanoma cell lines (Fig. 6C,D). While mutation of the F1 site reduced reporter expression to 78.5% ±18.6% (A375) and 82.1% ±19.4% (SKMEL-28) of the PAX3pm levels, this reduction was not significant. However, if the F2 or both F1 and F2 sites were mutated, the reporter levels dropped to 51.1% ±13.8% and 45.6% ±11.6% for A375 cells and 32.4% ±9.8% and 37.5% ±10.2% of the PAX3pm vector levels, respectively. This supports that FOXD3 can drive PAX3 expression from both the F1 and F2 enhancers, but the F2 enhancer is the site utilized in melanoma cells.
Fig. 6.
The PAX3 1.6kb proximal promoter is active in melanoma cells, and is bound to FOXD3 in melanoma cells. (A,B) A reporter construct containing 1.6 Kb of PAX3 promoter (PAX3pm) expresses luciferase in melanoma cells. pGL2-luciferase construct without (white bars) or with (black bars) a segment of the PAX3 promoter were transfected into A375 (A) and SKMEL-28 (B) cells. (C,D) The F1 and F2 PAX3 promoter sites are active in melanoma cells, with the F2 element being a major driver of expression. PAX3 promoter constructs with or without mutations in putative FOX sites were transfected into A375 and SKMEL-28 cells. For %PAX3pm levels, values were calculated by the measurement of arbitrary light units produced by each reporter divided by the PAX3pm levels, and multiplied by 100. For luciferase assays shown in A-D, each bar represents n=9, with standard error of the mean as shown. (E,F) FOXD3 is located on the endogenous PAX3 promoter in A375 (E) and SKMEL-28 (F) cells. Chromatin immunoprecipitation (ChIP) analysis was performed with primers specific for the PAX3 promoter region (top gels) or exon 4 of the beta tubulin gene (bottom gels, negative control). Proteins were precipitated with antibodies against FOXD3 (lane 1) or normal mouse IgG (negative control, lane 2). DNA from the cell lysate was tested as a positive PCR control (lane 3), and no DNA template as a negative control (lane 4).
FOXD3 REGULATES PAX3 EXPRESSION
ChIP assays were performed to determine if endogenous FOXD3 could bind to the PAX3 promoter within melanoma cells. A PCR amplicon was generated utilizing primers specific for the PAX3 promoter region when protein was precipitated with antibodies specific for FOXD3 in A375 and SKMEL28 cells (Fig. 6E,F, lane 1). A PCR product was not generated for the following negative controls: when the immunoprecipitation was performed with normal mouse IgG, when the primers were sequence specific for the coding region of the beta-tubulin gene that does not possess FOX enhancer sites, or when no ChIP sample template was added (PCR water blank). As a positive control of the input sample, DNA was isolated from the cell lysate before immunoprecipitation and utilized as a template for the PCR reactions (Fig. 6E,F lane 3). No PCR product was generated using PAX3 promoter-specific primers and an antibody against PAX3 protein (data not shown). Although these findings do not exclude the auto-regulation by PAX3, we did not find any evidence of this in these experiments. These findings support that FOXD3 is located on the PAX3 promoter in melanoma cells.
In summary, we delineate a model where FOXD3 directly binds to two consensus motifs within the PAX3 promoter from which it promotes protein expression. While both of these binding motifs are sufficient for FOXD3 to activate the PAX3 promoter in 293T cells, FOXD3 utilizes only the F2 site within melanoma cells. Further, we find that while FOXD3 overexpression results in an increase in PAX3 expression, loss of FOXD3 function does not alter PAX3 protein levels in melanoma cell lines. These data support that FOXD3 is sufficient but not necessary for PAX3 expression in melanoma cells.
DISCUSSION
THE IDENTIFICATION OF HIGHLY CONSERVED ENHANCER ELEMENTS IN THE PAX3 GENE
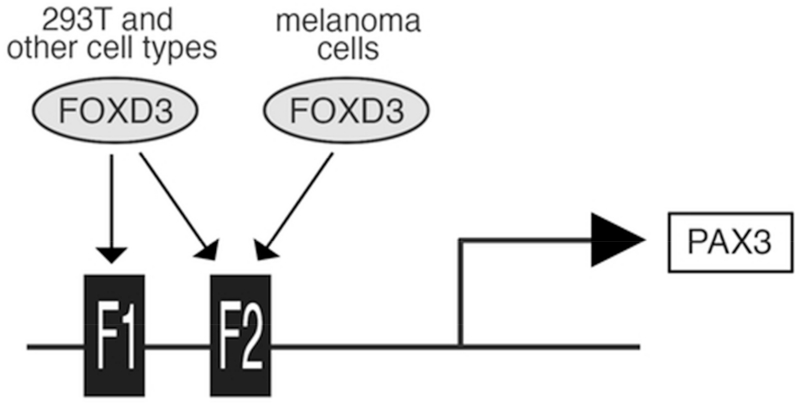
Transcription factors FOXD3 and PAX3 are both expressed in melanoma cells and exhibit a positive correlation between transcript levels (Fig. 1), suggesting a relationship between these two proteins. We identified two putative FOXD3 binding sites within conserved regions of the PAX3 gene, F1 and F2 (Fig. 4). FOXD3 is capable of binding to both of these sites in vitro (Fig. 5B). These sites appear to be redundant in driving the PAX3 element in 293T cells (Fig. 5C,D), while the F2 element was necessary in the two different melanoma cell lines tested (Fig. 6). These data suggest a mechanism where both of the FOX binding motifs may be utilized in some cells types but melanoma cells only utilize the F2 site (Fig. 7). The reason for this is not known, but it likely due to differences in cell types as well as the presence or absence of transcriptional co-factors. FOXD3 is known to bind to a variety of proteins in neural crest and melanocytes, including PAX3 [Thomas and Erickson, 2009]. Other FOXD3-interacting factors include Oct4 and GRG4/TLE4 [Guo et al., 2002; Yaklichkin et al., 2007]. F1 and F2 are situated within two separate conserved enhancer islands (NCE1 and NCE2, shown schematically in Fig. 4A), and the regulation of PAX3 by FOXD3 may differ during normal development due to the genetic element, cell type, and developmental stage. While the role of FOXD3 in the regulation of PAX3 during normal neural crest development, in melanoblasts, and in melanoma is not known, our work presented here supports that FOXD3 upregulates PAX3 in the melanoma disease state. It is predicted that FOXD3 regulates PAX3 through the F1 and F2 sites in both parallel and independent pathways in these different cell populations.
Fig. 7.
FOXD3 activates the PAX3 promoter from alternate FOX binding motifs in different cells lines. FOXD3 utilizes both FOX binding motifs, F1 and F2, in 293T cells. FOXD3 preferentially uses the F2 binding site in melanoma cells.
While the relationship between FOXD3 and PAX3 is not well defined in melanoma cells, here we discover FOXD3 response elements in the PAX3 locus. In addition, FOXD3 protein interacts with PAX3, and this leads to an inhibition of PAX3-dependent activation of MITF in neural crest cells [Thomas and Erickson, 2009]. It is not known if this pathway is also active in melanoma cells, although MITF levels are often independent of PAX3 expression status [He et al., 2011; Mascarenhas et al., 2010]. One intriguing possibility is that FOXD3 promotes PAX3 expression but selectively inhibits PAX3 activity in promoting MITF gene expression. Indeed, while PAX3 and FOXD3 expression are linked with invasion and drug resistance, so are lower MITF levels [Basile et al., 2012; Bonvin et al., 2012; Konieczkowski et al., 2014; Liu et al., 2013; Muller et al., 2014]. The possibility that FOXD3 functions in this manner in melanoma cells will be a focus of future studies.
FOXD3 IS SUFFICIENT BUT NOT NECESSARY FOR PAX3 EXPRESSION IN MELANOMA CELLS
We discovered that while overexpression of FOXD3 resulted in an increase of PAX3 protein levels (Fig. 2), inhibition of FOXD3 transcription factor activity was not able to reduce PAX3 levels in melanoma cells (Fig. 3), showing that FOXD3 is sufficient but not necessary for PAX3 expression in melanoma. FOXD3 is not the sole driver of PAX3 expression in melanoma but it does contribute to PAX3 expression and thus to some of melanoma phenotypes including growth, survival, and resistance to differentiation. That FOXD3 drives PAX3 expression may be especially important in the acquired resistance of melanoma to the mutant B-RAF inhibitor, PLX4032 or Vemurafenib. FOXD3 levels were found elevated in these cell death-resistant cells and knock-down of FOXD3 in these cells returned the drug-sensitivity to these cells [Abel and Aplin, 2010; Basile et al., 2012]. A similar trend is seen for PAX3 in Vemurafenib-resistant melanoma cells. Here, PAX3 levels are elevated, but cells become vulnerable to the drug again if PAX3 expression is blocked [Liu et al., 2013]. Since FOXD3 drives PAX3 expression in melanoma, it is possible that the overexpression of FOXD3 in these resistant cells is responsible, in part, for the increase in PAX3 levels, conferring resistance to cell death. Whether part of the mechanism through which increased FOXD3 expression confers PLX4032-resistance to melanoma cells is through the upregulation of PAX3 expression could be a focus of future research.
ACKNOWLEDGMENTS
We thank Patricia Labosky for providing for FOXD3 expression construct. The monoclonal PAX3 antibody was developed by C.P. Ordahl and obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242.
Grant sponsor: American Cancer Society; Grant number: RSG-CSM-121505; Grant sponsor: Friends of Dermatology-University of Chicago; Grant sponsor: Wendy Will Case Foundation; Grant sponsor: National Institutes of Health; Grant numbers: NIH R01CA130202, NIH R01CA184001.
Abbreviations
- ChIP
Chromatin immunoprecipitation
- DN
Dominant negative
- EMSA
Electrophoretic Mobility Shift Assay
- FOXD3
Forkhead box protein D3
- F1
FOX binding site 1
- F2
FOX binding site 2
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- GST
Glutathione S-transferase
- NCE
neural crest enhancer
- PAX3
Paired box protein 3
- qPCR
quantitative real-time polymerase chain reaction
- UTR
untranslated region
Footnotes
The authors declare no conflict of interest.
REFERENCES
- Abel EV, Aplin AE. FOXD3 is a mutant B-RAF-regulated inhibitor of G(1)-S progression in melanoma cells. Cancer Res. 2010;70:2891–900. doi: 10.1158/0008-5472.CAN-09-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basile KJ, Abel EV, Aplin AE. Adaptive upregulation of FOXD3 and resistance to PLX4032/4720-induced cell death in mutant B-RAF melanoma cells. Oncogene. 2012;31:2471–9. doi: 10.1038/onc.2011.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvin E, Falletta P, Shaw H, Delmas V, Goding CR. A phosphatidylinositol 3-kinase-Pax3 axis regulates Brn-2 expression in melanoma. Mol Cell Biol. 2012;32:4674–83. doi: 10.1128/MCB.01067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Stankunas K, Shang C, Kao SC, Twu KY, Cleary ML. Pbx1 functions in distinct regulatory networks to pattern the great arteries and cardiac outflow tract. Development. 2008;135:3577–86. doi: 10.1242/dev.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–20. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Dong L, Li Y, Cao J, Liu F, Pier E, Chen J, Xu Z, Chen C, Wang RA, Cui R. FGF2 regulates melanocytes viability through the STAT3-transactivated PAX3 transcription. Cell Death Differ. 2012;19:616–22. doi: 10.1038/cdd.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–38. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- Guo Y, Costa R, Ramsey H, Starnes T, Vance G, Robertson K, Kelley M, Reinbold R, Scholer H, Hromas R. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc Natl Acad Sci U S A. 2002;99:3663–7. doi: 10.1073/pnas.062041099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Li CG, Slobbe L, Glover A, Marshall E, Baguley BC, Eccles MR. PAX3 knockdown in metastatic melanoma cell lines does not reduce MITF expression. Melanoma Res. 2011;21:24–34. doi: 10.1097/CMR.0b013e328341c7e0. [DOI] [PubMed] [Google Scholar]
- Katiyar P, Aplin AE. FOXD3 regulates migration properties and Rnd3 expression in melanoma cells. Mol Cancer Res. 2011;9:545–52. doi: 10.1158/1541-7786.MCR-10-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, Frederick DT, Barzily-Rokni M, Straussman R, Haq R, Fisher DE, Mesirov JP, Hahn WC, Flaherty KT, Wargo JA, Tamayo P, Garraway LA. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014;4:816–27. doi: 10.1158/2159-8290.CD-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–79. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech Dev. 1998;76:185–90. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- Lang D, Epstein JA. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum Mol Genet. 2003;12:937–45. doi: 10.1093/hmg/ddg107. [DOI] [PubMed] [Google Scholar]
- Li J, Chen F, Epstein JA. Neural crest expression of Cre recombinase directed by the proximal Pax3 promoter in transgenic mice. Genesis. 2000;26:162–4. doi: 10.1002/(sici)1526-968x(200002)26:2<162::aid-gene21>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Li J, Liu KC, Jin F, Lu MM, Epstein JA. Transgenic rescue of congenital heart disease and spina bifida in Splotch mice. Development. 1999;126:2495–503. doi: 10.1242/dev.126.11.2495. [DOI] [PubMed] [Google Scholar]
- Liu F, Cao J, Wu J, Sullivan K, Shen J, Ryu B, Xu Z, Wei W, Cui R. Stat3-targeted therapies overcome the acquired resistance to vemurafenib in melanomas. J Invest Dermatol. 2013;133:2041–9. doi: 10.1038/jid.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JB, Littlejohn EL, Wolsky RJ, Young KP, Nelson M, Salgia R, Lang D. PAX3 and SOX10 activate MET receptor expression in melanoma. Pigment Cell Melanoma Res. 2010;23:225–37. doi: 10.1111/j.1755-148X.2010.00667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski RC, Chi NC, Li J, Brown C, Lu MM, Epstein JA. Identification of minimal enhancer elements sufficient for Pax3 expression in neural crest and implication of Tead2 as a regulator of Pax3. Development. 2004;131:829–37. doi: 10.1242/dev.00975. [DOI] [PubMed] [Google Scholar]
- Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Foppen MH, Kemper K, Goding CR, McDermott U, Blank C, Haanen J, Graeber TG, Ribas A, Lo RS, Peeper DS. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun. 2014;5:5712. doi: 10.1038/ncomms6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli TA, Ellsworth MK, Wu C, Gross KW, Pruitt SC. Positive and negative DNA sequence elements are required to establish the pattern of Pax3 expression. Development. 1997;124:617–26. doi: 10.1242/dev.124.3.617. [DOI] [PubMed] [Google Scholar]
- Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14:2755–66. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer RS, Shea CR, Nelson M, Powell SK, Freeman DM, Dan CP, Lang D. PAX3 expression in primary melanomas and nevi. Mod Pathol. 2008;21:525–30. doi: 10.1038/modpathol.3801019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt SC, Bussman A, Maslov AY, Natoli TA, Heinaman R. Hox/Pbx and Brn binding sites mediate Pax3 expression in vitro and in vivo. Gene Expr Patterns. 2004;4:671–85. doi: 10.1016/j.modgep.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ferras O, Coutaud B, Djavanbakht Samani T, Tremblay I, Souchkova O, Pilon N. Caudal-related homeobox (Cdx) protein-dependent integration of canonical Wnt signaling on paired-box 3 (Pax3) neural crest enhancer. J Biol Chem. 2012;287:16623–35. doi: 10.1074/jbc.M112.356394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit DJ, Smith AG, Parsons PG, Muscat GE, Sturm RA. Domains of Brn-2 that mediate homodimerization and interaction with general and melanocytic transcription factors. Eur J Biochem. 2000;267:6413–22. doi: 10.1046/j.1432-1327.2000.01737.x. [DOI] [PubMed] [Google Scholar]
- Sutton J, Costa R, Klug M, Field L, Xu D, Largaespada DA, Fletcher CF, Jenkins NA, Copeland NG, Klemsz M, Hromas R. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J Biol Chem. 1996;271:23126–33. doi: 10.1074/jbc.271.38.23126. [DOI] [PubMed] [Google Scholar]
- Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–24. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas AJ, Erickson CA. FOXD3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing MITF through a non-canonical mechanism. Development. 2009;136:1849–58. doi: 10.1242/dev.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaklichkin S, Steiner AB, Lu Q, Kessler DS. FoxD3 and Grg4 physically interact to repress transcription and induce mesoderm in Xenopus. J Biol Chem. 2007;282:2548–57. doi: 10.1074/jbc.M607412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Li Y, Nishimura EK, Xin H, Zhou A, Guo Y, Dong L, Denning MF, Nickoloff BJ, Cui R. Inhibition of PAX3 by TGF-beta modulates melanocyte viability. Mol Cell. 2008;32:554–63. doi: 10.1016/j.molcel.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Zhu BK, Pruitt SC. Determination of transcription factors and their possible roles in the regulation of Pax3 gene expression in the mouse B16 F1 melanoma cell line. Melanoma Res. 2005;15:363–73. doi: 10.1097/00008390-200510000-00004. [DOI] [PubMed] [Google Scholar]