Summary
Bacterial Type VI Secretion Systems (T6SS) function as contractile nanomachines to puncture target cells and deliver lethal effectors. In the ten years since the discovery of the T6SS, much has been learned about the structure and function of this versatile protein secretion apparatus. Most of the conserved protein components that comprise the T6SS apparatus itself have been identified and ascribed specific functions. In addition, numerous effector proteins that are translocated by the T6SS have been identified and characterized. These protein effectors usually represent toxic cargoes that are delivered by the attacker cell to a target cell. The field is beginning to better understand the lifestyle or physiology that dictates when bacteria normally express their T6SS. In this Chapter, we consider what is known about the structure and regulation of the T6SS, the numerous classes of antibacterial effector T6SS substrates, and how the action of the T6SS relates to a given lifestyle or behavior in certain bacteria.
Introduction
The type VI secretion system (T6SS), a Gram-negative secretion pathway (1, 2), delivers effectors upon direct contact with a target cell (3, 4). Death of the target cell is the primary outcome that follows the delivery of the lethal effectors, which are translocated from the attacker cell cytoplasm into the periplasm of the target cell via a T6S apparatus in a contact-dependent process (5, 6). It is well known that bacteria release bactericidal agents such as bacteriocins and antibiotics into the extracellular environment as a means to indiscriminately eliminate bacterial competitors (7). In addition, it is now understood that many Gram-negative bacteria also use the T6SS to directly antagonize bacteria in close proximity (5). Direct antagonism of neighboring cells can provide a selective advantage for bacteria in their natural habitat in dense biofilm communities or during a multicellular life-style that requires direct contact and cooperativity (8, 9). As a consequence, bacteria benefit from a specific mechanism that depends on cell-cell contact to discriminate between one another and eliminate non-self bacteria, potential cheaters, or competitors from the population. In particular, lethal action of the T6 contractile puncturing device provides a specific advantage for bacteria that possess the T6SS to discriminate, recognize, and kill competitors.
Many secretion systems are present in Gram-negative bacteria that function to translocate proteins. The T6SS is the most recently identified secretion system; the gene clusters that encode the system were named the T6SS following a genetic screen in Vibrio cholerae (2). Interestingly, genes or proteins that belong to or are dependent on the T6SS had been previously identified, mainly as virulence (or symbiosis) factors (10–13). In Rhizobium leguminosarum, a gene cluster later to be appreciated as encoding a T6SS was identified that played a role in symbiosis and was required for protein secretion (11). A proteomics study examining secreted proteins of Edwardsiella tarda also identified a large conserved gene cluster with noted similarity to the one identified in R. leguminosarum that was proposed to encode a novel protein secretion system (13). The widespread presence of T6SS genes were also noted prior to functional classification and discovery of the actual T6SS machinery (14–16). The T6SS has now been studied extensively in numerous organisms. Here, we consider much of what has been well-characterized since the T6SS was discovered nearly a decade ago.
T6SS Structural Components
Hcp and VgrG
The first discovered and most studied components of the T6SS are Hcp and VgrG (2), which are both secreted and required for T6SS activity (1, 2). These two components comprise the hollow tube and puncturing tip that is delivered by the action of the T6SS. Bioinformatic analysis (3) and structural analysis show that VgrG is an excellent match with the cell puncturing T4 bacteriophage tail spike (17). This structural study provides compelling evidence that Hcp and VgrG assemble to form a membrane puncturing device. While direct evidence that VgrG is indeed the puncturing tip is lacking, work on Hcp has demonstrated that it forms a tube made of hexameric “stacked rings” (1, 18). Additional studies have predicted that the Hcp tube would form a complex with a trimeric VgrG tip (17, 19, 20). It is now apparent that the T6SS functions in a manner analogous to an inverted phage tail and tube (21).
VipA and VipB
As predicted, the T6SS is known to contain proteins that assemble to form a sheath structure. The sheath proteins, VipA and VipB (or TssBC), have been shown to form tube structures or polysheaths (22, 23). It was also noted that the VipA and VipB polysheaths appeared to disassemble, an event dependent on ClpV-mediated ATP hydrolysis (22). The appearance of these tubules is remarkably consistent with T4 phage tail sheaths and suggests a common molecular mechanism between bacteriophage and T6SS function (24–26). When the bacteriophage tail or a T6SS make contact with target cells, contraction of the respective sheath delivers a puncturing device that facilitates delivery of phage DNA or T6SS effectors, respectively (Figure 1). The contraction event is presumed to coincide with effector secretion into target cells (27–30).
Figure 1. Comparison between bacteriophage and the T6SS.
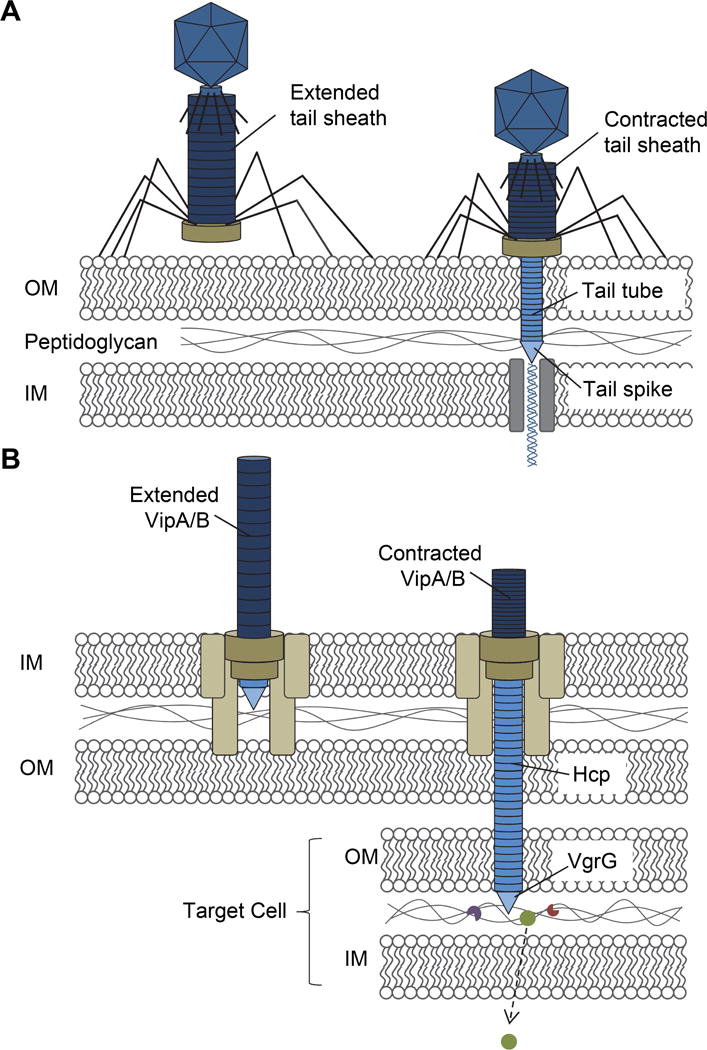
(A) Bacteriophage possess tail fibers that attach to lipopolysaccharides of target bacterial cells. During reversible binding, the tail fibers bring the base plate in contact with the cell surface. Once in contact, irreversible binding is initiated, the tail sheath contracts and the rigid tail tube and spike are forced though the outer membrane (OM), proteins of the spike degrade the peptidoglycan, and final interaction with the inner membrane (IM) initiates translocation of viral DNA into the cell. (B) The T6SS functions much like a bacteriophage with several proteins being structurally similar, depicted here in the same color. Formation of the T6SS base plate complex, which spans the IM, peptidoglycan, and OM initiates Hcp tube polymerization and sheath formation of VipA and VipB heterodimers. Upon contact with a target bacterial cell, this ‘ready to fire’ state is triggered causing the sheath to contract and deployment of the Hcp tube and VgrG spike into the target cell. Effector proteins are delivered into the periplasm, possibly degrading peptidoglycan for cytoplasmic effectors to gain further access into the cell. OM, outer membrane; IM, inner membrane.
Effector delivery module
The structure of the contractile sheath of the T6SS has been particularly well characterized. Cryo-electron microscopy and reconstruction of contracted VipA/B tubules have shown that the tubule is constructed of six protofilaments arranged in a right-handed helical arrangement (31). This symmetry has been described as an assembly of stacked hexameric rings analogous to the T4 tail sheath (31–33). An insightful result from these structural studies has shown that the ClpV recognition motif of VipB is buried in the protofilament of the elongated sheath and becomes exposed in the contracted sheath structure (31, 33). This distinct outer layer of the T6SS sheath is a key difference from the architecture of the bacteriophage tail sheath and facilitates interactions with the ClpV ATPase, which enables multiple rounds of sheath extension and contraction via a contraction-state-specific recycling mechanism (31, 33). This ability of the T6SS sheath to be re-used is in contrast to the bacteriophage tail sheath that only can be contracted once. In support of this, T6SS sheath recycling and activity has been visualized in real-time using fluorescent reporters fused to VipA (Figure 2) (8, 29). Collectively, evidence has led to a parallel assembly pathway between T6SS and bacteriophage morphogenesis. A proposed mechanism to form the T6SS effector delivery module is VgrG attracts Hcp that forms a hollow inner tube of stacked hexamers and Hcp tube formation then recruits and facilitates assembly of the outer tubule sheath structure (34).
Figure 2. Infiltration of resistant and sensitive opposing swarms by P. mirabilis HI4320 expressing VipA::sfGFP.
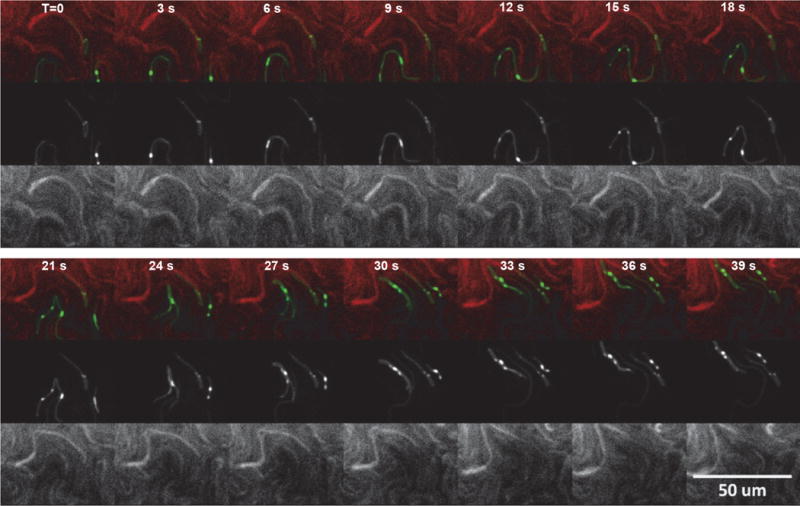
Agar plate inoculated with P. mirabilis HI4320 VipA::sfGFP opposing strains HI4320 or mutant 9C1 (mutation in immunity protein PefE) expressing dsRED. Infiltrating Proteus mirabilis HI4320 expressing VipA::sfGFP opposing strains HI4320 or mutant 9C1 expressing dsRED. Elapsed time (T) is indicated in seconds (reprinted with permission from Alteri CJ et al. 2013. PLoS Pathog 9: e1003608).
Envelope-spanning complex
The phage-like T6SS tail or effector delivery module is anchored to the cell membrane by a trans envelope complex (35). These additional structural components of the T6SS are TssL, TssM, and TssJ, which form the envelope-spanning complex (36, 37). The minimal core set of conserved membrane proteins required for T6SS function are the inner membrane proteins TssL and TssM as well as the outer membrane lipoprotein TssJ (35–38). These proteins are connected by interactions between TssM and TssL, and TssM and TssJ (35, 36, 39, 40). This envelope spanning complex has been shown to be assembled by the sequential addition of the three subunits, TssJ, TssM and TssL (41). The structure of the fully assembled envelope complex was determined by negative-stain electron microscopy and was shown form a large base in the cytoplasm (41). The envelope spanning complex was also observed to extend into the periplasm to form a double-ring structure containing the carboxy-terminal domain of TssM and TssJ that is anchored in the outer membrane (41). This structural study of the T6SS envelope complex suggests that conformational changes allow passage of the puncturing device made of the Hcp tube and VgrG spike through a transient pore in the outer membrane (41). Additionally, the cytoplasmic protein TssK has been shown to interact with Hcp, TssL, and TssC and is proposed to link the sheath-Hcp-VgrG complex to the envelope-spanning complex (42). Thus, it is not unreasonable that the assembled effector delivery module interacts with the membrane-spanning complex via TssK.
T6SS Effectors
The delivery module of the T6SS can translocate a diversity of protein effectors into the target cell. It is notable that the core T6SS genes, which encode structural components, are highly conserved across bacterial species (Figure 3), in contrast to the clear diversity among effectors identified from these same organisms. These T6SS effectors have been identified using a number of techniques and the toxic activities for many of the identified effectors have been biochemically characterized. T6SS effectors include VgrG, PAAR domain containing proteins that associate with VgrG, Rhs repeat proteins, and effector proteins that may be delivered within the hollow Hcp inner tube. The known and suspected activities of T6SS effectors include amidases, lipases, nucleases, and chaperone functions.
Figure 3. Conservation of the Type 6 Secretion System effector operons among Proteus mirabilis, Vibrio cholerae, and Pseudomonas aeruginosa.
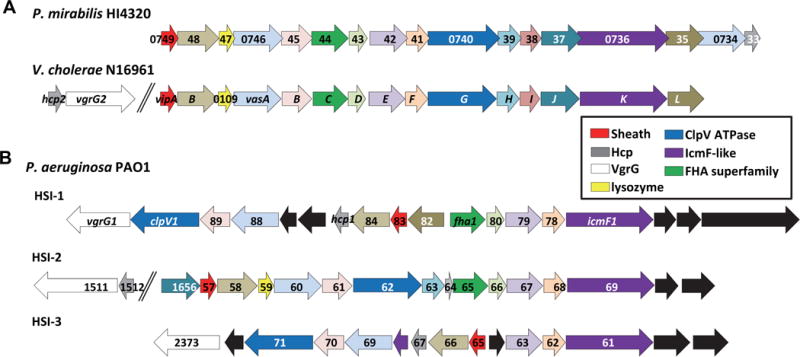
(A) The 17 genes that encode the Proteus mirabilis HI4320 T6SS are highly conserved with the well characterized T6SS genes from V. cholerae N16961 and have identical gene order in their respective chromosome and (B) comparison to the three known T6SSs (HIS-1, HIS-2, HIS-3) encoded by the genome of P. aeruginosa PA01. The arrows are color-coded based upon known or predicted gene function. Black arrows represent genes that are not found in either P. mirabilis HI4320 or V. cholerae N16961 T6SS gene locus (reprinted with permission from Alteri CJ et al. 2013. PLoS Pathog 9: e1003608).
Identification of effectors
Both biochemical and bioinformatics approaches have been successfully used to identify T6SS effectors. One study to identify T6SS effectors used bioinformatic analyses based on known P. aeruginosa effectors; this heuristic approach led to the identification of a superfamily of amidase effectors, which has greatly expanded the known diversity of effectors exhibiting amidase activity (43). This work also noted a potential for substantial diversity among T6SS effectors because these analyses failed to identify effectors that have been identified in other bacteria. (43). An alternative bioinformatics strategy to identify T6SS effectors is to scan the genome in areas immediately downstream of orphan hcp-vgrG gene pairs (8, 44, 45). Genes downstream of these hcp-vgrG pairs likely encode effectors in other organisms.
Biochemical approaches have also been employed to identify T6SS effectors. One key approach that led to the discovery of the first non-structural effectors, is to compare proteins secreted by a wild-type and T6SS mutant bacteria (5). The use of whole cell proteomics to identify proteins destabilized in the absence of Hcp has also been a successful approach to discover T6SS effectors (46).
VgrG family
The first T6SS effectors to be appreciated are members of the VgrG family, which are thought to act as a puncturing tip of the delivered Hcp tube. In V. cholera (3) and Pseudomonas aeruginosa (20), there is evidence that VgrG forms a trimer. It is notable that some organisms only have a single copy of VgrG, while other bacteria can have upwards of 30 distinct VgrG proteins. This indicates that delivery of different effectors may require a specifically adapted VgrG protein (18). These findings suggest that heterotrimeric VgrG complexes could be loaded and deliver multiple effectors in the context of a single puncturing tip. Aside from the proposed role as a structural component of the effector delivery module, VgrG can also be classified as an effector because some VgrG proteins contain additional functional domains beyond the minimal trimeric structure required for T6SS function. In V. cholerae, the carboxy-terminus of VgrG3 has muramidase activity (47) and VgrG-1 contains an actin crosslinking domain (3).
PAAR proteins
Another group of gene products, PAAR (proline alanine alanine arginine) domain-containing proteins have been shown to be T6SS effectors. It has been shown that PAAR proteins can interact with the tip of the VgrG trimer (48). PAAR proteins are associated with a myriad of predicted effector functions, suggesting that numerous PAAR proteins encoded within bacterial genomes are likely T6SS effectors (48). It has been speculated that PAAR proteins might complex with any given VgrG trimer; however, other data suggest specific interactions between VgrG proteins and effectors (20). One example of a PAAR effector is the nuclease, RhsA, which has been identified as T6SS-dependent in Dickeya dadantii (44). This nuclease requires one of two VgrG genes, demonstrating that this PAAR domain effector can be delivered by T6SS effector tubes capped with different VgrG puncturing tips. As mentioned, there are often multiple PAAR domain-containing proteins in bacteria that also encode a T6SS.
Rhs-repeat proteins
As noted above, another group of T6SS effectors are members of the Rhs-repeat-containing protein family (44). Structural analysis has shown that the Rhs-repeat domain forms a structure that completely encloses and protects the folded enzymatically active effector domain (49). Many Rhs-repeat proteins also contain PAAR domains in addition to effector domains (48). In P. aeruginosa PA14, the H2-T6SS encodes an Rhs effector protein that may contribute to internalization by eukaryotic cells (50). In addition, this group also found that VgrG1 of the H1-T6SS is responsible for delivering an Rhs effector protein to target cells (51). It is noteworthy that Rhs domain proteins function as antibacterial toxins in contact-dependent inhibition (Cdi) systems (44), which suggests Rhs proteins have evolved to be delivered by both the T6SS and two-partner secretion systems.
Non-VgrG effectors
The T6SS also delivers effectors that lack PAAR domains. This could suggest that the Hcp/VgrG puncturing tube may act as a conduit through which effectors are delivered to target cells (1, 52). Since the inner diameter of the Hcp tube is approximately 40Å (1), this requires that many effector proteins would have to be delivered in an unfolded state (53). Consistent with this notion, there is one example of a T6SS effector operon that encodes a small molecular mass thioredoxin that could presumably function to fold effector proteins upon delivery to the target cell (8). Alternatively, or in addition, a thioredoxin protein could also serve to maintain effectors in an unfolded state prior to being loaded into an Hcp/VgrG puncturing tube.
Hcp tubes may also serve a chaperone function during delivery of T6SS effectors. Hcp-effector interactions were initially noted during biochemical analysis of the T6SS in Edwardsiella tarda that showed the secreted effector EvpP interacted with Hcp (54). Additional biochemical approaches have shown that T6SS effectors can bind to residues exposed on the inner face of the tube forming Hcp hexamers and can even stabilize effectors; thus, Hcp could be considered a molecular chaperone for effectors (55). Collectively, these experiments suggest that effectors can be loaded into the puncturing tube during assembly of the T6SS structure and may be delivered into target cells concomitantly with the puncturing device.
Numerous non-VgrG effectors have been identified and the list of these effectors is continually growing (5, 43). In fact, T6SS effectors exhibit a broad range of functions capable of targeting both eukaryotic and prokaryotic cells. Effector activities include actin cross-linking (3, 56), muramidases and peptidases that hydrolyze peptidoglycan (5, 6, 43, 57–59), nucleases (44), and lipases (60). In P. mirabilis, there are numerous effectors that exhibit lethal activity through an unknown mechanism (8). In all cases, bacteria that produce these T6SS effectors also encode immunity proteins to protect against their cognate effectors (5, 8, 47, 58, 61).
Peptidoglycan-degrading effectors
One class of T6SS effectors targets the cell wall peptidoglycan. In P. aeruginosa, Tse1 and Tse3, among the first T6SS effectors to be characterized, both exhibit lytic activity (6). These effectors target peptide and sugar portions of the peptidoglycan. Characterization of Tse1 by several groups revealed crystal structures that indicate an amidase with a common cysteine protease fold (53, 57, 62). Peptidoglycan is a polymer composed of alternating N-acetylmuramic acid and N-acetylglucosamine residues. The muramic acid residues are modified with peptides that provide structural integrity by cross-linking with adjacent peptides. Tse1 cleaves between the second and third residues of the peptide modification (6). The specificity of Tse1 suggests that the effector could attack at sites of peptidoglycan synthesis (57). This is consistent with the observation that Tse1 is required for the P. aeruginosa T6SS-dependent lysis of target bacteria (63).
It is now appreciated that Tse1 belongs to a group of effectors that has been termed the type VI amidase effector (Tae) superfamily (43). Tae effectors appear to belong to four divergent groups of cell wall-degrading enzymes (64). Further evidence indicates the Tae proteins are functional analogs as tae genes are located adjacent to genes encoding immunity determinants. It appears that all members of the Tae family function as amidases that hydrolyze peptidoglycan of Gram-negative bacteria (64). Studies examining effectors of these Tae family members have shown that T6SS activity is required for their action on target cells (6, 43, 58).
In contrast to Tse1, Tse3 acts on the sugar backbone rather than on the peptide bonds contained within peptidoglycan (6). The β1,4 bonds between muramic acid and N-acetylglucosamine are common targets of a multitude of bacteriocidal enzymes, including lysozyme. Indeed, these enzymes share the lysozyme muramidase fold, but have an active site tyrosine residue and exhibit N-acetylglucosaminidase activity (6, 65). The VgrG protein of the puncturing device can also exhibit peptidoglycan degrading activity. The C-terminal domain of V. cholerae VgrG3 has a predicted muramidase fold, can degrade peptidoglycan, and can cause lysis when expressed in the periplasm (47). Furthermore, the presence of a cognate immunity gene suggests that the VgrG3 protein is itself a T6SS effector (61).
Lipases
T6SS effectors can also act as lipases to disrupt cell membranes. A group of phospholipase effectors have been identified that hydrolyze plasma membrane lipids (60). These lipase effectors degrade membranes by attacking different bonds in phospholipids (64). T6SS lipase effectors have been experimentally demonstrated to cleave phospholipids at three bonds (60). Certain lipase effectors demonstrate preference for specific head group moieties. One lipase effector in P. aeruginosa exhibits specificity for phosphatidylethanolamine, which is a primary phospholipid found in the bacterial membrane (60). Because phospholipids are ubiquitous in nature, that is, found in both bacterial and eukaryotic membranes, this raises the obvious possibility that T6SS lipase effectors would target both bacterial and host cell membranes. Consistent with this notion, genetic disruption of lipase effector-encoding genes in P. aeruginosa and V. cholerae creates fitness defects in infection models (60, 61).
Nucleases
Nucleases are another class of T6SS-dependent effectors. Recent studies of Rhs domain proteins show that subsets of these proteins are also T6SS nuclease effectors (44, 66). In D. dadantii, RhsA and RhsB, contain endonuclease effector domains that are dependent on neighboring VgrG proteins for delivery to target cells. When produced in target cells, these domains result in DNA degradation, growth inhibition, and a loss of nucleic acid staining (44). In Agrobacterium tumefaciens, a soil bacterium that triggers tumorigenesis in plants, produces a family of T6SS DNase effectors that are distinct from previously known polymorphic toxins and nucleases. These effectors exhibit an antibacterial DNase activity that relies on a conserved motif and can be counteracted by a cognate immunity protein (67).
T6SS Regulation
It is assumed that elaboration and usage of the T6SS nanomachine is energetically costly to the bacterial cell. It follows that expression and assembly of this structure would be tightly regulated. In some cases, there is evidence for transcriptional regulation of the T6SS via quorum sensing (68–70). In other cases, T6SS expression is regulated during biofilm formation (71, 72), by iron-limitation (73, 74), is temperature-dependent (70, 75), and may react to both osmolarity changes (70) and stress responses (76). There is also evidence for independent regulation for the expression of the structural components of the T6SS and the expression of effectors.
RpoN and VasH
In V. cholerae, the primary T6SS gene cluster encodes a regulator, VasH, which acts as an activator via effects on RpoN. In V. cholerae, both RpoN and VasH are required for T6SS activity (2, 77, 78). In V. cholerae, environmental cues likely stimulate the transcription of the major cluster so that VasH is produced, which subsequently activates the transcription of the hcp operons by RpoN (18, 79). However, this control is limited to the transcription of two hcp effector operons and not the T6SS itself (79). Finding that RpoN and VasH only control the hcp operons suggests additional regulators of the T6SS remain to be elucidated (18). The hcp operons encode the secreted T6SS effectors, while the main T6SS locus encodes structural components of the apparatus itself. This has been postulated as evidence for a two-tiered regulation that may be important for maintaining different levels of expression for components that can be recycled versus those that are released to target cells (18). It is notable that pandemic V. cholerae strains tightly regulate the expression of the T6SS (68), while non-pandemic serotype O37 strain V52 and environmental isolates constitutively express the T6SS (80). It is possible that this difference results from increased interbacterial competition in the environment and/or as a defense mechanism against predation.
Quorum sensing
Differences in quorum-sensing-mediated transcriptional control expression of vas genes could be partially responsible for T6SS variability among V. cholerae strains. There is evidence for a strong link between HapR and Hcp expression among a number of pandemic isolates (81). Further work showed that a deletion of luxO was able to induce T6SS expression in two O1 serotype strains (68, 81). It was also noted that T6SS activity was not fully induced despite transcriptional activation in the absence of LuxO (68). Thus, while quorum sensing represses T6SS expression, it appears that complete T6SS activation in pandemic V. cholerae involves additional inputs such as high osmolarity and possibly low temperature (82).
Iron regulation
Iron limitation controls the expression of the T6SS in Edwardsiella spp. and enteroaggregative E. coli. In these bacteria, the T6SS is repressed directly at the transcriptional level by Fur (73, 74). It was shown that Fur confers iron-dependent repression of production and export of an Hcp homolog (EvpC) in Edwardsiella tarda. and that the Fur protein binds directly to a Fur box sequence upstream of evpP, the first gene in the T6SS gene cluster (74). In enteroaggregative E. coli, the Sci-1 T6SS is required for biofilm formation and is regulated by iron availability through a pathway involving Dam methylation and Fur repression (73). Two Fur binding sites and three Dam methylation sites reside upstream of the Sci-1 T6SS gene cluster. Further, one of the Fur-binding sites overlaps a Dam methylation site. In the absence of iron, Fur dissociates and allows RNA polymerase to bind and initiate transcription. Similarly, the loss of Fur also permits methylation at the site, which inhibits the binding by Fur (73). Thus, iron limitation results in stable expression of the T6SS.
H-NS regulation
Regulation of T6SS gene clusters by members of the H-NS family of regulators has also been reported. These repressor proteins function as global regulators by controlling the expression of a large number of genes throughout the genome (83). Many genes and entire gene clusters that have been horizontally acquired are silenced by H-NS (84, 85). Indeed, this is what occurs for several T6SS gene clusters, like that in Salmonella enterica (86). In P. aeruginosa, the H-NS-like protein MvaT represses the HSI-2 and HSI-3 T6SS gene clusters (87).
RetS signaling pathway
Control of T6SS activity in P. aeruginosa is well characterized, however, much of what is described is based upon studies involving bacteria harboring a retS mutation, which leads to constitutive expression of the T6SS. The RetS regulon controls a range of virulence and fitness factors and is involved in the reciprocal control of traits important during acute and chronic phases of lung infection, respectively (88, 89). RetS is a hybrid sensor kinase that acts upstream of the Gac/Rsm pathway in P. aeruginosa (88). Along with regulation of the T6SS, this regulatory pathway control numerous traits related to antagonism or social behavior in Pseudomonads (90). It should be noted that wild-type P. aeruginosa poorly express the T6SS in liquid medium, however the T6SS is expressed by wild-type bacteria during surface growth (91) and in response to cell lysate (92).
Post-translational control of T6SS activity
P. aeruginosa also controls its T6SS through a post-translational regulatory system termed the threonine phosphorylation pathway (93). In this system, the T6SS requires a T6SS-encoded forkhead-associated protein, Fha, to be phosphorylated by the kinase PpkA (93). The activity of the kinase requires the lipoprotein TagQ and an outer membrane protein termed TagR (91). Studies have shown that TagQ is required for proper localization of TagR, and TagR likely interacts directly with the periplasmic domain of PpkA (94, 95). It has also been shown that two proteins, TagS and TagT, form a membrane-associated complex that is required for full activation of PpkA (95). In this scheme, the T6SS is shut down following dephosphorylation of Fha by the phosphatase PppA (93). Consistent with this, phosphatase mutants exhibit greater levels of T6SS activity (27, 93).
Studies aimed at dissecting the post-translational regulation of the T6SS in P. aeruginosa has led to a defensive T6SS hypothesis (27). This work, using a retS mutant strain, showed that when cultured on a solid surface, a P. aeruginosa bacterium that is attacked by a neighboring cell will strike back with a retaliatory T6SS counterattack (18, 27). This phenomenon has been named ‘T6SS dueling’ (28). The threonine phosphorylation pathway plays a role in this dueling phenomenon (27). Despite finding that bacteria lacking PppA have high levels of T6SS activity, the dueling response appeared defective (27). This result suggested that dephosphorylation of Fha might somehow control the positioning of the T6SS in this scenario (18, 27). This would be consistent with the observation that surface growth, with greater numbers of neighboring cells in contact, correlates with increased PpkA activity (95).
T6SS Activity and Actions
Mechanisms to trigger T6SS-mediated killing
The defensive nature of the T6SS activity in P. aeruginosa is supported by other studies. When cocultured with T6SS+ V. cholerae and A. baylyi, P. aeruginosa killed both species in a TagT-dependent manner (27). This killing was not observed when the attacking bacteria were lacking a functional T6SS. A similar differentiation between T6SS+ and T6SS− Burkholderia thailandensis has also been observed (63). Using the retS background in P. aeruginosa, it has also been shown that a variety of conditions that mimic direct antagonistic contact can trigger firing of the T6SS. For example, during conjugation, E. coli cells expressing the plasmid RP4 T4SS were highly sensitive to killing by P. aeruginosa (30). These studies further showed that DNA transfer was not required, but genes involved in pilus and mating pair formation were (30). The antimicrobial peptide polymyxin B also was shown to induce T6SS activity (30). These observations suggested that membrane perturbation may be the elusive signal that triggers defensive T6SS counter-attacks in P. aeruginosa (18).
Recent work from the Mougous Laboratory has provided a more complete understanding of P. aeruginosa response to antagonism (92). Their findings demonstrate that the response does not require the threonine phosphorylation pathway and involves the release of a diffusible signal from lysed P. aeruginosa sibling bacteria (92). This work goes on to show that the signal from lysed cells leads to post-transcriptional upregulation of the T6SS via the aforementioned Gac/Rsm pathway (92). Future studies are needed to better understand why P. aeruginosa would use the T6SS solely as a defensive weapon, or to identify the natural conditions under which the bacteria might deploy the T6SS in a preemptive manner. Other species such as Proteus mirabilis and Serratia marcescens do not require an attack by neighboring cells to stimulate the T6SS (8, 96). In enteroaggregative E. coli, it has been shown that a similar diffusible signal and counter-attack mechanism might exist (97). This T6SS activity observed in E. coli appeared to spread within a population of cells (97).
Proteus mirabilis swarming and Dienes line formation
The opportunistic pathogen P. mirabilis undergoes a characteristic developmental process to coordinate multicellular swarming behavior and discriminates itself from another Proteus isolate during swarming, resulting in a visible boundary termed a Dienes line (98, 99). The formation of this demarcation is dependent on the activity of the T6SS (8) (Figure 4). Further experiments using the P. mirabilis model to study T6SS demonstrated that all five identified T6SS hcp/vgrG effector operons, found in the single prototype strain, were transcriptionally active only during active swarming (8). The regulatory pathways that control this specific regulation of T6SS in P. mirabilis could involve the regulator MrpJ (100). The observation that P. mirabilis T6SS is controlled by swarming was also observed for the identity of self (ids) effector operon (8, 101). The implication from these studies is that P. mirabilis is preemptively deploying the T6SS during a growth phase, swarming, that requires close and direct contact across a large population of bacteria.
Figure 4. Contact-dependent preemptive antagonism is dependent on the T6SS in Proteus mirabilis.
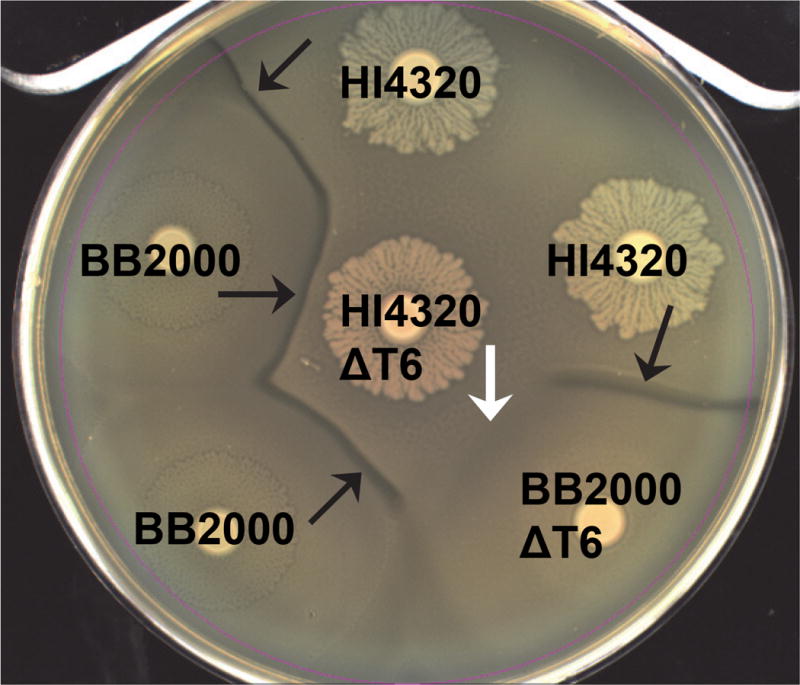
A Dienes line (black arrows) forms between two different wild-type isolates, HI4320 and BB2000 (strain A and B kill each other). Loss of the T6SS (ΔT6) in either isolate by disruption of PMI0742 does not affect the discriminatory Dienes line (strain A kills strain B or strain B kills strain A). Loss of the T6SS in both isolates allows non-identical swarms to merge and the lack of T6SS-dependent killing appears as “recognition” (white arrow) (reprinted with permission from Alteri CJ et al. 2013. PLoS Pathog 9: e1003608).
Interbacterial antagonism
The T6SS is a macromolecular machine that delivers protein effectors into both prokaryotic and eukaryotic cells, thus playing important roles during interbacterial competition and virulence in the host (102). In addition to the versatility, the genes encoding the T6SS are widely distributed and found in approximately one-third of sequenced Gram-negative bacterial genomes (103). Numerous studies have revealed that the activity of the T6SS is important in bacterial communities, either by promoting biofilm formation or by competing with neighboring bacteria (64). It is now appreciated that the T6SS is used to deliver antibacterial toxins directly inside neighboring bacterial cells. The role for the T6SS during interbacterial competition was first noted in P. aeruginosa (5). Subsequently, a similar function has been shown in B. thailandensis, V. cholerae, S. marcescens, enteroaggregative E. coli, Citrobacter rodentium, Acinetobacter baumanii, and P. mirabilis (8, 76, 104–107). The T6SS also plays an important role in intraspecies competition, that is, different isolates of the same species produce different sets of toxin effectors (8, 108). As noted this intraspecies warfare is responsible for the formation of macroscopic boundaries between non-isogenic strains in several species, most notably in swarming P. mirabilis (8, 109).
Biofilm formation
Several bacterial species use the T6SS during biofilm formation. Mutations in the enteroaggregative E. coli Sci-1 T6SS or avian pathogenic E. coli T6SS genes decrease the ability to form biofilms (38). Additionally, a second T6SS gene cluster in enteroaggregative E. coli, Sci-2 T6SS, and the P. aeruginosa H1-T6SS are upregulated under biofilm-forming conditions (93). Much like during cooperative swarming motility in P. mirabilis, coordination of T6SS production with biofilm determinants may help to promote formation of monospecies biofilms or create a protective isogenic zone within a mixed community.
Host-microbe interactions
There are also studies demonstrating that the T6SS is expressed during interactions with host cells (110–113). Closer inspection reveals a limited number studies that show a direct role for the T6SS in bacterial pathogenesis. The V. cholerae T6SS is required to escape predation and for toxicity towards phagocytes (2). In a mouse model of infection, V. cholerae causes inflammation and increased recovery of bacteria from the intestine (4). The P. aeruginosa H1- and H3-T6SS may influence interactions with host cells through enhanced invasion (113–115). It is important to note that most of the T6SS effectors characterized to date are antimicrobial. Thus, it must be considered that the observed upregulation of the T6SS in the host, or fitness defects of T6SS mutant bacteria in infection models, reflects an indirect relationship to pathogenesis. For example, T6SS antibacterial antagonism was recently revealed to be contribute to efficient colonization of V. cholerae in an animal model (116). That study clearly showed that bacteria lacking immunity against sister cells had a significant fitness defect during infection. Collectively, the limited numbers of studies that show a direct role for the T6SS in bacterial pathogenesis suggest the T6SS has evolved as a weapon used during interbacterial competition.
Competition in the host
Often, for bacteria to establish an infection, pathogens must be able to overcome colonization resistance created by the natural microbiome. This could be important for enteric bacteria that compete against the intestinal microbiome community (117). In support of this, a number of pathogens that colonize the intestine produce a T6SS, including Salmonella enterica, C. rodentium, Aeromonas hydrophila and enteroaggregative E. coli (38, 118–120). The T6SS can be envisioned to provide bacteria with a competitive advantage in other host sites. For example, opportunistic pathogens that cause wound infections, such as A. baumanii, P. aeruginosa, S. marcescens and P. mirabilis, all are known to produce at least one T6SS with antibacterial activity (5, 8, 106, 107). Once these opportunistic pathogens establish tissue colonization, the T6SS could provide a mechanism to prevent colonization by another strain or species. For P. aeruginosa, it is known that diversity within the cystic fibrosis lung decreases over time (121). Indeed, isolates of P. aeruginosa from cystic fibrosis patients have an active T6SS and these patients produce antibodies against Hcp (64, 72).
Additionally, there is a substantial amount of genomic evidence to support the primary role for the T6SS during interbacterial warfare. Immunity proteins are maintained in the genomes of bacteria that lack cognate T6SS effector proteins (43). The presence of orphan immunity proteins suggests that there is positive selection imposed by T6SS attacks (43). The observation that effector proteins are accompanied by immunity proteins is evidence for the selection against self-intoxication. This highlights the fitness cost that is associated with effector intoxication in the absence of immunity. It also appears that bacteria with a lifestyle that precludes interbacterial competition tend to lack a T6SS. For example, in Burkholderia mallei, the loss of interbacterial T6SS has been proposed to coincide with the evolution from a free-living opportunistic pathogen to an obligate pathogen (104).
Intraspecies antagonism
Bacteria that are highly related, for example, the same species or close relatives, are most likely to directly compete with one another for the exact same niche or resources. Although strains will cooperate and form mixed communities, in many cases, growth of one strain precludes growth of another (8, 98, 122–124). Antagonism between isolates has been well studied, the most obvious example is bacteriocin production and targeting of closely related competitiors (7). Another mechanism of intraspecies antagonism that has been recently characterized is known as contact dependent inhibition and requires direct contact like the T6SS (125, 126).
Intraspecies competition using the T6SS has been shown for many bacterial species. T6SS-mediated antagonism was observed between isolates of S. marcescens and in V. cholerae (80, 106). Additionally, bacteria such as P. aeruginosa, V. cholerae, and P. mirabilis all possess variable complements of T6SS effector and immunity pairs, which would result in competition between isolates of the species (8, 108). An exceptionally clear example of intraspecies antagonism using the T6SS can be observed between swarming populations of P. mirabilis. Macroscopic boundaries form between non-identical isolates upon contact during swarming (Figure 4). Upon initiation of swarming differentiation, the T6SS is assembled and appears to fire when opposing swarms meet (8). Boundary formation, underlying killing, and immunity to killing, can be assigned to specific T6SS genes and effector proteins. It is now becoming clear that the ability for P. mirabilis to self-recognize is a result of being immune to T6SS attack (8) (Figure 5). Indeed, the P. mirabilis recognition system appears to depend largely on diversity of hcp/vgrG effector operons and variation between otherwise identical effector operons (8, 127).
Figure 5. Immunity against T6SS is the basis for self-recognition.
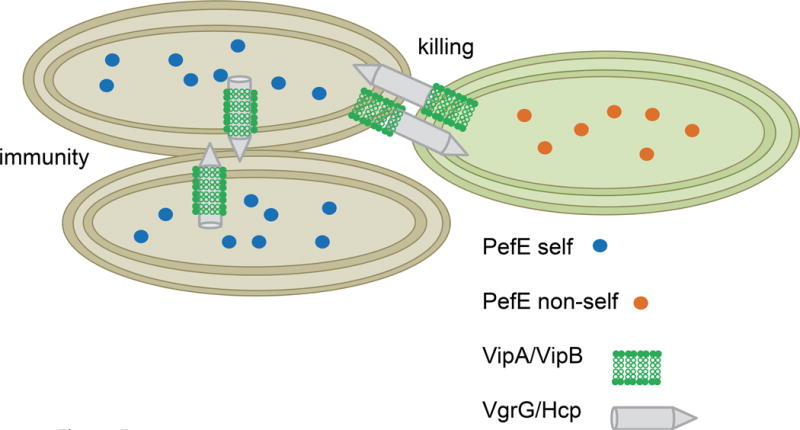
If two strains of Proteus mirabilis synthesize the same immunity protein then both strains are immune to killing by the T6SS and recognize as “self”. However, if two strains have divergent immunity proteins, then both strains are subject to killing by each of the other strain’s T6SS.
Intraspecies competition is consistent with the presence of diverse repertoires of T6SS effector and immunity pairs in many other bacterial species. Selection to maintain diversity could occur when a bacterium that lacks immunity to specific T6SS effector that is present in its neighbor, is rapidly killed (64). This further provides an explanation for why a given organism would translocate closely related effector proteins, which can differ in immune recognition and thus, would be non-redundant in antagonistic function (58, 122). Studying bacterial antagonism has led to the hypothesis that antibacterial activities could be preferentially produced in response to competition-induced stress (128). As discussed for P. aeruginosa and the Fha phosphorylation cascade, both intra- and inter-bacterial interactions of the T6SS in one cell can stimulate the system in neighboring cells (27, 63). This recognition is sufficiently integral to the activity of the P. aeruginosa H1-T6SS that inactivation of the T6SS of a competitor organism confers resistance to intoxication (27, 28).
The conditions that control T6SS activity can provide valuable insights into its physiological relevance of T6SS function. As noted, the H1-T6SS of P. aeruginosa and the T6SS of V. cholerae O1 serotypes, are repressed by quorum sensing (81, 129). Repression of the T6SS by quorum sensing indicates that these systems are active when cells are at relatively low cell density and may function to displace unrelated or more distantly related bacteria in a densely populated mixed community. Other systems, such as the H2-T6SS of P. aeruginosa, are induced under conditions of high cell density (72, 129). This might indicate a role for that T6SS during high cell density of an isogenic population or a role during intraspecies antagonism. As discussed, other bacteria control their T6SS in response to environmental cues, such as temperature, pH or iron availability (47, 70–77). These cues may indicate the movement of bacteria into a condition where competitor bacteria are likely to be encountered.
Concluding Remarks
There has been remarkable progress toward understanding the T6SS and its role in numerous bacterial species. The structural details of T6SS assembly and the effector delivery module has been elucidated. The diversity of effectors that elicit numerous antagonistic activities upon interaction with target cells are now appreciated. Studies by many research groups have provided substantial clues to answer the question of why bacteria would use contact-dependent delivery of effectors to eliminate one’s neighbors. It is becoming clear that bacteria can benefit from the T6SS to discriminate, “recognize”, and kill competitors rather than indiscriminately secrete bactericidal agents when competing for resources in their natural habitats. It is evident that T6SS-dependent interactions are involved in developmental processes such as multicellularity or organized communities (8). For bacteria that cooperate to achieve a behavior such as swarming, the T6SS could prevent cheaters from benefiting from the cooperative behavior (130, 131). In other instances, T6SS killing of neighboring cells can facilitate the release of genetic material for horizontal gene transfer between closely related species (132). Studies like these are likely to focus greater attention on the biological relevance of the T6SS in bacteria. As community behaviors of microbes are beginning to be more thoroughly appreciated, it is expected that T6SS will be a key determinant in shaping these microbial communities.
Acknowledgments
We thank Stephanie Himpsl for many helpful discussions and for assistance in creating figures. Our work on the T6SS in P. mirabilis is supported by grant AI059722 from the National Institute of Allergy and Infectious Disease.
References
- 1.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A. 2006;103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma AT, Mekalanos JJ. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A. 2010;107:4365–4370. doi: 10.1073/pnas.0915156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe. 2010;7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature. 2011;475:343–U392. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol. 2002;56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 8.Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HL. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog. 2013;9:e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotter P. Microbiology: Molecular syringes scratch the surface. Nature. 2011;475:301–303. doi: 10.1038/475301a. [DOI] [PubMed] [Google Scholar]
- 10.Folkesson A, Lofdahl S, Normark S. The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res Microbiol. 2002;153:537–545. doi: 10.1016/s0923-2508(02)01348-7. [DOI] [PubMed] [Google Scholar]
- 11.Bladergroen MR, Badelt K, Spaink HP. Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact. 2003;16:53–64. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- 12.Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, Elkins KL. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao PS, Yamada Y, Tan YP, Leung KY. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol. 2004;53:573–586. doi: 10.1111/j.1365-2958.2004.04123.x. [DOI] [PubMed] [Google Scholar]
- 14.Das S, Chaudhuri K. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 2003;3:287–300. [PubMed] [Google Scholar]
- 15.Pallen M, Chaudhuri R, Khan A. Bacterial FHA domains: neglected players in the phosphothreonine signalling game? Trends Microbiol. 2002;10:556–563. doi: 10.1016/s0966-842x(02)02476-9. [DOI] [PubMed] [Google Scholar]
- 16.Schlieker C, Zentgraf H, Dersch P, Mogk A. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol Chem. 2005;386:1115–1127. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- 17.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci U S A. 2008;105:3733–3738. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jove D, Filloux A. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J Biol Chem. 2011;286:12317–12327. doi: 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc Natl Acad Sci U S A. 2009;106:4160–4165. doi: 10.1073/pnas.0900044106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 2009;28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lossi NS, Manoli E, Forster A, Dajani R, Pape T, Freemont P, Filloux A. The HsiB1C1 (TssB-TssC) complex of the Pseudomonas aeruginosa type VI secretion system forms a bacteriophage tail sheathlike structure. J Biol Chem. 2013;288:7536–7548. doi: 10.1074/jbc.M112.439273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonemann G, Pietrosiuk A, Mogk A. Tubules and donuts: a type VI secretion story. Mol Microbiol. 2010;76:815–821. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- 25.Filloux A. The type VI secretion system: a tubular story. EMBO J. 2009;28:309–310. doi: 10.1038/emboj.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Records AR. The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol Plant Microbe Interact. 2011;24:751–757. doi: 10.1094/MPMI-11-10-0262. [DOI] [PubMed] [Google Scholar]
- 27.Basler M, Ho BT, Mekalanos JJ. Tit-for-Tat: Type VI Secretion System Counterattack during Bacterial Cell-Cell Interactions. Cell. 2013;152:884–894. doi: 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basler M, Mekalanos JJ. Type 6 secretion dynamics within and between bacterial cells. Science. 2012;337:815. doi: 10.1126/science.1222901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–U178. doi: 10.1038/nature10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho BT, Basler M, Mekalanos JJ. Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science. 2013;342:250–253. doi: 10.1126/science.1243745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kube S, Kapitein N, Zimniak T, Herzog F, Mogk A, Wendler P. Structure of the VipA/B type VI secretion complex suggests a contraction-state-specific recycling mechanism. Cell Rep. 2014;8:20–30. doi: 10.1016/j.celrep.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 32.Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH. Atomic structure of T6SS reveals interlaced array essential to function. Cell. 2015;160:940–951. doi: 10.1016/j.cell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudryashev M, Wang RY, Brackmann M, Scherer S, Maier T, Baker D, DiMaio F, Stahlberg H, Egelman EH, Basler M. Structure of the type VI secretion system contractile sheath. Cell. 2015;160:952–962. doi: 10.1016/j.cell.2015.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunet YR, Henin J, Celia H, Cascales E. Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 2014;15:315–321. doi: 10.1002/embr.201337936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aschtgen MS, Gavioli M, Dessen A, Lloubes R, Cascales E. The SciZ protein anchors the enteroaggregative Escherichia coli Type VI secretion system to the cell wall. Mol Microbiol. 2010;75:886–899. doi: 10.1111/j.1365-2958.2009.07028.x. [DOI] [PubMed] [Google Scholar]
- 36.Felisberto-Rodrigues C, Durand E, Aschtgen MS, Blangy S, Ortiz-Lombardia M, Douzi B, Cambillau C, Cascales E. Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 2011;7:e1002386. doi: 10.1371/journal.ppat.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E. Architecture and assembly of the Type VI secretion system. Biochim Biophys Acta. 2014;1843:1664–1673. doi: 10.1016/j.bbamcr.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 38.Aschtgen MS, Bernard CS, De Bentzmann S, Lloubes R, Cascales E. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol. 2008;190:7523–7531. doi: 10.1128/JB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma LS, Narberhaus F, Lai EM. IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J Biol Chem. 2012;287:15610–15621. doi: 10.1074/jbc.M111.301630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma LS, Lin JS, Lai EM. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol. 2009;191:4316–4329. doi: 10.1128/JB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durand E, Nguyen VS, Zoued A, Logger L, Pehau-Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A, Roussel A, Cambillau C, Cascales E, Fronzes R. Biogenesis and structure of a type VI secretion membrane core complex. Nature. 2015;523:555–560. doi: 10.1038/nature14667. [DOI] [PubMed] [Google Scholar]
- 42.Zoued A, Durand E, Bebeacua C, Brunet YR, Douzi B, Cambillau C, Cascales E, Journet L. TssK is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type VI secretion system. J Biol Chem. 2013;288:27031–27041. doi: 10.1074/jbc.M113.499772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe. 2012;11:538–549. doi: 10.1016/j.chom.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyata ST, Kitaoka M, Brooks TM, McAuley SB, Pukatzki S. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun. 2011;79:2941–2949. doi: 10.1128/IAI.01266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA, Cunningham DA, Tran BQ, Low DA, Goodlett DR, Hayes CS, Mougous JD. Genetically distinct pathways guide effector export through the type VI secretion system. Mol Microbiol. 2014;92:529–542. doi: 10.1111/mmi.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem. 2013;288:7618–7625. doi: 10.1074/jbc.M112.436725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Busby JN, Panjikar S, Landsberg MJ, Hurst MR, Lott JS. The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature. 2013;501:547–550. doi: 10.1038/nature12465. [DOI] [PubMed] [Google Scholar]
- 50.Jones C, Hachani A, Manoli E, Filloux A. An rhs gene linked to the second type VI secretion cluster is a feature of the Pseudomonas aeruginosa strain PA14. J Bacteriol. 2014;196:800–810. doi: 10.1128/JB.00863-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hachani A, Allsopp LP, Oduko Y, Filloux A. The VgrG proteins are “a la carte” delivery systems for bacterial type VI effectors. J Biol Chem. 2014;289:17872–17884. doi: 10.1074/jbc.M114.563429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and regulation of the type VI secretion system. Annu Rev Microbiol. 2012;66:453–472. doi: 10.1146/annurev-micro-121809-151619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benz J, Sendlmeier C, Barends TR, Meinhart A. Structural insights into the effector-immunity system Tse1/Tsi1 from Pseudomonas aeruginosa. PLoS One. 2012;7:e40453. doi: 10.1371/journal.pone.0040453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng J, Leung KY. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol. 2007;66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- 55.Silverman JM, Agnello DM, Zheng H, Andrews BT, Li M, Catalano CE, Gonen T, Mougous JD. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell. 2013;51:584–593. doi: 10.1016/j.molcel.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J Bacteriol. 2010;192:155–168. doi: 10.1128/JB.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chou S, Bui NK, Russell AB, Lexa KW, Gardiner TE, LeRoux M, Vollmer W, Mougous JD. Structure of a peptidoglycan amidase effector targeted to Gram-negative bacteria by the type VI secretion system. Cell Rep. 2012;1:656–664. doi: 10.1016/j.celrep.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. New secreted toxins and immunity proteins encoded within the Type VI secretion system gene cluster of Serratia marcescens. Mol Microbiol. 2012;86:921–936. doi: 10.1111/mmi.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whitney JC, Chou S, Russell AB, Biboy J, Gardiner TE, Ferrin MA, Brittnacher M, Vollmer W, Mougous JD. Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J Biol Chem. 2013;288:26616–26624. doi: 10.1074/jbc.M113.488320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature. 2013;496:508–512. doi: 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong TG, Ho BT, Yoder-Himes DR, Mekalanos JJ. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A. 2013;110:2623–2628. doi: 10.1073/pnas.1222783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding J, Wang W, Feng H, Zhang Y, Wang DC. Structural insights into the Pseudomonas aeruginosa type VI virulence effector Tse1 bacteriolysis and self-protection mechanisms. J Biol Chem. 2012;287:26911–26920. doi: 10.1074/jbc.M112.368043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LeRoux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc Natl Acad Sci U S A. 2012;109:19804–19809. doi: 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li L, Zhang W, Liu Q, Gao Y, Gao Y, Wang Y, Wang DZ, Li Z, Wang T. Structural Insights on the bacteriolytic and self-protection mechanism of muramidase effector Tse3 in Pseudomonas aeruginosa. J Biol Chem. 2013;288:30607–30613. doi: 10.1074/jbc.C113.506097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poole SJ, Diner EJ, Aoki SK, Braaten BA, de Roodenbeke CT, Low DA, Hayes CS. Identification of Functional Toxin/Immunity Genes Linked to Contact-Dependent Growth Inhibition (CDI) and Rearrangement Hotspot (Rhs) Systems. Plos Genetics. 2011;7 doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma LS, Hachani A, Lin JS, Filloux A, Lai EM. Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe. 2014;16:94–104. doi: 10.1016/j.chom.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A. 2010;107:21128–21133. doi: 10.1073/pnas.1014998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol. 2011;193:6471–6482. doi: 10.1128/JB.05414-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salomon D, Gonzalez H, Updegraff BL, Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One. 2013;8:e61086. doi: 10.1371/journal.pone.0061086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aubert DF, Flannagan RS, Valvano MA. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect Immun. 2008;76:1979–1991. doi: 10.1128/IAI.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol. 2011;13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 73.Brunet YR, Bernard CS, Gavioli M, Lloubes R, Cascales E. An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 2011;7:e1002205. doi: 10.1371/journal.pgen.1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chakraborty S, Sivaraman J, Leung KY, Mok YK. Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J Biol Chem. 2011;286:39417–39430. doi: 10.1074/jbc.M111.295188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pieper R, Huang ST, Robinson JM, Clark DJ, Alami H, Parmar PP, Perry RD, Fleischmann RD, Peterson SN. Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology. 2009;155:498–512. doi: 10.1099/mic.0.022160-0. [DOI] [PubMed] [Google Scholar]
- 76.Gueguen E, Durand E, Zhang XY, d’Amalric Q, Journet L, Cascales E. Expression of a Type VI Secretion System Is Responsive to Envelope Stresses through the OmpR Transcriptional Activator. PLoS One. 2013;8:e66615. doi: 10.1371/journal.pone.0066615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bernard CS, Brunet YR, Gavioli M, Lloubes R, Cascales E. Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J Bacteriol. 2011;193:2158–2167. doi: 10.1128/JB.00029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng J, Ho B, Mekalanos JJ. Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One. 2011;6:e23876. doi: 10.1371/journal.pone.0023876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dong TG, Mekalanos JJ. Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Research. 2012;40:7766–7775. doi: 10.1093/nar/gks567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Unterweger D, Kitaoka M, Miyata ST, Bachmann V, Brooks TM, Moloney J, Sosa O, Silva D, Duran-Gonzalez J, Provenzano D, Pukatzki S. Constitutive type VI secretion system expression gives Vibrio cholerae intra- and interspecific competitive advantages. PLoS One. 2012;7:e48320. doi: 10.1371/journal.pone.0048320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One. 2009;4:e6734. doi: 10.1371/journal.pone.0006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ishikawa T, Sabharwal D, Broms J, Milton DL, Sjostedt A, Uhlin BE, Wai SN. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun. 2012;80:575–584. doi: 10.1128/IAI.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 84.Grainger DC, Hurd D, Goldberg MD, Busby SJ. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–4652. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 86.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Castang S, McManus HR, Turner KH, Dove SL. H-NS family members function coordinately in an opportunistic pathogen. Proc Natl Acad Sci U S A. 2008;105:18947–18952. doi: 10.1073/pnas.0808215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 89.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci U S A. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 91.Silverman JM, Austin LS, Hsu F, Hicks KG, Hood RD, Mougous JD. Separate inputs modulate phosphorylation-dependent and -independent type VI secretion activation. Mol Microbiol. 2011;82:1277–1290. doi: 10.1111/j.1365-2958.2011.07889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.LeRoux M, Kirkpatrick RL, Montauti EI, Tran BQ, Peterson SB, Harding BN, Whitney JC, Russell AB, Traxler B, Goo YA, Goodlett DR, Wiggins PA, Mougous JD. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife. 2015;4 doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 94.Hsu F, Schwarz S, Mougous JD. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol Microbiol. 2009;72:1111–1125. doi: 10.1111/j.1365-2958.2009.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Casabona MG, Silverman JM, Sall KM, Boyer F, Coute Y, Poirel J, Grunwald D, Mougous JD, Elsen S, Attree I. An ABC transporter and an outer membrane lipoprotein participate in posttranslational activation of type VI secretion in Pseudomonas aeruginosa. Environ Microbiol. 2013;15:471–486. doi: 10.1111/j.1462-2920.2012.02816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fritsch MJ, Trunk K, Diniz JA, Guo M, Trost M, Coulthurst SJ. Proteomic identification of novel secreted antibacterial toxins of the Serratia marcescens type VI secretion system. Mol Cell Proteomics. 2013;12:2735–2749. doi: 10.1074/mcp.M113.030502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E. Imaging type VI secretion-mediated bacterial killing. Cell Rep. 2013;3:36–41. doi: 10.1016/j.celrep.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 98.Dienes L. Reproductive processes in Proteus cultures. Proc Soc Exp Biol Med. 1946;63:265–270. doi: 10.3181/00379727-63-15570. [DOI] [PubMed] [Google Scholar]
- 99.Dienes L. Further observations on the reproduction of bacilli from large bodies in Proteus cultures. Proc Soc Exp Biol Med. 1947;66:97. doi: 10.3181/00379727-66-15994. [DOI] [PubMed] [Google Scholar]
- 100.Bode NJ, Debnath I, Kuan L, Schulfer A, Ty M, Pearson MM. Transcriptional analysis of the MrpJ network: modulation of diverse virulence-associated genes and direct regulation of mrp fimbrial and flhDC flagellar operons in Proteus mirabilis. Infect Immun. 2015;83:2542–2556. doi: 10.1128/IAI.02978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gibbs KA, Wenren LM, Greenberg EP. Identity Gene Expression in Proteus mirabilis. J Bacteriol. 2011;193:3286–3292. doi: 10.1128/JB.01167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010;18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Durand E, Cambillau C, Cascales E, Journet L. VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 2014;22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 104.Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010;6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A. 2010;107:19520–19524. doi: 10.1073/pnas.1012931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol. 2011;193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carruthers MD, Nicholson PA, Tracy EN, Munson RS., Jr Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One. 2013;8:e59388. doi: 10.1371/journal.pone.0059388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun. 2014;5:3549. doi: 10.1038/ncomms4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wenren LM, Sullivan NL, Cardarelli L, Septer AN, Gibbs KA. Two independent pathways for self-recognition in Proteus mirabilis are linked by type VI-dependent export. MBio. 2013;4 doi: 10.1128/mBio.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bernard CS, Brunet YR, Gueguen E, Cascales E. Nooks and crannies in type VI secretion regulation. J Bacteriol. 2010;192:3850–3860. doi: 10.1128/JB.00370-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shalom G, Shaw JG, Thomas MS. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology. 2007;153:2689–2699. doi: 10.1099/mic.0.2007/006585-0. [DOI] [PubMed] [Google Scholar]
- 112.Parsons DA, Heffron F. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect Immun. 2005;73:4338–4345. doi: 10.1128/IAI.73.7.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chugani S, Greenberg EP. The influence of human respiratory epithelia on Pseudomonas aeruginosa gene expression. Microb Pathog. 2007;42:29–35. doi: 10.1016/j.micpath.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, Engel J, Filloux A, Bleves S. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J Biol Chem. 2012;287:27095–27105. doi: 10.1074/jbc.M112.376368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe. 2014;15:600–610. doi: 10.1016/j.chom.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 116.Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe. 2013;14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yurist-Doutsch S, Arrieta MC, Vogt SL, Finlay BB. Gastrointestinal microbiota-mediated control of enteric pathogens. Annu Rev Genet. 2014;48:361–382. doi: 10.1146/annurev-genet-120213-092421. [DOI] [PubMed] [Google Scholar]
- 118.Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics. 2009;10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Petty NK, Bulgin R, Crepin VF, Cerdeno-Tarraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron A, Clark L, Toribio AL, Parkhill J, Dougan G, Frankel G, Thomson NR. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J Bacteriol. 2010;192:525–538. doi: 10.1128/JB.01144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Suarez G, Sierra JC, Sha J, Wang S, Erova TE, Fadl AA, Foltz SM, Horneman AJ, Chopra AK. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb Pathog. 2008;44:344–361. doi: 10.1016/j.micpath.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A. 2012;109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gibbs KA, Urbanowski ML, Greenberg EP. Genetic determinants of self identity and social recognition in bacteria. Science. 2008;321:256–259. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu Rev Microbiol. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- 124.Vos M, Velicer GJ. Social conflict in centimeter-and global-scale populations of the bacterium Myxococcus xanthus. Curr Biol. 2009;19:1763–1767. doi: 10.1016/j.cub.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aoki SK, Pamma R, Hernday AD, Bickham JE, Braaten BA, Low DA. Contact-dependent inhibition of growth in Escherichia coli. Science. 2005;309:1245–1248. doi: 10.1126/science.1115109. [DOI] [PubMed] [Google Scholar]
- 127.Cardarelli L, Saak C, Gibbs KA. Two Proteins Form a Heteromeric Bacterial Self-Recognition Complex in Which Variable Subdomains Determine Allele-Restricted Binding. MBio. 2015;6:e00251. doi: 10.1128/mBio.00251-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 129.Lesic B, Starkey M, He J, Hazan R, Rahme LG. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology. 2009;155:2845–2855. doi: 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.West SA, Gardner A. Altruism, spite, and greenbeards. Science. 2010;327:1341–1344. doi: 10.1126/science.1178332. [DOI] [PubMed] [Google Scholar]
- 131.Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 132.Borgeaud S, Metzger LC, Scrignari T, Blokesch M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 2015;347:63–67. doi: 10.1126/science.1260064. [DOI] [PubMed] [Google Scholar]


